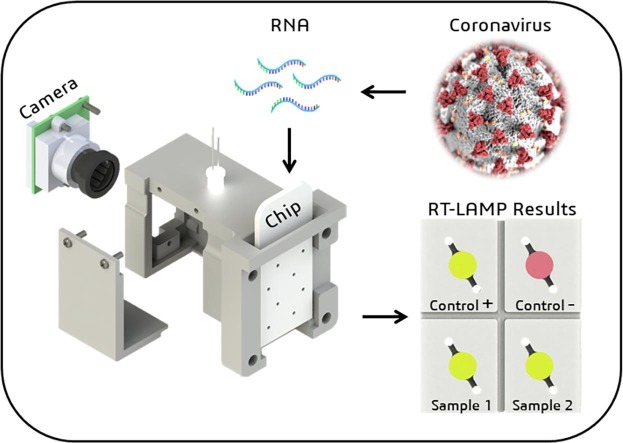
Graphical abstract
Keywords: LAMP-on-a-chip, SARS-CoV-2, Raspberry Pi, COVID-19, diagnostic, OpenCV
Abstract
This work describes the development of a Point-of-Care (POC) Lab-on-a-Chip (LOC) instrument for diagnosis of SARS-CoV-2 by Reverse-Transcription Loop-mediated isothermal amplification (RT-LAMP). The hardware is based on a Raspberry Pi computer ($35), a video camera, an Arduino Nano microcontroller, a printed circuit board as a heater and a 3D printed housing. The chips were manufactured in polymethyl methacrylate (PMMA) using a CO2 laser cutting machine and sealed with a PCR optic plastic film. The chip temperature is precisely controlled by a proportional–integral–derivative (PID) algorithm. During the RT-LAMP amplifications the chip was maintained at ∼ (65.0 ± 0.1) °C for 25 minutes and 5 minutes cooling down, totaling a 30 minutes of reaction .The software interpretation occurs in less than a second. The chip design has four 25 µL chambers, two for clinical samples and two for positive and negative control-samples. The RT-LAMP master mix solution added in the chip chambers contains the pH indicator Phenol Red, that is pink (for pH ∼ 8.0) before amplification and becomes yellow (pH ∼ 6.0) if the genetic material is amplified. The RT-LAMP SARS-CoV-2 diagnostic was made by color image recognition using the OpenCV machine vision software library. The software was programmed to automatically distinguish the HSV color parameter distribution in each one of the four chip chambers. The instrument was successfully tested for SARS-CoV-2 diagnosis, in 22 clinic samples, 11 positives and 11 negatives, achieving an assertiveness of 86% when compared to the results obtained by RT-LAMP standard reactions performed in conventional PCR equipment.
1. Introduction
The COVID-19 pandemic caused by the SARS-CoV-2 virus is a novel threat to global health and resulted in economic and social changes in the world since December-2019 [1]. To minimize the transmissibility of the virus, fast and accurate methods for detection of this virus are essential [2]. The gold standard method used to detect SARS-CoV-2 is the quantitative Real Time Polymerase Chain Reaction (RT-qPCR) [3], [4]. In general, samples from the upper respiratory tract (nasopharyngeal swab) are collected from suspects for RNA extraction followed by amplification of specific genomic regions. Despite the high sensitivity and specificity of the method, there are limitations to its use, such as complex and expensive laboratory instruments, reagents and highly-trained professionals [3], [5].
To decrease costs and complexity of COVID-19 diagnosis, approaches such as the Reverse Transcriptase Loop Mediated Isothermal Amplification (RT-LAMP) have been applied [6], [7]. RT-LAMP relies on auto-cycling strand displacement DNA synthesis resulting in exponential amplification, this reaction usually occurs in 30-40 minutes under isothermal conditions (65°C) using four to six sets of primers [8]. When the reaction is combined with pH indicators, it offers a rapid and simple colorimetric detection, demonstrating its potential as a point-of-care (POC) testing for COVID-19 [6]. When the reaction occurs, the dNTPs release ions into the solution and make it more acidic, thus changing the pH and causing the pH indicator to change its color to yellow.
Lab-on-a-chip (LOC) instruments may have advantages over other standardised methods, such as lateral flow tests and RT-qPCR, due to low sample consumption and portability, highlighting their potential for applications in point-of-care systems [9]. Traditional molecular biology techniques can be performed in chip devices, including the Loop Mediated Isothermal Amplification (LAMP) [10], [11]. Ganguli et al for example developed a 3D printed LOC device and a smartphone-based reader to detect COVID-19 [12]. Papadakis et al have reported a POC device with a miniature camera for the analysis of SARS-CoV-2 using conventional PCR tube with specificity and sensitivity comparable to the RT-qPCR [13]. Rodriguez-Mateos et al. [14], for example, developed a polymethyl methacrylate (PMMA) chip able to perform the RNA extraction and the RT-LAMP amplification reaction on the same chip with the use of a block heater and a magnet to move the reagents through the chip. Rodriguez-Manzano et al. [15], similarly, designed a complete handheld system but for SARS-CoV-2, with detection in under 20 minutes with a real time visualization of results. Song et al. [16] leads the thought that portable instruments are more competitive for its accessibility, high sensitivity, and lower costs.
LOC devices can be manufactured by several technologies and in many types of materials such as silicon, glass, and plastic substrates. For plastic substrate-based instruments, like polymethyl methacrylate (PMMA), there are methods of mass production, which include solvent imprinting, hot embossing, thermal bonding, injection molding, and laser ablation [17]. The CO2 laser ablation offers versatility, scalability, and cost-effectiveness in manufacturing [18]. Commercially available laser cutters can be used to generate a wide variety of microfluidic designs for different applications and accelerates the prototyping process. Adjusting the laser characteristics (as power and engraving speed) allows control of the depth and width of the chip [19]. The ablation process has the disadvantage of generating high roughness of the surface, which can reduce the applicability for some devices [19].
There are different methods for sealing PMMA chips, such as solvent bonding, chemical bonding, adhesive bonding, and thermal bonding [20]. Thermal bonding is also a commonly used method for manufacturing PMMA chips, allowing a simple and uniform surface bonding, however, this method may lead to channel deformation [21]. The chemical sealing method requires low pressure and low temperature, however, it involves several process steps [22]. Another example of sealing method is the solvent bonding, but some solvents can dissolve PMMA, leading to clogging the channels [23]. Sealing adhesives are commonly used and do not require complex steps but depending on its material and how it is applied. It can also lead to clogging of the channels [21].
Microfluidics-based LAMP chips have already been proposed in previous studies [24]. Fang et al. [25], for example, established a highly specific multiplex LAMP able to identify 3 human influenza viruses, with a significant cost/time-effective ratio and simple operational methodology. Zika virus has also been detected through RT-LAMP in a microfluidic chip and a chemically heated cup by Song et al. [26], with results observed in less than 40 minutes by naked-eyes.
In this work, we describe the development and validation of a point-of-care equipment and a sealed PMMA chip, able to perform colorimetric RT-LAMP reactions. We successfully standardised the POC diagnostic system by detecting SARS-CoV-2 in a chip, using machine vision combined with a colorimetric approach. The innovative approach proposed here was to demonstrate the feasibility of using simple components to develop a cheap and efficient POC device able to rapidly detect SARS-CoV-2 even in unfavorable environments. The device’s automatic colorimetric interpretation was also successfully implemented in order to avoid false positive/negative results by the human naked eye.
2. Material and methods
2.1. Chip manufacturing
The chip was manufactured in polymethyl methacrylate (PMMA) and designed using the software CorelDRAW (CorelDRAW Corel Corporation, Ottawa, USA) [27]. A CO2 laser cutting machine VS3020C (Visutec; Brazil) was used to fabricate the chip with external dimensions of 53 x 24 mm2 in a 2 mm thickness substrate, with four chambers of 25 µL volume each.
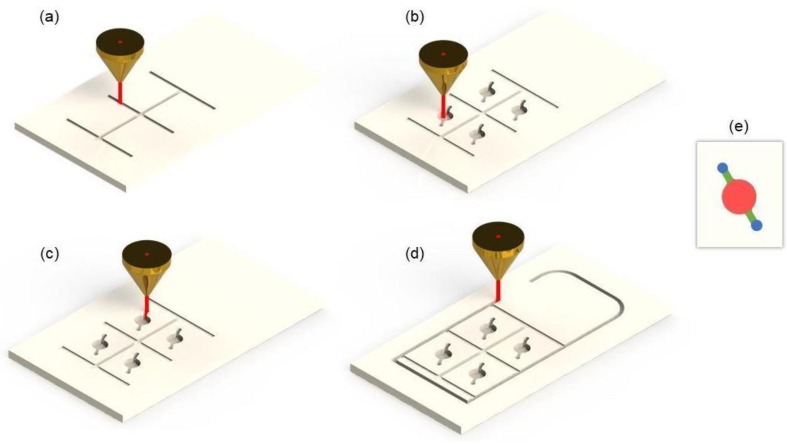
The CO2 laser cutting machine has a maximum laser power of 40 W, which may be adjusted from 1% to 100%. To cut the chip there were four steps: (1) engrave the grooves (Fig.1 .a); (2) engrave the chambers and channels (Fig.1.b); (3) cut the holes (Fig.1.c) and (4) cut out the chip (Fig.1.d). The first step required 12% of the CO2 maximum laser power combined with a speed of 160 mm/s to create a depth of 0.1 mm. This groove increases the adherence of the plastic film onto the chip when forced inside the groove using a spatula or a ballpoint pen. The next step was to manufacture the chambers and the channels with 29% of the CO2 laser power and a speed of 200 mm/s, resulting in a 25 µL chamber. To cut the hole, 100% of CO2 laser power was used and 20 mm/s of speed. In the final step the chip was cut out using 100% of the laser power and 20 mm/s speed. The chip’s project can be accessed in the supplementary material through a GitHub link (https://github.com/geovanimendonca/Pegasus).
Fig. 1.
PMMA Chip device fabrication through a laser ablation process. (a) Engraving the grooves. (b) Engraving the chambers and channels. (c) Cutting the holes. (d) Cutting the chip limits. (e) Chamber in detail.
Once the chip was cut, it followed these steps: (1) wash the chip with neutral detergent using a soft brush and tap water; (2) add the chip in a beaker with ultrapure water and sonicate it for 15 minutes (repeat the process twice); (3) dry it in a laboratory oven at 40°C for 30 minutes. After that a PCR optic plastic sealing film (Roche® 480 LightCycler®) [28] was applied to seal the front part of the chip, where the chambers are located. For better sealing, the optic plastic was pressed against the chip grooves with the assistance of a spatula or a ballpoint pen. Next, the chip was flipped to insert the sample (25 µL) on the chambers using a micropipette. The sample is inserted through the inlet holes (Fig.1.e, in blue), moves through the channel (Fig.1.e, in green), fills up the 25 µl reaction chamber (Fig.1.e, in red), and moves through the outlet channel. This design helps to introduce the sample with a conventional pipette and avoids bubbles formation because the plastic film is already in place. After inserting the sample, the entire back surface was also sealed with the same PCR optic plastic sealing film.
2.2. Samples and ethical statement
The SARS-CoV-2 clinical samples were acquired from 22 symptomatic patients from Erasto Gaertner Hospital (Curitiba - Brazil) with the Local Ethics Committee approval (CAAE 31592620.4.3001.5248 and 31592620.4.1001.0098). Sample’s manipulation and experiments were carried out in accordance with ANVISA (National Health Surveillance Agency), the Brazilian guidelines, and all recruited patients have written a consent. Two nasal and one oral rayon-swabs were collected in 3 mL of 1x Phosphate-Buffered Saline (PBS 1x) in 15 mL centrifuge tubes for each patient. Samples were stored at -20°C. The RNA extraction was performed with QIAmp Viral RNA Mini Kit (Qiagen) as described by the manufacturers [29] and the RNA was diluted in ultrapure water to avoid it from interfering with the colorimetric reaction.
2.3. Chip performance validation with SARS-CoV-2 samples
The RT-LAMP primers subjected for the chip validation were previously tested by our research group [6], [7]. These primers were designed by Rabe and Cepko and have four thymidine residues inserted in the middle of FIP and BIP primers [30] (Supplementary Table S1). Reactions were performed with WarmStart® Colorimetric LAMP 2X Master Mix (NEB, England) containing Phenol Red for pH detection, in a final volume of 25 μL with 6 μL of primer mix (FIP and BIP at 1.6 μM each, FOP and BOP at 0.2 μM each and FL and BL at 0.4 μM each) and 5 μL of RNA sample. The reactions were incubated at 65 °C for 25 minutes using our RT-LAMP on-a-chip instrument. Right after finishing the reaction the chip was cooled to 25 °C and a photo was taken for further analysis. For SARS-CoV-2 samples, two separate chambers were used for colorimetric RT-LAMP controls. As positive control (105 SARS-CoV-2 copies), we used RNA extracted from supernatants of SARS-CoV-2 cultured in Vero cells using QIAamp® RNA viral Mini Kit (Qiagen, Germany), following the manufacturer’s instructions. As non-template control (NTC) 5 μL of nuclease-free water was also used.
2.4. Software and image processing
The instrument’s software was written in Python 3 [31] and the firmware was written in C++. The accessed support libraries were the OpenCV version 4.1.0 (Open-Source Computer Vision Library) [32] for image processing, the color calibration was executed by removing the color parameters present in 96 samples and using them as delimiters for classifying the samples. The open-source framework for Python, Kivy version 2.0.0 [33], was used for developing the custom graphical user interface (GUI). All information is shown on a 5” Raspberry Pi display touch screen. The installed camera has a OV5647 sensor (OmniVision Technologies, China), a manual adjustable focal length of 3.6mm, a viewing angle of 76° and 5MP of resolution. The software and codes can be accessed in the Supplementary Material in a GitHub link (https://github.com/geovanimendonca/Pegasus). The software interpretation accuracy was calculated based on the comparison of previous colorimetric RT-LAMP results by our research group [6], [7].
2.5. Hardware
The motherboard circuit was manufactured using the LPKF Circuit Pro S103 PCB milling (LPKF Laser & Electronics, Germany); the electronic design with the software DipTrace 4.0.0.5 [34], and the heater with the Autodesk AutoCAD Student software (Autodesk, Inc. USA) [35]. The 35 μm copper FR4 PCB board heater was milled into thin tracks and used as a heating element, with a total resistance of ∼ (5.7 ± 0.5) Ω and a layout area over 22 x 27.5 mm2. The instrument was powered by an AC/DC power adapter (W-T5000 input 100-240V - 50/60 Hz - output 12V/5A). The main board contains a Raspberry Pi 3 model B+, single board computer (Raspberry Pi Foundation, United Kingdom), an Arduino Nano, microcontroller (Arduino, Italy), and a step-down module (LM2596 module) to lower the voltage from 12V to 5V. The Raspberry Pi was used to make the graphical interface (GUI) and communicate with the database (Firebase). Real time information of temperature was obtained through the analog 10k negative temperature coefficient sensor soldered in the heating board. The Arduino Nano controls the temperature with a proportional-integral-derivative controller (PID) algorithm library [36] by measuring the temperature from the sensor and receiving the setpoint (target) from the Raspberry Pi. The motherboard contains one IRL MOSFET 3103S to turn ON|OFF the heater. Similarly, two BC817 transistors were used to turn ON|OFF the cooler fan and the LED to light up the chip. The schematic diagram of the instrument is shown in Supplementary Figure 1.
2.6. Housing
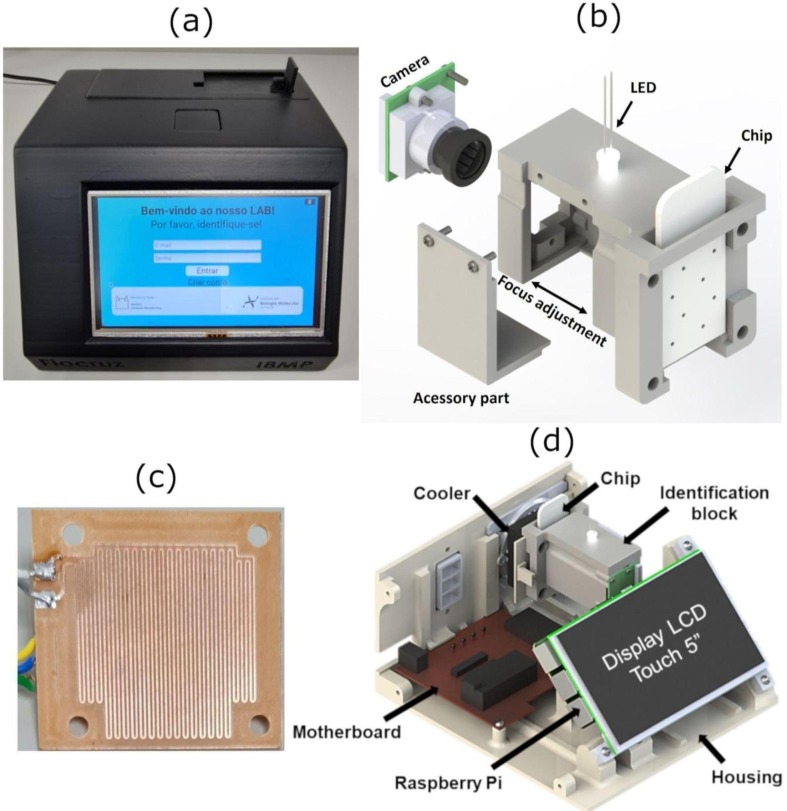
The instrument’s housing was completely produced by additive manufacturing (3D printing) using an Ender 3 Pro 3D printer (Creality, China) [37]. The identification block (ID) (Fig.2 .b) was also 3D printed. The ID block supports the heater, the area where the chip is inserted, the camera, and the white LED to light up the chip. The camera is aligned to the center of the chip to capture the four (4) chambers at the same time. The white LED was placed in the top-center of the ID.
Fig. 2.
Lab-on-a Chip RT-LAMP Instrument parts. (a) LAMP-on-a-Chip instrument. (b) Identification block. (c) Heating board. (d) Rendered image of the equipment showing its main components.
3. Results and Discussion
3.1. Lab-on-a Chip RT-LAMP instrument
The LAMP-on-a-chip instrument has four main parts: “housing” (Fig.2.a), “identification block” (Fig.2.b), “heating board“ (Fig.2.c) and ”motherboard“ (Fig.2.d). The identification block is used to hold the chip, to cool down the temperature, illuminate the chip chambers, and to hold the camera that records the chip image. The camera is aligned to the center of the samples, and a side cover can be removed to adjust the camera focus. The heating board is responsible for heating the chip to 65 °C. To control all system parameters, a motherboard was created. The motherboard has a microcontroller Arduino Nano used as a communication interface with the temperature sensor and the actuators (fan and heater). A Raspberry Pi 3 model B+ (low-cost computer) was used to create the user interface (HMI) through a 5” touch display and to communicate with the database.
As can be seen, the equipment hardware is relatively simple, that is the main goal of this project.
3.2. Heating/Temperature Control
In order to calibrate the chip temperature we compared the instrument temperature sensor (positioned near the heating board) with the temperature inside the chip (using a thermocouple). It was observed that the temperature in the sensor was always lower than the temperature on the chip. Figure 2 in supplementary materials shows the calibration curve, the linear dependence between them, and the equation TReal = (1.48).Tsensor - (11.4°C) created to automatically correct the values on the microcontroller firmware. The k-type thermocouple was measured in a Novus 1200 temperature controller (Novus International, Inc., EUA).
Our equipment takes 3 minutes to heat up the sample to the desired temperature of 65°C. In comparison, Pardy et al. [39] created a chip to perform LAMP reactions that reaches the target temperature in 5 minutes. They also created a heating system that does not need any electronic control but is fixed to a single temperature. Our equipment however has the flexibility to control the reaction’s temperature and time.
The main temperature applied was 65.0 °C due to the previous optimization of colorimetric RT-LAMP detection of SARS-CoV-2 by our research group [6], [7]. The RT-LAMP temperature was adjusted through a PID control set in the Arduino program, being responsible for stabilising the temperature within a given range of 0.1°C. A PID controller operates in a closed-loop system and uses the error measured at the system output as feedback to control the future actions that will be entered in the system input.
At the end of the amplification process, the instrument turns OFF the heating board while the fan is turned ON, to reach near room temperature, around 25°C. Only after reaching this temperature the image of the chambers is taken. The instrument takes approximately 5 minutes to cool down the temperature to 25°C. Although working, at the end of the project we concluded that new versions of this instrument should be designed using a Peltier device to cool the instrument faster. We noticed after several tests that as cold as the temperature reaction is (under 25°C), higher will be the color differences between an “amplified” and a “not amplified” sample.
3.3. Image Processing
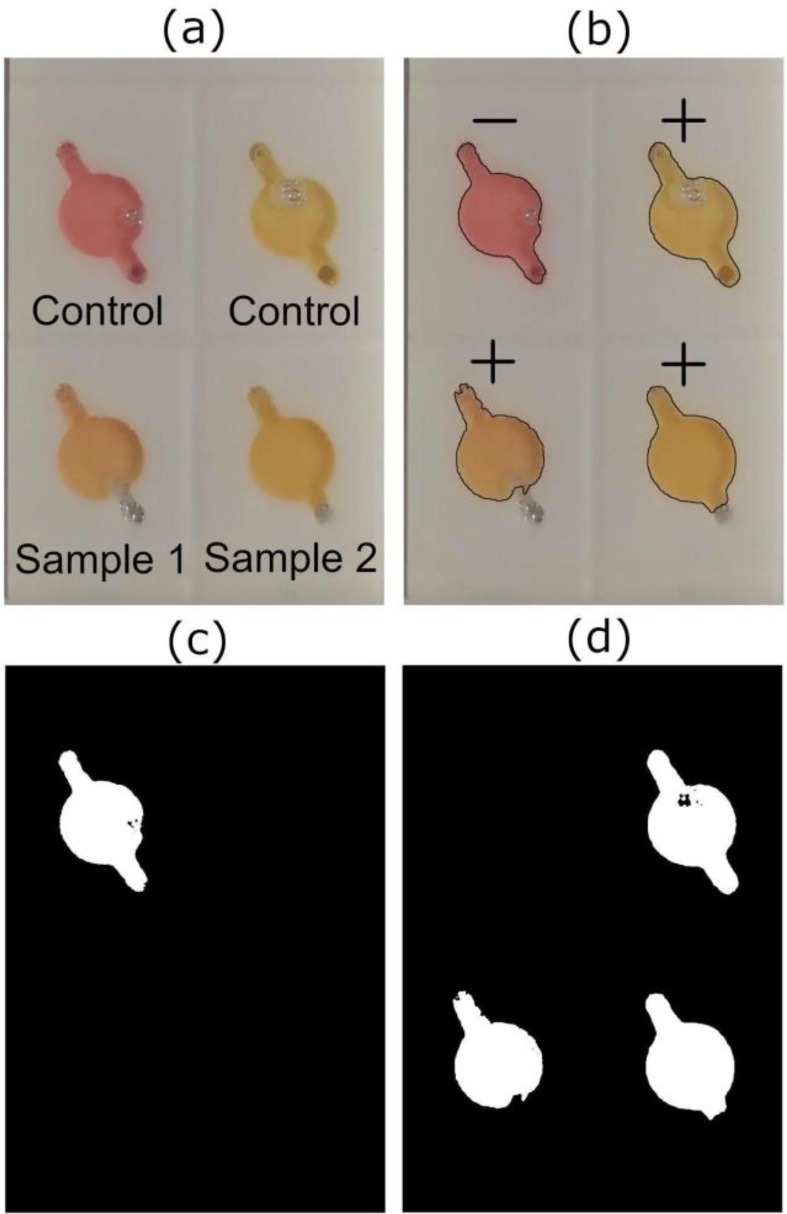
The process to identify the colors of the reagent has been previously performed by Pengfe et al. [40], in which a mobile app was created and calibrated to identify different reaction’s colors in tubes and classify them as “positive” or “negative”. We performed a very similar algorithm, but for identifying reagent color on our chips. The algorithm developed by our group for this instrument was named “Reagent Chamber Color Identification” (RCCI) and was designed to detect the color of the chip chamber after the RT-LAMP for SARS-CoV-2 occurs. The algorithm analyses the color (in HSV scale) of each chamber, using the OpenCV computer vision library. The positive control and negative temperature coefficient sensor were used as validation parameters, determining whether the sample's result was positive (yellow), negative (pink) or invalid. Invalid results occur when one or both controls do not behave as expected, invalidating the test. To start the identification process, the camera takes and saves a photo of the entire chip (Fig.3 .a). The reaction is finished, and when the chip is at room temperature (25 °C), the photo is loaded into the RCCI algorithm. The first step of the algorithm is to change the color model of the photo from RGB (Red, Green, Blue) to an alternative model, the HSV (Hue, Saturation, Value). The HSV model represents the hue in just one variable and can be used as a linear parameter to identify the color of an image. The RCCI creates two masks corresponding to positive (+) and negative (-) results (Fig.3.c, d). The parameters shown in Table S2 were extracted to calibrate the RCCI. A total of 96 samples were used (tested in 24 chips) to calibrate the artificial intelligence (A.I) of the instrument.
Fig. 3.
Lamp-on-a-Chip Image recognition process: (a) Photo taken after the RT-LAMP reaction of 2 positive samples of SARS-CoV-2, plus a positive and a negative control. (b) Diagnostic result showing the negative and positive controls on the top left and right of the chip respectively and the two positive results below to sample 1 and 2. (c and d) Masks created during the identification process to identify negative and positive samples respectively.
The masks are two binarized images (black/white) (Fig.3.c, d) that were analysed separately. Both images were subjected to noise reduction algorithms, performed by the OpenCV library, to avoid identification mistakes. The algorithm looks for contours in the image that match with the chambers, and once they are found, the algorithm classifies this area as positive (+) or negative (-) (Fig.3.b), depending on the analysed mask. The cartesian position of the samples were also analysed. Since the position of positive control and NTC are always in the same chambers, the software can classify the test as “valid” or “invalid”. For identification accuracy, 27 images containing four samples each were used, totalling 108 samples. A software interpretation accuracy of more than 96% was obtained when compared to results of colorimetric RT-LAMP analysed only by human naked eye. According to this data we can conclude that the developed instrument has the potential to detect as many colors as needed, as long as the “positive” and “negative” color controls are known, and their information is implemented into the algorithm.
3.4. Chip Validation
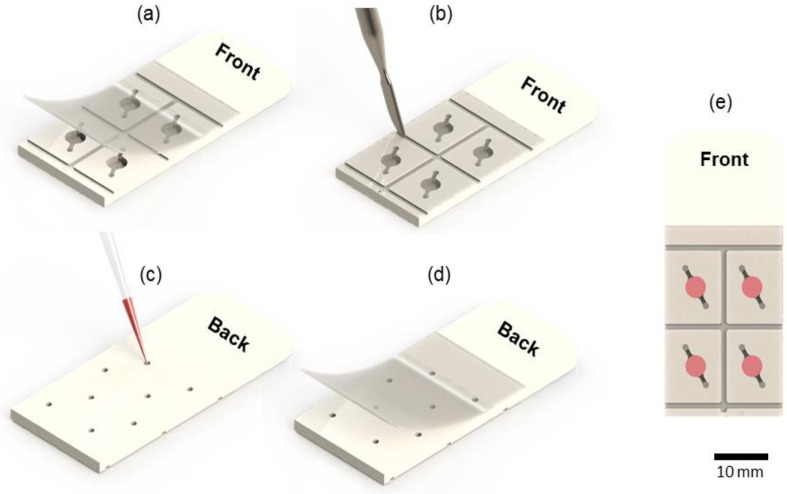
The first step to perform a RT-LAMP test is to seal the chip (Fig.4 .a). A PCR optic plastic sealing film Roche® 480 LightCycler® [28] was used. We tested both optical and non-optical sealing plastics. Non-optical sealing films influenced the reaction changing all results to false positive, whereas optical films did not influence the reaction result. This sealing material was tested on our instrument under the temperature of 65°C for 25 minutes (RT-LAMP standard conditions). It was verified that for temperatures above 75°C, the water vapour pressure formed inside the chip is higher than the plastic film adherence causing leaks. Therefore, the proposed chip design (Fig.4) can be used only for LAMP reactions but not for conventional PCR reactions, which is performed under higher temperatures [41].
Fig. 4.
Chip sealing and loading process. (a) Sealing the front of the chip with PCR optic plastic sealing film. (b) Fixing the sealing film in the grooves. (c) inserting samples. (d) Sealing the back side. (e) chip ready for reaction.
The use of a non-cutting tool, such as a spatula or a ballpoint pen tip can be used to force the plastic inside the grooves (Fig.4.b) improving the sealing. Each chip chamber is filled with the respective sample (Fig.4.c) and sealed on the back (Fig.4.d).
The process of manufacturing the chip through the laser ablation resulted in a rough surface (RMS 30,84µm) of chambers and lateral channels, as we can see in the Scanning Electron Microscopy (SEM) image of the chamber (Figure 3.a in supplementary material) and the grooves (Figure 3.b in supplementary material) (amplification 30X, and inclination of 45° from top view). No chemical treatment was made on the chip to minimize this roughness (RMS), and as far as we know, it did not significantly interfere in the tests.
A kit that also uses a microchip to detect COVID-19 was created by MiCo BioMed Co. Ltd. but the difference is that it was made for Real-time PCR reactions, taking 55 minutes to obtain the result, and testing only one sample at a time [42]. The chip and the equipment developed by us, on the other hand, achieves the result in 25 minutes with a capacity of two samples at a time.
3.5. Validation of SARS-CoV-2 molecular diagnosis using the LAMP-on-a-chip instrument
The SARS-CoV-2 detection by colorimetric RT-LAMP was performed in a conventional thermocycler instrument using nasal and throat swabs samples to establish a gold-standard tool for the chip and colorimetric RT-LAMP validation. Twenty-two clinical samples from Erasto Gaertner Hospital patients were analysed with RT-qPCR and resulted in 11 positive samples and 11 negative samples. All the 11 positive samples had a Ct under 30 for RT-qPCR. Using these pre-tested samples, the RT-LAMP experiments were performed using our LOC instrument.
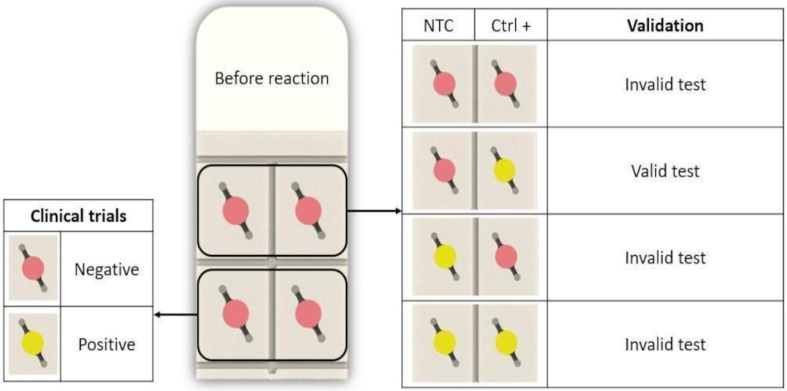
The chip was designed with four (4) chambers, two (2) for the clinical samples and the other two (2) as positive/negative control. On the upper left side, the NTC (Non-Template Control) was performed and on the upper right side the positive control was carried out using SARS-CoV-2 RNA (Fig.5 ). Reaction tests were considered effective for the presence or absence of SARS-CoV-2, the following three cumulative conditions were meet 1) all reactions were pink (pH ∼ 8.0) before incubation, 2) the positive control turned into yellow after incubation (pH 6.0), and 3) the negative control remained pink after the incubation (Fig.5).
Fig. 5.
RT-LAMP Diagnostic verification. The chip is considered “valid” when the negative control (NTC) remains pink after reaction and the positive control (Ctrl +) changes its color to yellow. The sample is considered “negative” when the color is pink and “positive” when color changes to yellow
The overall sensitivity and specificity of colorimetric RT-LAMP using our instrument was 100% and 77% respectively, when compared with RT-qPCR results. There were 3 samples which were negative for RT-qPCR but resulted in false positives samples in our device (Table S4). The colorimetric evaluation was made by our RCCI software when the instrument and the chip reached 25°C.
We also evaluated the limit of detection (LOD) of RT-LAMP for SARS-CoV-2 in our instrument by diluting the virus RNA in ultrapure water to avoid interfering with the colorimetric reaction as previously explained, from 20.000 copies/µl to 2 copies/µl (Table S3). The tests were performed six times for each concentration as shown in Supplementary Table S3. According to the obtained results, the lowest RNA copies number detected by the instrument was 200 copies/µl. This concentration agrees with the previously established RT-LAMP study published by our research group [6] and with Lu et al [43]. The authors obtained the same results targeting the N gene from SARS-CoV-2.
The sensitivity of 200 copies/µl was already expected due to the pH indicator, phenol red. This limitation has been mentioned by previous published studies by our research group [6], [7]. The colorimetric sensitivity depends directly on the viral load of each sample. When a clinical sample has a low viral load, the amplification of the SARS-CoV-2 RNA is not significant enough to change the reaction pH, therefore, the pH indicator is unable to detect it. Colorimetric RT-LAMP has high sensitivity performance with samples which have Ct under 30 for the gold standard method RT-qPCR. For this reason, colorimetric RT-LAMP is advised to be used only in acute cases (first five days of symptoms).
One of the most defiant aspects for POC instruments to merge into a commercial product is to establish a high analytical and diagnostic sensitivity, which must be comparable to gold standard methods. Consequently, emerging LOC devices require large numbers of clinical samples and approval from regulatory agencies, leading to delays in the authorization process [44]. After testing 22 clinical samples our results have already shown promising outcomes. We successfully validated a RT-LAMP reaction within 25 minutes, faster than conventional RT-LAMP for SARS-CoV-2 detection found in literature [38], [45], with the advantage of an automated result interpretation instead of visual diagnostics. Moreover, the instrument’s camera allows the real time observation of the reaction, which may also be an approach to study RT-LAMP reaction’s characteristics regardless of pathogen detection.
The COVID-19 pandemics asserted the demand of LOC instruments, stimulating the development of different devices for screening and prevention of many diseases. Despite this influx of LOC and POC approaches, most of these instruments are not deeply integrated and/or fully automated, showing that there is still space for research and development of new devices [46]. The instrument developed in this study has all the necessary requirements to be considered a POC device and brings new perspectives regarding LOC instrument’s manufacture and development.
4. Conclusions
A LAMP-on-a-Chip instrument was created using a Raspberry Pi 3B+, an Arduino Nano, a camera, a heating board, and machine vision software. PMMA chips (53 x 24 mm2) were fabricated using laser ablation technique and can be manufactured in industrial scale costing less than US$∼0,10 each. To seal our developed chip, no extra glue was required, just a PCR optic plastic sealing film. The chips were successfully tested in our instrument, with samples containing SARS-CoV-2 RNA in at least 200 copies/µl. A PID control system was implemented combined with a temperature calibration system, to precisely control the temperature (65.0 ± 0.1) °C. An image detection software, with machine vision technology, was created for the sample colorimetric detection with an accuracy of 96%. The final instrument was able to heat the samples and to maintain its temperature, while displaying the time reaction, the temperature in real time and the image of the chip on a user’s interface. In the user’s interface it is also possible to easily configure the main settings of the instrument. The results are comparable with other LOC LAMP instruments found in the literature [9]. Our proposed chip proved itself efficient and once that the main model is set, its design can be easily changed, alongside with the instrument temperature range (up to 75 °C). The LOC RT-LAMP instrument is effective, portable, and easy to handle. The instrument works with a 12V external power supply. Although this instrument was designed to operate with our chip containing two controls and two samples, the same principles here applied can be used to run other chip designs. Costing nearly US$∼182 nowadays (in parts), our LOC RT-LAMP instrument was tested successfully in the detection of the SARS-CoV-2 virus, achieving an assertiveness of 86% when analysed twenty-two samples, eleven positive and eleven negative. The overall sensitivity and specificity of our instrument was 100% and 77% respectively, when compared with conventional RT-LAMP results. Thus, the technology presented in this article is a viable alternative for diagnostics in laboratories, hospitals, and local health centers with low financial resources world wise. We also shared the software, firmware, electronic design, and chip design on GitHub (https://github.com/geovanimendonca/Pegasus), repository for open-source projects.
CRediT authorship contribution statement
Geovani Torezin Mendonça: Conceptualization, Data curation, Formal analysis, Methodology, Investigation, Validation, Writing – original draft. Mateus Cassaboni Stracke: Conceptualization, Investigation, Methodology, Writing – original draft. Bruna de Oliveira Coelho: Data curation, Formal analysis, Investigation, Writing – original draft. Heloisa Bruna Soligo Sanchuki: Formal analysis, Validation, Writing – original draft. Viviane Klassen de Oliveira: Formal analysis, Validation, Writing – original draft. Fabricio Klerynton Marchini: Project administration, Writing – original draft. Dalila Lucíola Zanette: Formal analysis, Supervision, Writing – original draft. Mateus Nóbrega Aoki: Formal analysis, Supervision, Writing – original draft. Emilson Ribeiro Viana: Conceptualization, Methodology, Supervision, Writing – original draft. Lucas Blanes: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank to Fiocruz, Fiotec (Brazilian Foundation for the Scientific and Technological Development in Health Grants. VPPIS-004-FIO-18–40 and VPPIS-005-FIO-20–2-19), Brazilian National Council for Scientific and Technological Development (CNPq) (Grant Number 442329/2019–9)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.microc.2022.107600.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Notes and references
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cojocaru R., Yaseen I., Unrau P.J., Lowe C.F., Ritchie G., Romney M.G., Sin D.D., Gill S., Slyadnev M. Microchip RT-PCR detection of nasopharyngeal SARS-CoV-2 Samples. J. Mol. Diagn. 2021;23:683–690. doi: 10.1016/J.JMOLDX.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., Maeng J.-S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B., Thakor A. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 Pandemic. Biology. 2020;9(8):182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki M.N., de Oliveira Coelho B., Góes L.G.B., Minoprio P., Durigon E.L., Morello L.G., Marchini F.K., Riediger I.N., do Carmo Debur M., Nakaya H.I., Blanes L. Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-88506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Oliveira Coelho B., Sanchuki H.B.S., Zanette D.L., Nardin J.M., Morales H.M.P., Fornazari B., Aoki M.N., Blanes L. Essential properties and pitfalls of colorimetric reverse transcription loop-mediated isothermal amplification as a point-of-care test for SARS-CoV-2 diagnosis. Mol Med. 2021;27(1) doi: 10.1186/s10020-021-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Fohlerová Z., Pekárek J., Basova E., Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020;153 doi: 10.1016/J.BIOS.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safavieh M., Kanakasabapathy M.K., Tarlan F., Ahmed M.U., Zourob M., Asghar W., Shafiee H. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomater. Sci. Eng. 2016;2:278–294. doi: 10.1021/ACSBIOMATERIALS.5B00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Ma Z., Qin J., Shen Z., Liu Q., Chen X., Wang H., An Z., Liu W., Li M. A versatile loop-mediated isothermal amplification microchip platform for Streptococcus pneumoniae and Mycoplasma pneumoniae testing at the point of care. Biosens. Bioelectron. 2019;126:373–380. doi: 10.1016/J.BIOS.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., de Ramirez S.A.S., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:22727–22735. doi: 10.1073/PNAS.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papadakis G., Pantazis A.K., Fikas N., Chatziioannidou S., Michaelidou K., Pogka V., Megariti M., Vardaki M., Giarentis K., Heaney J., Nastouli E., Karamitros T., Mentis A., Agelaki S., Gizeli E. Real-time colorimetric LAMP methodology for quantitative nucleic acids detection at the point-of-care. BioRxiv. 2020 doi: 10.1101/2020.07.22.215251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Mateos P., Ngamsom B., Walter C., Dyer C.E., Gitaka J., Iles A., Pamme N. A lab-on-a-chip platform for integrated extraction and detection of SARS-CoV-2 RNA in resource-limited settings. Anal. Chim. Acta. 2021;1177 doi: 10.1016/J.ACA.2021.338758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Manzano J., Malpartida-Cardenas K., Moser N., Pennisi I., Cavuto M., Miglietta L., Moniri A., Penn R., Satta G., Randell P., Davies F., Bolt F., Barclay W., Holmes A., Georgiou P. Handheld Point-of-Care System for Rapid Detection of SARS-CoV-2 Extracted RNA in under 20 min. ACS Cent. Sci. 2021;7:307–317. doi: 10.1021/ACSCENTSCI.0C01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Q.I., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21(9):1634–1660. doi: 10.1039/d0lc01156h. [DOI] [PubMed] [Google Scholar]
- 17.E. Team, Materials for microfluidic chips fabrication : a review 2017, Elveflow. (2021). https://www.elveflow.com/microfluidic-reviews/general-microfluidics/materials-for-microfluidic-chips-fabrication-a-review-2017/ (accessed September 28, 2021).
- 18.Hong T.-F., Ju W.-J., Wu M.-C., Tai C.-H., Tsai C.-H., Fu L.-M. Rapid prototyping of PMMA microfluidic chips utilizing a CO2 laser. Microfluid Nanofluid. 2010;9(6):1125–1133. [Google Scholar]
- 19.Matellan C., del Río Hernández A.E. Cost-effective rapid prototyping and assembly of poly(methyl methacrylate) microfluidic devices. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-25202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathur A., Roy S.S., Tweedie M., Mukhopadhyay S., Mitra S.K., McLaughlin J.A. Characterisation of PMMA microfluidic channels and devices fabricated by hot embossing and sealed by direct bonding. Curr. Appl Phys. 2009;9:1199–1202. doi: 10.1016/J.CAP.2009.01.007. [DOI] [Google Scholar]
- 21.Trinh K.T.L., Thai D.A., Chae W.R., Lee N.Y. Rapid fabrication of poly(methyl methacrylate) devices for lab-on-a-chip applications using acetic acid and UV treatment. ACS Omega. 2020;5:17396–17404. doi: 10.1021/ACSOMEGA.0C01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen T., Jung S.H., Lee M.S., Park T.-E., Ahn S., Kang J.H. Robust chemical bonding of PMMA microfluidic devices to porous PETE membranes for reliable cytotoxicity testing of drugs. Lab Chip. 2019;19:3706–3713. doi: 10.1039/C9LC00338J. [DOI] [PubMed] [Google Scholar]
- 23.Ng S.P., Wiria F.E., Tay N.B. Low distortion solvent bonding of microfluidic chips. Procedia Eng. 2016;141:130–137. doi: 10.1016/J.PROENG.2015.09.212. [DOI] [Google Scholar]
- 24.Zhang H., Xu Y., Fohlerova Z., Chang H., Iliescu C., Neuzil P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. TrAC, Trends Anal. Chem. 2019;113:44–53. doi: 10.1016/J.TRAC.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang X., Chen H., Yu S., Jiang X., Kong J. Predicting viruses accurately by a multiplex microfluidic loop-mediated isothermal amplification chip. Anal. Chem. 2010;83:690–695. doi: 10.1021/AC102858J. [DOI] [PubMed] [Google Scholar]
- 26.Song J., Mauk M.G., Hackett B.A., Cherry S., Bau H.H., Liu C. Instrument-free point-of-care molecular detection of zika virus. Anal. Chem. 2016;88:7289–7294. doi: 10.1021/ACS.ANALCHEM.6B01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graphic Design, Illustration and Technical Software | CorelDRAW, (n.d.). https://www.coreldraw.com/en/?link=wm (accessed September 30, 2021).
- 28.LightCycler® 480 Sealing Foil, (n.d.). https://lifescience.roche.com/en_br/products/lightcycler14301-480-sealing-foil.html (accessed September 30, 2021).
- 29.QIAamp Viral RNA Kits, (n.d.). https://www.qiagen.com/us/products/diagnostics-and-clinical-research/sample-processing/qiaamp-viral-rna-kits/ (accessed September 28, 2021).
- 30.Rabe B.A., Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. 2020;117:24450–24458. doi: 10.1073/PNAS.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welcome to Python.org, (n.d.). https://www.python.org/ (accessed September 28, 2021).
- 32.Home - OpenCV, (n.d.). https://opencv.org/ (accessed September 28, 2021).
- 33.Kivy: Cross-platform Python Framework for NUI Development, (n.d.). https://kivy.org/#home (accessed September 28, 2021).
- 34.DipTrace - Schematic and PCB Design Software, (n.d.). https://diptrace.com/ (accessed September 28, 2021).
- 35.AutoCAD Software | Get Prices & Buy Official AutoCAD 2022, (n.d.). https://www.autodesk.com/products/autocad/overview?term=1-YEAR&tab=subscription (accessed September 28, 2021).
- 36.PID - Arduino Reference, (n.d.). https://www.arduino.cc/reference/en/libraries/pid/ (accessed September 28, 2021).
- 37.Creality Ender-3 Pro, (n.d.). https://www.creality.com/br/goods-detail/ender-3-pro-3d-printer (accessed September 30, 2021).
- 38.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/SCITRANSLMED.ABC7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardy T., Tulp I., Kremer C., Rang T., Stewart R., Isalan M. Integrated self-regulating resistive heating for isothermal nucleic acid amplification tests (NAAT) in Lab-on-a-Chip (LoC) devices. PLoS ONE. 2017;12(12):e0189968. doi: 10.1371/journal.pone.0189968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. Liu, W. Zhang, J. Qiu, Q. Hu, K. He, Reagent color recognition model for android platform based on OPENCV and machine learning, Proceedings - 2019 International Conference on Machine Learning, Big Data and Business Intelligence, MLBDBI 2019. (2019) 270–273. https://doi.org/10.1109/MLBDBI48998.2019.00060.
- 41.Petrillo S., Carrà G., Bottino P., Zanotto E., De Santis M.C., Margaria J.P., Giorgio A., Mandili G., Martini M., Cavallo R., Barberio D., Altruda F. A novel multiplex qRT-PCR assay to detect SARS-CoV-2 infection: high sensitivity and increased testing capacity. Microorganisms. 2020;8(7):1064. doi: 10.3390/microorganisms8071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21:1634–1660. doi: 10.1039/D0LC01156H. [DOI] [PubMed] [Google Scholar]
- 43.Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. C, A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21(8):2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vashist S.K., Luppa P.B., Yeo L.Y., Ozcan A., Luong J.H.T. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015;33:692–705. doi: 10.1016/J.TIBTECH.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Klein S., Müller T.G., Khalid D., Sonntag-Buck V., Heuser A.-M., Glass B., Meurer M., Morales I., Schillak A., Freistaedter A., Ambiel I., Winter S.L., Zimmermann L., Naumoska T., Bubeck F., Kirrmaier D., Ullrich S., Barreto Miranda I., Anders S., Grimm D., Schnitzler P., Knop M., Kräusslich H.-G., Dao Thi V.L., Börner K., Chlanda P. SARS-CoV-2 RNA Extraction Using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12(8):863. doi: 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C., Liu M., Wang Z., Li S., Deng Y., He N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today. 2021;37 doi: 10.1016/J.NANTOD.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.