Abstract
Surface-enhanced Raman scattering (SERS)-based assays have been recently developed to overcome the low detection sensitivity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SERS-based assays using magnetic beads in microtubes slightly improved the limit of detection (LoD) for SARS-CoV-2. However, the sensitivity and reproducibility of the method are still insufficient for reliable SARS-CoV-2 detection. In this study, we developed a SERS-based microdroplet sensor to dramatically improve the LoD and reproducibility of SARS-CoV-2 detection. Raman signals were measured for SERS nanotags in 140 droplets passing through a laser focal volume fixed at the center of the channel for 15 s. A comparison of the Raman signals of SERS nanotags measured in a microtube with those measured for multiple droplets in the microfluidic channel revealed that the LoD and coefficient of variation significantly improved from 36 to 0.22 PFU/mL and 21.2% to 1.79%, respectively. This improvement resulted from the ensemble average effects because the signals were measured for SERS nanotags in multiple droplets. Moreover, the total assay time decreased from 30 to 10 min. A clinical test was performed on patient samples to evaluate the clinical efficacy of the SERS-based microdroplet sensor. The assay results agreed well with those measured by the reverse transcription-polymerase chain reaction (RT-PCR) method. The proposed SERS-based microdroplet sensor is expected to be used as a new point-of-care diagnostic platform for quick and accurate detection of SARS-CoV-2 in the field.
Keywords: Surface-enhanced Raman scattering, Microdroplet sensor, Magnetic bead, SERS nanotag, SARS-CoV-2
1. Introduction
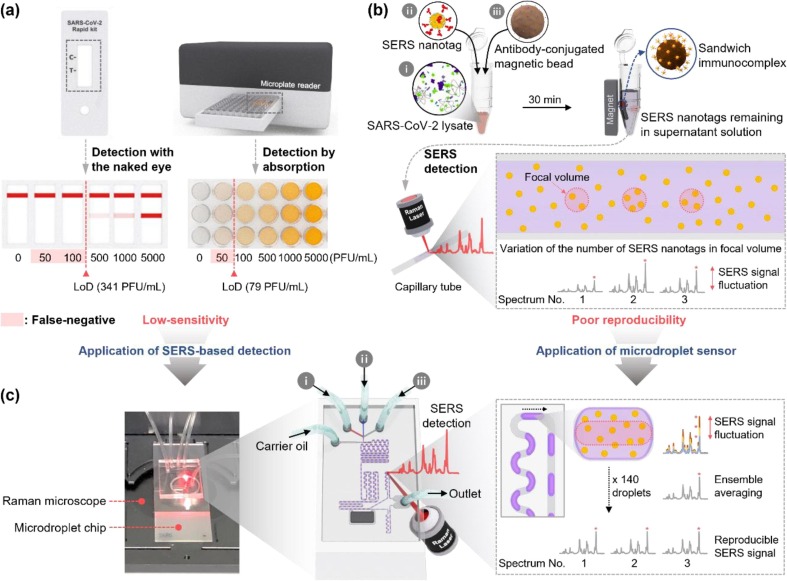
The worldwide spread of coronavirus disease 2019 (COVID-19) pandemic, a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in enormous economic, social, and cultural losses over the past two years [1], [2], [3], [4]. The reverse transcription-polymerase chain reaction (RT-PCR) method, which extracts and amplifies viral ribonucleic acid (RNA) and subsequently detects it using the fluorescence analysis, has been accepted as a standard method for SARS-CoV-2 detection. RT-PCR exhibits a reasonable sensitivity and specificity; however, it requires 3–4 h for sample pre-treatment, amplification, and detection [5], [6], [7]. Therefore, many studies have been conducted to reduce the diagnosis time using modern technologies, such as isothermal PCR [8], [9], genome editing [10], [11], and photonic PCR [12], [13]. Nevertheless, achieving a significant diagnosis time reduction using molecular diagnostic technologies is challenging. The lateral flow assay (LFA) strip that enables self-diagnosis based on immunoassays has been commercialized [14], [15], [16]; however, this point-of-care (POC) test is not considered a reliable diagnostic tool owing to its low sensitivity. Currently, the limit of detection (LoD) of an LFA strip is in the range of 300–500 PFU/mL (left, Scheme 1 a), leading to false-negative results for early-stage patients with low virus concentrations or asymptomatic infected patients [17], [18]. An enzyme-linked immunosorbent assay (ELISA) is another immunoassay method used for SARS-CoV-2 testing. Although the LoD of an ELISA kit is approximately 80 PFU/mL (right, Scheme 1 a), the proportion of false-negative diagnosis of this method remains high [19], [20]. Consequently, the critical issue in immunoassays for SARS-CoV-2 is the improvement of sensitivity.
Scheme 1.
(a) LFA strips and ELISA using a 96-well plate, (b) magnetic bead-based SERS assay platform in a microtube, and (c) microdroplet SERS sensor for the immunodiagnostic test of SARS-CoV-2. In the SERS assay in a microtube, the Raman signal of SERS nanotags in the focal volume is measured after transferring the supernatant solution to the capillary tube. The number of particles present in the focal volume varies along the measurement area. Hence, the reproducibility decreases owing to the signal intensity fluctuations. Reproducibility is significantly improved in the microdroplet sensor owing to the ensemble average effects because the Raman signal of SERS nanotags, contained in 140 droplets that continuously pass through the focal volume of the laser, is measured.
Many studies have recently demonstrated that the surface-enhanced Raman scattering (SERS) technique can be used for high-sensitivity biomedical detection of SARS-CoV-2 based on localized surface plasmon effects [21], [22], [23]. Raman signals are amplified using the electromagnetic enhancement when Raman reporter molecules present in the hot spot are exposed to a focal volume of an excitation laser beam. This amplification can resolve the low sensitivity issue inherent in absorbance or fluorescence detection, extensively used in immunoassays [24], [25], [26]. Hence, a magnetic bead-based SERS assay platform in a microtube for SARS-CoV-2 detection was developed [27], [28], [29]. In this assay, magnetic beads and SERS nanotags are used as capture substrates and detection probes, respectively. Then, sandwich immunocomplexes for the target SARS-CoV-2 antigen are formed on the surface of magnetic beads. Finally, the sandwich immunocomplexes are separated using a magnetic bar. Subsequently, the decrease in signal intensity from SERS nanotags in the supernatant solution is measured for quantitative analysis (Scheme 1 b). The magnetic bead-based SERS assay platform in a microtube could successfully detect SARS-CoV-2; however, there is room for improvement because of i) insufficient sensitivity due to the limited number of SERS nanotags in a focal volume, ii) poor reproducibility owing to fluctuation of the number of SERS nanotags in repeated measurements, and iii) difficulty of high throughput and POC diagnosis owing to large reagent consumption and labor-required experimental processes.
In this study, we developed a SERS-based microdroplet sensor for rapid, sensitive, and reproducible SARS-CoV-2 detection [30], [31], [32]. There have been many reports of a SERS detection method for diagnosing SARS-CoV-2 [33], [34], [35], but very few papers about high-sensitivity diagnosis using a SERS-based microfluidic platform. The supernatant solution containing SERS nanotags was divided into multiple droplets of equal volume in a microfluidic channel. Subsequently, SERS signals were measured for a certain number of droplets (Scheme 1 c) to improve the detection reproducibility. In particular, the detection sensitivity of this sensor significantly improved owing to ensemble average effects because the Raman signal was measured for multiple microdroplets containing SERS nanotags. We carefully optimized the SERS-based microdroplet sensor, and thus succeeded in SARS-CoV-2 detection at a low concentration of 0.22 PFU/mL and a coefficient of variation (CV) of 1.79%. These values are considerably lower than those of LFA strip, ELISA kits, and typical magnetic bead-based SERS assay platform in a microtube. The improved sensitivity and reproducibility of the sensor yield an accurate detection of SARS-CoV-2. A clinical test was performed on patient samples to evaluate the clinical efficacy of the SERS-based microdroplet sensor. The microdroplet sensor's assay results were compared with those measured by the RT-PCR method. Moreover, the sensing performance of the SERS-based microdroplet sensor was evaluated using a portable Raman system within 10 min. This proves the practical applicability of the current technique in the POC detection platform for SARS-CoV-2. The proposed SERS-based microdroplet sensor is expected to be used as a high-throughput POC diagnostic platform for various infectious diseases. Furthermore, this technology can be extended to multiple bio targets by changing receptors, multiplexed by increasing channels, and automated by integrating devices.
2. Experimental section
2.1. Reagent and materials
Gold(III) chloride trihydrate (HAuCl4·3H2O), sodium citrate dihydrate (99%), bovine serum albumin (BSA), thiol-PEG-COOH (HS-PEG-COOH, MW ∼3500), cobalt(II) chloride hexahydrate, 1H,1H,2H,2H-perfluorooctanol (PFO, 97%), perfluorodecalin (PFD, 95%), trichloro(1H,1H,2H,2H-perfluorooctyl)silane (97%), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS), and fluorescein sodium salt (FSS) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and used without further purification. Magic™ mouse anti-SARS-CoV-2 nucleoprotein monoclonal antibody (clone 0659, CABT-C S075) and Magic™ mouse anti-SARS-CoV-2 nucleoprotein monoclonal antibody (clone 0658, CABT-C S076) were purchased from Creative Diagnostics (Shirley, NY, USA). SARS-CoV-2 lysates were supplied by the Korea Research Institute of Bioscience and Biotechnology (KRIBB). Carboxylate magnetic beads, phosphate-buffered saline (PBS; 10X, pH 7.4), and malachite green isothiocyanate (MGITC) were purchased from Invitrogen Corporation (Eugene, OR, USA). The average diameter of magnetic beads was 1 μm. PBS solutions containing 0.05% Tween-20 (v/v) at a pH of 7.4 were prepared using standard protocols. Polydimethylsiloxane (PDMS, Sylgard 184 silicone elastomer kit) was purchased from Dow Corning (Midland, MI, USA). FC-40 (a mixture of perfluoro-tri-n-butylamine and perfluoro-di-n-butylmethylamine) and FC-70 (perfluorotripentylamine) were purchased from 3M (Maplewood, MN, USA). Ultrapure deionized (DI) water was obtained using the Arium Comfort Lab Water System (Sartorius, Goettingen, Germany), and used in all experiments. A nickel-coated neodymium (ND) magnet (4 × 1.9 × 0.7 mm3) was purchased from the LG magnet (Seoul, Korea). Gyeongsang National University College of Medicine provided 6 clinical samples (4 SARS-CoV-2 positives and 2 negatives). The nasopharyngeal aspirates samples were collected from patients using flocked nasopharyngeal swabs and placed into the virus transport media (Copan Diagnostics Inc., Murrieta, CA, USA). All samples were stored at −70 °C until use. The protocol for this study was reviewed and approved by the Institutional Review Board of Gyeongsang National University College of Medicine, Jinju, Korea (IRB approval number: 2020-10-002).
2.2. Preparation of detection antibody-conjugated hollow gold nanospheres
Cobalt nanoparticles (CoNPs) were synthesized based on a previously proposed method [36], [37]. During the synthesis of CoNPs, 0.1 M sodium citrate solution was deoxygenated using ultrapure N2 gas for 60 min to guarantee oxygen purging. Subsequently, 140 μL of 1.0 M NaBH4 and 100 μL of 0.4 M CoCl2 solution were rapidly added while stirring. The color of the mixture changed from colorless to dark brown, indicating the formation of CoNPs. The mixture was treated with N2 gas flow for 60 min to hydrolyze the remaining NaBH4. Subsequently, 50 μL of 0.1 M HAuCl4 solution was added nine times at intervals of 80 s. The flow of N2 gas stopped after the solution was stirred for 10 min. This completed the formation of a gold shell and exposed the remaining CoNPs to an oxidizing environment. The color of the solution changed to dark purple when the reaction was complete. The transmission electron microscopy (TEM) image demonstrates that the average size of the hollow gold nanospheres (HGNs) and Au shell thicknesses are 44 and 8 nm, respectively (Fig. S1 a). HGNs were washed by centrifugation (1,200 rcf, 30 min) at 25 °C before using to remove byproducts, such as chloride ions and CO2 generated in the process of oxidizing CoNPs. Three microliters of 10-4 M MGITC were adsorbed on the surface of HGNs to act as a Raman reporter when the HGNs were used as SERS-active probes. Sixty microliters of 10 µM HS-PEG-COOH linkers were added to this solution to form Au-S bonds on the surface of HGNs. PEGylated HGNs were created after 3 h of reaction at 25 °C, and washed twice with DI water. Moreover, 10 µL of 2.5 mM EDC and 10 µL of 2.5 mM NHS were sequentially added to activate the -COOH group at the end of the linker. The remaining EDC/NHS was washed with DI water after 30 min. Twenty microliters of 0.1 mg/mL detection antibodies were added to NHS-activated HGNs and incubated overnight at 4 °C for antibody conjugation. The surface of HGNs, to which antibodies were not attached, was blocked by adding 10 µL of 10% w/v BSA aqueous solution. The remaining proteins were removed by centrifugation at 1,200 rcf for 30 min. The pellets were re-dispersed in PBS after discarding the supernatant.
2.3. Preparation for capturing antibody-conjugated magnetic beads
To conjugate capture antibodies on the surfaces of magnetic beads, 500 μL of 0.5 mg/mL carboxyl-functionalized magnetic beads were suspended in DI water. Carboxylic groups on the surfaces of magnetic beads were activated by adding 15 μL of 0.1 M EDC and NHS dissolved in distilled water for 60 min. Subsequently, magnetic beads were separated using a magnet and washed with DI water to remove any unreacted molecules. After magnetic beads were re-suspended using 320 μL of DI water, 180 μL of 2.5 mg/mL capture antibodies were added to the magnetic bead solution and reacted overnight at 4 ℃. Furthermore, the capture antibody-conjugated magnetic beads were washed three times to remove nonspecific antibodies. The final product was stored in PBS buffer solution for future use.
2.4. Fabrication of magnetic bar-embedded droplet microfluidic sensor
A positive SU-8 50–100 photoresist (MicroChem Corp.) mold was fabricated using standard photolithography. Subsequently, the PDMS pre-polymer and curing agent (Sylgard 184, Dow Corning) were mixed in a ratio of 10:1 w/w, degassed under vacuum, and decanted onto the SU-8 mold. An ND magnet (4 × 1.9 × 0.7 mm3) was inserted into the magnet groove of the SU-8 mold. The PDMS layer with a channel structure was peeled from the master mold after curing at 70 °C for 2 h in an oven. Subsequently, the inlet and outlet holes were punched, and the structured PDMS substrate was bonded to a glass slide using the oxygen plasma. For hydrophobic treatment of the inner surface of the channel, 2% v/v trichloro(1H,1H,2H,2H-perfluorooctyl)silane in a PFD solution was filled in the channel and placed on a hotplate at 70 °C. The width of the main channel was 200 μm, whereas that of branch channels were 140 and 78 μm. The depth of all channels was 128 μm. A 10:20:3 v/v mixture of FC-40, FC-70, and 1H,1H,2H,2H perfluorooctanol (PFO) was used as the carrier oil.
2.5. Magnetic bead-based immunoassays in a microtube and SERS detection
Sandwich immunoassays for SARS-CoV-2 nucleoprotein were performed using SARS-CoV-2 lysates at six different concentrations. Furthermore, 30 µL of SERS nanotags, 30 µL of SARS-CoV-2 lysates, and 30 µL of antibody-conjugated magnetic beads were mixed and allowed to react under constant stirring. The reaction tube was placed on a magnet for 3 min, and the supernatant solution was collected after 30 min of incubation. The magnetic beads that formed sandwich immunocomplexes were washed three times using PBS, and the immunocomplexes were re-suspended in PBS. The separated supernatant solution and immunocomplexes were transferred to a capillary tube for Raman measurements. SERS signals were collected using a Renishaw inVia Raman microscope (Renishaw, New Mills, UK). A 10 mW He−Ne laser, operating at a wavelength of 632.8 nm, was used as the excitation source. A 20× objective lens, with a numerical aperture of 0.4, was used to focus the laser beam at the center of the capillary tube. The exposure time was 10 s with five averages, and the focal laser spot size was 1.93 μm. The Renishaw WiRE 4.0 software was used for data acquisition and control.
2.6. Fluorescence imaging of droplet generation/splitting and on-chip SERS detection
An Olympus IX71 inverted fluorescence microscope (Olympus, Japan) equipped with a high-speed camera (PCO AG, Germany) was used to record the droplet generation and splitting, and to measure droplet volumes and velocities. The samples were injected into the droplet channel using precision syringe pumps (PHD 2000, Harvard Apparatus, USA), 1 mL Norm-Ject plastic syringes (Henke-Sass Wolf GmbH, Germany), 23G needles (KOVAX NEEDLE, Korea Vaccine Co., Ltd., Korea), and TYGON tubing (ID = 0.02 IN/OD = 0.06 IN, Saint-Gobain PPL Corp., France). The SERS signals from droplets were collected using a portable Raman microscope (NS220-I Nanoscope Systems, Inc., Daejeon, Korea). A 16 mW He−Ne laser, operating at a wavelength of 633 nm, was used as the excitation source. A 20× objective lens, with a numerical aperture of 0.45, was used to focus the laser beam at the center of the microchannel. The exposure time was 5 s with three averages, and the focal laser spot size was 1.68 μm. The NS-Raman (Ver. 4.04) software was used for data acquisition and control. In addition, flow rates were simultaneously controlled using two microsyringe pumps.
3. Results and discussion
3.1. SERS-based magnetic assays of SARS-CoV-2 in a microtube
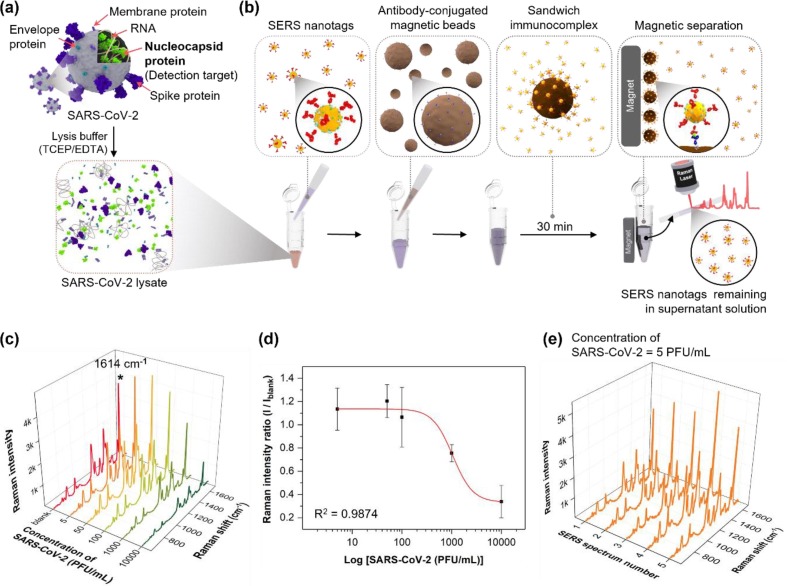
Fig. 1 illustrates the SERS-based assay for detecting SARS-CoV-2 using magnetic beads in a microtube. SARS-CoV-2, which was lysed using a TCEP/EDTA solution (Fig. 1 a), includes envelope proteins, membrane proteins, spike proteins on the virus surface, RNA, and nucleocapsid protein (N-protein). Here, N-protein was used as a target for SARS-CoV-2 detection because this protein is abundant inside SARS-CoV-2. Therefore, it is the most commonly used target in commercially available LFA strips [38], [39]. Most SARS-CoV-2 mutations occur in the spike protein, but the proposed SERS-based microdroplet sensor in this study targets the N-protein inside SARS-CoV-2, and the binding affinity of N-protein is less affected by mutation than spike protein. A SERS-based immunoassay for detecting SARS-CoV-2 was performed using SERS nanotags and magnetic beads in a microtube (Fig. 1 b). Detection antibody-conjugated HGNs were used as SERS nanotags to create electromagnetic enhancements. HGNs have multiple pinholes on the particle surface that intensify the incident light inside the holes of narrow nanogaps. Hence, sensitive assays with high reproducibility can be achieved at the single-particle level. Fig. S1 illustrates (a) transmission electron microscope (TEM) images, (b) dynamic light scattering (DLS) distribution data, and (c) UV-vis absorption spectra of HGNs and SERS nanotags. The TEM images illustrate that HGNs have an average diameter of 44 nm and a wall thickness of approximately 8 nm. Fig. S1 b demonstrates that the diameter increases by approximately 17 nm when the detection antibodies are conjugated on the HGN surface. The change in spectral data owing to conjugation is presented in the UV-vis spectra in Fig. S1 c. The absorption band was redshifted by 5 nm from 573 to 578 nm when the detection antibodies were conjugated on the surface. Therefore, we validated the successful synthesis of SERS nanotags.
Fig. 1.
SERS-based assay for detecting SARS-CoV-2 using magnetic beads in a microtube. (a) Lysis of SARS-CoV-2 using a TCEP/EDTA solution. (b) SERS-based immunoassay process of SARS-CoV-2 using SERS nanotags and magnetic beads in a microtube. (c) SERS spectra of the supernatant solution in a capillary tube measured in the SARS-CoV-2 concentration range of 0–10,000 PFU/mL. (d) Calibration curve for the immunoassay of SARS-CoV-2 determined using the four-parameter sigmoidal fitting equation. (e) SERS spectra measured for a supernatant solution at five different positions of the capillary tube after magnetic bead-based SERS assay was performed on a SARS-CoV-2 concentration of 5 PFU/mL in a microtube.
The other critical issue of SERS nanotags is their stability in solution. Raman reporter molecules (MGITCs) dissociate from the surface of the HGNs when exposed to high NaCl concentrations or harsh pH conditions. To resolve this issue, PEGylated HGN-MGITC (HGN-MGITC-PEG) has been developed to prevent the desorption of MGITC molecules and the adsorption of external species. The stability of HGN-MGITC and HGN-MGITC-PEG was ested under various pH and NaCl concentrations. As shown in Fig. S2, the UV/vis absorption intensity was significantly decreased with increased salt concentrations in HGN-MGITC (Fig. S2 a) since particles started to aggregate at a concentration of 40 mM NaCl. However, the UV/vis absorption intensity was not significantly changed for HGN-MGITC-PEG (Fig. S2), even under high NaCl concentrations. Fig. S2 b also shows the changes in the UV/vis absorption intensity when the pH of the solution was changed. In the case of HGN-MGITC, the UV/vis absorption intensity was significantly decreased at low pH conditions because hydrogen ions quickly neutralized the negative surface charge. However, for the HGN-MGITC-PEG, the decreasing UV/vis absorption intensity trend was much slower than that of the HGN-MGITC at low pH conditions. Therefore, we can conclude that the HGN-MGITC-PEG maintains good stability under extreme NaCl and pH conditions.
Moreover, 1-μm capture antibody-conjugated magnetic beads were used as support substrates for SERS-based assays. The N-proteins were selectively captured by SERS nanotags when a SARS-CoV-2 lysate reacted with them. The capture antibody-conjugated magnetic beads were added to the microtube and allowed to react for 30 min. Subsequently, sandwich immunocomplexes were formed on the surface of magnetic beads. Fig. S3 a presents the TEM images obtained after performing a SERS-based assay in the presence (1,000 PFU/mL) and absence of the target SARS-CoV-2. This figure demonstrates that SERS nanotags form sandwich immunocomplexes on the surface of magnetic beads only when the target SARS-CoV-2 is present. The number of immunocomplexes produced was proportional to the concentration level of SARS-CoV-2. Immunocomplexes were separated using a magnetic bar on the outer wall of the microtube. Then, they were washed with PBS buffer using a micropipette and re-dispersed in PBS buffer. The re-dispersed magnetic immunocomplexes were transferred to a capillary tube, and the SERS signal was measured. The number of SERS nanotags formed on the magnetic bead surface increased, enhancing the SERS signal intensity and increasing the concentration of SARS-CoV-2. Raman spectra for magnetic beads with SERS nanotags were measured and compared with those of magnetic beads only in Fig. S3 b. In the case of off-mode, the overall SERS peak intensity is relatively weak, but when SERS nanotags form sandwich immunocomplexes (on-mode), it shows a strong SERS intensity. Fig. S3 c compares the characteristic Raman peak intensity at 1614 cm-1 of SERS nanotags for SARS-CoV-2 between off- and on- modes.
In the second method, magnetic immunocomplexes were separated from the solution, and the remaining supernatant solution was directly transferred to a capillary tube to measure the SERS signal. The SERS signal intensity decreased with an increase in the SARS-CoV-2 concentration. However, both methods exhibited an insignificant reproducibility because the number of SERS nanotags in the focal volume of the capillary tube was not uniform. Fig. 1 c illustrates the SERS spectra measured using the second method in a SARS-CoV-2 concentration in the range of 0–10,000 PFU/mL. Fig. 1 d shows the calibration curve determined using the three-parameter sigmoidal fitting equation. The variations in the characteristic Raman peak intensity at 1614 cm-1 of SERS nanotags for the concentration of SARS-CoV-2 lysate is plotted in Fig. 1 d. The error bars represent standard deviations determined using five measurements. As shown in Fig. 1 d, significant deviations in the error bars were observed over the entire concentration range owing to variations in the number of SERS nanotags in the focal detection volume of the laser beam. Fig. 1 e illustrates the SERS spectra measured for a supernatant solution at five different positions of the capillary tube after the magnetic bead-based SERS assay was performed on a SARS-CoV-2 concentration of 5.0 PFU/mL in a microtube. Fig. 1 e demonstrates that the Raman peak intensity at 1614 cm-1 significantly fluctuates for each measurement.
Raman spectra were measured by re-dispersing magnetic immunocomplexes in PBS and transferring them to a capillary tube, as shown in Fig. S4 a. The calibration curve is determined using the method illustrated in Fig. S4 b. Therefore, this assay technique marginally improved the correlation coefficient for the SARS-CoV-2 lysate. However, reproducibility was low because of large standard deviations for each concentration. Moreover, the LoD determined using the standard deviation was slightly improved to 36 PFU/mL because the number of SERS nanotags in a focal detection volume is limited. Therefore, a new analytical technique to improve the sensitivity and reproducibility of SARS-CoV-2 detection is necessary.
3.2. Mixing/splitting efficiency of SERS-based microdroplet sensor
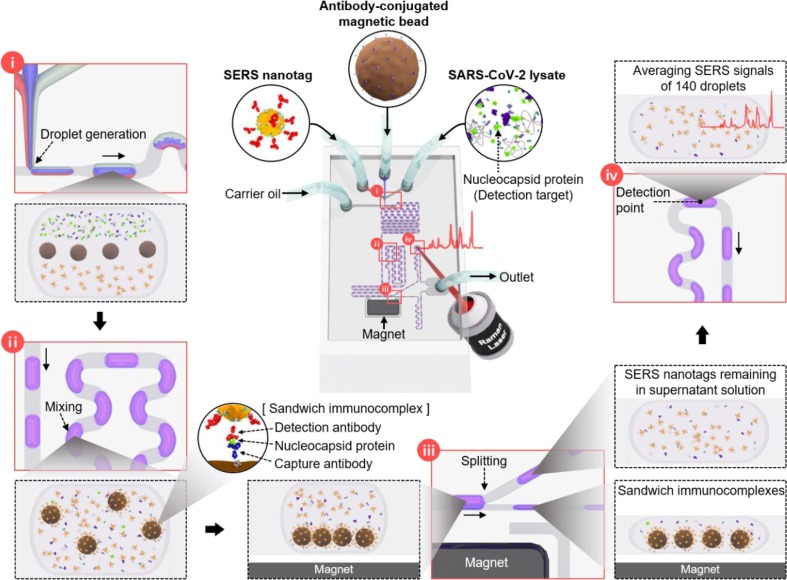
Fig. 2 illustrates the schematic design of the microdroplet sensor used to detect SARS-CoV-2. This microfluidic channel consists of four compartments: (i) droplet generation, (ii) droplet mixing, (iii) droplet splitting, and (iv) optical signal measurements. In particular, (i) SERS nanotags, capture antibody-conjugated magnetic beads, and SARS-CoV-2 lysate, all in aqueous phases, were injected from three different inlets to form a laminar flow. Subsequently, the carrier oil was injected through the inlet perpendicular to the aqueous phase to generate aqueous phase droplets at the T-junction. Herein, the inner surface of the microfluidic channel was treated with 2% v/v trichloro(1H,1H,2H,2Hperfluorooctyl) silane in a perfluorodecalin solution. Therefore, the interaction between N-protein and PDMS sensor does not occur because the immunoassay occurs in the aqueous phase droplet flowing along with the carrier oil. (ii) The laminar flow generated in the droplet passes through the winding channel, resulting in turbulent mixing that actively promotes the reaction. (iii) SERS nanotags, SARS-CoV-2 lysates, and capture antibody-conjugated magnetic beads forming immunocomplexes were separated using a magnetic bar embedded in the channel and moved to the lower narrow channel. The supernatant solution was moved to the upper wide channel. (iv) Raman signals of the SERS nanotags remaining in the supernatant solution were measured. One hundred and forty droplets passed through the focal volume at a fixed laser beam position, and their Raman signals were measured for 15 s.
Fig. 2.
Schematic design of the microdroplet channel used to detect SARS-CoV-2. This microfluidic channel consists of four compartments: (i) droplet generation, (ii) droplet mixing, (iii) droplet splitting, and (iv) optical signal measurements.
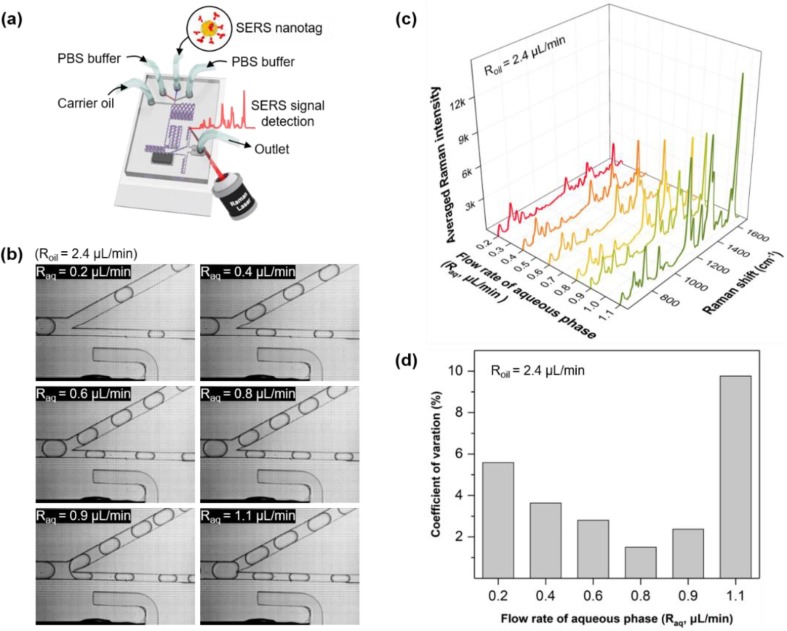
Microdroplet experiments were performed by exclusively injecting SERS nanotags and PBS buffer through the inlets to determine the optimal conditions for driving the chip, as shown in Fig. 3 a. Subsequently, flow rates of the aqueous and oil phases were varied using two syringe pumps. CVs of SERS intensities were analyzed to determine the optimum flow rate of the aqueous phase (Raq). Table 1 lists the variations of Raq in the range of 0.2–1.1 μL/min when the oil flow rate (Roil) was maintained at 2.4 μL/min. Fig. 3 b presents the photographic images of six different Raqs when each droplet passes through the droplet splitting junction, as shown in Fig. 2(iii). With an increasing flow rate, the size of droplets increases and the spacing between droplets decreases. After droplet splitting, regular intervals between droplets are maintained up to a flow rate of 0.8 μL/min. However, the intervals become irregular at flow rates higher than 0.9 μL/min because of changes in the channel hydrodynamic resistance [40], [41]. The resistance inside the channel varies after aqueous droplets pass through the splitting junction at a high flow rate. This change affects the movement of the next droplet entering the split intersection. This movement change results in an irregular spacing between droplets. However, constant droplet spacings are maintained at a low flow rate owing to the low hydrodynamic resistance and negligible feedback effect. The SERS signal intensity is determined by the ensemble average of droplets passing through the focal volume of the laser beam for a certain time. Hence, the reproducibility of the signal is poor because of the irregular spacing between droplets at a fast flow rate. Fig. 3 c illustrates the SERS spectra measured for six different flow rates for 15 s. The number of droplets passing through the focal volume fixed at the center of the channel during a specified measurement time increases, corresponding to an increase in flow rate. This flow rate effect leads to a rise in SERS signal intensity. Fig. 3 d demonstrates variations of the CV values corresponding to the SERS intensity for changes in Raq. Moreover, CV is determined using the values measured five times for each flow rate. The CV value decreases from 5.58% to 1.51%; however, it sharply increases to 9.77%, and the flow rate increases. The value of CV increases owing to the irregular spacing between droplets in the high flow rate region. Therefore, the flow rate was fixed at Raq = 0.8 μL/min for all experiments because the CV value was the lowest at this flow rate.
Fig. 3.
Effect of flow rates on the reproducibility of SARS-CoV-2 assays. (a) Introduction of SERS nanotags and PBS buffer into the inlets to establish optimal conditions for driving the chip. (b) Photographic images of six different Raqs when each droplet passes through the droplet splitting junction. (c) SERS spectra measured for six different flow rates in 15 s. (d) Variations in the CV values corresponding to the SERS intensity in (c) with respect to the variations of Raq.
Table 1.
Controlled flow rates for oil and aqueous phases to determine the optimal measurement condition.
| Flow rate conditions | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Roil (µL/min) | 2.4 | 2.4 | 2.4 | 2.4 | 2.4 | 2.4 |
| Raq (µL/min) | 0.2 | 0.4 | 0.6 | 0.8 | 0.9 | 1.1 |
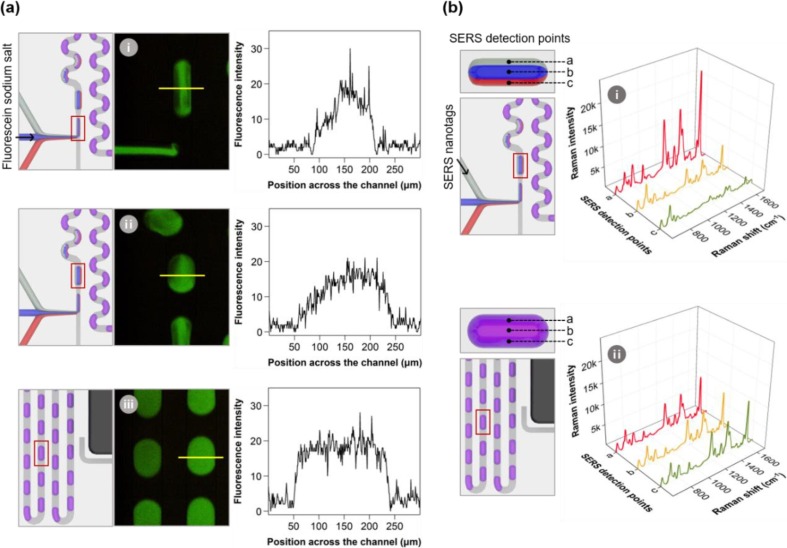
Additionally, we calculated the mixing and splitting efficiencies of the SERS-based microdroplet sensor. Magnetic beads and PBS buffer solutions were injected into the droplet channels through the central and other two inlets at both ends. Subsequently, the photographic image of each part of the microfluidic channel was captured to analyze the accuracy of the mixing and splitting of magnetic beads at the optimal flow rate, as shown in Fig. S5. Three laminar flows are formed in the sample injection part, and microdroplets are formed when the oil phase is vertically injected in the droplet generation compartment. In the droplet-mixing compartment, magnetic beads are evenly distributed throughout the droplet when droplets pass through the winding channels. As shown in the figure, the dispersed magnetic beads pass through the splitting junction embedded with a magnetic bar, and they are drawn toward the magnet and concentrate at the black dot. Subsequently, the droplets containing magnetic beads flow into the narrow channel, and the supernatant solution flows into the wide channel. Magnetic beads and supernatant solution droplets are separated into two branches. Furthermore, magnetic beads are re-distributed as they pass through multiple winding channels, and accumulate in the final outlet reservoir. Fig. 4 demonstrates that magnetic beads are separated effectively from the supernatant solution at an optimal flow rate.
Fig. 4.
(a) Fluorescence images and (b) SERS spectra of single droplet measured at different positions of microdroplet channel to evaluate the mixing efficiency of the laminar flow.
Fig. 4 presents fluorescence images and SERS spectra of droplets measured at different positions on the chip to evaluate the mixing efficiency of laminar flow when the chip is driven at an optimal flow rate. Fluorescence images of droplets at three different positions on the chip are shown in Fig. 4 a. Fluorescein sodium salt, exhibiting green fluorescence, was injected through the central inlet, and the PBS buffer solution was injected through the other two inlets on both ends. The laminar flow generated by the three flows in the sample injection compartment is not mixed. Therefore, the fluorescence intensity of the dye is strongly distributed in the central region. However, it decreases toward both ends, as demonstrated from the fluorescence intensity profiles measured in the vertical direction of the droplet. We analyzed the fluorescence intensity profile of droplets in the mixing compartment after passing through multiple winding channels and observed that mixing the laminar flow occurs actively. Hence, we observed uniform fluorescence intensity in the droplet mixing compartment. This mixing phenomenon was validated by Raman signal detection, as shown in Fig. 4 b. Subsequently, SERS nanotags were injected through the inlet at one end, and the PBS buffer solution was injected through the other two inlets. The high SERS signal intensity of the Raman reporter molecules was observed in the first laminar flow (position a), containing SERS nanotags before passing through the winding channel. However, moderate Raman signal intensities were observed at three positions (a, b, and c) after passing through the winding tracks. This experiment demonstrates the efficiency of the microdroplet channel.
3.3. SERS-based magnetic assays for detecting SARS-CoV-2 using a microdroplet channel
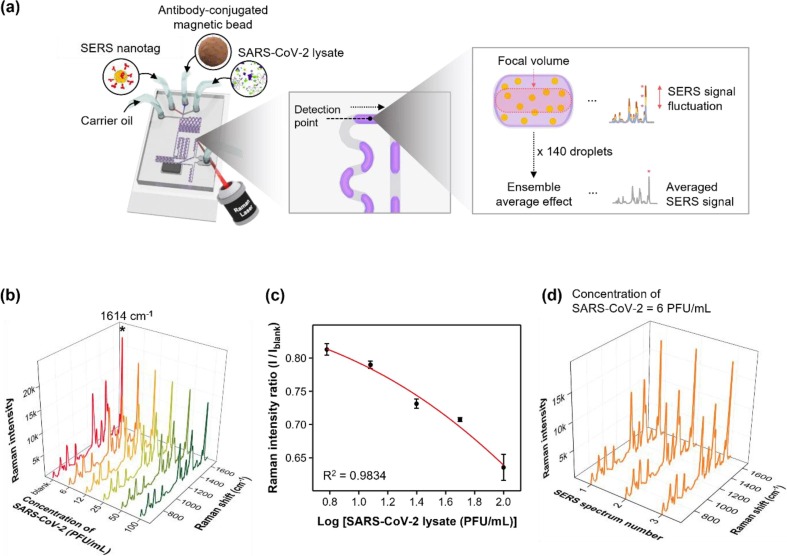
Fig. 5 illustrates the detection of SARS-CoV-2 lysate using a SERS-based microdroplet sensor. The immunoreaction was performed inside aqueous microdroplets. The droplet was split into two at a magnetic bar-embedded junction, and the SERS signal from droplets containing the supernatant solution was measured for 15 s. The number of magnetic immunocomplexes increased with an increase in the concentration of SARS-CoV-2. Therefore, the concentration of SERS nanotags is reduced in the supernatant solution, decreasing the measured SERS signal intensity. SARS-CoV-2 assays in a concentration range of 0–100 PFU/mL were performed using a SERS-based microdroplet sensor. The laser beam was focused on the center of the channel to detect Raman signals for SERS nanotags in microdroplets, as shown in Fig. 5 a. Fig. 5 b illustrates the on-chip detection SERS spectra for six different concentrations of SARS-CoV-2. Fig. 5 c shows the calibration curve for SARS-CoV-2 determined by the three-parameter sigmoidal fitting equation. The characteristic Raman peak intensity variations of SERS nanotags at 1614 cm-1 are plotted for the concentration of the SARS-CoV-2 lysate in Fig. 5 c. Error bars represent standard deviations determined through three measurements. Fig. 5 c demonstrates that the error bar for each concentration was significantly smaller than that for the magnetic bead-based assay performed in the microtube, as shown in Fig. 1 d. Fig. 5 d demonstrates the SERS spectra measured three times for droplets containing the supernatant solution for 15 s after the magnetic bead-based SERS assay was performed in a SARS-CoV-2 concentration of 6 PFU/mL in a microdroplet channel. Fig. 5 d illustrates that the Raman peak intensity at 1614 cm-1 is more consistent than the magnetic assay in the microtube. Moreover, the LoD determined from the calibration curve exhibits an extremely low value of 0.22 PFU/mL. Therefore, the probability of a false-negative detection for SARS-CoV-2 is significantly decreased. The LoD was determined using the three-parameter sigmoidal equation described in Fig. S6.
Fig. 5.
Detection of SARS-CoV-2 lysate using a SERS-based microdroplet sensor. (a) SERS-based microdroplet sensor for detecting SARS-CoV-2 lysate. The signal of 140 droplets passing through the focal volume was measured and averaged to improve its reproducibility. (b) On-chip detection SERS spectra for six different concentrations of SARS-CoV-2. (c) Calibration curve for SARS-CoV-2 lysate determined by the three-parameter sigmoidal fitting equation. (d) SERS spectra measured three times for droplets containing the supernatant solution for 15 s after magnetic bead-based SERS assay was performed on a SARS-CoV-2 concentration of 6 PFU/mL in a microdroplet channel.
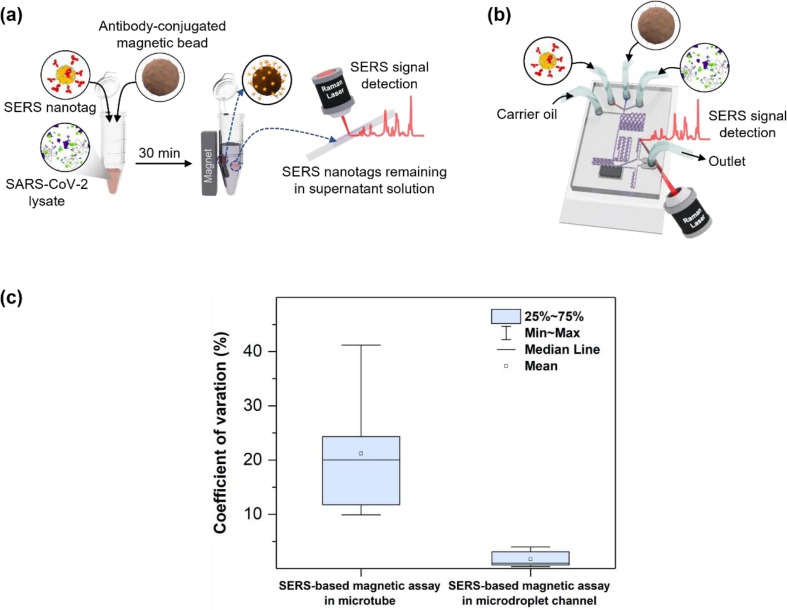
Fig. 6 presents a comparison of SERS-based magnetic assays for SARS-CoV-2 performed in a microtube and microfluidic channel. As for the SERS assay in a microtube, the supernatant solution was transferred to a capillary tube after assays, and SERS signals were measured. Therefore, the number of SERS nanotags in the focal volume is inconsistent at each measurement, and fluctuations in the SERS signal intensity result in a low reproducibility, as shown in Fig. 6 a. However, the microdroplet sensor significantly improves reproducibility owing to the ensemble average effects (Fig. 6 b). The signals from 140 droplets passing through the focal volume were measured and averaged. In Fig. 6 c, CV values from the assay performed using the two methods for the detection of SARS-CoV-2 are compared. CV values for SERS-based magnetic assay performed in the microtube and microfluidic channels were averages of five and three measurements, respectively. Fig. 6 c demonstrates that the average CV value is 21.2% for the microtube channel and 1.79% for the microdroplet channel. Therefore, the reproducibility improves by approximately 12 times in the SERS-based microdroplet sensor. Moreover, the Raman intensity in the microdroplet sensor was significantly enhanced because signals from SERS nanotags, contained in 140 droplets, were averaged in this case. Furthermore, the sensitivity of detection considerably improved using the SERS-based microdroplet sensor. A selectivity test was also performed for the SARS-CoV-2 (104 PFU/mL), influenza A/H1N1 (4032 HAU/mL), influenza B (2366 HAU/mL), and respiratory syncytial virus (RSV) (10 μg/mL) viruses. Corresponding SERS spectra and comparison of the Raman peak intensity ratio for different respiratory viruses were displayed in Fig. S7 a and S7b, respectively. Error bars in the figure denote the average deviations from five measurements. The intensity of the SERS signals significantly increased in the presence of SARS-CoV-2, however, weak signals were obtained in the presence of other viruses. These data suggest that the proposed SERS-based microdroplet sensor can accurately detect the virus due to the high specificity of the selected antibody pair against SARS-CoV-2.
Fig. 6.
Comparison of SERS-based assay for SARS-CoV-2 detection performed in (a) a microtube and (b) a microdroplet sensor. In the assay using a microtube, SERS signals were measured at five different positions of the capillary tube. The signal of 140 droplets passing through the focal volume was measured and averaged in the microdroplet sensor. (c) Comparison of CV values of the assays performed using two different methods for the detection of SARS-CoV-2.
Table 2 presents a comparison between the LoD values and detection times for the LFA strip, ELISA kit, SERS-based magnetic assay in a microtube, and SERS-based microdroplet sensor used for SARS-CoV-2 detection. The LoD is expressed in both tissue culture infectious doses (TCID50/mL) and PFU/mL units. TCID50 was determined by culturing host cells in a well plate titer and adding diluted virus fluids to the wells to determine the infection rate for each virus fluid. Commercial LFA, ELISA kits, and SERS-based magnetic bead assays exhibited LoDs of 341, 79.3, and 36.0 PFU/mL, respectively. Therefore, the sensitivity of these methods is insufficient to reduce the number of false-negative diagnosis for a patient having early signs of infection with low virus concentrations. Moreover, virus detection requires more than 3 h using ELISA kits, making rapid diagnosis impossible. The detection time in the case of the SERS-based assay performed in a microtube is approximately 30 min. However, this method is not convenient or feasible for SARS-CoV-2 detection at POCs because the assay needs to be performed manually using a micropipette. The proposed SARS-CoV-2 assay using the SERS-based microdroplet sensor exhibits an extremely low LoD of 0.22 PFU/mL. Thus, the possibility of a false-negative diagnosis for patients with early infection signs is significantly decreased. Moreover, the proposed method provides a rapid and convenient detection at POCs because its SARS-CoV-2 detection time is approximately 10 min.
Table 2.
Comparison of LoDs and detection times for four different SARS-CoV-2 assay methods.
| Lateral flowassay strip | Enzyme-linked immunosorbent assay | SERS-based magnetic assay in microtube | SERS-based magnetic assay in microdroplet channel | ||
|---|---|---|---|---|---|
| LoD | TCID50/mL | 494 | 115 | 52.2 | 0.32 |
| PFU/mL | 341 | 79.3 | 36.0 | 0.22 | |
| Detection time (min) | ≤ 30 | ≤ 180 | ≤ 30 | ≤ 10 | |
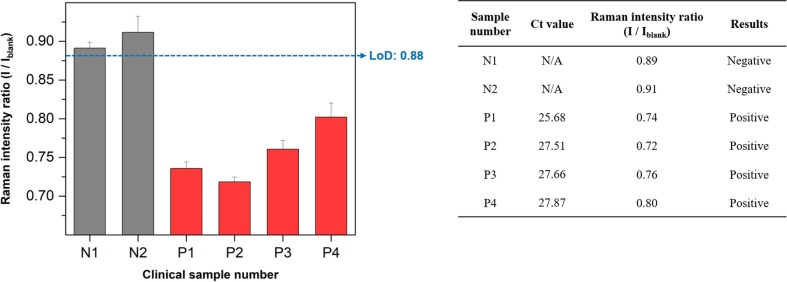
Finally, we performed a clinical assay using the proposed SERS-based microfluidic sensor for nasopharyngeal swab samples from six patients (4 SARS-CoV-2 positives and 2 negatives). RT-PCR assay results for the RNA marker ORF1 of SARS-CoV-2 were used as standard control data. In the calibration curve of Fig. 5 c, the LoD for I/Iblank was estimated to be 0.88. Therefore, a higher Raman intensity ratio than 0.88 is determined to be negative, and a lower intensity ratio than 0.88 is estimated to be positive. As a result of testing with six clinical samples, two negative samples (N1, N2) based on RT-PCR Ct values showed a Raman intensity ratio higher than 0.88, thus, it was judged negative. Conversely, all the positive samples (P1∼P4) showed a Raman intensity ratio lower than 0.88, resulting in a positive result as shown in Fig. 7 . Although the number of clinical samples tested in this work is limited, the proposed SERS-based microdroplet sensor shows a strong potential for clinical samples' positive/negative discrimination.
Fig. 7.
Clinical assay results for six nasopharyngeal swab clinical samples (4 SARS-CoV-2 positives and 2 negatives). RT-PCR assay results for the RNA marker ORF1 of SARS-CoV-2 were used as standard control data. A higher Raman intensity ratio than 0.88 is determined to be negative, and a lower intensity ratio than 0.88 is estimated to be positive.
4. Conclusion
The reproducibility of SERS-based magnetic bead assays in a microtube is poor (CV = 21.2%) owing to the limited number of SERS nanotags in the focal volume. Therefore, we developed a microdroplet sensor for rapid, reliable, and reproducible detection of SARS-CoV-2. This sensor separated the magnetic immunocomplex and supernatant solution droplets using a magnetic bar embedded in the channel. Subsequently, we measured the ensemble average signal of SERS nanotags containing 140 droplets passing through a fixed laser spot for 15 s. When the SARS-CoV-2 assay was performed using a microdroplet sensor, LoD and CV were improved from 36 to 0.22 PFU/mL and from 21.2% to 1.79%, respectively, compared to those of the assay performed in a microtube. Moreover, the detection time significantly decreased to less than 10 min. Therefore, the number of false-negative diagnosis in early-stage infected or asymptomatic patients with a low SARS-CoV-2 concentration decreased considerably. In addition, the cost required to manufacture one microdroplet chip, including PDMS, film mask, SYLGARD, base and curing agent, and slide glass, is estimated to be $ 1.50 per piece. The proposed SERS-based microdroplet sensor can be used as an efficient POC diagnostic platform for a safe and rapid detection of SARS-CoV-2 in the field because the proposed device can be easily integrated with a portable Raman spectrophotometer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that influence the work reported in this research.
Acknowledgements
This research was supported by the National Research Foundation of Korea (grant numbers 2019R1A2C3004375, 2020R1A5A1018052, and 2021M3E5E3080379) and KRIBB Research Initiative Program (1711134081).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2022.137085.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.He X.I., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 2.Nouvellet P., Bhatia S., Cori A., Ainslie K.E.C., Baguelin M., Bhatt S., Boonyasiri A., Brazeau N.F., Cattarino L., Cooper L.V., Coupland H., Cucunuba Z.M., Cuomo-Dannenburg G., Dighe A., Djaafara B.A., Dorigatti I., Eales O.D., van Elsland S.L., Nascimento F.F., FitzJohn R.G., Gaythorpe K.A.M., Geidelberg L., Green W.D., Hamlet A., Hauck K., Hinsley W., Imai N., Jeffrey B., Knock E., Laydon D.J., Lees J.A., Mangal T., Mellan T.A., Nedjati-Gilani G., Parag K.V., Pons-Salort M., Ragonnet-Cronin M., Riley S., Unwin H.J.T., Verity R., Vollmer M.A.C., Volz E., Walker P.G.T., Walters C.E., Wang H., Watson O.J., Whittaker C., Whittles L.K., Xi X., Ferguson N.M., Donnelly C.A. Reduction in mobility and COVID-19 transmission. Nat. Commun. 2021;12:1090. doi: 10.1038/s41467-021-21358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon S., Joshi A.D., Lo C.-H., Drew D.A., Nguyen L.H., Guo C.-G., Ma W., Mehta R.S., Shebl F.M., Warner E.T., Astley C.M., Merino J., Murray B., Wolf J., Ourselin S., Steves C.J., Spector T.D., Hart J.E., Song M., VoPham T., Chan A.T. Association of social distancing and face mask use with risk of COVID-19. Nat. Commun. 2021;12:3737. doi: 10.1038/s41467-021-24115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368:6936. doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen M., Zhou Y., Ye J., Abdullah Al-maskri A.A., Kang Y., Zeng S., Cai S., S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-NCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bwire G.M., Majigo M.V., Njiro B.J., Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-Analysis. J. Med. Virol. 2021;93:719–725. doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohammadniaei M., Zhang M., Ashley J., Christensen U.B., Friis-Hansen L.J., Gregersen R., Lisby J.G., Benfield T.L., Nielsen F.E., Henning Ramussen J., Pedersen E.B., Olinger A.C.R., Kolding L.T., Naseri M., Zheng T., Wang W., Gorodkin J., Sun Y. A non-enzymatic, isothermal strand displacement and amplification assay for rapid detection of SARS-CoV-2 RNA. Nat. Commun. 2021;12:5089. doi: 10.1038/s41467-021-25387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabe B.A., Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. U.S.A. 2020;117(39):24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahas A., Wang Q., Marsic T., Mahfouz M.M. A novel miniature CRISPR-Cas13 system for SARS-CoV-2 diagnostics. ACS Synth. Biol. 2021;10(10):2541–2551. doi: 10.1021/acssynbio.1c00181. [DOI] [PubMed] [Google Scholar]
- 12.Kang B.-H., Lee Y., Yu E.-S., Na H., Kang M., Huh H.J., Jeong K.-H. Ultrafast and real-time nanoplasmonic on-chip polymerase chain reaction for rapid and quantitative molecular diagnostics. ACS Nano. 2021;15(6):10194–10202. doi: 10.1021/acsnano.1c02154. [DOI] [PubMed] [Google Scholar]
- 13.Cheong J., Yu H., Lee C.Y., Lee J.-u., Choi H.-J., Lee J.-H., Lee H., Cheon J. Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat. Biomed. Eng. 2020;4(12):1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghodake G.S., Shinde S.K., Kadam A.A., Saratale R.G., Saratale G.D., Syed A., Elgorban A.M., Marraiki N., Kim D.-Y. Development and implementation of diagnostic tools and surveillance measures. Biosens. Bioelectron. 2021;177 doi: 10.1016/j.bios.2021.112969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y.e., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L., Li D., Ramadan S., Li Y., Klein N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020;170 doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mak G.C.K., Cheng P.K.C., Lau S.S.Y., Wong K.K.Y., Lau C.S., Lam E.T.K., Chan R.C.W., Tsang D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domenico M.D., de Rosa A., Boccellino M. Detection of SARS-CoV-2 proteins using an ELISA test. Diagnostics11. 2021:698. doi: 10.3390/diagnostics11040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H., Park S.-G., Choi N., Kwon H.-J., Kang T., Lee M.-K., Choo J. Sensitive detection of SARS-CoV-2 using SERS-based aptasensor. ACS Sen. 2021;6:2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Wang X., Cheng L.u., Ding S., Wang G., Choo J., Chen L. SERS-based test strips: Principles, designs and applications. Biosens. Bioelectron. 2021;189:113360. doi: 10.1016/j.bios.2021.113360. [DOI] [PubMed] [Google Scholar]
- 22.Xu K., Zhou R., Takei K., Hong M. Toward flexible surface-enhanced Raman scattering (SERS) sensors for point-of-care diagnostics. Adv. Sci. 2019;6:1900925. doi: 10.1002/advs.201900925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H., Das A., Bi L., Choi N., Moon J.-I., Wu Y., Park S., Choo J. Recent advances in surface-enhanced Raman scattering-based microdevices for point-of-care diagnosis of viruses and bacteria. Nanoscale. 2020;12(42):21560–21570. doi: 10.1039/d0nr06340a. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Zong S., Wu L., Zhu D., Cui Y. SERS-activated platforms for immunoassay: probes, encoding methods, and applications. Chem. Rev. 2017;117:7910–7963. doi: 10.1021/acs.chemrev.7b00027. [DOI] [PubMed] [Google Scholar]
- 25.Baumberg J.J., Aizpurua J., Mikkelsen M.H., Smith D.R. Extreme nanophotonics from ultrathin metallic gaps. Nat. Mater. 2019;18(7):668–678. doi: 10.1038/s41563-019-0290-y. [DOI] [PubMed] [Google Scholar]
- 26.Dang H., Park S.-G., Wu Y., Choi N., Yang J.-Y., Lee S., Joo S.-W., Chen L., Choo J. Reproducible and sensitive plasmonic sensing platforms based on Au-nanoparticle-internalized nanodimpled substrates. Adv. Funct. Mater. 2021;32:2105703. [Google Scholar]
- 27.Yu Q., Wu Y., Kang T., Choo J. Development of SERS-based immunoassay platforms using hollow Au nanostars for reliable SARS-CoV-2 diagnosis. Bull. Kor. Chem. Soc. 2021;42:1699–1705. doi: 10.1002/bkcs.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chon H., Lee S., Son S.W., Oh C.H., Choo J. Highly sensitive immunoassay of lung cancer marker carcinoembryonic antigen (CEA) using surface-enhanced Raman scattering of hollow gold nanospheres. Anal. Chem. 2009;81:3029–3034. doi: 10.1021/ac802722c. [DOI] [PubMed] [Google Scholar]
- 29.Chon H., Lee S., Yoon S.-Y., Lee E.K., Chang S.-I., Choo J. SERS-based competitive immunoassay of troponin I and CK-MB markers for early diagnosis of acute myocardial infarction. Chem. Commun. 2014;50(9):1058–1060. doi: 10.1039/c3cc47850e. [DOI] [PubMed] [Google Scholar]
- 30.Choi N., Lee J., Ko J., Jeon J.H., Rhie G.-E., deMello A.J., Choo J. Integrated SERS-based microdroplet platform for the automated immunoassay of F1 antigens in Yersinia pestis. Anal. Chem. 2017;89(16):8413–8420. doi: 10.1021/acs.analchem.7b01822. [DOI] [PubMed] [Google Scholar]
- 31.Gao R., Cheng Z., deMello A.J., Choo J. Wash-free magnetic immunoassay of the PSA cancer marker using SERS and droplet microfluidics. Lab Chip. 2016;16(6):1022–1029. doi: 10.1039/c5lc01249j. [DOI] [PubMed] [Google Scholar]
- 32.Chung E., Gao R., Ko J., Choi N., Lim D.W., Lee E.K., Chang S.-I., Choo J. Trace analysis of mercury (II) ions using aptamer-modified Au/Ag core-shell nanoparticles and SERS spectroscopy in microdroplet channel. Lab Chip. 2013;13:260–266. doi: 10.1039/c2lc41079f. [DOI] [PubMed] [Google Scholar]
- 33.Moitra P., Chaichi A., Abid Hasan S.M., Dighe K., Alafeef M., Prasad A., Gartia M.R., Pan D. Probing the mutation independent interaction of DNA probes with SARS-CoV-2 variants through a combination of surface-enhanced Raman scattering and machine learning. Biosens. Bioelectron. 2022;208 doi: 10.1016/j.bios.2022.114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim W., Kim S., Han J., Kim T.G., Bang A., Choi H.W., Min G.E., Shin J.-H., Moon S.W., Choi S. An excitation wavelength-optimized, stable SERS biosensing nanoplatform for analyzing adenoviral and AstraZeneca COVID-19 vaccination efficacy status using tear samples of vaccinated individuals. Biosens. Bioelectron. 2022;204 doi: 10.1016/j.bios.2022.114079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paria D., Kwok K.S., Raj P., Zheng P., Gracias D.H., Barman I. Label-free spectroscopic SARS-CoV-2 detection on versatile nanoimprinted substrates. Nano Lett. 2022;22(9):3620–3627. doi: 10.1021/acs.nanolett.1c04722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartzberg A.M., Oshiro T.Y., Zhang J.Z., Huser T., Talley C.E. Improving nanoprobes using surface-enhanced Raman scattering from 30-nm hollow gold particles. Anal. Chem. 2006;78(13):4732–4736. doi: 10.1021/ac060220g. [DOI] [PubMed] [Google Scholar]
- 37.Wang H., Goodrich G.P., Tam F., Oubre C., Nordlander P., Halas N.J. Controlled texturing modifies the surface topography and plasmonic properties of Au nanoshells. J. Phys. Chem. B. 2005;109(22):11083–11087. doi: 10.1021/jp051466c. [DOI] [PubMed] [Google Scholar]
- 38.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bokelmann L., Nickel O., Marivic T., Paeaebo S., Meyer M., Borte S., Riesenberg S. Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat. Commun. 2021;12:1467. doi: 10.1038/s41467-021-21627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glawdel T., Elbuken C., Ren C. Passive droplet trafficking at microfluidic junctions under geometric and flow asymmetries. Lab Chip. 2011;11:3774–3784. doi: 10.1039/c1lc20628a. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y., Fu T., Zhu C., Lu Y., Ma Y., Li H.Z. Asymmetrical breakup of bubbles at a microfluidic T-junction divergence: feedback effect of bubble collision. Microfluid. Nanofluid. 2012;13(5):723–733. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.