Summary
Introduction
Secondary bacterial infections have been reported in majority of patients hospitalized with coronavirus disease 2019 (COVID-19). A study of the antimicrobial susceptibility profiles of these bacterial strains revealed that they were multidrug resistant, demonstrating their resistance to at least three classes of antimicrobial agents including beta-lactams, fluoroquinolones and aminoglycosides. Bacterial co-infection remains as an important cause for high mortality in patients hospitalized with COVID-19.
Methods
In our study, we conducted a retrospective comparative analysis of bacterial co-infections and the antimicrobial resistance profile of bacterial isolates obtained from inpatients admitted in COVID-19 and non-COVID-19 intensive care units. The goal was to obtain the etiology and antimicrobial resistance of these infections for more accurate use of antimicrobials in clinical settings. This study involved a total of 648 samples collected from 356 COVID-19 positive patients and 292 COVID-19 negative patients admitted in the intensive care unit over a period of six months from May to October 2020.
Results
Among the co-infections found, maximum antimicrobial resistance was found in Acinetobacter species followed by Klebsiella species in both the ICU’s. Incidence of bacterial co-infection was found to be higher in COVID-19 intensive care patients and most of these isolates were multidrug resistant strains.
Conclusion
Therefore, it is important that co-infections should not be underestimated and instead be made part of an integrated plan to limit the global burden of morbidity and mortality during the SARS-CoV-2 pandemic and beyond.
Key words: Novel corona virus, Hospital acquired infections, SARS-CoV-2, COVID-19, Secondary bacterial infections, Multidrug resistance, Anti-microbial resistance, COVID ICU, Non-COVID ICU
Introduction
The Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) first appeared in Wuhan, China in December 2019. It has now become a global pandemic affecting most of the countries round the world. Globally as on 3rd May 2021, the total confirmed cases and deaths due to COVID-19 disease reported to WHO include 152,387,917 and 3,195,624 respectively. India reported 19,925,604 confirmed cases of COVID-19 disease including 218,959 deaths to WHO [1]. Also, during the second wave, between 1st March and 3rd May, India has reported 8,813,363 confirmed cases and 61,802 deaths, thereby accounting for a total of 44.23% confirmed cases and 28.23% mortalities.
Antibiotics remain ineffective in treating COVID-19 but they are used to treat patients suspected of COVID-19. This is because of the difficulty in ruling out the Secondary bacterial infection and remains as a dangerous and common complication in hospitalized patients, especially with COVID-19 disease. They occur at an approximate incidence of 10-15% [2, 3]. Also, COVID-19 patients with secondary bacterial infections have a higher mortality rate of around 50% [4]. Therefore, early initiation of the appropriate antimicrobial therapy can help to tackle the life threatening secondary bacterial infections. Current recommendations are extrapolated from the data available to treat other viral pneumonias as there are no proper controlled clinical trials available to support the use of empirical antimicrobial agents for the treatment of COVID-19 disease [5]. Studies have recommended empirical antimicrobial therapy like 3rd generation cephalosporins to treat all COVID-19 disease associated secondary bacterial infections [6, 7]. However, irrational use of broad-spectrum antibiotics to treat hospitalized patients can lead to the emergence of multidrug resistant isolates [8, 9]. Clear understanding of COVID-19 patients with secondary bacterial infection and the etiological agents will help to treat the COVID-19 patients promptly in order to assure the controlled use of antibiotics and to reduce the adverse effects of antibiotic overuse.
Therefore, based on the existing microbiological data, antibiotics ought to be judiciously used to treat COVID-19 patients with confirmed secondary bacterial infections. Currently there is no pre-existing data about the etiology or spectrum of secondary bacterial infections in patients with COVID-19 disease [10-12].
In the current study, a retrospective comparative analysis of secondary bacterial infections was made in critically ill patients with and without COVID-19 disease. The aim was to obtain the etiology and antimicrobial resistance profile of bacterial isolates causing secondary bacterial infections in COVID-19 patients admitted in the ICU and also to compare the susceptibility profile of bacterial isolates obtained from critically ill COVID-19 negative patients. This can lead to a more accurate and effective antimicrobial use.
Materials and methods
This was a retrospective observational study carried out at Chettinad Hospital and Research Institute, a tertiary care hospital situated in Kelambakkam, Tamilnadu after obtaining due approval from the Institutional Human Ethics Committee.
Sample size: This study included a total of 648 patients which comprised of 356 COVID positive patients and 292 COVID negative patients admitted in other intensive care units (ICU). The study was conducted over a period of six months from May to October 2020.
The COVID-19 positive patients included in the study had tested positive for SARS CoV-2 infection by Real time RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) as per Indian Council of Medical Research (ICMR) guidelines using SD-BIOSENSOR Real Time detection kit. SARS Cov2 RNA extraction for these samples was done using Qiacube, an automated nucleic acid extractor. The clinical samples from these patients, like blood, urine and sputum were obtained and processed for bacterial culture and sensitivity according to standard microbiological procedures [13]. The bacterial isolates were subjected to anti-microbial susceptibility testing (AST) by Kirby Bauer disc diffusion method as per Clinical and Laboratory Standards Institute (CLSI) Guidelines 2021. According to Centres for Disease Control and Prevention (CDC) definition the bacterial strains were identified as multidrug resistant if they were resistant to 1 drug each in at least 3 of the categories of drugs like extended spectrum cephalosporin, fluoroquinolones, aminoglycosides, carbapenems, and piperacillin tazobactam.
Results
Total of 648 patients were included in the study. Out of which 356 patients (54.94%) from the COVID ICU and 292 patients (45.06%) from the non-COVID ICUs.
Total respiratory samples processed was 316, out of which 178 (56.33%) samples were from COVID ICU and 138 (43.67%) samples were from other ICU’s. Of the total 316 samples processed, 113 samples yielded pathogenic bacterial isolates, out of which 76 (67.26%) samples were from COVID ICU and 37 (32.74%) samples were from other ICU’s.
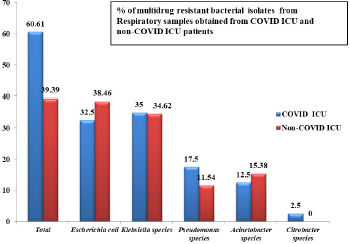
Among the 113 samples that showed growth, 66 samples yielded multidrug resistant bacterial isolates. 40 (60.61%) samples were from COVID ICU and 26 (39.39%) samples were from other ICU’s.
Bacteriological profile of the multidrug resistant isolates obtained from respiratory samples from COVID ICU and other ICU’s includes Escherichia coli, Klebsiella species, Pseudomonas species, Acinetobacter species and Citrobacter species (Fig. 1). Multidrug resistant strains of Escherichia coli and Klebsiella species were found to be more prevalent in COVID ICU (32.5% & 35% respetively) and other ICU’s (38.46% & 34.62% respectively).
Fig. 1.
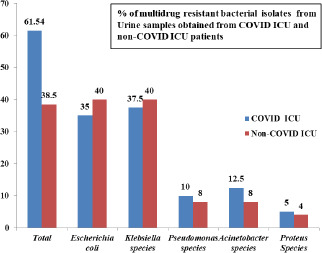
A total of 320 urine samples were analyzed. Of the total 320 samples processed, 123 samples yielded growth, out of which 74 (60.16%) samples were from COVID ICU and 49 (39.84%) samples were from other ICU’s. Among the 123 samples that yielded growth, 65 samples yielded multidrug resistant isolates. 40 (61.54%) Multi-drug Resistant (MDR) isolates were from COVID ICU and 25 (38.46%) MDRO’s were from other ICU’s.
The bacteriological profile of the multidrug resistant bacterial urine isolates from COVID ICU and other ICU’s includes Escherichia coli, Klebsiella species, Pseudomonas species, Acinetobacter species and Proteus species (Fig. 2) Multidrug resistant Escherichia coli and Klebsiella species were found to be more prevalent in COVID ICU (35% and 37.5% respectively) and other ICU’s (40% each).
Fig. 2.
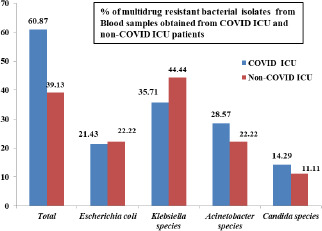
Total blood samples processed was 175, out of which 106 (60.57%) samples were from COVID ICU and 69 (39.43%) samples were from other ICU’s. Of the total 175 samples processed, 76 samples yielded growth, out of which 53 (69.74%) samples were from COVID ICU and 23 (30.26%) samples were from other ICU’s. Among the 76 samples that showed growth, 23 yielded multidrug resistant isolates. 14 (60.87%) samples were from COVID ICU and 9 (39.13%) samples from other ICU’s.
The bacteriological profile of the MDRO’s in blood samples obtained from COVID ICU and other ICU’s included Escherichia coli, Klebsiella species, Acinetobacter species and Candida albicans (Fig. 3). MDR Klebsiella species were found to be more prevalent in COVID ICU (35.71%) and other ICU’s (44.44%).
Fig. 3.
An overview on the total profile of various bacterial isolates and multidrug resistant isolates isolated from respiratory, urine and blood samples of COVID ICU and other ICU patients are demonstrated (Tab. I).
Tab. I.
An overview on the total profile of various bacterial isolates and drug resistant isolates isolated from various clinical samples.
| Sample type | No of Organisms | Urine | Blood | Respiratory | |||
|---|---|---|---|---|---|---|---|
| Type of ICU | COVID ICU | Other ICU | COVID ICU | Other ICU | COVID ICU | Other ICU | |
| Escherichia coli | TOTAL | 28 | 22 | 9 | 7 | 23 | 17 |
| MDR | 14 | 10 | 3 | 2 | 13 | 10 | |
| % MDR | 50 | 45.45 | 33.33 | 28.57 | 56.52 | 58.82 | |
| Klebsiella species | TOTAL | 21 | 17 | 9 | 11 | 21 | 16 |
| MDR | 15 | 10 | 5 | 4 | 14 | 9 | |
| % MDR | 71.42 | 58.82 | 55.55 | 36.36 | 66.66 | 56.25 | |
| Pseudomonas species | TOTAL | 9 | 7 | 0 | 0 | 14 | 9 |
| MDR | 4 | 2 | 0 | 0 | 7 | 3 | |
| % MDR | 44.44 | 28.57 | 0 | 0 | 50.00 | 33.33 | |
| Acinetobacter species | TOTAL | 5 | 3 | 5 | 2 | 7 | 5 |
| MDR | 5 | 2 | 4 | 2 | 5 | 4 | |
| % MDR | 100.00 | 66.67 | 80.00 | 100.00 | 71.43 | 80.00 | |
| Citrobacter species | TOTAL | 0 | 0 | 0 | 0 | 1 | 0 |
| MDR | 0 | 0 | 0 | 0 | 1 | 0 | |
| % MDR | 0 | 0 | 0 | 0 | 100 | 0 | |
| Proteus species | TOTAL | 4 | 3 | 0 | 0 | 0 | 0 |
| MDR | 2 | 1 | 0 | 0 | 0 | 0 | |
| % MDR | 50 | 33.33 | 0 | 0 | 0 | 0 | |
| Coagulase Negative Staphylococcus species | TOTAL | 0 | 0 | 13 | 5 | 0 | 0 |
| MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| % MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| Candida albicans | TOTAL | 0 | 0 | 7 | 4 | 0 | 0 |
| MDR | 0 | 0 | 2 | 1 | 0 | 0 | |
| % MDR | 0 | 0 | 28.57 | 25 | 0 | 0 | |
| Candida species (non-albicans) | TOTAL | 2 | 2 | 0 | 0 | 0 | 0 |
| MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| % MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| Staphylococcus aureus | TOTAL | 0 | 0 | 2 | 2 | 0 | 0 |
| MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| % MDR | 0 | 0 | 0 | 0 | 0 | 0 | |
In our study highest resistance was demonstrated among Acinetobacter species (n = 22) (Fig. 4). Of these, 14 (63.6 %) were from COVID ICU and 8 (36.3%) were from other ICU patients.
Fig. 4.
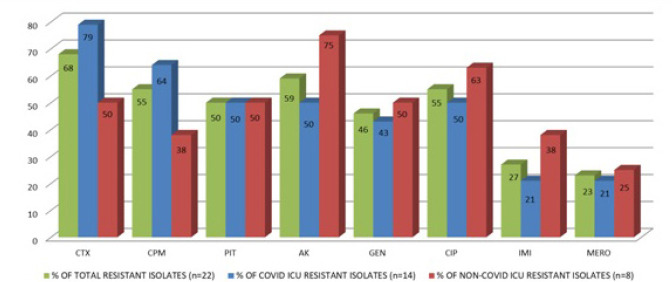
Acinetobacter Species - Antibiotic resistance profile.
Among the 14 strains from COVID ICU, 11 (78.6%) strains were resistant to cefotaxime, 9 (64.3%) resistant to cefepime, 7 each (50%) were resistant to piperacillin tazobactam, ciprofloxacin and amikacin, 6 (42.9%) to gentamicin and 3 each (21.4%) to imipenem and meropenem.
Of the 8 Acinetobacter strains from other ICUs, 4 each (50%) showed resistance to gentamicin, cefotaxime and to piperacillin tazobactam, 3 each (37.5%) to cefepime and imipenem, 6 (75%) to amikacin, 5 (62.5%) to ciprofloxacin and 2 (25%) to meropenem.
The total resistance pattern of Acinetobacter isolates from both the ICU’s (COVID and non- COVID) is as follows: 68.2% (n = 15/22) to cefotaxime, 54.5% (n = 12/22) to cefipime, 50% (11/22) to piperacillin tazobactam, 59.1% (13/22) to amikacin, 45.5% (n = 10/22) to gentamicin, 54.5% (n = 12/22) to ciprofloxacin, 27.3% (6/22) to imipenem and 22.7% (n = 5/22) to meropenem.
In our study next highest resistance was demonstrated among Klebsiella species (n = 57) (Fig. 5). Of the total 57 isolates of Klebsiella species, 34 were (59.6%) were isolated from COVID ICU and 23 (40.3%) were from other ICU patients.
Fig. 5.
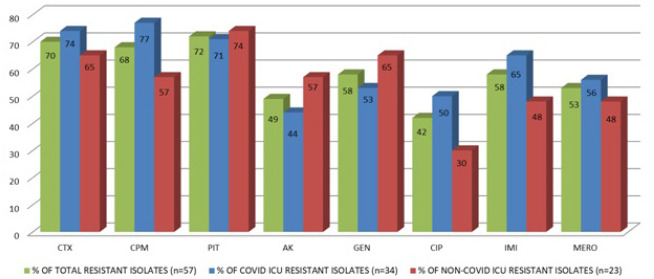
Klebsiella species - Antibiotic resistance profile.
Among the 34 strains from COVID ICU, 25, (73.5%) were resistant to cefotaxime, 26 (76.5%) to cefepime, 24 (70.6%) to piperacillin tazobactam, 15 (44.1%) to amikacin, 18 (52.9%) to gentamicin, 17 (50%) to ciprofloxacin, 22 (64.7%) to imipenem and 19 (55.9%) to meropenem.
Of the 23 strains from other ICUs, 15 (65.2%) were resistant to cefotaxime, 13 (56.5%) to cefepime, 17 (73.9%) to piperacillin tazobactam, 13 (56.5%) to amikacin, 15 (65.2%) to gentamicin, 7 (30.4%) to ciprofloxacin, 11 (47.8%) to imipenem and meropenem.
Therefore, the total resistance pattern of Klebsiella species from COVID and non-COVID ICU is as follows: 70.2% (n = 40/57) to cefotaxime, 68.4% (n = 39/57) to cefepime, 71.9% (n = 41/57) to piperacillin tazobactam, 49.1% (n=28/57) to amikacin, 57.9% (n = 33/57) to gentamicin, 42.1% (n = 24/57) to ciprofloxacin, 57.9% (n = 33/57) to imipenem and 52.6% (n = 30/57) to meropenem.
Next highest resistance was demonstrated among Escherichia coli (n = 52). Of these, n = 30/52 isolates were from COVID ICU and n = 22/52 were from other ICU patients. (Fig. 6)
Fig. 6.
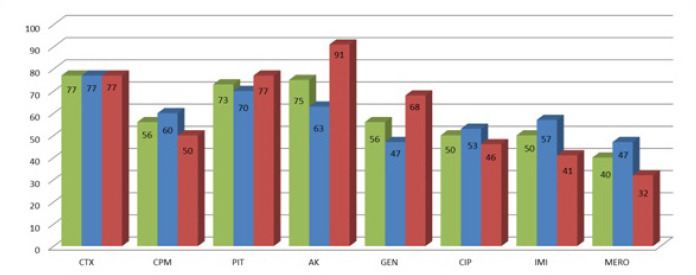
Escherichia coli - Antibiotic resistance profile.
Among the 30 strains from COVID ICU, 23 (76.7%) showed resistance to cefotaxime, 18 (60%) to cefepime, 21 (70%) to piperacillin tazobactam, 19 (63.3%) to amikacin, 17 (56.7%) to imipenem, 14 (46.7%) to gentamicin and meropenem, 16 (53.3%) to ciprofloxacin.
Of the 22 strains from other ICUs, 17 (77.3%) showed resistance to cefotaxime, 11 (50%) to cefepime, 17 (77.3%) to piperacillin tazobactam, 20 (90.9%) to amikacin, 15 (68.2%) gentamicin, 10 (45.5%) to ciprofloxacin, 9 (40.9%) to imipenem and 7(31.8%) to meropenem.
Therefore, the total resistance pattern of Escherichia coli from both the ICUs is as follows: 76.9% (n = 40/52) of the strains showed resistance to cefotaxime, 55.8% (n = 29/52) to cefepime, 73.1% (n = 38/52) to piperacillin tazobactam, 75% (n = 39/52) to amikacin, 55.8% (n = 29/52) to gentamicin, 50% (n = 26/52) to ciprofloxacin, 50% (n = 26/52) to imipenem and 40.4%(n = 21/52) to meropenem.
Also the resistance profile of Pseudomonas species, (n = 16) was analyzed. Of these, n = 11/16 isolates were from COVID ICU and n = 5/16 were from other ICU patients. (Fig. 7)
Fig. 7.
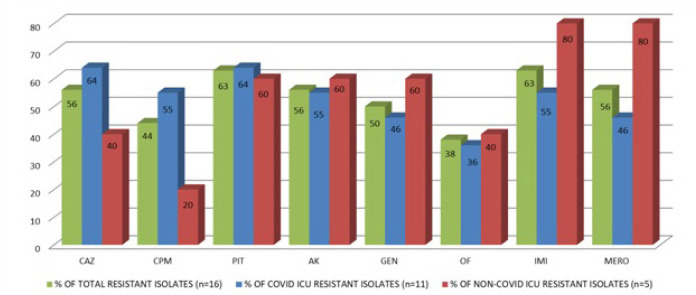
Pseudomonas species - Antibiotic resistance profile.
Among the 11 strains from COVID ICU, 7 (63.6%) showed resistance to ceftazidime and piperacillin tazobactam, 6 (54.5%) to imipenem, cefepime and amikacin, 5 (45.5%) to gentamicin and meropenem, 4 (36.4%) to ofloxacin.
Of the 5 strains from other ICU’s, 2 (40%) strains showed resistance to ofloxacin and ceftazidime, 1 (20%) to cefepime, 3 (60%) to piperacillin tazobactam, amikacin and gentamicin, 4 (80%) each to imipenem and meropenem.
Therefore, the total resistance pattern of Pseudomonas from both the ICUs is as follows: 56.3% (n = 9/16) to ceftazidime, 43.8% (n = 7/16) to cefepime, 62.5% (n = 10/16) to piperacillin tazobactam, 56.3% (n = 9/16) to amikacin, 50% (n = 8/16) to gentamicin, 37.5% (n = 6/16) to ofloxacin, 62.5% (n = 10/16) to imipenem and 56.3% (n = 9/16) to meropenem.
In this study, 3 out of 11 Candida albicans isolates demonstrated 27.27% resistance, out of which 2 (66.66%) of the isolates were from COVID ICU patients and 1(33.33%) was from patients admitted in other ICUs. All the three resistant strains were blood isolates.
Among the 2 strains isolated from COVID ICU, no strains (0%) showed resistance to amphotericin-B and nystatin, 1 strain (50%) showed resistance to ketoconazole and both strains (100%) showed resistance to clotrimazole, fluconazole and itraconazole. The single strain isolated from other ICU’s, showed no resistance to amphotericin-B, fluconazole and itraconazole, and showed resistance to clotrimazole, ketoconazole and nystatin.
The total resistance pattern of Candida albicans from both the ICUs is as follows: 0% (n = 0/3) to amphotericin-B, 100% (n = 3/3) to clotrimazole, 66.67% (n = 2/3) to fluconazole, itraconazole and ketoconazole and 33.33% (n = 1/3) to nystatin.
In our study we also isolated other Candida species which include three strains of Candida tropicalis and one strain of Candida parapsilosis obtained from urine samples and these isolates did not demonstrate any antifungal resistance.
Discussion
Bacterial co-infections occurring in patients hospitalized with COVID-19 are a leading cause of mortality. A meta-analysis done by Langford BJ et.al states that bacterial co-infection remains very common among COVID-19 infected ICU patients than critically ill patients belonging to other ICU settings [14].
Among the Gram-negative bacteria, Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii are the most prevalent bacterial isolates and the anti-microbial resistance rates among these bacteria were found to be very high.
A study conducted by Russell et al., showed that among the Gram negative bacteria, members belonging to the family Enterobacteriaceae, like Escherichia coli commonly are associated blood stream infections [15]. In our study Escherichia coli and Klebsiella species are the predominant isolates causing blood stream infections affected with COVID-19.
In our study, we included 356 COVID positive patients admitted in the intensive care unit and 292 COVID negative patients admitted in other non-COVID intensive care units. We found that COVID patients admitted in the ICU were more prone to bacterial co-infections and also the resistant bacterial strains were found to be more prevalent in COVID patients admitted in the ICU (Figg. 1-3).
Higher resistance was found in Acinetobacter spp 81.48% (n = 22/27), out of which 63.63% (n = 14/22) of the bacterial strains were isolated from COVID ICU and 36.26% (n = 8/22) were from other ICU patients.
This finding is similar to the study done by Jie Le et al. [9]. However, the study conducted by Mahmoudi and another study conducted by Surbhi Khurana et al. [16, 17] showed higher resistance among Klebsiella species, followed by Acinetobacter species. In our study, Acinetobacter species showed higher resistance followed by Klebsiella species. A study by Ehsan Sharifipour et al. also showed that a wide spread resistance was contributed by Acinetobacter species [18]. Another study conducted by Sama Rezasoltani et al. showed more resistance among Staphylococcus aureus (MRSA) strains [19].
Klebsiella species was the next most resistant organism isolated (60%), out of which 59.64% (n = 34/57) were from COVID ICU patients and 40.35% (n = 23/57) were from other ICU patients. A study by Sreenath K et.al states that Klebsiella species is the common organism that usually complicates treatment options in COVID-19 affected patients admitted in ICU setting [20]. Another study by Arcari et al. shows that multidrug resistant Klebsiella species remain as a common pathogen in critically ill COVID-19 patients [21].
Other bacterial isolates like Escherichia coli showed 49.05% resistance (n = 52/106), out of which 57.69% (n = 30/52) were obtained from COVID ICU patients 42.31% (n = 22/52) were from non-COVID ICU patients. A study by Mahmoudi et al. shows that Escherichia coli obtained by COVID-19 patients showed greater resistance to cephalosporins, co-trimoxazole and piperacillin-tazobactam [16]. Howerver, in our study many isolates were resistant to cephalosporins and few were also resistant to imipenem and meropenem.
Drug resistance was found to be 42.85% (n = 3/7) in Proteus spps isolates, out of which 66.66% (n = 2/3) were from COVID ICU patients and 33.33% (n = 1/3) were from non-COVID ICU patients.
Pseudomonas spps showed 41.02% resistance (n = 16/39), out of which 68.75% (n = 11/16) were obtained from COVID ICU patients and 31.25% (n = 5/16) were obtained from patients admitted in non-COVID ICU’s. A study by Qu et al. states that Pseudomonas aeruginosa can remain as an important co-infecting pathogen in critically ill COVID-19 patients with a greater capacity of producing biofilm and thereby conferring antibiotic resistance [22].
In our study we isolated a single strain of Citrobacter spps (n = 1/1) that was found to be multidrug resistant. That strain was isolated from a respiratory sample in COVID ICU.
Other gram-positive bacterial isolates like Coagulase negative staphylococci (n = 18) and Staphylococcus aureus (n = 4) obtained from blood culture did not show any multi drug resistance.
In our study we respiratory samples from COVID-19 patients remain predominant as lower respiratory tract bacterial co-infections remain common in these patients.
The underlying pathogenesis of bacterial co-infection in severe and critical COVID-19 cases is due to the host-pathogen interactions, which include the virulence factors of the pathogens, dysregulations of immune responses and disturbed microbial flora during viral pneumonia [23, 24].
Viral pneumonia and bacterial co-infection act as mutual reinforcing factor to promote the progression of COVID-19 disease. Severe cases of COVID-19 cause multiple damages in the lungs, thereby decreasing the oxygen and carbon dioxide diffusion capacities. Surfactant disruption and sloughing of cells into the airways provide access and rich source of nutrients, thereby leading to rapid bacterial invasion. Both the factors like changes in microbial flora and bacterial virulence can alter the immune responses to SARS-CoV-2, resulting in the rebound of viral titres and high mortality in severe and critically ill patients [25].
The strengths of the present study include, we analysed significant number of samples from critically ill COVID-19 patients and we were able to present an appropriate microbiological and antibiotic resistance profile. However, the study also has weakness that has to be addressed in the future. This study is a retrospective study and therefore systemic patient review was not performed. Also this study was conducted in a single centre and not a multicentric study because the microbiological and antibiotic profile can vary with respect to the different geographical location.
Conclusion
An effective antimicrobial regimen remains the key step for the successful treatment of COVID-19. Also, due to the scarcity of data available on the empirical use of antimicrobials to treat COVID-19 secondary infections and the irrational use of antimicrobials to treat these patients, has led to the emergence of multidrug resistance [25].
Therefore, molecular approaches like meta genomic next-generation sequencing can help in the detection of a broad range of pathogens and their antimicrobial resistance which further contribute to the appropriate antibiotic stewardship programmes. In addition, molecular understanding of the causes and consequences of bacterial co-infections in COVID-19 patients aid the development of novel therapeutic interventions influencing targets with high efficacy and safety during co-infections [26].
To conclude, it is important that the bacterial co-infections in critically ill patients should not be underestimated. It should be taken as a key factor in order to reduce the morbidity and mortality rate in COVID-19 patients globally. Investigation of bacterial co-infections and antibiotic profile can further help in improved health of COVID-19 patients and also will help us understand the viral and bacterial pathogen interaction within the host [26]. Ethical and rational use of antibiotics is highly recommended.
Acknowledgement
We thank Chettinad Hospital and Research Institute for the support to carry out this study.
Figures and tables
Footnotes
Conflict of interest statement
No conflicts of interest.
Authors’ contributions
PS and SR designed the study. SR collected and analysed the data. SR, PS and APSR participated in the manuscript revision. All authors of the manuscript gave their complete approval.
References
- [1].World Health Organization. WHO Coronavirus disease (COVID-2019) dashboard. Available at: https://covid19.who.int
- [2].Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. https://doi.org/10.1016/S0140-6736(20)30183-5 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054-62. https://doi.org/10.1016/S0140-6736(20)30566-3 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol 2017;8:1041. https://doi.org/10.3389/fmicb.2017.01041 10.3389/fmicb.2017.01041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alhazzani W, Møller MH, Arabi YM, Loeb M, Gong MN, Fan E, Oczkowski S, Levy MM, Derde L, Dzierba A, Du B, Aboodi M, Wunsch H, Cecconi M, Koh Y, Chertow DS, Maitland K, Alshamsi F, Belley-Cote E, Greco M, Laundy M, Morgan JS, Kesecioglu J, McGeer A, Mermel L, Mammen MJ, Alexander PE, Arrington A, Centofanti JE, Citerio G, Baw B, Memish ZA, Hammond N, Hayden FG, Evans L, Rhodes A. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med 2020;46:854-87. https://doi.org/10.1007/s00134-020-06022-5 10.1007/s00134-020-06022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, Han Y, Hu B, Hu F, Li BH, Li YR, Liang K, Lin LK, Luo LS, Ma J, Ma LL, Peng ZY, Pan YB, Pan ZY, Ren XQ, Sun HM, Wang Y, Wang YY, Weng H, Wei CJ, Wu DF, Xia J, Xiong Y, Xu HB, Yao XM, Yuan YF, Ye TS, Zhang XC, Zhang YW, Zhang YG, Zhang HM, Zhao Y, Zhao MJ, Zi H, Zeng XT, Wang YY, Wang XH, for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team, Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM). A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 2020;7:4. https://doi.org/10.1186/s40779-020-0233-6 10.1186/s40779-020-0233-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Critical CR, Medicine C, Hospital YC, Medicine AO. Chinese experts’ consensus on diagnosis and treatment of severe and critical coronavirus disease 2019 (revised edition). Zhonghua wei zhong bing ji jiu yi xue 2020;32:269-74. https://doi.org/10.3760/cma.j.cn121430-20200218 10.3760/cma.j.cn121430-20200218 [DOI] [PubMed] [Google Scholar]
- [8].Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases of coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 2020;71:769. https://doi.org/10.1093/cid/ciaa272 10.1093/cid/ciaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li J, Wang J, Yang Y, Cai P, Cao J, Cai X, Zhang Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control 2020;9:153. https://doi.org/10.1186/s13756-020-00819-1 10.1186/s13756-020-00819-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507-13. https://doi.org/10.1016/S0140-6736(20)30211-7 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dong X, Cao YY, Lu XX, Zhang JJ, Du H, Yan YQ, Akdis CA, Gao YD. Eleven faces of coronavirus disease 2019. Allergy 2020;75:1699-709. https://doi.org/10.1111/all.14289 10.1111/all.14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, Satlin MJ, Campion TR, Jr, Nahid M, Ringel JB, Hoffman KL, Alshak MN, Li HA, Wehmeyer GT, Rajan M, Reshetnyak E, Hupert N, Horn EM, Martinez FJ, Gulick RM, Safford MM. Clinical Characteristics of COVID-19 in New York City. N Engl J Med 2020;382:2372-4. https://doi.org/10.1056/NEJMc2010419. 10.1056/NEJMc2010419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mackie TJ, Collee J. G. McCartney, Mackie & McCartney practical medical microbiology 14 th edition. New York:Churchill Livingstone; 2006. [Google Scholar]
- [14].Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, Soucy JP, Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 2020;26:1622-9. https://doi.org/10.1016/j.cmi.2020.07.016 10.1016/j.cmi.2020.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Russell CD, Fairfield CJ, Drake TM, Turtle L, Seaton RA, Wootton DG, Sigfrid L, Harrison EM, Docherty AB, de Silva TI, Egan C, Pius R, Hardwick HE, Merson L, Girvan M, Dunning J, Nguyen-Van-Tam JS, Openshaw PJM, Baillie JK, Semple MG, Ho A; ISARIC4C investigators. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet Microbe 2021;2:e354-65. https://doi.org/10.1016/S2666-5247(21)00090-2 10.1016/S2666-5247(21)00090-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mahmoudi H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS Hyg Infect Control 2020;15. https://doi.org/10.3205/dgkh000370 10.3205/dgkh000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Khurana S, Singh P, Sharad N, Kiro VV, Rastogi N, Lathwal A, Malhotra R, Trikha A, Mathur P. 2021. Profile of co-infections & secondary infections in COVID-19 patients at a dedicated COVID-19 facility of a tertiary care Indian hospital: Implication on antimicrobial resistance. Indian J Med Microbiol 2021;39:147-53. https://doi.org/10.1016/j.ijmmb.2020.10.014 10.1016/j.ijmmb.2020.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sharifipour E, Shams S, Esmkhani M, Khodadadi J, Fotouhi-Ardakani R, Koohpaei A, Doosti Z, Ej Golzari S. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis 2020;20:1-7. https://doi.org/10.1186/s12879-020-05374-z 10.1186/s12879-020-05374-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rezasoltani S, Yadegar A, Hatami B, Aghdaei HA, Zali MR. Antimicrobial Resistance as a Hidden Menace Lurking Behind the COVID-19 Outbreak: The Global Impacts of Too Much Hygiene on AMR. Front Microbiol 2020;11:590683. https://doi.org/10.3389/fmicb.2020.590683 10.3389/fmicb.2020.590683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sreenath K, Batra P, Vinayaraj EV, Bhatia R, SaiKiran KV, Singh V, Singh S, Verma N, Singh UB, Mohan A, Bhatnagar S, Trikha A, Guleria R, Chaudhry R. Coinfections with other respiratory pathogens among patients with COVID-19. Microbiol Spectr 2021;9:e00163-21. https://doi.org/10.1128/Spectrum.00163-21 10.1128/Spectrum.00163-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arcari G, Raponi G, Sacco F, Bibbolino G, Di Lella FM, Alessandri F, Coletti M, Trancassini M, Deales A, Pugliese F, Antonelli G, Carattoli A. Klebsiella pneumoniae infections in COVID-19 patients: a 2-month retrospective analysis in an Italian hospital. Int J Antimicrob Agents 2021;57:106245. https://doi.org/10.1016/j.ijantimicag.2020.106245 10.1016/j.ijantimicag.2020.106245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Qu J, Cai Z, Liu Y, Duan X, Han S, Liu J, Zhu Y, Jiang Z, Zhang Y, Zhuo C, Liu Y, Liu Y, Liu L, Yang L. Persistent Bacterial Coinfection of a COVID-19 Patient Caused by a Genetically Adapted Pseudomonas aeruginosa Chronic Colonizer. Front Cell Infect Microbiol 2021;11:641920. https://doi.org/10.3389/fcimb.2021.641920 10.3389/fcimb.2021.641920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lucien MAB, Canarie MF, Kilgore PE, Jean-Denis G, Fénélon N, Pierre M, Cerpa M, Joseph GA, Maki G, Zervos MJ, Dely P, Boncy J, Sati H, Rio AD, Ramon-Pardo P. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int J Infect Dis 2021;104:250-4. https://doi.org/10.1016/j.ijid.2020.12.087 10.1016/j.ijid.2020.12.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vaillancourt M, Jorth P. The unrecognized threat of secondary bacterial infections with COVID-19. MBio 2020;11(4). https://doi.org/10.1128/mBio.01806-20 10.1128/mBio.01806-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rawson TM, Moore LS, Castro-Sanchez E, Charani E, Davies F, Satta G, Ellington MJ, Holmes AH. COVID-19 and the potential long-term impact on antimicrobial resistance. J Antimicrob Chemother 2020;75:1681-4. https://doi.org/10.1093/jac/dkaa194 10.1093/jac/dkaa194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bengoechea JA, Bamford CG. SARS-CoV-2, bacterial co-infections, and AMR: the deadly trio in COVID-19? EMBO Mol Med 2020;12:e12560. https://doi.org/10.15252/emmm.202012560 10.15252/emmm.202012560 [DOI] [PMC free article] [PubMed] [Google Scholar]


