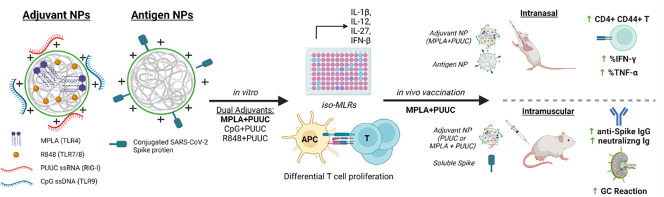
Abstract
Despite success in vaccinating populations against SARS-CoV-2, concerns about immunity duration, continued efficacy against emerging variants, protection from infection and transmission, and worldwide vaccine availability remain. Molecular adjuvants targeting pattern recognition receptors (PRRs) on antigen-presenting cells (APCs) could improve and broaden the efficacy and durability of vaccine responses. Native SARS-CoV-2 infection stimulates various PRRs, including toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors. We hypothesized that targeting PRRs using molecular adjuvants on nanoparticles (NPs) along with a stabilized spike protein antigen could stimulate broad and efficient immune responses. Adjuvants targeting TLR4 (MPLA), TLR7/8 (R848), TLR9 (CpG), and RIG-I (PUUC) delivered on degradable polymer NPs were combined with the S1 subunit of spike protein and assessed in vitro with isogeneic mixed lymphocyte reactions (isoMLRs). For in vivo studies, the adjuvant-NPs were combined with stabilized spike protein or spike-conjugated NPs and assessed using a two-dose intranasal or intramuscular vaccination model in mice. Combination adjuvant-NPs simultaneously targeting TLR and RIG-I receptors (MPLA+PUUC, CpG+PUUC, and R848+PUUC) differentially induced T cell proliferation and increased proinflammatory cytokine secretion by APCs in vitro. When delivered intranasally, MPLA+PUUC NPs enhanced CD4+CD44+ activated memory T cell responses against spike protein in the lungs while MPLA NPs increased anti-spike IgA in the bronchoalveolar (BAL) fluid and IgG in the blood. Following intramuscular delivery, PUUC NPs induced strong humoral immune responses, characterized by increases in anti-spike IgG in the blood and germinal center B cell populations (GL7+ and BCL6+ B cells) in the draining lymph nodes (dLNs). MPLA+PUUC NPs further boosted spike protein-neutralizing antibody titers and T follicular helper cell populations in the dLNs. These results suggest that protein subunit vaccines with particle-delivered molecular adjuvants targeting TLR4 and RIG-I could lead to robust and unique route-specific adaptive immune responses against SARS-CoV-2.
Keywords: COVID-19 protein subunit vaccine, SARS-CoV-2 spike protein, Combination adjuvant, Monophosphoryl lipid A, Intranasal versus intramuscular vaccination, Adaptive immune response
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by SARS-CoV-2 virus, has elicited a global scientific effort to develop vaccines and therapies at an unprecedented rate. Though mRNA-based vaccines against SARS-CoV-2 spike protein have been approved by the FDA, much work remains to develop long-term immunity and understand how both local immunity in the lung and systemic immunity protect against emerging SARS-CoV-2 variants. The incorporation of molecular adjuvants in vaccines, specifically protein subunit vaccines, is a potential strategy to induce robust immune responses against SARS-CoV-2 through the targeting of receptors on antigen-presenting cells (APCs) [1,2]. Aluminum-containing adjuvants (e.g., alum) have been included in vaccines to enhance immunogenicity since the 1930s [3]. Traditional adjuvants like alum have been developed and tested empirically, but in recent years vaccine design strategy has shifted to a more rational approach where each component elicits a defined immunological pathway to modulate the immune response. Pathogen-associated molecular patterns (PAMPs), including lipids, carbohydrates, peptides, and nucleic acids commonly expressed by pathogens, are currently investigated as adjuvants because they specifically bind to pattern recognition receptors (PRRs) on APCs and induce maturation [2]. Mature APCs, namely dendritic cells (DCs), initiate antigen-specific adaptive immune responses by activating naïve T cells, which differentiate into effector cells, such as cytotoxic T cells (CD8+) and T helper cells (CD4+) [4]. T helper cells (type 2, Th2) mediate the differentiation of B cells into antibody-producing plasma cells [5,6].
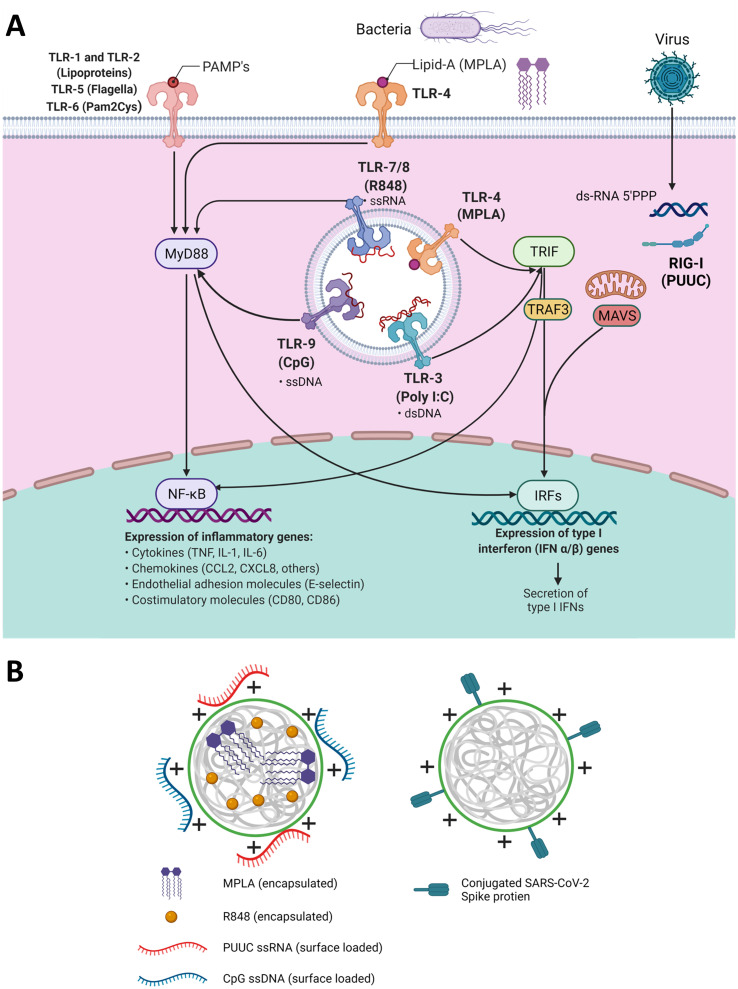
Two classes of PRRs are toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors. RIG-I receptors are in the cytosol and recognize short double-stranded RNA (dsRNA), a replication intermediate for RNA viruses, that exhibit viral motifs such as an uncapped 5′ diphosphate or triphosphate end [7]. The poly-uridine core of Hepatitis C virus (HCV), poly-U/UC (PUUC), activates RIG-I and triggers potent anti-HCV responses [8]. The most studied TLR and ligand combination is TLR4 and the Gram-negative bacterial cell wall component lipopolysaccharide (LPS), which includes monophosphoryl lipid A (MPLA), a molecule used in FDA-approved adjuvant systems (AS01 and AS04, GlaxoSmithKline) [[9], [10], [11]]. Endosomal TLRs 7 and 8 recognize single-stranded RNA and can be activated by synthetic imidazoquinolines including resiquimod (R848) [12]. Endosomal TLR9 recognizes single-stranded DNA, and current understanding is that TLR9 recognizes unmethylated CpG motifs common to bacteria and viruses to discriminate from self-DNA [13]. Most TLRs (TLR4, TLR7/8, TLR9) signal through the MyD88 pathway, which activates NF-κB to induce production of proinflammatory cytokines (i.e., IL-1β) and IRF7 to induce type I interferon secretion [[14], [15], [16]]. When trafficked from the plasma membrane to endosomes, TLR4 can signal through the TRIF pathway and induce expression of IFN-β (Fig. 1A) [16,17].
Fig. 1.
TLR and RIG-I signaling pathways intersect and can be activated by nanoparticles with encapsulated and surface-loaded pathogen-associated molecular patterns. A) Schematic of signaling pathways initiated by TLRs 1–9, and RIG-I receptors including adaptor proteins MyD88, TRIF, TRAF3, and MAVS and transcription factors NF-κB and various IRFs which regulate the transcription of proinflammatory genes. B) Left: Depiction of poly(lactic-co-glycolic acid)-polyethyleneimine nanoparticles (PLGA-PEI NPs) with encapsulated hydrophobic molecules (R848 or MPLA) and surface-loaded nucleic acids (CpG or PUUC) for adjuvant delivery during vaccination. Right: PLGA-PEI NPs with surface-loaded SARS-CoV-2 spike protein for antigen delivery during vaccination.
A single pathogen can express multiple PAMPs which concurrently stimulate multiple PRRs on immune cells. For instance, the live-attenuated yellow fever vaccine activates TLR2, TLR7, TLR8 and TLR9 on different DC subsets. Similarly, the live-attenuated Bacillus Calmette-Guérin (BCG) tuberculosis vaccine signals through TLR2, TLR4 and TLR9, among other PRRs [18]. Positive-sense single-stranded RNA viruses like SARS-CoV-2 interact with TLR7 and TLR8, and produce dsRNA replication intermediates that are recognized by RIG-I and TLR3 [[19], [20], [21], [22]]. TLR2 (dimerized with TLR1 or TLR6) and TLR4 have also exhibited potential to recognize viral proteins; in fact, SARS-CoV-2 spike protein reportedly induces TLR4 signaling [[23], [24], [25]]. Studies have shown that combinations of PAMPs result in synergistic, complementary, and antagonistic effects on innate and adaptive immunity. We have recently reported co-delivery of MPLA and CpG DNA on poly(lactic-co-glycolic acid)-polyethyleneimine (PLGA-PEI) particles induces prolonged IRF5 phosphorylation, leading to synergistic increases in proinflammatory cytokine secretion in APCs [26]. Further, we have demonstrated that intramuscular vaccination with R848, PUUC and H1N1 hemagglutinin on PLGA-PEI particles elevates CD8+ T cell populations in the lung and antigen-specific CD4+ T cell populations in the spleen in mice [27].
To evaluate the effects of PRR-targeted adjuvants on SARS-CoV-2 protein subunit vaccination, we first formulated PLGA-PEI nanoparticles (NPs) with pairings of MPLA (TLR4), R848 (TLR7/8), or CpG DNA (TLR9) plus PUUC (RIG-I), and then combined the NPs with SARS-CoV-2 spike S1 subunit (containing the ACE-2 receptor binding domain) for in vitro studies. Adjuvant-NPs were combined with full stabilized spike protein or spike-conjugated PLGA-PEI NPs (spike-NPs) for in vivo studies. The various formulations were tested in vitro using isomixed leukocyte reactions (isoMLRs) and in vivo using two-dose intranasal and intramuscular vaccination models in mice. SARS-CoV-2 is an airborne virus that is mainly transmitted through contact with respiratory droplets from the nose and throat of an infected person [28,29], resulting in severe lung infection and subsequent systemic infection (i.e., viremia) [30]. Therefore, we explored intranasal (I.N.) vaccination to induce local immune memory in the lung for prevention against SARS-CoV-2 infection and transmission, as well as intramuscular (I.M.) vaccination (the current method for all FDA-approved SARS-CoV-2 vaccines). We show that I.N. vaccination with MPLA+PUUC NPs and spike-NPs induced CD4+CD44+ memory T cell responses in the lung. Overall, I.N. vaccination did not produce strong humoral responses; however, MPLA NPs plus soluble spike protein generated increases in anti-spike protein IgA and IgG in broncho-alveolar lavage (BAL) fluid and serum, respectively. PUUC NPs, when delivered I.M. with various doses of soluble spike protein, produced strong humoral responses characterized by increases in anti-spike IgG and neutralizing antibodies as well as germinal center B cell expansion in draining lymph nodes (dLNs). I.M. vaccination with MPLA+PUUC NPs plus soluble spike protein generated lower total anti-spike IgG compared to PUUC NPs, but a higher neutralizing antibody response. Our results show route-specific immune polarization induced by SARS-CoV-2 protein subunit vaccines adjuvanted with NPs carrying MPLA and PUUC, specifically localized T cell responses in the lungs after I.N. vaccination and systemic humoral responses after I.M. vaccination.
2. Materials and methods
2.1. Synthesis of PUUC (RIG-I agonist)
The RIG-I agonist poly-U/UC (PUUC) is based on a Hepatitis C Virus (HCV) RNA sequence [8]. PUUC RNA was transcribed from custom DNA templates (Integrated DNA Technologies, Custom PAGE-purified Ultramer oligos) with the MEGAshortscript™ T7 Transcription Kit (Invitrogen, #AM1354) as previously described [17]. The templates were: 5′-TAA TAC GAC TCA CTA TAG GCC ATC CTG TTT TTT TCC CTT TTT TTT TTT CTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTC TCC TTT TTT TTT CCT CTT TTT TTC CTT TTC TTT CCT TT-3′ (forward) and 5′-AAA GGA AAG AAA AGG AAA AAA AGA GGA AAA AAA AAG GAG AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AGA AAA AAA AAA AGG GAA AAA AAC AGG ATG GCC TAT AGT GAG TCG TAT TA-3′ (reverse). PUUC was purified by chilled ethanol precipitation followed by centrifugation and resuspension in nuclease-free water (Boston BioProducts, #R-100DR) at 0.5 to 1.0 mg/mL. Yields were quantified using the Nucleic Acid Quantification workflow on a Synergy HT plate reader (BioTek) with Gen5 software. PUUC was stored at −80°C.
2.2. PLGA-PEI nanoparticle synthesis and adjuvant loading
PLGA NPs were synthesized using a double-emulsion and solvent evaporation method previously reported [26,27,31]. PLGA (50:50, MW: 7000–17,000, Resomer RG 502H, Sigma-Aldrich, #719897) was dissolved in dichloromethane (DCM, Sigma-Aldrich, #270997) in a 1:20 w/v ratio. Endotoxin-free water was added in a 1:4 v/v ratio to the mixture to form the first water-oil emulsion. For particles with encapsulated adjuvant, either R848 (STEMCELL Technologies, #73784) or MPLA PHAD® (1 μg/mg PLGA, Avanti Polar Lipids, #699800P) was dissolved in the DCM added to the first water-oil emulsion. The first emulsion was sonicated at 65% power for 2 min at RT, and then added to 5% PVA (MW: 31,000–50,000) in water at a 5:16 v/v ratio to form a second water-oil-water emulsion. The second emulsion was sonicated at 65% power for 5 min at RT. DCM was evaporated by magnetically stirring the second emulsion at RT for 3 h. Large PLGA aggregates were removed by centrifugation at 2000 x g for 10 min. The supernatant was ultracentrifuged at 80,000 x g for 20 min to pellet the PLGA NPs. NPs were washed with DI water using ultracentrifugation, resuspended in DI water, and lyophilized for 48 h.
Branched PEI (Polysciences, #06090) was coated onto the PLGA NPs by EDC (Thermo Scientific, #22980) and sulfo-NHS (Thermo Scientific, #PG82071) chemistry to produce cationic NPs. PLGA NPs were resuspended in 0.1 M aqueous MES (Sigma-Aldrich, #M5287) and 40 M excess of EDC and 25 M excess of sulfo-NHS were added to the suspension. After end-to-end rotation for 2 h, a 1:2 v/v bPEI solution in 0.2 MES was added to the particle suspension. After stirring for 2 h, particles were ultracentrifuged at 80,000 x g twice in 1 M NaCl and once in DI water. PLGA-PEI NPs were resuspended in DI water and lyophilized for 48 h. Nanoparticle size and surface zeta potential were measured with a Zetasizer Nano ZS (Malvern). For particles electrostatically loaded with nucleic acid adjuvants, either Class B CpG ODN 1826 (Invitrogen, #tlrl-1826) or PUUC was incubated with particles in 10 mM sodium phosphate buffer (made with nuclease-free water) under end-to-end rotation for 24 h.
Adjuvant concentrations (μg adjuvant per mg NP, Table 1 ) were selected based on data from previous work evaluating the loading efficiencies of TLR agonists on PLGA-PEI particles (MPLA, CpG), the effects of adjuvant density on synergistic TLR signaling (MPLA, CpG), and the effects of dosing ratio (R848, PUUC) on interferon production by murine and human pDCs in vitro [26,27,32]. R848 loading was determined by dissolving particles in sterile-filtered DMSO (Tocris, #3176) followed by absorbance readings against a standard curve at 324 nm. The target adjuvant concentration for R848 was 6 μg per mg PLGA; however, due to batch-to-batch variation, encapsulation of R848 in PLGA NP ranged between 4.8 and 6.67 μg per mg PLGA NP. MPLA loading estimation via GC–MS, LC-MS, and surrogate fluorometry has been previously described [26]. Incorporation of MPLA in PLGA NP was assumed to be 100% based on a previous study [33]. To quantify the loading efficiency of PUUC and CpG, PLGA-PEI NPs were centrifuged at 10,000 x g for 20 min. Drops of supernatants were added to the BioTek Take3 microvolume plate and measured for unbound RNA (PUUC) or ssDNA (CpG) using the Nucleic Acid Quantification feature of the Gen5 software on a Synergy HT plate reader. The loading efficiency of CpG and PUUC was confirmed to be 100%.
Table 1.
Single and combination adjuvant loading on PLGA NP and doses for GM-CSF and FLT3L BMDCs in isoMLR assays. Size, PDI and zeta measurements were taken for all NPs prior to electrostatically loading adjuvants CpG or PUUC. PDI is polydispersity index. *The target adjuvant concentration for R848 was 6 μg per mg PLGA. Due to batch-to-batch variation, encapsulation of R848 in PLGA NP ranged between 4.8 and 6.67 μg per mg NP and the adjuvant dose ranged between 80 to ~110 ng per 500,000 BMDCs.
| PLGA-PEI NP Formulation | Size (nm) | PDI | Zeta (mV) | Targeted adjuvant concentration |
Adjuvant dose for BMDCs |
|---|---|---|---|---|---|
| (per mg NP) | (ng/5e5 cells/mL) | ||||
| CpG NP | 269.4 ± 41.1 | 0.45 | 31.7 ± 0.7 | 60 μg CpG | 500 ng CpG |
| MPLA NP | 246.0 ± 69.26 | 0.134 | 25.0 ± 4.92 | 6 μg MPLA | 100 ng MPLA |
| PUUC NP | 269.4 ± 41.1 | 0.45 | 31.7 ± 0.7 | 6 μg PUUC | 100 ng PUUC |
| R848 NP | 253.2 ± 61.10 | 0.37 | 29.1 ± 2.3 | 4.8 μg R848 | 100 ng R848 |
| CpG+PUUC NP | 269.4 ± 41.1 | 0.45 | 31.7 ± 0.7 | 30 μg CpG and 6 μg PUUC |
500 ng CpG and 100 ng PUUC |
| MPLA+CpG NP | 246.0 ± 69.26 | 0.134 | 25.0 ± 4.92 | 6 μg MPLA and 60 μg CpG |
50 ng MPLA and 500 ng CpG |
| MPLA+PUUC NP | 246.0 ± 69.26 | 0.134 | 25.0 ± 4.92 | 6 μg MPLA and 6 μg PUUC |
100 ng MPLA and 100 ng PUUC |
| R848+PUUC NP | 253.2 ± 61.10 | 0.37 | 29.1 ± 2.3 | 6 μg R848* and 6 μg PUUC |
100 ng R848* and 100 ng PUUC |
2.3. Spike-conjugated nanoparticle synthesis
To conjugate SARS-CoV-2 spike to PLGA-PEI NPs (spike-NPs), free amines of particles were converted to thiols via 2-iminothiolane (Traut's reagent). Initially, PLGA-PEI NPs were dispersed in 100 mM phosphate buffer with 2 mM EDTA (pH = 8.0) and mixed with excess amount of Traut's reagent (Sigma-Aldrich, #I6256). After continuous rotation overnight at RT, thiolated PLGA-PEI (PLGA-PEI-SH) NPs were purified by centrifugation, washing, and lyophilization overnight. Thiol groups on particles were estimated by Ellman's reagent (G Biosciences, #BC87) using cysteine (Sigma-Aldrich, #168149) as the standard, yielding a 60–70% thiolation efficiency. Until spike loading, PLGA-PEI-SH NPs were stored at −20°C. Prior to in vivo experiments, a stock solution of NHS-PEG-SPDP (bifunctional crosslinker, Sigma-Aldrich #803499) was prepared in anhydrous DMSO (10 mg/mL, Sigma-Aldrich, #276855). Further, excess NHS-PEG-SPDP (200 μg) crosslinker was added to well-dispersed PLGA-PEI-SH NPs solution in PBS (5 mg/mL, pH = 7.6) with stabilized SARS-CoV-2 spike glycoprotein (10 μg/mg NP, with Avi Tag, BEI Resources, #NR-53524). Following rotation for 6 h at RT, NPs were centrifuged at 21,000 x g for 10 min, and supernatant was saved (first wash). NPs were washed with nuclease-free water, centrifuged, and supernatant was also saved (second wash). Supernatant washes were filtered through a 100,000 MWCO Ultra-4 Amicon centrifugal filter (Millipore Sigma, #UFC810096) to retain unconjugated spike glycoprotein and separate remaining NHS-PEG-SPDP crosslinker. Unconjugated spike protein was measured via micro-BCA (Boster Bio, #AR1110). The loading efficiency of spike protein on nanoparticles ranged from 70 to 75%. The concentration of spike-NPs was adjusted to match the dose of soluble spike protein in all in vivo experiments.
2.4. Bone marrow derived dendritic cell (BMDC) culture
Female Balb/cJ (5–10 weeks old, Jackson Labs) were euthanized with CO2. PBS was flushed through tibiae and femurs to isolate bone marrow, which was filtered through a 40 μm strainer to create single-cell suspensions. Bone marrow cells were centrifuged (500 x g, 5 min), resuspended in RBC lysis buffer (Thermo Fisher, #00-4333-57), and incubated at RT for 10 min. Cells were centrifuged and resuspended in complete RPMI medium (with l-glutamine, Gibco, #11875119) comprised of 10% characterized fetal bovine serum (GE Healthcare, #SH30071.03), 1% penicillin-streptomycin (Corning, #30-002-CI), 55 μM β-mercaptoethanol (Thermo Fisher, #21985023), and 1 mM sodium pyruvate (Thermo Fisher, #11360070). To generate GM-CSF BMDCs, cells were concentrated at 1 million cells/ mL in petri dishes (20 mL per dish) and medium was supplemented with 20 ng/mL murine GM-CSF (Peprotech, #315-03). On days 2 and 4, 10 mL of medium was carefully removed from each petri dish and replaced with fresh medium (40 ng/mL GM-CSF). On day 6, 10 mL of medium was added (60 ng/mL GM-CSF) without any medium removal. Experiments with GM-CSF BMDCs were conducted on day 7 of culture. To generate FLT3L BMDCs, cells were concentrated at 2 million cells/mL in 6-well plates (5 mL per well) and media was supplemented with 200 ng/mL human FLT3 ligand (PeproTech, #300-19) instead of GM-CSF. Experiments with FLT3L BMDCs were conducted on day 9 of culture (no media changes). All BMDCs were cultured at 37°C with 5% CO2.
2.5. Characterization of APC subsets in BMDC culture
FLT3L and GM-CSF BMDCs were stained with 1:1000 Zombie Green Fixable Viability Kit (BioLegend, #423111) and were blocked with anti-mouse CD16/32 (BioLegend, #101320) and True-Stain Monocyte Blocker (BioLegend, #426102). After blocking, cells were stained with anti-mouse CD11b (BUV395, BD, #563553), CD11c (Brilliant Violet 421, Biolegend, #117343), B220 (APC, Biolegend, #103212), Ly-6C (Brilliant Violet 711, Biolegend, #128037), Ly-6G (Brilliant Violet 785, Biolegend, #127645), CD64 (PE, Biolegend, #139304), F4/80 (PE/Cy5, Biolegend, #123112), and MHC-II (APC/Cy7, Biolegend, #107628). Cells were fixed with BD Cytofix (#554655) and analyzed with a BD LSRFortessa flow cytometer. FlowJo Software (BD) was used to generate t-distributed stochastic neighbor embedding (t-SNE) plots.
2.6. In vitro activation of BMDCs and iso-mixed lymphocyte reaction (iso-MLR)
Adjuvant-loaded PLGA-PEI NPs (adjuvant-NPs) and the S1 subunit of the SARS-CoV-2 spike protein (Novus Biologicals, #NBP2-90985, 100 ng protein/500,000 cells/mL RPMI medium) were added to GM-CSF or FLT3L BMDCs in a U-bottom 96 well plate. Each well contained 100,000 BMDCs in 200 mL complete RPMI medium and adjuvant doses are outlined in Table 1. The concentration of PLGA-PEI NPs was adjusted to match the adjuvant dose and the NP-only (i.e., Blank) control was matched to the highest PLGA-PEI NP concentration. The cells were incubated with S1 protein and adjuvant-NPs for 24 h before addition of T cells. To isolate T cells, spleens from female Balb/cJ mice were dissociated in 2 mg/mL Collagenase D (Sigma-Aldrich, #11088882001) in Opti-MEM media (Gibco, #11058021) and filtered through a 40 μm cell strainer. T cells were isolated using the Mouse Pan-T Cell Isolation Kit II (Miltenyi Biotec, #130-095-130), labeled with CellTrace™ CFSE (Thermo Fisher, #C34554), and resuspended in complete RPMI medium at 1 million cells/mL. BMDCs in U-bottom plates were centrifuged (500 x g, 5 min) and supernatants were collected and frozen at −20°C. 200,000 T cells were added to each well containing BMDCs. After 72 h, T cells were stained with Zombie UV (Biolegend, #423107) to exclude dead cells, and anti-mouse CD3 (Brilliant Violet 786, Biolegend), anti-mouse CD4 (APC, Biolegend) and anti-mouse CD8a (PE, Biolegend) to identify CD4+ and CD8+ T cells. A well containing only CSFE-stained T cells was included as a non-proliferative control. Percentages of proliferating T cells were calculated by gating on the reduced FITC signal (Fig. S1). DuoSet ELISA kits (R&D Systems) were used to quantify IFN-β and IFN-λ3 from BMDC supernatants. IL-12p70, IL-1β, and IL-27 were quantified using customized Luminex® assays (R&D systems). Supernatants were diluted 4-fold for all cytokines except for IL-1β, in which case they were diluted 8-fold. Supernatants were analyzed in triplicate for each experimental condition.
2.7. In vivo intranasal vaccination
On days 0 (1st dose) and 28 (2nd dose), 4 mg adjuvant-NPs were combined with 1 μg spike protein antigen in 60 μL saline (0.9% sodium chloride, Hanna Pharmaceutical, #0409488810) and administered to 9- to 10-week-old female BALB/cJ mice dropwise in the bilateral nares. IVIS® fluorescence imaging (PerkinElmer) confirmed adjuvant-NPs are present in the trachea and all mouse lung lobes 24 h after I.N. administration (Fig. S2). For seven of the treatment groups, mice received 1 μg of unformulated (i.e., soluble) stabilized SARS-CoV-2 spike protein (R&D Systems, #10549-CV-100) and 4 mg adjuvant-NPs with total adjuvant doses of MPLA (24 μg), CpG (20 μg), PUUC (17 μg), CpG+PUUC (20 μg and 17 μg), MPLA+PUUC (20 μg and 17 μg), R848+PUUC (20 μg and 20 μg), or MPLA+CpG (24 μg and 20 μg). For two of the treatment groups, mice received 1 μg of antigen on spike-NPs and 4 mg adjuvant-NPs with total adjuvant doses of PUUC (17 μg) or MPLA+PUUC (20 μg and 17 μg). An additional treatment group received 2 μg of unformulated stabilized SARS-CoV-2 spike protein administered with 4 mg adjuvant-NPs containing 20 μg MPLA and 17 μg PUUC. Two antigen-only control groups included mice receiving 1 μg antigen unformulated or on spike-NPs in 60 μL saline. An untreated group received only 60 μL saline. Half of each group (3 mice) was euthanized with sodium pentobarbital on day 27 to collect blood using cardiac puncture and bronchoalveolar lavage (BAL) fluid using methods described in Hoecke et al. [34] The remaining 3 mice in each group were euthanized on day 35, and blood, BAL fluid, and lungs were collected. All blood samples were clotted for 30–60 min at RT in serum separator tubes (BD, #365967), and sera were isolated by centrifugation at 4000 x g for 15 min at 4°C. Sera were heat inactivated at 56°C for 30 min in a water bath to inhibit complement binding and then stored at −80°C. BAL fluid samples were centrifuged to remove cells and stored at −80°C.
2.8. Ex vivo lung cell restimulation
Harvested lungs from I.N. vaccinated mice were processed into single cell suspensions with a gentleMACS™ Octo Dissociator and Lung Dissociation Kit (Miltenyi Biotec) according to manufacturer's instructions including RBC lysis. Cells were centrifuged (500 x g, 5 min), resuspended at 10 million cells/mL in complete RPMI medium, plated in a U-bottom 96-well plate (2.4 million cells per well), and incubated at 37°C with 5% CO2 overnight. Lung cells were centrifuged and resuspended in fresh RPMI complete medium containing 20 μL/mL of PeptTivator® SARS-CoV-2 Prot_S (Miltenyi Biotec) and 5 μg/mL Brefeldin A (BioLegend). After 6 h, cells were stained for 30 min at RT with Zombie Green™ Fixable Viability Kit (BioLegend) and blocked with anti-mouse CD16/32 (BioLegend) and True-Stain Monocyte Blocker™ (BioLegend). Following blocking, cell surfaces were stained for 30 min at 4°C with anti-mouse CD45 (BD, BUV395), CD44 (Biolegend, BV711), CD69 (Biolegend, BV785), CD103 (Biolegend, PE-Dazzle 594), CD8a (Biolegend, PE/Cy5), and CD4 (Biolegend APC). Surface-stained cells were fixed and permeabilized for 30 min at 4°C with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Then cells were stained with anti-mouse IFN-γ (PE), Granzyme B (Pacific Blue), and TNF-α (PE/Cy7). Cell fluorescence was quantified using a BD LSRFortessa™ flow cytometer.
2.9. In vivo intramuscular vaccination and analysis of popliteal lymph nodes
Four mg of adjuvant-NPs (4 mg/mouse) loaded with MPLA (24 μg), PUUC (20 μg) or MPLA+PUUC (24 μg, 20 μg) was combined with variable doses (80 ng, 200 ng, 1000 ng) of unformulated stabilized SARS-CoV-2 spike glycoprotein (BEI Resources, #NR-52397) in 100 μL saline. 50 μL was injected into the left and 50 μL into the right anterior tibialis muscle of 9- to 10-week-old female BALB/cJ mice on day 0 (1st dose) and day 28 (2nd dose). On day 26, blood was sampled from all mice via the jugular veins (no euthanasia) and serum was separated using methods previously outlined. On day 36, mice were euthanized with CO2 and blood, bilateral popliteal lymph nodes, and spleens were collected. Both popliteal lymph nodes from each mouse were combined and passed through a 40 μm cell strainer to generate single cell suspensions, which were centrifuged (500 x g, 5 min) and washed with PBS. Lymph node cells were stained with Zombie Green™ (Biolegend) and blocked with anti-mouse CD16/32 as above. After blocking, cell surfaces were stained for 30 min at 4°C with anti-mouse B220 (Biolegend, APC), GL7 (Biolegend, PE/Cy7), and CXCR5 (Biolegend, BV421). Surface-stained lymph node cells were then fixed, permeabilized and intracellularly stained with anti-mouse BCL6 (Biolegend, PE) in permeabilization buffer. Cell fluorescence was quantified using a BD FACSymphony™ A5 Cell Analyzer. A parallel I.M. vaccination study was conducted with the same adjuvant-NP and control groups, except antigen was delivered on spike-NPs instead of unformulated.
2.10. Ex vivo splenocyte restimulation
Harvested spleens from I.M. vaccinated mice were processed into single cell suspensions with a gentleMACS™ Octo Dissociator and Spleen Dissociation Kit (Miltenyi Biotec). Splenocytes were centrifuged (500 x g, 5 min) and resuspended in complete RPMI medium at a concentration of 10 million cells per mL. Cells were plated in a U-bottom 96-well plate (2 million cells per well) and left to culture overnight at 37°C in 5% CO2. Cells were centrifuged and resuspended in fresh RPMI complete medium containing 40 μL/mL of PeptTivator® SARS-CoV-2 Prot_S (Miltenyi Biotec, #130-126-700), 1 μL/mL monensin (BioLegend), and 5 μg/mL Brefeldin A (BioLegend). After 6 h of incubation, splenocytes were stained for 30 min at RT with Zombie Green™ Fixable Viability Kit (BioLegend) and blocked with anti-mouse CD16/32 (BioLegend). Following blocking, cell surfaces were stained for 30 min at 4°C with anti-mouse CD8a (Biolegend, PE/Cy5), CD4 (Biolegend, APC), CD3 (BD, BUV395), CD44 (Biolegend, BV711). Surface-stained cells were fixed and permeabilized for 30 min at 4°C with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Then cells were stained with anti-mouse IFN-γ (Biolegend, PE), Granzyme B (Biolegend, BV421), IL-4 (Biolegend, PE/Dazzle 594) and TNF-α (Biolegend, PE/Cy7). Cell fluorescence was quantified using a BD FACSymphony™ A5 Cell Analyzer.
2.11. ELISA assay for quantifying anti-spike antibodies in serum and BAL fluid
SARS-CoV-2 stabilized spike protein (BEI Resources, #NR-52397) was diluted 1 μg/mL in 0.05 M carbonate-bicarbonate buffer (pH 9.6) and adsorbed onto Nunc™ MaxiSorp™ ELISA Plates by incubating 100 ng/well overnight at 4°C. Antigen-coated plates were washed three times with wash PBST (0.01 M PBS + 0.05% Tween-20), and plates were blocked for 6 to 8 h at 4°C with PBSTBA (PBST + 1% BSA + 0.02% NaN3). Blocked plates were incubated overnight at 4°C with 102- to 106-fold diluted BAL fluid or sera from I.N. vaccinated mice and sera from I.M. vaccinated mice. Plates were washed three times with PBST. A secondary biotinylated anti-mouse IgA, IgG, IgG1, or IgG2a antibody (SouthernBiotech) was diluted 5000-fold in 5-fold diluted PBSTBA and was added to plates for 2 h at RT. Plates were washed three times with PBST. 5000-fold diluted streptavidin-conjugated horseradish peroxidase (strep-HRP, ThermoFisher) was incubated for 2 h at RT. Plates were washed six times with PBST. Ultra TMB-ELISA Substrate Solution (ThermoFisher) was added to wells and plates were incubated for 15 to 25 min at RT. 2 N sulfuric acid was added to each well, and absorbance was measured at 450 and 630 nm on a Synergy HT plate reader (BioTek) with Gen5 software.
2.12. Assay for quantifying spike protein-neutralizing antibodies in serum
Spike-neutralizing antibodies were quantified using a modified ELISA assay in a 384-well UltraCruz® ELISA high-binding plate (Santa Cruz Biotechnology). Diluted spike protein in 0.05 M carbonate-bicarbonate buffer (1 μg spike/mL, pH 9.6) was incubated in wells of a 384-well plate (50 ng/well) overnight at 4°C. Plates were washed three times with PBST, and plates were blocked in PBSTBA for 6–8 h at 4°C. Blocked plates were incubated overnight at 4°C with 102- to 106-fold diluted sera from I.M. vaccinated mice. Plates were washed three times with PBST. Plates were incubated for 2 h at RT with 500 ng/mL (25 ng/well) recombinant biotinylated human ACE-2 (R&D Systems, #BT933-020) diluted in PBSTBA. Plates were washed three times with PBST, and 5000-fold diluted strep-HRP (ThermoFisher) was incubated for 2 h at RT. Plates were washed six times with PBST, and incubated with Ultra TMB-ELISA Substrate Solution (ThermoFisher) for 20 min. The reaction was stopped with 2 N sulfuric acid, and absorbance was measured at 450 and 630 nm on a Synergy HT plate reader (BioTek) with Gen5 software. Absorbances were normalized by row to correct for 384-well plate effects; specifically, absorbances of samples at 450 nm were divided by absorbances of blank wells at 450 nm in each row.
2.13. Statistical analysis
All flow cytometry FCS files were analyzed with FlowJo (v10, BD). Statistical analyses were performed with GraphPad Prism 9. Normality was assessed with the Kolmogorov-Smirnov test with Dallal-Wilkinson-Lilliefors p value. For more than two comparisons, statistical differences between normally distributed datasets were determined with a one-way ANOVA with Tukey's post-hoc test for multiple comparisons. Nonparametric datasets were evaluated with the Kruskal-Wallis test and Dunn's post-hoc test. For antibody titer quantifications with ELISA, titers were defined as the highest fold dilution with an absorbance outside the cutoff value. For total IgG, IgG1, and IgG2a titers, the cutoff value was defined as the mean plus three times the sample standard deviation of the absorbance values of the least-diluted saline groups. For neutralizing titers, the cutoff was defined as the mean minus four times the sample standard deviation of the normalized absorbances of the least-diluted saline groups [35].
3. Results
3.1. TLR- and RIG-I-targeted combination adjuvants differentially induce GM-CSF BMDC proinflammatory cytokine secretion and FLT3L BMDC activation of T cells when delivered with SARS-CoV-2 S1 subunit protein
Hydrophobic R848 or MPLA were encapsulated into PLGA NP using a w/o/w emulsion-solvent evaporation method previously published. Blank particles without adjuvants were also prepared as a negative control. The average diameter of PLGA NPs prior to PEI modification ranged from 240 to 270 nm [26,27,31]. To electrostatically load CpG DNA and PUUC RNA, PLGA NPs were modified with surface bPEI to produce cationic particles with an approximate zeta potential of 30 mV [26,27,31]. To assess if adjuvants are more immunostimulatory when paired with another or delivered alone, combination adjuvants MPLA+PUUC, CpG+PUUC, and R848+PUUC on NPs as well as single-adjuvant and unformulated (Blank) control NPs were mixed with SARS-CoV-2 S1 subunit in media and incubated with murine BMDCs generated using either GM-CSF or FLT3L cytokines in a 96 well plate (doses in Table 1). After 24 h, supernatants were collected to quantify BMDC proinflammatory cytokine secretion, and then T cells were added for another 72 h with activated BMDCs before quantifying T cell proliferation using CellTrace CFSE.
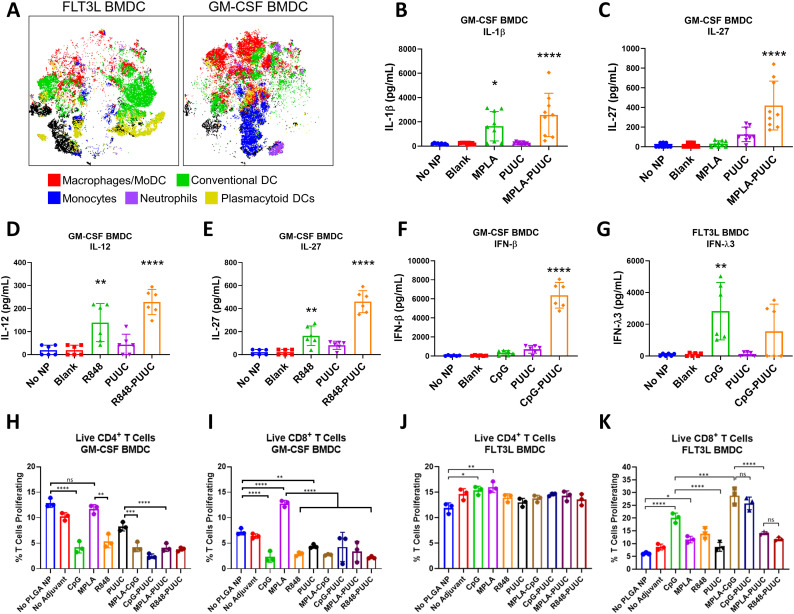
Flow cytometry and tSNE analysis revealed Day 9 FLT3L-derived BMDC cultures were composed of conventional dendritic cells (cDCs; CD11c+MHCIIhiCD64lo/−F4/80lo/−) and plasmacytoid dendritic cells (pDCs; B220+Ly6C+CD11c+MHCIIloLy6Glo), whereas Day 7 GM-CSF-derived BMDCs were primarily monocytes (Ly6ChiCD11c−) and monocyte-derived DCs/macrophages (MoDC; CD64+F4/80+). There is some macrophage survival in FLT3L culture, and neutrophil (Ly6GhiLy6C+) survival in GM-CSF culture (Fig. 2A). MPLA+PUUC NPs statistically increased IL-1β and IL-27 secretion, R848+PUUC NPs upregulated IL-12p70 and IL-27, and CpG+PUUC NPs increased production of IFN-β in GM-CSF BMDC culture compared to single-adjuvant NP, blank NP, and antigen-only controls (Fig. 2B–F). In FLT3L BMDC culture, however, CpG NPs increased and CpG+PUUC NPs antagonistically decreased IFN-λ3 secretion (Fig. 2G).
Fig. 2.
GM-CSF and FLT3L BMDCs secrete different cytokine profiles and activate CD8+ T cells in response to S1 protein with combination adjuvants. A) t-SNE plots of GM-CSF and FLT3L BMDCs with labeled clusters of APC subsets. Macrophages/Mo-DCs are CD64+ F4/80−, Conventional DCs are CD11c+ MHCII+ CD64lo F4/80lo, Monocytes are Ly6Chi CD11c−, Neutrophils are Ly6Ghi Ly6C+, plasmacytoid DCs are B220+ Ly6C+ CD11c+ MHCIIlo. B–G) Cytokine concentrations (pg/mL) of IL-1β, IL-27, IL-12p70, IFN-β, and IFN-λ3 in supernatants of BMDC culture after incubation with adjuvanted NPs for 24 h. H–I) Percentage of live CD3+CD4+ T cells or CD3+CD8+ T cells proliferating in presence of GM-CSF BMDCs, gated on diminished CFSE signal (Fig. S1). “No Adjuvant” condition is blank NPs. J, K) Percentage of live CD3+CD4+ T cells or CD3+CD8+ T cells proliferating in presence of FLT3L BMDCs. *p < 0.05. **p < 0.01, ***p < 0.001, ****p < 0.0001 based on a One-Way ANOVA and Tukey Test for multiple comparison.
Next, we evaluated the ability of combination adjuvant-activated BMDCs to stimulate murine T cell proliferation in isomixed lymphocyte (isoMLR) reactions. After 72 h there were no significant changes in CD4+ T cell proliferation with MPLA- or PUUC-stimulated GM-CSF BMDCs while CpG, R848, MPLA+CpG, CpG+PUUC, MPLA+PUUC and R848+PUUC-stimulated GM-CSF BMDCs induced two-fold decreases in proliferation compared to antigen-only controls (Fig. 2H). Alternatively, there was a two-fold increase in CD8+ T cell proliferation with MPLA-stimulated GM-CSF BMDCs (Fig. 2I). CD4+ T cell proliferation significantly increased when mixed with CpG NP- and MPLA NP-stimulated FLT3L BMDCs (Fig. 2J). CD8+ T cell proliferation significantly increased when mixed with FLT3L BMDCs stimulated with CpG, MPLA+CpG, and CpG+PUUC NPs (Fig. 2K). Combination adjuvants synergistically or additively induced proinflammatory cytokine secretion by APCs in vitro while select adjuvants, including MPLA NP with GM-CSF BMDCs and CpG-single and dual NP formulations with FLT3L BMDCs, increased CD8+ T cell proliferation in vitro. We were motivated to evaluate whether these in vitro studies would predict how combination and single TLR- and RIG-I targeted adjuvants would improve SARS-CoV-2 subunit vaccination in vivo.
3.2. MPLA-PUUC NPs increase lung T cell responses in mice following intranasal vaccination with SARS-CoV-2 stabilized spike protein
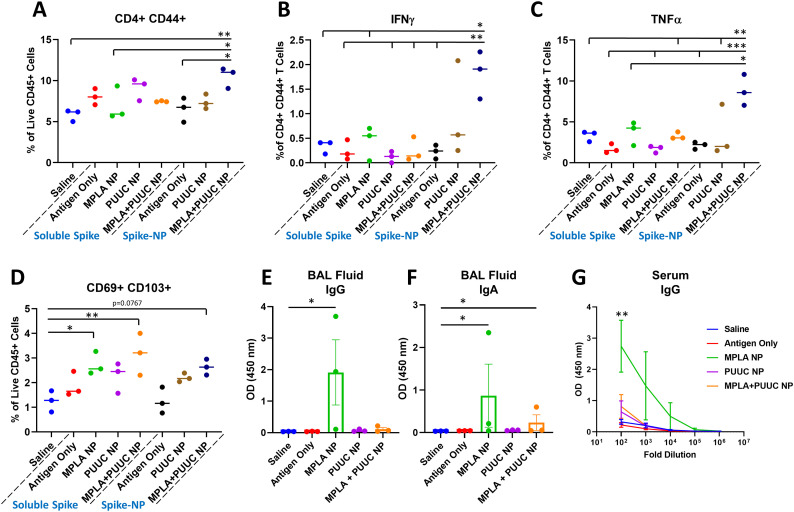
Given the protective role of mucosal immunity against respiratory virus SARS-CoV-2 [36], we vaccinated mice I.N. with single- and dual-adjuvant-loaded NPs combined with either soluble spike protein or NP-conjugated spike protein (spike-NP). Mice were immunized I.N. with the 1st dose on Day 0 and 2nd dose on Day 28, and lungs were harvested on Day 35 for restimulation with spike peptide pools. Lungs from mice immunized with spike-NPs plus MPLA+PUUC NPs had increased CD4+CD44+ T cell populations with significant increases in IFNγ and TNFα secretion after antigen restimulation (Fig. 3A–C). CD69+CD103+ tissue-resident memory T cell populations were highest in mice vaccinated with MPLA NPs or MPLA+PUUC NPs plus soluble spike protein (Fig. 3D). These cells exhibited a double negative CD4−CD8− phenotype, suggesting they could be γδ cells, a subset of T cells enriched in epithelial and mucosal tissues that are activated in an MHC-independent manner. I.N. vaccination with CpG, CpG+PUUC, R848+PUUC, or MPLA+CpG NPs and soluble spike protein failed to generate significant increases in T cells producing IFNγ or TNFα (Fig. S3A–K). In mice immunized with MPLA NPs and soluble spike protein, anti-spike BAL fluid IgG and IgA and serum IgG levels were higher compared to mice in PUUC NP and MPLA+PUUC NP groups ( Fig. 2E–G).
Fig. 3.
MPLA+PUUC NPs increase T cell responses in the lung when delivered intranasally with spike protein. On days 0 (1st dose) and 28 (2nd dose), female BALB/c mice were immunized I.N. with unformulated (i.e., soluble) or NP-conjugated spike protein (1 μg) and PLGA-PEI NPs (4 mg) loaded with MPLA (24 μg), PUUC (17 μg), and MPLA+PUUC (20 μg, 17 μg). Mice were euthanized and lungs were collected on day 35, one week after the 2nd dose. Lung cells were restimulated with spike peptide pools for 6 h and stained for analysis by flow cytometry. Percentages of cells expressing A) CD4+CD44+ out of CD45+ cells, B) IFNγ+ out of CD4+CD44+ cells, C) TNFα+ out of CD4+CD44+ cells, D) CD69+CD103+ (tissue resident memory T cells) out of CD45+ cells. BAL fluid from vaccinated mice using soluble spike antigen was assayed for anti-spike E) IgG and F) IgA with ELISA. G) Sera were assayed for anti-spike IgG with ELISA (error bars represent the SEM). p values are *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 calculated using A–D) One-way ANOVA with Tukey post-hoc test, E, F) Kruskal-Wallis with Dunn's post-hoc test for nonparametric data, or G) Two-way ANOVA with Tukey post-hoc test.
3.3. PUUC NPs increase humoral responses in mice following intramuscular vaccination with various doses of SARS-CoV-2 stabilized spike protein
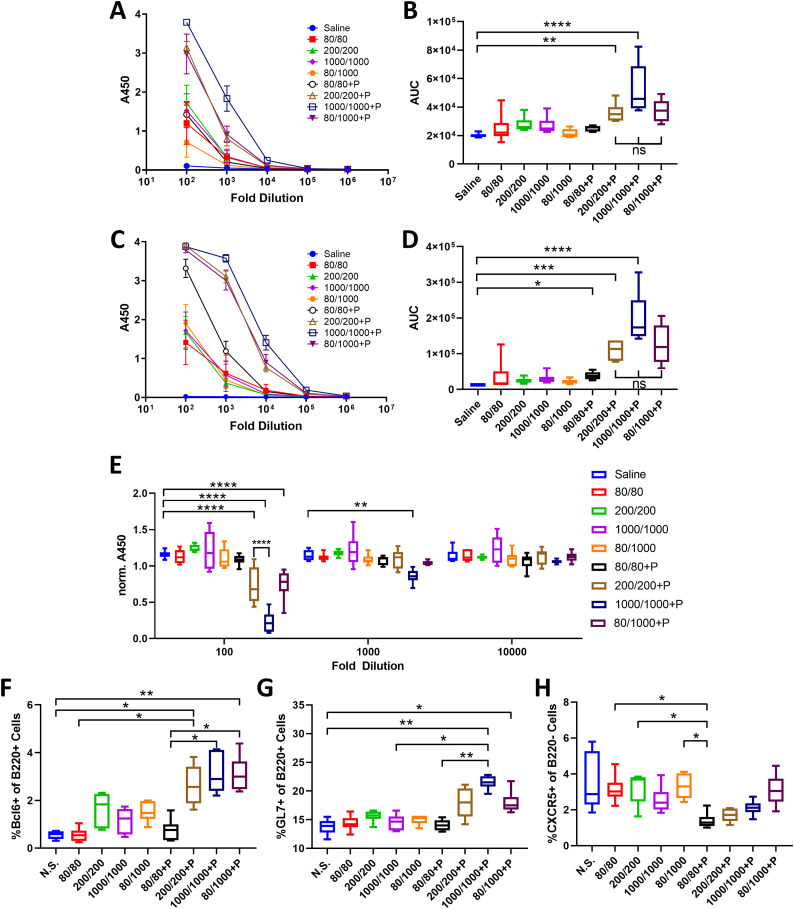
Because authorized COVID vaccines in the US are administered I.M. and antigen doses have been shown to impact T cell responses [37], we evaluated the ability of PUUC NPs to improve SARS-CoV-2 I.M. protein subunit vaccines with different doses of spike protein. On Days 0 and 28, mice were immunized with soluble spike protein at doses of 80, 200, or 1000 ng with or without PUUC NPs. A cohort of mice received a mixed dose of 80 ng on Day 0 and 1000 ng on Day 28 (80/1000). Blood was drawn via the jugular vein of mice on Day 26 and cardiac puncture on Day 36 to quantify levels of antigen-specific IgG after the 1st and 2nd doses, which were higher in mice with PUUC NP-adjuvanted vaccines (Fig. 4A–D). Anti-spike IgG1 and IgG2a titers after the 2nd dose were higher for mice that received the 200/200, 1000/1000, and 80/1000 ng (1st dose/2nd dose) of antigen I.M. with PUUC NPs (Fig. S4A–D) compared to antigen-only and saline control mice. Up to a 1000-fold dilution, neutralization of spike protein was significantly higher (ACE-2 A450 absorbance signal lower) in the sera of mice that received 1000/1000 ng antigen mixed with PUUC NP (Fig. 4E). Graphs of antibody titers are shown in Fig. S8.
Fig. 4.
PUUC NPs delivered intramuscularly with spike protein enhance humoral responses. Female BALB/c mice were immunized I.M. into both tibialis anterior muscles at day 0 (1st dose) with soluble spike protein at doses of 80 ng, 200 ng, 1000 ng with or without adjuvant-NPs (4 mg) loaded with PUUC (+P, 20 ng PUUC dose). Peripheral blood was sampled on day 26. On day 28, mice received a 2nd dose of protein subunit vaccines. Mice received the same formulations, except for two groups that received 80 ng spike protein as a 1st dose received 1000 ng spike protein as a 2nd antigen dose (80/1000 and 80/1000 +P). Mice were euthanized on day 36 for to collect blood and popliteal LNs. A) Anti-spike IgG in post-1st dose sera at various dilutions measured by absorbance at 450 nm during ELISA assays and B) comparison of area under the curve (AUC). C–D) Anti-spike IgG in post-2nd dose sera measured by absorbance at 450 nm and comparison of AUC. E) ACE-2 signal measured by absorbance at 450 nm in spike protein neutralization assay with post-2nd dose sera. Absorbance was normalized to a blank well in each row of a 384 well plate to correct for plate effects. Lower absorbance values indicate higher spike-neutralizing antibody levels in sera. Percentages of cells expressing F) Bcl6+ out of B220+ cells, G) GL7+ out of B220+ cells and H) CXCR5+ out of B220− cells from combined popliteal lymph nodes. B,D) Normality was assessed with the Kolmogorov-Smirnov test. Statistical significance was determined with the Kruskal-Wallis test and Dunn's post-hoc test for multiple comparisons. E–H) Statistical significance calculated with One-Way ANOVA and Tukey post-hoc test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 for all graphs.
By Day 36, percentages of germinal center (GC) BCL6+B220+ cells in popliteal lymph nodes significantly increased in the 200/200, 1000/1000, and 80/1000 ng antigen mice immunized with PUUC NPs compared to antigen-only and saline control mice (Fig. 4F). Similarly, GL7+B220+ cell populations were significantly increased in mice immunized with 1000/1000 and 80/1000 ng antigen plus PUUC NPs compared to controls (Fig. 4G). PUUC NPs with 80/80 ng antigen significantly decreased populations of B220− CXCR5+ cells (T follicular helper, Tfh cells) in the dLNs (Fig. 4H). In the spleen, PUUC NPs did not significantly change CD4 T cell population percentages (Fig. S5A) nor IFN-γ, IL4, or TNF-α secretion by CD4 T cells (Fig. S5B–D). CD8+ T cell populations and secretion of Granzyme B and TNF-α also did not change (Fig. S5I, J, L, M, N, P). Interestingly, CD8+ T cell secretion of IFNγ decreased in mice receiving antigen with PUUC NPs (Fig. S5K and O).
3.4. Combination adjuvant MPLA+PUUC NPs do not increase humoral responses to intramuscular vaccination with SARS-CoV-2 spike protein compared to single-adjuvant PUUC NPs
I.N vaccination with MPLA+PUUC NPs plus spike-NPs increased lung T cell responses and antigen-specific IgG levels were not significantly different between the mice vaccinated I.M. with 1000/1000 and 80/1000 ng (1st dose/2nd dose) soluble spike protein and PUUC NPs. Therefore, we evaluated if MPLA+PUUC NPs mixed with 80/1000 ng soluble spike protein would enhance immune responses compared to single-adjuvant NPs. Mice were immunized with antigen plus MPLA NPs, PUUC NPs, or MPLA+PUUC NPs on Days 0 and 28. Antigen-specific IgG levels most significantly increased in mice vaccinated with PUUC NPs after the 1st and 2nd doses (Fig. S6A–D). Neutralization of spike protein was detectable in mouse sera after the 2nd dose of PUUC NPs up to a 100-fold dilution and MPLA+PUUC NP-vaccinated mice up to a 10,000-fold dilution (Fig. S6E). In the draining popliteal LNs, B220+ cell percentages increased approximately 1.5× in the MPLA NP group (Fig. S6F). Out of B220+ cells, BCL6+ and GL7+ percentages significantly increased in the MPLA, PUUC, and MPLA+PUUC NP groups relative to the saline control group, and in the PUUC NP group relative to the antigen-only control group (Fig. S6G–H). Out of B220− cells, the CXCR5+ population significantly increased in the PUUC and MPLA+PUUC NP groups compared to the saline control and in the MPLA+PUUC NP group relative to the antigen-only control group (Fig. S6I). For mice vaccinated with spike-NP (1000/1000 ng spike protein, 1st dose/2nd dose), MPLA+PUUC NPs increased anti-spike total IgG in serum after the 1st dose, but not after the 2nd dose (Fig. S7). I.M. vaccination with MPLA+PUUC NPs did not generate systemic T cell responses, similar to previous observations with PUUC NPs (Fig. S5).
4. Discussion
In response to studies revealing that PAMP combinations synergistically and complementarily enhance immunity, we investigated the singular and combined effects of nanoparticle-delivered TLR and RIG-I agonists on cellular and humoral immune responses against SARS-CoV-2 spike protein. Our data suggest that the RIG-I agonist PUUC has the potential to be an effective adjuvant for SARS-CoV-2 protein subunit vaccines when combined with TLR4 agonist MPLA on NPs and delivered I.N. with spike protein-conjugated NPs or I.M. with soluble spike protein (Fig. 5 ). MPLA NPs delivered with soluble spike protein also have the capability of inducing systemic and localized humoral responses when administered I.N. (Fig. 5). This data is supported by evidence that the SARS-CoV-2 virus interacts with both TLR4 and RIG-I during natural infection [38,39].
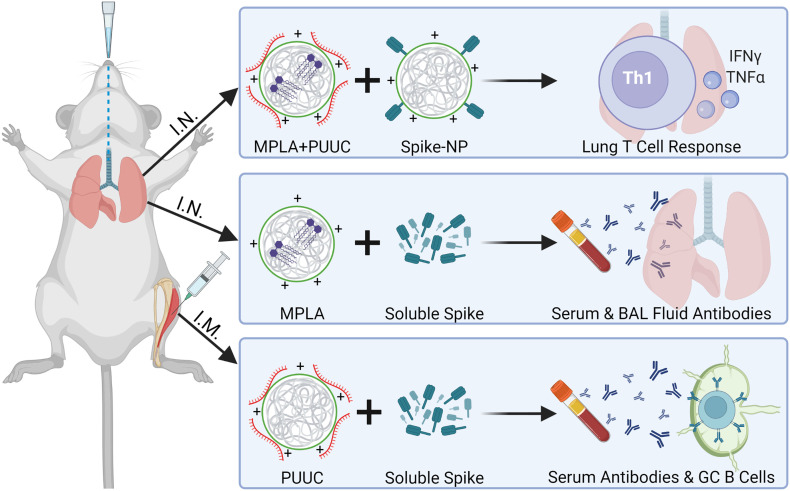
Fig. 5.
Key takeaways from intranasal and intramuscular two-dose vaccination models. Top: MPLA-PUUC NPs administered with spike-NPs intranasally induce local T cell responses, characterized by increases in IFNγ+ and TNFα+ CD4 T cells and tissue resident memory cells after lung cell restimulation with spike peptide pools. Middle: MPLA NPs administered with soluble spike protein intranasally induce systemic and localized humoral responses, characterized by increases in serum and BAL fluid antibodies. Bottom: PUUC NPs injected with soluble spike protein intramuscularly induce systemic humoral responses, characterized by increases in serum antibodies and germinal center B cells in the popliteal lymph nodes.
In vitro data show that MPLA+PUUC, R848+PUUC, and CpG+PUUC PLGA-PEI NPs stimulate more GM-CSF BMDC proinflammatory cytokine secretion (IL-1β, IL-12p70, IFN-β) than single-adjuvant controls, and MPLA NPs and CpG-containing NPs (i.e., CpG, CpG+PUUC) maintain or induce CD8+ T cell proliferation with GM-CSF or FLT3L BMDCs, respectively. GM-CSF BMDCs and FLT3L BMDCs, both widely used for evaluating APC maturation in vitro, had contradicting responses to TLR and RIG-I agonists most likely because the cultures are comprised of different APC populations (monocytes, monocyte-derived macrophages, and DCs). Consistent with previous reports, we found that GM-CSF BMDCs were primarily composed of a heterogeneous population of monocytes, monocyte-derived macrophages, monocyte-derived DCs, and neutrophils [40]. FLT3L BMDCs contained higher percentages of conventional DCs and plasmacytoid DCs [41]. Interestingly, cells derived from the Ly6Chi monocyte lineage are known to secrete IL-27 in vivo in response to subunit vaccination with multiple TLR adjuvants, which may explain higher IL-27 production by GM-CSF BMDCs compared to FLT3L BMDCs [42].
Due to observed increases in proinflammatory cytokine production in vitro, we predicted that MPLA+PUUC NPs, R848+PUUC NPs, and CpG+PUUC NPs would elicit stronger cellular and humoral immune responses in vivo to SARS-CoV-2 spike protein subunit vaccines compared to single-adjuvanted or non-adjuvanted vaccines. Previous studies have reported delivering antigens and adjuvants in separate PLGA particles can improve or attain the same immune responses compared to co-delivered antigens and adjuvants. Therefore, we opted to deliver antigen and adjuvant separately for these studies [33,43]. Also, because APCs innately recognize the particulate state of microbes, we tested adjuvants with both NP-conjugated (spike-NP) and soluble SARS-CoV-2 spike protein antigens [44]. CpG+PUUC and R848+PUUC NPs delivered with soluble spike protein failed to generate significant increases in percentages of T cells producing IFNγ or TNFα. A previous study showed that I.M. vaccination with R848+PUUC NPs induced lymphopenia, possibly due to the overproduction of type I IFN [27]. Studies utilizing CpG 1018 or TLR7/8 agonists in SARS-CoV-2 spike protein subunit vaccines combine the TLR agonists with alum, which alone traditionally induces a Th2-type immune response [[45], [46], [47]].
I.N. vaccination of mice with MPLA+PUUC NPs plus spike-NPs increased CD44+CD4+ T cell populations with IFN-γ and TNF-α responses in lung cells during restimulation with SARS-CoV-2 spike protein peptide pools. CD44 is a marker that distinguishes effector and memory T cells from naïve subsets [48]. Because these CD44+CD4+ T cells were enriched for intracellular IFN-γ and TNF-α, our results suggest that these cells were polarized towards a Th1 effector phenotype, which may be essential for controlling SARS-CoV-2 infection in humans [[49], [50], [51]]. Cell populations bearing tissue-resident memory T cell markers (CD69+CD103+) also increased. Because these T cells were CD4−CD8− we speculate they could be γδ T cells, a subset of T cells enriched in epithelial and mucosal tissues that are activated in an MHC-independent manner [52,53]. Unexpectedly, in post-booster BAL fluid and sera, antigen-specific IgG and IgA were present following I.N. vaccination with MPLA NPs plus soluble spike protein, but not with PUUC NPs or MPLA+PUUC NPs.
There are a few relevant TLR- and RIG-I-targeted SARS-CoV-2 protein-subunit vaccine studies that serve as a benchmark for our I.N. vaccination data. Routhu et al. administered I.N. a SARS-CoV-2 RBD trimer subunit vaccine with alum plus 3M-052 (TLR7/8 agonist) in rhesus macaques and also observed a Th1-biased response, but no detectable antigen-specific CD8+ T cell response [46]. A preclinical study by Jangra et al. investigating a three-dose nanoemulsion +/− RIG-I agonist (IVT DI) adjuvanted SARS-CoV-2 S1 subunit I.N. vaccine observed a systemic Th1 response, characterized by an increase in IFNγ secreted by splenocytes and dLN cells [54]. Jangra et al. did observe IgG in the BAL fluid and sera but it is worth noting they administered three doses of 15-μg S1 subunit protein, compared to our two doses of 1-μg spike protein, which may be necessary to induce humoral responses. The addition of RIG-I agonist to their nanoemulsion adjuvant also did not enhance humoral responses against S1 subunit protein [54].
I.M. vaccination with spike protein plus MPLA, PUUC, or MPLA+PUUC NPs induced different antigen-specific immune responses compared to I.N. vaccination. The antigen-specific IgG response in mice immunized I.M. with spike-NPs was minimal; IgG was only detectable at a 1:100 serum dilution in formulations without adjuvant after the 2nd dose (Fig. S7). Conversely, I.M. vaccines with soluble spike protein and PUUC NPs generated significant systemic antigen-specific IgG responses in mice after receiving one or both doses. Sera from mice immunized with soluble spike protein (after two doses) also contained antibodies neutralizing spike protein, meaning ACE-2 binding was reduced. These results are noteworthy because SARS-CoV-2 enters respiratory endothelial cells via endocytosis following ACE-2 binding [55,56]. A study by Powell et al. observed strong neutralizing antibody titers after a single dose of SARS-CoV-2 spike protein delivered on self-assembling ferritin nanoparticles (S-Fer) adjuvanted with MPLA and Quil-A (another TLR-4 agonist) [57]. However, this protein subunit vaccine was delivered subcutaneously with an antigen dose of 10-μg.
We also observed I.M. vaccination with soluble spike protein and PUUC NPs increased GC BCL6+ and GL7+ B cells in the dLNs. Because the GC reaction produces high-affinity isotype switched antibodies by both plasma cells and memory B cells [27,58], we suspect that the increased GC response associated with PUUC NPs explains the increase in antigen-specific-spike IgG and neutralizing antibodies. This GC reaction may be a response to early innate inflammation associated with PUUC, such as endothelial cell activation which has been reported with RIG-I ligation in viral models [[59], [60], [61]], and the APC activation which we have observed in our in vitro studies. Because both IgG1 and IgG2a were increased following I.M. vaccination with PUUC NPs and soluble spike protein, we do not attribute the overall increase in antibodies to either a Th1- or Th2-mediated bias [62]. This finding contrasts with previous studies that have shown a Th2 bias with RIG-I-based adjuvants following intraperitoneal vaccination with influenza VLPs [63].
Zhou et al. showed that the inclusion of alum in SARS-CoV spike protein subunit vaccines reduced the effective antigen dose tenfold. In mice, the alum-adjuvanted 5-μg spike protein subunit vaccine produced twice as many neutralizing antibodies as the nonadjuvanted 50-μg spike protein subunit vaccine [64]. Interestingly in our studies, the 80-ng 1st dose and 1000-ng spike protein 2nd dose produced similar increases in GC B cell populations compared to the 1000-ng 1st dose and 1000-ng 2nd dose, indicating an opportunity for antigen dose sparing with the inclusion of PUUC as an adjuvant. MPLA+PUUC NPs plus soluble spike protein reduced antigen-specific IgG following the first injection and performed equally to PUUC NPs following the 2nd dose. MPLA+PUUC NPs and PUUC NPs increased BCL6+ and GL7+ B cell populations. MPLA+PUUC NPs significantly induced more neutralizing antibody activity than the saline control up to a 10,000-fold dilution of serum, and increased B220−CXCR5+ cell populations (Tfh markers).
Compared to I.N. trials with spike-NP and I.M. trials with soluble spike protein, it is interesting that MPLA+PUUC NPs failed to induce strong anti-spike cellular or humoral immune responses when delivered I.M. with spike-NPs. This result could be explained by a hindered ability of spike protein to encounter B cells when conjugated to NP, as particles >200 nm in diameter drain less efficiently through lymphatic vessels [65]. Additionally, given that particle surface topology alone can trigger innate immune responses, particulate antigen might be sufficiently immunogenic enough that when combined with PUUC or MPLA+PUUC NPs, lymphocytes become anergic, resulting in a lower adaptive immune response [66].
5. Conclusion
Our results demonstrate that polymer-NP delivery of RIG-I agonist PUUC and TLR4 agonist MPLA increases immune responses to SARS-CoV-2 spike protein subunit vaccines compared to non-adjuvanted vaccines. MPLA NPs, PUUC NPs, and MPLA+PUUC NPs elicited differential cellular and humoral responses against SARS-CoV-2 spike protein depending on the APCs encountered (GM-CSF versus FLT3L-derived), route of administration (I.N. versus I.M.), and antigen delivery platform (soluble versus NP-conjugated). We show that I.N. administration with MPLA+PUUC NPs induces Th1 and tissue-resident memory T cell responses in the lung while MPLA NPs induce local IgA responses in the lung. In contrast, I.M. vaccination with soluble spike protein and PUUC NPs induces robust systemic humoral responses characterized by increases in anti-spike IgG and neutralizing antibodies. Future investigations should examine whether a combination of I.N. and I.M. vaccination can produce balanced systemic and lung-specific protective immunity against SARS-CoV-2 challenge and improve vaccine durability and protection against infection and transmission.
CRediT authorship contribution statement
Alexandra Atalis: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Visualization, Supervision, Project administration. Mark C. Keenum: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Visualization, Supervision, Project administration. Bhawana Pandey: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Visualization. Alexander Beach: Validation, Formal analysis, Investigation, Resources, Data curation. Pallab Pradhan: Conceptualization, Methodology, Investigation, Supervision. Casey Vantucci: Investigation, Resources, Writing – review & editing. Laura O'Farrell: Investigation, Resources, Supervision. Richard Noel: Investigation, Resources, Supervision. Ritika Jain: Investigation, Resources. Justin Hosten: Investigation, Resources. Clinton Smith: Investigation, Resources. Liana Kramer: Investigation, Resources. Angela Jimenez: Investigation, Resources. Miguel Armenta Ochoa: Investigation, Resources. David Frey: Investigation, Resources. Krishnendu Roy: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
There are no conflicts of interest to disclose.
Acknowledgments
We wish to acknowledge the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for the use of their shared equipment, services, and expertise. These facilities include: the Biopolymer Characterization Core for the preparation of PLGA particles, the Engineered Biosystems Building Physiological Research Laboratory for animal experiments, and the Cellular Analysis and Cytometry Core for flow cytometry experiments. Research reported in this publication was supported in part by the Pediatrics/Winship Flow Cytometry Core of Winship Cancer Institute of Emory University, Children's Healthcare of Atlanta and National Institutes of Health - National Cancer Institute (NIH/NCI) under award number P30CA138292. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was partially funded by National Institutes of Health - National Institute of Allergy and Infectious Disease (NIH/NIAID) grant U01-AI124270-02 to KR, funds from the Georgia Tech Foundation to KR, the National Science Foundation Graduate Research Fellowship to AA, the NIH T32 Cellular and Tissue Engineering training fellowship (National Institutes of Health (NIH) T32 Cellular and Tissue Engineering Training Fellowship grant T32-GM0843) to AA and AB, and the Robert A. Milton Chaired Professorship to KR.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2022.05.023.
Appendix A. Supplementary data
Supplementary material
References
- 1.R&D Blue Print . 2021. COVID-19 Vaccine Tracker and Landscape. [Google Scholar]
- 2.Awate S., Babiuk L.A.B., Mutwiri G. Mechanisms of action of adjuvants. Front. Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed S.G., Orr M.T., Fox C.B. Key roles of adjuvants in modern vaccines. Nat. Med. 2013;19:1597–1608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 4.Saravia J., Chapman N.M., Chi H. Helper T cell differentiation. Cell. Mol. Immunol. 2019;16:634–643. doi: 10.1038/s41423-019-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maddur M.S., Bayry J. B cells drive Th2 responses by instructing human dendritic cell maturation. Oncoimmunology. 2015;4 doi: 10.1080/2162402X.2015.1005508. e1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ionescu L., Urschel S. Memory B cells and long-lived plasma cells. Transplantation. 2019;103:890–898. doi: 10.1097/TP.0000000000002594. [DOI] [PubMed] [Google Scholar]
- 7.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnell G., Loo Y.-M., Marcotrigiano J., Gale M., Jr. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002839. e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y.-C., Yeh W.-C., Ohashi P.S. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Pålsson-McDermott E.M., O’Neill L.A. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113:153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casella C.R., Mitchell T.C. Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell. Mol. Life Sci. 2008;65:3231–3240. doi: 10.1007/s00018-008-8228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmi H., et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88–dependent signaling pathway. Nat. Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 13.Ashkar A.A., Rosenthal K.L. Toll-like receptor 9, CpG DNA and innate immunity. Curr. Mol. Med. 2002;2:545–556. doi: 10.2174/1566524023362159. [DOI] [PubMed] [Google Scholar]
- 14.Warner N., Núñez G. MyD88: a critical adaptor protein in innate immunity signal transduction. J. Immunol. 2013;190:3–4. doi: 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda, K. & Akira, S. Seminars in Immunology. 3–9 (Elsevier). [DOI] [PubMed]
- 17.Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moliva J.I., Turner J., Torrelles J.B. Immune responses to bacillus Calmette–Guérin vaccination: why do they fail to protect against Mycobacterium tuberculosis? Front. Immunol. 2017;8:407. doi: 10.3389/fimmu.2017.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lund J.M., et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kell A.M., Gale M., Jr. RIG-I in RNA virus recognition. Virology. 2015;479:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirato K., Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06187. e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y., et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021:1–3. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradhan P., et al. TRAF6-IRF5 kinetics, TRIF, and biophysical factors drive synergistic innate responses to particle-mediated MPLA-CpG co-presentation. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abd4235. eabd4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toy R., et al. TLR7 and RIG-I dual-adjuvant loaded nanoparticles drive broadened and synergistic responses in dendritic cells in vitro and generate unique cellular immune responses in influenza vaccination. J. Control. Release. 2021;330:866–877. doi: 10.1016/j.jconrel.2020.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. 2020;117:11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., et al. SARS-CoV-2: mechanism of infection and emerging technologies for future prospects. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2168. e2168. [DOI] [PubMed] [Google Scholar]
- 30.Trougakos I.P., et al. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J. Biomed. Sci. 2021;28:1–18. doi: 10.1186/s12929-020-00703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leleux J.A., Pradhan P., Roy K. Biophysical attributes of CpG presentation control TLR9 signaling to differentially polarize systemic immune responses. Cell Rep. 2017;18:700–710. doi: 10.1016/j.celrep.2016.12.073. [DOI] [PubMed] [Google Scholar]
- 32.Madan-Lala R., Pradhan P., Roy K. Combinatorial delivery of dual and triple TLR agonists via polymeric pathogen-like particles synergistically enhances innate and adaptive immune responses. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-02804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasturi S.P., et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoecke L.V., Job E.R., Saelens X., Roose K. Bronchoalveolar lavage of murine lungs to analyze inflammatory cell infiltration. J. Vis. Exp. 2017:55398. doi: 10.3791/55398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nybo K. Immunology and immunochemistry: ELISA. BioTechniques. 2018;49 doi: 10.2144/000113475. [DOI] [Google Scholar]
- 36.Russell M.W., Moldoveanu Z., Ogra P.L., Mestecky J. Mucosal immunity in COVID-19: a neglected but critical aspect of SARS-CoV-2 infection. Front. Immunol. 2020;11:3221. doi: 10.3389/fimmu.2020.611337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billeskov R., Beikzadeh B., Berzofsky J.A. The effect of antigen dose on T cell-targeting vaccine outcome. Hum. Vaccines Immunotherapeutics. 2019;15:407–411. doi: 10.1080/21645515.2018.1527496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya M., et al. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect. Genet. Evol. 2020;85:104587. doi: 10.1016/j.meegid.2020.104587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada T., et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021:1–9. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 40.Helft J., et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+ MHCII+ macrophages and dendritic cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Brasel K., De Smedt T., Smith J.L., Maliszewski C.R. Generation of murine dendritic cells from flt3-ligand–supplemented bone marrow cultures. Blood J. Am. Soc. Hematol. 2000;96:3029–3039. [PubMed] [Google Scholar]
- 42.Kilgore A.M., et al. IL-27p28 production by XCR1+ dendritic cells and monocytes effectively predicts adjuvant-elicited CD8+ T cell responses. ImmunoHorizons. 2018;2:1–11. doi: 10.4049/immunohorizons.1700054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazzaz J., et al. Encapsulation of the immune potentiators MPL and RC529 in PLG microparticles enhances their potency. J. Control. Release. 2006;110:566–573. doi: 10.1016/j.jconrel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Moyer T.J., Zmolek A.C., Irvine D.J. Beyond antigens and adjuvants: formulating future vaccines. J. Clin. Invest. 2016;126:799–808. doi: 10.1172/JCI81083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo T.-Y., et al. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-77077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Routhu N.K., et al. SARS-CoV-2 RBD trimer protein adjuvanted with Alum-3M-052 protects from SARS-CoV-2 infection and immune pathology in the lung. Nat. Commun. 2021;12:1–15. doi: 10.1038/s41467-021-23942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brewer J.M., et al. Aluminium hydroxide adjuvant initiates strong antigen-specific Th2 responses in the absence of IL-4-or IL-13-mediated signaling. J. Immunol. 1999;163:6448–6454. [PubMed] [Google Scholar]
- 48.Schumann J., Stanko K., Schliesser U., Appelt C., Sawitzki B. Differences in CD44 surface expression levels and function discriminates IL-17 and IFN-γ producing helper T cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132479. e0132479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raphael I., Nalawade S., Eagar T.N., Forsthuber T.G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine. 2015;74:5–17. doi: 10.1016/j.cyto.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moderbacher C.R., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.038. 996-1012. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang S., et al. Tissue resident memory γδT cells in murine uterus expressed high levels of IL-17 promoting the invasion of trophocytes. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.588227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Do J.-S., et al. γδ T cells coexpressing gut homing α4β7 and αE integrins define a novel subset promoting intestinal inflammation. J. Immunol. 2017;198:908–915. doi: 10.4049/jimmunol.1601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jangra S., et al. A combination adjuvant for the induction of potent antiviral immune responses for a recombinant SARS-CoV-2 protein vaccine. Front. Immunol. 2021;12:729189. doi: 10.3389/fimmu.2021.729189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 57.Powell A., et al. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 2021;7:183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gatto D., Brink R. The germinal center reaction. J. Allergy Clin. Immunol. 2010;126:898–907. doi: 10.1016/j.jaci.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 59.da Conceicao T.M., et al. Essential role of RIG-I in the activation of endothelial cells by dengue virus. Virology. 2013;435:281–292. doi: 10.1016/j.virol.2012.09.038. [DOI] [PubMed] [Google Scholar]
- 60.Kell A.M., Hemann E.A., Turnbull J.B., Gale M., Jr. RIG-I-like receptor activation drives type I IFN and antiviral signaling to limit Hantaan orthohantavirus replication. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008483. e1008483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asdonk T., et al. Endothelial RIG-I activation impairs endothelial function. Biochem. Biophys. Res. Commun. 2012;420:66–71. doi: 10.1016/j.bbrc.2012.02.116. [DOI] [PubMed] [Google Scholar]
- 62.Holdsworth S.R., Kitching A.R., Tipping P.G. Th1 and Th2 T helper cell subsets affect patterns of injury and outcomes in glomerulonephritis. Kidney Int. 1999;55:1198–1216. doi: 10.1046/j.1523-1755.1999.00369.x. [DOI] [PubMed] [Google Scholar]
- 63.Beljanski V., et al. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J. Virol. 2015;89:10612–10624. doi: 10.1128/JVI.01526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z., et al. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine. 2006;24:3624–3631. doi: 10.1016/j.vaccine.2006.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schudel A., Francis D.M., Thomas S.N. Material design for lymph node drug delivery. Nat. Rev. Mater. 2019;4:415–428. doi: 10.1038/s41578-019-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baranov M.V., Kumar M., Sacanna S., Thutupalli S., Van Den Bogaart G. Modulation of immune responses by particle size and shape. Front. Immunol. 2020;11:3854. doi: 10.3389/fimmu.2020.607945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material