Abstract
The bacteriophage-host sensitivity patterns of 16 strains of Lactococcus lactis originally isolated from a mixed strain Cheddar cheese starter culture were determined. Using phages obtained from cheese factory whey, four of the strains were found to be highly phage resistant. One of these isolates, Lactococcus lactis subsp. cremoris HO2, was studied in detail to determine the mechanisms responsible for the phage insensitivity phenotypes. Conjugal transfer of plasmid DNA from strain HO2 allowed a function to be assigned to four of its six plasmids. A 46-kb molecule, designated pCI646, was found to harbor the lactose utilization genes, while this and plasmids of 58 kb (pCI658), 42 kb (pCI642), and 4.5 kb (pCI605) were shown to be responsible for the phage resistance phenotypes observed against the small isometric-headed phage φ712 (936 phage species) and the prolate-headed phage φc2 (c2 species). pCI658 was found to mediate an adsorption-blocking mechanism and was also responsible for the fluffy pellet phenotype of cells containing the molecule. pCI642 and pCI605 were both shown to be required for the operation of a restriction-modification system.
Bacteriophage infection of lactococcal starter cultures is undoubtedly a persistent problem of the dairy fermentation industry. Prolonged production schedules, partial or complete process failure, and substantial economic losses are particular hallmarks of phage-related problems. The exploitation of naturally occurring lactococcal bacteriophage defense systems, through their conjugal transfer to phage-sensitive starter strains, for example (6, 7, 20, 27, 29, 30, 44, 50), can increase the level of insensitivity towards phages that are commonly encountered in cheese factories. Four principal mechanisms have been distinguished in Lactococcus species following intensive studies of phage-host interactions, adsorption inhibition, phage DNA penetration blocking, restriction-modification, and abortive infection (for recent reviews, see references 9, 10, 16 and 22). These mechanisms are generally encoded by plasmid-located genes, which has proved to be advantageous in terms of their characterization and for conjugal dissemination to other strains. As these are naturally occurring lactococcal plasmids with GRAS (generally recognized as safe) status, their introduction into other lactococcal hosts by conjugation does not require regulatory clearance. It has been demonstrated that the stacking of several insensitivity mechanisms which target a wide variety of phages at different points in the lytic cycle can improve the inherent resistance of a strain (7, 12, 13, 28, 32, 49, 51, 55). This ongoing challenge to effectively counter phage relies on the availability of broad-spectrum, highly efficient mechanisms which are well characterized at both the phenotypic and genotypic levels.
This study reports the identification and characterization of novel lactococcal bacteriophage resistance plasmids. A screening program involving 16 lactococcal strains from the University College Cork (UCC) Culture Collection was undertaken, and transmissible phage resistance systems were identified in four of these strains following conjugation and curing experiments. For the purpose of this study, one of the strains, designated Lactococcus lactis subsp. cremoris HO2, was selected for further study. This strain, which originated from a mixed strain Cheddar cheese starter culture, harbors six endogenous plasmids, four of which were found to either confer or influence bacteriophage resistance mechanisms of different strengths and specificities.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
The bacterial strains and bacteriophages used in this study are listed in Tables 1 and 2. Lactococcal strains were grown at 30°C in M17 medium (56) containing 0.5% glucose or lactose as required. Streptomycin (Sm; 600 μg ml−1) was added when appropriate. Escherichia coli V517 was grown at 37°C with aeration in Luria-Bertani medium (43). Stocks of all cultures were maintained at −20°C in 40% glycerol. Bacteriophages were propagated on their homologous hosts at 30°C in GM17/LM17. Phages used for the selection of L. cremoris MG1363-derived transconjugants were the homologous small isometric-headed φ712 (936 phage species) and prolate-headed φc2 (c2 species), while the lytic prolate-headed φ952 (c2 species) was employed for selection of L. cremoris 952-derived transconjugants (Table 1). Additional phages used for sensitivity or resistance assays were either initially isolated from a cocktail which contained isolates specific for dairy strains or were derived from factory whey which had previously been shown to cause inhibition of the test strains (Table 2).
TABLE 1.
L. lactis strains screened for phage resistance mechanisms and bacteriophages used for transconjugant selection
| Lactococcus sp. | Plasmid content (kb)a | Source or comment |
|---|---|---|
| Strains | ||
| UL033 | ND | UCC culture collection |
| UC301 | ND | UCC culture collection |
| UC161 | ND | UCC culture collection |
| UC073 | ND | UCC culture collection |
| UC021-2 | ND | UCC culture collection |
| UC147 | ND | UCC culture collection |
| UC109 | ND | UCC culture collection |
| 107 | ND | UCC culture collection |
| UC103 | ND | UCC culture collection |
| UC029 | ND | UCC culture collection |
| HO2 | 58, 46, 42, 22.5, 8.9, 4.5 | UCC culture collection |
| H15 | ND | UCC culture collection |
| I23 | ND | UCC culture collection |
| UC107 | ND | UCC culture collection |
| F31 | ND | UCC culture collection |
| F35 | ND | UCC culture collection |
| MG1363 | Plasmid free | Derivative of L. cremoris 712 (17) |
| Tc-AF021 Lac+ | 58, 46 | Lac+; phage-resistant transconjugant of HO2 × MG1363 mating |
| Tc-AF021 Lac− | 58 | Lac−; phage-resistant cured derivative of Tc-AF021 Lac+ |
| Tc-AF030 Lac+ | 46, 42, 22.5, 4.5 | Lac+; phage-resistant transconjugant of HO2 × MG1363 mating |
| Tc-AF030 Lac− 1 | 42, 22.5, 4.5 | Lac−; phage-resistant cured derivative of Tc-AF030 Lac+ |
| Tc-AF030 Lac− 3 | 22.5, 4.5 | Lac−; phage-sensitive cured derivative of Tc-AF030 Lac+ |
| Tc-AF030 Lac− 4 | 42, 22.5 | Lac−; phage-sensitive cured derivative of Tc-AF030 Lac+ |
| Tc-AF009 Lac+ | 46 | Lac+; phage-resistant transconjugant of HO2 × MG1363 mating |
| L. lactis subsp. cremoris 952 | 55, 8, 3.6, 2.6 | Chr. Hansens Ltd. culture collection, Hørsholm, Denmark |
| E. coli V517 | 55.9, 7.5, 5.8, 5.3, 4.1, 3.1, 2.1 | Source of size reference plasmids (35) |
| Phages | ||
| φc2 (c2 species) | Prolate-headed, lytic phage for MG1363 | |
| φ712 (936 species) | Small isometric-headed phage for MG1363 | |
| φ952 (c2 species) | Prolate-headed phage for 952 |
ND, not determined.
TABLE 2.
Factory-derived phages isolated against L. lactis strains
| Phage designation | Strain | Plaque description | Plaque size (mm) | Titer (PFU ml−1) |
|---|---|---|---|---|
| φI07-1 | I07 | Clear | 3 | 1 × 107 |
| φI07-2 | I07 | Clear | 2 | 5 × 108 |
| φ147-1 | UC147 | Clear | <1 | 7 × 106 |
| φ147-2 | UC147 | Clear, lysin halo | 5 | 7 × 106 |
| φ103-1 | UC103 | Clear, lysin halo | 4 | 2 × 108 |
| φ103-2 | UC103 | Clear, lysin halo | 2 | 5 × 106 |
| φ029 | UC029 | Clear | 0.5 | 1 × 104 |
| φ161-1 | UC161 | Clear | 3 | 5 × 108 |
| φ161-2 | UC161 | Clear | 1 | 1 × 106 |
| φ021 | UC021-2 | Clear | <1 | 5 × 106 |
| φI23-1 | I23 | Clear | 1 | 6 × 103 |
| φI23-2 | I23 | Pinpoint, hazy | <0.5 | 2.5 × 105 |
| φ063 | UC063 | Clear | 1 | 1.2 × 108 |
| φ109-1 | UC109 | Clear | 2.5 | 1.4 × 108 |
| φ109-2 | UC109 | Clear | 1 | 1.4 × 108 |
Bacterial conjugation experiments.
Conjugal transfer of plasmid DNA was performed on GM17 agar or skimmed milk agar based on the method of McKay (37) or on that of Harrington and Hill (20). Donor and recipient cultures (pregrown at 30°C for 4 h) were mixed in a 1:2 ratio, and 0.2 ml of the mixture was plated on nonselective solid agar surfaces. The mating mix was harvested with 2 ml of Ringers solution and plated either on lactose indicator agar (LIA) (36) containing 600 μg of Sm ml−1 or fast slow differential agar (26) to select for any resulting Lac+ MG1363- or 952-derived transconjugants, respectively.
Bacteriophage plaque assays.
Bacteriophage plaque assays were performed with φ712 and φc2 at 21, 30, and 37°C. Assays conducted with φ712 at 37°C involved the substitution of 1 M calcium-boro-gluconate for 0.185 M CaCl2 to allow plaques to be more clearly visualized and enumerated. Levels of adsorption by phages to sensitive and insensitive hosts was determined by the method described by Lucey et al. (34) with a control sample with no culture added. Phage adsorption was calculated by using the following formula: % adsorption = [(control titer − residual titer)/(control titer)] × 100. Efficiency of plaquing (EOP) of φ712 on its nonrestricting homologous host L. cremoris MG1363 and on the restricting test hosts was assayed according to the method described by Sanders and Shultz (46). A decrease in the EOP on the test culture relative to the nonrestricting homologous host was evidence of restriction activity, while modification was recognized by a restoration of full plaquing ability following one cycle of growth through the restricting test host. Cell survival was estimated by the procedure of Behnke and Malke (3). Cells which survived phage infection were enumerated as CFU milliliter−1, and the percentage of the population which died was calculated as [(CFU milliliter−1 in cultures without phage) − (CFU milliliter−1 in cultures with phage)/(CFU milliliter−1 in cultures without phage)] × 100.
Plasmid curing.
Cured variants of selected transconjugants were isolated based on the method of Sinha (53). Strains were subcultured four times in M17 broth with and without the buffering agent β-glycerophosphate (52) at 30 and 37°C. Individual Lac− isolates recovered from LIA were restreaked for purity and analyzed for phage resistance patterns and plasmid content.
Plasmid preparation and analysis.
Lactococcal plasmid DNA was isolated as described by Anderson and McKay (2). Plasmid profiles were analyzed by electrophoresis on 0.7% vertical agarose gels with TAE buffer (40 mM Tris-acetate, 2 mM EDTA [pH 8.0]). Gels stained with ethidium bromide (0.5 μl ml−1) were viewed under UV light and photographed by using a Polaroid type 667 film or a UVP Imagestore 5000 gel documentation system (UV Products Ltd., Cambridge, United Kingdom). Plasmid sizes were estimated based on size reference plasmids isolated from E. coli V517 (Table 1).
Southern blotting and hybridization analysis.
DNA was transferred from agarose gels to nylon membranes (Hybond N+; Amersham International, Bucks, United Kingdom) by the method of Southern (54) as modified by Wahl et al. (57). DNA was labelled by using the enhanced chemiluminescence gene detection system. Probe labelling, hybridization conditions, and washing steps were performed according to the instructions provided by the manufacturer.
Intracellular phage DNA replication.
Replication of phage DNA within the sensitive and resistant hosts was compared by the method described by Hill et al. (23). Phages were used to infect cells at a multiplicity of infection greater than 1, and samples were taken at specific time intervals after infection until the sensitive host had lysed. Extracted DNA samples were digested with EcoRI (φc2) or HindIII (φ712) and electrophoresed on 0.7% agarose gels, followed by Southern blotting and hybridization with the test phage DNA.
PCR.
Lactococcal template DNA for PCRs was extracted by physically disrupting the cells by shaking in the presence of glass beads (106 μm; Sigma Corp., Poole, United Kingdom). Oligonucleotide primers (Table 3) specific for lactococcal abi genes were synthesized with a PCR-MATE DNA synthesizer (Applied Biosystems Inc., Foster City, Calif.). PCR reagents were purchased from Promega (Madison, Wis.), and reactions were executed with an Omnigene thermal cycler (Hybaid Ltd., Middlesex, United Kingdom). The annealing temperatures for PCR programs varied according to the melting temperatures of the specific primers used.
TABLE 3.
Sequences of lactococcal abi primers used in PCRs and expected product sizes in each case
| Primera | Sequence | Size (bp) | Refer-ence(s) |
|---|---|---|---|
| abiA (Fwd) | 5′-TATGTCTGTTAATGCTG-3′ | ||
| abiA (Rev) | 5′-ACAAGAGTTATGAGTGC-3′ | 219 | 8, 25 |
| abiB (Fwd) | 5′-ATGGATATTGAAGTTGA-3′ | ||
| abiB (Rev) | 5′-TCAAATGTCTTTTAGCT-3′ | 753 | 5 |
| abiC (Fwd) | 5′-TATAGGAAATGGGCTGG-3′ | ||
| abiC (Rev) | 5′-CCAGTCCCAAGCCCCTC-3′ | 341 | 11 |
| abiD (Fwd) | 5′-GGTAGAAACTAGAACAG-3′ | ||
| abiD (Rev) | 5′-CCGACTTGTAGCTCTCC-3′ | 399 | 39 |
| abiD1 (Fwd) | 5′-CAATGGACTACTAATC-3′ | ||
| abiD1 (Rev) | 5′-CGGTTATCCAGCTTTTC-3′ | 616 | 1 |
| abiEi (Fwd) | 5′-TTCAGCAGAAATACAGC-3′ | ||
| abiEi (Rev) | 5′-TACTGTTTCTATTTGTG-3′ | 257 | 15 |
| abiEii (Fwd) | 5′-AGATGATGTTTCCTGATG-3′ | ||
| abiEii (Rev) | 5′-TGAAAATCGTAATAATC-3′ | 240 | 15 |
| abiF (Fwd) | 5′-GAACAGAGAGTAAAACC-3′ | ||
| abiF (Rev) | 5′-TGTAGGTTTGATTTGGC-3′ | 527 | 15 |
| abiG (Fwd) | 5′-AAGCCTCTAGATGGTAGC-3′ | ||
| abiG (Rev) | 5′-AACTGGCATAAGGAATC-3′ | 842 | 40 |
| abiH (Fwd) | 5′-AATCAACTAGCAATTCG-3′ | ||
| abiH (Rev) | 5′-TACAGGCTCTATAAAGC-3′ | 1,338 | 42 |
Fwd, forward; Rev, reverse.
RESULTS
Sensitivity patterns of lactococcal strains to lactococcal phages.
Sixteen lactococcal donor strains from the UCC Culture Collection were assessed for their phage sensitivity patterns. Bacteriophages were isolated against a number of the strains by challenging them with a phage cocktail by a spot test. Wherever a positive reaction was obtained, a sample from the lysed area was removed and propagated on the sensitive host. The lysate was then plaque assayed on the same host, and different phage types were isolated on the basis of morphological differences. These individual isolates were then repropagated through the relevant culture. A total of 15 phage types were identified based on plaque morphology, and sensitivity of 9 of the 16 lactococcal strains was examined (Table 2). The 15 bacteriophages were reacted against all 16 strains in order to determine the host range of each phage. Strains I07, UC103, and UC029 were strongly inhibited by most or all of the phages, while in contrast, six strains, designated UL033, HO2, H15, I23, F31, and F35, appeared to be particularly phage resistant. The sensitivity of the intended conjugal recipient strain L. lactis subsp. cremoris MG1363 was also examined, and this strain was infected by most of the phages, somewhat surprisingly, considering it is not commercially used (Table 4).
TABLE 4.
Host ranges of bacteriophages isolated against Lactococcus cultures
| Strain | Reaction against indicated bacteriophagea
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| φ103b | φ107-1 | φ107-2 | φ021 | φ109b | φ063 | φ161b | φ029 | φ123b | φ147b | |
| UL033 | − | − | − | − | − | − | − | − | − | − |
| UC301 | − | +− | − | +− | − | − | − | + | − | − |
| UC161 | +− | +− | +− | − | ++ | − | ++ | − | − | − |
| UC073 | − | − | − | − | − | − | − | − | − | − |
| UC021-2 | − | − | ++ | ++ | − | ++ | − | − | − | − |
| UC147 | − | − | − | − | ++ | − | − | − | − | ++ |
| UC109 | ++ | − | ++ | − | ++ | − | − | − | − | ++ |
| I07 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ |
| UC103 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ |
| UC029 | +− | ++ | − | +− | ++ | ++ | − | ++ | − | − |
| HO2 | − | − | − | − | − | ++ | − | − | − | − |
| H15 | − | − | − | − | − | − | − | − | − | − |
| I23 | − | − | − | − | − | − | − | − | ++ | − |
| UC107 | +− | +− | +− | +− | +− | +− | +− | +− | − | +− |
| F31 | − | − | − | − | − | − | − | +− | − | − |
| F35 | − | − | − | − | − | − | − | − | − | − |
| MG1363 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ |
+, very hazy plaque, no phage propagated from plaque; ++, clear plaque, good lysis, sensitive; −, no clearing, not sensitive to phage.
Mixtures of φ103-1 and 2, φ109-1 and 2, φ161-1 and 2, φ123-1 and 2, and φ147-1 and 2, respectively, were used in experiments.
Conjugal mating experiments were performed with the 16 strains as donors to determine if genetic determinants for phage resistance could be cotransferred to L. cremoris MG1363 with those for lactose metabolism. While a number of the strains were able to transfer lactose at frequencies ranging from 1 × 10−7 to 3.5 × 10−2 (Table 5), only transconjugants derived from four of these (I07, F31, I23, and HO2) exhibited concomitant transfer of insensitivity against φ712 and/or φc2. The latter three strains were also previously shown to be strongly phage resistant as described in Table 4. L. lactis subsp. cremoris HO2 was selected for further study, which involved the genetic localization and the characterization of its phage resistance systems.
TABLE 5.
Conjugal transfer of lactose metabolism and phage resistance determinants from 16 lactococcal strains to L. lactis subsp. cremoris MG1363
| Donor strain | Frequency of Lac+ transconjugants/recipient | % phage resistancea |
|---|---|---|
| UL033 | 5.2 × 10−4 | ND |
| UC301 | <10−9 | ND |
| UC161 | <10−9 | ND |
| UC073 | 4 × 10−6 | ND |
| UC021-2 | <10−9 | ND |
| UC147 | <10−9 | ND |
| UC109 | <10−9 | ND |
| I07 | 10−7 | 60 |
| UC103 | <10−9 | ND |
| UC029 | 3 × 10−7 | ND |
| HO2 | 2.5 × 10−4 | 36 |
| H15b | ND | |
| I23 | 3.5 × 10−2 | 2 |
| UC107 | 4 × 10−2 | ND |
| F31 | 2.2 × 10−6 | 41 |
| F35 | 1.2 × 10−6 | ND |
ND, no phage resistance detected.
Strain was resistant to streptomycin at 600 μg ml−1.
Identification of phage resistance plasmids in L. cremoris HO2-derived transconjugants.
Three individual transconjugant types, based on the extent of phage resistance they exhibited, were derived following the mating between L. cremoris HO2 and L. cremoris MG1363. These were designated Tc-AF021 Lac+, Tc-AF030 Lac+, and Tc-AF009 Lac+. Tc-AF021 Lac+ had acquired complete insensitivity to the small isometric-headed φ712 and partial insensitivity to the prolate-headed φc2, both of which are lytic for the recipient strain L. cremoris MG1363 (Table 6). Tc-AF021 Lac+ also displayed a soft fluffy pellet following centrifugation. Tc-AF030 Lac+ exhibited partial resistance to both phages manifested by a reduction in plaque size and EOP, and Tc-AF009 Lac+ conferred very slight insensitivity to φ712 by decreasing the plaque size and the EOP and to φc2 by reducing the plaque size (Table 6).
TABLE 6.
Reactions of φ712 and φc2 on L. lactis subsp. cremoris MG1363 and L. lactis subsp. cremoris HO2-derived transconjugants
| Strain (plasmid) | φ712
|
φc2
|
||
|---|---|---|---|---|
| EOP | PD (mm) | EOP | PD (mm) | |
| MG1363 (plasmid free) | 1.0 | 0.75–1.0 | 1.0 | 3–4 |
| Tc-AF021 Lac+ (pCI658, pCI646) | NVPb | NVP | 5 × 10−1 | 1–2 (hazy) |
| Tc-AF030 Lac+ (pCI646, pCI642, pCI623, pCI605) | 7.9 × 10−4 | Pinpoint–1.0 | 2.7 × 10−2 | 1–2 |
| Tc-AF030 Lac− 1 (pCI642, pCI623, pCI605) | 3.3 × 10−2 | Pinpoint–1.0 | 2.7 × 10−2 | 1–2 |
| Tc-AF030 Lac− 3 (pCI623, pCI605) | 1.8 | 0.75–1.0 | 1.04 | 3–4 |
| Tc-AF030 Lac− 4 (pCI642, pCI623) | 1.5 | 0.75–1.0 | 9 × 10−1 | 3–4 |
| Tc-AF009 Lac+ (pCI646) | 4.4 × 10−1 | Pinpoint–0.5 | 9 × 10−1 | 1.5–2.5 |
PD, plaque diameter.
NVP, no visible plaques.
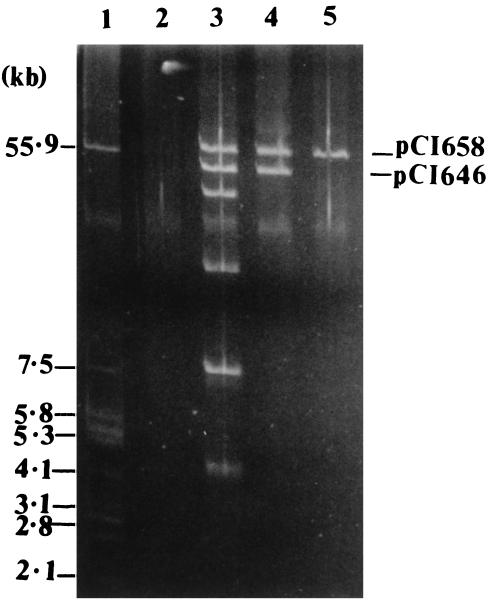
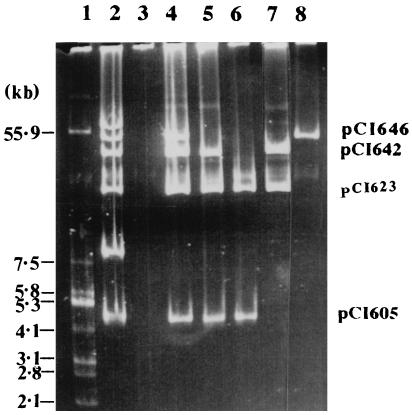
Analysis of the various transconjugants showed that they had acquired a number of plasmids which were also detected in the donor strain. Tc-AF021 Lac+ harbored two plasmids, of 58 and 46 kb, designated pCI658 and pCI646, respectively (Fig. 1, lane 4). A Lac− cured derivative of Tc-AF021 Lac+ which lacked pCI646 was generated (Fig. 1, lane 5), implying that this plasmid encoded lactose utilization. This cured derivative, Tc-AF021 Lac−, retained pCI658 and still exhibited the phage resistance and fluffy pellet phenotypes, demonstrating that this plasmid was likely to be responsible for both characteristics. Plasmid profiles revealed that Tc-AF030 Lac+ possessed, in addition to the lactose plasmid pCI646, three plasmids, of 42, 22.5, and 4.5 kb (designated pCI642, pCI623, and pCI605) (Fig. 2, lane 4). Plasmid-cured variants which lacked one or more of the plasmids present in the parent transconjugant Tc-AF030 Lac+ were obtained. Derivative Tc-AF030 Lac− 1 (Fig. 2, lane 5) was cured of the Lac plasmid pCI646 and maintained its insensitivity to phage infection, although the level of resistance against φ712 was up to 40 times less powerful as demonstrated by differences in EOP values between the strains. Variants Tc-AF030 Lac− 3 and 4 (Fig. 2, lanes 6 and 7) had also lost plasmids pCI642 and pCI605, respectively, and both these strains were found to succumb to phage attack. Tc-AF009 Lac+, which carried only the Lac plasmid pCI646 (Fig. 2, lane 8) was only partially resistant to phage infection. (The EOP values and plaque diameters for all strains tested are given in Table 6.)
FIG. 1.
Plasmid profile analysis of L. lactis subsp. cremoris HO2, L. lactis subsp. cremoris MG1363, and Tc-AF021 Lac+ and Lac− derivatives. Lanes: 1, E. coli V517 (source of size reference plasmids); 2, L. lactis subsp. cremoris MG1363 (plasmid-free, phage-sensitive recipient); 3, L. lactis subsp. cremoris HO2 (phage-resistant donor); 4, Tc-AF021 Lac+ (Lac+, phage-resistant transconjugant); 5, Tc-AF021 Lac− (Lac−, phage-resistant cured derivative of Tc-AF021 Lac+).
FIG. 2.
Plasmid profile analysis of L. lactis subsp. cremoris HO2, L. lactis subsp. cremoris MG1363, Tc-AF030 Lac+ and Lac− cured derivatives, and Tc-AF009 Lac+. Lanes: 1, E. coli V517 (source of size reference plasmids); 2, L. lactis subsp. cremoris HO2 (phage-resistant donor strain); 3, L. lactis subsp. cremoris MG1363 (plasmid-free, phage sensitive recipient); 4, Tc-AF030 Lac+ (Lac+, phage-resistant transconjugant); 5, Tc-AF030 Lac− 1 (Lac−, phage-resistant cured derivative of Tc-AF030 Lac+); 6, Tc-AF030 Lac− 3 (Lac−, phage-sensitive cured derivative of Tc-AF030 Lac+); 7, Tc-AF030 Lac− 4 (Lac−, phage-sensitive cured derivative of Tc-AF030 Lac+); 8, Tc-AF009 Lac+ (Lac+, phage-resistant transconjugant).
Characterization of the phage resistance mechanism in Tc-AF021.
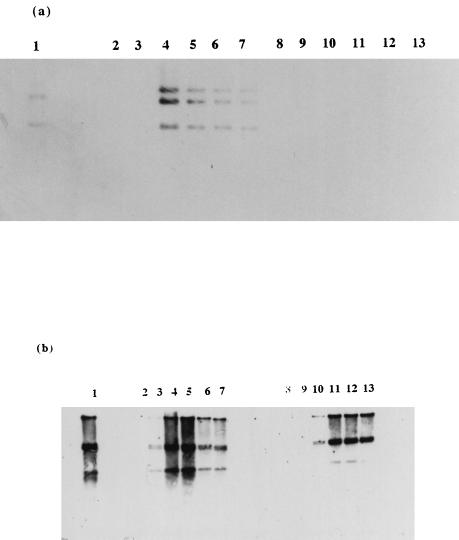
Transconjugant Tc-AF021 Lac− harboring the phage resistance plasmid pCI658 was found to adsorb φ712 and φc2 at reduced levels compared with the control host L. cremoris MG1363 (Table 7), suggesting that the plasmid confers an adsorption-blocking phenotype. In addition, none of the Tc-AF021 Lac− cells died upon infection with φ712, and only a minor proportion of the population was killed following φc2 infection (Table 7). It was also demonstrated that while normal replication of φ712 occurred in the MG1363 host, φ712 DNA was not detected at all in the phage-resistant transconjugant Tc-AF021 Lac− (Fig. 3a), consistent with observations that adsorption of φ712 to this strain is dramatically inhibited. Infection with φc2 revealed that levels of phage DNA increased at a slower-than-normal rate in extracts up to 60 min postinfection (Fig. 3b), probably as a result of the decreased ability of the phage to adsorb to the pCI658-containing cell.
TABLE 7.
Percent phage adsorption to and percent cell death of L. lactis subsp. cremoris MG1363 and L. lactis subsp. cremoris HO2-derived transconjugants following infection with φ712 and φc2
| Strain | % Adsorption
|
% Cell death
|
||
|---|---|---|---|---|
| φ712 | φc2 | φ712 | φc2 | |
| MG1363 | 99 | 87 | 83.3 | 91.5 |
| Tc-AF021 Lac− | 14.7 | 57 | 0 | 20.5 |
| Tc-AF030 Lac− 1 | 99.9 | 98 | 0 | 65.4 |
FIG. 3.
φ712 (a) and φc2 (b) DNA replication in L. lactis subsp. cremoris MG1363 (control) and phage-resistant transconjugant Tc-AF021 Lac−. φ712 DNA samples were digested with HindIII; φc2 DNA samples were digested with EcoRI. (a) Lanes: 1, φ712 DNA restricted with HindIII (control); 2, MG1363 sample extracted at 0 min without added φ712; 3 to 7, MG1363 samples taken at 15, 30, 45, 60, and 75 min postinfection; 8, Tc-AF021 Lac− sample at extracted 0 min without added φ712; 9 to 13, Tc-AF021 Lac− samples extracted at 15, 30, 45, 60, and 75 min. (b) Lanes: 1, φc2 DNA digested with EcoRI (control); 2, MG1363 sample taken at 0 min without added φc2; 3 to 7, MG1363 samples extracted 10, 20, 30, 40, and 50 min postinfection; 8, Tc-AF021 Lac− sample taken at 0 min without added φc2; 9 to 13, Tc-AF021 Lac− samples extracted at 10, 20, 30, 40, and 50 min.
Phage resistance in Tcs-AF030 and AF009.
While transconjugant Tc-AF030 Lac+ adsorbed φ712 and φc2 normally, experimental evidence presented in Table 8 suggested that restriction-modification (R/M) may be the mechanism of phage defense in this strain (EOP reduction of 10−3 for φ712). Again it was recognized that Tc-AF030 Lac− 1, which does not harbor the Lac plasmid pCI646, was more susceptible to infection by φ712. Also noteworthy was the plaque size demonstrated by φ712 and φc2 on both Tc-AF030 Lac+ and Tc-AF030 Lac− 1 variants, where a mixture of small and large plaques (ranging from pinpoint to 1 mm for φ712 and from 1 to 2 mm for φc2) was evident. The reduced plaque size may represent the operation of an additional resistance system within the strain. While cell death was negligible following infection of Tc-AF030 Lac− 1 with φ712, this value remained relatively high (65.4%) following φc2 attack. In addition, phage DNA internalization experiments revealed that replication of φ712 and φc2 within Tc-AF030 Lac− 1 was significantly less efficient compared to that within the sensitive host MG1363 (data not shown). Notably, an examination for phage sensitivity of cured derivatives Tc-AF030 Lac− 3 and 4, from which pCI642 and pCI605, respectively, had been eliminated, indicated that both acquired a fully sensitive phenotype equivalent to that of MG1363. This indicated that both plasmids were involved in conferring the R/M traits observed in Tc-AF030 Lac− 1. Tc-AF009 Lac+ exhibited slightly reduced sensitivity to φ712 as seen by a decrease in the plaque diameter range to between pinpoint and 0.5 mm and a reduction in the EOP to 4.4 × 10−1. This result supports the hypothesis that the Lac plasmid pCI646 may also play a role in phage resistance. However, this strain did not conform to a classical R/M pattern (Table 8), so it seems likely that an alternative intracellular defense mechanism is responsible for the phenotype.
TABLE 8.
Comparison of the EOP of φ712 on L. lactis subsp. cremoris MG1363 and phage-resistant transconjugants Tc-AF030 Lac+, Tc-AF030 Lac− 1, and Tc-AF009 Lac+
| Phage host strain | EOP
|
||
|---|---|---|---|
| φ712 | φ712.Ba | φ712.Bb | |
| MG1363 | 1.0 | 1.0 | 1.0 |
| Tc-AF030 Lac+ | 2.6 × 10−3 | 7.2 × 10−1 | 5.3 × 10−3 |
| Tc-AF030 Lac− 1 | 1.9 × 10−2 | 3.1 | 5.8 × 10−2 |
| Tc-AF009 Lac+ | 4.5 × 10−1 | 1.0 | 1.4 |
φ712 passaged through the restrictive test host.
φ712 passaged through its nonrestrictive host, MG1363.
The defense mechanisms operating against φc2 within transconjugants Tc-AF021 Lac− and Tc-AF030 Lac− 1 remained unaffected by incubation at 37°C. It was not possible to observe an effect on φ712 due to the absence of plaque formation on any of the strains, despite the substitution of 1 M calcium-boro-gluconate for 0.185 M CaCl2.
Identification of a homolog of the abortive infection gene, abiB, on pCI642.
Oligonucleotide primers were created based on 9 of 14 lactococcal abortive infection (Abi) genes for which DNA sequence data were available. With template DNA from L. cremoris HO2, a PCR product was observed only with primers specific for the AbiB system from L. lactis subsp. lactis IL416 (5), which has been shown to be responsible for arresting phage development through the dramatic decay of sensitive phage RNA transcripts (41). The 753-bp PCR product obtained was found to emanate from pCI642, and analysis of its sequence revealed that it was almost indistinguishable from the original IL416-derived abiB sequence, having 97.1 and 93.2% identity at the nucleotide and amino acid levels, respectively. Subsequent experiments indicated that the pCI642-located abiB homolog was expressed at low levels and did not appear to induce degradation of φ712 RNA transcripts (data not shown).
Conjugal transfer of pCI658 to a commercial cheese starter culture.
Strategies to transfer the adsorption-blocking plasmid pCI658 to a spectrum of commercial lactococcal starter cultures were undertaken by using phage resistance as the primary selection. The acquisition of the characteristic fluffy pellet phenotype by putative transconjugants was used as a second selective marker. Initial transfer efforts with L. lactis subsp. cremoris 952 as a recipient were accomplished relatively easily (frequency of transfer, 1.5 × 10−4 per recipient). Plasmid analysis of a transconjugant (Tc-903) exhibiting partial resistance to its homologous prolate-headed φ952 (hazy plaques of reduced size) and a fluffy pellet morphology revealed that it had acquired pCI658. This was subsequently confirmed by using Southern hybridization experiments. Unfortunately, transfer of the plasmid to 10 other cheese manufacturing cultures could not be detected.
DISCUSSION
Sixteen lactococcal cultures from the UCC Culture Collection were screened for transmissible phage resistance phenotypes, and four strains, designated I07, F31, I23, and HO2, demonstrated these properties. The latter three strains were shown to possess potent phage insensitivity mechanisms based on their resistance patterns to 15 phages, which had previously been found to inhibit 9 of the original 16 cultures. This report describes the identification and characterization of the phage resistance mechanisms originating in one of the strains, L. lactis subsp. cremoris HO2. A combination of conjugation and curing experiments, with L. lactis subsp. cremoris MG1363 as a host, allowed the detection of three transconjugant types, based on the extent of resistance to the small isometric-headed φ712 (936 phage species) and the prolate-headed φc2 (c2 species). Transconjugant Tc-AF021 Lac+ contains a 46-kb lactose plasmid, pCI646, and a larger, 58-kb plasmid, pCI658, which is linked to a soft, easily suspended, fluffy pellet phenotype and encodes complete and partial insensitivities against φ712 and φc2, respectively. Tc-AF030 Lac+ possesses two plasmids involved in phage resistance, pCI642 and pCI605, in addition to the Lac plasmid pCI646. pCI642 and pCI605 exert a cooperative effect in the production of phage defense against φ712 and φc2 in that both are required for maintenance of the phage resistance phenotype. Tc-AF009 Lac+ harbors pCI646 and displays only slight phage insensitivity. It was shown that the phage resistance mechanisms functional against φc2 in transconjugants Tc-AF021 Lac− and Tc-AF030 Lac− 1 (i.e., derivatives of Tc-AF021 Lac+ and Tc-AF030 Lac+ cured of the lactose plasmid) remained unaffected by incubation at 37°C, which is significant in the context of Cheddar cheese manufacturing processes.
A comprehensive examination of the phage-resistant transconjugants was performed in order to determine the underlying mode of action. It was found that the phage resistance phenotype exhibited by Tc-AF021 Lac− was the result of a striking decrease in the ability of the cell to adsorb phages compared to that of the sensitive strain MG1363. The reduction in phage adsorption was more pronounced for φ712 (from 99 to 14.7%) than for φc2 (87 to 57%). The difference in resistance levels was also reflected during phage internalization experiments, when no φ712 DNA could be detected in Tc-AF021 Lac−, while levels of φc2 DNA did increase, although more slowly than normal, with the time lag most likely resulting from the adsorption-blocking mechanism mediated by the pCI658-containing derivative. The difference in adsorption levels for both phages is most probably due to differences in their receptor sites and/or the mode of adsorption of the phages. Budde-Niekiel and Teuber (4) speculated that isometric-headed phages have a more restricted specificity of adsorption than prolate-headed phages. Evidence for adsorption inhibition in Tc-AF021 was further substantiated by the observation of a loose fluffy pellet following centrifugation of the strain, indicating the presence of an extracellular coating which probably masks phage receptors, thereby providing an effective shield against infection (data not shown). The phenomenon of cell surface alteration was also previously reported for the adsorption-blocking plasmids pSK112 (48) and pCI528 (34) and for the phage-resistant variant L. lactis subsp. cremoris 398 (19).
It was established that φ712 and φc2 had the capacity to adsorb normally to transconjugant Tc-AF030 Lac+. R/M was indicated as the mechanism of defense functioning within this derivative. The R/M system of Tc-AF030 Lac+ displayed a level of restriction which was greater for φ712 than for φc2, which may be related to the relative genome sizes of the infecting phage (31). However, the level of insensitivity expressed against φ712 was weaker in the absence of the native lactose plasmid pCI646, suggesting that this molecule is intrinsically involved in phage resistance. Higgins et al. (21) recorded the opposite effect, whereby the R/M phenotype encoded by pTN20 was actually suppressed when the lactose plasmid pTR1040 was coresident. It was found during this study that the existence of pCI646 alone in Tc-AF009 Lac+ did confer a small degree of phage insensitivity, indicating that the plasmid probably possesses inherent resistance which is as yet uncharacterized. Furthermore, a highly interesting feature of the Tc-AF030 Lac− 1 system was that resistance to φ712 and φc2 was eliminated when either plasmid pCI642 or pCI605 was removed from the strain; the presence of both plasmids was essential for the generation of R/M-mediated phage insensitivity.
Tc-AF030 Lac+ and Tc-AF030 Lac− 1 displayed reduced plaque size following infection with φ712 and φc2. In addition, cell death was negligible following infection of Tc-AF030 Lac− 1 with φ712, while a relatively high percentage of the population was killed following infection with φc2. The results of phage replication experiments showed that the abilities of φ712 and φc2 to proliferate in Tc-AF030 Lac− 1 were considerably weaker than that of the corresponding sensitive host, MG1363. These observations could reflect the presence of R/M and/or other additional activities, such as abortive infection, which could affect intracellular phage development within the strain. Indeed, R/M systems have been distinguished in many lactococcal strains harboring other complementary defense mechanisms (11, 14, 18, 21, 24, 33, 38, 47, 58), but the demonstration of the combined existence of R/M and Abi activities within a strain can prove difficult, since the effects of one system may overshadow those of the other (11, 24, 38). In the case of Tc-AF030 Lac− 1, the results of PCR experiments allowed the detection on pCI642 of a homolog of the abortive infection gene abiB (5), previously demonstrated by Parreira et al. (41) to be responsible for phage RNA degradation. However, the pCI642-derived abiB appeared to be poorly expressed and did not seem to promote degradation of φ712 RNA transcripts (data not shown).
The availability of the substantial amount of information regarding phage defense systems in lactococci, both at the genetic and mechanistic levels, has provided exciting opportunities for the development of phage-resistant starter cultures. Conjugal transfer of phage resistance plasmids, which is a food grade technology, has been used to enhance the resistance properties of commercial starter cultures (6, 7, 20, 27, 29, 30, 44, 50). During the course of this work, several experiments were undertaken to transfer the adsorption-blocking plasmid pCI658 to a broad range of lactococcal starter cultures; however, only one experiment was successful. A transconjugant of L. lactis subsp. cremoris 952 was generated which contained pCI658 and which exhibited the fluffy pellet phenotype characteristic of this plasmid. In addition, the strain had acquired partial resistance to its homologous φ952 (c2 phage species). L. cremoris 952 was previously shown at UCC to promote conjugal transfer of plasmids between lactococcal strains (29) and thus was employed as an intermediate strain to facilitate the transfer of pCI658 to other cultures. However, in this study, transfer of pCI658 to 10 other starter strains could not be achieved. In the construction of phage-resistant strains, several considerations should be made. According to Sanders (45), factors such as low transfer frequency, reverse conjugation (which confuses recovery of desired transconjugants), lack of antibiotic resistance markers, the uncertainty of the use of phage resistance as a selective marker, and phenotypic and genotypic instability of the phage defense system in different backgrounds are of prime importance. Any one (or a combination) of these criteria may have been responsible for the difficulties that were encountered in attempts to introduce pCI658 to strains with a wide variety of diverse backgrounds.
In conclusion, L. cremoris HO2 acts as an excellent reservoir of phage resistance plasmids, four of which were shown to be responsible for mediating the phage insensitivity phenotypes observed. Adsorption inhibition was associated with pCI658, R/M was expressed through the combined presence of pCI642 and pCI605, and the Lac plasmid pCI646 was also found to exert a degree of phage resistance. It is possible that these plasmids could be manipulated to improve the range of phage-resistant starter cultures available to the cheese manufacturing industry.
ACKNOWLEDGMENTS
This work was supported by the Department of Agriculture, Food and Forestry (DAFF), Dublin, Ireland, under the Food Industry Sub-Programme of EU Structural Funds, 1994–9, and by the Irish Co-operative Organisation Society (ICOS) Ltd., Dublin, Ireland.
We also acknowledge Ruth Davis and Judy Casey, who performed the initial culture screening program, Aidan Coffey for his advice, and Gro Johannessen for her contribution to the R/M studies.
REFERENCES
- 1.Anba J, Bidnenko E, Hillier A, Ehrlich D, Chopin M-C. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J Bacteriol. 1995;177:3818–3823. doi: 10.1128/jb.177.13.3818-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behnke D, Malke H. Bacteriophage interference in Streptococcus pyogenes. 1. Characterization of prophage-host systems interfering with the virulent phage A25. Virology. 1978;85:118–128. doi: 10.1016/0042-6822(78)90416-6. [DOI] [PubMed] [Google Scholar]
- 4.Budde-Niekiel A, Teuber M. Electron microscopy of the adsorption of bacteriophages to lactic acid streptococci. Milchwissenschaft. 1987;42:551–553. [Google Scholar]
- 5.Cluzel P-J, Chopin A, Ehrlich S D, Chopin M-C. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an iso-ISS1 element. Appl Environ Microbiol. 1991;57:3547–3551. doi: 10.1128/aem.57.12.3547-3551.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coakley M, Fitzgerald G F, Ross R P. Application and evaluation of phage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl Environ Microbiol. 1997;63:1434–1440. doi: 10.1128/aem.63.4.1434-1440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffey A G, Fitzgerald G F, Daly C. Identification and characterization of a plasmid encoding abortive infection from Lactococcus lactis subsp. lactis UC811. Neth Milk Dairy J. 1989;43:229–244. [Google Scholar]
- 8.Coffey A G, Fitzgerald G F, Daly C. Cloning and characterization of the determinant for abortive infection from the lactococcal plasmid pCI829. J Gen Microbiol. 1991;143:1355–1362. doi: 10.1099/00221287-137-6-1355. [DOI] [PubMed] [Google Scholar]
- 9.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteriophage resistance. Antonie Leeuwenhoek. 1996;70:99–110. doi: 10.1007/BF00395928. [DOI] [PubMed] [Google Scholar]
- 10.Dinsmore P K, Klaenhammer T R. Bacteriophage resistance in Lactococcus. Mol Biotechnol. 1995;4:297–314. doi: 10.1007/BF02779022. [DOI] [PubMed] [Google Scholar]
- 11.Durmaz E, Higgins D L, Klaenhammer T R. Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis subsp. lactis ME2. J Bacteriol. 1992;174:7463–7466. doi: 10.1128/jb.174.22.7463-7469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durmaz E, Klaenhammer T R. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defenses in a single Lactococcus lactis strain. Appl Environ Microbiol. 1995;61:1266–1273. doi: 10.1128/aem.61.4.1266-1273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emond E, Hollier B J, Boucher I, Vandenbergh P A, Vedamuthu E R, Kondo J K, Moineau S. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl Environ Microbiol. 1997;63:1274–1283. doi: 10.1128/aem.63.4.1274-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Froseth B R, Harlander S K, McKay L L. Plasmid-mediated phage insensitivity in Streptococcus lactis KR5. J Dairy Sci. 1988;71:275–284. doi: 10.3168/jds.S0022-0302(88)79555-7. [DOI] [PubMed] [Google Scholar]
- 15.Garvey P, Fitzgerald G F, Hill C. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4160–4166. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defence systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 17.Gasson M J. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier M, Chopin M-C. Plasmid-determined systems for restriction and modification activity and abortive infection in Streptococcus cremoris. Appl Environ Microbiol. 1987;53:923–927. doi: 10.1128/aem.53.5.923-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gopal P K, Crow V L. Characterization of loosely associated material from the cell surface of Lactococcus lactis subsp. cremoris E8 and its phage-resistant variant strain 398. Appl Environ Microbiol. 1993;59:3177–3182. doi: 10.1128/aem.59.10.3177-3182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington A, Hill C. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid pNP40. Appl Environ Microbiol. 1991;57:3405–3409. doi: 10.1128/aem.57.12.3405-3409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins D L, Sanozky-Dawes R B, Klaenhammer T R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988;170:3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill C. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:87–108. [Google Scholar]
- 23.Hill C, Massey I J, Klaenhammer T R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991;57:283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill C, Pierce K, Klaenhammer T R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+) Appl Environ Microbiol. 1989;55:2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill C, Miller L A, Klaenhammer T R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990;56:2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huggins A R, Sandine W E. Differentiation of fast and slow acid producing strains of lactic streptococci. J Dairy Sci. 1984;67:1674–1679. [Google Scholar]
- 27.Jarvis A W. Conjugal transfer in lactic streptococci of plasmid-encoded insensitivity to prolate- and small isometric-headed bacteriophages. Appl Environ Microbiol. 1988;54:777–784. doi: 10.1128/aem.54.3.777-783.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josephsen J, Klaenhammer T R. Stacking of three different restriction and modification systems in Lactococcus lactis by co-transformation. Plasmid. 1990;23:71–75. doi: 10.1016/0147-619x(90)90046-f. [DOI] [PubMed] [Google Scholar]
- 29.Kelly W, Dobson J, Jorck-Ramberg D, Fitzgerald G F, Daly C. Introduction of bacteriophage resistance plasmids into commercial Lactococcus starter cultures. FEMS Microbiol Rev. 1990;87:P63. [Google Scholar]
- 30.Klaenhammer T R. Development of bacteriophage-resistant strains of lactic acid bacteria. Biochem Soc Trans. 1991;19:675–681. doi: 10.1042/bst0190675. [DOI] [PubMed] [Google Scholar]
- 31.Klaenhammer T R. Plasmid directed mechanisms for bacteriophage defence in lactic streptococci. FEMS Microbiol Rev. 1987;46:313–325. [Google Scholar]
- 32.Klaenhammer T R. Genetic characterization of multiple mechanisms of phage defence from a prototype phage insensitive strain of Lactococcus lactis ME2. J Dairy Sci. 1989;72:3429–3443. [Google Scholar]
- 33.Laible N J, Rule P L, Harlander S K, McKay L L. Identification and cloning of plasmid deoxyribonucleic acid coding for abortive phage infection from Streptococcus lactis subsp. diacetylactis KR2. J Dairy Sci. 1987;70:2211–2219. [Google Scholar]
- 34.Lucey M, Daly C, Fitzgerald G F. Cell surface characteristics of Lactococcus lactis harbouring pCI528, a 46 kb plasmid encoding inhibition of bacteriophage adsorption. J Gen Microbiol. 1992;138:2137–2143. [Google Scholar]
- 35.Macrina F L, Kopecko D J, Jones K R, Ayers D J, McCowen S M. A multiple plasmid-containing Escherichia coli strain: a convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 36.McKay L L, Baldwin K A, Zottola E A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972;23:1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKay L L, Baldwin K A, Walsh P M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980;40:84–91. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKay L L, Bohanon M J, Polzin K M, Rule P L, Baldwin K A. Localization of separate genetic loci for reduced sensitivity towards small isometric-headed bacteriophage sk1 and prolate-headed bacteriophage c2 on pGBK17 from Lactococcus lactis subsp. lactis KR2. Appl Environ Microbiol. 1989;55:2702–2709. doi: 10.1128/aem.55.10.2702-2709.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLandsborough L A, Kolaetis K M, Requena T, McKay L L. Cloning and characterization of the abortive infection genetic determinant abiD isolated from pBF61 of Lactococcus lactis subsp. lactis KR5. Appl Environ Microbiol. 1995;61:2023–2026. doi: 10.1128/aem.61.5.2023-2026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor L, Coffey A, Daly C, Fitzgerald G F. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol. 1996;62:3075–3082. doi: 10.1128/aem.62.9.3075-3082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parreira R, Valyasevi R, Ehrlich S D, Chopin M-C. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol Microbiol. 1996;19:221–230. doi: 10.1046/j.1365-2958.1996.371896.x. [DOI] [PubMed] [Google Scholar]
- 42.Prevots F, Daloyau M, Bonin O, Dumont X, Tolou S. Cloning and sequencing of the novel abortive infection gene abiH of Lactococcus lactis subsp. lactis biovar. diacetylactis S94. FEMS Microbiol Lett. 1996;142:295–299. doi: 10.1111/j.1574-6968.1996.tb08446.x. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sanders M E, Leonhard P J, Sing W D, Klaenhammer T R. Conjugal strategy for the construction of fast acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986;52:1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders M E. Phage resistance in lactic acid bacteria. Biochimie. 1988;70:411–421. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- 46.Sanders M E, Shultz J W. Cloning of phage resistance genes from Lactococcus lactis subsp. cremoris. J Dairy Sci. 1990;73:2044–2053. [Google Scholar]
- 47.Schouler C, Clier F, Lerayer A L, Erhlich S D, Chopin M-C. A type IC restriction-modification system in Lactococcus lactis. J Bacteriol. 1998;180:407–411. doi: 10.1128/jb.180.2.407-411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sijtsma L, Jansen N, Hazeleger W C, Wouters J T M, Hellingwerf K J. Cell surface characteristics of bacteriophage-resistant Lactococcus lactis subsp. cremoris SK110 and its bacteriophage-sensitive variant SK112. Appl Environ Microbiol. 1990;56:3230–3233. doi: 10.1128/aem.56.10.3230-3233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sing W D, Klaenhammer T R. Characterization of restriction-modification plasmids from Lactococcus lactis subsp. cremoris and their effects when combined with pTR2030. J Dairy Sci. 1991;74:1133–1144. [Google Scholar]
- 50.Sing W D, Klaenhammer T R. Conjugal transfer of bacteriophage resistance determinants on pTR2030 into Streptococcus cremoris strains. Appl Environ Microbiol. 1986;51:1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sing W D, Klaenhammer T R. A strategy for rotation of different bacteriophage defenses in a lactococcal single-strain starter culture system. Appl Environ Microbiol. 1993;59:365–372. doi: 10.1128/aem.59.2.365-372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha R P. Effects of buffering media with phosphates on antibiotic resistance of lactic streptococci. Appl Environ Microbiol. 1984;47:1175–1177. doi: 10.1128/aem.47.5.1175-1177.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha R P. Stability of plasmids in lactococci during extended incubation in growth media. Can J Microbiol. 1991;47:488–490. doi: 10.1139/m91-082. [DOI] [PubMed] [Google Scholar]
- 54.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 55.Steenson L R, Klaenhammer T R. Plasmid heterogeneity in Streptococcus cremoris M12R: effects on proteolytic activity and host dependent phage replication. Appl Environ Microbiol. 1986;50:851–858. doi: 10.3168/jds.S0022-0302(86)80661-0. [DOI] [PubMed] [Google Scholar]
- 56.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahl G, Stern M, Stark G R. Efficient transfer of large DNA fragments from agarose gels to diazobenzylomethyl paper and rapid hybridization using dextran sulphate. Proc Natl Acad Sci USA. 1979;76:3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward A C, Davidson B E, Hillier A J, Powell I B. Conjugally transferable phage resistance activities from Lactococcus lactis DRC1. J Dairy Sci. 1992;75:683–691. [Google Scholar]