Abstract
Root system architecture is an important determinant of below-ground resource capture and hence overall plant fitness. The plant hormone auxin plays a central role in almost every facet of root development from the cellular to the whole-root-system level. Here, using Arabidopsis as a model, we review the multiple gene signaling networks regulated by auxin biosynthesis, conjugation, and transport that underpin primary and lateral root development. We describe the role of auxin in establishing the root apical meristem and discuss how the tight spatiotemporal regulation of auxin distribution controls transitions between cell division, cell growth, and differentiation. This includes the localized reestablishment of mitotic activity required to elaborate the root system via the production of lateral roots. We also summarize recent discoveries on the effects of auxin and auxin signaling and transport on the control of lateral root gravitropic setpoint angle (GSA), a critical determinant of the overall shape of the root system. Finally, we discuss how environmental conditions influence root developmental plasticity by modulation of auxin biosynthesis, transport, and the canonical auxin signaling pathway.
In vascular plants, root systems form the interface between the plant body and the soil. They play a crucial role in basic functions such as anchorage and nutrient and water uptake, as well as in species-specific functions such as vegetative reproduction, minimizing underground competition with adjacent individuals, and maintaining abiotic and biotic interactions within the rhizosphere (Morris et al. 2017). Plants encounter myriad subterranean macro- and micro-environmental stresses during their lifetimes, including biotic stresses, such as pathogen attack, and abiotic stresses such as nutrient and water scarcity. Root systems must therefore retain the ability to perceive and respond to these stresses at multiple levels, ranging from individual cells to tissues and eventually whole organs. The ability of root systems to maintain a high degree of plasticity is therefore key to ensuring the survival, fitness, and adaptation of individual plants (Khan et al. 2016).
Roots can have one of two distinct origins: the embryo (embryonic, i.e., primary or seminal roots) or other roots and/or non-root tissues (postembryonic). Roots that arise from pre-existing roots are usually secondary or lateral roots (LRs), while those that arise from non-root tissues are adventitious or basal roots (for review, see Du and Scheres 2018). In dicot species, including Arabidopsis, the primary embryonic root is dominant and produces a number of secondary LRs. Over time, these LRs reiterate the formation of new LRs, which leads to the production of a highly ordered and spatially distinct root system termed as the “taproot” or allorhizic root system. In contrast, monocots form “homorhizic” root systems comprised of numerous adventitious roots that in turn produce additional secondary or LRs (Osmont et al. 2007; Bellini et al. 2014). An important distinction between the two types of root systems lies in the life span of the primary embryonic root: in taproot systems, the primary root remains dominant during the entire life span of the plant, while in homorhizic systems, embryonic roots are often short-lived and play a role in the establishment of the root systems at the seedling stage, before the development of adventitious roots, which dominate the root system (Bellini et al. 2014).
Root system architecture (RSA) describes the spatial distribution of the root system, capturing the specific deployment of root axes in the soil matrix or substrate upon which the plant grows (Smith and De Smet 2012; Khan et al. 2016). Macro RSA, the large-scale overall form of the root system, comprises three major parameters: length, degree of branching, and angle of growth. At a smaller scale, the “micro” parameters of RSA that influence the extent of root surface area include root diameter and the formation of root hairs. These macro- and micro-scale components of RSA are a critical determinant of the plant's ability to capture the water and nutrients that are typically distributed heterogeneously within the soil (Smith and De Smet 2012; Morris et al. 2017; for review, see Rellán-Álvarez et al. 2016). RSA parameters can be modulated by multiple genetic, physiological, and environmental factors, allowing for an efficient capture of resources from the rhizosphere (for review, see Rellán-Álvarez et al. 2016). Understanding the molecular mechanisms that regulate RSA and its plasticity is therefore key to enhancing both the productivity and sustainability of agriculture through developing crop varieties that are better able to take up water and nutrients, reducing the need for supplementary fertilizer application and irrigation (Lynch 2013).
The plant hormone auxin is the master regulator of plant architecture (for review, see Benjamins and Scheres 2008; Overoode et al. 2010). Auxin is a small molecule that is dynamically distributed via a system of polar auxin transport. The resulting gradients, minima and maxima of auxin concentration that are formed to act to pattern, trigger developmental transitions, and control growth. Auxin can enter cells passively, but is also actively imported into cells by the influx carriers AUX1 and LIKE AUX1 (LAX) 1-3 (Swarup et al. 2005; Péret et al. 2012). Auxin must be exported from cells by the PIN-FORMED family of efflux transporter proteins as well as the ABCB and TWISTED DWARF1 (TWD1) proteins (for review, see Geisler et al. 2017; Zwiewka et al. 2019; Hammes et al. 2021). It is this requirement for active auxin efflux from cells that forms the basis of the directional control of auxin movement. Arabidopsis contains eight PIN proteins, of which PINs 1–4 and 7 are canonical “long PINs” targeted to the plasma membrane, while PINs 5 and 6 are short PINs targeted to the endoplasmic reticulum (ER). Although each of the long PINs have specific expression patterns and play distinct roles in root and shoot development, recent work has shown that all five canonical PINs share equivalent capacity to generate auxin maxima and regulate different aspects of root (and shoot) development (Zhang et al. 2020). Thus, coordinated regulation of the subcellular distribution of PIN proteins within the field of cells allows flows of auxin to be generated (Blilou et al. 2005). Spatiotemporal variation in auxin concentration is then translated into developmental control principally via the TIR1/AFB-Aux/IAA-ARF system of auxin-dependent transcriptional regulation (see Morffy and Strader 2021). Briefly, in this system auxin acts to regulate the abundance of Aux/IAA transcriptional corepressor proteins, which are targeted to auxin-regulated genes via their interaction with DNA-binding transcription factors called auxin response factors (ARFs) (for review, see Leyser 2018). Auxin promotes the formation of an auxin coreceptor complex comprising an F-box protein of the TIR1/AFB family and an Aux/IAA protein. This auxin-enhanced interaction catalyzes to ubiquitin-mediated proteolysis of the Aux/IAA, thereby de-repressing the ARF-bound loci previously transcriptionally silenced by the presence of the Aux/IAA (Gray et al. 2001; Dharmasiri et al. 2005; Kepinski and Leyser 2005). Here, we review the complex role of auxin in root development and its impact on RSA.
AUXIN IN PRIMARY ROOT DEVELOPMENT
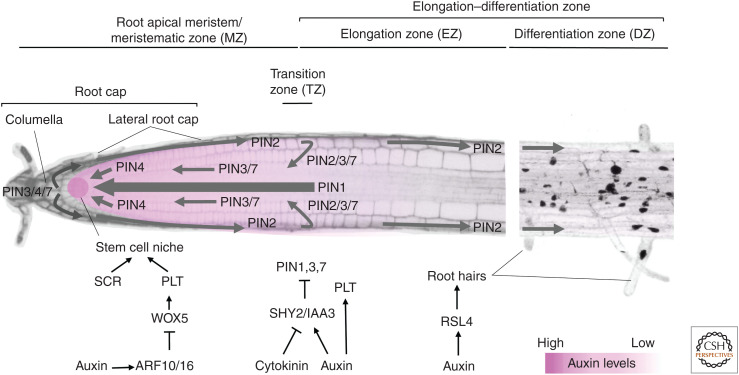
In the Arabidopsis root, tissues originate through the activity of the root meristem. Just above the columella, which constitutes the root tip, is the stem cell niche, which is composed of a group of slow-dividing cells that constitute the organizing center, also called quiescent center (QC), and the surrounding long-term stem cells (for review, see García-Gómez et al. 2021). These stem cells divide asymmetrically to short-term stem cells, which undergo multiple cell divisions with little expansion, in the proximal meristem (PM). The PM is enveloped by the lateral root cap (LRC), an important tissue for auxin transport and homeostasis in the root meristem. Cells are displaced away from the stem cell niche by subsequent cell divisions, until they reach the transition zone (TZ), where they stop dividing and start to differentiate. Cells then enter the elongation zone (EZ) within which cells undergo vast increases in length (up to 300% in 3 h) with virtually no division. The EZ is followed by the differentiation zone (DZ), where cells stop elongating and complete their differentiation gaining cell-type-specific features and functions (Verbelen et al. 2006; Markakis et al. 2012). Research has shown that some of the signaling processes that mediate differentiation begin while cells are still elongating (see below) (Datta et al. 2015). For this reason, the EZ and DZ have sometimes been referred to collectively as the elongation−differentiation zone (EDZ), a name that better captures the overlap in the processes of cell elongation and cell differentiation (Takatsuka and Umeda 2014).
T he patterning and coordination of the processes that maintain the constant self-renewal and growth of the root is complex. At first sight, the extent of auxin's involvement in so many of them seems quite remarkable. First, a localized, auxin maximum in the root apex acts to position and maintain the QC and stem cell niche (Sabatini et al. 1999; Grieneisen et al. 2007). This auxin maximum is specified during embryogenesis in the hypophyseal cell at the base of the embryo, where it first establishes the population of root stem cells. In both the embryo and postembryonic root, the formation of these auxin maxima is principally dependent on the expression and subcellular localization of members of the PIN family of auxin efflux transporters (see Fig. 1 and legend for details; Blilou et al. 2005; Michniewicz et al. 2007; Zourelidou et al. 2014). Auxin maintains stem cell organization by regulating the differentiation of distal stem cells (the cells that give rise to the columella) through a signaling pathway involving the transcription factors PLETHORA (PLTs) and the homeodomain transcription factor WUSCHEL RELATED HOMEOBOX 5 (WOX5) (Sabatini et al. 2003; for review, see Drisch and Stahl 2015). Through an ARF10/16-IAA17/AXR3 signaling pathway, local auxin levels are tightly regulated in the root tip by biosynthesis and transport, in turn repressing the expression of WOX5 and restricting it to the QC where it is required for the expression of PLTs (Ding and Friml 2010).
Figure 1.
Organization of the primary root in Arabidopsis. In the root apex, individual PINs are localized to specific cell membranes to generate a pattern of auxin distribution in the root tip. In the stele, several PINs but principally PIN1 are localized to rootward cell membranes leading to a strong tipward flux of auxin. In the columella cells of the root cap, PIN3, PIN4, and PIN7 distribute auxin laterally toward the lateral root cap, from where it is transported shootward through the epidermis by AUX/LAX proteins and PIN2 (Hu et al. 2021). At the transition zone, some auxin is also refluxed back into the rootward stream in the stele via lateral, inward transport by PIN2, PIN3, and PIN7 (Blilou et al. 2005). This topology of PINs proteins in the root apex creates both the maximum of auxin that positions the stem cell niche and through the gravity-responsive, asymmetric lateral redistribution of auxin from the columella, a means to steer the growth of the root with reference to gravity (for review, see García-Gómez et al. 2021). The identity of the stem cell niche is also dependent on the high levels of PLT expression, tightly regulated by complex coordinated hormone signaling pathways. Further shootward, antagonistic interaction between auxin and cytokinin maintain the identity of the meristematic and transition zones, partially through the regulation of levels of the Aux/IAA, SHY2/IAA3. At the transition zone PLT levels are intermediate and become further reduced in the elongation–differentiation zone, where cells begin to stop elongating and differentiate to acquire specific functions, such as root hairs.
Next, a complex network of hormone signaling pathways specify the boundaries of mitotic activity in the primary root apex, positioning the TZ and maintaining a meristem of sufficient size to sustain continuous root growth. At the TZ, auxin and cytokinin act antagonistically to regulate the expression of the PIN efflux transporters through modulation of the expression of the auxin signaling corepressor SHY2/IAA3. This, in turn, promotes auxin redistribution leading to cell differentiation. Conversely, auxin promotes SHY2 degradation thereby promoting cell division (Dello Ioio et al. 2008). Further insight into auxin-dependent TZ maintenance came from a recent study that combined computational and genetic approaches (Di Mambro et al. 2017). This work showed that cytokinin-dependent degradation of auxin, via conjugation by the enzymes of the GRETCHEN HAGEN (GH) family, in the LRC is required for the specification of an auxin minimum, which coincides with the TZ. The authors predicted, using mathematical modeling, that perturbation of this auxin minimum would lead to changes in TZ positioning and root meristem size and, conversely, showed that manipulation of GH expression level resulted in variations in root meristem size (Di Mambro et al. 2017). In addition, auxin also controls TZ positioning by regulating the graded distribution of PLTs (Aida et al. 2004; Mähönen et al. 2014; Durgaprasad et al. 2019). PLTs are transcribed in a narrower domain around the stem cell niche, and PLT proteins form a gradient through a mechanism that involves coordinated cell divisions and cell-to-cell transport. Auxin regulates PLT levels and distribution by both inducing PLT expression around the stem cell niche and by regulating cell division in the PM. PLT controls root zonation in a dose-dependent manner: high levels of PLT in the QC are necessary to maintain slow mitotic stem cell identity, intermediate PLT levels in the meristem induce rapid cell divisions, and low PLT levels at the TZ allow for the onset of cell expansion and differentiation (Galinha et al. 2007; Mähönen et al. 2014). A recent systems biology approach further revealed the complex interplay between auxin-cytokinin-PLT signaling pathways in root development. This study showed that PLT2 levels drop in the root TZ, leading to ARR12 activation. The resulting PLT2-ARR12 antagonism controls the growth of the root meristem in the initial phases of primary root growth. Subsequent activation of ARR1 further reduces PLT2 levels through induction of the cell-cycle repressor KRP2, thus setting final meristem size (Salvi et al. 2020).
Proximal to the meristem, auxin concentrations decrease as daughter cells repeatedly divide before they differentiate. Crucially, auxin concentrations rise again after the TZ, where it is needed for appropriate cell differentiation. In the DZ, auxin is an essential driver of developmental events in both proximodistal and radial axes (Marhava et al. 2018). For example, root protophloem sieve element (PPSE) cells accumulate higher levels of auxin and begin the process of differentiation while their neighbouring cells are still meristematic. A recent study has demonstrated the novel role of two plasma membrane proteins, BRX (BREVIS RADIX) and PAX (a D6PK-like protein kinase, member of the AGC protein family) in regulating this pathway. This work showed that BRX and PAX coordinately act at the rootward membrane of PPSE cells to inhibit PIN1-dependent auxin efflux. This causes a rise in auxin levels, which triggers PPSE differentiation. Following the localized increase in auxin levels, BRX is displaced from the membrane, which in turn activates PAX-dependent auxin efflux and leads to localized reduction in auxin levels. The authors propose that BRX and PAX act as a “molecular rheostat” to control the timing of PPSE differentiation in a multicellular context (Marhava et al. 2018).
Surprisingly, the role of auxin homeostasis and signaling networks in regulating root elongation still remains poorly understood. However, a large number of studies have demonstrated that the exogenous application of both natural and synthetic auxins inhibits cellular elongation and, therefore, root growth. Interestingly, recent work also has demonstrated that tissue-specific biosynthesis of auxin effects root growth. In this study, Hu et al. (2021) expressed the YUCCA and TAA1 auxin biosynthesis genes under the control of promoters that restricted their expression to specific root tissues: pWER—epidermis, pSCR—endodermis, pSHR—stele, pAPL—phloem (protophloem, companion cells, and metaphloem sieve elements), and pWOX5—QC. The authors showed that the ectopic expression of YUCCA-TAA1 in the epidermis, endodermis, and stele caused very short roots, while YUCCA-TAA1 expression in the phloem and QC had relatively mild effects (Hu et al. 2021).
The inhibition of root growth by auxin has been linked at least partially to auxin-dependent disruption of the auxin cytoskeleton (Rahman et al. 2007; for review, see Zhu and Geisler 2015). Auxin homeostasis during overall root development was also recently found to be dependent on the interplay between the dynamic regulation of the auxin oxidase AtDAO1 (DIOXYGENASE FOR AUXIN OXIDATION 1) and the conjugation enzyme GH3. Mellor et al. (2016) used a combination of mathematical modeling, biochemical, and physiological approaches to show that at lower concentrations of auxin, AtDAO1 was expressed at higher levels to oxidize endogenous auxin, while higher levels of auxin led to expression of the conjugating enzyme GH3. Further work by Zhang et al. (2016) showed that Arabidopsis contains two distinct DAO genes, AtDAO1 and AtDAO2, and loss-of-function dao mutants had elongated primary roots, increased LR density, and longer root hairs (Porco et al. 2016a; Zhang et al. 2016) compared to wild-type (WT) seedlings. All of these phenotypes are attributed to elevated IAA levels in dao mutants.
Auxin also regulates epidermal cell patterning. The Arabidopsis root epidermis is alternately spaced with files consisting of smaller trichoblast or hair cells, which generate root hair, and longer atrichoblast or non-hair cells (Dolan 2001; Löfke et al. 2013). Interestingly, at least two auxin transporters are known to be differentially expressed within these distinct cell files. First, the activity of the auxin influx carrier AUX1 mediates auxin transport through atrichoblast cells and is necessary to maintain the identity of root hair cells, and promote root hair elongation. Surprisingly, AUX1 itself is not present in root hair cells, suggesting that auxin is transported longitudinally through canals in epidermal tissue (Jones et al. 2009). Second, PIN2 is also targeted to the plasma membrane of non-hair cells at higher levels than the hair cell files, likely by differential rates of trafficking to lytic vacuoles within these files (Löfke et al. 2015). Taken together, these data suggest that distinctly different downstream auxin-dependent processes occur in these two cell types.
Downstream of PIN/AUX1 auxin transport-dependent patterning, auxin regulates hair cell growth by modulating the expression of a basic helix-loop-helix transcription factor RSL4 (ROOT HAIR DEFECTIVE 6 LIKE 4). RSL4 is sufficient to promote postmitotic hair cell elongation and, accordingly, rsl4 mutants have very short root hairs (Yi et al. 2010). RSL4 is an auxin-inducible gene (Yi et al. 2010) and more recently it has been shown that root hair elongation is directly proportional to the amplitude of a pulse of RSL4 expression that precedes formation of the root hairs in the DZ (Datta et al. 2015). The intensity of this transient increase in RSL4 expression can be modulated by external environmental signals such as low phosphate, providing another mechanism for the integration of environmental cues to modulate plant development (Datta et al. 2015).
AUXIN IN LATERAL ROOT DEVELOPMENT
In Arabidopsis, LRs arise from single-layered pericycle cells. Radially, the pericycle is surrounded by the endodermal, cortical, and epidermal cell layers, and itself surrounds the central root vasculature. The pericycle is composed of two distinct cell types: phloem pole pericycle (PPP) and xylem pole pericycle (XPP). LRs arise exclusively from XPP cells, also known as pericycle founder cells. Crucially, these cells are thought to retain semi-meristematic properties based on their ultrastructure (small vacuoles, dense cytoplasm, and ribosomes) and maintain the ability to undergo cell divisions upon activation by remaining in the G2 phase of the cell cycle for long periods.
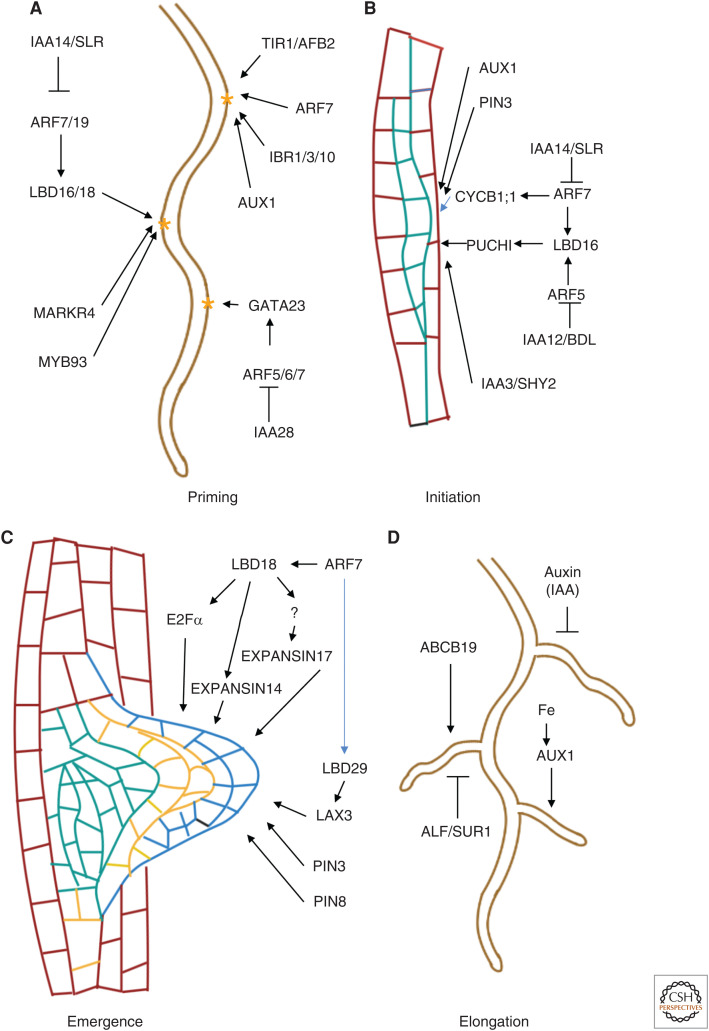
LR development occurs in distinct steps, including (1) pericycle priming and specification of founder cells; (2) initiation, defined as the first round of anticlinal divisions of the founder cells; (3) primordium formation, defined as the sum of divisions leading to the formation of a dome-shaped primordium; and (4) emergence, defined as the protrusion of the newly formed LR through overlying ground and epidermal tissue (Fig. 2; covered in detail in Cavallari et al. 2021).
Figure 2.
Auxin signaling networks in lateral root development. Visual summary of genes and networks acting both upstream and downstream of the canonical TIR1/AFB-Aux/IAA-ARF auxin signaling pathway during the multiple stages (A) priming, (B) initiation, (C) emergence, and (D) elongation of lateral root development in Arabidopsis. Yellow asterisks in A denote priming sites. Arrowheads denote up-regulation of expression of target genes, while bars denote target down-regulation.
Priming
Early patterning events that determine the regular spacing of LRs coincide with the oscillation of auxin-dependent transcriptional mechanism. In Arabidopsis, LRs emerge from the DZ of the primary root at regularly spaced intervals. When roots are observed in a 2D growth system, such as a petri dish, LRs emerge specifically at the outside edges of waves or curves. An auxin-dependent root clock transcriptional signal is one of the earliest drivers of early LR priming and, therefore, patterning as confirmed by several studies (De Smet et al. 2007; Laskowski et al. 2008; Moreno-Risueno et al. 2010; Xuan et al. 2015, 2016). Crucially, these studies first demonstrated the oscillatory expression of the auxin-responsive promoter DR5 driving the expression of two different markers, β-glucuronidase (GUS) and luciferase (LUC). The oscillations have an amplitude of ∼6 h and occur within the basal meristem and elongation zones, together termed the oscillation zone (OZ). However, the periodicity of the root clock can be modified by environmental signals or auxin treatment (Moreno-Risueno et al. 2010; Kircher and Schopfer 2016). In standard conditions, however, over time, a portion of cells within the OZ maintained stable high expressions of DR5:LUC signal. These cells then began to initiate LRs as they reached the DZ in the elongating root and were defined as LR pre-branch sites. Interestingly, the authors found large numbers of genes that oscillate both in phase (2000) and in antiphase (1500) of DR5:LUC. Also, the application of exogenous auxin was not sufficient to induce LR priming. Taken together, these data suggest that priming is a result of fluctuations not only in auxin levels, but in the expression of a significant number of genes.
Studies using of loss-of-function mutants have reiterated the importance of auxin perception and signaling in LR priming and pre-branch site formation. The arf7, tir1, and afb2 mutants all display aberrant DR5 oscillatory patterns and have markedly lower numbers of LR pre-branch sites (for review, see Du and Scheres 2018). Recently, ARF7 and its auxin-sensitive inhibitor POTENT/IAA18 were found to regulate gene oscillation in the OZ required for the formation of LR pre-branch sites by developing a negative regulatory loop circuit that was predicted to be influenced by environmental cues (Perianez-Rodriguez et al. 2021). Interestingly, an indole-3-butyric acid (IBA) to indole acetic acid ([IAA], the most common form of active auxin) conversion pathway acting specifically within the LRC seems to play a significant role in the generation of stable auxin fluxes required for pre-branch site establishment. Mutations in three INDOLE-3-BUTYRIC ACID RESPONSE genes (IBR1, IBR3, and IBR10) lead to reduced frequency of pre-branch sites (Fig. 2A; Xuan et al. 2015). In a follow-up study, the authors demonstrated the significance of local fluctuations in auxin levels in pre-branch site establishment (Xuan et al. 2016). LRC cells reaching the TZ displayed a gradual reduction in DR5:VENUS signal just prior to their apoptosis and act as a source of auxin for neighboring stele cells, consistent with DR5 activity oscillation. This work also showed that AUX1 expression in the LRC was required for correct DR5 oscillation amplitude and LR patterning.
Interestingly, several studies have demonstrated that not all pre-branch sites showing static DR5 signal go on to become lateral root founder cells (LRFCs) or, indeed, form LRs. One of the earliest molecular markers for LRFC formation is the auxin regulatory GATA23 transcription factor. GATA23 is activated at specific pre-branch sites through an IAA28-ARF 6/7/8/19-dependent pathway in the basal meristem (Fig. 2A). XPP cells leaving the basal meristem displays broad peaks in GATA23 expression and both gata23 gain- and loss-of-function mutants show altered numbers and spacing of LRs (De Rybel et al. 2010). Moreover, LBD16/ASL18 and other redundantly expressed LBDs, expressed in the adjacent cells of the XPPs undergoing asymmetric cell division downstream of an SLR14-ARF7/19 signaling module, are required for founder cell specification (Fig. 2A; Goh et al. 2012). Additional molecular genetic studies also identified MARKR4 as another candidate gene in LRFC specification. markr4 mutants show fewer LRs, but have unaltered numbers of pre-branch sites, suggesting that MARKR4 is required for the conversion of pre-branch sites into LRFCs (Fig. 2A). MARKR4 was identified as a downstream molecular component of the IBA to IAA conversion pathway (Xuan et al. 2016). Another potential candidate regulator is MYB93, a member of the R2R3 MYB transcription factor family, which is strongly induced by auxin in the basal meristem (Gibbs et al. 2014).
A known positional cue that tightly regulates LR initiation is curvature, either induced mechanically or by gravity. Several studies have convincingly demonstrated that LRs preferentially originate from the convex side of a curved root. Several in silico and mechanotransduction studies have attempted to explain this phenomenon by considering factors such as the differences in geometry of XPP cells on convex and concave sides of the bend, and heterogenous changes in of Ca2+ levels in stretched XPP cells. However, experimental work recently showed that curvature alone is not sufficient to specify positioning of the LR, as even if curvature frequency increases, the frequency of LR initiation remains the same (Kircher and Shopfer 2016). Thus, while further work is required to fully understand the signaling pathways that regulate unilateral LR initiation after LRFC specification (Laskowski et al. 2008; Richter et al. 2009), it is evident that auxin transport from the LRC, via the influx transporter AUX1, also plays a role in specifying LR initiation sites. Roots of the Arabidopsis aux1 mutant display a coiled growth phenotype, with LRs emerging mainly from the convex side of the bend, in complete contrast to the normal wavy growth patterns of WT roots (Marchant et al. 2002; De Smet et al. 2007). Further, aux1 mutants also have fewer LRs and display lower levels of DR5 oscillatory activity in the basal meristem. However, targeted expression of AUX1 to the LRC and epidermal cells restores both the agravitropic root and aberrant LR branching patterns in the aux1 mutant (Swarup et al. 2005).
Studies using a heat-shock-inducible system to generate clonal sectors of XPP cells with local high IAA levels (iaaM) were sufficient to enhance LR production in pericycle cells (Dubrovsky et al. 2008). In silico approaches proposed that existing LRFCs were able to inhibit new LRFCs in the distal region of the root, by depleting auxin levels in the surrounding cells (Laskowski et al. 2008) These local auxin gradients are formed by the action of influx and efflux carriers. Conversely, molecular genetic studies showed that roots of polar auxin transport mutants (e.g., pin2pin3pin7 and weak alleles of gnom) and loss- (e.g., mp/arf5) or gain-of-function (e.g., shy2, bdl) signaling mutants show altered branching patterns, with closely grouped, fused, or fewer LRP/LRs (Laskowski et al. 2008; Geldner 2009; De Smet et al. 2010; Goh et al. 2012; Okumura et al. 2013).
Other regulatory genes/modules also interact with auxin to induce and regulate LRFC patterning. For example, the PLT triple loss-of-function plt357 mutant has several clustered LR primordia (Hofhuis et al. 2013). Finally, auxin-responsive small terminal peptide RALF34, which is expressed around the flanks of the pericycle cells undergoing divisions to form LRPs, seems to be required for spatial distribution of LRs. Indeed, two loss-of-function insertion mutants of RAFLF4 showed a threefold increase in clustered LRPs (Murphy et al. 2016). Another auxin-repressed peptide signaling module CEP5/CEPR has a similar phenotype in loss-of-function background (Roberts et al. 2016).
LR Initiation and Primordium Formation
The first step in LR initiation involves the formation of an auxin maximum in specified LRFCs (for review, see Torres-Martínez et al. 2019). This maximum is dependent on the influx carrier AUX1 and the activation of YUCCA4-mediated auxin biosynthesis by plant-specific B3 transcription factors FUSCA3 and LEC2 (Tang et al. 2017). During primordium formation, the first cell division occurs anticlinally to form a series of cells that undergo further periclinal division (for review, see Du and Scheres 2018). This step is preceded by nuclear migration within the XPP cells toward the center of the LR-initiation site. This is followed by asymmetric cell division, which is again regulated by BDL-ARF5- and SLR14-ARF7-dependent LBD16 expression in the XPP cells (Fig. 2B; Goh et al. 2012; Lee et al. 2015). LBD16 is required for LR initiation and subsequent cell fate re-specification after the first round of asymmetric divisions (Goh et al. 2012). LBD16 in turn up-regulates PUCHI, an auxin-inducible transcription factor expressed within newly developed LRPs following the first round of asymmetric divisions (Fig. 2A; Goh et al. 2019). Premature induction of PUCHI during the preinitiation phase disrupts LR primordium formation.
The divisions necessary to initiate LR formation require pericycle cells to reenter the cell cycle and start dividing actively. This first series of XPP divisions depends on mechanical feedback from the overlying endodermal cells. A SHY2/IAA3-dependent signal triggers loss of endodermal cell volume and turgor pressure by relinquishing their tight junction-like diffusion barriers (Vermeer et al. 2014). These biophysical changes trigger CYCB1;1 expression in the XPP cells, which regulates G2-M transition. Again, the SLR-ARF7/19 signaling module seems to be required for this pathway and slr and arf7/19 mutants have almost no LRs and show reduced expression of CYCB1;1, likely through an LBD16/LBD18 signaling module (Fig. 2B; Vanneste et al. 2005; Okushima et al. 2007). In addition to triggering changes in endodermal cell shape, auxin signaling in pericyle cells is also required to specify the plane of division. tir1/afb mutants show periclinal XPP cell divisions that are not competent to form primordia. This auxin-dependent input into cell plane division specification relies on a dynamic microtubule cytoskeleton (for review, see Van Norman et al. 2013).
PIN3 is transiently expressed in the endodermal cells overlying the LRFC, until just after the first round of anticlinal divisions. Interestingly, pin3 mutants have increased numbers of founder cells but reduced numbers of LRPs, suggesting that transient endodermal PIN3 expression is required for the transition from founder cell to primordium stage. PIN3 is initially distributed in a nonpolar manner in the endodermal cells in the DZ. However, during the course of LRP formation, PIN3 becomes polarized to the inner cell wall (inner lateralization) (Marhavý et al. 2013, 2016) facilitating auxin efflux into the LRFCs for the first round of anticlinal divisions.
LR Emergence
LRs originate from pericycle cells deep within the parental root tissue and must emerge through three distinct cell layers (endodermis, cortex, and epidermis). These cell layers undergo cell wall loosening and hydraulic separation via multiple auxin-dependent signaling modules. Particularly within the endodermis, the dynamic opening and resealing of lignified and suberized barriers are tightly regulated for LR emergence, and to prevent leakage of nutrients and pathogen entry from the soil. This endodermal remodeling is also critical for root system plasticity (Barberon et al. 2016). A new transcriptomic study identified specific members of GDSL-type esterases/lipases (GELPs), proteins that are expressed sequentially downstream of SHY2 and regulate auxin-dependent desuberization during LR emergence, and consequently resuberization of the endodermal barrier post-LR emergence (Ursache et al. 2021). A study from a few years ago, which used a combination of mathematical modeling and cutting-edge light sheet fluorescence microscopy, showed that while the first division is tightly regulated, subsequent divisions do not occur in a rigid sequence as the primordium center grows, but instead their orientations (anticlinal or periclinal) depend on cell geometry and follow the “shortest wall” rule. The orientations of these division planes, the apical growth of the primordium due to cell proliferation, and accommodation from the overlying cell layers are all key factors that determine the emergence of the characteristic layers that make up the dome-shaped primordium (von Wangenheim et al. 2016).
Interestingly, recent data has suggested that the Arabidopsis noncanonical ARF ETTIN/ARF3 (ETT), which lacks the PB1 DNA-binding domain, may play an important role in LR emergence (Stoeckle et al. 2018). Loss-of-function and auxin-insensitive mutants have increased numbers of emerged LRs, suggesting that auxin can act directly through ETT to negatively regulate LR emergence (Simonini et al. 2017).
Downstream of an ARF7/19 module, LBD transcription factors play multiple roles in LR initiation and emergence. An LBD18/33 dimer are other direct downstream targets of this module and themselves regulate the expression of E2Fα, which encodes a transcriptional activator of cell-cycle genes (Berckmans et al. 2011). Further, LBD18 also directly up-regulates EXPANSIN14, and indirectly regulates EXPANSIN17, required for loosening of the cell wall during LR emergence (Fig. 2C).
The auxin influx transporters, AUX1 and LAX3, play a crucial role in regulating spatial separation of adjacent overlying cell files as the newly formed LRP emerges. Initially, AUX1 facilitates the loading of shoot-derived IAA into the vascular system to ensure that the auxin required for cell wall loosening is channeled to the overlying tissue via the LRPs (Marchant et al. 2002; Swarup et al. 2005; Péret et al. 2013). At later developmental stages, the primordium itself becomes the source of auxin required in the overlying tissue layers to trigger the expression of cell wall remodeling enzymes required for emergence. Several in silico and in vivo studies have also demonstrated that sequential expression of PIN3 and LAX3 in turn in the two adjacent cell files overlying the LRP is crucial to ensure that the auxin-dependent separation of cells occurs solely along the shared walls. LAX3 is a downstream target of LBD29, which is a direct target of ARF7 (Fig. 2C; Porco et al. 2016a,b).
The intracellular auxin homeostasis required during LR emergence is maintained by the short PIN, PIN8. PIN8 is expressed in the phloem cells adjected to the primordium at a later stage of development. Although the mechanisms by which PIN8 promotes LR emergence is not fully understood, pin8 mutants show significantly reduced numbers of emerged LRs that are arrested at the primordium stage and have reduced expression levels of GATA23 and LBD genes (Lee et al. 2020).
LR Elongation
Surprisingly, there is little information available on the effect of auxin and the role of auxin signaling networks on LR elongation. In Arabidopsis, at least, several studies have reported that auxin inhibits the growth of LRs in young seedlings. Interestingly, however, in tomato, Muday and Haworth (1994) reported that auxin promoted LR growth, suggesting that the growth responses of LRs to auxin are likely to be species specific (for review, see Waidmann et al. 2020).
Gain- and loss-of-function studies using Arabidopsis mutants have identified a small number of additional genes that regulate LR elongation. Celenza et al. (1995) reported that the constitutively active allele of alf1/sur displays hyperproliferation of LRs (Fig. 2D). Whereas Boerjan et al. (1995) established that this mutation leads to an overproduction of endogenous IAA, the ALF gene is yet to be mapped. Moreover, Wu et al. (2007) demonstrated that the loss-of-function mutants of the ABCB14/PGP19/MDR1 gene show arrested and/or impaired LR growth rates and a 40% reduction of acropetal auxin transport within the LRs. In contrast, no differences in LR growth rates have been reported between WT and aux1 mutants in normal conditions (Linkohr et al. 2002; Giehl et al. 2012). However, localized iron supply was found to promote LR elongation via altering AUX1-mediated auxin distribution, suggesting that environmental triggers are able to influence LR growth rates via the modulation of auxin transport and/or signaling components (Fig. 2D; see the “Environmental Regulation” section).
GRAVITROPIC SETPOINT ANGLE (GSA)
Perhaps the most significant parameter that determines overall root architecture is root angle. As described previously, secondary roots in most plant species grow at oblique angles in the soil. These angles, and indeed all angles that plant organs may grow at are known as gravitropic setpoint angles (GSAs). GSAs are defined as the angle at which a plant organ grows to maintain equilibrium with gravity (Digby and Firn 1995). Spatiotemporal developmental and environmental cues regulate the setting and maintenance of these root angles within the soil, and can therefore shape root architectures that can contrastingly range from shallow, widely distributed roots, to narrow deeper rooting systems. The distribution of the root system within the soil is key to maximizing the capture of heterogenously distributed soil minerals and water. For example, phosphate and nitrate are often found while water is found in the deeper layers.
In Arabidopsis, as described above, LRs emerge from the pericycle layer and emerge through overlying tissues at a near horizontal orientation (stage I). Following emergence, LRs undergo a brief period of downward growth, characterized by asymmetric distribution of auxin (as visualized by the reporter DR5v2) as well as the efflux transporter PIN3 (stage II) (Rosquete et al. 2013). Subsequently, LRs undergo stable angled growth, progressively transitioning through increasingly vertical states to adopt near vertical GSAs (stages III–V). Importantly, LRs actively maintain GSAs from stage II onward (i.e., they retain the capacity to reorient themselves both downward and upward) when moved from the GSA.
Several studies have recently sought to explain the molecular mechanisms that underpin the active setting and maintenance of these GSAs, most often using Arabidopsis roots as a model system. One set of studies proposes that LRs grow nonvertically due to the onset of sequential expressions of PIN3, PIN7, and PIN4 in the root columella cells of stage II, III, and IV LRs, respectively. This is in direct contrast to primary root tips where PIN3, 4, and 7 are highly expressed within the columella initials (Guyomarc'h et al. 2012; Rosquete et al. 2013; Ruiz Rosquete et al. 2018; Ogura et al. 2019). In this model, LRs grow at nonvertical angles because they lack the molecular machinery required for full gravitropic competence. However, both these studies do not explain how LRs not only grow obliquely, but also maintain the capacity to bend upward to return to their original GSAs upon reorientation. Other work proposes that nonvertical GSAs are a result of balanced auxin fluxes across the upper and lower sides of the root that act antagonistically. In this model, the authors define a gravitropic auxin flux that increases in magnitude when an LR is reoriented above its GSA, to promote downward bending. This increase in downward auxin flux occurs in an angle-dependent manner: the larger the displacement above GSA, the greater the increase in the magnitude of downward auxin flux. However, when the LR is reoriented below its GSA, the magnitude of this gravitropic auxin flux decreases. In this scenario, the relative magnitude of the antagonistic “antigravitropic offset” flux increases, and the root bends upward toward its original GSA (for review, see Roychoudhry and Kepinski 2015). Interestingly, another recent study discovered that the cytokinin reporter TCSn:GFP is expressed asymmetrically in higher levels toward the upper flank of stage II LRs. The authors proposed that asymmetric cytokinin signaling inhibits downward bending at the upper flank of LRs, and cytokinin therefore functions as an “antigravitropic” signal (Waidmann et al. 2019).
In the last few years, several studies have described the role of multiple members of the LAZY gene family in regulating gravitropism and root (and indeed shoot) angles in multiple plant species. Uga et al. first described DEEPER ROOTING 1 (DRO1), a QTL that controlled root angle in rice. Rice accessions that contained a truncated version of the DRO1 allele, termed DRO1-NIL, had shallow rooting angles and demonstrated impaired root gravitropism in young seedlings. DRO1 was found to be expressed in the root EZ and was negatively regulated by auxin (Uga et al. 2011). Subsequently, DRO1 was found to be orthologous to LAZY4 in Arabidopsis (Hollender and Dardick 2015). Subsequent studies showed that LAZY4 was part of a larger gene family in Arabidopsis, termed as the IGT family, which also contains TAC1 (Guseman et al. 2017). LAZY proteins are expressed in the root tip and EZ, and share a conserved 14 amino acid carboxyl terminus (termed as CCL domain), which seems to be important for directional graviresponse in primary roots (Taniguchi et al. 2017). Several single and multiple loss-of-function mutants of LAZYs have multiple phenotypes including shallow LR growth angles and delayed primary root gravitropism. Interestingly, a recent study showed that primary roots of a lazy234 multiple mutant are negatively gravitropic and bend upward (Ge and Chen 2016). Whether this phenotype is the result of an actively maintained GSA, or loss of the capacity to respond to gravity, remains to be investigated.
While the precise roles of LAZYs in regulating root angle are not yet fully understood, what is evident is that PIN3 polarity is altered in LR columella cells of lazy mutants. Several lines of evidence suggest that the shallow LR GSA phenotype in lazy loss-of-function mutants is due to increased localization of PIN3 to the upper membrane of LR columella cells (Taniguchi et al. 2017; Furutani et al. 2020). Another recent study showed that LAZY proteins may themselves be polarized in LR columella cells and likely recruit PIN3 to the plasma membrane by interacting with the BRX domain of Regulator of Chromosome Condensation 1 (RCC1)-Like proteins (RLD proteins), identified as novel regulators of PIN polarity (Furutani et al. 2020).
ENVIRONMENTAL REGULATION
Vascular plants have evolved complex 3D root architectures to maximize the volume of soil they can explore and interact with. This is to acquire numerous highly heterogeneously distributed resources within multiple soil strata. The ability of the root system to adapt its genetically determined developmental parameters to multiple environmental signals, such as nutrient and water availability, has been a key factor determining the success of land plants (for review, see Morris et al. 2017). This adaptive behavior is known as developmental plasticity.
Phosphate Availability
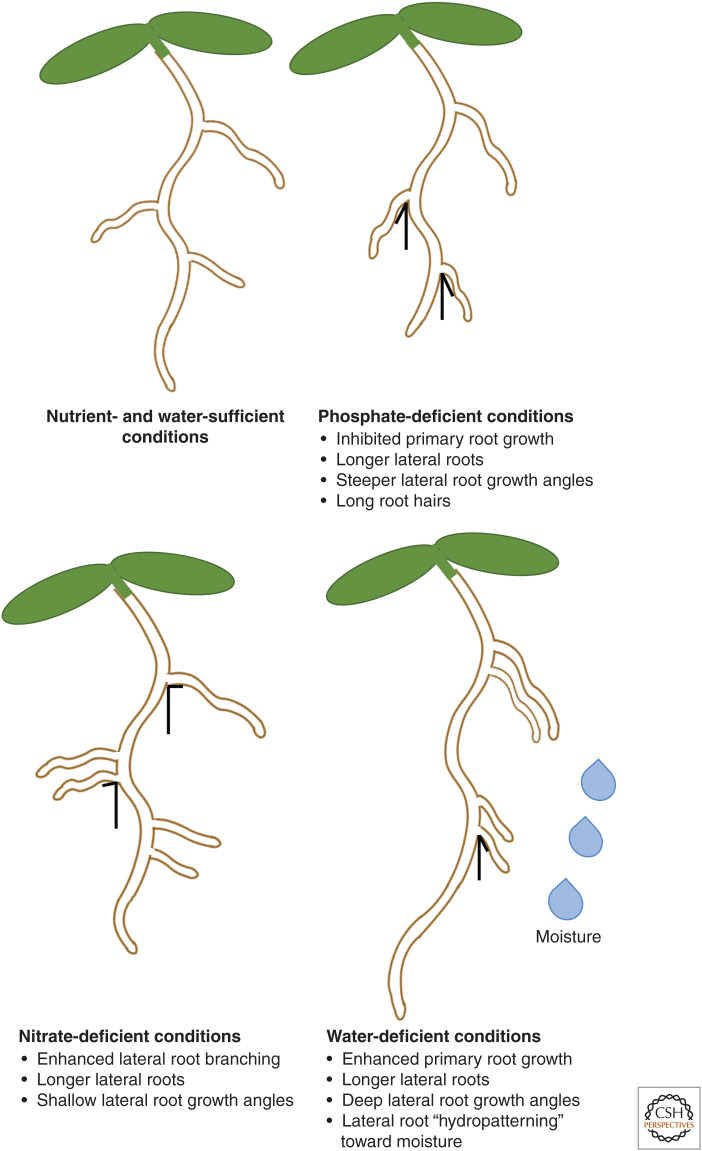
Diverse environmental signals can have dramatic and often diametrically opposite effects on RSA. Phosphates and nitrates are both examples of plant macronutrients that are often limiting in soil (Morgan and Connolly 2013). Phosphorus, specifically, is crucial in the building of nucleic acids, ATP, and other molecules, and is acquired by the plant in its inorganic forms of phosphates. Phosphates are highly immobile in soil and often form complexes with soil components or can be converted to organic phosphate, leading to reductions in the availability of soil Pi for the plants (Brady and Weil 2008). In Arabidopsis, phosphate deficiency leads to PR length inhibition, increases in LR length, and deeper root angle (Fig. 3). These effects are due to a signaling pathway that enhances sensitivity to auxin via up-regulation of the TIR1 auxin receptor (López-Bucio et al. 2002; Ma et al. 2003; Pérez-Torres et al. 2008; Roychoudhry et al. 2017). Interestingly, these effects of low phosphate appear to be root class specific. In bean, growth under phosphorus deficiency had been demonstrated to induce shallower root angles in basal (adventitious) roots in contrast to the effect of low phosphorus on Arabidopsis LR growth angle (Bonser et al. 1996). Further examination of the response to low phosphate in bean showed that, as in Arabidopsis, bean LRs become more vertical while, as previously shown, basal roots adopt a shallower growth habit (Roychoudhry et al. 2017). The physiological basis of this root class-specific response is not yet understood.
Figure 3.
Environmental regulation of root system architecture (RSA) plasticity. Effect of phosphate, nitrate, and water deficiency on different parameters of RSA. Phosphate deficiency leads to inhibition of primary root growth, with contrasting increases in lateral root and root hair length along with steeper lateral root growth angles in Arabidopsis. In contrast, nitrate deficiency has little to no effect on primary root length, but leads to increases in lateral root length, and shallower growth angles. Water deficiency broadly enhances primary and lateral root growth, causes steeper lateral root growth angles and stimulates “hydropatterning” of lateral roots (i.e., induces enhanced lateral root branching toward the side of the primary root that is in contact with higher moisture levels).
Nitrate Availability
Nitrogen, a crucial macronutrient required for the production of amino acids and proteins, is primarily acquired by the root system in its inorganic form from nitrates and ammonia, supplied as fertilizers. In smaller quantities, nitrogen may also be taken up in its organic forms (urea, amino acids, and peptides). Depending on its source, nitrogen availability may have different effects on RSA (Fig. 3). For example, ammonium application limits PR growth (Liu et al. 2013). However, local application of ammonium enhances LR length and branching, likely due to increased expression of the ammonium transporter AMT (AMMONIUM TRANSPORTER) 1;3 in LRs, which enhances nitrogen uptake (Lima et al. 2010). In contrast to these findings, several studies have demonstrated that localized nitrogen supply either in the form of nitrate or L-glutamate inhibits LR elongation, while nitrate deficiency enhances LR elongation and branching (Zhang and Forde 1998; Zhang et al. 1999; Linkohr et al. 2002; Tian et al. 2009). Nitrogen availability or deprivation seems to have a relatively small effect on PR length (for review, see Waidmann et al. 2020). Thus, depending on the source of nitrogen, the root system has distinct responses in terms of LR branching and elongation, suggesting that there are several additional mechanisms/hormone-signaling pathways that fine-tune the uptake of N. A recent study has also demonstrated that nitrate deficiency resulted in shallow LR growth angles in Arabidopsis (Roychoudhry et al. 2017). However, the effect of localized nitrate supply on LR growth angle has not yet been explored in detail.
Light
The effect of light on RSA has remained largely unexplored, as the root systems of most terrestrial plants remain in darkness, underground. Interestingly, however, several studies have demonstrated that multiple photoreceptors are expressed in root tissues, enabling them to detect ambient light and launch appropriate tropic and developmental regimes (for review, see van Gelderen et al. 2018). Broadly, total darkness inhibits root growth because of the reduction in basipetal auxin transport, due to depletion of PIN proteins from the plasma membrane (Laxmi et al. 2008). However, when only root systems are placed in darkness, root growth is largely enhanced in Arabidopsis (Zhang et al. 2019). In contrast, light exposure limits root growth and promotes negative phototropism, via a signaling pathway that depends on the photoreceptors PHOT1 and PHOT2, with receptors CRY1/CRY2 and PHYA playing minor roles. Downstream of the direction of light perception, polar auxin transport is required for directional root growth. The coordinated rootward polarities of PIN1 in the stele and PIN2 in root epidermal cells as well as the lateral relocalization of PIN3 in the columella cells are required for the formation of the auxin gradient (Wang et al. 2012; Zhang et al. 2013). During gradient formation, PIN1 polarity in the stele is also influenced by flavonols, which were found to accumulate on the side of the root exposed to light and promote cell elongation at this side (Buer and Muday 2004; Silva-Navas et al. 2015). Another recent study demonstrated that UV-B light inhibits LR growth through a signaling pathway that involved the UV-B receptor UVR8, which antagonistically regulates auxin-dependent gene expression. Upon UV-B irradiation, UVR8 interacts with the MYB73 transcription factor within the nucleus, thereby inhibiting its DNA-binding activity and resulting in direct repression of target auxin-responsive genes (Yang et al. 2020).
Water
The effect of water on overall RSA has not been extensively studied to date (for review, see Dietrich 2018). Root systems with deeper, soil-penetrating architectures have enhanced resistance to drought, because of their ability to capture available water from deep within the soil. In this light, it is not surprising perhaps that a recent study showed that water deficit promoted vertical growth in Arabidopsis LRs (Fig. 3; Rellán-Álvarez et al. 2016). Interestingly, it was found that in addition to regulating growth angles, water availability around the circumferential axis of the root also acts as a positional cue to specify the patterning of LRs. This phenomenon was shown to occur before the initiation step and was defined as “hydropatterning.” It seems that high water levels promote patterning through a pathway that creates a local LR inductive signal through the biosynthesis (dependent on TRYPTOPHAN AMINOTRANSFERASE1) and transport (PIN3-dependent) of auxin (Bao et al. 2014). In addition, it has been shown that ARF7 promotes LR initiation by triggering asymmetric transcription of LBD16 at the circumference of the root that is in contact with water. This asymmetric induction of LBD16 was regulated by the preferential posttranslational modification of ARF7 by the small ubiquitin-like modifier (SUMO) protein. SUMOylation of ARF7 leads to an increased association with the repressor IAA3/SHY2, presumably at the “dry” circumference of the root, and consequent inhibition of LBD16 transcription (Orosa-Puente et al. 2018).
Similar to LRs, primary roots in several species also develop hydrotropic curvatures in response to water availability. Initially, several lines of evidence suggested that this hydrotropic response was antagonistic to gravitropic bending (for review, see Takahashi et al. 2009), but this response appears to be species-specific. For instance, in cucumber, the root cap is required for normal hydrotropism, suggesting that the interactions between the two tropisms may be more complex. In Arabidopsis, while auxin transport is not required for the hydrotropic response, a functional auxin response seems to be needed. For example, studies showed that treatment with auxin response inhibitors led to contrasting results. Whereas treatment with p-chloro-phenoxy-isobutyl acetic acid (PCIB), an inhibitor of Aux/IAA protein degradation (Oono et al. 2003), produced a decrease in hydrotropic response, addition of auxinole or α-(phenylethyl-2-oxo)-indole acetic acid (PEO-IAA), a TIR1 antagonist (Hayashi et al. 2012), accelerated the response (Kaneyasu et al. 2007; Shkolnik et al. 2016). These contrasting results could be due to different modes of action and target specificities of inhibitors. Thus, currently, additional experiments with multiple auxin-responsive mutants are required to confirm the effect of auxin signaling on the hydrotropic response.
CONCLUSIONS
The number of processes governed by auxin during root, and indeed most aspects of plant development seems extraordinary. This versatility rests in part on the fact that auxin can be moved within tissues to create and maintain gradients of concentration. Equally important is capacity of the auxin signaling system to generate specific outputs relevant to distinct and diverse developmental phenomena. These context-specific responses to auxin are a property of the existence of different TIR1/AFB-Aux/IAA-ARF signaling modules, which can then be wired to downstream genes to effect appropriate modes of developmental control. Nevertheless, even with this rationalization of how so much of development can, often simultaneously, depend on one simple molecule, the sheer robustness of auxin-regulated development is remarkable. A conspicuous example of this robustness is seen in the control of LR formation. This is a process with multiple, distinct, auxin-regulated steps and yet, in the face of high concentrations of auxin, the result is simply an increase in the number of LR primordia, each initiating into perfectly formed LRs. Here, the fact that the development of individual LRs is so well buffered against large changes in auxin concentrations may well stem from the fact that successive Aux/IAA-ARF signaling modules are activated consecutively in time and space. Understanding this molecular genetic basis of auxin's capacity for self-organization is one of the frontiers of auxin research and one that is at the interface of genetics, biophysics, and mathematical modeling.
This review has been very focused on the dicot model Arabidopsis and within that system, principally on auxin's role in primary and LR growth. This leaves out the unsurprisingly central role of auxin in adventitious roots and also in the regulation of root development in monocot species. Nevertheless, many of the paradigms established in Arabidopsis have been found to be translatable to cereal species such as maize, rice, and wheat in particular, albeit with interesting differences (for review, see McSteen 2010; Balzan et al. 2014; Steffens and Rasmussen 2016; Waidmann et al. 2020). Understanding these differences and maximizing the potential application of our understanding of auxin's control of root biology will be essential if we are to make rapid progress in developing crop varieties that perform better in more sustainable farming systems in which inputs such as water, nitrogen, and phosphorous must be reduced. Indeed, there was an extent to which root performance as a trait became a casualty of post-Green Revolution breeding programs based on high levels of agricultural inputs. While this was an appropriate technological response to alleviating food insecurity at that point in history, we are now way beyond the stage at which food production needs to be radically decarbonized. Reducing the use of nitrogen fertilizer derived from the environmentally damaging Haber–Bosch process and conserving the world's finite supply of phosphorous are high on the list of targets for crop improvement. It is here that our deep understanding of the networks of auxin response controlling root growth and system architecture can be brought to bear in a way that can accelerate the development of new varieties in time to meet our commitments to limiting global warming and feeding a global population predicted to peak at 9–10 billion people just 30 years from now.
Whereas overall tremendous progress has been made in understanding the role of auxin and its signaling networks in regulating multiple facets of root and, indeed, overall plant development, many questions still remain unanswered. Whereas a large number of gene networks that regulate different steps of both primary and LR development have now been studied, at least partially, many still remain to be discovered. Specific examples of these include exceptionally rapid auxin-dependent signaling pathways that regulate root elongation (Fendrych et al. 2016, 2018) and those that underpin the mechanisms of LR growth angle setting and maintenance. Following on from this, how multiple environmental signals can fine tune these myriad auxin-dependent developmental responses remains to be fully understood. Perhaps the most basic question of all relates to the evolution of such complex systems of auxin response and how the proliferation and subfunctionalization of the three gene families in auxin signaling, including the development of specific preferential interactions between pairs and multimers of different Aux/IAAs, ARFs, and TIR1/AFBs, may have underpinned the evolution of increasingly sophisticated developmental phenomena. Interdisciplinary advances in the multiple fields of biology, physics, and mathematical modeling will enable researchers to answer these and other fundamental, yet exciting, questions in the coming decade.
ACKNOWLEDGMENTS
We apologize to those authors whose work we could not cover in this review. We thank Marta Del Bianco for critical reading of the manuscript and The Leverhulme Trust (RPG-2018-137) and BBSRC (BB/N010124/1) for their support of relevant projects in the Kepinski laboratory.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Balzan S, Johal GS, Carraro N. 2014. The role of auxin transporters in monocots development. Front Plant Sci 5: 393. 10.3389/fpls.2014.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Aggarwal P, Robbins NE 2nd, Sturrock CJ, Thompson MC, Tan HQ, Tham C, Duan L, Rodriguez PL, Vernoux T, et al. 2014. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc Natl Acad Sci 111: 9319–9324. 10.1073/pnas.1400966111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Vermeer JE, De Bellis D, Wang P, Naseer S, Andersen TG, Humbel BM, Nawrath C, Takano J, Salt DE, et al. 2016. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164: 447–459. 10.1016/j.cell.2015.12.021 [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I. 2014. Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666. 10.1146/annurev-arplant-050213-035645 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. 2008. Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465. 10.1146/annurev.arplant.58.032806.103805 [DOI] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, Magyar Z, Alvim Kamei CL, Koncz C, Bögre L, et al. 2011. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683. 10.1105/tpc.111.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. 1995. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonser A, Lynch J, Snapp S. 1996. Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytol 132: 281–288. 10.1111/j.1469-8137.1996.tb01847.x [DOI] [PubMed] [Google Scholar]
- Brady NC, Weil RR. 2008. The nature and properties of soil, 14th ed. Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- Buer CS, Muday GK. 2004. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205. 10.1105/tpc.020313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cavallari N, Artner C, Benkova E. 2021. Auxin-regulated lateral root organogenesis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL Jr, Grisafi PL, Fink GR. 1995. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142. 10.1101/gad.9.17.2131 [DOI] [PubMed] [Google Scholar]
- Datta S, Prescott H, Dolan L. 2015. Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat Plants 1: 15138. 10.1038/nplants.2015.138 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706. 10.1016/j.cub.2010.09.007 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei Dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci 107: 2705–2710. 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. 2005. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119. 10.1016/j.devcel.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Dietrich D. 2018. Hydrotropism: how roots search for water. J Exp Bot 69: 2759–2771. 10.1093/jxb/ery034 [DOI] [PubMed] [Google Scholar]
- Digby J, Firn RD. 1995. The gravitropic set-point angle (GSA): the identification of an important developmentally controlled variable governing plant architecture. Plant Cell Environ 18: 1434–1440. 10.1111/j.1365-3040.1995.tb00205.x [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey P, Busch W, Novak O, Ljung K, De Paola L, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci 114: E7641–E7649. 10.1073/pnas.1705833114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci 107: 12046–12051. 10.1073/pnas.1000672107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. 2001. How and where to build a root hair. Curr Opin Plant Biol 4: 550–554. 10.1016/S1369-5266(00)00214-4 [DOI] [PubMed] [Google Scholar]
- Drisch RC, Stahl Y. 2015. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front Plant Sci 6: 505. 10.3389/fpls.2015.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. J Exp Bot 69: 155–167. 10.1093/jxb/erx223 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105: 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgaprasad K, Roy MV, Venugopal A, Kareem A, Raj K, Willemsen V, Mähönen AP, Scheres B, Prasad K. 2019. Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep 29: 453–463.e3. 10.1016/j.celrep.2019.08.099 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Hirano Y, Nishimura T, Nakamura M, Taniguchi M, Suzuki K, Oshida R, Kondo C, Sun S, Kato K, et al. 2020. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat Commun 11: 76. 10.1038/s41467-019-13729-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- García-Gómez ML, Garay-Arroyo A, García-Ponce B, Sánchez MP, Álvarez-Buylla ER. 2021. Hormonal regulation of stem cell proliferation at the Arabidopsis thaliana root stem cell niche. Front Plant Sci 12: 628491. 10.3389/fpls.2021.628491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Chen R. 2016. Negative gravitropism in plant roots. Nat Plants 2: 16155. 10.1038/nplants.2016.155 [DOI] [PubMed] [Google Scholar]
- Geisler M, Aryal B, di Donato M, Hao P. 2017. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol 58: 1601–1614. 10.1093/pcp/pcx104 [DOI] [PubMed] [Google Scholar]
- Geldner N. 2009. Cell polarity in plants: a PARspective on PINs. Curr Opin Plant Biol 12: 42–48. 10.1016/j.pbi.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Voß U, Harding SA, Fannon J, Moody LA, Yamada E, Swarup K, Nibau C, Bassel GW, Choudhary A, et al. 2014. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol 203: 1194–1207. 10.1111/nph.12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, Lima JE, von Wirén N. 2012. Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 24: 33–49. 10.1105/tpc.111.092973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H. 2012. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893. 10.1242/dev.071928 [DOI] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Yamaguchi N, Okamoto Y, Uehara T, Kaneko S, Takebayashi Y, Kasahara H, Ikeyama Y, Okushima Y, et al. 2019. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol 224: 749–760. 10.1111/nph.16065 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. 10.1038/nature06215 [DOI] [PubMed] [Google Scholar]
- Guseman JM, Webb K, Srinivasan C, Dardick C. 2017. DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J 89: 1093–1105. 10.1111/tpj.13470 [DOI] [PubMed] [Google Scholar]
- Guyomarc'h S, Léran S, Auzon-Cape M, Perrine-Walker F, Lucas M, Laplaze L. 2012. Early development and gravitropic response of lateral roots in Arabidopsis thaliana. Philos Trans R Soc Lond B Biol Sci 367: 1509–1516. 10.1098/rstb.2011.0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hammes UZ, Murphy AS, Schwechheimer C. 2021. Auxin transporters—a biochemical view. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H. 2012. Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem Biol 17: 590–598. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. 2013. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol 23: 956–962. 10.1016/j.cub.2013.04.048 [DOI] [PubMed] [Google Scholar]
- Hollender CA, Dardick C. 2015. Molecular basis of angiosperm tree architecture. New Phytol 206: 541–556. 10.1111/nph.13204 [DOI] [PubMed] [Google Scholar]
- Hu Y, Omary M, Hu Y, Doron O, Hoermayer L, Chen Q, Megides O, Chekli O, Ding Z, Friml J, et al. 2021. Cell kinetics of auxin transport and activity in Arabidopsis root growth and skewing. Nat Commun 12: 1657. 10.1038/s41467-021-21802-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HM, Grierson CS. 2009. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11: 78–84. 10.1038/ncb1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneyasu T, Kobayashi A, Nakayama M, Fujii N, Takahashi H, Miyazawa Y. 2007. Auxin response, but not its polar transport, plays a role in hydrotropism of Arabidopsis roots. J Exp Bot 58: 1143–1150. 10.1093/jxb/erl274 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Khan MA, Gemenet DC, Villordon A. 2016. Root system architecture and abiotic stress tolerance: current knowledge in root and tuber crops. Front Plant Sci 7: 1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P. 2016. Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. J Exp Bot 67: 1411–1420. 10.1093/jxb/erv541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. 10.1371/journal.pbio.0060307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R. 2008. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510. 10.1371/journal.pone.0001510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Cho C, Kim J. 2015. Lateral Organ Boundaries Domain16 and 18 Act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol 168: 1792–1806. 10.1104/pp.15.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Ganguly A, Lee RD, Park M, Cho HT. 2020. Intracellularly localized PIN-FORMED8 promotes lateral root emergence in Arabidopsis. Front Plant Sci 10: 1808. 10.3389/fpls.2019.01808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wirén N. 2010. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22: 3621–3633. 10.1105/tpc.110.076216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760. 10.1046/j.1365-313X.2002.01251.x [DOI] [PubMed] [Google Scholar]
- Liu Y, Lai N, Gao K, Chen F, Yuan L, Mi G. 2013. Ammonium inhibits primary root growth by reducing the length of meristem and elongation zone and decreasing elemental expansion rate in the root apex in Arabidopsis thaliana. PLoS ONE 8: e61031. 10.1371/journal.pone.0061031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Dünser K, Kleine-Vehn J. 2013. Epidermal patterning genes impose non-cell autonomous cell size determination and have additional roles in root meristem size control. J Int Plant Biol 55: 864–875. 10.1111/jipb.12097 [DOI] [PubMed] [Google Scholar]
- Löfke C, Scheuring D, Dünser K, Schöller M, Luschnig C, Kleine-Vehn J. 2015. Tricho- and atrichoblast cell files show distinct PIN2 auxin efflux carrier exploitations and are jointly required for defined auxin-dependent root organ growth. J Exp Bot 66: 5103–5112. 10.1093/jxb/erv282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. 2002. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256. 10.1104/pp.010934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot 112: 347–357. 10.1093/aob/mcs293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. 2003. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390. 10.1104/pp.012161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597. 10.1105/tpc.010354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Bassukas AEL, Zourelidou M, Kolb M, Moret B, Fastner A, Schulze WX, Cattaneo P, Hammes UZ, Schwechcheimer C, et al. 2018. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. 10.1038/s41586-018-0186-z [DOI] [PubMed] [Google Scholar]
- Marhavý P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benková E. 2013. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 32: 149–158. 10.1038/emboj.2012.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Montesinos JC, Abuzeineh A, Van Damme D, Vermeer JE, Duclercq J, Rakusová H, Nováková P, Friml J, Geldner N, et al. 2016. Targeted cell elimination reveals an auxin-guided biphasic mode of lateral root initiation. Genes Dev 30: 471–483. 10.1101/gad.276964.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis MN, De Cnodder T, Lewandowski M, Simon D, Boron A, Balcerowicz D, Doubbo T, Taconnat L, Renou JP, Höfte H, et al. 2012. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol 12: 208. 10.1186/1471-2229-12-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. 2010. Auxin and monocot development. Cold Spring Harb Perspect Biol 2: a001479. 10.1101/cshperspect.a001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor N, Band LR, Pěnčík A, Novák O, Rashed A, Holman T, Wilson MH, Voß U, Bishopp A, King RJ, et al. 2016. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci 113: 11022–11027. 10.1073/pnas.1604458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. 2007. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. 10.1016/j.cell.2007.07.033 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311. 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morffy N, Strader LC. 2021. Structural aspects of auxin signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JB, Connolly EL. 2013. Plant–soil interactions: nutrient uptake. Nature Education Knowledge 4: 2. [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP, et al. 2017. Shaping 3D root system architecture. Curr Biol 27: R919–R930. 10.1016/j.cub.2017.06.043 [DOI] [PubMed] [Google Scholar]
- Muday GK, Haworth P. 1994. Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32: 193–203. [PubMed] [Google Scholar]
- Murphy E, Vu LD, Van den Broeck L, Lin Z, Ramakrishna P, van de Cotte B, Gaudinier A, Goh T, Slane D, Beeckman T, et al. 2016. RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J Exp Bot 67: 4863–4875. 10.1093/jxb/erw281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Goeschl C, Filiault D, Mirea M, Slovak R, Wolhrab B, Satbhai SB, Busch W. 2019. Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178: 400–412.e16. 10.1016/j.cell.2019.06.021 [DOI] [PubMed] [Google Scholar]
- Okumura K, Goh T, Toyokura K, Kasahara H, Takebayashi Y, Mimura T, Kamiya Y, Fukaki H. 2013. GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for Arabidopsis lateral root initiation. Plant Cell Physiol 54: 406–417. 10.1093/pcp/pct018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H. 2003. p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147. 10.1104/pp.103.027847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Ann Rev Plant Biol 58: 93–113. 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- Overoode P, Fukaki H, Beeckman T. 2010. Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. 2012. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. 10.1105/tpc.112.097766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Middleton AM, French AP, Larrieu A, Bishopp A, Njo M, Wells DM, Porco S, Mellor N, Band LR, et al. 2013. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol Syst Biol 9: 699. 10.1038/msb.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 12: 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perianez-Rodriguez J, Rodriguez M, Marconi M, Bustillo-Avendaño E, Wachsman G, Sanchez-Corrionero A, De Gernier H, Cabrera J, Perez-Garcia P, Gude I, et al. 2021. An auxin-regulable oscillatory circuit drives the root clock in Arabidopsis. Sci Adv 7: eabd4722. 10.1126/sciadv.abd4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rashed A, Voß U, Casanova-Sáez R, Bishopp A, Golboeweska A, Bhosale R, Swarup R, Swarup K, et al. 2016a. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci 113: 11016–11021. 10.1073/pnas.1604375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. 2016b. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143: 3340–3349. [DOI] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI. 2007. Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J 50: 514–528. 10.1111/j.1365-313X.2007.03068.x [DOI] [PubMed] [Google Scholar]
- Rellán-Álvarez R, Lobet G, Dinneny J. 2016. Environmental control of root system biology. Ann Rev Plant Biol 67: 619–642. 10.1146/annurev-arplant-043015-111848 [DOI] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiol 151: 1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I, Smith S, Stes E, De Rybel B, Staes A, van de Cotte B, Njo MF, Dedeyne L, Demol H, Lavenus J, et al. 2016. CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J Exp Bot 67: 4889–4899. 10.1093/jxb/erw231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, von Wangenheim D, Marhavý P, Barbez E, Stelzer EH, Benková E, Maizel A, Kleine-Vehn J. 2013. An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr Biol 23: 817–822. 10.1016/j.cub.2013.03.064 [DOI] [PubMed] [Google Scholar]
- Roychoudhry S, Kepinski S. 2015. Shoot and root branch growth angle control—the wonderfulness of lateralness. Curr Opin Plant Biol 23: 124–131. 10.1016/j.pbi.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Roychoudhry S, Kieffer M, Del Bianco M, Liao CY, Weijers D, Kepinski S. 2017. The developmental and environmental regulation of gravitropic setpoint angle in Arabidopsis and bean. Sci Rep 7: 42664. 10.1038/srep42664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Rosquete M, Waidmann S, Kleine-Vehn J. 2018. PIN7 auxin carrier has a preferential role in terminating radial root expansion in Arabidopsis thaliana. Int J Mol Sci 19: 1238. 10.3390/ijms19041238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. 10.1016/S0092-8674(00)81535-4 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358. 10.1101/gad.252503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi E, Rutten JP, Di Mambro R, Polverari L, Licursi V, Negri R, Dello Ioio R, Sabatini S, Ten Tusscher K. 2020. A self-organized PLT/auxin/ARR-B network controls the dynamics of root zonation development in Arabidopsis thaliana. Dev Cell 53: 431–443.e23. 10.1016/j.devcel.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Shkolnik D, Krieger G, Nuriel R, Fromm H. 2016. Hydrotropism: root bending does not require auxin redistribution. Mol Plant 9: 757–759. 10.1016/j.molp.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Silva-Navas J, Moreno-Risueno MA, Manzano C, Pallero-Baena M, Navarro-Neila S, Téllez-Robledo B, Garcia-Mina JM, Baigorri R, Gallego FJ, del Pozo JC. 2015. D-Root: A system for cultivating plants with the roots in darkness or under different light conditions. Plant J 84: 244–255. 10.1111/tpj.12998 [DOI] [PubMed] [Google Scholar]
- Simonini S, Bencivenga S, Trick M, Østergaard L. 2017. Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell 29: 1864–1882. 10.1105/tpc.17.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Phil Trans R Soc B 367: 1441–1452. 10.1098/rstb.2011.0234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Rasmussen A. 2016. The physiology of adventitious roots. Plant Phys 170: 603–617. 10.1104/pp.15.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle D, Thellmann M, Vermeer JE. 2018. Breakout—lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol 41: 67–72. 10.1016/j.pbi.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065. 10.1038/ncb1316 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N. 2009. Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69: 489–502. 10.1007/s11103-008-9438-x [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M. 2014. Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65: 2633–2643. 10.1093/jxb/ert485 [DOI] [PubMed] [Google Scholar]
- Tang LP, Zhou C, Wang SS, Yuan J, Zhang XS, Su YH. 2017. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phyt 213: 1740–1754. 10.1111/nph.14313 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Furutani M, Nishimura T, Nakamura M, Fushita T, Iijima K, Baba K, Tanaka H, Toyota M, Tasaka M, et al. 2017. The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 29: 1984–1999. 10.1105/tpc.16.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian QY, Sun P, Zhang WH. 2009. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol 184: 918–931. 10.1111/j.1469-8137.2009.03004.x [DOI] [PubMed] [Google Scholar]
- Torres-Martínez HH, Rodríguez-Alonso G, Shishkova S, Dubrovsky JG. 2019. Lateral root primordium morphogenesis in angiosperms. Front Plant Sci 10: 206. 10.3389/fpls.2019.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Okuno K, Yano M. 2011. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J Exp Bot 62: 2485–2494. 10.1093/jxb/erq429 [DOI] [PubMed] [Google Scholar]
- Ursache R, De Jesus Vieira Teixeira C, Dénervaud Tendon V, Gully K, De Bellis D, Schmid-Siegert E, Grube Andersen T, Shekhar V, Calderon S, Pradervand S, et al. 2021. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat Plants 7: 353–364. 10.1038/s41477-021-00862-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Pierik R. 2018. Light signaling, root development, and plasticity. Plant Physiol 176: 1049–1060. 10.1104/pp.17.01079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050. 10.1105/tpc.105.035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. 2013. To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140: 4301–4310. 10.1242/dev.090548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen JP, Cnodder TDe, Le J, Vissenberg K, Baluška F. 2006. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal Behav 1: 296–304. 10.4161/psb.1.6.3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. 2014. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183. 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A. 2016. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr Biol 26: 439–449. 10.1016/j.cub.2015.12.047 [DOI] [PubMed] [Google Scholar]
- Waidmann S, Ruiz Rosquete M, Schöller M, Sarkel E, Lindner H, LaRue T, Petřík I, Dünser K, Martopawiro S, Sasidharan R, et al. 2019. Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat Commun 10: 3540. 10.1038/s41467-019-11483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann S, Sarkel E, Kleine-Vehn J. 2020. Same same, but different: growth responses of primary and lateral roots. J Exp Bot 71: 2397–2411. 10.1093/jxb/eraa027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng D, Shi Y, Miao N, Bian Y, Yin Z. 2012. Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Mol Biol Rep 39: 2401–2415. 10.1007/s11033-011-0991-z [DOI] [PubMed] [Google Scholar]
- Wu G, Lewis DR, Spalding EP. 2007. Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19: 1826–1837. 10.1105/tpc.106.048777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. 2015. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388. 10.1016/j.cub.2015.03.046 [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Moller BK, Skorzinki N, Njo MF, et al. 2016. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387. 10.1126/science.aad2776 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Chen P, Liang L, Li X, Liu H. 2020. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39: e101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42: 64–267. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. 10.1126/science.279.5349.407 [DOI] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci 96: 6529–6534. 10.1073/pnas.96.11.6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KX, Xu HH, Yuan TT, Zhang L, Lu YT. 2013. Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J 76: 308–321. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lin JE, Harris C, Pereira FCM, Wu F, Blakeslee JJ, Peer WA. 2016. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci 113: 11010–11015. 10.1073/pnas.1604769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang C, Xu H, Shi X, Zhen W, Hu Z, Huang J, Zheng Y, Huang P, Zhang KX, et al. 2019. HY5 contributes to light-regulated root system architecture under a root-covered culture system. Front Plant Sci. 10: 1490. 10.3389/fpls.2019.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodriguez R, Li L, Zhang X, Friml J. 2020. Functional innovations of PIN auxin transporters mark crucial evolutionary transitions during rise of flowering plants. Sci Adv 6: eabc8895. 10.1126/sciadv.abc8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Geisler M. 2015. Keeping it all together: auxin-actin crosstalk in plant development. J Exp Bot 66: 4983–4998. 10.1093/jxb/erv308 [DOI] [PubMed] [Google Scholar]
- Zourelidou M, Absmanner B, Weller B, Barbosa IC, Willige BC, Fastner A, Streit V, Port SA, Colcombet J, de la Fuente van Bentem S, et al. 2014. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. 10.7554/eLife.02860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M, Bilanovičová V, Seifu YW, Nodzyński T. 2019. The nuts and bolts of PIN auxin efflux carriers. Front Plant Sci 10: 985. 10.3389/fpls.2019.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]