Abstract
We have investigated the pressure- and temperature-induced conformational changes associated with the low complexity domain of hnRNP A1, an RNA-binding protein able to phase separate in response to cellular stress. Solution NMR spectra of the hnRNP A1 low-complexity domain fused with protein-G B1 domain were collected from 1 to 2,500 bar and from 268 K to 290 K. While the GB1 domain shows the typical pressure-induced and cold temperature-induced unfolding expected for small globular domains, the low-complexity domain of hnRNP A1 exhibits unusual pressure and temperature dependences. We observed that the low-complexity domain is pressure sensitive, undergoing a major conformational transition within the prescribed pressure range. Remarkably, this transition has the inverse temperature dependence of a typical folding-unfolding transition. Our results suggest the presence of a low-lying extended, and fully solvated state(s) of the low-complexity domain that may play a role in phase separation. This study highlights the exquisite sensitivity of solution NMR spectroscopy to observe subtle conformational changes and illustrates how pressure perturbation can be used to determine the properties of metastable conformational ensembles.
Introduction
High-pressure NMR spectroscopy has emerged as a powerful technique to characterize the stability of globular proteins,1–4 their mechanisms of folding,5–7 and presence of low-lying intermediate states.8–10 According to Le Chatelier’s principle an increase in pressure will shift the thermodynamic equilibrium toward states of lower molar volume. Since the application of pressure leads (over most of the accessible temperature range) to protein unfolding, it signifies that the volume change upon unfolding is negative (ΔVF→U <0), i.e. the molar volume of the unfolded state is smaller than that of the folded state.11 The magnitude of ΔVF→U values measured for globular proteins typically lies around 50 to 100 ml/mol, which represent only 0.5% to 2% of the protein’s molar volume.12 The physical origin of ΔVF→U has been the subject of much debate but strong experimental and computational evidences point toward an imperfect balance between negative contributions (i.e. elimination of cavities and void volume13–14) and positive contributions (i.e. volume changes due to exposure of hydrophobic side chain upon unfolding15). The mechanisms of pressure unfolding have also been thoroughly investigated by molecular simulations and experimental approaches. It is generally believed that under the influence of high pressure, water molecules penetrate into internal cavities of the protein core and destabilize hydrophobic interactions.16–18 The volume difference upon unfolding is known to be strongly temperature dependent. Indeed, because the thermal expansivity of the unfolded states is larger than that of the folded state, ΔVF→U is more negative at lower temperatures and it’s magnitude decreases as temperature increases. ΔVF→U can even become positive at high enough temperatures.19–21
Only a handful of studies have examined the effects of pressure on protein unfolded states and intrinsically disordered proteins and peptides.22–24 In the case of α-synuclein, no major effect was observed besides non-specific pressure-induced chemical shift changes and a small decrease in 3JHNHα couplings.23 Separately, it has been reported that folding of small helical motifs can be promoted under high-pressure conditions due to preferential hydration of helical structure.25–27 These evidences suggest that unfolded protein chains may not remain entirely featureless at high-pressure.
Low-complexity (LC) domains found in RNA-binding proteins associated with liquid-liquid phase separation form a specific class of intrinsically disordered domains. LC domains have distinct amino acid compositions; they are enriched in polar amino acids (especially Ser and Gly), and feature conserved patterns of aromatic residues.28 In many cases, these disordered LC domains are necessary and sufficient for driving phase separation.29,30 Liquid condensates are believed to be the result of multivalent weak interactions between multiple interacting motifs in LC domains, including electrostatic, cation-π and π-π interactions.31–35 Yet, the relative contribution of these interactions to phase separation is largely unknown. The effects of pressure on phase separated systems have recently been investigated revealing a strong pressure dependence that can lead to complete dissolution of liquid condensates over a few hundred bars.36–38
In the present study we investigated the effects of pressure on the LC domain of the isoform A of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1-A). hnRNP A1-A is a 34 kDa protein composed of 320 amino acids and three separate protein domains, two RNA recognition motifs (RRMs) and a disordered C-terminal domain (LCDA1) rich in glycine and arginine (Fig. 1A). The two RRMs form the nucleic acid binding domain, referred to as Unwinding Protein-1 (UP1), and work in tandem to bind RNA and DNA substrates.39 The C-terminal domain engages in protein interactions that are necessary for the protein to carry out its nucleic acid processing activities.40 Many of these processing activities are regulated by post-translational modification of LCDA1, such as methylation, phosphorylation, and SUMOylation.41 LCDA1 also contains a 38 amino acid nuclear localization signal (NLS) responsible for shuttling the protein between the nucleus and the cytoplasm, with the shuttling activity being mediated between interactions with Transportin 1 and Transportin 2.42,43 The NLS mediates protein interactions, primarily through aromatic residues present in this region.44 Given the multitude of protein-protein and protein-RNA interactions the LCDA1 forms, it is imperative to examine the biophysical properties of the domain as well as the nature of the chemical interactions it is capable of engaging in with other proteins and nucleic acids.
Figure 1.
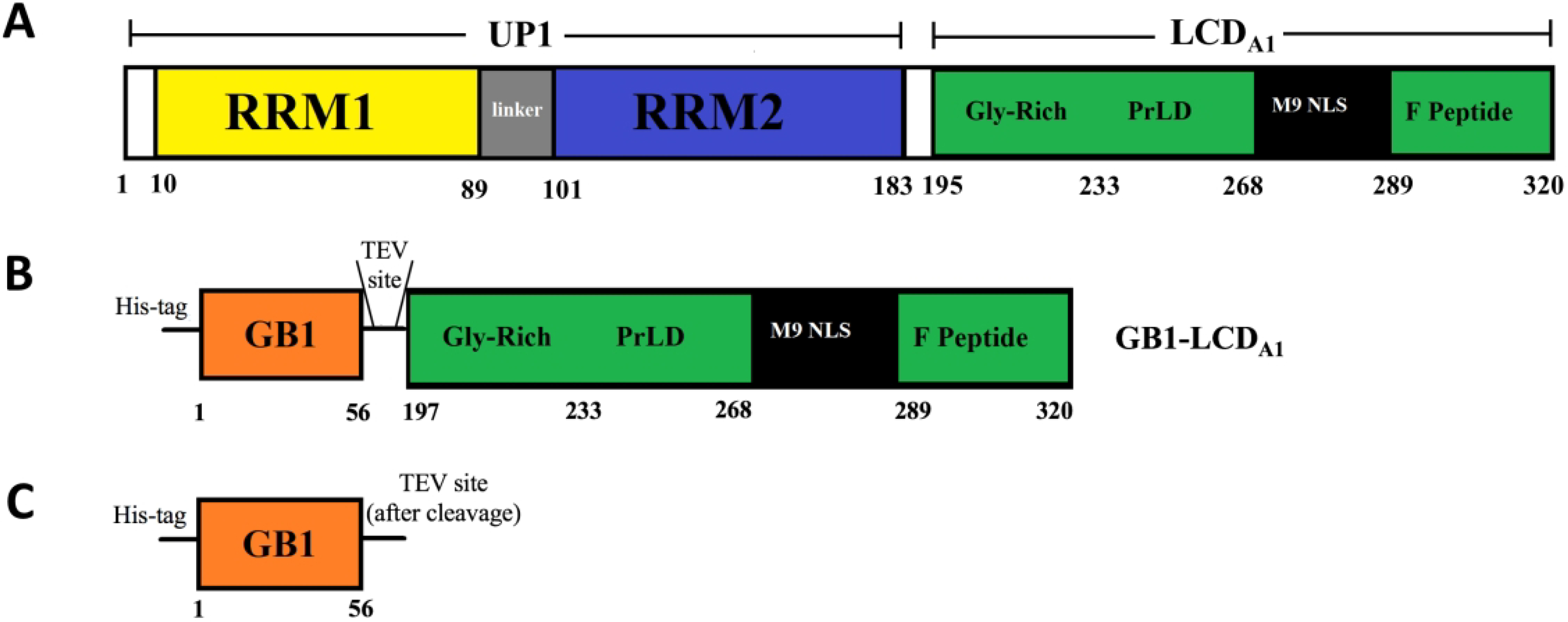
Schematic representation of the sequence of (A) hnRNP A1-A with the N-terminal Unwinding Protein-1 domain (UP1) encompassing two RNA Recognition Motifs (RRM1 and RRM2) and the C-terminal Low-Complexity Domain (LCDA1) encompassing the glycine-rich region (Gly-rich), prion-like domain (prLD), M9 nuclear localization signal (M9 NLS), and phosphopeptide (F Peptide). (B) GB1-LCDA1 construct composed of GB1 domain fused with hnRNP A1-A Low-Complexity Domain. This construct encompasses an N-terminal His-tag and a TEV recognition site between the GB1 domain and the LCDA1 domain. (C) Isolated GB1 domain obtained after TEV cleavage of GB1-LCDA1.
hnRNP A1 has shown the ability to form liquid condensates in response to cellular stress and other stimuli.29 These stress granules are formed from pools of stalled, untranslated mRNAs, with assembly often occurring through interactions of the LC domains of various RNA binding proteins.45 Although RNA binding to the UP1 domain is known to promote granule assembly, the isolated LC domain has the ability to self-assemble into granules.29,46,47 The arginines present in the RGG box within the Gly-rich region were shown to be necessary for stress granule assembly in hnRNP A1.48 The details of this mechanism has been investigated for hnRNP A2, where it was found that arginine methylation of the RGG boxes reduces phase separation by disrupting interactions between the charged arginines and the aromatic residues in the region preceding the prion-like domain.49
The LCDA1 domain used in the present study encompasses amino acids 197–320 of hnRNP A1-A (Fig. 1A). We examined the behavior of the LCDA1 under pressure using solution NMR spectroscopy in solution phase (no liquid condensate) with the aim of characterizing low-lying conformational states that may play a role in triggering phase separation. For this purpose, we used a chimeric construct consisting of protein G B1 domain (GB1) fused at its C-termini to LCDA1 (Fig. 1B). The GB1 domain aided protein purification and sample stability. Its presence, along with the sample buffer conditions, prevents phase separation even at the high sample concentration required for NMR experiments and low temperature range sampled here (268 K to 290 K). Therefore, fusion of LCDA1 with a soluble domain such as GB1 allows the characterization of the conformational ensemble of LCDA1 in fully homogeneous solution phase without formation of liquid condensates. This construct also offers the advantage of presenting within the same chain two structurally different domains: a well folded globular domain (GB1) and a low complexity disordered domain (LCDA1). By designing this construct, we intended to use GB1 domain as an internal control to help interpret the effects of pressure measured on LCDA1.
Materials and Methods
Sample preparation –
Constructs for the isolated GB1 domain and GB1-LCDA1 fusion protein were cloned from gBlock gene fragments (IDT) into pMCSG plasmid.50 The following sequence was used for the GB1-LCDA1 construct: MHHHHHHSSGVDLQYKLALNGKTLKGETTTEAVDAATAEKVFKQYANDNGVDGEWTYDDATKTFTVTEGTENLYFQSNIMRSGSGNFGGGRGGGFGGNDNFGRGGNFSGRGGFGGSRGGGGYGGSGDGYNGFGNDGSNFGGGGSYNDFGNYNNQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGYGGSSSSSSYGSGRRF (the TEV recognition site separating the GB1 sequence to the LCDA1 sequence is underlined). Overexpression was carried out in BL21 (DE3) cells (NEB) in minimal media supplanted with 15NH4Cl. Cells were induced at OD600 ~1.0 with 0.2 mM IPTG. Cells were harvested after overnight expression. For GB1-LCDA1, cell pellet was resuspended in lysis buffer (20 mM Na2HPO4 pH 7=5, 1 M NaCl, 20mM imidazole, 0.25 mM EDTA, 1 mM PMSF, 10% Glycerol, protease inhibitor cocktail tablet). Cells were lysed by sonication, cell debris was spun down, and lysate was applied to a HiTrap Chelating HP column (GE Healthcare) charged with NiSO4. Protein was washed with lysis buffer, then eluted with elution buffer (20 mM Na2HPO4 pH=7.5, 1 M NaCl, 250mM imidazole, 0.25 mM EDTA, 1 mM PMSF, 10% Glycerol). Eluted protein was further purified by FPLC gel filtration with a Sephacryl S-100 column in gel filtration buffer (100 mM HEPES pH=7.5, 350 mM NaCl, 0.5 mM EDTA, 10% glycerol). Fractions containing protein were pooled and washed into NMR Buffer through Amicon centrifugal filtration. Purification of GB1 from GB1-peptide was conducted the same as GB1-LCDA1, except before the FPLC gel filtration, the peptide was cleaved off with TEV protease and separated from GB1 by another run through the HiTrap Chelating HP column.
NMR –
1H-15N TROSY-HSQC spectra were recorded with uniformly 15N labeled samples on a Bruker 700 MHz Avance II spectrometer, equipped with a z-shielded gradient triple resonance cryoprobe. A total of 100 × 1048 complex points were collected, for acquisition times of 104 and 121 ms in the 15N and 1H dimensions, respectively, using an interscan delay of 1.5 s. 1H-13C HSQC spectra were recorded with triple labeled 2H-15N-13C samples for a total of 128 × 512 complex points on a Bruker 800 MHz spectrometers equipped with a z-shielded gradient triple resonance cryoprobe. resonance cryoprobe. All the experiments were recorded using 150 μM protein in buffer 100 mM HEPES pH=7.5, 350 mM NaCl, 0.1 mM leupeptin, 0.5 mM EDTA, 10% 2H2O, 10% glycerol. A commercial ceramic high-pressure NMR cell and an automatic pump system (Daedalus Innovations, Philadelphia, PA) were used to vary the pressure in the 1 bar to 2.5 kbar range. The spectra were processed using NMRPipe51 and displayed with SPARKY.52
SAXS-
GB1-LCDA1 for SAXS was prepared the same as NMR, but overexpressed in TB broth instead of minimal media. SEC-SAXS experiments were performed on a 650 μM GB1-LCDA1 sample at BioCAT (beamline 18-ID, Advanced Photon Source). Data was collected as previously described.53 Molecular weight calculations and data analysis were performed in PRIMUS54 from the ATSAS suite of programs.55
Results
Pressure-induced unfolding of isolated GB1 domain.
We first studied the stability of the isolated GB1 domain under pressure at both 290 K and 277 K (Fig. 1C). The intensity of amide crosspeaks was monitored from series of 15N-1H heteronuclear single-quantum coherence (HSQC) spectra collected every 500 bar (Fig. S1 A and B). The intensity profiles measured for each residue followed a sigmoidal curve, which makes reasonable the assumption of a two-state equilibrium between folded (F) and unfolded (U) states (Fig. S1 C and D). Assuming a negligible difference in compressibility between F and U, one can express the change of the equilibrium constant Keq as a function of pressure as:
| (1) |
Here ΔGF→U and ΔG0F→U are the Gibbs-free energy changes from F to U at pressures p and p0 (= 0.1 MPa), respectively; ΔVF→U is the molar volume change between F and U; R is the gas constant, and T is the absolute temperature.
Using NMR spectroscopy observables such as amide crosspeak intensities, the equilibrium constant can be written as:
| (2) |
where, IF represents the maximal intensity of the folded crosspeaks, IU the intensity of the same crosspeaks when the protein is fully unfolded (IU usually converges to 0) and I the intensity of the crosspeak at a given pressure-temperature condition. Combining equation (1) with equation (2) gives:
| (3) |
We noticed that the peak widths of the folded-state amide resonances did not broaden as a function of pressure, which means that native crosspeak intensity can be reliably used as a proxy to monitor the relative folded population at a residue level. Nevertheless, it should be acknowledged that these residue-level ΔVF→U values must be considered as apparent and not as pure thermodynamic parameters.56
The average ΔVF→U measured for the isolated GB1 domain at 290 K is −34.1 ± 1.2 ml/mol (Table 1 and Fig. S1 E), which is similar to volume differences measured for other small globular protein domains such as ubiquitin, CI2, and CspB.11 We found that the average ΔVF→U values were slightly more negative at 277 K compared to 290 K (−37.4 ± 1.6 vs. −34.1 ± 1.2 ml/mol) (Table 1). This indicates, as expected for globular proteins, that the unfolded states of GB1 has a larger thermal expansivity than that of its folded states.
Table 1.
Thermodynamic parameters measured for the isolated GB1 domain, GB1 domain in GB1-LCDA1 construct (GB1-LCDA1), and LCDA1 domain in the GB1-LCDA1 construct (GB1-LCDA1). Measured ΔV values correspond to the volume change upon unfolding in the case of GB1 (ΔVF→U) and to the volume change associated with LP-to-HP transition in the case of LCDA1 (ΔVLP→HP). Values in the table represents the average and standard deviation calculated over all residue-specific ΔV values.
| ΔV (ml/mol) | |
|---|---|
| GB1 (290 K) | −34.1 ± 1.2 |
| GB1 (227 K) | −37.4 ± 1.6 |
| GB1−LCDA1 (290 K) | −32.9 ± 1.1 |
| GB1−LCDA1 (277 K) | −37.5 ± 1.7 |
| GB1-LCDA1 (290 K) | −43.6 ± 2.9 |
| GB1-LCDA1 (277 K) | −32.2 ± 1.2 |
GB1-LCDA1 is monomeric in solution and does not phase separate at low temperature or high pressure.
We then determined the oligomeric state of the chimeric protein GB1-LCDA1 in conditions similar to that of the NMR experiments. Figure S2 shows the profile of GB1-LCDA1 eluting from a Superdex 200 10/300 GL gel filtration column with an injected sample concentration of 150 μM. The protein sample and column were kept at 277 K. A single peak is observed with an elution profile similar to isolated GB1, as the presence of LCDA1 does not alter its SEC profile. No peak corresponding to higher molecular weight oligomers was observed confirming that GB1-LCDA1 is predominantly monomeric in the NMR experiments conditions. Further evidence for the monomeric state of the construct was found by small-angle x-ray scattering (SAXS) experiments run at 298K and 650 μM. The molecular weight of GB1-LCDA1 was calculated using a five different concentration-independent determination Bayesian method.57 The results ranged from 16–23 kDa, in-line with the expected weight of 21 kDa for the monomeric protein (Table 2). Visual analysis of the NMR spectra also indicates that GB1-LCDA1 does not form liquid condensate over the range of temperature and pressure sampled here. Liquid-liquid phase separation leads to drastic broadening of peak linewidths which was observed in none of the 2D NMR spectra collected from 268 K to 290 K and 1 bar to 2.5 kbar.
Table 2.
Concentration-independent molecular weight determination for GB1-LCDA1 calculated using the PRIMUS program.
| Method | Results |
|---|---|
| MMQp | qmax[A−1]=0.26626 MW=15294 Da |
| MoW | qmax[A−1]=0.30025 V[A3]=22104 MW=18237 Da |
| Volume of Correlation | qmax[A−1]=0.30025 VC=234 MW=16918 Da |
| Size & Shape | MW=22974 Da |
| Bayesian Inference | MW=18050 Da MW Probability=43.6% Credibility Interval=15800–19000 Da Interval Probability=96.83% |
GB1-LCDA1 ensemble is compact and disordered in solution.
Kratky plots are commonly used for determining the degree of structural disorder of protein ensembles in solution.58 The Kratky plot generated for GB1-LCDA1 shows an initial parabolic peak is followed by a plateaued baseline (Fig. 2A) as expected for a multi-domain protein combining the features of folded and disordered domains.58,59 Guinier analysis of the SAXS data revealed a radius of gyration, Rg of 26.2 ± 0.1 Å (Fig. 2B), which shows that the chimeric protein is highly compact in solution (i.e. the theoretical Rg of GB1-LCDA1 in a fully extended state is ~46.5 Å).60,61
Figure 2.
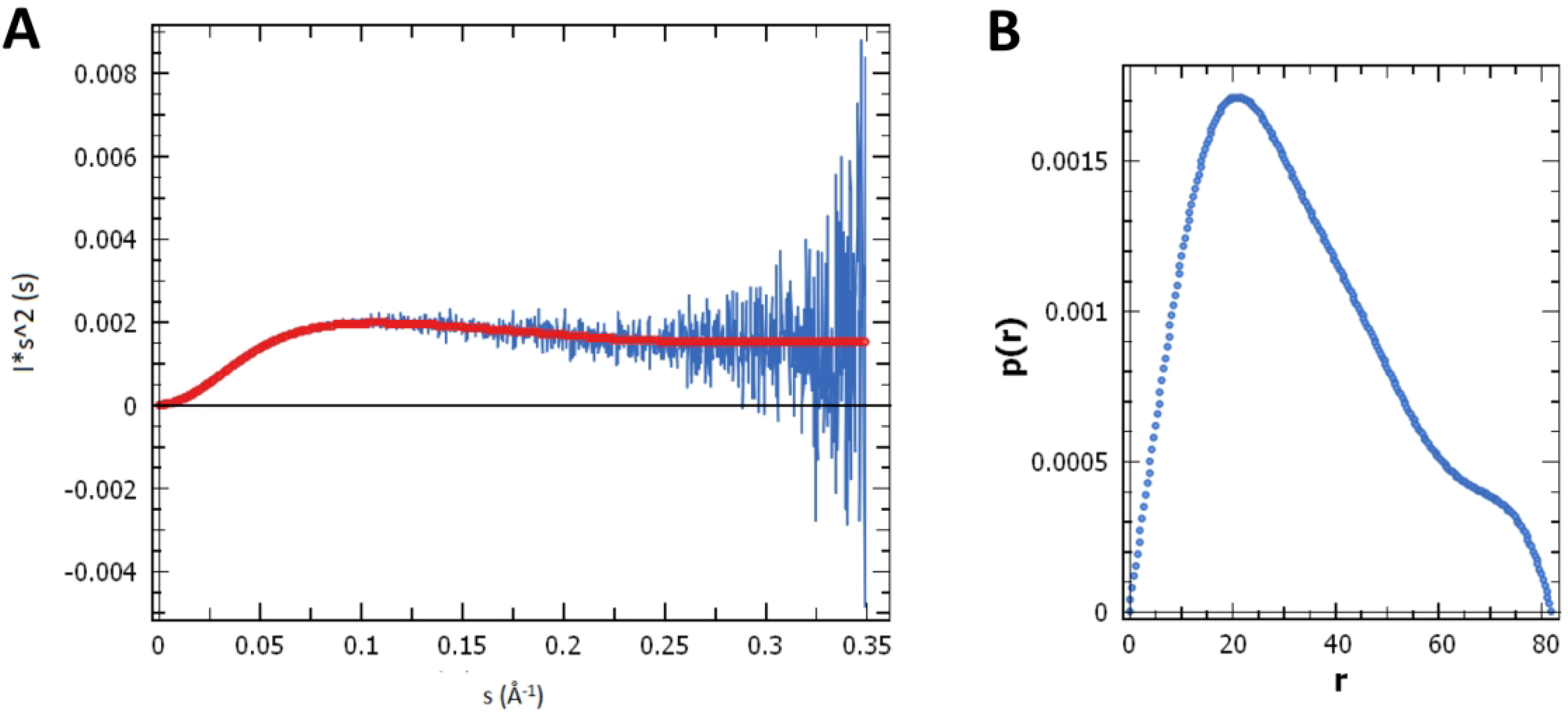
Small-angle X-ray scattering (SAXS) profile of GB1-LCDA1 collected at a protein sample concentration of 650 μM. (A) Analysis of the Kratky plot shows that GB1-LCDA1 is a multi-domain protein composed of both globular and disordered domains. (B) Pair-wise distance distribution function P(r). A radius of gyration estimated of 26.2 ± 0.1 Å was calculated for GB1-LCDA1 in solution.
Pressure-induced unfolding of GB1 in GB1-LCDA1.
To probe the effect of pressure on the GB1-LCDA1, we first examined the stability under pressure of the GB1 domain in the chimeric construct and compared it with the isolated GB1 domain described above. Comparison of the spectra recorded for isolated GB1 domain and GB1-LCDA1, show an almost perfect overlap of GB1 domain crosspeaks in both constructs (Fig. S3). In addition, ΔVF→U values measured for GB1 crosspeaks in the GB1-LCDA1 construct were remarkably similar to those measured for the isolated GB1 domain at both 290 K and 277 K (Fig. 3 and Table 1). These results demonstrate that GB1 structure and folding/unfolding thermodynamics remain unchanged when fused to LCDA1 domain.
Figure 3.
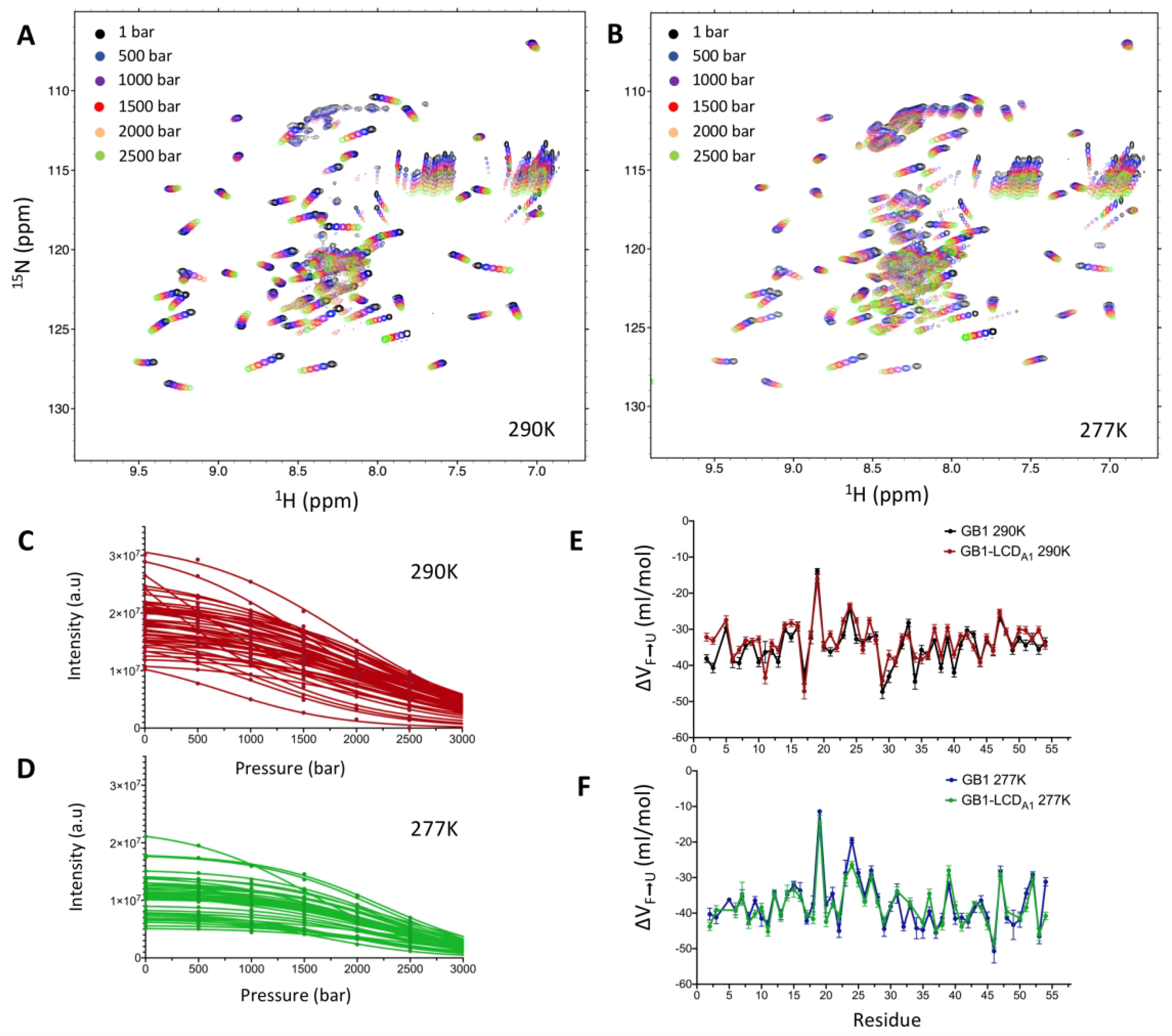
Pressure-induced unfolding of GB1 domain in GB1-LCDA1 construct. 1H-15N HSQC spectra of GB1-LCDA1 were collected at 290 K (A) and 277 K (B) at pressure varying from 1 bar to 2,500 bar. Intensity profiles of individual GB1 crosspeaks in GB1-LCDA1 were measured as a function of pressure at 290 K (C) and 277 K (D) and fitted to equation (3) (solid line) to obtain residue-specific ΔVF→U values. Comparison of ΔVF→U values as a function of GB1 sequence at 290 K (E) and 277 K (F). ΔVF→U values measured for GB1 in GB1-LCDA1 construct are compared here with those measured for the isolated GB1 domain (see Fig. S1).
Pressure-induced transition of LCDA1 in GB1-LCDA1.
While the GB1-LCDA1 construct presents the advantage of preventing phase separation at low temperature and high protein concentration, which is essential for this study, it seemingly causes significant exchange-induced linewidth broadening of a large number of LCDA1 crosspeaks. As apparent from the 2D 1H-15N spectrum of GB1-LCDA1 collected in standard conditions (i.e. 290 K, 1 bar), less than twenty crosspeaks can be confidently assigned to the LCDA1 domain (Fig. S3). Similarly, comparison of 1H-13C HSQCs spectra of GB1 and GB1-LCDA1 shows that only about twelve crosspeaks can be attributed to the LCDA1 (Fig. S4). These experiments suggest that conformational exchange over unfavorable time scale rather than extensive solvent exchange is the main factor limiting the number of LCDA1 resonances that can be detected. Since residue-specific chemical shift assignment is impossible in such conditions, we chose 11 non-overlapping crosspeaks in the 2D 1H-15N spectrum (indicated with arrows in Fig. S3) as internal probes to characterize the overall pressure-induced transitions experienced by LCDA1.
Remarkably, when monitoring the intensity of the selected LCDA1 crosspeaks over the 1–2.5 kbar pressure range, we also observed a significant pressure-induced loss of intensity that can be fitted to the same two-state model described by equations (1–3) (Fig. 4 A and B). Since the pressure-induced transition observed for LCDA1 crosspeaks is manifestly not a simple “folded-to-unfolded” transition, we will define the corresponding volume changes as ΔVLP→HP, where “LP” corresponds to main state populated by LCDA1 at atmospheric pressure and “HP” the state promoted in high-pressure conditions. ΔVLP→HP values measured at 290 K were large and negative with an average value of −43.6 ± 2.9 ml/mol (Table 1 and Fig. 4C). We found that the magnitude of ΔVLP→HP values significantly decreased at 277 K with an average of −32.2 ± 1.2 ml/mol (Table 1 and Fig. 4C). This result indicates that the “HP” state of LCDA1 has a smaller thermal expansivity than its “LP” state, strongly suggesting that the LP↔HP transition is indeed fundamentally different from a simple Folded↔Unfolded transition such as that observed in the GB1 domain. Indeed, in the case of GB1 unfolding transition, the thermal expansivity of the unfolded states is larger than that of the folded states (as expected for any globular protein) (Table 1).
Figure 4.
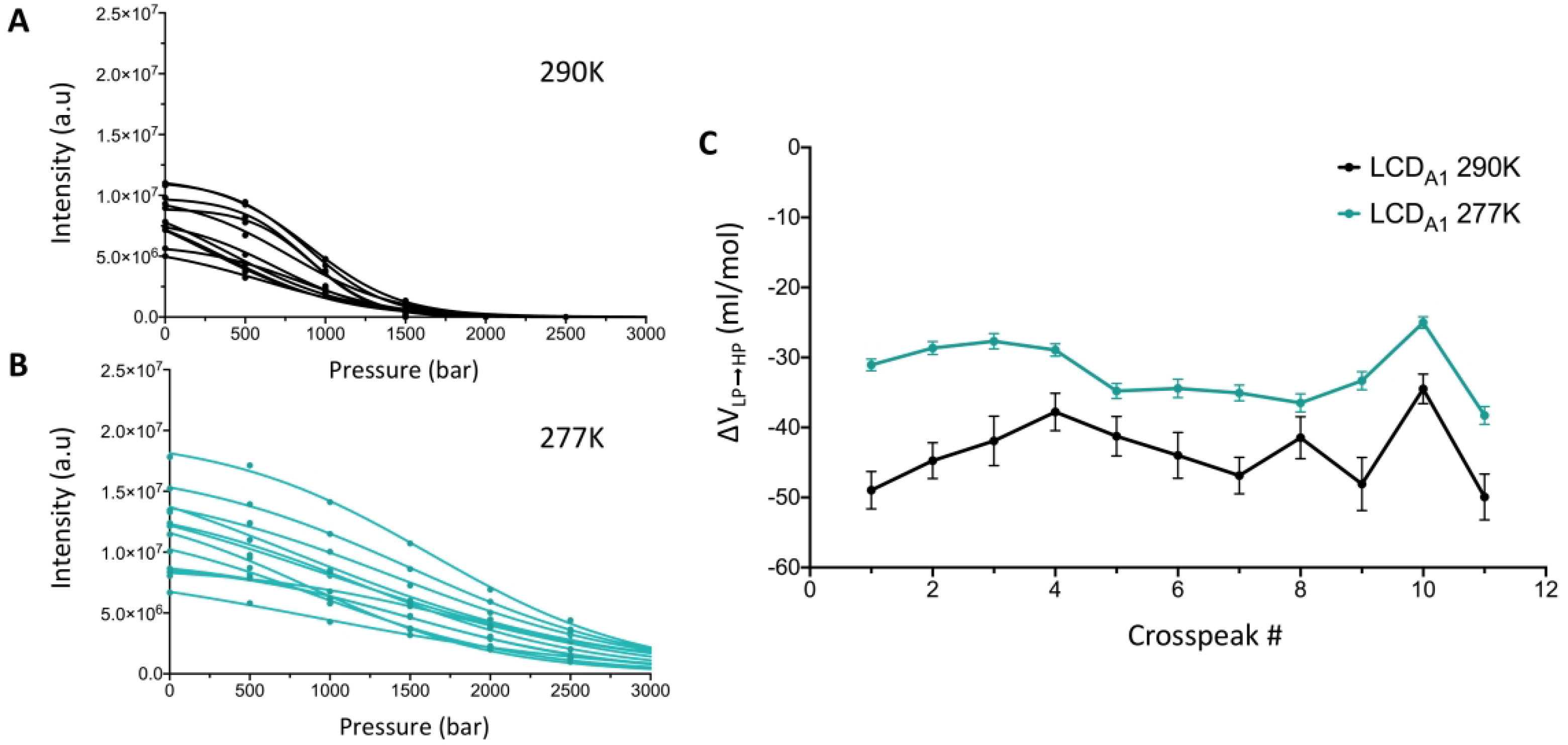
Pressure-induced conformational changes of LCDA1 in GB1-LCDA1. Individual LCDA1 crosspeak intensities were monitored as a function of pressure at 290 K (A) and 277 K (B) and fitted to equation (3) (solid line) to obtain individual ΔVLP→HP values. (C) Comparison of ΔVLP→HP values measured for 11 LCDA1 crosspeaks in GB1-LCDA1 at 290 K (black line and dots) and 277 K (cyan line and dots).
In order to map the pressure-temperature landscape of GB1-LCDA1 construct we recorded additional spectra at extreme low temperature and extreme high-pressure. The relative population of the folded state, in the GB1 domain, and “LP” state in the LCDA1 were calculated for each condition based on average crosspeak intensities (Fig. 5). By mapping the relative population of each state over a broad range of pressure and temperature conditions, one can clearly observe that the folded population of GB1 domain decreases as pressure increases, and decreases even further with lower temperatures (green circle in Fig. 5). On the other hand, the main state populated by LCDA1 (“LP” state) in standard conditions also decreases as pressure increases but its relative population increases at lower temperatures (blue rectangle in Fig. 5).
Figure 5.
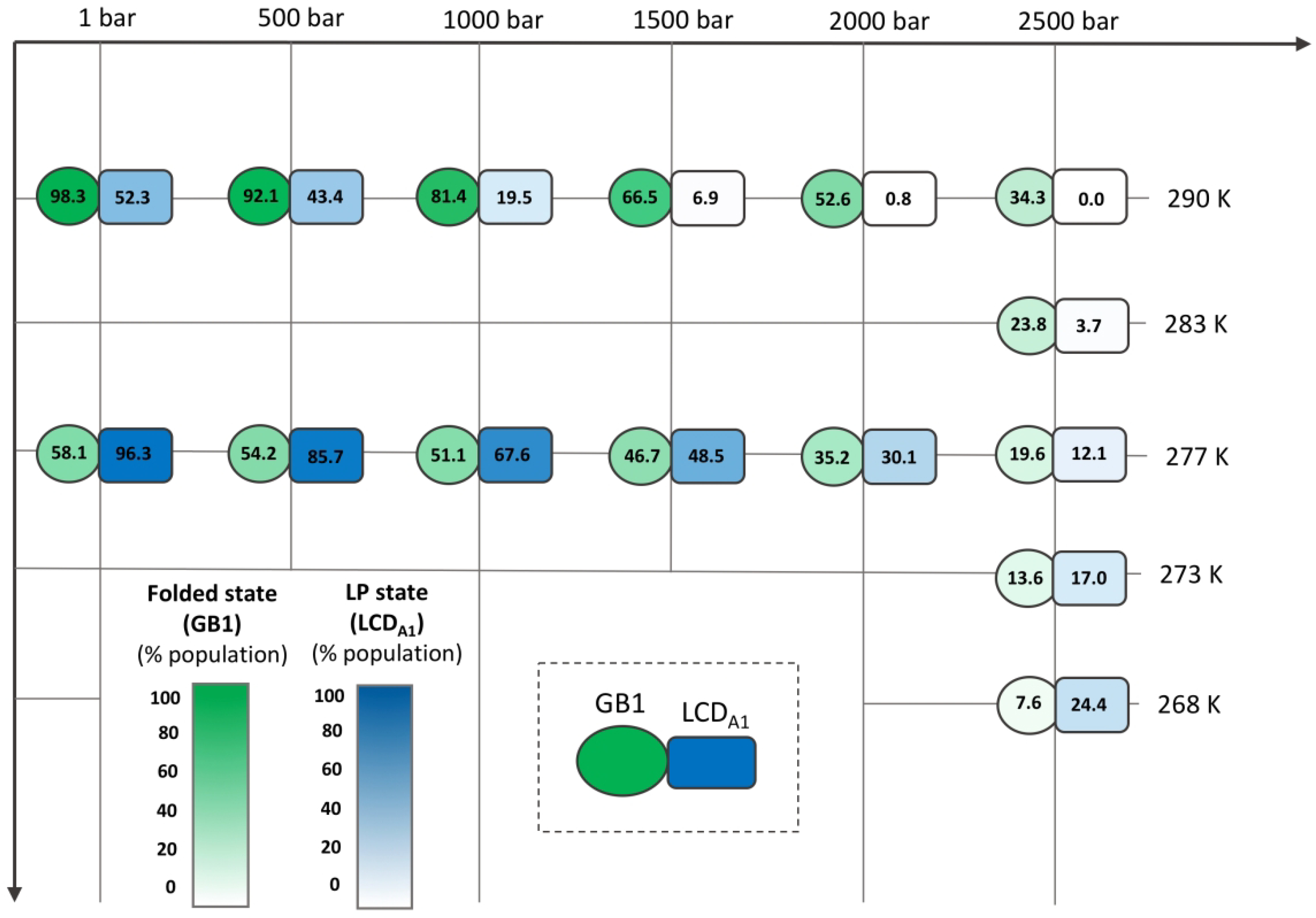
Pressure-temperature mapping of the relative folded population of GB1 in GB1-LCDA1 (green circle) and relative LP population of LCDA1 in GB1-LCDA1. The theoretical maxima (100%) were calculated for both GB1 folded state LCDA1 LP state from the fit of the pressure denaturation profiles (see Fig. 3 C and D for GB1 and Fig. 3 A and B for LCDA1) using equation (3) (i.e. IF in equation (3)). Numbers shown here correspond to the average of the relative population calculated over all individual crosspeaks.
Discussion.
The use of GB1 domain as a fusion tag to overcome solubility and sample stability issues has become a standard tool for biomolecular NMR studies.62 Fusion of GB1 with a low complexity domain such as LCDA1 presents the additional advantage of preventing phase separation. The high salt buffer conditions of the experiments also prevented phase separation. The structure and thermodynamics of the GB1 has been extensively studied by NMR spectroscopy,63,64 and the influence of pressure on GB1 native chemical shifts65 as well as on its equilibrium and kinetic unfolding at acidic pH have been previously described.66 Here we found that GB1 domain, either isolated or as part of the GB1-LCDA1 construct, exhibits the typical pressure-temperature dependence that has been observed for many other small globular proteins: i) negative ΔVF→U values (i.e. GB1 unfolds as pressure increases); ii) a decrease in ΔVF→U magnitude as temperature increases (i.e. the thermal expansivity of GB1 unfolded states is larger than the expansivity of its folded states); and iii) the folded population of GB1 decreases at lower temperature (i.e. indicative of cold denaturation).11
In contrast, LCDA1 exhibits unique pressure-temperature dependence properties, that are distinct from both well-structured proteins (e.g. GB1 domain) and intrinsically disordered proteins (e.g. α-synuclein23). Indeed, the results presented here demonstrate that LCDA1 undergoes conformational changes as pressure increases (called here “LP↔HP” transition), characterized by negative ΔVF→U values. Our data also show that the “HP” state has a smaller thermal expansivity than that of the “LP” state. In addition, the “LP” state is stabilized at lower temperatures. To reconcile all above observations, we propose a model in which the “LP↔HP” transition corresponds to transient formation and stabilization of structural motifs with concomitant exposure of polar and charged residues to solvent (Fig. 6). In this model, we hypothesize that LCDA1 adopts a structurally disordered but compact conformation in standard conditions (i.e. atmospheric pressure) stabilized by a mesh-like network of transient polar and electrostatic interactions. This model conforms with recent experiments showing the LC domain of hnRNP A2 to be largely disordered, yet compact.49 Exposure of polar and charged side chain to solvent is accompanied by a significant negative volume change as the density of water molecules is higher around polar and charged moieties. The sum of negative contribution resulting from exposure of these side chains would explain the large and negative ΔVLP→HP values measured for LCDA1, as pressure disrupts the network of transient interactions present in the LP state. Since pressure can promote the formation of small helical motifs,25–27 we also hypothesize that the HP state is not entirely disordered but rather encompasses short structural motifs. The higher degree of secondary structure in the HP state compared to the LP state would explain the negative change in thermal expansivity measured upon LP-to-HP transition. Cold denaturation of these motifs would explain why the LP state is favored at low temperature (Fig. 5). Similar cold denaturation is expected for short β-strand motifs that have been identified recently in the low complexity domains of hnRNPA1 and hnRNPA2.67,68 It should be noted that the significant increase in water density from 290 K to 268 K could also play a role in modulating the free-energy of LCDA1 sub-ensembles at subfreezing temperatures. While we recognize that assessing the nature of these structural motifs is important, the small number of LCDA1 crosspeaks observable with the present construct prevents us to fully assign the backbone chemical shifts of the low complexity domain with triple resonance NMR experiments.
Figure 6.
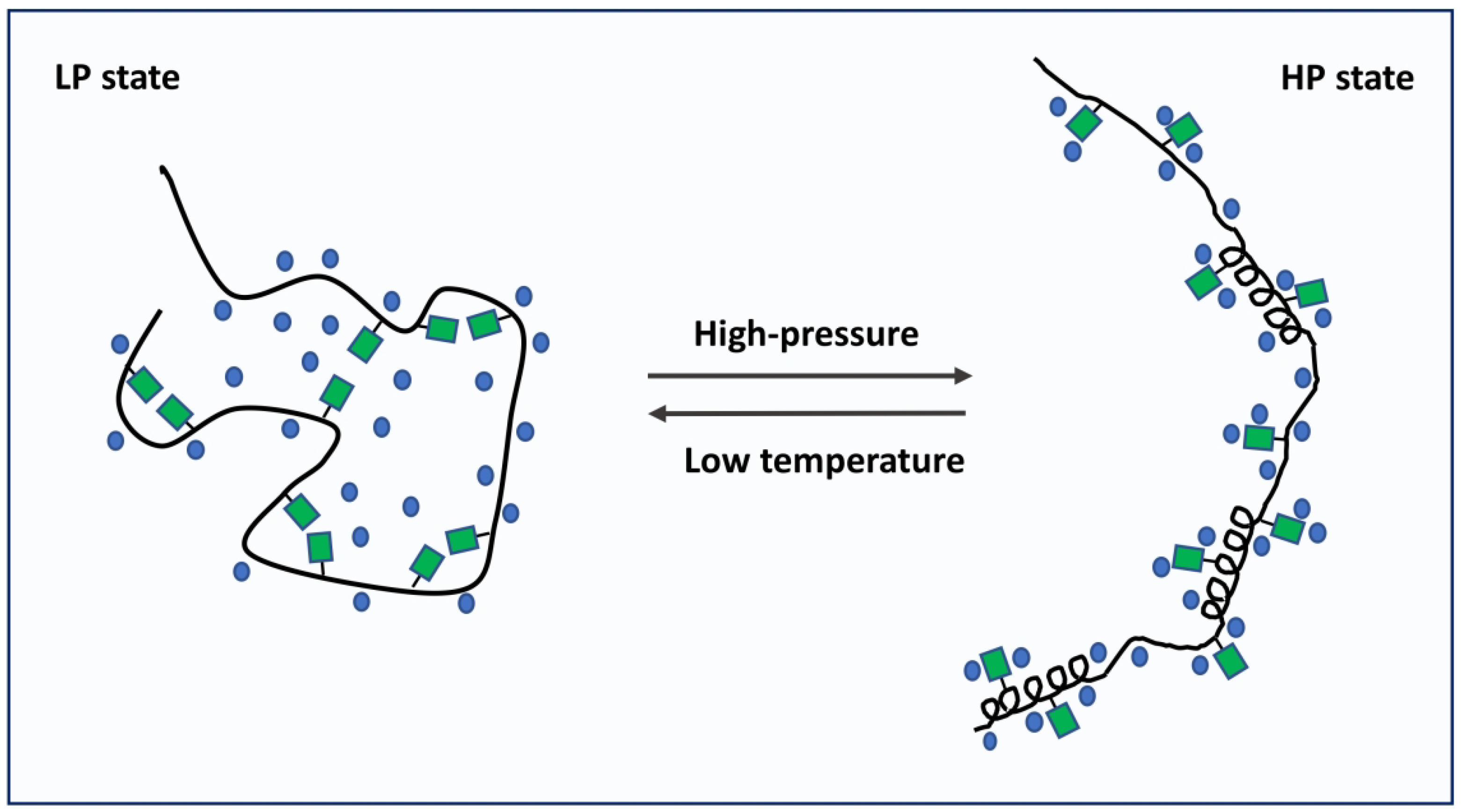
Model of the conformational changes experienced by LCDA1 under high-pressure and low temperature conditions. Our data suggest that in standard conditions (atmospheric pressure), LCDA1 adopts a compact conformation stabilized by a mesh-like network of polar and electrostatic interactions (LP state). High-pressure will promote the exposure of polar and charged side chains to solvent, therefore breaking the network of interactions present in the LP state. We hypothesize that the HP state also contains structural motifs that are denaturated at very low temperature.
Phase separation is generally believed to be triggered by multivalent weak interactions including electrostatic, cation-π, and π-π interactions.31–35 A recent study on hnRNP A1 has demonstrated that valence of aromatic residues plays a major role in determining the temperature dependence of chain compaction.69 In the case of TDP-43, phase separation has been shown to be mediated by an α-helix in its LC domain.70,71 The data presented here suggest that a similar mechanism may take place for hnRNP A1. Indeed, the low-lying HP state identified in our study may contain short helical motifs able to mediate interactions between LC domains. While this mechanism is certainly not the sole thermodynamic factor driving hnRNP A1 phase separation, it may constitute a non-negligible contribution to the formation of liquid condensates.
Conclusion
We have shown that the low-complexity domain of hnRNP A1 exhibits unique temperature and pressure dependences. We measured volume differences associated with pressure-induced conformational changes, characterizing the transition between a disordered, compact state and a fully solvated, extended state. In view of these results, we hypothesize that the extended state of LCDA1 contains helical motifs that may play a role in driving the formation of liquid condensates. This study illustrates how high-pressure NMR can be used to describe low lying populations in a structurally disordered ensemble.
Supplementary Material
Acknowledgements
We thank Dr. Vincenzo Venditti for critical reading of the manuscript. We thank Srinivas Chakravarthy for performing the SAXS experiments. J.R. acknowledges the Iowa State University College of Liberal Arts and Sciences and the Roy J. Carver Charitable Trust for their support. B.T. acknowledges NIH U54AI150470.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Nucci NV, Fuglestad B, Athanasoula EA, Wand AJ. Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proc Natl Acad Sci USA 2014;111:13846–13851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeno A et al. Cavity as a source of conformational fluctuation and high-energt state : high-pressure NMRstudy of a cavity-enlarged mutant of T4 lysozyme. Biophys J 2015;108:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roche J et al. Remodeling of the folding free energy landscape of staphylococcal nuclease by cavity-creating mutations. Biochemistry 2012;51:9535–9546 [DOI] [PubMed] [Google Scholar]
- 4.Nisius L, Grzesiek S Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network. Nat Chem 2012;4: 711–717 [DOI] [PubMed] [Google Scholar]
- 5.Roche J et al. Effect of internal cavities on folding rates and routes revealed by real-time pressure-jum NMR spectroscopy. J Am Chem Soc 2013;135:14610–14618 [DOI] [PubMed] [Google Scholar]
- 6.Charlier C et al. Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample tube. Proc Natl Acad Sci USA 2018;115: E4169–E4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korzhnev DM. et al. Probing the transition state ensemble of a protein folding reaction by pressure-dependent NMR relaxation dispersion. J Am Chem Soc 2006;128:5262–5269 [DOI] [PubMed] [Google Scholar]
- 8.Akasaka K, Li H. Low-lying excited states of proteins revealed from nonlinear pressure shifts in 1H and 15N NMR. Biochemistry 2001;40:8665–8671 [DOI] [PubMed] [Google Scholar]
- 9.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: ubiquitin at 30 bar-3 kbar. J Mol Biol 2005;347:277–285 [DOI] [PubMed] [Google Scholar]
- 10.Tugarinov V, Libich DS, Meyer V, Roche J, Clore GM. The energetics of a three-state protein folding system probed by high-pressure relaxation dispersion NMR spectroscopy Angew Chem Int E. Engl 2015;54:11157–11161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roche J, Royer CA. Lessons from pressure denaturation of proteins. J R Soc Interface 2018;15:pii 20180244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royer CA. Revisiting volume changes in pressure-induced protein unfolding. Biochim Biophys Acta 2002;1595:201–209 [DOI] [PubMed] [Google Scholar]
- 13.Roche J et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA;109:6945–6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouget JB. et al. Size and sequence and the volume change of protein folding. J Am Chem Soc 2011;133:6010–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CR, Makhatadze GI. Molecular determinant of the effects of hydrostatic pressure on protein folding stability. Nat Commun 2017;8:14561–14570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummer G, Garde S, Garcia AE. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA 1998;95:1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh T, Garcia AE, Garde S. Molecular dynamics simulations of pressure effects on hydrophobic interactions. J Am Chem Soc 2001;123:10997–11003 [DOI] [PubMed] [Google Scholar]
- 18.Oliveira GAP, Silva JL. A hypothesis to reconcile the physical and chemical unfolding of proteins. Proc Natl Acad Sci USA 2015;112:E2775–E2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seemann H, Winter R, Royer CA. Volume, expansivity and isothermal compressibility changes associated with temperature and pressure unfolding of Staphylococcal nuclease J Mol Biol 2001;307:1091–1102 [DOI] [PubMed] [Google Scholar]
- 20.Mitra L, Rouget JB, Garcia-Moreno B, Royer CA, Winter R. Towards a quantitative understanding of protein hydration and volumetric properties. Chemphyschem 2008;9:2715–2721 [DOI] [PubMed] [Google Scholar]
- 21.Chen CR, Makhatadze GI. Molecular determinants of temperature dependence of protein volume change upon unfolding. J Phys Chem B 2017;121:8300–8310 [DOI] [PubMed] [Google Scholar]
- 22.Schroer MA. et al. High-pressure SAXS study of folded and unfolded ensembles of proteins. Biophys J 2010;99:3430–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roche J, Ying J, Maltsev AS, Bax A. Impact of hydrostatic pressure on an intrinsically disordered protein: a high-pressure NMR study of alpha-synuclein. Chembiochem 2013;14:1754–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes CA, Robertson AJ, Louis JM, Anfinrud P, Bax A. Observation of β-amyloid peptide oligomerization by pressure-jump NMR spectroscopy. J Am Chem Soc 2019;141:13762–13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekiyo T, Shimizu A, Kato M, Taniguch Y. Pressure tuning FT-IR spectroscopic study on the helix-coil transition of Ala-rich oligopeptide in aqueous solution. Biochim Biophys Acta 2005;1750:1–4 [DOI] [PubMed] [Google Scholar]
- 26.Neumaier S, Buttner M, Bachmann A, Kiefhaber T. Transition state and ground state properties of the helix-coil transition in peptides deduced from high-pressure studies. Proc Natl Acad Sci USA 2013;110:20988–20993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Best RB, Miller C, Mittal J. Role of solvation in pressure-induced helix stabilization. J Chem Phys 2014;141:22D522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin EW, Mittag T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry 2018;57:2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molliex A et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrilization. Cell 2015;163:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pak CW. et al. Sequence determinants of intracellular phase separation by complex coarcervation of a disordered protein. Mol Cell 2016;63:72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qamar S et al. FUS Phase separation is modulated by a molecular chaperone and methylation of arigine cation-π interactions. Cell 2018;173:720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YH, Forman-Kay JD, Chan HS. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry 2018;57:2499–2508 [DOI] [PubMed] [Google Scholar]
- 33.Vernon RM. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. elife 2018;7:pii 231486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alshareedah I et al. Interplay betwee short-range attracttion and lang-range repulsion controls reeentrant liquid condensation of ribonucleoprotein-RNA complexes. J Am Chem Soc 2019;141:14593–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J et al. A molecular grammar goverining the driving forces for phase separation of prion-like RNA binding proteins. Cell 2018;174:688–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cinar S, Cinar H, Chan HS, Winter R Pressure-sensitive and osmolyte-modulated liquid-liquid phase separation of eye-lens γ-crystallins. J Am Chem Soc 2019;141:7347–7354 [DOI] [PubMed] [Google Scholar]
- 37.Cinar H, Cinar S, Chan HS, Winter R. Pressure-induced dissolution and reentrant formation of condensed, liquid-liquid phase-separated elastomeric α-elastin. Chemistry 2018;24:8286–8291 [DOI] [PubMed] [Google Scholar]
- 38.Cinar H et al. Temperature, hydrostratic pressure and osmolyte effects on liquid-liquid phase separation in protein condensates: physical chemistry and biological implications. Chemistry 2019;25:13049–13069 [DOI] [PubMed] [Google Scholar]
- 39.Levengood JD, Tolbert JS. Idiosyncrasies of hnRNP A1-RNA recognition: can binding mode influence function. Semin Cell Dev Biol 2019;86:150–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci 2013;14:18999–19024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in helath and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci 2013;56:436–446 [DOI] [PubMed] [Google Scholar]
- 42.Pollard VW. et al. A novel receptor-mediated nuclear protein import pathway. Cell 1996;86:985–994 [DOI] [PubMed] [Google Scholar]
- 43.Izaurralde E et al. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol 1997;1317:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartegni L et al. hnRNP A1 selectively interacts through its gly-rich domain with different RNA-binding proteins. J Mol Biol 1996;259:337–348 [DOI] [PubMed] [Google Scholar]
- 45.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol 2016;26:668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato M et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012;149:753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Y Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 2015;60:208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wall ML, Lewis SM. Methylarginines within the RGG-motif region of hnRNP A1 affects its IRES trans-acting factor activity and are requires for hnRNP A1 stress hranule localization and formation. J Mol Biol 2017;429:295–307 [DOI] [PubMed] [Google Scholar]
- 49.Ryan VH. et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell 2018;69:465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stols L et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus proteas cleavage site. Protein Expr Purif 2002;25:8–15 [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F et al. NMRPipe : a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995;5:277–293 [DOI] [PubMed] [Google Scholar]
- 52.Goddard TD, Kneller DG. Sparky 3, University of California San Francisco, San Francisco, CA: (2010) [Google Scholar]
- 53.Morgan CE. et al. The First Crystal structure of the UP1 domain of hnRNP A1 bound to RNA reveals a new look for an old RNA binding protein. J Mol Biol 2015;427:3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konarev PV. et al. PRIMUS: a windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 2003;36:1277–1282 [Google Scholar]
- 55.Franke D ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 2017;50:1212–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen LM, Roche J. High-pressure NMR techniques for the study of protein dynamics, folding and aggregation. J Magn Reson 2017;277:179–185 [DOI] [PubMed] [Google Scholar]
- 57.Hajizadeh NR. et al. Consensus Bayesian assessment of protein molecular masss from solution X-ray scattering data. Sci Rep 2018;8:7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Putnam CD. et al. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 2007;40:191–285 [DOI] [PubMed] [Google Scholar]
- 59.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011;95:559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernardó P, Svergun DI. Structural analysis of intrinsically disordered protein by small-angle X-ray scattering. Mol Biosyst 2012;8:151–167 [DOI] [PubMed] [Google Scholar]
- 61.Bernardó P, Blackledge M. A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering. Biophy J 2009;97:2839–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou P, Wagner G. Overcoming the solubility limit with solubility-enhancement tags successful applications in biomolecular NMR studies. J Magn Res 2010;46:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gronenborn AM et al. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 1991;253:657–661 [DOI] [PubMed] [Google Scholar]
- 64.Ding K, Louis JM, Gronenborn AM. Insights into conformation and dynamics of protein GB1 during folding and unfolding by NMR. J Mol Biol 2004;335:1299–1307 [DOI] [PubMed] [Google Scholar]
- 65.Wilton DJ et al. Pressure-induced changes in the solution structure of the GB1 domain of protein G. Proteins 2008;71:1432–1440 [DOI] [PubMed] [Google Scholar]
- 66.Dreydoppel M et al. Equilibrium and kinetic unfolding of GB1: stabilization of the native state by pressure. J Phys Chem B 2018;122:8846–8852 [DOI] [PubMed] [Google Scholar]
- 67.Gui X et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their roles in stress granule assembly. Nat Commun 2019;10: doi: 10.1038/s41467-019-09902-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu J et al. CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nat Commun 11: doi: 10.1038/s41467-020-17905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin EW. et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020;367:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 2016;24:1537–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conicella AE. et al. TDP-43 α-helical structure tunes liqui-liquid phase separation and function. Proc Natl Acad Sci USA 2020;117:5883–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


