Abstract
Introduction
Acute myeloid leukemia (AML) is a heterogeneous clonal disorder of hematopoietic progenitor cells, and the AML cells are differentiation retarded which results in the hyperproliferation of those malignant tumor cells. To stop the uncontrollable proliferation, inducing the AML cell differentiation is one highly expected therapy because it can bring relatively low systematic side effects compared to conventional chemotherapies; however, there are few options of inductive therapeutics in the clinical applications so far. This study aims to investigate the differentiation-induction effects of lab-developed hydrophilic nanocrystals of As4S4 (ee-As4S4).
Methods
In this work, ee-As4S4 was applied upon a refractory mouse model co-expressing AML1-ETO and HyC-KITD816V as well as a related human AML cell line, Kasumi-1, to investigate whether the nanocrystals can break the retardation of differentiation and drive the cells undergo apoptosis.
Results
It was shown that ee-As4S4 induced the upregulation of surface markers CD11b, CD235a, and CD41a, which indicate granulocytic, erythroid, and megakaryocytic differentiation respectively, leading to the multiple-lineage differentiation and post-differentiation apoptosis, and the inhibition of histone deacetylase activity was largely involved with the differentiation-induction effects. In the AML mice, orally administered ee-As4S4 increased the level of Ter119, CD11b, and CD41 in bone marrow-derived leukemia cells while reducing the percentage of leukemic cells in the bone marrow. Also, ee-As4S4 improved the hemogram and relieved the hepatomegaly and splenomegaly of the AML mice. As a result, the survival of the AML mice was significantly prolonged. Importantly, ee-As4S4 did not cause acute or chronic toxicity in healthy mice.
Conclusion
In conclusion, ee-As4S4 induced effective and multiple-lineage differentiation and apoptosis of AML cells in the refractory AML mouse model and cell line, suggesting that it holds promising potential as a novel inductive agent in differentiation therapy of AML.
Keywords: As4S4, nanocrystals, acute myeloid leukemia, differentiation therapy, histone deacetylation
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous hematopoietic malignancy that originates from differentiation blocks at various stages of commitment and maturation within the myeloid, which leads to the accumulation of immature myeloid blasts in the bone marrow.1,2 Conventional chemotherapies have been a chief strategy in the AML treatment, which killed proliferating cells and generally succeed in debulking the disease.3 However, the treatment regimens based on cytotoxic mechanisms often lead to intolerable side effects and result in a very poor 5-year survival rate due to drug resistance and relapse. Hence, more efficacious and tolerable therapies are urgently demanded.
It has been well known that differentiation blockade is the main characteristic of leukemic cells, which results in uncontrollable proliferation.4 Therefore, induction of differentiation represents a promising strategy for leukemia treatment. Different from cytotoxic therapies, differentiation therapies can accelerate the maturation of non-clonogenic leukemic cells and decrease the fraction of cells endowed with excessive proliferation,5,6 therefore is an alternative way to treat leukemia with much lower systematic toxicity. The hallmark success of the differentiation therapy regimen is the combination of retinoic acid (RA) and arsenic trioxide, which has made one of the subtypes of AML, acute promyelocytic leukemia (APL) highly curable.7 However, APL is less than 10% in AML patients, and more than 90% of AML patients are not able to obtain benefits from the ATRA-based differentiation therapy.
Several recent studies have demonstrated that epigenetic changes associated with regulation of chromatin structure and gene expression are largely involved in the pathogenesis of AML.8 In the past few years, it has been known that AML blasts generally have reduced acetylation of histones compared to normal white blood cells,9,10 and histone deacetylase (HDAC) plays a central role in regulating the acetylation of histones. Inhibition of HDAC can reverse oncogene-induced transcriptional repression, therefore had predicted a major impact on AML. Some HDAC inhibitors have been reported to elicit AML cell differentiation ex vivo or in vivo,11–13 suggesting that the inhibition of HDAC may provide an alternative approach to induce AML cell differentiation.
Realgar (arsenic sulfide, As4S4) is a kind of crystal substance with large lattice energy, therefore makes it poor solubility in water and the acidic solution. Even after ground with a traditional purification technique named Shuifei,14 the particle size of realgar is still tens of microns and referred as raw realgar (r-As4S4).15 Several composite formulations containing r-As4S4 have shown certain clinical therapeutic effects on some blood cancers,16–19 however, the bioavailability is very low when orally administrated15 and the effects are needed to increase. We previously developed a solid dispersion formulation by using hot-melting extrusion technique, in which r-As4S4 was processed into nanosized crystalline with the amphiphilic polymer-coating (referred to as ee-As4S4) and the bioavailability is largely improved,15 at the same time, some new effects have been observed due to the nano-size.20 It was found out that ee-As4S4 was able to induce chronic myeloid leukemia (CML) cells undergo erythroid differentiation through downregulating the intracellular ROS and inducing autophagic degradation of BCR-ABL fusion protein, which has not been observed with the raw As4S4.20 Besides, ee-As4S4 was able to inhibit HDAC activity, therefore inducing megakaryocytic differentiation in the CML cells.21 The differentiation-induction and HDAC inhibition effects of ee-As4S4 on the CML cells shed the light that the hydrophilic nanocrystal of As4S4 may hide an attractive capacity for inducing AML cell differentiation.
In this study, we applied ee-As4S4 to a well-established refractory AML mouse model22 and human AML cell line Kasumi-1, both are featured with t (8:21) translocation and AML1/ETO fusion protein.23 We showed that ee-As4S4 could effectively induce multi-lineage differentiations including erythroid, megakaryocytic, and granulocytic ones, and consequently post-differentiation apoptosis of the AML cells through inhibiting the activity of HDAC. These effects collectively in turn prolonged the survival of the AML mice that received the oral administration of ee-As4S4.
Materials and Methods
Cell Culture
Kasumi-1, a t (8;21)-positive M2b-type of the French-American-British (FAB) classification AML cell line was kindly provided by Professor Min Wang from the Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Tianjin, China) and cultured in RPMI 1640 medium (Hyclone, Cytiva, Marlborough, MA, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Scientific, Rockford, IL, USA), 100 U/mL penicillin, and 100 U/mL streptomycin (Hyclone, Cytiva, Marlborough, MA, USA) in a humidified atmosphere of 5% CO2 at 37 °C.
Histone Deacetylase Activity Assay
The ee-As4S4 was prepared and characterized as described previously.18 HDAC activity was detected according to the manufacturer’s protocol using a Colorimetric HDAC Activity Assay Kit (Bio Vision, San Francisco, CA, USA). Briefly, Kasumi-1 cells were collected and lysed with a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) after being treated with ee-As4S4. The nuclear protein of 85 μL (50 μg), assay buffer of 10 μL HDAC, and HDAC substrate of 5 μL were mixed and incubated at 37 °C for 1 h. Afterward, the reaction was stopped by adding 10 μL lysine developer and mixed well, and then incubated for 30 min at 37 °C. The absorbance was read at 405 nm with a Synergy H1 microplate spectrophotometer (BioTek Instruments, Winooski, VT, USA).
Quantitative Real-Time PCR (q-RT PCR)
Total RNA was extracted by Trizol reagent (Merck, Darmstadt, Germany) and reverse transcribed to cDNA. Quantitative real-time PCR (q-RT PCR) was performed using SYBR probe (Takara Bio, Kusatsu, Japan), and fluorescence was detected with a CFX96 Connect Real-Time PCR Detection System (Biorad, Hercules, CA, USA). The primers used in the present study were purchased from Sangon Biotech (Shanghai, China), and the sequences were as below:
GAPDH forward primer: 5ʹ-GGTCACCAGGGCTGCTTTTA-3ʹ
GAPDH reverse primer: 5ʹ-GAGGGATCTCGCTCCTGGA-3ʹ
CEBPA1 forward primer: 5ʹ-GGTGGACAAGAACAGCAACG-3ʹ
CEBPA1 reverse primer: 5ʹ-GGTCAGCTCCAGCACCTTCT-3ʹ
GATA1 forward primer: 5ʹ-GCCTTCATCACTCCCTGTCC-3ʹ
GATA1 reverse primer: 5ʹ-AGGCGTTGCATAGGTAGTGG-3ʹ
SPI1 forward primer: 5ʹ-GTGCCCTATGACACGGATCTA-3ʹ
SPI1 reverse primer: 5ʹ-AGTCCCAGTAATGGTCGCTAT-3ʹ
Cell Differentiation Assay
Kasumi-1 cells were seeded in 24 well plates at a density of 2 × 105 cells/mL. After incubated with 0.5–2.0 mg/L ee-As4S4 for 24 h or 72 h, cells were collected and rinsed with PBS. Then cells were then labeled with PE-conjugated antibodies against CD235a (ebioscience, ThermoFisher Scientific, Rockford, IL, USA), FITC-conjugated antibodies against CD41a (ebioscience, ThermoFisher Scientific, Rockford, IL, USA), and FITC-conjugated antibodies against CD11b (BioLegend, San Diego, CA, USA), respectively. The antibody-labeled cells were subsequently analyzed by flow cytometry. Post-differentiation apoptosis was assessed by co-staining with APC-conjugated Annexin V (Dojindo, Kumamoto, Japan) and differentiation markers, followed by flow cytometry analyses.
Western Blot
After incubated with ee-As4S4, Kasumi-1 cells were collected and lysed in RIPA cell lysis buffer (Solarbio life sciences, Beijing, China) supplemented with 1 mM PMSF (Solarbio life sciences, Beijing, China) and protein phosphatase inhibitor (Applygen, Beijing, China). The lysates were then centrifuged at 12,000 rpm for 15 min at 4°C. The protein was quantified by a Pierce™ BCA Protein Assay Kit according to the manufacturer’s instructions. After denatured by boiling in loading buffer (Applygen, Beijing, China), an equal amount of protein (20 µg) was loaded and separated on 12% glycine SDS-PAGE gel, and transferred to polyvinylidene difluoride (PVDF) membranes (0.45 µm; Millipore, Merck, Darmstadt, Germany). Then the membrane was incubated with primary antibodies at 4°C overnight, including anti-β-actin, anti-pro-caspase 3, and anti-caspase 3 (Cell Signaling Technology, CST, Boston, MA, USA). The membrane was incubated with Horseradish Peroxidase (HRP)-conjugated secondary antibodies and visualized using an automatic chemiluminescence image analysis system (Tanon, Shanghai, China) with HRP substrate luminol reagent and peroxide solution (Merck, Darmstadt, Germany). The relative expression of the protein was analyzed using Image J software.
Caspase 3 Activity Assay
Caspase 3 activity was detected according to the manufacturer’s protocol using a Colorimetric Caspase 3 Activity Assay Kit (Dojindo, Kumamoto, Japan). Briefly, Kasumi-1 cells were collected and lysed with lysis buffer after being treated with ee-As4S4. After being incubated for 10 min at 4°C and centrifuge for 10 min at 10,000 rpm, the supernatant was collected and quantified by Bradford and adjusted to 1 mg/mL before being mixed with assay buffer. The protein of 50 μL (50 μg), assay buffer of 10 μL, and substrate of 10 μL were mixed and incubated at 37°C for 2 h. The absorbance was read at 405 nm with a Synergy H1 microplate spectrophotometer.
In vivo Therapeutic Effect of ee-As4S4
Six-week-old, female C57 mice were bred in the Experimental Animal Center of the Institute of Basic Medical Sciences (Beijing, China) under specific-pathogen-free conditions. The transplantable AML mouse model was constructed as described previously.22,24 In brief, GFP labeled leukemic cells, which express AML1-ETO fusion gene and HyCKITD816V were injected intravenously to sub-lethally irradiated (450 cGy) mice (1 × 106 cells per mouse). On day-7 after transplantation, the percentage of GFP-positive (GFP+) cells in peripheral blood was detected, and the mice were randomly grouped according to the percentage of GFP+ cells. After grouping, the mice were intragastrically administered with distilled water (control), r-As4S4 (0.5 mg/day), and ee-As4S4 (containing 0.5 mg ee-As4S4 in it). The overall survival was measured from transplantation until the occurrence of the last death (n = 7).
To evaluate the histological changes after different treatments, the AML mice were intragastrically administered with distilled water, r-As4S4, and ee-As4S4 for 7 days, and sacrificed on day-15 after transplantation. White blood cell (WBC), red blood cell (RBC), and platelet (PLT) count of the peripheral blood were measured by a hematology analyzer (Mindray Bio-Medical Electronics Co., Shenzhen, China). Femurs were collected for further study. One of the femurs of each mouse were flushed, and the bone marrow mononuclear cells were collected to detect the percentages of GFP+ leukemia cells (n = 5). Another femur of each mouse was fixed and subjected to immunofluorescent staining for the detection of Ter119, CD41, CD11b, and caspase-3. The expression of differentiation markers Ter119, CD41, and CD11b were also detected in the bone marrow mononuclear cells (n = 4). The antibodies used in flow cytometry and immunofluorescent staining were as follows: APC-conjugated antibodies against mouse Ter119 (CAT#: 17-5921-82, ebioscience, ThermoFisher Scientific, Rockford, IL, USA), PE-conjugated antibodies against mouse CD41 (CAT#:133906, BioLegend, San Diego, CA, USA), PE-conjugated antibodies against mouse CD11b (CAT#: 101208, BioLegend, San Diego, CA, USA); mouse CD11b antibodies (CAT#: GB11058, Servicebio, Wuhan, China), mouse CD41 antibodies (CAT#: PA5-79526, Invitrogen, ThermoFisher Scientific, Rockford, IL, USA), mouse Ter119 antibodies (CAT#: ab91113, Abcam, Cambridge, UK).
In vivo Biological Safety Evaluation
Healthy C57BL/6 mice (n = 6) were orally administered with or without ee-As4S4 for 14 days. The body weights were measured every two days and the peripheral blood was collected every two days and analyzed by a hematology analyzer (Mindray Bio-Medical Electronics Co.).
Statistical Analysis
All of the quantitative data are expressed as the mean ± SEM and the statistical analysis was carried out by SPSS26.0. Homogeneity of variance was tested by the Levene test. Unpaired two-tailed Student’s t-test was used for the analysis between two groups defined by one factor. One-way ANOVA was used to compare three or more groups defined by one factor, and two-way ANOVA was used to compare the groups that involve two factors. Tukey’s multiple comparison test was used as a follow-up to ANOVA to compare the mean of different groups. P-value less than 0.05 was considered statistically significant.
Results
The ee-As4S4 Induced Multi-Lineage Differentiation and Post-Differentiation Apoptosis in vitro
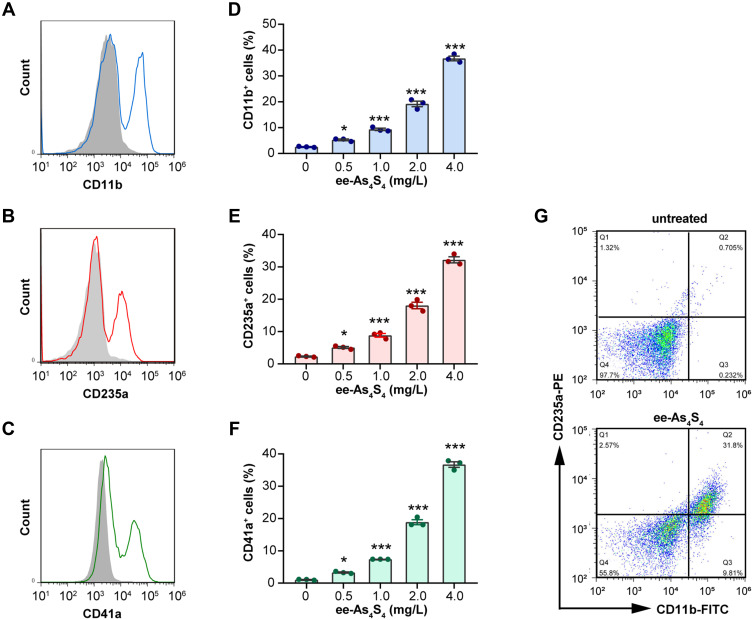
In normal physiological conditions, immature myeloid cells hold the potential to undergo granulocytic differentiation, erythroid differentiation, and megakaryocytic differentiation to generate mature granulocytes, red blood cells, and platelets. Kasumi-1 cells are featured with t (8:21) translocation and AML1/ETO fusion protein, which was classified as M2b subtype of the FAB classification and defined as AML with t (8;21) (q22;q22) according to the WHO classification and belongs to the category of AML with recurrent genetic abnormalities. The expression of AML1/ETO fusion protein leads to the hyperplasia of the primitive and immature myeloid cells. By ee-As4S4 incubation for 72h, surface markers CD11b, CD235a, and CD41a were significantly increased on the cells, which indicate granulocytic, erythroid, and megakaryocytic differentiation, respectively (Figure 1A–C). The portion of markers positive cells increased in a dose-dependent manner, and the percentage of CD11b+, CD235a+, and CD41a+ cells reached 36.77%, 32.19%, and 36.72% after incubation with 4 mg/L ee-As4S4 (Figure 1D–F). In the same way, the mean fluorescence intensity (MFI) of cells also increased gradually (Figure S1A–C). Overall, these results indicated that ee-As4S4 induced multi-lineage myeloid differentiation of Kasumi-1 cells.
Figure 1.
The ee-As4S4 induced multi-lineage differentiation of Kasumi-1 cells. (A–C) Overlap distribution of CD11b, CD235a, and CD41a. The filled gray peak indicated the control group, and the concentration of ee-As4S4 In (A–C and G) is 4.0 mg/L. (D–F) The portion of differentiation marker positive cells. (G) Plot distribution of Kasumi-1 cells defined by CD11b and CD235a antibodies. The incubation time for all experiments is 72 h. *P<0.05, ***P<0.001.
After being incubated with ee-As4S4, the cells were double-stained with CD11b and CD235a antibodies to investigate whether the differentiation to multiple lineages was dependent. Results showed that the portion of double-negative cells decreased from 97.7% to 55.8% after the ee-As4S4 treatment, indicating that 44.2% of the cells underwent either erythroid or myeloid differentiation or both. Among them, 41.6% of the cells were CD11b positive, and 34.4% of the cells were CD235a positive, while 31.8% of the cells were double-positive (Figure 1G), which suggested that the cells undergoing differentiation were partially overlapped and partially independent.
Wright-Giemsa staining was conducted to observe the morphological changes of Kasumi-1 cells. It was shown that some of the cells exhibited typical megakaryocyte-like (Figure S2A-II), granulocyte-like (Figure S2A-III), and erythrocyte-like (Figure S2A-IV) phenotypes. Besides, the cells became bigger (Figure S2A-I). We also compared the FSC value in flow cytometry. Results showed that after incubation with ee-As4S4, the FSC value of the cells was increased from 5.12 × 106 to 5.54 × 106 (Figure S2B), indicating an increase in the cellular size, which is consistent with the differentiation occurrence.
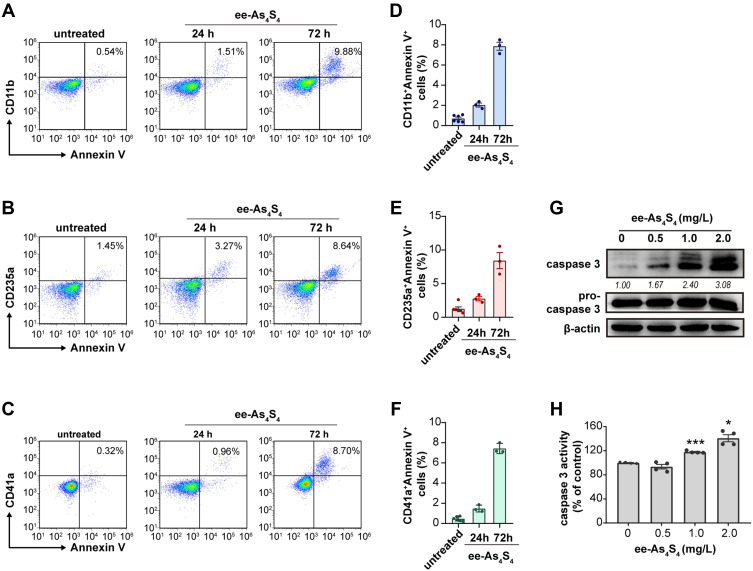
The differentiated cells are more likely to undergo apoptosis compared with immature leukemic cells.25,26 It was shown that after incubated with ee-As4S4 for 24 h or 72 h, most of the Kasumi-1 cells were double-stained with lineage markers and Annexin V conjugated with APC, indicating that the differentiated cells (CD11b+, CD235a+ or CD41a+ cells) were undergoing apoptosis (Figure 2A–F). Besides, both the intracellular content and the activity of caspase-3 were elevated significantly in a dose-dependent manner (Figure 2G and H), which clearly showed the effect of apoptosis induction of ee-As4S4. Taken above together, the ee-As4S4 induced differentiation and apoptosis in Kasumi-1 cells.
Figure 2.
The ee-As4S4 induced post-differentiation apoptosis of Kasumi-1 cells. (A–C) Representative plots distribution of the Kasumi-1 cells incubated with ee-As4S4 at 2.0 mg/L for 72 h after co-staining with Annexin V and antibodies against differentiation markers. (D–F) Proportion of Annexin V and differentiation marker double-positive cells. (G) Western blot of pro-caspase 3 and caspase 3 after incubation with ee-As4S4 for 72 h. (H) Caspase-3 activity after incubation with ee-As4S4 for 72 h. *P<0.05, ***P<0.001.
ee-As4S4 Induced Differentiation Through Reducing the Activity of HDAC
Histone deacetylases (HDAC) inhibit the acetylation of histones and repress gene expression through chromatin remodeling, inappropriate repression of genes has been linked to several types of cancer,11,27 and aberrant histone acetylation is known to play a key role in differentiation block in AML. It was reported that inhibition of HDAC resulted in transcriptional activation of genes that participated in hematopoietic differentiation,25 therefore activating these transcript factors could be a candidate strategy for differentiation therapy of leukemia.
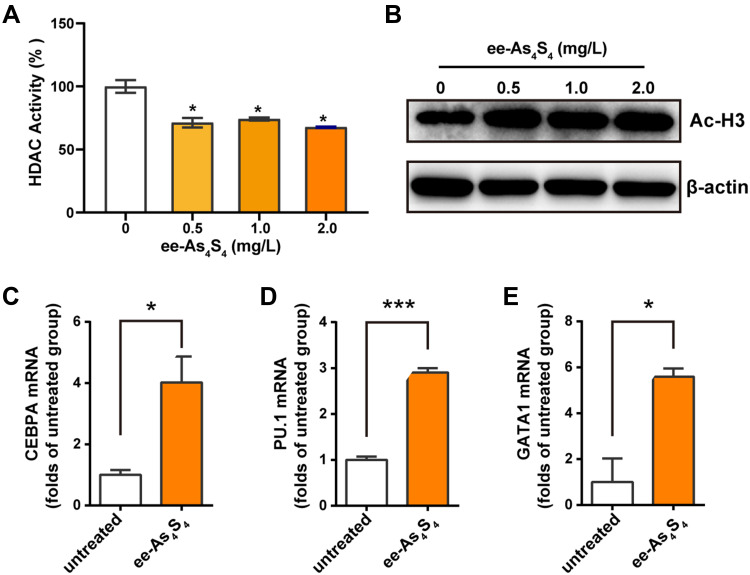
It was shown that the activity of HDAC of Kasumi-1 cells was reduced to about 70% of the untreated group after incubation with ee-As4S4 (Figure 3A); meanwhile, the acetylated histone H3 was increased along with the down-regulation of HDAC activity (Figure 3B). The inhibition of HDAC activity resulted in the transcriptional activation of genes that participated in hematopoietic differentiation, including CEBPA and PU.1, the regulators of granulocytic differentiation28,29 (Figure 3C and D), and GATA1, (Figure 3E) the master transcription factor of megakaryopoiesis and erythropoiesis.30,31
Figure 3.
Ee-As4S4 inhibited the activity of HDAC in Kasumi-1 cells. (A) HDAC activity in lysate Kasumi-1 cell that incubated with ee-As4S4 for 72 h. (B) The expression of acetylated histone H3 determined by Western Blotting in Kasumi-1 cell that incubated with ee-As4S4 for 72 h. (C–E) Relative mRNA levels of CEBPA, PU.1, and GATA1 of Kasumi-1 cells incubated with ee-As4S4 at 2.0 mg/L for 48 h. *P<0.05 ***P<0.001.
Oral Administered ee-As4S4 Induced Differentiation and Apoptosis of Leukemic Cells in AML Mice
The in vivo differentiation induction effect was investigated using a transplantable AML mouse model co-expressing AML1-ETO fusion protein and mutant HyC-KITD816V, the leukemic cells were also constitutively expressed GFP reporter gene.22 The FAB classification and WHO category and definition of the mouse model are the same as Kasumi-1 cells. Moreover, co-occurrence of kit mutation and the t (8;21) translocation is associated with a poor outcome and a higher risk of relapse, making it a refractory AML mouse model.
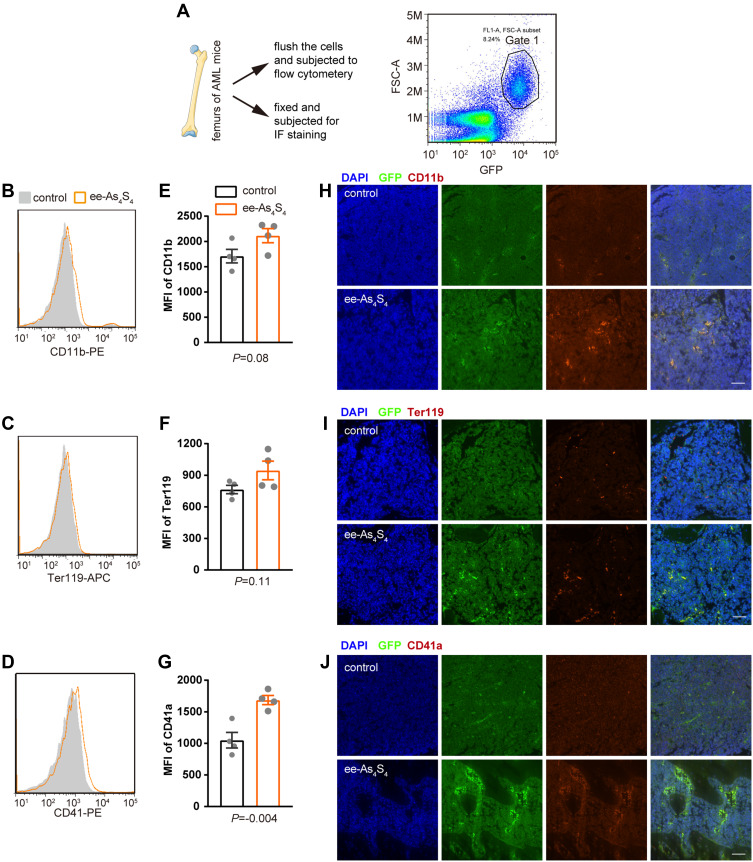
The AML mice were orally administered with ee-As4S4 once a day for 7 days and sacrificed and the femurs were collected. Bone marrow mononuclear cells were flushed from one of the femurs of each mouse and subjected to flow cytometry to analyze differentiation-associated surface markers. Ter119 and CD41 severed as an erythroid and megakaryocytic marker in mice, respectively. The AML cells were gated in Gate 1 according to the expression of GFP (Figure 4A). It was observed that ee-As4S4 increased the expression of CD11b, Ter119, and CD41 on the AML cells (Figure 4B–D). The MFI of the three markers for the leukemia cells separated from the bone marrow of AML mice were all elevated after the ee-As4S4 treatment (Figure 4E–G). The other femurs of each mouse were collected and fixed for immunofluorescence staining. The results showed that ee-As4S4 increased the expression of the three differentiation markers in the bone marrow cells (Figure 4H–J), which is consistent with the results derived from flow cytometry. Together, these results demonstrated that ee-As4S4 induced leukemia cell differentiation in vivo.
Figure 4.
The ee-As4S4 induced multi-lineage differentiation in vivo. (A) Illustration of the bone marrow assay and the gated scheme, cells in Gate 1 are leukemia cells. (B–D) Overlap distribution of CD11b, Ter119, and CD41 expression in the leukemia cells in the control group and the ee-As4S4-treated group. (E–G) Mean fluorescence intensity of the three markers in leukemia cells. (n=4) (H–J) Immunofluorescence staining of the three markers in femurs of mice from control and ee-As4S4 group.
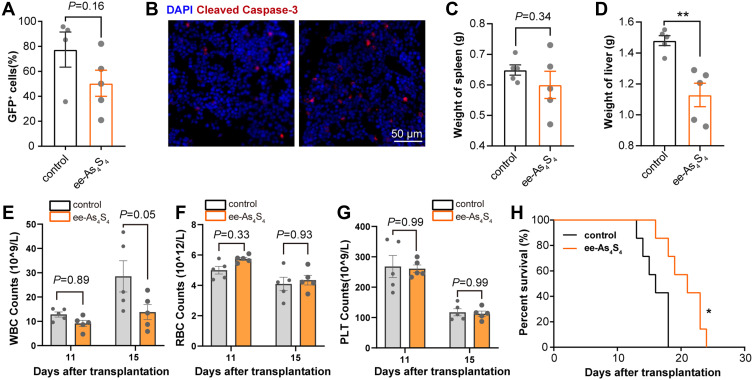
The in vivo apoptosis induction effect was next investigated. Results showed that the percentage of GFP+ cells in the bone marrow decreased from 77.4% in the control group to 53.0% with ee-As4S4 treatment (Figure 5A). Furthermore, caspase-3 was increased in the femur of ee-As4S4 treated mice, indicating that ee-As4S4 induced significant apoptosis in vivo (Figure 5B). In summary, these results indicated that ee-As4S4 induced differentiation and apoptosis in vivo, and inhibited the hyperproliferation of GFP+ leukemic cells in vivo.
Figure 5.
The ee-As4S4 induced apoptosis in vivo. (A) The percentage of GFP+ leukemia cells in the bone marrow determined by flow cytometry. (B) Immunofluorescence staining of cleaved caspase-3 in the femur of mice from control and ee-As4S4 group. (C and D) The weight of spleen and liver of AML mice (n=5). (E–G). The count of WBC, RBC, and PLT (n=5). (H) Survival curve of AML mice (n=7). *P < 0.05, **P<0.01.
Hepatomegaly and splenomegaly are typical symptoms resulting from leukemia cell extramedullary infiltration, which was relieved after the AML mice received ee-As4S4 treatment as the spleen weight was slightly reduced by the mean although not statistically (Figure 5C). The average weight of the livers was reduced to 1.13 g in the ee-As4S4 group, while that was 1.48 g in the control group (Figure 5D). It was noticed that the relief effect of splenomegaly was not as strong as that of hepatomegaly; this could be due to that ee-As4S4 was administered orally and likely to be metabolized through the liver in priority, therefore showing a stronger effect on hepatomegaly.
The peripheral hemogram of acute leukemia can be characterized by an obvious increase of white blood cells (WBC), and higher WBC counts in AML patients were associated with an increased mortality rate.32 Besides, anemia and thrombocytopenia, ie, reduction of red cell count (RBC) and platelet (PLT), were also observed in AML patients. When comparing the blood cell counts detected on Day 11 and Day 15 after transplantation, it was found that the WBC increased and the RBC and PLT decreased in the control group. Due to the apoptosis-induction effect, the peripheral WBC was significantly reduced by ee-As4S4 administration in the mice, suggesting the remission of AML (Figure 5E). The composition of WBC on Day 15 was shown in Figure S5A since there was a significant difference between the two groups. The differentiated counts of WBC including neutrophils, lymphocytes, monocytes, eosinophils, and basophils were all decreased along with the decrease of total WBC counts.
When compared with the control group, the ee-As4S4 treated group showed the tendency of decreased WBC. The RBC in the ee-As4S4 group was 5.74×109/L and 4.34 ×109/L while that was 4.99×109/L and 4.09 ×109/L in the control group on Day 11 and Day 15, revealing an anemia-alleviating effect (Figure 5F). However, the ee-As4S4 did not affect the PLT count (Figure 5G). Due to the reduced leukemic cells, relief of extramedullary infiltration, and improvement of hemogram, ee-As4S4 treatments prolonged the survival of AML mice compared with the control group and the r-As4S4 group (Figures 5H, S3 and S4). The median survival was 21 days for the ee-As4S4 group, while that was about 16 days for the control group.
In vivo Safety of Multiple Administration of ee-As4S4
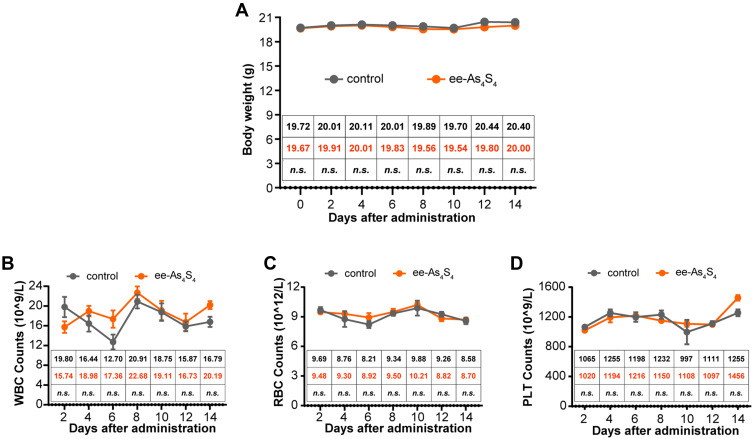
To assess the in vivo safety of ee-As4S4, healthy C57BL/6 mice were orally administered with ee-As4S4 or ddH2O twice a day for 14 days. Results showed that there were no significant differences detected in the bodyweight in the two groups (Figure 6A). Besides, the counts of WBC as well as its composition, RBC, and PLT were at a similar level among the different groups (Figures 6B–D and S5B), indicating that the oral administration of ee-As4S4 had no significant toxicity in the healthy mice, exhibiting good biosafety as a therapeutic agent.
Figure 6.
The in vivo safety of ee-As4S4 in healthy mice. (A) The bodyweight of mice for each group (n=6). (B–D) The counts of WBC (B), RBC (C), and PLT (D) were analyzed after the administration for 14 days (n=6). Mean of body weight, WBC, RBC, and PLT count were listed in the inserted table, the mean of the control group was black-colored, and that of the ee-As4S4 treated group is Orange-colored. “n.s.” indicates no significance between the two groups in the same day.
Discussion
Although existing chemotherapies can help to reach complete remission for initial treatment of AML, relapse and drug resistance still take about six in ten patients within five years.33,34 Therefore, developing alternative medicines is still an unmet health demand in this field. Differentiation therapy is an attractive and promising strategy for AML, which gained the eyesight of researchers since 1970s.5 Great success has been obtained in APL, and research interests are also increased in the treatment of other cancer types, especially leukemia; however, clinically available options are still in lack.
Nanoparticle-based drug delivery systems have shown great potentials in the development of novel therapeutics against blood cancer.35 For example, Vyxeos (CPX-351) is a liposomal formulation of cytarabine and daunorubicin, which demonstrate a striking higher response rate with improved overall survival among secondary AML (including MRC-AML) and therapy-related AML compared to the traditional 3 + 7 regimen.36,37 The therapeutic effects of Vyxeos come from the synergistic cytotoxicity of two chemotherapeutic drugs. In the present study, by utilizing nanoparticle-based technology, ee-As4S4 was prepared and it displayed the novel function of differentiation induction in the AML cells in vitro and in vivo. Moreover, the mouse model used in this study is a refractory one, which was unable to benefit from cytosine arabinoside treatment.38 These results strongly suggested that ee-As4S4 could serve as a therapeutic for refractory AML patients.
The differentiation induction effect of the ee-As4S4 was first noted in chronic myeloid leukemia cells K562, as the erythroid differentiation of K562 cells was induced through the autophagic elimination of BCR-ABL,20 and the megakaryocytic differentiation was induced through the inhibition of HDAC activity and transcriptional activation of RUNX1.21 The preference of ee-As4S4 beyond the r-As4S4 was clearly shown in this work, indicating that the nanoformulation may lead to new therapeutic opportunities. In the mechanistic study, we showed that ee-As4S4 was able to activate the expression of myeloid differentiation-associated transcript factors and induce the differentiation of AML cells, meanwhile the activity of HDAC was reduced, which implied that ee-As4S4 might serve as a potent treatment option for different subtypes of AML.39
The experimental evidence both in vitro and in vivo also showed that ee-As4S4 was capable of inducing multi-lineage differentiation of M2b subtypes of AML cells. We would like to address that the in vitro and in vivo effects of differentiation induction were consistent, though the proportion of the cells undergoing differentiation was different. The difference between the in vitro and in vivo data could be attributed to two reasons: 1. the cells were of different species origin, and the performance might be different; 2. unlike in vitro culture, cells undergoing differentiation could be rapidly cleared by phagocytes due to the apoptosis, resulting in lower expression of differentiation markers in the harvested cells.
Conclusion
In conclusion, ee-As4S4 was able to induce multi-lineage differentiation including granulocytic, erythroid, and megakaryocytic differentiation through reducing the activity of HDAC, which led to apoptosis of the AML cells. The oral administration of ee-As4S4 prolonged the survival of the refractory AML mice by inducing the in situ leukemia cells in the bone marrow as well as the extramedullary infiltrating AML cells to undergo differentiation and apoptosis.
Funding Statement
This work was supported by the National Key R&D Program of China (2017YFA0205504).
Ethics Approval
The in vivo study was performed in accordance with the regulations of Chinese Academy of Medical Sciences Standing Committee on animal experiments and was approved by the Institutional Animal Care and Use Committee (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences, and Peking Union Medical College, Beijing, China). The use of the Kasumi-1 cells was approved by the research ethics committee of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences.
Disclosure
The authors declare no competing interest in this work.
References
- 1.Bose P, Konopleva MY. ORY-1001: overcoming the differentiation block in AML. Cancer Cell. 2018;33(3):342–343. doi: 10.1016/j.ccell.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 2.Dhall A, Zee BM, Yan F, Blanco MA. Intersection of epigenetic and metabolic regulation of histone modifications in acute myeloid leukemia. Front Oncol. 2019;9:432. doi: 10.3389/fonc.2019.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiNardo CD, Cortes JE. Mutations in AML: prognostic and therapeutic implications. Hematology Am Soc Hematol Educ Program. 2016;2016(1):348–355. doi: 10.1182/asheducation-2016.1.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sykes DB, Kfoury YS, Mercier FE, et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell. 2016;167(1):171–186.e15. doi: 10.1016/j.cell.2016.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de The H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18(2):117–127. doi: 10.1038/nrc.2017.103 [DOI] [PubMed] [Google Scholar]
- 6.Sell S. Leukemia: stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev. 2005;1(3):197–205. doi: 10.1385/scr:1:3: [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Liu L, Jin J, Lou Y, Mills K. All-trans retinoic acid plus arsenic trioxide versus all-trans retinoic acid plus chemotherapy for newly diagnosed acute promyelocytic leukemia: a meta-analysis. PLoS One. 2016;11(7):e0158760. doi: 10.1371/journal.pone.0158760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai SF, Levine RL. Genetic and epigenetic determinants of AML pathogenesis. Semin Hematol. 2019;56(2):84–89. doi: 10.1053/j.seminhematol.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal-Singh S, Isken F, Agelopoulos K, et al. Genome-wide analysis of histone H3 acetylation patterns in AML identifies PRDX2 as an epigenetically silenced tumor suppressor gene. Blood. 2012;119(10):2346–2357. doi: 10.1182/blood-2011-06-358705 [DOI] [PubMed] [Google Scholar]
- 10.Sauer T, Arteaga MF, Isken F, et al. MYST2 acetyltransferase expression and Histone H4 Lysine acetylation are suppressed in AML. Exp Hematol. 2015;43(9):794–802.e4. doi: 10.1016/j.exphem.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 11.Göttlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. doi: 10.1038/35106079 [DOI] [PubMed] [Google Scholar]
- 13.Ungerstedt JS. Epigenetic modifiers in myeloid malignancies: the role of histone deacetylase inhibitors. Int J Mol Sci. 2018;19(10):3091. doi: 10.3390/ijms19103091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Xu H. Development of realgar nanotherapeutics for cancer treatments. In: Xu H, Gu N, editors. Nanotechnology in Regenerative Medicine and Drug Delivery Therapy. Springer Singapore; 2020:421–454. [Google Scholar]
- 15.Ma Q, Wang C, Li X, et al. Fabrication of water-soluble polymer-encapsulated As4S4 to increase oral bioavailability and chemotherapeutic efficacy in AML mice. Sci Rep. 2016;6(1):29348. doi: 10.1038/srep29348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu DP, Qiu JY, Jiang B, et al. Tetra-arsenic tetra-sulfide for the treatment of acute promyelocytic leukemia: a pilot report. Blood. 2002;99(9):3136–3143. doi: 10.1182/blood.V99.9.3136 [DOI] [PubMed] [Google Scholar]
- 17.Zhu HH, Huang XJ, Arsenic O. Retinoic acid for non-high-risk acute promyelocytic leukemia. New Engl J Med. 2014;371(23):2239–2241. doi: 10.1056/NEJMc1412035 [DOI] [PubMed] [Google Scholar]
- 18.Zhu HH, Liu YR, Jia JS, Qin YZ, Zhao XS, Lai YY. Oral arsenic and all-trans retinoic acid for high-risk acute promyelocytic leukemia. Blood. 2018;131(26):2987–2989. doi: 10.1182/blood-2018-02-834051 [DOI] [PubMed] [Google Scholar]
- 19.Zhu HH, Wu DP, Du X, et al. Oral arsenic plus retinoic acid versus intravenous arsenic plus retinoic acid for non-high-risk acute promyelocytic leukaemia: a non-inferiority, randomised Phase 3 trial. Lancet Oncol. 2018;19(7):871–879. doi: 10.1016/S1470-2045(18)30295-X [DOI] [PubMed] [Google Scholar]
- 20.Wang T, Wen T, Li HM, et al. Arsenic sulfide nanoformulation induces erythroid differentiation in chronic myeloid leukemia cells through degradation of BCR-ABL. Int J Nanomed. 2019;14:5581–5594. doi: 10.2147/Ijn.S207298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia M, Wang T, Xu S, et al. Arsenic sulfide nanoformulation induces megakaryocytic differentiation through histone deacetylase inhibition. Adv Ther. 2019;3(5):1900151. [Google Scholar]
- 22.Meng J, Ge Y, Xing H, et al. Synthetic CXCR4 antagonistic peptide assembling with nanoscaled micelles combat acute myeloid leukemia. Small. 2020;16(31):e2001890. doi: 10.1002/smll.202001890 [DOI] [PubMed] [Google Scholar]
- 23.Larizza L, Magnani I, Beghini A. The Kasumi-1 cell line: a t (8;21)-kit mutant model for acute myeloid leukemia. Leuk Lymphoma. 2005;46(2):247–255. doi: 10.1080/10428190400007565 [DOI] [PubMed] [Google Scholar]
- 24.Yan D, Wei H, Lai X, et al. Co-delivery of homoharringtonine and doxorubicin boosts therapeutic efficacy of refractory acute myeloid leukemia. J Control Release. 2020;327:766–778. doi: 10.1016/j.jconrel.2020.09.031 [DOI] [PubMed] [Google Scholar]
- 25.Altucci L, Rossin A, Hirsch O, et al. Rexinoid-triggered differentiation and tumor-selective apoptosis of acute myeloid leukemia by protein kinase A-mediated desubordination of retinoid X receptor. Cancer Res. 2005;65(19):8754–8765. doi: 10.1158/0008-5472.CAN-04-3569 [DOI] [PubMed] [Google Scholar]
- 26.Martin SJ, Bradley JG, Cotter TG. HL-60 cells induced to differentiate towards neutrophils subsequently die via apoptosis. Clin Exp Immunol. 1990;79(3):448–453. doi: 10.1111/j.1365-2249.1990.tb08110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anastas JN, Zee BM, Kalin JH, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36(5):528–544.e10. doi: 10.1016/j.ccell.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 28.Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPalpha is required for induction of granulocytic differentiation. Blood. 2003;102(4):1267–1275. doi: 10.1182/blood-2003-02-0477 [DOI] [PubMed] [Google Scholar]
- 29.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170 [DOI] [PubMed] [Google Scholar]
- 30.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–3376. doi: 10.1038/sj.onc.1205326 [DOI] [PubMed] [Google Scholar]
- 31.Shin E, Jeong JG, Chung H, et al. The Gata1(low) murine megakaryocyte-erythroid progenitor cells expand robustly and alter differentiation potential. Biochem Biophys Res Commun. 2020;528(1):46–53. doi: 10.1016/j.bbrc.2020.04.143 [DOI] [PubMed] [Google Scholar]
- 32.Ram R, Amit O, Zuckerman T, et al. Venetoclax in patients with acute myeloid leukemia refractory to hypomethylating agents—a multicenter historical prospective study. Ann Hematol. 2019;98(8):1927–1932. doi: 10.1007/s00277-019-03719-6 [DOI] [PubMed] [Google Scholar]
- 33.Molica M, Breccia M, Foa R, Jabbour E, Kadia TM. Maintenance therapy in AML: the past, the present and the future. Am J Hematol. 2019;94(11):1254–1265. doi: 10.1002/ajh.25620 [DOI] [PubMed] [Google Scholar]
- 34.Short NJ, Konopleva M, Kadia TM, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506–525. doi: 10.1158/2159-8290.Cd-19-1011 [DOI] [PubMed] [Google Scholar]
- 35.Wan Z, Sun R, Moharil P, et al. Research advances in nanomedicine, immunotherapy, and combination therapy for leukemia. J Leukoc Biol. 2021;109(2):425–436. doi: 10.1002/jlb.5mr0620-063rr [DOI] [PubMed] [Google Scholar]
- 36.Alfayez M, Kantarjian H, Kadia T, Ravandi-Kashani F, Daver N. CPX-351 (vyxeos) in AML. Leuk Lymphoma. 2020;61(2):288–297. doi: 10.1080/10428194.2019.1660970 [DOI] [PubMed] [Google Scholar]
- 37.Lancet JE, Uy GL, Cortes JE, et al. CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J Clin Oncol. 2018;36(26):2684–2692. doi: 10.1200/jco.2017.77.6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S, Zhang M, Fang X, et al. A novel CD123-targeted therapeutic peptide loaded by micellar delivery system combats refractory acute myeloid leukemia. J Hematol Oncol. 2021;14(1):193. doi: 10.1186/s13045-021-01206-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27(32):5459–5468. doi: 10.1200/jco.2009.22.1291 [DOI] [PubMed] [Google Scholar]