Abstract
The unique functionality of Akkermansia muciniphila in gut microbiota indicates it to be an indispensable microbe for human welfare. The importance of A. muciniphila lies in its potential to convert mucin into beneficial by-products, regulate intestinal homeostasis and maintain gut barrier integrity. It is also known to competitively inhibit other mucin-degrading bacteria and improve metabolic functions and immunity responses in the host. It finds a pivotal perspective in various diseases and their treatment. It has future as a promising probiotic, disease biomarker and therapeutic agent for chronic diseases. Disease-associated dysbiosis of A. muciniphila in the gut microbiome makes it a potential candidate as a biomarker for some diseases and can provide future theranostics by suggesting ways of diagnosis for the patients and best treatment method based on the screening results. Manipulation of A. muciniphila in gut microbiome may help in developing a novel personalized therapeutic action and can be a suitable next generation medicine. However, the actual pathway governing A. muciniphila interaction with hosts remains to be investigated. Also, due to the limited availability of products containing A. muciniphila, it is not exploited to its full potential. The present review aims at highlighting the potential of A. muciniphila in mucin degradation, contribution towards the gut health and host immunity and management of metabolic diseases such as obesity and type 2 diabetes, and respiratory diseases such as cystic fibrosis and COVID-19.
Keywords: Akkermansia muciniphila, Biomarker, COVID-19, Gut microbiome, Host immunity, Mucus degradation, Obesity, Probiotic, Therapeutic, Type 2 diabetes
Introduction
Akkermansia muciniphila (A. muciniphila) is a recently discovered member of commensal gut microbiota and constitutes a new genus of the phylum Verrucomicrobia (Derrien et al. 2004). It is oval, strictly anaerobic, non-motile and Gram-negative bacteria that do not form endospores. It has circular genome of 2,664,102 base pairs, sharing 29% gene similarity with phylum Verrucomicrobia (van Passel et al. 2011). As unveiled by whole-genome sequencing, its proteome consists of 5644 unique proteins (Guo et al. 2017). A. muciniphila colonizes the gastrointestinal tracts at an early stage through human milk and accounts for 1–4% of total gut microbiota (Collado et al. 2008). The abundance of A. muciniphila in caecum is ubiquitous in infants and healthy adults. Besides the large intestine, it is also found in the lining of the lungs and saliva.
The importance of A. muciniphila lies in its potential to degrade mucin, the significant component in mucus. It consumes mucin as a carbon and nitrogen source during its life cycle and metabolism. The optimum temperature and pH for its growth are 37 °C and 6.5 respectively. A recent study showed that despite being a strict anaerobe, it could sustain lower amounts of oxygen (Ouwerkerk et al. 2016). This property is quite similar to other microbes in the intestine, especially anaerobes, such as Bifidobacterium adolescentis and Bacteroides fragilis, which can tolerate ambient amounts of oxygen for 48 h (Ouwerkerk et al. 2016). A. muciniphila is known to competitively inhibit other mucin-degrading bacteria and improve metabolic functions and immunity responses in the host, making it a suitable candidate as a probiotic (Belzer and de Vos 2012). A. muciniphila was even found to be effective in treatment of inflammatory bowel diseases and cancer (Png et al. 2010; Chen et al. 2020a). The present review aims at studying the potential of A. muciniphila in mucin degradation, contribution towards the gut health and host immunity, and management of metabolic diseases such as obesity and type 2 diabetes, and respiratory diseases such as cystic fibrosis and COVID-19.
A. muciniphila in gut microbiome
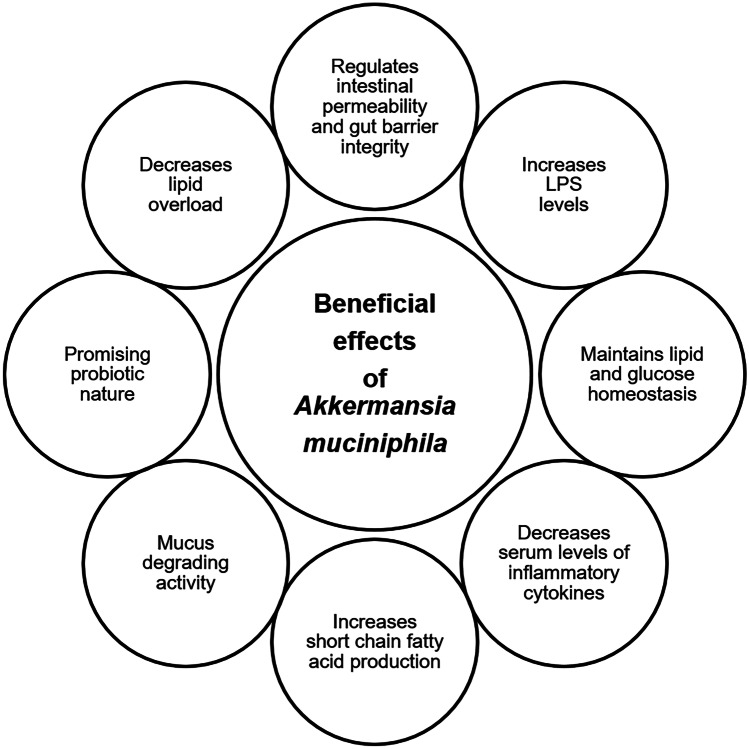
The prevalence of A. muciniphila has been connected with a healthy gut, and therefore, its richness is inversely linked to numerous disease conditions (Jakobsson et al. 2015). An investigation of its relationships with the hosts revealed A. muciniphila to enhance the intestinal barrier function in mice (Shin et al. 2014). Colonization by A. muciniphila culminated in transcriptome alterations, leading to a rise in the genetic expression linked with immunogenicity. Outer membrane proteins of A. muciniphila were discovered to play a function in controlling immunological responses. One of the outer membrane proteins was recently discovered (Amuc-1100) (Ottman et al. 2017b). The work demonstrated that the outer membrane pili-like protein is essential in immunological modulation and the increase of trans-epithelial resistance. A. muciniphila performed a function in regulating metabolic endotoxemia and adipose tissue metabolism. Several investigations have consciously or inadvertently discovered the existence of Akkermansia-like spp. in regions of the human body other than the colon, where A. muciniphila could also have vital activities. The physiology and environmental factors of Akkermansia-like spp. in distinct anatomic locations of the digestive tract allow us to evaluate the ability of A. muciniphila to colonize and be productive at all these niches. Various beneficial effects of A. muciniphila in the human microbiome are presented in Fig. 1.
Fig. 1.
Beneficial effects of A. muciniphila in human microbiome
Mucin-degrading activity of A. muciniphila
Mucus consists of heavily glycosylated mucin-2 (MUC2), an oligosaccharide composed of various amino sugars and monosaccharide sugars, including N-acetyl-D-galactosamine (GalNAc), N-acetyl-D-glucosamine (GlcNAc), D-galactose and L-fucose (Ottman et al. 2017a). In many cases, these sugars are further substituted with acetate, phosphate and sulfate groups. Mucin has defining roles like a lubricant for food transport over membranes and provides selective permeability that allows the flow of nutrients to epithelial cells. It also acts as the first line of defence against mechanical damage, pathogens, and toxins and provides a surface layer to bacteria for its growth, adhesion and protection (Cone 2009; Johansson et al. 2013). However, some bacterial species of human microbiota release inflammatory toxins, which increase the permeability of the mucus layer and ultimately decrease its barrier property (Jakobsson et al. 2015). It has been concluded that bacterial colonies reside only at the outer layer of the intestinal tract. In contrast, the inner layer intends to keep the bacteria at bay from the epithelial cells to enforce immune tolerance to the guts by transporting IgA and antimicrobial proteins (Johansson et al. 2008, 2011).
It is observed that A. muciniphila can maintain an exciting microbial relationship in the host intestine by converting mucin into beneficial by-products (Ottman et al. 2017a, b). Recent studies showed that A. muciniphila could be grown on a synthetic medium where mucin can be replaced with media containing glucose, threonine, peptone and GlcNAc (Plovier et al. 2016). The amino group of sugars promotes the growth of bacteria in the presence of casitone, tryptone, yeast extract and peptone. One of the essential factors that account for the proliferation of A. muciniphila is glucose-6-phosphate, one of the constituents of mucin known to promote the adaptation of mucosal niche (van der Ark et al. 2018). In order to study substrate uptake abilities of A. muciniphila, few studies were conducted on a genome-scale metabolic model to demonstrate amino acids auxotrophy, sugar degrading capacities and vitamin biosynthesis (Ottman et al. 2017a). These experiments have also been validated through in vivo experiments in which A. muciniphila has been shown to proficiently utilize mucin-derived monosaccharide sugars and amino sugars. It has been found that the uptake of mucin-derived sugars and non-mucin sugar glucose by A. muciniphila is enhanced in a mucin-rich environment which indicates the need of mucin-derived components for the optimal growth of bacteria. In vivo experiments have also suggested that A. muciniphila may have galactose metabolism; however, mucin-derived components are necessary for its growth.
Transcriptomic analysis of A. muciniphila under mucin-rich and mucin-depleted conditions showed differential gene expression suggesting a global change in cellular functions (Shin et al. 2019). Out of 1126 differentially expressed genes (DEGs), 583 genes were upregulated while 543 were downregulated in mucin-rich conditions as compared to mucin-depleted conditions. The upregulated genes were significantly related to hydrolase activity acting on glycosidic bonds and their transporters, thereby confirming the activity of mucin-degrading genes under mucin-rich conditions. Thus, the genes that encode mucin-degrading enzymes, such as sulfatases, galactosidases, acetyl-glucosaminidase, neuraminidases and L-fucosidase transporters were upregulated in mucin-rich conditions. Furthermore, their downregulation in mucin-depleted medium determined the importance of its role in mucin-degradation. The catabolic glycolysis pathway is also correlated with mucin-degradation pathways (Shin et al. 2019). Thus, under mucin-depleted conditions, genes involved in glycolysis and energy metabolism, such as NADH dehydrogenase, succinate dehydrogenase and ATP synthase, either showed similar expression levels or were upregulated significantly. At the same time, there were few exceptions, including one ATP-dependent 6-phosphofructokinase gene (Amuc_1481), two enolase genes (Amuc_844, Amuc_1184) and one dihydrolipoyl dehydrogenase gene (Amuc_1689).
A. muciniphila in host immunity and probiotic nature
The symbiotic relationship between the gut microbiota and host determines the normal physiology, immunity and pathogen susceptibility of an individual. The interplay between host and gut microbiome that helps in pathogen displacement, regulating immune response and anti-inflammatory pathways, is a vital phenomenon. There is abundant evidence in the literature on mucin-utilizing A. muciniphila conferring immunity (Tummler and Puchelle 1997; Plovier et al. 2016; Ottman et al. 2017a). Many mucin degradation pathways regulate the host pathway by signalling through tumour necrosis factor α (TNF-α), interferon γ, interleukins-10 (IL-10) and interleukins-4 (IL-4) (Derrien et al. 2011; Andersson et al. 2012; Collado et al. 2012). Decreased levels of anti-inflammatory cytokines (IL-10 and IL-4) induced interleukins, while increased proinflammatory cytokines (TNF-α and IFN-γ) causing rapid proliferation of A. muciniphila. An increase in 2-arachidonoylglycerol levels was noted post A. muciniphila treatment, reducing inflammation (Gunderson and Kopito 1994; Everard et al. 2013). The secreted proteins from bacteria interact with host immune cells to induce signalling pathways that exhibit anti-inflammatory and immunomodulatory activity (Sánchez et al. 2008, 2010; Bernardo et al. 2012; Ruiz et al. 2014). The extracellular material secreted from it activates the downstream signalling pathway like toll-like receptors 2 (TLR2) (Ottman et al. 2017b). Amuc_110, a specific protein in the outer membrane, recapitulates the effect of bacteria on TLR2 activation and improves the barrier integrity of intestines (Dean and Annilo 2005; Belzer and de Vos 2012; Plovier et al. 2016; Ottman et al. 2017b). However, it is still unknown how Amuc_110 protein is regulated in the presence of a dynamic mucosal environment. The gene encoding Amuc_110 protein is highly regulated in a mucin-depleted environment (Plovier et al. 2016). Amuc_110 has also shown its ability to exert a probiotic effect on diet-induced obesity and was present in the extracellular proteins of A. muciniphila as well (Plovier et al. 2016). To conclude, A. muciniphila is inversely correlated with inflammatory conditions and helps in epithelial barrier integrity by stimulating anti-inflammatory pathways (Gunderson and Kopito 1994; Shin et al. 2014; Cantarel et al. 2015; Caesar et al. 2015; Schneeberger et al. 2015).
The composition and functioning of the human gut microbiota are primarily proportional to nutritional accessibility of microbiota either obtained from a host or food (Zoetendal et al. 2012; Nicholson et al. 2012; Salonen and de Vos 2014). A. muciniphila is one of the good bacteria of the human gut microbiota. The presence of A. muciniphila in the intestinal mucus layer indicates it to be involved in gut regulation and host metabolism. It exists in a symbiotic relationship with the mucosal layer, and its abundance is greatly affected by the nutrients present in the mucus layer located around the intestinal epithelial cells. Its presence also supports other beneficial bacteria in the gut microbiome. A. muciniphila catabolizes mucins and turns them into short-chain fatty acids (SCFAs), including acetate, which other beneficial bacteria exploit, such as Firmicutes, to produce butyrate, a vital source of energy for the cells lining the gut. The production of SCFAs from the breakdown of mucin supplies energy to the goblet cells, which are responsible for secreting mucins. Furthermore, the consumption of specific dietary fibres can increase the abundance of this friendly bacteria, which helps thicken the mucus lining the gut. This strengthens the gut lining and improves gut barrier function and may, in turn, help in preventing weight gain. Chelakkot et al. demonstrated the role of Amuc_1100, isolated from A. muciniphila, in AMP-activated protein kinase (AMPK) activation mechanism, thereby improving gut integrity (Chelakkot et al. 2018). Amuc_1100 has been implicated in enhancing the expression of tight junction protein-1 (Tjp-1) and occludin (Li et al. 2016), thereby contributing to the gut barrier function. Thus, the presence of A. muciniphila in the mucus of the intestine regulates intestinal homeostasis and its integrity barrier through host signalling pathways (Derrien et al. 2004; Ottman et al. 2017a).
A. muciniphila dysbiosis associated with disease states and its management
The gut microbiome of healthy people is quite diverse. Gut microflora through microbial antigens and metabolites is the master regulator of innate and adaptive immunity. Disease associated dysbiosis results in induction, training and function of immune system. The disturbance of the mucus layer by any means may lead to inflammation and increase the risk of infection. Even slight variance in intestinal flora may be associated with sensitivity and severity of a disease. Therefore, gut microbes find use as potential diagnostic biomarkers by studying their abundance in different diseases.
A. muciniphila has been linked with wide range of diseases and disorders such as type 2 diabetes (Tilg and Moschen 2014), alcoholic steatohepatitis (ASH) (Grander et al. 2018), appendicitis (Swidsinski et al. 2011), obesity (Dao et al. 2016), atopic diseases (Drell et al. 2015), colorectal cancer (Weir et al. 2013), autism (Wang et al. 2011), inflammatory bowel disease (Png et al. 2010), cystic fibrosis (Hayden et al. 2019), and COVID-19 (Yeoh et al. 2021) (Table 1). Various studies demonstrated A. muciniphila to be negatively correlated with inflammatory bowel diseases (Png et al. 2010; Rajilić-Stojanović et al. 2013), appendicitis (Swidsinski et al. 2011), obesity (Karlsson et al. 2012; Dao et al. 2016) and type 2 diabetes (Tilg and Moschen 2014). Lower abundance of A. muciniphila is commonly observed in the majority of metabolic disorders while higher abundance is seen in few cases like colorectal cancer (Yu et al. 2017).
Table 1.
Altered abundance of A. muciniphila in various disease states in humans
| Disease | A. muciniphila abundance | Detection method | Sample type | References |
|---|---|---|---|---|
| Allergic asthma | Reduced | qPCR | Faeces | (Demirci et al. 2019) |
| Asthma | Reduced | 16S rRNA sequencing | Faeces | (Michalovich et al. 2019) |
| Alcoholic steatohepatitis (ASH) | Reduced | 16S rRNA sequencing | Faeces | (Grander et al. 2018) |
| Atopy | Reduced | 16S rRNA sequencing | Faeces | (Candela et al. 2012) |
| Atopy | Reduced | Pyrosequencing | Faeces | (Drell et al. 2015) |
| Autism | Elevated | bTEFAP | Faeces | (De Angelis et al. 2013) |
| Autism | Reduced | qPCR | Faeces | (Wang et al. 2011) |
| Clostridium difficile infection | Elevated | qPCR | Faeces | (Vakili et al. 2020) |
| Colorectal cancer | Elevated | 16S rRNA sequencing | Faeces | (Weir et al. 2013) |
| Colorectal cancer | Elevated | qPCR | Tissue biopsy | (Mira-Pascual et al. 2015) |
| Crohn’s disease | Reduced | qPCR | Tissue biopsy | (Png et al. 2010) |
| Crohn’s disease | Reduced | 16S rDNA pyrosequencing | Faeces | (Opstelten et al. 2016; Malham et al. 2019) |
| Cystic fibrosis | Reduced | 16S-based tag-encoded FLX amplicon pyrosequencing (bTEFAP) | Faeces | (Hoffman et al. 2014) |
| Cystic fibrosis | Reduced | Metagenomic sequencing | Faeces | (Hayden et al. 2019) |
| COVID-19 | Elevated | Shotgun sequencing | Faeces | (Yeoh et al. 2021) |
| Oesophageal cancer | Elevated | 16S rRNA sequencing | Tissue biopsy | (Snider et al. 2019) |
| Hyperlipidaemia | Reduced | 16S rRNA sequencing | Faeces | (Gargari et al. 2018) |
| Microscopic colitis | Reduced | 16S rDNA pyrosequencing | Faeces | (Fischer et al. 2015) |
| Multiple system atrophy | Elevated | Metagenomic sequencing | Faeces | (Wan et al. 2019) |
| Obesity | Reduced | qPCR | Faeces | (Marvasti et al. 2020) |
| Obesity | Elevated | qPCR | Faeces | (Remely et al. 2015) |
| Parkinson’s disease | Elevated | qPCR | Faeces | (Unger et al. 2016) |
| Parkinson’s disease | Elevated | Shotgun sequencing | Faeces | (Bedarf et al. 2017) |
| Prediabetes | Reduced | 16S rRNA sequencing | Faeces | (Allin et al. 2018) |
| Psoriasis | Reduced | 16S rDNA pyrosequencing | Faeces | (Tan et al. 2018) |
| Pulmonary arterial hypertension | Reduced | Shotgun sequencing | Faeces | (Kim et al. 2020) |
| Pulmonary tuberculosis | Reduced | Metagenomic sequencing | Faeces | (Hu et al. 2019) |
| Schizophrenia | Elevated | 16S rRNA sequencing | Faeces | (Xu et al. 2020a) |
| Schizophrenia | Elevated | Shotgun sequencing | Faeces | (Zhu et al. 2020) |
| Spleen deficiency syndrome | Reduced | qPCR | Faeces | (Peng et al. 2020) |
| Type 1 diabetes | Reduced | qPCR | Faeces | (Fassatoui et al. 2019) |
| Type 2 diabetes | Elevated | Metagenomic sequencing | Faeces | (Chelakkot et al. 2018) |
| Type 2 diabetes | Elevated | Shotgun sequencing | Faeces | (Qin et al. 2012) |
| Type 2 diabetes | Reduced | 16S rRNA sequencing | Urine | (Liu et al. 2017) |
| Type 2 diabetes | Reduced | Metagenomic sequencing | Faeces | (Zhong et al. 2019) |
| Type 2 diabetes | Reduced | qPCR | Faeces | (Fassatoui et al. 2019) |
| Ulcerative colitis | Reduced | qPCR | Tissue biopsy | (Png et al. 2010) |
| Ulcerative colitis | Reduced | MiSeq sequencing | Faeces | (Bajer et al. 2017) |
| Ulcerative colitis | Reduced | 16S rRNA sequencing | Faeces | (Malham et al. 2019) |
Recently, an association between A. muciniphila and metastasis of lymph nodes in lung adenocarcinoma was reported (Chen et al. 2020a). The relative abundance of A. muciniphila was observed to be greater in metastasis cohort (0.057) than the non-metastasis cohort (0.023) pointing towards its potential as a promising biomarker to predict lymph node metastasis. Likewise, a study to investigate whether A. muciniphila could enhance the antitumor effect of cisplatin (cis-diamminedichloroplatinum; CDDP) was conducted (Chen et al. 2020b). It was found that when A. muciniphila was combined with cisplatin, the growth of tumour volume slowed and the changes of tumour pathomorphology significantly improved. At molecular level, upregulation of factor-associated suicide (Fas) proteins and downregulation of p53, ki-67 and Fas ligand (FasL) proteins were also observed. Several proinflammatory factors (TNF-α, IFN-γ and IL-6) were induced while the expression of CD4+CD25+Foxp3+ Treg was suppressed indicating the role of A. muciniphila in regulating immune inflammatory microenvironment in reversion of tumour growth and tumour immune escape. Also, the levels of IFI27l2 and IGFBP7, two most differentially expressed genes in lung cancer, were found to be increased because of the combined treatment with A. muciniphila and CDDP. Signalling pathways such as JAK-STAT, FOXO, cytokine-cytokine receptor interaction, PI3K-Akt, Th17 and cell differentiation were associated with the antitumor effect of A. muciniphila and CDDP. The study suggested that A. muciniphila combined with CDDP provides good symbiotic environment to achieve maximum therapeutic efficacy of antitumor drugs. The human gut microbiome could therefore, be responsible for differences in drug response of individuals paving way for personalized therapeutics to significantly improve human health care. Thus, by regulating the abundance of A. muciniphila in the gut microbiome in a personalized manner, early treatment of related diseases may be facilitated. The microbiome in general, and A. muciniphila in particular, can act as a biomarker of these diseased states and provide future theranostics by suggesting ways of disease diagnosis and treatment (Morgan and Huttenhower 2012).
A. muciniphila and metabolic diseases
Decreased abundance of A. muciniphila in obesity
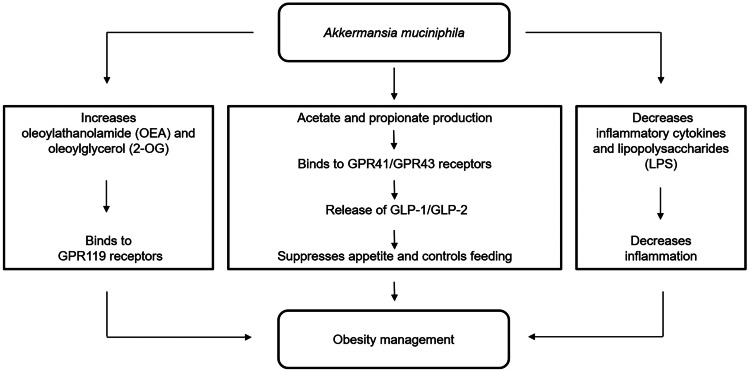
World Health Organization (WHO) defines obesity as abnormal fat accumulation leading to health implications like type 2 diabetes, fatty liver disease and hypertension. Being a multifactorial disorder, not only does it result in fatal complications like cardiovascular diseases and psychological effects but also challenges one to perform everyday tasks with ease. It is a low-grade inflammatory metabolic disorder resulting in higher levels of inflammatory cytokines such as IL-6, TNF-α and hypersensitive C-reactive protein (hs-CRP). Inflammation in obesity leads to significant changes in gut microbiota, for example, increase in the abundance of Firmicutes, Bifidobacterium spp. and Lactobacillus gasseri while decrease in the abundance of Bacteroidetes (Ley et al. 2005; Wang and Jia 2016). A. muciniphila was inversely associated with obesity and found to be more abundant in lean individuals than overweight individuals (Remely et al. 2016). The various mechanisms by which A. muciniphila helps in obesity management are shown in Fig. 2. A. muciniphila maintains the intestinal immunity, gut barrier integrity and permeability by reducing inflammatory cytokines, thereby achieving metabolic homeostasis (Ottman et al. 2017b). Also, lipopolysaccharide (LPS) found in cell wall of Gram-negative bacteria is a potential proinflammatory molecule and is associated with onset of inflammation. Imbalance in LPS levels leads to activation of proinflammatory signalling pathways causing increased secretion of IL-6 and TNF-α. The inherent property of A. muciniphila of SCFAs production signals G-protein coupled receptor (GPCR) activation and histone deacetylase (HDAC) inhibition to maintain energy homeostasis and appetite sensation (Lukovac et al. 2014). SCFAs production facilitate increased production of glucagon-like peptide 1 (GLP-1) and glucose-dependent insulin tropic polypeptide (GIP) upon binding to GPR41/GPR43 receptors present in L-cells in the intestinal mucosa. This results in improvement in insulin resistance and glucose tolerance thereby, suppressing appetite through metabolic signalling. Furthermore, A. muciniphila decreases inflammation by increasing the levels of oleoylathanolamide (OEA), 2-palmitoylglycerol (2-PG), 2-acylglycerol (2-AG) and 2-oleoylglycerol (2-OG) which bind to GPR119 receptors, stimulating release of GLP-1 (Everard et al. 2013). In a recent study, oral supplementation of A. muciniphila in obese mice through high fat diet (HFD) has been demonstrated to reduce the intestinal endotoxin levels, hence reducing inflammation (Cani and de Vos 2017; Fuke et al. 2019). It helped restore gut barrier dysfunction through symbiotic relationships with other beneficial microbes like Bacteroidetes, Euryarchaeota, Firmicutes and Actinobacteria and improved intestinal permeability through the inhibition of claudin 3 (Cldn3), cannabinoid receptor 1 (Cnr1) and occludin like tight junction proteins or lowering of flavin-containing monooxygenase 3 (FMO3) expression (Dao et al. 2016).
Fig. 2.
Role of A. muciniphila in obesity management
In humans, both live and pasteurized A. muciniphila were found to be safe for oral consumption by heavy body weight individuals (Plovier et al. 2016). In a clinical trial (NCT02637115), oral administration of A. muciniphila in obese patients for 3 months was found to be an effective and safe treatment. Similarly, in a randomized, double-blind human study based on A. muciniphila supplementation, a decrease in body weight along with improvement in liver dysfunction and inflammation in patients was observed (Depommier et al. 2019). Also, supplementation with prebiotic containing oligofructose helped restore A. muciniphila abundance. But since A. muciniphila does not grow in vitro on oligofructose-enriched media, it can be concluded that complex cross-feeding interactions might be involved. While it is clear that the human mucus colonizer maintains gut barrier integrity and homeostasis during obesity, the role of human gut microbiome on etiology of obesity remains to be investigated. The study of Akkermansia-obesity relationship could provide better insights to microbe-based treatments. Since A. muciniphila regulates the energy metabolism of host, its therapeutic intervention in obesity could be explored (Xu et al. 2020b). The reduction in fat-mass ratio in obesity can be studied by urinary metabolomics profile of A. muciniphila, making it a suitable biomarker for obesity (Png et al. 2010). Furthermore, the risk of obesity and associated metabolic diseases can be reduced by proper management of gut microbial profile.
Decreased abundance of A. muciniphila in type 2 diabetes
According to WHO, 1 out of 11 people suffer from diabetes with over 1.5 million deaths globally. Type 2 diabetes is a silent killer marked by the body’s inability to utilize insulin produced by pancreatic β-cells. A study conducted by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) suggests that factors such as sedentary lifestyle and genetic conditions may contribute to early onset of type 2 diabetes. The increased levels of blood sugar, i.e. hyperglycaemia results in nervous, circulatory and immune system–related complications. Insulin resistance as a result of type 2 diabetes affects the gut microbial diversity and metabolite production. Alterations in the abundance of various gut microbes during the onset and progression of type 2 diabetes have also been observed. Many studies have reported negative correlation of genus Akkermansia, Bacteroides, Faecalibacterium, Roseburia and Bifidobacterium, while positive correlation of genus Ruminococcus, Blautia and Fusobacterium with type 2 diabetes (Sedighi et al. 2017; Gao et al. 2018). Patients with normal glucose tolerance exhibited higher abundance of A. muciniphila as compared to pre-diabetic or type 2 diabetes patients. Interestingly, several anti-diabetic drugs like metformin, dapagliflozin and liraglutide were found to favour the abundance of A. muciniphila (Shin et al. 2014; Wang et al. 2018; Lee et al. 2018). Furthermore, a study conducted on the administration of an antidiabetic drug, metformin revealed that A. muciniphila further enhanced its anti-diabetic effects (Shin et al. 2014). Mice fed on HFD, when treated with metformin, showed increased abundance of A. muciniphila and improved blood sugar levels. Moreover, improved tolerance to glucose was observed upon oral administration of A. muciniphila but not metformin.
A. muciniphila is known to protect the intestinal barrier function by maintaining normal blood sugar levels. The intrinsic ability of A. muciniphila to catabolize complex carbohydrates further assists in inhibition of α-glucosidase and reduction of postprandial hyperglycaemia (Everard et al. 2013). Both obesity and type 2 diabetes may be ameliorated by increasing fatty acid oxidation and energy expenditure but reducing fatty acid biosynthesis (Everard et al. 2013; Gurung et al. 2020). Thus, fatty acid oxidation in adipose tissues and adipocyte differentiation may be promoted by administration of A. muciniphila, and other bacteria such as Lactobacillus gasseri, Bacteroides acidifaciens and thus SCFAs. This is correlated to increased levels of 2-PG, 2-AG and 2-OG in the adipose tissue. Furthermore, A. muciniphila can be regulated by increasing circulation of tryptophan metabolites through dietary intake (Cronin et al. 2021). Interestingly, A. muciniphila positively influences the host’s glucose metabolism by inducing IL-10, thus protecting from ageing-related insulin resistance (Wang et al. 2015; Greer et al. 2016). It fights against diabetic oxidative damage and improves resistance to gluco/lipotoxicity by decreasing hepatic glycogen levels and increasing HDL-C levels (Zhang et al. 2018). Moreover, it reduces inflammation by inhibiting the expression of TNF-α and lipid oxidative damage by lowering malondialdehyde levels in diabetic animals (Zhang et al. 2018). Increased paracellular gut permeability and gut barrier disruptions result in inflammation and metabolic diseases by increased absorption of LPS. A. muciniphila maintains glucose homeostasis and fat mass storage by controlling host mucus turnover and higher L-cell activity. Therefore, prebiotic treatment could help restore A. muciniphila abundance and counter metabolic endotoxemia in type 2 diabetes and obesity. Therefore, it becomes necessary to study the host-gut microbe interactions for better understanding of governing mechanisms in development of type 2 diabetes. A. muciniphila, through the alteration of the gut microbiota, could help to control and manage type 2 diabetes in the near future.
A. muciniphila and respiratory diseases
Decreased abundance of A. muciniphila in cystic fibrosis
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene located on chromosome 7. CFTR protein functions as an important anion-selective ion channel responsible for epithelial fluid secretion and intra-luminal hydration (Welsh and Smith 1993). Defective CFTR protein leads to accumulation of mucus in the lungs and intestine largely affecting the pulmonary and intestinal microbiota (Price and O’Toole 2021). As the mucus gets more and more viscous, the mucociliary clearance mechanism (MCC) becomes unable to clear the mucus resulting in manifestation of opportunistic microbial infections. Dysfunctional CFTR results in an altered intestinal condition including dysbiosis of gut microbiota due to physiological and biochemical imbalance. It includes changes in intestinal pH, inflammation, malabsorption and gut barrier disruptions (Meeker et al. 2020). Various factors such as prolonged antibiotics intake, immunosuppressive mediations and high-calorie diet further shape the CFTR microbiome. Specifically, in delF508 mutations, bacteria such as E. coli and Eubacterium biforme were found in higher abundance while Bifidobacterium and Faecalibacterium species were in lower abundance (Schippa et al. 2013). Recent studies on the gut microbiota of CFTR-deficient mice reported an increase in abundance of Enterobacteriaceae, Mycobacteria and Bacteroides while a decrease in abundance of Lactobacilli, Acinetobacter lwoffii and A. muciniphila (Thomsson et al. 2002; Bazett et al. 2016). Through integrated metagenomics and metabolomics study on CFTR gut microbiota, an increase in the expression of associated metabolites such as propylbutyrate, γ-aminobutyric acid (GABA), ethanol, choline and pyridine was observed while there was reduction in expression levels of 4-methylphenol, methylacetate, uracil, sarcosine, acetate, phenol, benzaldehyde and glucose (Vernocchi et al. 2018). The multi-omics-based model pointed out the correlation of microbial and metabolite variations caused by CFTR functional defects. Therefore, it becomes important to study the relationship between host gut, microbes and associated metabolites to investigate CF and its biomarkers. Major evidence of their relationship is the study on the administration of CFTR potentiator Ivacaftor which resulted in an increase in A. muciniphila. It could be explained by the release of bicarbonates from CFTR post Ivacaftor treatment that provided the optimal environment for mucin degraders (Ambort et al. 2012; Schütte et al. 2014). Post treatment with Ivacaftor, a significant reduction in stool calprotectin, a protein released by neutrophils, but no change in M2 pyruvate kinase (M2-PK) was observed. The reduction of calprotectin indicated that intestinal inflammation could be improved in CF patients upon restoration of intestinal milieu. Also, there was selective loss of pathogens like those of Enterobacteriaceae family which was positively correlated with stool calprotectin level (Manor et al. 2016). It led to an increase in the expression of an antimicrobial peptide (Reg III), which has direct metabolic activity in the intestine against Gram positive bacteria. In CF, A. muciniphila accounted for normal stool M2-PK concentration and decreased amount of Enterobacter. Thus, the increased abundance of A. muciniphila supports its potential as a biomarker for the gut and it may be used for the microbe based therapy in CF (Pang et al. 2014). Many gut microbes including A. muciniphila could be associated with CF and hold future as its promising therapeutic intervention. By regulating the gut profile of A. muciniphila through personalized nutrition and supplementation, host immunity can be improved, which could serve as one of the prophylactic ways by which the severity of CF could be minimized.
Increased abundance of A. muciniphila in COVID-19
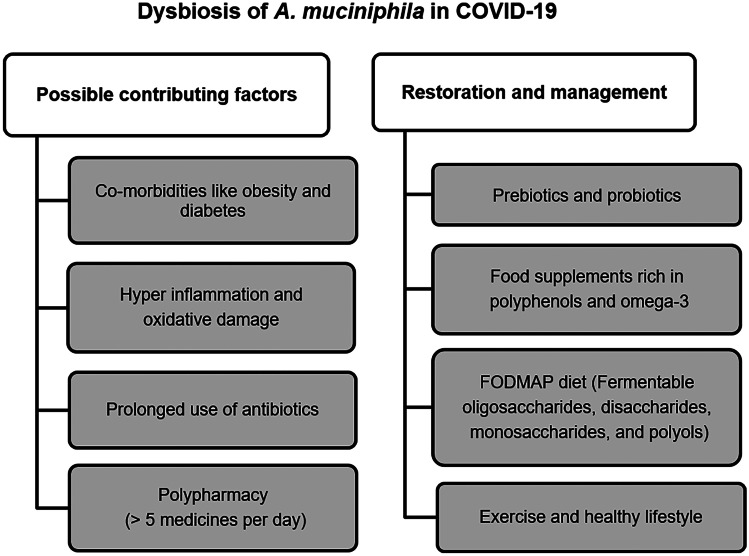
On 11th March 2020, COVID-19 caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) was declared as a pandemic. Over 180 countries were affected globally leading to nationwide lockdowns. The virus primarily attacked the respiratory system causing high-grade fever, severe cough, shortness of breath and pneumonia in severe cases. Several people infected with the virus experienced neurological and gastrointestinal (GI) manifestations with or without respiratory symptoms. On the other hand, some people were asymptomatic or symptom-free. The viral infection was also correlated with gut-lung-brain axis and microbiome imbalance. Significant reduction in the abundance of beneficial microbes was associated with inflammation and pathogenesis in COVID-19 (Hussain et al. 2021). Altered microbiome composition could further weaken body’s immunity and may play a role in SARS-CoV-2 infection. Recent studies have elucidated the relationship between gut and lung microbiota in COVID-19 and potential as prognostic markers (Wang et al. 2021). Although there is no direct evidence of specific interaction between resident gut microbe and COVID-19, some studies suggest that the gut microbiome in COVID-19 could be a key player in modulating host response and disease severity (Hussain et al. 2021; Yamamoto et al. 2021; Yeoh et al. 2021). GI symptoms were accompanied by gut dysbiosis during the early phase causing changes in gut microbiome and increase in inflammatory cytokines. Recently, proinflammatory cytokine storm due to significant increase in levels of IL-6 and IL-10 was reported to be predictive of COVID-19 severity (Han et al. 2020). Moreover, the presence of SARS-CoV-2 RNA was reported in faecal samples suggesting gut to be a viral replication site (Xiao et al. 2020). According to a study performed on SARS-CoV-2 recovered patients, A. muciniphila along with B. dorei was found to be elevated in the COVID-19 patients (Yeoh et al. 2021). Moreover, these bacteria were positively correlated with inflammatory cytokines, namely IL-1β and IL-6 and proinflammatory cytokine C-X-C motif ligand 8 (CXCL8). On the other hand, Faecalibacterium prausnitzii, Eubacterium rectale and some species of Bifidobacteria were found in lower abundance. A recent faecal metabolomics studies through machine learning approach suggested that particular set of gut microbiota could be used to predict proteomic risk score based on 20 blood proteomic biomarkers for COVID-19 severity (Gou et al. 2021). The gut microbial profile was used as a tool for prediction of the blood molecular signatures, indicating amino acid related pathways such as aminoacyl-tRNA biosynthesis, arginine biosynthesis and valine, leucine and isoleucine biosynthesis to be a possible link between inflammation and gut microbiota. It is well known that the gut microbiome renders beneficial effects on pulmonary mucosal immunity and host defence, thus safeguarding against respiratory infections (Gray et al. 2017). Downregulation of angiotensin-converting enzyme II (ACE2) involved in amino acid transport, tryptophan and antimicrobial peptide metabolism upon binding with viral spike protein might affect gut microbial ecology leading to dysbiosis in COVID-19 (Kuba et al. 2005). The possible factors contributing to A. muciniphila dysbiosis in COVID-19 along with strategies to restore its normal abundance are shown in Fig. 3. Co-morbidities such as diabetes, obesity and cardiovascular diseases largely influence the risk of infection and severity of disease. A lot of stress and consumption of fat and carbohydrate rich foods during the quarantine period was observed, leading to reduced CD8 + T cell response which could be linked to higher risk of infection (Mattioli et al. 2020). On the contrary, non-pharmacological measure such as reduction in consumption of fast food and increased emphasis on a healthy balanced diet also helped mitigate severe health conditions. Lifestyle habits, including diet and physical exercise, can profoundly influence the composition of the microbiome and consequently host metabolism and well-being. With the regulation of dietary habits, the optimum abundance of this friendly bacteria can be achieved in COVID-19 (Dhar and Mohanty 2020). Food supplements rich in polyphenols, omega-3 and FODMAP (Fermentable oligosaccharides, disaccharides, monosaccharides and polyols) are well known to increase A. muciniphila. Thus, by consuming polyphenol-rich foods, like fruits and vegetables, the abundance of A. muciniphila in the gut can be enhanced. Probiotic supplementations along with standard therapies as a prophylactic measure could also help mitigate the increased risk of comorbidities and move towards effective treatment. It becomes necessary to manage gut microbiome during and post disease recovery. The gut microbiome is asserted to critically impact the severity of infection as well as host immunity in COVID-19. By monitoring the GI symptoms, early diagnosis and treatment of COVID-19 can be facilitated. Therefore, it becomes important to study interaction between coronavirus and intestinal microbiome to develop novel treatment approaches.
Fig. 3.
Possible factors contributing to A. muciniphila dysbiosis in COVID-19 along with strategies to restore its normal abundance
Conclusion and future perspectives
A. muciniphila is a key mucus degrading bacteria in host immunity and infection response. The correlated metabolites and pathways are closely associated with inflammation, post-infection severity and recovery. A. muciniphila fortifying the intestinal mucus layer promotes several health-mediating effects. It regulates intestinal homeostasis and helps in maintaining epithelial barrier integrity by stimulating anti-inflammatory pathways. A. muciniphila presents itself as a powerful gut microbe having many metabolic interventions and as a promising therapeutic agent. The close relation of intestinal anti-inflammatory and protective effects of A. muciniphila emphasizes on its promising probiotic role (Neef and Sanz 2013). On the basis of cross-talk elucidated across gut-lung axis, alterations in the gut microbiota through administration of SCFAs, probiotics or micronutrients could act as potential therapeutic strategies. A specific protein in the outer membrane of A. muciniphila, Amuc-110, which recapitulates the effect of bacteria on TLR2 activation and improves the barrier integrity of intestines, could serve as a strong candidate for drug production in future. Thus, by unveiling the inter-relationships between host factors such as diet, lifestyle habits, clinical markers and A. muciniphila in the human gut microbiome, interventions and clinical trials may be designed. Through extensive investigation, new dimensions of the impact of A. muciniphila in the microbiome on human health may be explored in obesity, type 2 diabetes, cystic fibrosis and COVID-19 by examining its dysbiosis in patients as compared to healthy individuals and adopting suitable strategies for its restoration (Fig. 4). The study of the relationship between A. muciniphila in gut microbiome and personalized medicine can be one of the most attractive aspects of future research, which can provide significant perspectives for the treatment of metabolic diseases like type 2 diabetes and obesity, and even respiratory diseases such as cystic fibrosis and COVID-19. To conclude, potential opportunities exist for targeted interventions to modify the composition of A. muciniphila in the gut microbiome to improve host health.
Fig. 4.
Dysbiosis and restoration of A. muciniphila in obesity, type 2 diabetes, cystic fibrosis and COVID-19
Acknowledgements
The authors are grateful to the Delhi Technological University, New Delhi, India, for providing support to conduct this study.
Author contribution
Conceptualization: Vidushi Aggarwal and Sushant Sunder. Writing — original draft: Vidushi Aggarwal and Sushant Sunder. Writing — review & editing: Vidushi Aggarwal, Sushant Sunder and Smita Rastogi Verma. Supervision: Smita Rastogi Verma.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allin KH, Tremaroli V, Caesar R, et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810–820. doi: 10.1007/s00125-018-4550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambort D, Johansson MEV, Gustafsson JK, et al. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci. 2012;109:5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson KE, Axling U, Xu J, et al. Diverse effects of oats on cholesterol metabolism in C57BL/6 mice correlate with expression of hepatic bile acid-producing enzymes. Eur J Nutr. 2012;52:1755–1769. doi: 10.1007/s00394-012-0479-1. [DOI] [PubMed] [Google Scholar]
- Bajer L, Kverka M, Kostovcik M, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23:4548. doi: 10.3748/wjg.v23.i25.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett M, Bergeron ME, Haston CK. Streptomycin treatment alters the intestinal microbiome, pulmonary T cell profile and airway hyperresponsiveness in a cystic fibrosis mouse model. Sci Rep. 2016;6:1–13. doi: 10.1038/srep19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedarf JR, Hildebrand F, Coelho LP, et al. Erratum to: functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017;9:61. doi: 10.1186/s13073-017-0451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C, de Vos WM. Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo D, Sánchez B, Al-Hassi HO, et al. Microbiota/host crosstalk biomarkers: regulatory response of human intestinal dendritic cells exposed to lactobacillus extracellular encrypted peptide. PLoS ONE. 2012;7:e36262. doi: 10.1371/journal.pone.0036262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Tremaroli V, Kovatcheva-Datchary P, et al. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M, Rampelli S, Turroni S, et al. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, de Vos WM. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Waubant E, Chehoud C, et al. Gut microbiota in multiple sclerosis. J Investig Med. 2015;63:729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelakkot C, Choi Y, Kim D-K, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med. 2018;50:e450–e450. doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Song S, Zhao Y, et al. 146P A new biomarker of microbe: Akkermansia muciniphila in lung adenocarcinoma tissues may predict lymph node metastasis in lung adenocarcinoma. Ann Oncol. 2020;31:S297–S298. doi: 10.1016/j.annonc.2020.08.267. [DOI] [Google Scholar]
- Chen Z, Qian X, Chen S, et al. Akkermansia muciniphila enhances the antitumor effect of cisplatin in lewis lung cancer mice. J Immunol Res. 2020;2020:1–13. doi: 10.1155/2020/2969287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. 2012;72:77–85. doi: 10.1038/pr.2012.42. [DOI] [PubMed] [Google Scholar]
- Cone RA. Barrier properties of mucus. Adv Drug Deliv Rev. 2009;61:75–85. doi: 10.1016/j.addr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Cronin P, Joyce SA, O’Toole PW, O’Connor EM. Dietary fibre modulates the gut microbiota. Nutrients. 2021;13:1655. doi: 10.3390/nu13051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- De Angelis M, Piccolo M, Vannini L, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Annilo T. Evolution of the atp-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- Demirci M, Tokman HB, Uysal HK, et al. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol (madr) 2019;47:365–371. doi: 10.1016/j.aller.2018.12.009. [DOI] [PubMed] [Google Scholar]
- Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Van Baarlen P, Hooiveld G, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011 doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Dhar D, Mohanty A. Gut microbiota and COVID-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drell T, Larionova A, Voor T, et al. Differences in gut microbiota between atopic and healthy children. Curr Microbiol. 2015;71:177–183. doi: 10.1007/s00284-015-0815-9. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassatoui M, Lopez-Siles M, Díaz-Rizzolo DA, et al. Gut microbiota imbalances in Tunisian participants with type 1 and type 2 diabetes mellitus. Biosci Rep. 2019 doi: 10.1042/BSR20182348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Holst E, Karlsson F, et al. Altered microbiota in microscopic colitis. Gut. 2015;64:1185–1186. doi: 10.1136/gutjnl-2014-308956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke N, Nagata N, Suganuma H, Ota T. Regulation of gut microbiota and metabolic endotoxemia with dietary factors. Nutrients. 2019;11:2277. doi: 10.3390/nu11102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Zhu C, Li H, et al. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity. Obesity. 2018;26:351–361. doi: 10.1002/oby.22088. [DOI] [PubMed] [Google Scholar]
- Gargari G, Deon V, Taverniti V, et al. Evidence of dysbiosis in the intestinal microbial ecosystem of children and adolescents with primary hyperlipidemia and the potential role of regular hazelnut intake. FEMS Microbiol Ecol. 2018 doi: 10.1093/femsec/fiy045. [DOI] [PubMed] [Google Scholar]
- Gou W, Fu Y, Yue L, et al. Gut microbiota, inflammation, and molecular signatures of host response to infection. J Genet Genomics. 2021;48:792–802. doi: 10.1016/j.jgg.2021.04.002. [DOI] [PubMed] [Google Scholar]
- Grander C, Adolph TE, Wieser V, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891–901. doi: 10.1136/gutjnl-2016-313432. [DOI] [PubMed] [Google Scholar]
- Gray J, Oehrle K, Worthen G, et al. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med. 2017;9:eaaf9412. doi: 10.1126/scitranslmed.aaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer RL, Dong X, Moraes ACF, et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat Commun. 2016;7:13329. doi: 10.1038/ncomms13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson KL, Kopito RR. Effects of pyrophosphate and nucleotide analogs suggest a role for ATP hydrolysis in cystic fibrosis transmembrane regulator channel gating. J Biol Chem. 1994;269:19349–19353. doi: 10.1016/s0021-9258(17)32174-9. [DOI] [PubMed] [Google Scholar]
- Guo X, Li S, Zhang J, et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genomics. 2017 doi: 10.1186/s12864-017-4195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M, Li Z, You H, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden H, Eng A, Pope C, et al. P314 Fecal dysbiosis is associated with growth failure in infants with cystic fibrosis: a multicentre study. J Cyst Fibros. 2019;18:S146. doi: 10.1016/S1569-1993(19)30607-1. [DOI] [Google Scholar]
- Hoffman LR, Pope CE, Hayden HS, et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis. 2014;58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Feng Y, Wu J, et al. The gut microbiome signatures discriminate healthy from pulmonary tuberculosis patients. Front Cell Infect Microbiol. 2019 doi: 10.3389/fcimb.2019.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I, Cher GLY, Abid MA, Abid MB. Role of gut microbiome in COVID-19: an insight into pathogenesis and therapeutic potential. Front Immunol. 2021 doi: 10.3389/fimmu.2021.765965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Rodríguez‐Piñeiro AM, Schütte A et al (2015) The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16:164–177. 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed]
- Johansson MEV, Holmén Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson CLJ, Önnerfält J, Xu J, et al. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity. 2012;20:2257–2261. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- Kim S, Rigatto K, Gazzana MB, et al. Altered gut microbiome profile in patients with pulmonary arterial hypertension. Hypertension. 2020;75:1063–1071. doi: 10.1161/HYPERTENSIONAHA.119.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DM, Battson ML, Jarrell DK, et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol. 2018;17:62. doi: 10.1186/s12933-018-0708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin S, Vanhoutte PM, et al. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe −/− mice. Circulation. 2016;133:2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- Liu F, Ling Z, Xiao Y et al (2017) Dysbiosis of urinary microbiota is positively correlated with Type 2 diabetes mellitus. Oncotarget 8:3798–3810. 10.18632/oncotarget.14028 [DOI] [PMC free article] [PubMed]
- Lukovac S, Belzer C, Pellis L, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. Mbio. 2014 doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malham M, Lilje B, Houen G, et al. The microbiome reflects diagnosis and predicts disease severity in paediatric onset inflammatory bowel disease. Scand J Gastroenterol. 2019;54:969–975. doi: 10.1080/00365521.2019.1644368. [DOI] [PubMed] [Google Scholar]
- Manor O, Levy R, Pope CE, et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep. 2016;6:22493. doi: 10.1038/srep22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvasti FE, Moshiri A, Sadat Taghavi M et al (2020) The first report of differences in gut microbiota composition between obese and normal weight iranian subjects. Iran Biomed J 24:148–154. 10.29252/ibj.24.3.148 [DOI] [PMC free article] [PubMed]
- Mattioli AV, Sciomer S, Cocchi C, et al. Quarantine during COVID-19 outbreak: changes in diet and physical activity increase the risk of cardiovascular disease. Nutr Metab Cardiovasc Dis. 2020;30:1409–1417. doi: 10.1016/j.numecd.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker SM, Mears KS, Sangwan N, et al. CFTR dysregulation drives active selection of the gut microbiome. PLoS Pathog. 2020;16:e1008251. doi: 10.1371/journal.ppat.1008251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalovich D, Rodriguez-Perez N, Smolinska S, et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat Commun. 2019;10:5711. doi: 10.1038/s41467-019-13751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira-Pascual L, Cabrera-Rubio R, Ocon S, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef A, Sanz Y. Future for probiotic science in functional food and dietary supplement development. Curr Opin Clin Nutr Metab Care. 2013;16:679–687. doi: 10.1097/MCO.0b013e328365c258. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Opstelten JL, Plassais J, van Mil SWC, et al. Gut microbial diversity is reduced in smokers with Crohnʼs disease. Inflamm Bowel Dis. 2016;22:2070–2077. doi: 10.1097/MIB.0000000000000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman N, Davids M, Suarez-Diez M, et al. Genomescale model and omics analysis of metabolic capacities of Akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.01014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman N, Reunanen J, Meijerink M, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE. 2017 doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk JP, van der Ark KCH, Davids M, et al. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol. 2016;82:6983–6993. doi: 10.1128/AEM.01641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T, Leach ST, Katz T, et al. Fecal biomarkers of intestinal health and disease in children. Front Pediatr. 2014 doi: 10.3389/fped.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Zhang S, Liu Z, et al. Gut microbiota and Chinese medicine syndrome: altered fecal microbiotas in spleen (Pi)-deficient patients. J Tradit Chin Med = Chung i Tsa Chih Ying Wen Pan. 2020;40:137–143. [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2016;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Price CE, O’Toole GA (2021) The gut-lung axis in cystic fibrosis. J Bacteriol 203 [DOI] [PMC free article] [PubMed]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- Remely M, Hippe B, Geretschlaeger I, et al. Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wien Klin Wochenschr. 2015;127:394–398. doi: 10.1007/s00508-015-0755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remely M, Hippe B, Zanner J, et al. Gut microbiota of obese, type 2 diabetic individuals is enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after weight loss. Endocrine, Metab Immune Disord - Drug Targets. 2016;16:99–106. doi: 10.2174/1871530316666160831093813. [DOI] [PubMed] [Google Scholar]
- Ruiz L, Hevia A, Bernardo D, et al. Extracellular molecular effectors mediating probiotic attributes. FEMS Microbiol Lett. 2014;359:1–11. doi: 10.1111/1574-6968.12576. [DOI] [PubMed] [Google Scholar]
- Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci Technol. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- Sánchez B, Bressollier P, Urdaci MC. Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol Med Microbiol. 2008;54:1–17. doi: 10.1111/j.1574-695x.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- Sánchez B, Urdaci MC, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa–bacteria interactions. Microbiology. 2010;156:3232–3242. doi: 10.1099/mic.0.044057-0. [DOI] [PubMed] [Google Scholar]
- Schippa S, Iebba V, Santangelo F, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) allelic variants relate to shifts in faecal microbiota of cystic fibrosis patients. PLoS ONE. 2013;8:e61176. doi: 10.1371/journal.pone.0061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger M, Everard A, Gómez-Valadés AG, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte A, Ermund A, Becker-Pauly C, et al. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci. 2014;111:12396–12401. doi: 10.1073/pnas.1407597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi M, Razavi S, Navab-Moghadam F, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- Shin J, Noh JR, Chang DH, et al. Elucidation of akkermansia muciniphila probiotic traits driven by mucin depletion. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N-R, Lee J-C, Lee H-Y, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- Snider EJ, Compres G, Freedberg DE, et al. Alterations to the esophageal microbiome associated with progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2019;28:1687–1693. doi: 10.1158/1055-9965.EPI-19-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- Tan L, Zhao S, Zhu W, et al. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp Dermatol. 2018;27:144–149. doi: 10.1111/exd.13463. [DOI] [PubMed] [Google Scholar]
- Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, et al. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucα1-2 glycosyltransferase. Biochem J. 2002;367:609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- Tummler B, Puchelle E. CFTR: a multifaceted epithelial molecule. Trends Cell Biol. 1997;6(7):250–251. doi: 10.1016/S0962-8924(97)01062-3. [DOI] [PubMed] [Google Scholar]
- Unger MM, Spiegel J, Dillmann K-U, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Vakili B, Fateh A, Asadzadeh Aghdaei H, et al. Characterization of gut microbiota in hospitalized patients with clostridioides difficile infection. Curr Microbiol. 2020;77:1673–1680. doi: 10.1007/s00284-020-01980-x. [DOI] [PubMed] [Google Scholar]
- van der Ark KCH, Aalvink S, Suarez-Diez M, et al. Model-driven design of a minimal medium for Akkermansia muciniphila confirms mucus adaptation. Microb Biotechnol. 2018;11:476–485. doi: 10.1111/1751-7915.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MWJ, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE. 2011 doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernocchi P, Del CF, Russo A, et al. Gut microbiota signatures in cystic fibrosis: Loss of host CFTR function drives the microbiota enterophenotype. PLoS ONE. 2018;13:e0208171. doi: 10.1371/journal.pone.0208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Zhou X, Wang C, et al. Alterations of the gut microbiota in multiple system atrophy patients. Front Neurosci. 2019 doi: 10.3389/fnins.2019.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H, Sun Y, et al. Potential associations between microbiome and COVID-19. Front Med. 2021 doi: 10.3389/fmed.2021.785496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jia H. Metagenomewide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- Wang J, Tang H, Zhang C, et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015;9:1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ, et al. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in Feces of children with autism. Appl Environ Microbiol. 2011;77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Saha S, Van Horn S, et al. Gut microbiome differences between metformin- and liraglutide-treated T2DM subjects. Endocrinol Diabetes Metab. 2018;1:e00009. doi: 10.1002/edm2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-R. [DOI] [PubMed] [Google Scholar]
- Xiao F, Sun J, Xu Y, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Wu B, Liang J, et al. Altered gut microbiota and mucosal immunity in patients with schizophrenia. Brain Behav Immun. 2020;85:120–127. doi: 10.1016/j.bbi.2019.06.039. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang N, Tan H-Y, et al. Function of Akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Saito M, Tamura A, Prawisuda D, Mizutani T, Yotsuyanagi H. The human microbiome and COVID-19: a systematic review. PLoS ONE. 2021;16(6):e0253293. doi: 10.1371/journal.pone.0253293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh YK, Zuo T, Lui GC-Y, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- Zhang L, Qin Q, Liu M, et al. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog Dis. 2018 doi: 10.1093/femspd/fty028. [DOI] [PubMed] [Google Scholar]
- Zhong H, Ren H, Lu Y, et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine. 2019;47:373–383. doi: 10.1016/j.ebiom.2019.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Ju Y, Wang W, et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat Commun. 2020;11:1612. doi: 10.1038/s41467-020-15457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, van den Bogert B, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]