SUMMARY
Microglia, the resident immune cells of the brain, have emerged as crucial regulators of synaptic refinement and brain wiring. However, whether the remodeling of distinct synapse types during development is mediated by specialized microglia is unknown. Here, we show that GABA-receptive microglia selectively interact with inhibitory cortical synapses during a critical window of mouse postnatal development. GABA initiates a transcriptional synapse remodeling program within these specialized microglia, which in turn sculpt inhibitory connectivity without impacting excitatory synapses. Ablation of GABAB receptors within microglia impairs this process and leads to behavioral abnormalities. These findings demonstrate that brain wiring relies on the selective communication between matched neuronal and glial cell types.
In brief
GABA-receptive microglia selectively prune inhibitory synapses in development and disruption of this microglial response leads to behavioral abnormalities highlighting a critical function of selective communication between microglia and neuronal cell types.
Graphical Abstract

INTRODUCTION
Brain function relies on interactions among diverse cell types. Microglia are the primary brain macrophages and play diverse roles in tissue defense during infection and injury (Ransohoff and Perry, 2009). In the healthy developing brain, microglia regulate a plethora of processes that impact the organization of neural circuits, including synapse pruning (Thion et al., 2018; Bohlen et al., 2019; Neniskyte and Gross, 2017; Wilton et al., 2019). Synapses exhibit a striking molecular and functional heterogeneity, the best example of which is the dichotomy between excitatory and inhibitory synapses that possess distinct molecular components and properties (Favuzzi and Rico, 2018; Vogels and Abbott, 2009). These fundamental differences have profound implications for circuit function (Sohal and Rubenstein, 2019). However, whether microglia are generic effectors of synapse pruning or specialized microglia are able to discriminate between distinct synapse types is unknown.
Our understanding of microglia diversity in both development and disease has been greatly enhanced by the examination of their transcriptomic differences at the single cell level (Hammond et al., 2019; Keren-Shaul et al., 2017; Krasemann et al., 2017; Li et al., 2019; Masuda et al., 2019; Matcovitch-Natan et al., 2016). Such analysis led to the discovery of disease-associated microglia (DAM), which act as universal immune sensors of neurodegeneration (Deczkowska et al., 2018). However, whether variations in microglial transcriptomes map onto differences in function in the healthy developing brain remains poorly understood. We explored the hypothesis that functional microglia diversity has evolved to ensure the selective pruning of excitatory versus inhibitory synapses. Thus far, the examination of microglia-mediated synaptic pruning has focused on excitatory synapses (Paolicelli et al., 2011; Schafer et al., 2012). An association between microglia and inhibitory synapses has been suggested in the adult and under pathological conditions (Chen et al., 2014; Lui et al., 2016, Liu et al., 2021), the latter of which are often characterized by an aberrant reactivation of developmental programs (Wilton et al., 2019). However, support for this hypothesis has been to date limited to studies in the embryonic brain, where prenatal immune challenges regulate the laminar positioning and connectivity of neocortical parvalbumin (PV) interneurons (Squarzoni et al., 2014; Thion et al., 2019). Here, we demonstrate that GABA-receptive microglia remodel inhibitory but not excitatory synapses during mouse postnatal cortical development. The selectivity revealed by this process identifies specialized microglia dedicated to remodeling distinct synapse types.
RESULTS
Microglia depletion during cortical development alters inhibitory and excitatory synapse connectivity
To investigate whether microglia are required for inhibitory synapse development, we examined inhibitory connectivity after depleting myeloid cells—including microglia—for the first two postnatal weeks. Daily injections of the colony-stimulating factor 1 receptor inhibitor PLX5622 efficiently depleted microglia beginning at postnatal day 4 (P4) (Figures 1A and 1B). We focused on the barrels in the mouse somatosensory (S1) cortex and its most abundant interneuron subtype, PV cells. At P15, PV inhibitory synapses onto excitatory neurons were increased in microglia-depleted mice compared to controls (Figure 1C). Of note, this phenotype was not due to changes in the distribution or number of PV interneurons (Figure S1A). Upon microglia depletion, significant changes in PV synapses were detected only after their initial assembly (P12) (Figures S1B and S1C), suggesting a role for microglia in the maturation or refinement rather than formation of these connections. Conversely, the depletion of microglia during the third and fourth postnatal weeks did not alter the density of PV synapses (Figures S1C and S1D). Synapse development in different neocortical areas follows common principles but is asynchronous (Pinto et al., 2013). Consistent with this, the increase in PV synapses upon microglia depletion was also observed in the visual cortex (V1), although the exact time window was shifted in accordance with its later development (Figure S1E). Notably, inhibitory synapses made by dendrite-targeting somatostatin (SST) interneurons were also increased in P15 microglia-depleted mice (Figure S1F). As for PV cells, the density of SST interneurons was unaltered (Figure S1G).
Figure 1. Microglia depletion during cortical development alters inhibitory and excitatory synapse connectivity.
(A) Schematic of microglia depletion experiment.
(B) Image and density of Iba1+ microglia in control and depleted mice (n = 3–8). ***p < 0.001, Student’s t test (P4) and one-way ANOVA, Sidak’s multiple comparisons test (P8-P15).
(C) Images, masks, and densityofSyt2+Gephyrin+ synapses made by PV cells onto excitatory neurons (NeuN) in P15control and depleted mice (n = 5). *p < 0.05, Student’s t test.
(D) Schematic ofoptogenetics experiment, IPSCtraces, and IPSCamplitude (n = 16cellsfrom 4 controlsand n = 16cellsfrom 3 depleted mice). *p < 0.05, Mann-Whitney test.
(E and F) Traces, frequency, and amplitude of mIPSCs (n = 22 cells from 3 control and n = 25 cells from 3 depleted mice) and mEPSCs (n = 25 cells from 3 control and n = 28 cells from 3 depleted mice). *p < 0.05, **p < 0.01, ns p > 0.05, Student’s t test.
(G) mEPSC/mIPSC ratio; n = 22 cells from 3 control mice (Ctl) and n = 25 cells from 3 depleted (Dpl) mice. ns p > 0.05, Mann-Whitney test.
(H) Schematic, images, masks, and density of VGlut2+Homer1+ synapses onto excitatory or PV cells in controls (n = 5–11) and depleted mice (n = 4–9) at P15. *p < 0.05, Student’s t test.
(I) Schematic and legend for experiment in (J) and (K).
(J) Images and density of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons in P30 control and depleted mice (n = 6) after microglia repopulation. *p < 0.05, Student’s t test.
(K) Density of VGlut2+Homer1+ synapses onto excitatory neurons in P30 control (n = 9) and depleted mice (n = 7) after microglia repopulation. *p < 0.05, Mann-Whitney test.
Full and empty arrowheads indicate colocalization and boutons not meeting criteria. Scale bars, 1 μm except in (B) where it equals 100 μm. Data are mean ± SEM. See also Figure S1.
We next asked whether the structural increase in PV synapses was paralleled by a functional increase in PV inhibition. While recording from excitatory neurons, we stimulated PV cells expressing Channelrhodopsin-2 (PVCre/+;Ai32) and found that the amplitude of optogenetically evoked inhibitory postsynaptic currents (IPSCs) was significantly increased in microglia-depleted animals compared to controls (Figure 1D). The increased inhibitory connectivity onto excitatory neurons in microglia-depleted mice was also confirmed by a higher frequency of miniature and spontaneous inhibitory synaptic currents (mIPSCs and sIPSCs) (Figures 1E and S1H). Together, these results demonstrate the existence of sequential and temporally restricted waves during which the maturation of cortical inhibitory circuits is regulated by microglia.
For comparison, we examined the impact of microglia depletion on glutamatergic connectivity. The frequency of miniature and spontaneous excitatory synaptic currents (mEPSCs and sEPSCs) was higher in P15 microglia-depleted mice compared to controls (Figures 1F and S11). Importantly, developmental microglia depletion did not significantly alter miniature or spontaneous EPSC/IPSC frequency ratios (Figures 1G and S1J). Consistently, structural synapse analyses showed that both PV and excitatory neurons received more thalamocortical synapses in microglia-depleted mice (Figures 1H and S1K–S1M). Taken together, these experiments suggest that microglia regulate the development of both excitatory and inhibitory synapses.
We next asked whether the exuberant connectivity recovers once microglia are allowed to repopulate the brain. Within 48 h after cessation of the P1-P15 depletion treatment, microglia sequentially repopulated subcortical and cortical regions. Cortical repopulation was complete by P21 (Figures 1I and S1N). At P30, the supernumerary inhibitory and excitatory synapses persisted (Figures 1J, 1K, and S1O). However, in the adult (P60), synapse density returned to control levels (Figure S1P). This indicates that depleting microglia during development causes long-lasting, albeit not permanent, defects in inhibitory and excitatory connectivity.
Microglia interact with inhibitory synapses during development
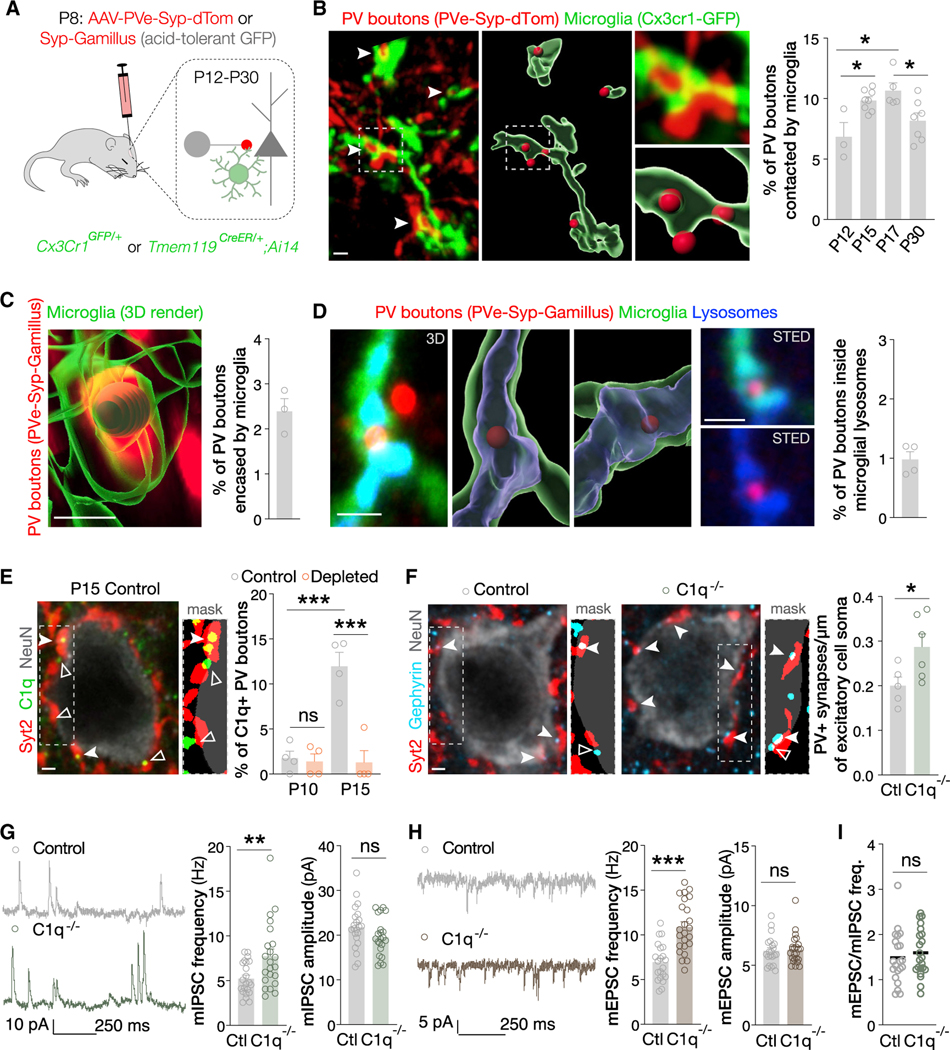
The previous experiments are consistent with microglia refining inhibitory synapses during development. Microglia-mediated synapse remodeling has been posited to depend on distinct and sequential processes: chemotaxis, target recognition, and phagocytosis (Neniskyte and Gross, 2017; Wilton et al., 2019). In response to various chemotactic signals, microglia are attracted to and interact with neurons and synapses (Badimon et al., 2020; Cserép et al., 2020; Madry and Attwell, 2015). To test whether microglia directly interact with inhibitory synapses during development, we used confocal and stimulated emission depletion (STED) super-resolution microscopy (Figure 2A). We generated mice expressing fluorescent reporters in both microglia and PV synaptic terminals. To this end, we injected Cx3cr1GFP/+ mice with adeno-associated viruses (AAVs) expressing synaptophysin-tdTomato under the control of a PV-specific enhancer (Vormstein-Schneider et al., 2020). At P15, microglia contacted 10% of PV boutons and their processes ensheathed these presynaptic terminals. Moreover, the fraction of PV boutons contacted by microglia increased between P12 and P15, peaked at P15-P17, and decreased by P30 (Figure 2B).
Figure 2. Microglia interact with inhibitory synapses during development.
(A) Schematic of experiments in (B) to (D).
(B) Images and 3D reconstruction of microglia processes (Cx3cr1GFP/+) contacting PV boutons (PVe-Syp-dTom: synaptophysin-tdTomato under the control of a PV-specific enhancer). Fraction of PV boutons contacted by microglia (n = 3–8 mice). *p < 0.05, one-way ANOVA, Holm-Sidak multiple comparisons test.
(C) 3D reconstruction and fraction of PV boutons (PVe-Syp-Gamillus) encapsulated by microglia (Tmem119CreER/+;AI 14) at P15 (n = 3 mice).
(D) Confocal image, 3D reconstruction, STED image and fraction of PV boutons (PVe-Syp-Gamillus) engulfed by microglial (Tmem119Cre/+;Ai14) lysosomes (CD68) at P15 (n = 4 mice).
(E) Image, mask, and fraction of C1q+ PV boutons (Syt2) in control and depleted mice (n = 4). ns, p > 0.05; ***p < 0.001, one-way ANOVA, Holm-Sidak multiple comparisons test.
(F) Images, masks, and density of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons (NeuN) in P15 control (n = 5) and C1q−/− (n = 6) mice. *p < 0.05, Student’s t test.
(G and H) Traces, frequency, and amplitude of mIPSCs (n = 22 cells from 3 control and n = 22 cells from 3 C1q−/− mice) and mEPSCs (n = 21 cells from 3 control and n = 24 cells from 3 C1q−/− mice). **p < 0.01, ***p < 0.001, ns p > 0.05, Student’s t test except for mEPSC amplitude where Mann-Whitney test was used. (I) mEPSC/mIPSC ratio (n = 21 cells from 3 control and n = 22 cells from 3 C1q−/− mice). ns, p > 0.05; Student’s t test.
Scale bars, 1 μm. Full and empty arrowheads indicate colocalization and boutons not meeting criteria. Data are mean ± SEM. See also Figures S2 and S3.
The developmental interactions of microglia with excitatory synapses involve their phagocytic engulfment and elimination (Neniskyte and Gross, 2017; Wilton et al., 2019). We therefore examined whether similar processes occur at inhibitory synapses. Because fluorescence quenching and protein degradation by lysosomal proteases may affect the detection of fluorescent proteins inside microglial lysosomes, we used the acid-tolerant monomeric green fluorescent protein Gamillus (Katayama et al., 2008; Shinoda et al., 2018a, 2018b). We injected AAVs expressing synaptophysin-Gamillus to label PV synaptic terminals in mice with genetically labeled microglia (Tmem119CreER/+;Ai14) (Figure 2A). At P15, a subset of these boutons was encapsulated within microglia and colocalized with microglial lysosomes (Figures 2C and 2D).
Classical complement proteins tag subsets of excitatory synapses for elimination by microglia (Schafer et al., 2012; Stevens et al., 2007). We examined complement C1q accumulation on PV boutons and found that C1q was deposited on 10% of PV synaptic terminals at P15 (Figure 2E). Next, we compared inhibitory and excitatory connectivity in controls versus C1qa knockout mice (C1q−/−). At P15, PV inhibitory synapses onto excitatory neurons were increased in C1q−/− mice (Figure 2F). The frequency of both mIPSCs and mEPSCs was also increased (Figures 2G and 2H). As with microglia depletion, there was no significant change in the ratio of mEPSCs/mIPSC frequency received by each cell (Figure 2I). Thus, C1q deficiency mimicked the defects observed in microglia-depleted mice, demonstrating that C1q is involved in regulating inhibitory connectivity.
GABA-receptive microglia preferentially interact with inhibitory versus excitatory synapses
To visualize real-time interactions, we performed in vivo two-photon imaging of microglia and PV boutons during the peak contact period (P15-P17) in S1 (Figures 3A–3D and S2). The imaging experiments revealed a bimodal distribution in microglia-PV synapse dynamics. Microglia either contacted few PV puncta (14%) or engaged in interactions with the majority of PV boutons (60%) in their vicinity (Figures 3C, S2C, and S2I; Videos S1 and S2). Of note, 1.8% of interactions involved microglia with phagocytic morphology (Figure S2G; Videos S3 and S4). Among those microglia that engaged more with PV boutons, most interactions were sustained for periods ranging from 12 to 15 min. By contrast, within the population that rarely contacted boutons, the interactions were of significantly shorter duration (Figures 3D, S2E, and S2K).
Figure 3. GABA-receptive microglia preferentially interact with inhibitory versus excitatory synapses.

(A) Schematic of in vivo imaging experiments and brain vasculature imaged through the cranial window. Scale bar, 500 μm.
(B) Time-lapse images from Video S1 showing microglia contacting PV boutons. Scale bar, 10 μm.
(C) Distribution of microglia contacting the indicated percentages of PV boutons over 20 min (n = 88 cells from 12 mice).
(D) Duration of contacts between microglia interacting with a minority (n = 24 cells) or majority (n = 49 cells) of local PV boutons. **p < 0.01, Mann-Whitney test.
(E) Images (left: single plane; right: 2-μm stack) of microglia (Cx3cr1GFP/+) expressing Gabbr1 and Gabbr2 mRNA at P15 (smFISH). Scale bars, 10 μm.
(F) Fraction of microglia expressing Gabbr1, Gabbr2, and both mRNAs at P15 (n = 7) in layer 4 of the primary somatosensory cortex (S1).
(G) 3D reconstruction and fraction of PV boutons (PVe-Syp-tdTom) contacted by Gabbr2+ and Gabbr2− microglia (Cx3cr1GFP/+) at P15 (n = 58 Gabbr2+ and 59 Gabbr2− cells from 4 mice). ***p < 0.001, Mann-Whitney test. Scale bar, 8 μm.
(H) 3D reconstruction and fraction of VGlut2+ boutons contacted by Gabbr2+ and Gabbr2− microglia (Cx3cr1GFP/+) at P15 (n = 23 Gabbr2+ and 36 Gabbr2− cells from 4 mice). **p < 0.01, Student’s t test. Scale bar, 10 μm.
Arrowheads indicate colocalization. Data are mean ± SEM. In (D), thick and thin lines are median and quartiles. See also Figures S2, S3, S4, S5 and Videos S1, S2, S3, and S4.
These differential interactions suggested the existence of at least two microglia subpopulations or states. We therefore mined published transcriptional data (Favuzzi et al., 2019; Matcovitch-Natan et al., 2016) to seek annotated ligand-receptor pairs expressed in microglia and inhibitory—but not excitatory—neurons during development (Figure S3A). GABA-GABAB receptors were among the leading candidates that could conceivably mediate signaling from inhibitory synapses to microglia. Such signaling has a strong precedent. In addition to its role as a neurotransmitter, GABA acts as a paracrine signal to control a myriad of developmental events. These include stem cell proliferation, migration, synaptogenesis, synapse pruning and astrocyte activity (Nagai et al., 2019; Oh et al., 2016; Wang and Kriegstein, 2009; Wu et al., 2012), most of which are mediated by GABAB receptors (Gaiarsa and Porcher, 2013; Mederos and Perea, 2019; Nagai et al., 2019). Moreover, previous work showed that a subset of microglia express GABAB receptors (GABABRs), and GABA elicits chemical, electrical, and morphological responses in microglia (Fontainhas et al., 2011; Kuhn et al., 2004).
For GABAB-responsiveness, the expression of both Gabbrl and Gabbr2 is required (Jones et al., 1998). Using used single molecule fluorescent in situ hybridization (smFISH), we determined that 50% of all microglia express Gabbrl, with only half of this population co-expressing Gabbr2. Hence, 25% of all microglia are double-positive for both GABABR subunits within barrels in S1 at P15 (Figures 3E, 3F, and S3B–S3E). The expression of GABABR subunits in microglia is upregulated at postnatal stages and exhibits regional differences (Matcovitch-Natan et al., 2016; Saunders et al., 2018) (Figures S3D, S3F, and S3G). To test the involvement of microglial GABABRs in the regulation of inhibitory synapse refinement, we combined smFISH with three-dimensional reconstruction. We found that inhibitory PV boutons were preferentially contacted and ensheathed by Gabbr2-expressing microglia (Figures 3G and S3H). In contrast, VGlut2+ excitatory synaptic terminals were preferentially contacted and ensheathed by microglia that did not express Gabbr2 (Figures 3H and S3I).
Removal of GABAB1Rs from microglia selectively impacts inhibitory connectivity
To directly test the idea that GABA-receptive microglia may be dedicated to remodel inhibitory synapses during development, we generated conditional mutant mice lacking GABAB1Rs in microglia using two distinct Cre-driver lines (cKO, Cx3cr1Cre/+; GABAB1Rfl/fl and Tmem119CreER/+;GABAB1Rfm). In each, we examined the impact of GABAB1R removal on microglia-PV synapse interactions. We found that cKO microglia contacted significantly fewer PV boutons compared to controls (Cx3cr1+/+; GABAB1Rfl/fl, Cx3cr1Cre/+;GABAB1R+/+, and Tmem119+/+; GABAB1Rfl/fl) (Figures 4A and S4A). In contrast, the proportion of contacted VGlut2+ synaptic terminals was not affected by the removal of GABAB1 Rs (Figures 4B and S4B). Consistent with these observations, in vivo two-photon imaging showed that cKO microglia interactions with PV boutons were no longer bimodal and more closely resembled those that interact less with PV synapses (Figures 4C–4E and S4C).
Figure 4. Removal of GABAB1Rs from microglia selectively impacts inhibitory connectivity.
(A) 3D reconstruction and fraction of PV boutons (PVe-Syp-tdTom) contacted by microglia (Iba1)inP15 control (n = 7) and GABAb1RcKO (n = 5) mice. **p < 0.01, Student’s t test. Scale bar, 4 μm.
(B) 3D reconstruction and fraction ofVGlut2+ boutons contacted by microglia (Iba1) in P15 control (n = 7) and GABAb1RcKO (n = 5) mice. ns, p > 0.05; Student’s t test. Scale bar, 4 μm.
(C) Schematic of in vivo imaging experiments in (D)-(F) and brain vasculature imaged through the cranial window. Scale bar, 500 μm.
(D) Time-lapse images showing cKO microglia contacting PV boutons. Scale bar, 20 μm.
(E) Distribution of microglia contacting the indicated percentages of PV boutons over 20 min in cKO mice (n = 62 cells from 6 mice). Control is from Figure 3C.
(F) Duration of contacts between microglia and PV boutons in cKO mice (n = 37 cells from 3 mice). Control is from Figure 3D.
(G) Schematic of experiments in (H) to (L).
(H) Images and density of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons in P15 control (n = 8) and cKO (n = 6) mice. ***p < 0.001, Mann-Whitney test. Scale bar, 2 μm.
(I) Images and density of VGlut2+Homer1+ synapses onto excitatory neurons in P15 control (n = 6) and cKO (n = 7) mice. ns p > 0.05, Student’s t test. Scale bar, 2 μm.
(J and K) Representativetraces,frequency, and amplitude ofmIPSCsand mEPSCs (n = 13cellsfrom 3 control and n = 14cellsfrom 3 cKO mice) at P15. *p < 0.05; ns, p > 0.05, Student’s t test.
(L) mEPSC/mIPSC ratio; n = 13 cells from 3 controls (Ctl) and n = 14 cells from 3 cKO mice (cKO). ns, * < 0.05; Student’s t test.
cKO, Cx3cr1Cre/+;GABAB1Rfl/fl. Full and empty arrowheads indicate colocalization and boutons not meeting criteria. Insets in (H) and (I) show masks. Data are mean ± SEM. See also Figures S4 and S5.
We reasoned that if GABABR signaling contributes to microglia-mediated remodeling of inhibitory synapses, cKO and microglia-depleted mice may display similar abnormalities in connectivity (Figures 1, 4F–4K, and S4D–S4K). To test this hypothesis, we quantified the density of PV inhibitory synapses contacting the soma of excitatory neurons in P15 mutants. We found that excitatory cells received significantly more PV synapses in GABABR mutants than were observed in control neurons (Figures 4G, S4D, and S4E). These data demonstrate that the removal of GABABRs from microglia phenocopies the changes in PV efferent connectivity observed when microglia are depleted. In contrast, no change in the density of excitatory inputs occurred (Figures 4H and S4F–S4H). We confirmed this selective effect on inhibitory connectivity by recording synaptic activity from P15 control and cKO mice. Analysis of mIPSCs and mEPSCs demonstrated that while inhibitory events were significantly increased in frequency, excitatory currents were unaffected (Figures 4I, 4J, S4I, and S4J). As a result, the mEPSC/mIPSC frequency ratio was decreased in cKO mice (Figures 4K and S4K). Together, these results reveal that removal of GABAB1Rs decouples the effects of microglia perturbation upon inhibitory versus excitatory synapses.
The increased PV innervation of mutants persisted at P30 (Figure S4L) and was detected in both V1 and in the dorsolateral striatum (Figure S4M). Moreover, disrupting GABABR signaling in microglia also affected the efferent connectivity of SST interneurons (Figure S4N). Of note, the presence of supernumerary synapses was not due to off-targets effects such as changes in the density of microglia or PV and SST interneurons, defects in interneuron survival or synapse assembly, changes in the fraction of PV cells surrounded by perineuronal nets (Crapser et al., 2020; Nguyen et al., 2020) or GABAB1R deletion from neurons (Figures S5A–S5G). In addition, mutant mice did not exhibit increased susceptibility to epilepsy (Figures S5H–S5J). Importantly, although excitatory synapses remained unaltered, inhibitory synapses were decreased in P60 cKO mice compared to controls (Figures S5K–S5O). Depleting microglia from P30 to P60 did not prevent this phenotype (Figure S5M), demonstrating that the late loss of inhibitory synapses in these mutants is not microglia-dependent. Together, these findings indicate that GABA-receptive microglia selectively mediate inhibitory synapse remodeling, and disrupting this process leads to permanent defects in inhibitory connectivity.
Ablation of GABAB1Rs within microglia alters genes involved in synapse remodeling
To explore the molecular mechanisms downstream of GABAB1 Rs in microglia, we isolated P15 wild-type (WT) (Cx3cr1+/+; GABAB1Rfl/fl) and cKO (Cx3cr1Cre/+;GABAB1Rfl/fl) microglia from the S1 cortex and performed single-cell RNA sequencing (scRNA-seq) (Figures 5 and S6; Table S1). To identify transcriptional changes due to the loss of one copy of Cx3cr1, we examined Cx3cr1Cre/+;GABAB1R+/+ mice as an additional control (Cre-Het).
Figure 5. Ablation of GABAB1Rs within microglia alters genes involved in synapse remodeling.
(A) UMAP plots of WT microglia showing 5 clusters and scaled expression of representative enriched genes.
(B) UMAP plots of WT and cKO integrated scRNA-seq dataset.
(C) Same as (B), showing 8 mixed clusters and representative enriched genes.
(D) Percentage of WT and cKO microglia composing each cluster.
(E) Mixed cluster contributions to total differentially expressed genes (DEGs) between WT and cKO microglia.
(F) Gene Ontology analysis of downregulated genes from cluster 4.
(G) Violin plots of normalized log-expression values for representative genes significantly downregulated in cKO microglia from cluster 4.
(H) Heatmap showing scaled expression of genes downregulated in cKO.
(I) Cluster 4 subclusters and percentage of WT and cKO microglia. Representative downregulated genes are highlighted.
Upon unsupervised clustering, WT microglia segregated in 5 major clusters of which two were notable (Figure 5A; Table S1). Cluster 1 cells exhibited higher levels of genes previously identified as transcriptional signatures of postnatal immature microglia (e.g., Tmsb4x) (Li et al., 2019; Masuda et al., 2019). Cells in Cluster 2 expressed higher levels of synapse pruning (e.g., C1qa [Stevens et al., 2007] and Trem2 [Filipello et al., 2018]) and homeostatic microglia core (e.g., P2ry12) genes. Of note, WT cells from our dataset could be integrated into a developmental trajectory with microglia ranging from embryonic to P30 (Hammond et al., 2019) (Figure S6A). All genetic alleles examined had comparable transcriptional states (Table S1; STAR Methods). However, consistent with the anti-inflammatory role of GABABRs in microglia (Kuhn et al., 2004), we found increases in cKO microglia related to DAM (Keren-Shaul et al., 2017) and with inflammatory profiles (clusters 2 and 5) (Figure S6D; Table S1).
We next combined WT and cKO microglia. The aligned dataset segregated into eight mixed clusters, most of which contained microglia from both experimental groups indicating that the loss of GABAB1Rs did not fundamentally alter the range of microglial states (Figures 5B–5D and S6E; Table S1). Clusters 1–3 shared transcriptional signatures of postnatal immature microglia. Cluster 4 had higher levels of synapse pruning and homeostatic microglia core genes. Cells in cluster 5 were transcriptionally related to DAM, and cluster 6 was composed of actively proliferating cells. Microglia in cluster 7 exhibited inflammatory and interferon-responsive profiles, whereas long non-coding RNAs were enriched in cluster 8. In agreement with the role of GABABRs in attenuating inflammation, cKO cells were more prevalent in clusters 5, 6, and 7 (Figures 5D and S6E). To query which genes are altered by the loss of GABAB1 Rs, we compared WT and cKO microglia in the mixed clusters. Only a few genes—involved in inflammation and proliferation—were differentially expressed in clusters 1–3,6, and 8. In contrast, cluster 4 harbored a large number of differentially expressed genes (Figure 5E; Table S2). The majority of genes downregulated in cluster 4 are involved in synaptic pruning and related processes such as chemotaxis and phagocytosis (Figures 5F and 5G; Table S2). Similar alterations were also observed in cKOs and Cre-Het microglia and hence are not the result of the loss of one Cx3cr1 allele (Figures S6F–S6J; Table S2). Thus, removal of microglial GABABRs impacts pruning genes selectively within more mature microglia (cluster 4).
As an alternative approach, we trained a classifier to identify genes predictive of WT versus cKO microglia across all mixed clusters and observed that the downregulation of pruning genes was a distinctive feature of mutant cells (Figure S6K). Further confirming the relevance of the transcriptional changes driven by cluster 4, pruning genes were also detected in a “pseudo-bulk” differential expression analysis (Table S2). Finally, analysis following denoising of gene expression (Tjärn-berg et al., 2020) yielded similar results (Figures S6L–S6N; Table S2). Importantly, we found that the downregulated genes were altered only in a subset of cluster 4 microglia, which segregated as a transcriptionally defined subgroup (Figures 5H and 5I; Table S2). Taken together, these data implicate several genes relevant to synapse remodeling as effectors of GABABR function in microglia.
The transcriptional changes observed in cKOs are restricted to GABA-receptive microglia
scRNA-seq technologies often fail to detect low-expressed mRNAs. As a consequence, Gabbr1 and Gabbr2—both low expressed genes in microglia—were incompletely detected in our scRNA-seq analysis. We therefore performed imaging-based single-cell gene expression profiling of WT and cKO microglia using multiplexed error-robust fluorescence in situ hybridization (MERFISH) (Chen et al., 2015b; Moffitt et al., 2018) (Figure 6A). To this end, we selected a 23-plex gene panel focused on assessing the co-expression of Gabbr1 and Gabbr2 with both microglia markers and pruning genes. Of note, Gabbr1 could be detected also in cKOs due to the presence of a non-deleted portion of the gene.
Figure 6. The transcriptional changes observed in cKOs are restricted to GABA-receptive microglia.
(A) Schematic of MERFISH experiment.
(B) UMAP plots of WT control and cKO microglia in the MERFISH dataset.
(C) Same as (B), showing 6 clusters.
(D) Same as (B), showing scaled expression of Gabbr1 and Gabbr2.
(E) Region imaged for MERFISH and cell maps on DAPI signal from a control slice. Scale bar, 100 μm.
(F) Layer distribution of microglia. Data are mean ± SD between slices.
(G) Split violin plots of normalized log-expression values for a representative pruning gene (C1qc) enriched in clusters 4 and 4GG control (ctl) cells.
(H) Split violin plots of normalized log-expression values for representative genes enriched in GABA-receptive control (ctl) cells.
(I) Volcano plot showing differentially expressed genes (DEGs) between control and cKO microglia for the MERFISH clusters. The negative log10-transformed p values are plotted against the log2 fold change. DEGs with an absolute log2 fold change higher than 0.25 and an adjusted p value <0.05 are depicted as opaque shapes with gene name, the rest is depicted with transparency. When close to the threshold, Gabbr-genes are also shown with opacity. Four data points with an adjusted p value >0.05 are outside the × axis limit.
(J) Images, masks, and fraction of C1q+ PV synaptic terminals (Syt2) in P15 control and cKO mice (n = 5 each). *p < 0.05, Student’s t test. Scale bar, 1 μm.
(K) Images, masks, and fraction of C1q+ VGlut2+ synaptic terminals in P15 control (n = 6) and cKO (n = 7) mice. ns, p > 0.05; Student’s t test. Scale bar, 1 μm.
Data in (J) and (K) are mean ± SEM. Full and empty arrowheads indicate colocalization and boutons not meeting criteria. See also Table S2.
Unsupervised clustering of microglia based on MERFISH profiling generated 6 cell groups (1,2GG, 3,3GG, 4, and 4GG), containing both WT and cKO microglia (Figures 6B and 6C; Table S2). Among the MERFISH groups, GABA-receptive microglia were highly enriched in three clusters (i.e., 2GG, 3GG, and 4GG) (Figure 6D; Table S2), most of which had a corresponding Gabbr-negative population. MERFISH cluster 4 and 4GG cells were distinguished from other microglia by their enhanced expression of homeostatic core genes, such as P2ry13, and were hence more closely related to the cluster 4 cells in our scRNA-seq data (Table S2). Notably, GABA-receptive microglia were uniformly distributed across cortical layers and anatomically interspersed with Gabbr-negative cells (Figures 6E and 6F). Pruning genes were enriched in both clusters 4 and 4GG at comparable levels (Figure 6G). In addition, GABA-receptive microglia had higher levels of transcripts encoding various transmembrane proteins. These include members of the tetraspanin family involved in cell motility, signaling, and membrane dynamics (Charrin et al., 2014) (Figure 6H; Table S2). Importantly, these same genes were downregulated in cKO microglia within cluster 4 of our scRNA-seq analysis (Table S2), suggesting a reduction of GABA-receptive microglia in cluster 4 rather than these molecules being obligatory effectors of GABABRs.
We next determined whether other gene expression changes observed in our scRNA-seq data were cell-autonomously restricted to GABA-receptive cells. Using the same MERFISH gene panel, we compared the expression of pruning genes in WT and cKO microglia. Genes encoding phagocytic receptors and those involved in microglial chemotaxis (C1qc, Trem2, Gpr34, and P2ry12) were only downregulated in cluster 4 GABA-receptive cells (i.e., selectively within 4GG). These genes were unaffected in GABA-receptive microglia within other clusters (Figure 6I; Table S2).
Among the genes selectively downregulated are those encoding the complement molecule C1q. Consistent with our transcriptomic results, C1q accumulation at PV synapses was significantly decreased in P15 cKO mice compared to controls (Figure 6J). In contrast, no change in C1q accumulation at VGlut2+ terminals was observed (Figure 6K). Taken together, our findings demonstrate that GABABRs activate a synapse remodeling program selectively in GABA-receptive microglia during development.
Loss of GABAB1Rs within microglia causes behavioral defects
To test if the loss of microglial GABAB1Rs results in behavioral abnormalities, we performed unsupervised analysis using motion sequencing (MoSeq). MoSeq identifies sub-second motifs (syllables) that compose mouse behavior (Markowitz et al., 2018; Wiltschko et al., 2015). We compared the usage of behavioral syllables in control versus GABAb1R cKO mice at P30 and P60. Compared to both WTs (Cx3cr1+/+;GABAB1Rfl/fl) and Cre-Hets (Cx3cr1Cre/+;GABAB1R+/+), high velocity syllables (e.g., jump and run) were less frequent in P30 cKOs (Cx3cr1Cre/+; GABAB1Rfl/fl), although this difference was more pronounced in males than females (Figures 7A, S7A, and S7B). In addition, P30 cKOs showed an overall decrease in spatial exploration (Figure 7B).
Figure 7. Loss of GABAB1Rs within microglia causes behavioral defects.
(A) Illustration of behavioral syllables enriched in P30 WTs or cKOs.
(B) Heatmap depicting the position of P30 WTs (n = 17) and cKOs (n = 29) during MoSeq.
(C) Expression probability of syllable usage (left) and syllable speed (right) in P60 WT control (n = 9), cKO (n = 9), and Cre-Het control (n = 3) female mice. *p < 0.05,z test on bootstrapped syllable usage/speed distribution corrected for false discovery rate (FDR). Data are mean ± SEM.
(D) Expression probability of syllable usage (left) and syllable speed (right) in P60 WT (n = 10), cKO (n = 5), and Cre-Het (n = 5) male mice. *p < 0.05, z test onbootstrapped syllable usage distribution corrected for FDR. Data are mean ± SEM.
(E) Illustration of syllables enriched in P60 WTs or cKOs.
(F) Heatmap depicting the position of P60 WTs (n = 19) and cKOs (n = 14) during MoSeq.
(G) Transition graphs depicting syllables (nodes) and transition probabilities (edges) in P60 WT and cKO mice. Node size proportional to syllable usage, edges weighted by bigram probability. Numbers correspond to syllables in Figure S7C.
(H) Transition graph depicting the difference in syllable usage and transition probability in WT and cKO mice. Low probability transitions were removed.
cKO: Cx3cr1Cre/+;GABAB1Rfl/fl. Syllable labels were assigned by a human observer. In (C) and (D), only relevant or significant syllables are shown. See also Figure S7 and Video S5.
In contrast to P30, P60 cKOs displayed high velocity syllables with greater frequency and executed behavioral motifs with increased rapidity compared to P60 WT and Cre-Het controls. Conversely, usage of low-velocity syllables (e.g., pause) was decreased in P60 cKOs (Figures 7C–7E, S7C, S7E, and S7G; Video S5). In addition, P60 cKO mice showed an overall increase in spatial exploration, although this effect was more pronounced in males than females (Figures 7F and S7D). Of note, P60 cKOs generated using either Cx3cr1Cre or Tmem119CreER drivers appeared comparable in their usage of high velocity syllables (Figure S7F). We also observed an overall decrease in syllabic transitions in P60 cKOs compared to WTs (Figure 7G and 7H). Of note, the few enhanced transitions observed in cKOs involved high-velocity syllables, further reaffirming their hyperactivity.
DISCUSSION
Multiple lines of evidence support that GABA-receptive microglia selectively remodel inhibitory synapses during development. (1) In vivo two-photon imaging indicates that a subset of microglia directly interacts with inhibitory synapses and these interactions are strongly attenuated in GABAB1R cKOs. (2) GABA-receptive microglia preferentially contact inhibitory versus excitatory synapses. (3) The contacts between microglia and inhibitory synapses are reduced upon ablation of GABABRs within microglia. (4) Orthogonal transcriptomic analyses demonstrate that the conditional removal of microglial GABABRs alters synapse pruning genes selectively within GABA-receptive microglia. (5) Removing GABABRs from microglia alters inhibitory connectivity without impacting excitatory synapses. (6) cKOs exhibit behavioral abnormalities that correlate with the inhibitory synaptic alterations.
A role for microglia in the wiring of inhibitory circuits
Distinct microglia-dependent mechanisms play a crucial role in shaping developing excitatory circuits. For example, both phagocytic and non-phagocytic microglia functions contribute to excitatory synapse refinement (Cheadle et al., 2020; Schafer et al., 2012). Our findings indicate that the same principles hold true for inhibitory synapses and that microglia-mediated synapse engulfment contributes to inhibitory synapse remodeling. Nonetheless, additional pruning mechanisms (e.g., release of factors that induce receptor endocytosis) may also play a role. Alternatively, or in addition to synapse elimination, microglia may retard the maturation of inhibitory synapses. As PV synapse density continues to increase between P12and P15 in S1, microglia depletion or GABAB1 R ablation may lead to supernumerary synapses.
GABA acts as a paracrine signal to initiate inhibitory synapse remodeling
Our results showed that the genes downregulated in cKOs are largely confined to cluster 4 GABA-receptive microglia. This suggests that microglia-mediated remodeling of inhibitory circuits requires three conditions to be met: (1) GABA release (Wu et al., 2012), (2) expression of GABABRs in microglia, and (3) the appearance of pruning competent microglia.
Our work adds to the evidence demonstrating a non-synaptic role of ambient GABA signaling through GABABRs in neural circuit development (Cellot and Cherubini, 2013; Gaiarsa and Porcher, 2013). We anticipate that GABA binding to microglial GABABRs triggers a complex cascade, which initiates a series of synapse-specific and activity-dependent processes. P2Y12 receptors play a crucial role in attracting microglia to cell bodies and synapses (Badimon et al., 2020; Cserep et al., 2020). Here, we showed that microglia lacking GABABRs contact fewer inhibitory synapses and that P2ry12 is downregulated in cKO GABA-receptive microglia. In addition, our MERFISH analysis identified transmembrane molecules enriched in GABA-receptive microglia. Some of these molecules may interact with partners selectively found at GABAergic terminals, thereby explaining the marked preference of GABA-receptive microglia for inhibitory synapses. In the adult, microglia are attracted to active synapses and dampen neuronal activity through the generation of adenosine (Badimon et al., 2020). An intriguing possibility is that a similar adenosine-mediated weakening of synapses triggers pruning during development.
Loss of microglial GABAB1Rs trigger a neurodevelopmental disorder-related behavioral phenotype
Our results show that disruption of GABA-signaling within microglia causes reduced activity at P30 but hyperactivity in adult animals. Hyperactivity is a hallmark symptom of neurodevelopmental disorders, including attention deficit/hyperactivity disorder (ADHD) (American Psychiatric Association, 2013). Abnormal development of synaptic contacts and an altered excitatory versus inhibitory synapse ratio underlie the pathophysiology of these disorders (Braat and Kooy, 2015; Chen et al., 2015a). Consistent with previous findings (Thion et al., 2019), in cortex and striatum, loss of microglial GABAB1Rs causes a selective increase in inhibitory synapses at P15and P30 that shifts to a selective decrease at P60. We showed that this change is not microglia-dependent and is therefore likely a compensatory effect. Finally, the behavioral phenotype probably reflects circuit anomalies distributed across many brain areas. Reduced cortical inhibition has been reported in ADHD (Edden et al., 2012; Gilbert et al., 2011). Nonetheless, synaptic alterations in the striatum are likely a crucial component. Accordingly, striatal dysfunction is associated with diverse neuropsychiatric conditions involving hyperactivity (Cubillo et al., 2012; Nagai et al., 2019).
Our work indicates that specialized microglia selectively remodel specific synapse types. Microglia express various neurotransmitter and neuromodulators receptors (Pocock and Kettenmann, 2007). Unveiling whether this allows them to target specific synapses will deepen our knowledge of brain function and dysfunction.
Limitations of study
Two additional aspects may contribute to the observed phenotypes: non-autonomous changes in microglial function not involving transcription and nonspecific effects of the enhanced pro-inflammatory microglia profiles.
Inhibitory synapses are preferentially, but not exclusively, contacted by GABA-receptive microglia, suggesting that additional subsets of microglia might also contribute to inhibitory synapse pruning.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by Lead Contact, Gord Fishell (Gordon_Fishell@hms.harvard.edu).
Materials availability
The AAV-PVe-Syp-Gamillus plasmid generated in this study is available from the corresponding author on request.
Data and code availability
scRNA-seq data have been deposited in NCBI GEO and assigned the following accession number GSE159947.
MERFISH raw and processed data are available at: https://drive.google.com/drive/folders/1TG4KkAW6-0HBmKnYqHCD4vfbBZXzSXkA
Should this link not be accessible, MERFISH processed data are also available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH while the raw data will be shared upon request in a manner determined on a case by case basis (e.g., hard drive, cloud server).
Scripts for synaptic analyses are available at: https://github.com/emiliafavuzzi/synaptic-analyses (https://doi.org/10.5281/zenodo.4899956)
Scripts reproducing scRNA-seq and MERFISH analyses are available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021 (https://doi.org/10.5281/zenodo.4899812)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cx3Cr1GFP/+ (Jackson Laboratories #005582), Tmem119CreER/+ (kind gift from G. Feng; Jackson Laboratories #031820) (Kaiser and Feng, 2019), Cx3Cr1Cre/+ (Jackson Laboratories #025524), Cx3Cr1Cre/+ (MMRRC #036395-UCD), Ai14 (Jackson Laboratories #007914), pvCre/+ (Jackson Laboratories #008069), Tmem119GFP (kind gift from G. Feng; Jackson Laboratories #031823) (Kaiser and Feng, 2019), SSTFlp/+ (Jackson Laboratories #028579), RCE:FRT (Jackson Laboratories #032038), C1qa−/− (kind gift from M. Botto), Ai34 (Jackson Laboratories#012570) (Bottoet al., 1998), SSTCre/+ (Jackson Laboratories#013044), VGlut1Cre/+ (Jackson Laboratories #023527) and Ai32 (Jackson Laboratories #024109) were maintained in a C57BL/6 background; GABAB1 floxed mice (kind gift from B. Bettler) (Haller et al., 2004) were maintained in a BALB/c background; RCE:loxP (Jackson Laboratories #032037) and Lhx6GFP (GENSAT, MMRRC #000246-MU) were maintained in a SWR/J background.
Animals were group housed and maintained under standard, temperature controlled laboratory conditions. Mice were kept on a 12:12 light/dark cycle and received water and food ad libitum. Both male and female mice were used for all experiments. Except for the behavioral experiments, similar results were obtained in both males and females.
For all experiments involving a comparison between conditions, littermates were used and they were always processed and analyzed together to minimize technical variability. For all experiments involving a comparison between controls (e.g., Cx3cr1+/+; GABAB1Rfl/fl) and GABAB1R cKO (e.g., Cx3cr1Cre/+; GABAB1Rfl/fl) mice, wild-type littermates (Cx3cr1+/+; GABAB1Rfl/fl) were used. When specified, Cre-Het controls (Cx3cr1Cre/+; GABAB1R+/+) were used as additional control.
Cx3cr1Cre mice from Jackson Laboratories were used in most of the experiments while Cx3cr1Cre mice from MMRRC were used to confirm most synaptic phenotypes (not shown, except in Figure S4E) and in the MERFISH experiment in Figure 6.
All animal maintenance and experimental procedures were conducted in accordance with the regulations established by the National Institutes of Health (NIH, USA) and were approved by the Institutional Animal Care and Use Committee at Harvard Medical School (protocol number IS00001269) and The Broad Institute (protocol number 0156–03-17).
METHOD DETAILS
Microglia Depletion
Microglia depletion was achieved by administering the CSF1R inhibitor PLX5622 (generously provided by Plexxikon Inc, Spangenberg et al., 2019). To deplete microglia in mice older than post-natal (P) day 18, animals were fed PLX5622-formulated AIN-76A diet (1200 mg PLX5622 per kilogram added to chow AIN-76A, Research Diets) ad libitum. Control mice received control diet (AIN-76A, Research Diets). To ablate microglia at early postnatal stages (P1-P18), PLX5622 was delivered to pups via intragastric (P1-P7) and intraperitoneal (P8-P18) injections. PLX5622 powder was provided by Plexxikon. A 20 × stock was made every week by dissolving PLX5622-FA in DMSO. The 20 × stock was stored at room temperature, protected from light. A diluent containing 0.5% hydroxypropyl methyl cellulose (HPMC, Sigma) and 1% Polysorbate 80 (PS80, Sigma) in deionized water was prepared. On each dosing day, a working solution containing 0.5 volumes of the PLX5622-DMSO 20 × stock (or DMSO 20 × as control) were dissolved to obtain a final concentration of 0.5% HPMC, 1% PS80, 5% DMSO in water. The mixture was placed in a sonicating water bath for 20–45 minutes to make a uniform suspension. Care was taken not to allow the compound to settle for more than 10 minutes before injection. P1-P18 pups received an intragastric or intraperitoneal injection of 50 mg/kg of PLX5622 every 24 hours. PLX5622 early postnatal treatment was well tolerated and resulted in more than 95% survival.
Perfusion and Immunohistochemistry
Animals were deeply anesthetized with sodium pentobarbital by intraperitoneal injection and transcardially perfused with PBS followed by 4% paraformaldehyde (PFA) in PBS. Brains were dissected out, postfixed for two hours at 4°C and cryoprotected in 30% sucrose-PBS solutions overnight at 4°C. Tissue was sectioned at 40 mm on a sliding microtome (Leica). Free-floating brain sections were incubated with 0.3% Triton X-100 in PBS for 4 × 15 minutes and then blocked for 2 hours (0.3% Triton X-100,10% Normal Goat and/or Donkey Serum), followed by incubation with primary antibodies in 0.3% Triton X-100, 5% Normal Goat and/or Donkey Serum overnight at 4°C. The next day, sections were rinsed 4 × 15 minutes in PBS, incubated with the appropriate secondary antibodies in 0.3% Triton X-100, 5% Normal Goat and/or Donkey Serum for 2 hours at room temperature, rinsed 4 × 15 minutes in PBS and then incubated with DAPI. The following primary antibodies were used: mouse anti-Synaptotagmin-2 (1:1000, ZFIN #ZDB-ATB-081002–25), mouse anti-gephyrin (1:500, Synaptic Systems #147 011), rabbit anti-NeuN (1:500, Millipore #ABN78), guinea-pig anti-NeuN (1:500, Millipore #ABN90P), guinea-pig anti-VGlut2 (1:2000, Millipore #AB2251), guinea-pig anti-VGlut2 (1:1000, Synaptic Systems #135404), guinea-pig anti-VGlut1 (1:1000, Millipore #AB5905), rabbit anti-Homer 1b/c (1:500, Synaptic Systems #160023), mouse anti-parvalbumin (1:1000, Sigma #P-3088), rabbit anti-DsRed (1:500, Clontech #632496), chicken anti-GFP (1:1000, Aves Lab #1020), rabbit anti-Iba1 (1:500, Wako Chemicals #019–19741), guinea-pig anti-Iba1 (1:500, Synaptic Systems #234 004), guinea-pig anti-parvalbumin (1:2000, Swant #GP72), rabbit anti-C1q (1:500, Abcam #ab182451), rat anti-CD68 (1:500, Bio-Rad #MCA1957), mouse anti-GABABR1 (1:500, NeuroMab #73–183), guinea-pig anti-GABABR2 (1:500, Millipore #AB2255), anti-WFA (1:500, L1516–2mg), mouse anti-GAD65 (1:500, Millipore #MAB351R), rabbit anti-SST (1:2,000, Peninsula Laboratories #T4103.0050). Of note, the following anti-GABABR1 antibody exhibited non-specific signal in GABAB1R cKO mice and we therefore discourage its use: Synaptic Systems # 322 102. Conversely, the following anti-GABABR2 antibody did not work in our hands: clone N81/2, NeuroMab # 73–124. Sections were mounted in Fluoromount-G (Southern Biotechnology, #100241–874), except for STED imaging where ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, #P36961) was used.
Generation of AAV Expression Vectors
The pAAV-PVe-Syp-tdTomato (synaptophysin-tdTomato under the control of the S5E2 PV-specific enhancer) was engineered as follows: thepAAV-S5E2-dTomato (Vormstein-Schneider et al., 2020) was used as backbone and the dTomato reporter was replaced by the Syp-dTomato reporter (Ai34, a gift from Hongkui Zeng, Addgene plasmid #34881) (Madisen et al., 2012) using Gibson Assembly (primers: Fw-TCC ACA GCG AGC TCG CTA GCG CCA CCA TGG ACG TGGT and Rv-TCC AGA GGT TGA TTA TCG ATA AGC TTC TAC TTG TAC AGC TCG TCC ATG CC). The pAAV-PVe-Syp-Gamillus was engineered as follows: the pAAV-PVe-Syp-tdTomato was used as backbone and the dTomato reporter was replaced by the acid-tolerant monomeric green fluorescent protein Gamillus (Gamillus/pcDNA3, a gift from Takeharu Nagai, Addgene plasmid #124837) (Shinoda et al., 2018a) using Gibson Assembly (primers: Fw-CTT CTC CAA TCA GAT GTC GCG AAT GGT GAG CAA GGG CGAG and Rv-TCC AGA GGT TGA TTA TCG ATA TTA CTT GTA CAG CTC GTC CAT GCC).
Cell Culture, transfection and AAV production
HEK293FT cells (Thermo Fisher Scientific, #R70007) were cultured in Dulbecco’s Modified Eagle’s medium with high glucose and pyruvate, GlutaMAX Supplement, 10% fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml). The cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. For AAV production, HEK293FT cells were seeded on 15-cm plates without antibiotics for 24 hours and co-transfected with the following plasmids using Polyethylenimine (100 μg/dish, Polysciences, #23966–1): pHGTI-helper (22 μg/dish), pAAV2/PHP.eB (cloned from pUCmini-iCAP-PHP.eB, a gift from Viviana Gradinaru, Addgene plasmid #103005; 9 μg/dish) and the AAV expression vector (12 μg/dish). 72 hours after transfection, transfected cells were harvested and lysed (150 mM NaCl, 20 mM Tris pH8.0) by three freeze-thaw cycles and Benzonase treatment (375 U/dish; Sigma, #E1014) for 15 minutes at 37°C. The supernatants were cleared by centrifugation at 4000 RPM for 20 minutes at 4°C, then transferred to Iodixanol gradients (OptiPrep Density Gradient Medium, Sigma, #D1556) for ultracentrifugation (VTi50 rotor, Beckman Coulter) at 50,000 RPM for 1.5 hours at 16°C. The 40% iodixanol fraction containing the AAVs was collected, underwent ultrafiltration with PBS in Amicon Ultra (15 ml, 100K, Millipore, #UFC910024) for 4 times, aliquoted and stored at −80°C. The number of genomic viral copies was determined by qPCR using the following primers against the WPRE sequence: Fw: AGC TCC TTT CCG GGA CTT TC and Rv: CAC CAC GGA ATT GTC AGT GC.
Intracranial injections
P8-P10 pups were anesthetized with 5% isoflurane and mounted on a stereotaxic frame. Isoflurane concentration during surgery was kept between 1%−2% and the body temperature was maintained at 37 °C using a heating pad. Surgery was performed using the ‘notouch’ sterile procedure, and all surgical tools were sterilized prior to surgery. The scalp was cleaned with betadine and ethanol (after shaving the skin for P10 injections) and cut open to expose the skull covering the somatosensory cortex. A small craniotomy (~1 mm) was opened overthe primary somatosensory cortex using a micro knife (Fine Science Tools). Then, 200 nL of AAV-PHP.B-PVe-Syp-tdTomato or AAV-PHP.B-PVe-Syp-Gamillus were unilaterally injected in the somatosensory cortex (anteroposterior −2.1/−2.8 mm, mediolateral +2.4/2.6 mm relative to Lambda; dorsoventral −0.34 and −0.44 mm relative to the pial surface) at a rate of 100 nl/minute using a Nanoject III Injector (Drummond Scientific, USA) followed by 2 additional minutes to allow diffusion. After surgery, mice were given Meloxicam (Metacam) subcutaneously at 5mg/kg of body weight (Boehringer-Ingelheim) and, upon recovery, were placed back in the home cage with the mother.
Tamoxifen Induction
Tamoxifen was dissolved in corn oil (20 mg/ml) at 37°C with constant agitation and stored at −80°C. Before injection, a 10 mg/ml dilution in corn oil was made. Microglia labeling in Tmem119CreER/+; Ai14 mice or GABAB1R removal in Tmem119CreER/+; GABAB1 Rfl/fl mice was achieved by performing intra-gastric tamoxifen injections in P2 and P3 post-natal mouse pups at a dose of 0.1mg/g of body weight for 3 consecutive days. Control mice (Tmem119+/+; GABAB1Rfl/fl) also received tamoxifen injections.
Single Molecule Fluorescent In Situ Hybridization Histochemistry
For single molecule fluorescent in situ hybridization (smFISH) combined with immunohistochemistry, mice were perfused and brains were fixed overnight in 4% PFAin PBS followed by cryoprotection in 30% sucrose PBS. Then, 16 μm or (for 3D reconstruction) 40 μm thick brain sections were obtained using a Leica cryostat or sliding microtome. Samples were processed according to the ACDBio Multiplex Flourescent v2 Kit protocol (ACDBio #323100). Briefly, tissue was pre-treated with a series of H2O2, antigen retrieval and protease IV steps before incubation with the probe for 1.5 hours at 40°C. Probes were purchased from ACDBio: RNAscope® Probes-Mm-Gabbr1 (#425181), Mm-Gabbr2 (#317971) and a custom-made probe to specifically detect Gabbr1 exons 7 and 8 deleted in GABAB1R cKOs (exon number is based on the exons used by both GABAB1a and GABAB1b isoforms). Three amplification steps were carried out prior to developing the signal with Cy3 fluorophore (Perkin-Elmer #NEL752001KT). Sections were then stained for GFP (chicken anti-GFP, 1:500, Aves lab #1020) or Iba1 (rabbit anti-Iba1,1:500, Wako Chemicals #019–1974) and tdTomato (rabbit anti-DsRed, 1:500, Clontech #632496) or VGlut2 (guinea-pig anti-VGlut2 (1:2000, Millipore #AB2251). Corresponding secondary antibodies were: 488 anti-Chicken (1:500, JIR #703–545-155), 488 anti-rabbit (1:500, Thermo Scientific #A-21206), 546 anti-rabbit (1:500, Thermo Scientific #A-10040) and 594 anti-guinea pig (1:500, JIR #706–585-148). Samples were counterstained with DAPI (5 μM, Sigma #D9542) and mounted using Prolong Gold antifade mounting medium (Molecular Probes #P369300).
Image Acquisition
For all analyses, images were taken in layer 4 of the somatosensory cortex (or visual cortex when specified), except for SST synapse analysis which was performed in layer 1, the main output layer of SST interneurons.
For analysis of synapse density, tissue samples were imaged on an upright ZEISS LSM 800 confocal using a 40X oil immersion objective, 1.4 NA, 2.5 digital zoom, 1024 × 1024 pixels (~0.22 μm resolution using 510 nm emission).
For analysis of microglia-synapse contacts and engulfment, tissue samples (layer 4 of S1 region) were imaged on an upright ZEISS LSM 800 confocal using a 40X oil immersion objective, 1.4 NA, 2.5 digital zoom, 1024 × 1024 pixels, 0.2 μm step size to produce confocal stacks of ~10–15 μm. For the analysis of contacts made by Gabbr2+ microglia, images were acquired with no zoom at 2048 × 2048 pixels. For the percentage of tdTomato+ PV synapses contacted or encapsulated by microglia, images were taken only in the center of the infection, where at least 95% of Syt2+ PV synapses were tdTomato+ (Vormstein-Schneider et al., 2020).
Stimulated Emission Depletion Microscopy (STED) images were acquired with a Leica SP8 Confocal/STED 3 × microscope with an oil-immersion 100 ×, 1.44-N.A. objective, at the Harvard Neurobiology Imaging Core. For contact and engulfment analysis, infection-rich areas were first scanned in confocal mode (zoom factor 4.70, pixel size 24.2 × 24.2 nm, speed 400 Hz) and confocal scans containing putative contacts or engulfed boutons were followed by sequential STED scans with gated detectors. PV synapse STED images were acquired using a similar approach but the region of interest was selected as a NeuN+ cell body. During STED scanning, Alexa Fluor 647, Alexa Fluor 546/555 and Alexa Fluor 488 signals were excited with 653-nm, 553-nm, and 499-nm white light lasers, respectively and in this particular order, and were depleted with 770 nm, 660 nm and 592 nm time-gated depletion lasers.
For cell colocalization and density, tissue samples were imaged on a ZEISS Axio Imager using or on an upright ZEISS LSM 800 confocal using 10X or 20X dry objectives (with or without tiling mode).
Seizure Susceptibility
The susceptibility to seizures was evaluated as described before with some modifications (Wamsley et al., 2018). Briefly, a 20 mg/ml stock of kainic acid (Sigma) was made in PBS 1X. Each mouse received an intraperitoneal injection of kainic acid at a dose of 20 mg/kg of mouse.
In Vivo Two-Photon Imaging
All in vivo two-photon imaging experiments were performed at P15-P17.
For in vivo two-photon imaging in controls, two sets of experiments were performed: Cx3cr1GFP/+ mice injected with PVe-Syp-tdTomato (6 mice, 3 males and 3 females) and Tmem119CreER/+;Ai14 mice injected with PVe-Syp-Gamillus (6 mice, 3 males and 3 females).
For in vivo two-photon imaging in GABAB1R cKOs, three sets of experiments were performed: Cx3cr1GFP/+;Tmem119CreER/+; GABAB1Rfl/fl injected with PVe-Syp-tdTomato (3 mice, 1 male and 2 females), Tmem119GFP; Cx3cr1Cre/+; GABAB1Rfl/fl injected with PVe-Syp-tdTomato (2 mice, 2 females), Tmem119CreER/+; RCE; GABAB1Rfl/fl injected with PVe-Syp-tdTomato (1 male).
Surgery
P15-P17 Cx3cr1GFP/+ or Tmem119CreER/+;Ai14 mice that had been previously injected with PVe-Syp-tdTomato or PVe-Syp-Gamillus AAVs (see intracranial injections above for details) were used. On the imaging day, mice were injected with dexamethasone (2 μg/g of body weight) to reduce brain swelling during surgery. Mice were then anesthetized with 2% isoflurane and held on a stereotaxic frame. Isoflurane concentration during surgery was kept between 1%−2% and the body temperature was maintained at 37 °C using a heating pad. The eyes were protected with a lubricant ointment (Systane) to prevent drying. Surgery was performed using the ‘notouch’ sterile procedure, and all surgical tools were sterilized prior to surgery. The skin was shaved, the scalp was cleaned with betadine and ethanol and cut open to expose the skull covering the somatosensory cortex. A 3% hydrogen peroxide solution (Sigma) was applied to the skull and the periosteal tissue surrounding the skull was gently scraped with forceps. Using a biopsy punch (VWR) and a micro knife (Fine Science Tools), a 3-mm craniotomy was opened over the previously injected region of the primary somatosensory cortex. Care was taken not to damage the dura mater and to absolutely avoid any bleeding as it clearly affected microglia behavior and motility (in addition to imaging quality). A glass window comprised of a 5 mm round cover glass (Warner Instruments, Harvard Bioscience, Inc.) and a 3 mm diameter cover glass (Warner Instruments, Harvard Bioscience, Inc.) attached to it with an ultraviolet curable adhesive (NOA 71) was placed over the craniotomy and its edges were sealed with Vetbond tissue adhesive (3M). A custom-made metal head-bar (Harvard Medical School Machine Shop) was attached over the right hemisphere using an ethyl-based instant gel adhesive (Loctite 409). Dental cement (Metabond, Parkell) was used to reinforce the attachment of the head-bar to the skull and to further seal the edges of the coverslip. The animal was allowed to recover for at least 1 hour in its home cage before starting imaging.
Imaging
Time lapse imaging of microglia and PV synapses was performed in layer 4 or deep layer 3 of the primary somatosensory cortex using a Bergamo II multiphoton microscope from ThorLabs (software ThorImageLS 3.2), equipped with a 8 kHz galvo-resonant scanner. For control Cx3cr1GFP/+; PVe-Syp-tdTomato experiments, the majority (64%) of the imaging was done in awake animals and a subset (36%) was performed under light anesthesia. For control Tmem119CreER/+;Ai14; PVe-Syp-Gamillus experiments, the majority (87.5%) of the imaging was performed under light anesthesia and a subset (12.5%) was performed in awake animals. For cKO experiments, 30% of the sessions were carried out in awake mice and the rest under light anesthesia. Mice were anesthetized with Equitisin because of the short half-life of this anesthetic mixture. A 1 mL solution containing 0.02 mg of magnesium sulfate, 0.32 mL of double-distilled sterile water, 0.1 mL of ethanol, 0.4 mL of propylene glycol and 0.18 mL of Nembutal was prepared and injected intraperitoneally (1.4 μl/g) While anesthetized, body temperature was maintained at 36.5–37.5°C using a heating pad. Imaging was done using a dual path Insight X3 laser (Spectra Physics). TdTomato was excited using a λ = 1045 nm fixed output while Gamillus and GFP were excited using the tunable output path tuned at λ = 930 nm, and signals were collected using two GaAsP PMTs. Imaging was performed using a 16 × /0.8 NA water-immersion objective (Nikon) at a zoom 8.2 ×. For Cx3cr1GFP/+; PVe-Syp-tdTomato control experiments and all cKO experiments, the field of view measured 99.22 × 99.22 μm (1024 × 1024 pixels) yielding a 0.097 pixel size. For Tmem119CreER/+::Ai14; PVe-Syp-Gamillus control experiments, the field of view measured 99.22 × 99.22 μm (512 × 512 pixels) yielding a 0.194 pixel size. Scan mode was set to unidirectional in averaging mode. Averaging of 5–7 frames resulted in a frame rate of 1.1 frames per seconds. The choice of the field of view was biased toward regions where we could detect some interactions between microglia and synapses prior to starting imaging. To image an entire microglia cell, z stacks of 10–15 μm were taken using a 1–3 μm step size using a piezo coupled to the objective (ThorLabs). Each imaging session lasted 20–30 minutes and multiple imaging sessions were performed in one day with a mouse recovery time of at least 60 minutes every hour. Based on the average contact duration observed in controls, all cKO imaging sessions lasted 20 minutes.
Patch-Clamp Recordings and Optogenetics
P15–28 mice were deeply anesthetized with isoflurane, and P60 mice were deeply anesthetized with sodium pentobarbital by intraperitoneal injection. In the case of P25–28 or P60 recordings, mice were first transcardially perfused with the slicing solution. Brains were removed and 300 mm coronal slices were cut using a vibratome (Leica). P15–28 brains were cut with ice-cold sucrose artificial cerebrospinal fluid (ACSF) of the following composition (in mM): 87 NaCl, 26 NaHCO3, 2.5 KCl, 1.25, NaH2PO4, 0.5 CaCl, 4 MgCl2,10 glucose, 75 sucrose, saturated with 95% O2,5% CO2 at pH 7.3–7.4. For P60 brains, the slicing solution was NMDG-ACSF (Ting et al., 2018) of the following composition (in mM): 92 NMDG, 2.5 KCl, 1.25 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, 3 Na-pyruvate, 0.5 CaCl2, and 10 MgSO4, pH 7.3–7.4. P15-P28 slices were transferred to a heated chamber at 34 C° with oxygenated ACSF recording solution, where they underwent recovery for 30 minutes. Slices were then moved to ACSF recording solution at room temperature, where they remained for at least an hour before recording. For P60 recordings, slices were recovered in oxygenated NMDG slicing solution at 34 C° for 25 minutes. Na+ was reintroduced (to 52mM) by gradually adding 2M NaCl-NMDG solution during recovery as previously described (Ting et al., 2018). Slices were then transferred to HEPES-holding solution of the following composition (in mM): 92 NaCl, 2.5 KCl, 1.25 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, 3 Na-pyruvate, 2 CaCl2, and 1 MgCl2, pH 7.3–7.4 at room temperature for at least an hour prior to recordings.
For recordings, slices were transferred to the recording chamber of an up-right microscope (Zeiss Axioskope II). All recordings were carried out at a constant temperature (30 C°). P15-P28 slices were perfused with ACSF of the following composition (in mM): 125 NaCl, 2.5 KCl, 1 MgCl2, 1.25 NaH2PO4, 25 NaHCO3, 20 Glucose, 2 CaCl2, saturated with 95% O2, 5% CO2 at pH 7.3–7.4. For P60 recordings, the ACSF had the following composition (in mM): 124 NaCl, 2.5 KCl, 1.25 NaH2PO4, 24 NaHCO3, 12.5 glucose, 5 HEPES, 2 CaCl2, and 1 MgCl2, saturated with 95% O2, 5% CO2 at pH 7.3–7.4.
Excitatory neurons were visualized with infrared-differential interference (IR-DIC) optics through a 40x water-immersion objective (Zeiss) and recorded in layer 4 or 5 of S1 barrel field. Recording pipettes were pulled from borosilicate glass (Harvard Apparatus) to obtain a tip resistance of 3–5 MΩ. For any experiment, only cells with access resistance < 30MΩ were accepted. Access resistance was monitored throughout the recording and any cells where access resistance deteriorated and changed more than 20% were discarded. Spontaneous excitatory and inhibitory postsynaptic currents (sEPSCs and sIPSCs) or miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs) were recorded by clamping the cells at either −70 mV to record EPSCs and at +0 mV to record IPSCs. For voltage-clamp recordings from pyramidal cells, the following internal solution was used (in Mm): 130 Cs-methanosulfonate, 5 CsCl, 10 HEPES, 0.2 EGTA, 4 MgATP, 0.3 Na-GTP, 8 Phosphocreatine-Tris, 5 QX-314-Cl, equilibrated with CsOH at pH 7.3). Data were acquired at a 20 kHz sampling rate using a MultiClamp 700B amplifier (Molecular Devices) and were filtered at 10 kHz.
Electrophysiology recordings were carried out at P15-P17 mice for all experiments measuring miniature synaptic currents (recorded with bath application of 1 μM TTX, Tocris #1069), P58–61 for P60 experiments and P25-P28 mice (depletion P1-P28) for optogenetics. For spontaneous synaptic currents, data from P15 mice (depletion P1-P15) are shown as pool with data from P26–28 mice (depletion P1-P28), as similar results were observed in the age-specific datasets.
Optogenetics
All experiments were conducted under wide-field photostimulation through a 40x water-immersion objective. The recorded neuron was centered in the field of view and a 470 nm LED was triggered to deliver a square-shaped pulse of 1 or 5 ms illumination at two different irradiance values (50% and 100% of the maximum stimulation intensity ~1 mW/mm2). Light pulses eliciting IPSCs were delivered every 15 s. The LED output was driven using a digital output from the Clampex software of the pCLAMP 9.0 program suite (Molecular Devices) controlling a BioLED controller (Mightex). Optogenetics experiments were performed in P26-P28 (depletion P1-P28) mice to allow functional expression of Channelrhodopsin-2 under the control of the late PVCre promoter. Results are shown only for the 5 ms pulse using 100% of the power, however similar results were observed for the 1 ms pulse using 50% of the power. To test monosynapticity, TTX (1 μM) and 4-aminopyradine (1 mM, Tocris #0940) were applied in some of the optogenetics recordings.
Tissue dissociation and microglia isolation
Microglia single cell suspensions for sequencing were generated as described previously with some modifications (Hammond et al., 2019). Centrifuges and tools were all prechilled at 4°C or on ice. P15 wild-type controls (Cx3cr1++; GABAB1Rfl/fl; N = 12), Cre-Het controls (Cx3cr1Cre/+; GABAB1R++; N = 10) and GABAB1R cKO (Cx3cr1Cre/+; GABAB1Rfl/fl; N = 13) mice were transcardially perfused using ice-cold Hank’s balanced salt solution (1X HBSS, Thermo Fisher Scientific) and the brains were removed and placed on ice-cold HBSS. Coronal slices encompassing the whole somatosensory region were cut and the S1 region was quickly dissected out using a scalpel and placed in ice-cold HBSS. For each experiment, tissue from at least 4 mice/genotype was pooled to obtain a good cell yield and Dounce homogenized in ice cold HBSS 15–20 times each with the loose and tight pestles while simultaneously rotating the pestle. The cell suspension was then transferred to prechilled 50 mL tubes and filtered using a pre-wet (with HBSS) 70 μm cell strainer. Cell suspensions were transferred into a prechilled 15mL tube and centrifuged at 500 g for 5 min at 4°C. The cell pellet was resuspended in 5.5 mLof HBSS1X. A 90% Percoll (Thermo Fisher Scientific) stock solution was made by adding 10% of HBSS 10xand 4.5 mLof the 90% Percoll stock solution were added to the cell suspension in order to obtain a 40% isotonic Percoll solution. The mix was centrifuged at 500 g for 1 hour at 4° C with full acceleration and braking. The top layers (myelin and Percoll) were removed by vacuum suction and the microglia pellet at the bottom of the 15mL tube was resuspended in 2 mL of ice-cold FACS buffer (2mM EDTAin 1XPBS), transferred to 2 mL tubes and centrifuged at 800 g for 10 minutes at 4°C to remove any excess of Percoll. The pellet was then resuspended in 200 μL of FACS buffer containing 4 μL of Mouse Fc Block (1:50, 10 μg/ml, BD Bioscience #553141) and blocked for 15 minutes on ice. Next, 300 μL of ice-cold FACS buffer as well as the following antibodies were added to the single cell suspension at a 1:200 final dilution and incubated for 20–30 minutes on ice: Cd11b-PE (BioLegend, #101208), CD45-APC-Cy7 (BioLegend, #103116), and Cx3cr1-APC (BioLegend, #149008). DAPI was also added to the mixture at a final dilution of 0.1 μg/ml and used to distinguish alive and dead cells. Samples were then washed in 1 mL of ice cold FACS buffer, centrifuged for 5 minutes at 800 g, resuspended in 500 μL of ice-cold FACS buffer containing RNase inhibitor at a concentration of 20 units/10 μL (RNaseOUT, Thermo Fisher Scientific #10777019) and transferred to pre-coated FACS tubes. Alive (low DAPI), individual microglia exhibiting high fluorescence levels of Cd11b and Cx3cr1 but low levels of Cd45 were purified from the suspension by fluorescence-activated cell sorting (FACS) using a 100 μm chip on a SH800S Sony sorter in purity mode. Of note, cells exhibiting high fluorescence levels of Cd45 were absent or extremely rare. Approximately 11,000–16,000 microglia were collected in each sorting experiment (experiments yielding fewer than 10,000 cells did not produce good quality libraries and were discarded). Samples were kept at 4°C before, during and after sorting.
Cells from a total of N = 12 wild-type controls (P15 Cx3cr1++; GABAB1Rfl/fl; 3 females and 9 males), N = 10 Cre-Het controls (P15 Cx3cr1Cre/+;GABAB1R+/+; 8 females and 2 males) and N = 13 GABAB1R cKOs (P15 Cx3cr1Cre/+; GABAB1Rfl/fl; 9 females and 4 males) were used for library preparation and sequencing. In order to minimize batch effects, (1) in each experiment wild-type control and GABAB1R cKO mice from the same litter were used and (2) each experiment consisted in parallel sorting of cells from both wild-type control and GABAB1R cKO mice. Cre-Het control microglia were purified using the same experimental procedure. However, these mice were not from the same litter as control or GABAB1R cKO mice and the experiment was performed separately. Nevertheless, this additional sample exhibited similar transcriptional states as the wild-type control microglia (see cluster annotation below) and was therefore used to identify genes expression changes that could be attributed to the loss of one copy of Cx3cr1.
Single cell library preparation & sequencing
After sorting, collected cells were centrifuged at 800 g for 5 minutes at 4°C and resuspended in 50–60 μL of PBS1x-BSA0.04% (aiming at obtaining 700 cells/μl). A small volume of cells was then diluted 1:10 and cells were counted using a hemocytometer to determine the exact number of cells/μl. Single-cell RNA-seq libraries were prepared on the 10x Genomics platform using the Chromium Single Cell 3′ Library and Gel Bead Kit v3.0 (PN-1000075), Chromium Single Cell 3′ Library Construction Kit v3.0 (PN-1000078), Chromium Chip B Single Cell kit (PN-1000154) and Chromium i7 Multiplex Kit (PN-120262) as instructed by the manufacturer. Libraries were sequenced using the Nova-Seq 100 cycle kit (Illumina) by Broad Institute Genomic Services.
Design of MERFISH encoding probes
In order to examine the co-expression of Gabbr1 and Gabbr2 with microglia markers, pruning genes, genes enriched in postnatal immature microglia (scRNA-seq clusters 1–3) and genes downregulated in cKO microglia within the scRNA-seq cluster 4, a MERFISH gene panel comprising 23 genes was designed. These were: Gabbr1, Gabbr2, Tmem119, Fcrls, P2ry13, P2ry12, Gpr34, Trem2, C1qc, Tmsb4x, Rps29, Ftl1, Cd164, Clec4a3, Ecscr, Laptm4a, Laptm5, Ppib, Selenok, Selenop, Tmem14c, Tspan3, Tspan4, Tspan7, mt_Nd2, mt_Co3. The Gabbr1 probe was designed to specifically avoid exons 7 and 8 deleted in GABAB1R cKOs (the exon number is based on the exons used by both GABAB1a and GABAB1b isoforms). C1qc, Tmsb4x, Rps29, Ftl1, mt_Nd2, mt_Co3 did not pass the RNA length and expression criteria to be part of the combinatorial single-molecule FISH (MERFISH) imaging run as described in previous MERFISH work (Chen et al., 2015b; Moffitt et al., 2018) and were therefore imaged in a non-combinatorial set of sequential smFISH imaging rounds with one single gene measured in each color channel per round. Thus, our23-plex MERFISH panel consisted of a 12-bit library for the combinatorial set (Cat #VZG114) and 6 genes imaged in the linear steps. All probes were produced by Vizgen.
Tissue sectioning, staining and imaging
P15 control (Cx3cr1+/+; GABAB1Rfl/fl) and GABAB1R cKO (Cx3cr1Cre/+; GABAB1Rfl/fl) mice were deeply anesthetized with isoflurane and the brains were removed, fresh-frozen in Tissue-Tek O.C.T. (Electron Microscopy Sciences, #62550–01) and stored at −80°C. Functionalized coverslips (Vizgen, #FCS01) for mounting the tissue sections for MERFISH imaging experiments were coated with fiducial beads (Polysciences, #17149–10) by incubation in PBS1Xfor 15 minutes and dried prior to tissue sectioning. 10 μm brain slices from the S1 region were sectioned on a cryostat (Leica) and placed onto the coverslip by dropping the glass onto the tissue slice with the bead coated side facing the tissue. Three brain slices from the same brain were mounted on each coverslip. The mounted brain slices were allowed to adhere to the coverslip for 5 minutes at −20°C. Slices were then incubated in fixation buffer (4% PFA in 1XPBS) for 15 minutes at room temperature and washed 3 times for 5 minutes with 1X PBS. Slices were incubated in 70% ethanol for 5 minutes and stored in ethanol at 4°C for up to a week prior to staining. MERFISH staining was carried out as instructed by the manufacturer (Vizgen). Briefly, coverslips with brain slices were washed in 2XSSC and incubated first in formamide wash buffer for 30 minutes at 37°C and then in ready-to-use Encoding Probe Hybridization Buffer Mix for 36–48 hours at 37°C. After a series of washes in 2XSSC and formamide wash buffer at 47°C, samples were gel embedded at room temperature for 1.5 hours. Next, tissue was cleared by adding proteinase K supplemented clearing solution to each sample and incubating it for 48h at 37°C. After a series of washes in 2XSSC, samples were incubated in hybridization buffer 1 from the Vizgen Imaging Reagent Kit (Vizgen, #IK-18) containing fluorescent probes for the first round of imaging (i.e., the first 3 bits of the codebook). Coverslips were then assembled into the imaging chamber and the bottom of the coverslip was cleaned with 100% methanol to ensure the optical imaging surface was entirely clean. MERFISH imaging of 11 brain slices from 2 controls (1 male and 1 female) and 8 slices from 2 cKOs (1 male and 1 female) was performed on an automated Vizgen Alpha Instrument with parameter files provided by the company and available at https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH. Stacks of seven images spaced by 1.5 μm in Z and comprising the whole S1 region were acquired with a Nikon 60X/1.4NA objective. Images were acquired in five colors, three encoding readout probes from the library, one serving as a fiducial marker for registration of the images during analysis and a nuclear staining (DAPI) to help downstream segmentation.
Motion Sequencing (MoSeq)
MoSeq experiments were carried out as described previously (Wiltschko et al., 2015; Markowitz et al., 2018). Briefly, P30 or P60 control, Cre-Het or GABAB1R cKO mice were placed in a circular open field in the dark (red light) and their 3D pose dynamics were measured during 60 minutes at 30 Hz through the use of a depth camera (Kinect 2, Microsoft). All mice were tested in the afternoon (i.e., 4–11 pm), when more active. For P60 MoSeq experiments, a total of N = 19 wild-type controls (Cx3cr1+/+; GABAB1Rfl/fl; 9 females and 10 males), N = 8 Cre-Het controls (Cx3cr1Cre/+; GABAB1R+/+; 3 females and 5 males), N = 14 GABAB1R cKOs (Cx3cr1Cre/+; GABAB1Rfl/fl; 9 females and 5 males) and N = 6 Tmem119-GABAB1R cKOs (Tmem119CreER/+; GABAB1Rfl/fl; 3 females and 3 males) were tested. For P30 MoSeq experiments, a total of N = 17 wild-type controls (Cx3cr1+/+; GABAB1Rfl/fl; 11 females and 6 males), N = 12 Cre-Het controls (Cx3cr1Cre/+; GABAB1R+/+; 5 females and 7 males) and N = 29 GABAB1R cKOs (Cx3cr1Cre/+; GABAB1Rfl/fl; 15 females and 14 males) were tested.
QUANTIFICATION AND STATISTICAL ANALYSIS
All of the statistical details of experiments can be found in the figure legends which include the statistical tests used, exact value of n, exclusion of any data, and what n represents.
All statistical analyses were performed using GraphPad Prism or R software. Unless otherwise stated, parametric data were analyzed by t test or one-way ANOVA followed by Holm-Sidak or Tukey post hoc analysis for comparisons of multiple samples. Non-parametric data were analyzed by the Mann-Whitney rank sum test or Kruskal-Wallis one-way analysis of variance on ranks followed by Holm-Sidak or Dunn post hoc analysis for comparisons of multiple samples. P values <0.05 were considered statistically significant.
Quantifications of Cell and Synapse Density
The quantification of synapse density during development is highly influenced by technical (e.g., perfusion and fixation, temperature during sectioning, antibody batch, etc.) as well as biological (e.g., maternal care, exact time of birth, etc.) factors. To minimize variability, littermates were used in all experiments involving a comparison in synapse density between conditions. Moreover, brains from the two conditions were strictly processed, imaged and analyzed together. When mice from multiple litters were pooled, they were collected simultaneously or within a short period of time (less than a month) and each collection always included both experimental and control groups. To confirm all main phenotypes, analyses were repeated and performed blindly.
Synapse density, contacts & engulfment
For PV and excitatory synapse analysis, single planes were analyzed using a custom script in Fiji (ImageJ) (Schneider et al., 2012) software as described before (Favuzzi et al., 2017). Briefly, processing of all channels included noise reduction and smoothing. All single channel images were converted to RGB. Next, a color threshold was automatically set to identify the cell body (or dendrite), its perimeter was automatically measured and a masked binary image with the cell body (or dendrite) only was created. For bouton segmentation, a watershed-based method was used such that boutons were separated based on the local minima of the pixel gray values. For the presynaptic boutons (Syt2, VGlut1 or VGlut2) or postsynaptic clusters (Gephyrin or Homer1), a color threshold was selected to segment boutons as isolated puncta (i.e., low enough to allow local minima detection and background exclusion but high enough to detect all putative presynaptic boutons or postsynaptic clusters). The comparison between the original images and the masks was used to guide the choice of the threshold value. The same criteria for each channel were applied to all images from the same experiment. The area of each presynaptic bouton was 0.2–0.8 μm2 whereas postsynaptic clusters measured 0.10.3 μm2. The “Analyze Particles” (where the minimum size for presynaptic and postsynaptic structures was 0.20 μm and 0.10μm, respectively) and “Watershed” tools were applied and a mask was generated. A merged image from all masks was created, converted to an 8-bit image and, using an automatic threshold, the overlap between presynaptic boutons, postsynaptic clusters and cell body was automatically detected as particles with a size greater than 0.05 μm2 in the “analyze particles” tool (corresponding to at least half of the smallest postsynaptic cluster overlapping with both presynaptic bouton and soma). Sample scripts for such analyses can be found at https://github.com/emiliafavuzzi/synaptic-analyses https://doi.org/10.5281/zenodo.4899956. A video showing an example of the workflow for synapse analysis can be found at https://www.youtube.com/watch?v=2WHMuz5oMAM&feature=youtu.be.
For C1q accumulation, the analysis was carried out as described above and the minimum size for the colocalization between C1q puncta and presynaptic boutons was 0.20 μm.
For the analysis of microglia-synapse contacts and engulfment, confocal stacks (~10–15 μm) were analyzed with IMARIS 9.3.1 or 9.5.0 software using a MATLAB script to automatize the analysis (an example can be found at https://github.com/emiliafavuzzi/synaptic-analyses) as described before (Favuzzi et al., 2019). All channels were subjected to background subtraction and Gaussian filtering. Then, three-dimensional isosurfaces were created for GFP+ microglia and volume was quantified automatically. A threshold was selected to include as much of the processes as possible while excluding any background and a 0.01 μm3 size filter was applied. The automatic threshold calculated in Imaris based on k-means statistical methods was used in the majority of analyses. Then, presynaptic tdTomato+ PV boutons were reconstructed as “spots” of 0.8 μm diameter (corresponding to the largest measured size of presynaptic boutons) and their total number was automatically calculated. Briefly, the built-in spot detection algorithm in Imaris first applies a 3D Mexican Hat filter using the spot size and then locates the spot centroid at the local maxima of the filtered image. Next, the number of spots located at no more than 0.4 mm from the microglia surface was automatically determined and indicated as contact. The 0.4 μm distance is calculated from the center of mass of the reconstructed “spot” and corresponds to its radius thus identifying as contact only those spots that are completely juxtaposed to the surface. In the representative images only contacted boutons are shown as reconstructed spots.
For the analysis of contacts made by Gabbr2+ microglia, the surface of all cells was reconstructed. Using the slice view, individual microglia were identified as Gabbr2-positive or Gabbr2-negative in single confocal planes. The surface corresponding only to the cell of interest was then duplicated and spots were reconstructed only within the region of interest containing the selected cell. Contact analysis was then carried out as described above.
Fluorescence quenching as well as protein degradation by lysosomal proteases may affect the detection of Syp-tdTomato (pKa 4.7) once inside microglial lysosomes (Katayama et al., 2008; Shinoda et al., 2018b). For this reason, instead of Syp-tdTomato, the acid-tolerant monomeric green fluorescent protein Gamillus was used for the encapsulation and engulfment analysis. Gamillus exhibits fluorescence stability at acidic pH (pKa 3.4) such as that of the lysosomes (pH ~4.5–5.0) as well as a proven resistance to proteolytic degradation in lysosomes (Katayama et al., 2008; Shinoda et al., 2018b). In particular, PVe-Syp-Gamillus was used to label PV boutons and Tmem119CreER/+;Ai14 mice (upon tamoxifen injection) were used to label microglia. For the encapsulation analysis, reconstruction was performed as described above and the “Split spots into surface” tool was used. The number of PV spots encapsulated inside microglia was then automatically quantified. For the engulfment analysis, three-dimensional isosurfaces were created for the lysosome marker CD68 inside tdTomato+ microglia. The subset of Gamillus+ PV boutons that had been detected as encapsulated within microglia was further subjected to the “Split spots into surface” tool using microglial CD68 as surface and the number of PV spots embedded was automatically determined.
For SST synapses using SSTCre;Ai34 mice, the analysis was performed in Fiji as described above except for the absence of a cell body. Synapses were analyzed in layer 1 and normalized by area.
For SST synapses in cKOs, since a Cre driver was already included for Gabbr1 genetic deletion, we used a four-allele genetic strategy to delete Gabbr1 in microglia while also labeling SST axons (Cx3cr1Cre/+; GABAB1RfUfl; SSTFlp/+;RCE:FRT) and performed the analysis as described before (Favuzzi et al., 2019). Briefly, 0.2 μm step size confocal stacks (~5–10 μm) were analyzed using IMARIS 9.3.1 or 9.5.0 and a MATLAB script to automatize the analysis. Briefly, SST axons (genetically labeled using SSTFlp/+;RCE:FRT mice) were reconstructed in 3D and GAD65+ presynaptic boutons contained within the axon were reconstructed as spots (size 0.6 μm) and selected as SST pre-synapses using the “split into surface” tool. Then, the number of GAD65+ presynaptic boutons within the axon colocalizing with Gephyrin+ postsynaptic clusters (size 0.3 μm) was automatically obtained and normalized per axon volume. The threshold for colocalization was 0.45 μm, corresponding to the sum of the radius of each spot and indicating that pre- and post-synapses are identified as such only when juxtaposed.
Cell colocalization and density
PV and microglia cell density was analyzed using a custom script in Fiji (ImageJ) software (an example can be found at https://github.com/emiliafavuzzi/synaptic-analyses). Briefly, a region of interest corresponding to the barrel field of the somatosensory cortex (or individual layers within it) was selected and the area was measured. Next, using an automatic threshold and a size filter of at least 10 μm in the “analyze particle” tool the total number of cells in the region was automatically determined. Due to the small soma size and highly ramified nature of microglia, microglia cell density was analyzed manually. Briefly, a region of interest corresponding to the barrel field of the somatosensory cortex was selected and the area was measured. Next, cells within this area were counted using the cell counter plugin in ImageJ (created by Kurt De Vos). For the colocalization of microglia expressing GABAB receptor mRNAs or protein, confocal stacks (~10–15 μm) were analyzed manually. Analysis was performed in individual confocal planes using the cell counter plugin. For a cell to be considered Gabbr1+ or Gabbr2+, mRNA clusters had to be present in at least three different focal planes. For the colocalization of microglia with GABAB1R and GABAB2R proteins, the signal in GABAB1R cKOs was used as baseline to determine background. Note that a reduction in GABAB2R protein has also been reported in GABAB1R cKOs (Haller et al., 2004). Using GABAB1R cKOs, we determined that clusters having an area higher than 0.15 μm were bona fide GABABRs. Images were analyzed using the same procedure described above for the synaptic analysis and a mask for each channel was created. Then, microglia expressing GABAB1R and GABAB2R proteins were identified manually in individual confocal planes using the cell counter plugin. Note that, due to the high antibody background, smFISH should be the preferred method for identification of GABA-receptive cells.
Seizure Susceptibility
A total of N = 7 controls (Cx3cr1+/+; GABAB1Rfl/fl; 1 P16, 3 P20 and 3 P26; 3 females and 4 males) and N = 7 GABAB1R cKOs (Cx3cr1Cre/+; GABAB1Rfl/fl; 3 P16, 2 P20 and 2 P26; 3 females and 4 males) was used. Since no significant difference or trend was observed in the susceptibility to seizures based on age or sex, data from all mice of the same genotype were pooled. Immediately after the kainic acid injection, seizure susceptibility was investigated according to a modified Racine Scale (Gehman et al., 2011; Racine, 1972): stage 0, normal behavior; stage 1, immobility; stage 2, mouth and facial movements; stage 3, head bobbing; stage 4, forelimb clonus; stage 5, rearing; stage 6, continuous rearing and falling (tonic-clonic seizures); stage 7, status epilepticus and/or death. Each animal was continuously inspected for up to 90 minutes from the time of kainic acid injection. The maximum score of each animal’s behavior every 5 minutes was used to determine the average score and standard deviation for both control and GABAB1R cKO mice. Of note, two-way ANOVA for repeated-measures revealed a significant effect of time but no significant effect of the genotype factor.
In Vivo Two-Photon Imaging
To correct for motion artifacts, image registration was carried out in ImageJ using a script created by Albert Cardona and Robert Bryson-Richardson (Parslow et al., 2014). To facilitate data analysis, four-dimensional data were projected into one plane using a standard maximum projection procedure, contrast was enhanced (saturation 0.35, normalize all) and a noise reduction step was performed using an edge-preserving Kuwahara-type filter developed by Wayne Rasband with a radius of 5. Contacts between microglia and presynaptic PV boutons were defined as an overlap between tdTomato+ PV boutons and GFP+ microglia (or Gamillus+ PV boutons and tdTomato+ microglia) fluorescence signals higher or equal to 30% of the synaptic bouton surface that lasted for more than 1 minute. Each contact was verified in at least one individual z plane for the time frame at which the corresponding contact started. For the quantification of the percentage of PV boutons contacted by one cell, the total number of PV boutons in a 20 μm radius around the microglia cell body was counted; next, the number of PV boutons contacted by microglia over a total of 20 minutes was calculated and expressed as a fraction of the total number of PV boutons in the area. For the quantification of the contact duration, each contact was identified and labeled, and the contact start as well as end time were recorded. tdTomato signal (for PV boutons or microglia) exhibited significant bleaching over time, preventing imaging sessions of 30–60 minutes to be included in the analysis. For the quantification of the percentage of PV boutons contacted by one cell, synapses or microglia processes that underwent significant bleaching over the 20–30 minute imaging session were still considered if contacted by a microglia while still visible. However, such synapses were not considered for the contact duration analysis as it was not possible to determine the end of the contact (because of bleaching or, occasionally, loss of focus). Conversely, the contact duration analysis included both synapses detected in a 20 μm radius around the microglia cell body and synapses contacted by microglia processes that did not have a cell body in the field of view. Of note, for Cx3cr1GFP/+; PVe-Syp-tdTomato experiments, occasional bleed-through of the GFP signal in thetdTomato channel was observed; however, in most of these cases, the GFP signal detected in thetdTomato channel was observed in microglia cell bodies while contacts were detected between boutons and microglia processes. cKO experiments using Cx3cr1GFP/+;Tmem119CreER/+; GABAB1Rfl/fl mice were analyzed as described above. In the cKO experiments using Tmem119GFP; Cx3cr1Cre/+; GABAB1Rfl/fl or Tmem119CreER/+; RCE; GABAB1Rfl/fl mice, the dimmer GFP signal required a higher laser power which caused extensive bleaching of the PVe-Syp-tdTomato boutons over time. This is why, when using those reporters, the analysis was carried out using only the first frames for PV boutons (static) and all imaging sessions where motion was not entirely corrected by registration were removed. Since a similar percentage of interactions was observed in these two sets, data were pooled. However, contact duration was significantly lower in the experiments using Tmem119GFP; Cx3cr1Cre/+; GABAB1Rfl/fl or Tmem119CreER/+; RCE; GABAB1Rflm mice and therefore only data from Cx3cr1GFP/+;Tmem119CreER/+; GABAB1Rfl/fl mice were included.
For Cx3cr1GFP/+; PVe-Syp-tdTomato control experiments, a total of 51 microglia cells and 222 contacts were analyzed from 6 P15-P17 mice (3 males and 3 females). For Tmem119CreER/+::Ai14; PVe-Syp-Gamillus control experiments, a total of 37 microglia cells and 115 contacts were analyzed from 6 P15-P17 mice (3 males and 3 females). For Cx3cr1GFP/+;Tmem119CreER/+; GABAB1Rfl/fl cKO experiments a total of 37 microglia cells and 99 contacts were analyzed from 3 P15-P17 mice (1 male and 2 females). For Cx3cr1GFP/+; Tmem119CreER/+; GABAB1Rfl/fl and Tmem119CreER/+; RCE; GABAB1Rfl/fl cKO experiments a total of 25 microglia cells and 85 contacts were analyzed from 3 P15-P17 mice (1 male and 2 females).
Data were visualized using the “ggplot2” R package. The peaks of the probability density function were found using the following coding lines in R: d < - density(x) and d$x[c(F, diff(sign(diff(d$y))) < 0)] where × was the vector containing all observations. The two populations of microglia contacting few or most boutons where split based on the local minimum of the Kernel density estimation identified using the following coding line in R (the interval was selected within the two peaks): optimize(approxfun(d$x,d$y), interval = c(12,65))$minimum. Data from L4 and L3 were pooled as no significant differences were detected in the parameters analyzed. As previously reported (Liu et al., 2019; Stowell et al., 2019), microglia appeared more active in anesthetized versus awake mice. Under light anesthesia, microglia exhibited a trend for contacting more PV boutons (although the difference did not reach significance). Moreover, the interactions had a small but significant duration increase in anesthetized versus awake mice. However, since similar conclusions could be reached in both the awake and anesthetized datasets with respect to microglia-PV bouton interactions (e.g., compare the PVe-Syp-tdTomato with PVe-Syp-Gamillus experiments) data from anesthetized versus awake imaging were pooled.
Patch-Clamp Recordings
Miniature synaptic currents were analyzed using the Minianalysis software (Synaptosoft, Decatur, GA) except for mEPSCs/mIPSCs from Cx3cr1Cre P15 cKO experiment and sEPSC/sIPSC experiments where a threshold search in Clampfit (Molecular Devices) was used. For the analysis using Minianalysis, the detection parameters were: Threshold 6.72 pA; local maximum period 8000 μs; baseline period 4000 μs; decay time period 8000 μs; decay time fraction 0.32; baseline average period 1000 ms; area threshold 11 pA; peak average points 1. For the analysis using Clampfit, a detection parameter of 4 pA for event threshold was used. The analysis was performed within the first 10 and 40 s of recording or within the 30 s period that showed the most stable baseline. In the case of optogenetically evoked IPSCsfrom pyramidal cells, monosynaptic currents were identified by the latency of the light mediated response. This was similar to the latency using TTX+4-AP to isolate monosynaptic responses (used in some of the recordings). Peak amplitude was identified using the threshold search in Clampfit. IPSC peak amplitudes were averaged across 10 trials at each irradiance level per cell. Average values per cell were then condensed into a grand average per group.
In silico Identification Of Ligand-Receptor Pairs
Developmental transcriptomic datasets from Matcovitch-Natan et al. (2016) and Favuzzi et al. (2019) were used to identify receptors (or cell adhesion and membrane proteins) within microglia that could interact with ligands (or cell adhesion and membrane proteins) expressed by interneurons but not pyramidal cells. First, the list of genes enriched in cluster 6 (upregulated in postnatal brain) and 7 (upregulated throughout development) from Matcovitch-Natan et al. (2016) were used as input for a Gene Ontology search in Panther. Genes belonging to the following categories were selected: transmembrane signal receptor, cell adhesion molecule, cell junction protein, defense immune protein, extracellular matrix protein, intercellular signal molecule, membrane traffic protein, transmembrane signal receptor (for protein class); cell junction, extracellular region part, extracellular region, membrane part, membrane, synapse part, synapse (for cellular component); biological adhesion, developmental process, immune system process, response to stimulus, signaling (for biological process). Duplicates were removed and the resulting list was further filtered by function and cellular location in The UniProt Consortium (2017) and only receptors, membrane and cell-adhesion proteins were kept (e.g., cytokines were removed). The resulting list of genes was used as input in BioGRID (Stark et al., 2006), to detect interactors. Intracellular interactions were manually removed and only inter-cellular interactors were kept. An exception was made for the gene Lmbr1l, whose BioGRID output of 939 interactors was filtered using Panther to remove intracellular interactions by keeping genes in the following GO cellular component categories and removing duplicates: cell junction, extracellular region part, extracellular region, membrane part, membrane, synapse part and synapse. Interactors not expressed in interneurons were removed using a cutoff of FPKM < 4 in P10 SST and PV interneurons. The expression in SST and PV interneurons was averaged to obtain the expression values for interneurons at P5 and P10. Then a specificity score was calculated as log10(average expression across P0, P5 and P10 interneurons/expression in P12 pyramidal cells) and genes with a score > 0.2 were considered enriched in interneurons as compared to pyramidal cells. The resulting list of interactors was then ranked by specificity score.
scRNA-seq data processing
Raw scRNA-seq data were processed using the standard CellRanger v3.0.0 pipeline (10x Genomics). Reads were aligned to the GRCm38 (mm10) Mus musculus genome. Doublets, cells with low quality or likely undergoing stress and putative non-microglia were excluded from the analysis. Criteria for exclusion were: fraction of reads mapping to mitochondrial genes greater than 10%, fewer than 800 genes detected, more than 20,000 total UMI detected and no reads mapping to Fcrls and Cx3cr1 genes. The almost complete absence of cells expressing Ms4a7 confirmed that CNS border-associated macrophages (BAMs) (Van Hove et al., 2019) were not contaminating the dataset. After these filtering steps, 6,235 wild-type control cells, 9,078 Cre-Het control cells and 9,752 GABAB1R KO cells were retained for further analysis.
scRNA-seq data clustering & visualization
Since previous work has shown that microglia gene expression follows a continuous rather than discrete distribution (Hammond et al., 2019; Li et al., 2019; Masuda et al., 2019), the Monocle3 R package was used to perform unsupervised clustering analysis on scRNA-seq data (Cao et al., 2019). Briefly, a gene by cell expression matrix was generated for wild-type and GABAB1R KO and annotated by sample (WT_1 and WT_2, KO_2 and KO_3 respectively), preprocessed using default parameters (normalized by log and size factor, and reduced to 50 dimensions with PCA), batch effect were removed with align_cds using sample as the alignment group and condition as residual_model_formula_str; UMAP was computed using reduce_dimension, clusters were computed using the Leiden community detection with cluster_cells. Cre-Het data were preprocessed in the same manner. The same procedure was carried out for subclustering with the only difference that cells belonging to the cluster of interest were used. All plots from the analysis were generated using Monocle3 or Scanpy (Amir et al., 2013; Wolf et al., 2018) built in functions as well as custom scripts in R.
To generate featureplots and heatmaps, data were log-transformed after adding a pseudocount and scaled to unit variance and zero mean. More specifically, UMI count values per cell were transformed by preprocess_cds in Monocle3, normalize_total and Freeman-Tukey transform (FTT) or sc.pp.log1p with Scanpy; they were then mean centered and scaled by genes with maximum and minimum values set to 3 and −3. These values were plotted using a custom R script and ggplot2.
For the analysis of the cell cycle phase, each cell was assigned a G2M score and an S score, based on its expression of G2/M and S phase markers using CellCycleScoring from Satija et al. (2015).
Single Cell RNA Sequencing Dataset Integration
For integrating the wild-type control and GABAB1R KO datasets, a gene by cell expression matrix was generated and annotated by sample (WT_1 and WT_2, KO_2 and KO_3) and condition (WT and KO). Data were then preprocessed using default parameters in Monocle3 (normalized by log and size factor, and reduced to 50 dimensions with PCA). Batch effect were removed with align_cds using condition as the alignment group. UMAP was computed using reduce_dimension and clusters were computed using the Leiden community detection with cluster_cells. To evaluate the validity of wild-type and cKO cell integration, the composition of each mixed cluster was assessed. Briefly, each cell from all integrated clusters (I1 to I8) was traced back to their corresponding wild-type control (C1 to C5) or GABAB1R cKO (GC1 to GC7) cluster. Proportions were then computed for each integrated cluster and are shown in the composition charts in Figure S6.
The Cre-Het control and GABAB1R KO datasets were generated separately and, as such, exhibited stronger batch effects than the wild-type control and GABAB1R KO datasets. For integrating the Cre-Het control and GABAB1R KO datasets, DEWÄKSS was used (Tjärnberg et al., 2020). Following standard pre-processing (normalization and log1p transformation), the optimal amount of principal components and k for the k-nearest neighbor graph was computed in DEWÄKSS over the BBKNN algorithm to integrate the conditions (Polański et al., 2020). Unsupervised clustering was performed using Leiden clustering with the adjacency matrix computed in DEWÄKSS. Denoised values and clusters were then imported in Seurat for differential expression analysis (see below).
To integrate our P15 wild-type microglia single-cell data with embryonic, early postnatal (P4-P5) and juvenile (P30) datasets from Hammond et al. (2019), Monocle3 was run with default parameters as described above. Due to the higher number of cells, the following modifications were introduced: num_dim set to 200 for preprocess_cds and residual_model_formula_str = ‘-sample + tp’ for align_cds (“sample” indicates each replicates and “tp” indicates each time point). A developmental trajectory was then computed using learn_graph.
Single Cell RNA Sequencing Cluster Annotation
Lists of genes enriched in each cluster were generated using the top_markers function in the Monocle3 R package (marker score ≥ 0.15). Additionally, clusters from Monocle3 were transferred to Seurat (Satija et al. (2015) and lists of genes enriched in each cluster were generated using the FindAllMarkers function in the Seurat R package with Wilcoxon rank sum tests (default parameters: absolute value of the average log2FC ≥ 0.25 and p value ≤ 0.01). The full lists of cluster-enriched genes for wild-type, Cre-Het, cKO and the integrated datasets are shown in Table S1. Confirming the soundness and reliability of our data and analysis, both the Seurat and Monocle analyses returned the same genes or genes with similar functions. The relative enrichment of gene groups involved in analogous processes as well as the expression of known marker genes was used for cluster identification. The cellular function of each gene was based on gene ontology (GO) terms and Medline search. The functional annotation of individual clusters was based on the roles of cluster-enriched genes in addition to a comparison with annotated clusters from previous work (including but not limited to references Hammond et al., 2019; Keren-Shaul et al., 2017; Li et al., 2019; Masuda et al., 2019).
Control WT, GABAB1R cKO & WT+KO integrated clusters
Unsupervised clustering of wild-type microglia returned 5 main clusters. Cells from cluster 1 exhibited higher levels of genes implicated in metal homeostasis (e.g., Fth1, Ftl1, Mt1), actin cytoskeleton dynamics (e.g., Tmsb4x, Pfn1, Cfl1) and ribosomal components (e.g., Rps26, Rps15a, Rpl32, Rps5), all of which have been previously identified as transcriptional signatures of postnatal immature microglia (Li et al., 2019; Masuda et al., 2019). Consistently, cells from cluster 1 also shared some transcriptional signatures (e.g., Ftl1, Cfl1, Mt1, Rpl30) with cluster 4 P4-P5 microglia from Hammond et al. (2019). Subclustering of cluster 1 further revealed an additional 22 subclusters of which two were notable. Microglia from subcluster 1.15 shared some of the transcriptional signatures of DAM (e.g., Cd63, Cd9, Lyz2) (Keren-Shaul et al., 2017). These are also shared by proliferative-region-associated microglia (Li et al., 2019). Subcluster 1.22 contained a small group of cells expressing higher levels of many established pro-inflammatory (e.g., Ccl12, Stat1) and interferon response (e.g., Ifitm3, Ifi44, Ifi27l2a) genes. These genes are typically induced in microglia upon injury or pathological conditions (Hammond et al., 2019). Cluster 2 was composed of cells expressing higher levels of homeostatic microglia core genes (e.g., Fcrls, P2ry13, Hexb), as well as genes involved in synapse pruning (e.g., C1qa (Stevens et al., 2007), Trem2 (Filipello et al., 2018)). Cluster 3 microglia are distinguished by their relatively high expression of Actb and the long non-coding RNA Gm42418. Cells belonging to cluster 3 appeared as a transitional state between cluster 1 and 2. They expressed intermediate levels of genes encoding ribosome components and microglia core genes but lower levels of synapse pruning genes. Cluster 4 cells are characterized by high expression of long non-coding RNAs with unknown function (e.g., Gm26870, Gm10800, Gm21738). Finally, cluster 5 was composed of actively proliferating microglia, as shown by their high expression of genes encoding cyclins (e.g., Ccnb2), proteins involved in chromosome condensation and segregation during mitosis (e.g., Cenpf, Smc2), DNA replication (e.g., Top2a, Mcm5) and proliferation markers (e.g., Mki67).
The cKO dataset was composed of 7 clusters. Alike microglia from the wild-type cluster 1, cKO microglia from cluster 1 were less distinctive except for a relatively higher expression of genes encoding ribosomal proteins (e.g., Rps15a, Rpl32, Rps5) and, therefore, resembled postnatal immature microglia. Cluster 2 was composed of microglia sharing some of the transcriptional signatures of disease-associated microglia (DAM [Keren-Shaul et al., 2017], e.g., Cd63, Cd9, Lyz2). These cells were also characterized by a higher expression of classical pro-inflammatory genes (e.g., Ccl12) or genes induced in microglia under pathological conditions (e.g., C3aR1) (Doolen et al., 2017). However, these microglia also exhibited an increased expression of the anti-inflammatory gene Mrc1, typically down-regulated by pro-inflammatory and up-regulated by anti-inflammatory cytokines (von Ehr et al., 2020; Zimmer et al., 2003). Cluster 2 did not seem to have an obvious wild-type counterpart although it shared some transcriptional signatures with subcluster 1.15. In addition, microglia from cKO cluster 2 was extremely similar to Cre-Het cells from subcluster 1.3 (see below). GABAB1R cKO microglia from cluster 3 resembled microglia from the wild-type cluster 2. These cells expressed higher levels of homeostatic microglia core genes (e.g., Fcrls, P2ry13, Hexb) and genes involved in synapse pruning (e.g., C1qa). Like microglia from the wild-type cluster 2, cKO cells from cluster 3 upregulated mitochondrial genes (e.g., mt-Co3, mt-Atp6, mt-Cytb) and downregulated numerous genes encoding ribosomal proteins. Cells belonging to cluster 4 appeared as expanding microglia from cluster 1. Like cells from cluster 1, they expressed higher levels of ribosomal subunit genes (e.g., Rps15a, Rpl32) but were distinct in that they expressed a higher level of genes involved in DNA replication and cell cycle progression (e.g., Hells, Cdca7). Cluster 5 was composed of cells expressing higher levels of many classical pro-inflammatory (e.g., Ccl12, Stat1) and interferon response (e.g., Ifitm3, Ifi44) genes. Notably, microglia from cluster 5 shared several gene expression signatures with cells collected from the white matter of adult mice exposed to a focal demyelinating injury caused by lysolecithin (LPC) injection (cluster 9 from Hammond et al., 2019). Cluster 5 resembled the wild-type subcluster 1.22, although it was considerably more abundant, consistent with the loss of the anti-inflammatory role of GABAB1Rs in microglia (Kuhn etal., 2004). Finally, microglia belonging to clusters 6 and 7 were easily identified as cells undergoing mitosis, as shown by the higher expression of genes encoding cyclins (e.g., Ccnb2), proteins involved in chromosome segregation during mitosis (e.g., Cenpf), DNA replication (e.g., Top2a) and proliferation markers (e.g., Mki67). In addition, cells from cluster 6 almost exclusively expressed high levels of several histone proteins (e.g., Hist1h1b).
The integrated dataset (wild-type+GABAB1 R cKO) was composed of 8 clusters that had a clear correspondence to the wild-type and cKO clusters. Microglia from clusters 1–3 resembled cells from wild-type or cKO clusters 1: immature postnatal microglia. In fact, these cells displayed a relative upregulation of genes implicated in metal homeostasis (Fth1, Ftl1, and Mt1), actin cytoskeleton dynamics (Tmsb4x, Pfn1, and Cfl1), and encoding ribosomal components (e.g., Rps26, Rps15a, Rpl32, and Rps5). Cells belonging to cluster 2 had a relative enrichment in Macf1, another gene involved in actin cytoskeleton dynamics and microglia from cluster 3 exhibited a small enrichment in ribosomal components and expressed higher levels of the long non-coding RNA Gm42418, resembling the wild-type cluster 3. Overall, microglia from clusters 2 and 3 were less distinctive than cells from cluster 1, although they exhibited slightly higher levels of homeostatic microglia core genes compared to cells from cluster 1. This last observation suggests that cells in clusters 2 and 3 might be transitioning from immature to homeostatic states. In contrast, microglia from cluster 4 were very distinctive. These cells expressed higher levels of homeostatic microglia core genes (e.g., Fcrls, P2ry13, Hexb) as well as genes involved in synapse pruning (e.g., C1qa, Trem2) and corresponded to microglia from wild-type and cKO clusters 2 and 3, respectively. Cluster 5 was the only cluster composed exclusively of GABAB1R cKO microglia and fully matched the cKO cluster 2 or Cre-Het subcluster 1.3 (see below). Like the wild-type cluster 5 and the cKO clusters 6 and 7, cluster 6 was composed of actively proliferating microglia as shown by the higher expression of genes involved in proliferation and DNA replication (e.g., Mcm5, Hells, Mki67) and by a cell cycle phase analysis. Microglia belonging to cluster 7 were characterized by a higher expression of classical pro-inflammatory genes (e.g., Ccl12, Stat1) and a large number of genes induced in microglia upon injury or pathological conditions (e.g., Ifi44, Ifi27l2a). These cells corresponded to the pro-inflammatory microglia from the cKO cluster 7 as well as the small wild-type subcluster 1.22. Consistent with this, GABAB1R cKO microglia were more abundant in cluster 7 than wild-type microglia, an expected result given the anti-inflammatory role of GABAB1 Rs in microglia (Kuhn et al., 2004). Finally, cells from cluster 8 (like wild-type cells from cluster 4) uniquely up-regulated a set of long non-coding RNAs whose exact function is yet unidentified (e.g., Gm26870, Gm10800, Gm21738).
Control (Cre-Het) clusters
Unsupervised clustering of Cx3cr1Cre/+ microglia returned 5 main clusters. Like microglia from wild-type or cKO cluster 1, Cre-Het microglia from cluster 1 shared transcriptional signatures of postnatal immature microglia (Li et al., 2019; Masuda et al., 2019). These cells exhibited a relative upregulation of genes implicated in metal homeostasis (e.g., Fth1, Ftl1), actin cytoskeleton dynamics (e.g., Tmsb4x, Pfn1, and Cfl1) and encoding ribosomal components (e.g., Rps26, Rps15a, Rpl32, and Rps5). However, like wild-type microglia, subclustering of Cre-Het cluster 1 revealed additional differences. In particular, subcluster 1.1 could be identified as prototypic postnatal immature microglia. In fact, the main transcriptional feature of microglia from subcluster 1.1 was a lower expression of genes that characterized subclusters 1.2 and 1.3. Like wild-type subcluster 1.22 or wild-type+cKO cluster 7, Cre-Het subcluster 1.2 contained cells expressing higher levels of pro-inflammatory (e.g., Ccl12, Stat1) and interferon response (e.g., Ifitm3, Ifi44, Ifi27l2a) genes typically induced in microglia upon injury or pathological conditions (Hammond et al., 2019). Similarly to microglia from wildtype subcluster 1.15, cKO cluster 2 or wild-type+cKO cluster 7, microglia belonging to subcluster 1.3 shared some of the transcriptional signatures of disease-associated microglia (DAM [Keren-Shaul et al., 2017], e.g., Cd63, Cd9, Lyz2). The higher expression of the anti-inflammatory gene Mrc1 further confirmed the resemblance of these microglia to cells from cKO cluster 2 or wild-type+cKO cluster 7. Like the wild-type cluster 2, Cre-Het clusters 2 and 3 were composed of cells expressing higher levels of homeostatic microglia core genes (e.g., Fcrls, P2ry13, Hexb), genes involved in synapse pruning (e.g., C1qa [Stevens et al., 2007], Trem2 [Filipello et al., 2018]). Like the wild-type cluster 3, cells belonging to Cre-Het cluster 4 appeared as a transitional state between clusters 1 and 2–3. These cells expressed intermediate levels of genes characterizing cluster 1 but - like wild-type microglia from cluster 3 - could be distinguished based on their relatively higher expression of Actb and the long non-coding RNA Gm42418. Finally, like wild-type cluster 5, Cre-Het cluster 5 was composed of actively proliferating microglia as shown by the higher expression of genes encoding cyclins (e.g., Ccnb2), proteins involved in chromosome condensation and segregation during mitosis (e.g., Cenpf), DNA replication (e.g., Top2a) and proliferation markers (e.g., Mki67).
Differential expression analyses
For Cre-Het versus wild-type analysis, all cells from wild-type (Cx3cr1+/+; GABAB1Rfl/fl) and Cre-Het (Cx3cr1Cre/+; GABAB1R+/+) controls were compared to identify genes expression changes that could be attributed to the loss of one copy of Cx3cr1. A differential expression analysis using all genes and all cells (no clusters) was performed using the FindMarkers function in the Seurat R package with Wilcoxon rank sum tests (log2 fold change = 0) and the identity class set on condition (wild-type or Cre-Het). Differentially expressed genes (DEGs) having an absolute value of the average log2 fold change ≥ 0.25 and an adjusted p value ≤ 0.01 were flagged as “Cre-Het DEGs” and used as additional filter applied to all differential expression analyses between wild-type and cKO microglia.
For each cluster composed of both wild-type and GABAB1R cKO microglia in similar proportions (i.e., clusters 1, 2, 3, 4, 6 and 8), a differential expression analysis was performed between wild-type and cKO cells. For this “within cluster” wild-type versus cKO differential expression analysis, first, clusters from Monocle were transferred to Seurat. Then, for cells belonging to each cluster, a differential expression analysis was performed using the FindMarkers function in the Seurat R package with Wilcoxon rank sum tests (absolute value of the average log2 fold change ≥ 0.25) and the identity class set on condition (wild-type or cKO). The list of differentially expressed genes (DEGs) for each cluster was then intersected with the Cre-Het DEG list and overlapping genes were removed. Finally, only DEGs with an adjusted p value ≤ 0.05 were used for further analysis. The fraction of DEGs was calculated dividing the number of DEGs in each cluster (or group of clusters) by the total number of DEGs found across all clusters. The same conclusions (cluster 4 having the highest proportion of DEGs) were reached with or without the filtering steps.
For the “pseudo-bulk” wild-type versus cKO analysis, a differential expression analysis using all genes and all cells (no clusters) was performed using the FindMarkers function in the Seurat R package with Wilcoxon rank sum tests (absolute value of the average log2 fold change ≥ 0) and the identity class set on condition (wild-type or cKO). Criteria for DEGs identification were an absolute value of the average log2 fold change ≥ 0.10 and an adjusted p value ≤ 0.01. A relatively low fold change was used in this analysis to account for dilution of differences occurring within specific clusters only. Cre-Het DEGs were filtered from the final list. Since pseudobulk analyses yielded a higher number of DEGs, a more stringent adjusted p value was used (i.e., 0.01 instead of 0.05).
For differential expression analyses using microglia from only males or females, cells were selected based on the expression of × (Xist) or Y (Ddx3y and Eif2s3y) chromosome genes. Identity was set as female for cells expressing Xist and exhibiting no Ddx3y and Eif2s3y expression while identity was set as male for cells expressing Ddx3y or Eif2s3y and exhibiting no Xist expression. For microglia from either males or females, DEGs between WT and cKO within each cluster were then computed in Seurat as described above.
Gene Ontology (GO) enrichment analysis of DEGs
A Gene Ontology (GO) enrichment analysis was performed in AmiGO using PANTHER Overrepresentation Test (released 20200407; annotation version and release date: GO Ontology database https://doi.org/10.5281/zenodo.3727280 Released 2020–03-23). The list of genes differentially expressed in cKO microglia within each cluster was used as input for a Gene Ontology enrichment analysis using all mouse genes as reference list. The annotation datasets used were: GO biological process complete, GO molecular function complete and GO cellular component complete. Enrichment was verified using Fisher’s Exact test with Bonferroni correction for multiple comparisons. The same analysis was carried out for downregulated genes, upregulated genes and for the combined list of up-regulated and downregulated genes.
Supervised discriminative gene identification via elastic net penalized regression
To identify genes predictive of control versus cKO microglia, a normalized expression matrix from the integrated cells composing clusters 1 to 4, 6 and 8 was used as input matrix. A hot-one-encoded response vector was used where wild-type was set to 0 and cKO to 1. 20% of the input matrix and response vector were randomly removed for each class. The remaining input matrix and response vector were used with cross validation (cv.glmnet, family set to Gaussian and ‘a’ ranging from 0 to 1) to determine optimal lambda and alpha hyper parameters. Model evaluation was run on the unseen 20% using the area under the curve and mean squared error. Once the best model was computed, coefficients were refitted using the whole input matrix. Genes were ranked based on their fitted coefficient value and further filtered on their median expression. Cre-Het genes were removed from the final list and a heatmap was generated as described above.
Denoising Expression data with a Weighted Affinity Kernel and Self-Supervision (DEWÄKSS)
Cells were processed using Scanpy and DEWÄKSS (Tjärnberg et al., 2020). Briefly, each sample was aggregated in a single expression matrix and then filtered as described above with the addition of a minimum number of counts required fora gene set to 10, cells were then normalized and transformed using Freeman-Tukey transform (FTT). DEWÄKSS was then run on FTT values (n_neighbors ranging from 5 to 250, n_pcs ranging from 10 to 300, use_global_err set to False, modest set to max, neighbor_alg set to bbknn using euclidean metric and samples as batch keys for neighbor_args). Denoised data were then used for further analysis. First, unsupervised clustering was performed using Leiden clustering with the adjacency matrix computed in DEWÄKSS. Then, a list of top markers for each cluster was generated using rank_genes_groups (method = ‘wilcoxon’, use_raw = False, layer = ‘Ms’) (Table S6). Next, a comparison of cell assignment to each cluster before and after denoising (DEWÄKSS clusters versus integrated Monocle clusters) was carried out. This comparison revealed an almost complete overlap between cells in cluster 4. Cells from cluster 4 were then selected and a differential expression analysis between denoised wild-type and cKO microglia was performed using rank_genes_groups as described above.
MERFISH image analysis and cell segmentation
Encoded MERFISH images were decoded using the MERlin pipeline (Emanuel and Babcock, 2020) available at https://github.com/emanuega/MERlin, using the codebook provided by Vizgen. Decoded data were used to generate binary mosaics of the tissue where transcripts were represented as a single pixel. A mask was applied to select only the S1 region and microglia were segmented using the signal of the established Tmem119 microglia marker. The segmentation script is available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH. Briefly, microglia were segmented as Tmem119 rich regions: sequential dilations and erosions on the Z projection of the binary mosaics representing Tmem119 allowed transcripts that were close together to merge into a larger object. Transcripts for each gene within these cells were then counted by quantifying the number of “on” pixels in their respective binary representation. For genes specific to or highly enriched in microglia, only the Z projection was used to count transcripts. For Gabbr1 and Gabbr2, which are also highly expressed in neurons, expression was assessed only in Z planes containing the microglia markers Tmem119, Fcrls and P2ry12 (TFP).
The expression of genes imaged in the non-combinatorial smFISH rounds was measured using a high pass filter followed by a peak-finding algorithm, also available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH. Briefly, transcripts were identified as local maxima on the high frequency images. The minimal intensity for one of these local maxima to be considered a transcript was automatically set for each gene as the 95th percentile of the intensity values of the filtered image. To avoid counting the same molecule twice, a minimal distance between two spots was set to 4 pixels (~0.44 μm).
In total, 6,886 microglia were detected. The total number of Z planes with a TFP count higher than 7 was recorded and 92 cells that did not reach this threshold in any Z plane were removed. 93 cells located at the edges of the S1 region mask were also removed.
Cell clustering and DE analysis of MERFISH data
After these filtering steps, a total of 3,563 control and 3,138 cKO microglia were retained for further analysis. However, as reported previously (Chen et al., 2015b; Moffitt et al., 2018), batch effects between MERFISH runs were observed. Since most of the genes in our panel may be affected in cKO microglia and based on the assumption that batch effects are largely not cell-specific, the influence of these batch effects was removed by calculating the total counts for all genes in all cells in each slice (i.e., not only microglia, and largely dominated by Gabbr neuronal signal). The total number of counts was calculated by summing the total spots on each image and the total number of cells in each brain slice was detected by binarizing the DAPI image and counting the number of objects. These average counts per cells were then used to normalize the raw RNA counts in microglia. As previously reported (Chen et al., 2015b; Moffitt et al., 2018), given the slice thickness, some cells were not imaged completely. To remove differences in RNA counts due to imaged cell volumes, RNA counts per cell were normalized by the imaged volume of each microglia. For both batch and volume correction, a scaling factor was used to ensure that the normalized expression values were on the same scale as the original counts.
Residual technical cell-specific biases that affected all genes equally were removed using the deconvolution strategy for scaling normalization (Lun et al., 2016a). This involved dividing all RNA density counts for each cell by a “deconvolved” cell-specific scaling “size factor.” The size factor is based on the pool-based total sum of counts across all genes for each cell and such that the mean size factor across all cells is equal to 1. Specifically, RNA density values were imported using the “SingleCellExperiment” R package (Amezquita et al., 2020) and scaling normalization by deconvolving size factors from cell pools was performed using the computeSumFactors function from the scran R package (Lun et al., 2016b). The logNormCounts function from the scater R package (McCarthy et al., 2017) was then used to compute the log-transformed normalized RNA density values for each cell which were used for downstream analyses. Of note, two mitochondrial genes (mt-Co3, mt-Nd2) imaged in the non-combinatorial rounds were too locally concentrated to count single transcripts. Gene expression analysis using integrated fluorescence intensity failed to clearly mark any defined cell group and therefore these two genes were removed from the subsequent analysis.
Given the low number of genes in our MERFISH panel, a reduction in the number of included genes or dimensionality reduction were not computationally necessary for clustering. Clustering of both control and cKO microglia was performed on the z-scores of the log-transformed normalized RNA densities per cell for all genes using the Jaccard-Louvain community-based detection with a k value of 20. This was implemented with the buildSNNGraph function in Scran while the Louvain method from the igraph package (Csardi and Nepusz, 2006) was used to identify communities. Next, data were imported in Seurat and cluster-enriched genes were identified using the FindMarkers function. For visualization, Uniform Manifold Approximation and Projection (UMAP) was used to embed cells in two dimensions.
A small cluster containing only 18 (15 control and 3 cKO) cells characterized by a low-level of pruning genes was not included for visualization or differential expression analysis. Of note, this cluster might correspond to the small fraction of WT cells in the scRNA-seq subcluster 4.2.
Lastly, differentially expressed genes between control and cKO microglia within each cluster were identified using the FindMarkers function in Seurat (absolute log-fold change threshold 0.25, adjusted p value lower than 0.05, ident cKO/control). Few Gabbr-negative cells were included in the Gabbr-enriched clusters and vice versa. For the differential expression analysis, these cells were removed by selecting only cells with a log-transformed normalized Gabbr1 and Gabbr1 RNA density higher than 1 (identified as local minimum in the Gabbr1 and Gabbr1 histogram distribution). A similar approach (removing few Gabbr-positive cells) was used for the non GABA-receptive clusters. Of note, similar conclusions (with, as expected, slightly lower fold changes) were reached with or without this last filtering step. Plots were generated using custom R scripts and ggplot2. For cell map visualization, cells were dilated with a disk of 13-pixel radius. The code for the downstream analysis is also available at https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH. https://doi.org/10.5281/zenodo.4899812.
Motion Sequencing (MoSeq)
Pre-processing code automatically identified the mouse in the arena, centered it in a 80 × 80 pixel square and aligned the mouse’s nose to the right and tail to the left. Next, principal component analysis (PCA) was automatically performed on the 80 × 80 pixel aligned movies of each mouse. Principal component scores for the data were then computed and fit to a generative model for mouse behavior through the use of computational inference techniques (Wiltschko et al., 2015; Markowitz et al., 2018). The model returned a set of behavioral syllables that characterizes the expressed behavior of those mice, and the statistics that govern the frequency and the order in which those syllables were expressed in the experiment. Syllable usage was calculated by summing the number of occurrences of each syllable and dividing by total syllable usage across a recording session, converting syllable usage into a percentage. Each syllable was assigned a label by a human observer. Note that out of all syllables discovered, only a subset is shown (including those with relevance for the identified phenotype and all syllables that were significantly different as assessed by a statistical test). Syllable speed was calculated by computing, for all mice within each group, the mean of the mouse centroid speeds (frames were labeled using the respective syllable label). Transition matrices were calculated by counting the total number of occurrences in which syllable A transitions into syllable B (for all syllables). For comparing significant syllables between experimental conditions, a z-test was run on bootstrapped syllable usage distributions (one test per syllable) followed by the non-negative Benjamini-Hochberg procedure with a 10% family-wise error rate to correct for multiple comparisons.
ADDITIONAL RESOURCES
We have developed a searchable website where users can examine microglia gene expression patterns in our P15 wild-type and cKO scRNA-seq datasets http://zoidberg.bio.nyu.edu/index.
Supplementary Material
Figure S1. Consequences of early postnatal microglia depletion for cortical connectivity, related to Figure 1 (A) Representative images and quantitation of the density and layer distribution of PV cells in control (n = 4) and microglia-depleted (n = 4) mice at P15 after P1-P15 microglia depletion. ns p > 0.05, Student’s t test (for density) and One-Way ANOVA followed by Sidak’s multiple comparisons test (for layer distribution). Scale bar equal 100 μm.
(B) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in P10 control (n = 4) or depleted (n = 4) mice after P1-P10 microglia depletion and in P12 control (n = 3) or depleted (n = 5) mice after P1-P12 microglia depletion. ns p > 0.05, One-Way ANOVA followed by Tukey’s multiple comparisons test.
(C) Representative images and quantitation of Iba1+ microglia density in control (n = 4) and microglia-depleted (n = 4) mice at P30 after P15-P30 microglia depletion. ***p < 0.001, Student’s t test. Scale bar equal 100 μm.
(D) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in P30 control (n = 4) and depleted (n = 3) mice after P15-P30 microglia depletion. One brain with incomplete depletion was excluded from the analysis. ns p > 0.05, Student’s t test.
(E) Representative images and quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in the primary visual cortex (V1) of P15 control (n = 6) and depleted (n = 9) mice after P1-P15 microglia depletion and of P25 control (n = 12) and depleted (n = 10) mice after P1-P25 microglia depletion. ns p > 0.05 and *p < 0.05, One-Way ANOVA followed by Tukey’s multiple comparisons test. Scale bar equal 2 μm.
(F) Representative images and quantitation of synapses made in L1 of S1 by SST+ cells (SSTCre;Ai34 Synaptophysin-tdTomato labeled SST+ presynaptic terminals colocalizing with Gephyrin) in P15 control (n = 8) and depleted (n = 6) mice after P1-P15 microglia depletion. *p < 0.05, Student’s t test. Scale bar equal 2 μm.
(G) Quantitation of the density of SST cells in P15 control (n = 6) and microglia-depleted (n = 5) mice after P1-P15 microglia depletion. ns p > 0.05, Mann-Whitney test.
(H) Traces, frequency and amplitude of sIPSCs (n = 14 cells from 3 control and n = 17 cells from 5 depleted mice). ***p < 0.001, ns p > 0.05, Student’s t test.
(I) Traces, frequency and amplitude of sEPSCs (n = 17 cells from 4 control and n = 18 cells from 5 depleted mice). ***p < 0.001, ns p > 0.05, Student’s t test.
(J) sEPSC/sIPSC ratio (n = 14 cells from 3 control and n = 15 cells from 3 depleted mice). ns p > 0.05, Mann-Whitney test.
(K) Schematic of synapses analyzed in L and M.
(L) Representative images and quantitation of VGlut2+Homer1+ synapses onto excitatory cell dendrites (TdTomato+, labeled by virus injection of pyramidalneurons) in P15 control (n = 6) and depleted (n = 4) mice. **p < 0.01, Student’s t test. Scale bar equal 2 μm.
(M) Quantitation of VGlut1+Homer1+ synapses made onto L4 excitatory cells (NeuN) in P15 control (n = 6) and depleted (n = 4) mice (left), onto PV cells in P15 control (n = 9) or depleted (n = 10) mice (middle) and onto excitatory cell dendrites (TdTomato+, labeled by virus injection of pyramidal neurons) in P15 control (n = 7) or depleted (n = 4) mice. ns p > 0.05, Mann-Whitney test for excitatory neurons and Student’s t test for PV cells and dendrites.
(N) Representative images of Iba1+ microglia in control and P1-P15 microglia-depleted brains at the indicated stages of repopulation and quantitation of microglia density in control and microglia-repopulated brains at P17, P19, P21, P25 and P30 (n = 3–4 mice per condition). ***p < 0.001, ns p > 0.05, One-Way ANOVA followed by Sidak’s multiple comparisons test. Scale bar equal 100 μm.
(O) Quantitation of synapses made in L1 of S1 by SST+ cells (SSTCre;Ai34 labeled presynaptic terminals colocalizing with Gephyrin) in P30 control (n = 4) and microglia-repopulated (n = 5) mice after P1-P15 microglia depletion. *p < 0.05, Student’s t test.
(P) Quantitation of Syt2+Gephyrin+ synapses (left), SSTCre;Ai34+Gephyrin+ synapses (middle) and VGlut2+Homer1+ synapses (right) made by PV, SST and thalamic cells in P60 control (n = 6–8) and microglia-repopulated (n = 4–7) mice after P1-P15 microglia depletion. ExCs: excitatory cell soma. ns p > 0.05, Student’s t test for SST+ and thalamic synapses and Mann-Whitney test for PV synapses.
All data are mean ± SEM, each data point represents one experimental animal except in F, I and J where it represents one cell. Arrowheads indicate colocalization.
Figure S2. Microglia-PV synapse interactions during development, related to Figure 2 (A) Masks showing the criteria for contact identification in the in vivo imaging experiments. Percentages indicate the fraction of bouton area contacted by microglia.
(B) Proportion of PV boutons contacted by one microglia during the in vivo imaging experiments in deep layer 3 (n = 15 cells contacting 68 boutons) or layer 4 (n = 36 cells contacting 154 boutons). Data are the same as in (C) and (I) where they are shown as pooled dataset. ns p > 0.05, Mann-Whitney test. Data are mean ± SEM, each data point represents one cell.
(C) Distribution of Cx3cr1GFP/+microglia contacting the indicated percentages of PV boutons in a 20 μmradius around the microglia cell body over 20 minutes (n = 51 cells from 6 mice) during the in vivo imaging experiments. PV boutons are labeled with PVe-Syp-tdTomato. The distribution is shown as density histogram and kernel density estimation. The first peak corresponds to 11.7% of PV boutons contacted by microglia and the second peak to 60.9%. The local minimum of the bimodal distribution is 26.4%. These data were pooled with data in (I) to generate the graph in Figure 3C.
(D) Distribution of microglia-PV boutons interactions for the indicated duration during the in vivo imaging experiments. The distribution is shown as density histogram and kernel density estimation of the proportion of Syp-tdTomato+ PV synaptic boutons contacted by microglia for the indicated duration (n = 222 PV boutons from 6 mice).
(E) Duration of contacts between microglia interacting with a minority (n = 11 cells) or the majority (n = 25 cells) of local PV boutons during the in vivo imaging experiments. *p < 0.01, Mann-Whitney test. These data represent the subset of data shown in (D) for which it was possible to visualize the microglia cell body and they were pooled with data in (K) to generate the graph shown in Figure 3D. Data were split based on the local minimum of the kernel density estimation in (C). Data are mean ± SEM, each data point represents one cell.
(F) Time-lapse images from Video S2 showing a Syp-tdTomato+ PV bouton being enwrapped by a microglia process (Cx3cr1GFP/+) and contacted by a second process over 10 minutes. Scale bar equal 10 μm.
(G) Time-lapse images from Video S3 showing a Cx3cr1GFP/+ microglial process that terminates into what resembles a phagocytic cup engulfing a Syp-tdTomato+ PV bouton over 20 minutes. Scale bar equal 10 μm.
(H) Time-lapse image from in vivo imaging of Syp-Gamillus+ PV boutons and microglia genetically labeled using Tmem119CreER/+;Ai14 mice. Scale bar equal 10 μm.
(I) Distribution of Tmem119CreER/+;Ai14 microglia contacting the indicated percentages of PV boutons (labeled with PVe-Syp-Gamillus) in a 20 μm radius around the cell body over 20 minutes (n = 37 cells from 6 mice). The distribution is shown as density histogram and kernel density estimation. The first peak corresponds to 18.1% of PV boutons contacted by microglia and the second peak to 57.4%. The local minimum of the bimodal distribution is 34.4%. These data were pooled with data in (C) to generate the graph in Figure 3C.
(J) Distribution of microglia-PV boutons interactions for the indicated duration. The distribution is shown as density histogram and kernel density estimation of the proportion of Syp-Gamillus+ PV synaptic boutons contacted by microglia for the indicated duration (n = 115 PV boutons from 6 mice).
(K) Duration of contacts between microglia interacting with a minority (n = 13 cells) or the majority (n = 23 cells) of local PV boutons. *p < 0.01, Student’s t test. These data represent the subset of data shown in (J) for which it was possible to visualize the microglia cell body and they were pooled with data in E to generate the graph shown in Figure 3D. Data are mean ± SEM, each data point represents one cell.
(L) Representative images of the same 3D Imaris space in two different orientations. Note that, in the visualization of a 3D volume, boutons can appear closer than they are depending on the orientation. In contrast, “real contacts” appear close regardless of the orientation. Scale bar equal 5 μm.
(M) Representative 3D reconstruction of a microglia process and surrounding PV boutons. Contacted PV boutons are reconstructed as red spots whereas PV boutons not contacted by microglia are reconstructed as light blue spots. Full and empty arrowheads indicate positive and negative (i.e., boutons that did not meet the contact criteria) examples, respectively. The yellow empty arrowhead indicates a bouton that is far from the process. In all relevant figures, only contacted boutons (see methods for criteria) have been highlighted as “reconstructed spots.” Scale bar equal 5 μm.
Figure S3. Expression of GABAb receptors in microglia, related to Figure 2 (A) Schematic of analysis and heatmaps showing ligands-receptor pairs that are leading candidates for mediating interneuron-microglia communication. The heatmap on the left is showing the expression (at the indicated developmental time points) of putative ligands enriched in interneurons as compared to pyramidal cells. For P5 and P10 interneurons, the corresponding values in SST and PV cells were averaged. The list is ranked according to specificity and developmental upregulation. For GABA, the expression of the GABA-synthetizing enzyme Gad1 is shown. The heatmap on the right is showing the expression (at the indicated developmental time points) of receptors expressed in microglia that are experimentally validated interactors of the ligands shown on the left. For the P3-P9 time point, values at P3 and P9 were averaged.
(B) Venn diagram depicting the fraction of microglia expressing Gabbr1+ mRNA, Gabbr2+ mRNA as well as double positive for both Gabbr1+ and Gabbr2+ mRNAs at P15 (smFISH: single molecule fluorescent in situ hybridization). The diagram is a visualization of the data shown in Figure 3F.
(C) Validation of the specificity of GABAB1R antibody. Representative images from P15 controls and VGlut1Cre;Gabbr1 cKO mice, lacking GABAB1R in cortical excitatory neurons. Note the absence of GABAB1R signal within NeuN+ excitatory neurons in VGlut1Cre cKO mice. Scale bar equal 10 μm.
(D) Left: quantitation of the percentage of microglia (Iba1+) expressing GABAB1R protein at P0, P10 and P15 in control and Cx3cr1Cre GABAB1R cKO mice (n = 3–6 each). ns p > 0.05, **p < 0.01, ***p < 0.001, One-Way ANOVA followed by Sidak’s multiple comparisons test. Note the higher expression of GABAB1R in controls at P15 compared to P0 or P10. Right: quantitation of the percentage of microglia (Iba1+) expressing GABAB1R protein at P15 in control and Tmem119CreER GABAB1R cKO mice (n = 4 each). *p < 0.05, Student’s t test. Data are mean ± SEM, each data point represents one experimental animal.(E) Representative images and quantitation of microglia (Iba1+) expressing both GABAb-|R and GABAB2R proteins in P15 control mice (n = 3) within layer 4 of S1. Scale bar equal 5 μm. See STAR Methods for details about noise in the IHC experiment versus smFISH. Arrowheads indicate colocalization. Data are mean ± SEM, each data point represents one experimental animal.(F) Gabbr1 and Gabbr2 expression in adult microglia/macrophages from different brain regions. Data and visualization are from Saunders et al. (2018). FC: frontal cortex, TH: thalamus, STR: striatum, PC: posterior cortex, CB: cerebellum, SN: substantia nigra, HC: hippocampus, GP: globus pallidus. The reported confidence intervals reflect statistical sampling noise.(G) Fraction of GABA-receptive microglia (defined as expressing both Gabbr1 and Gabbr2 mRNAs by single molecule fluorescent in situ hybridization) in layer 4 of S1 and the CA1 region of the hippocampus of Cx3cr1GFP/+ mice at P15. S1 data are the same as shown in Figure 3F. Data are mean ± SEM, each data point represents one experimental animal.(H) 3D reconstruction and fraction of PV boutons (PVe-Syp-tdTom) encased by Gabbr2+ and Gabbr2-microglia (Cx3cr1GFP/+) at P15 (n = 58 Gabbr2+ and 59 Gabbr2-cells from 4 mice). ***p < 0.001, Mann-Whitney test. Scale bar equal 1 μm. Data are mean ± SEM, each data point represents one cell.(I) 3D reconstruction and fraction ofVGlut2+ boutons encased by Gabbr2+ and Gabbr2-microglia (Cx3cr1GFP/+) at P15 (n = 23 Gabbr2+ and 36 Gabbr2-cells from 4 mice). *p < 0.05, Student’s t test. Scale bar equal 1 mm. Data are mean ± SEM, each data point represents one cell.
Figure S4. Synaptic phenotype of GABAB1R cKO mice, related to Figure 3 (A) Left: quantitation of PV boutons contacted by microglia at P15 in wild-type control mice (n = 7, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 8, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Tukey’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4A. Right: quantitation of PV boutons contacted by microglia at P15 in control (n = 4, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 6, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. *p < 0.05, **p < 0.01.(B) Left: quantitation of VGlut2 boutons contacted by microglia at P15 in wild-type control mice (n = 7, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 6, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Tukey’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4B. Right: quantitation of VGlut2 boutons contacted by microglia at P15 in control (n = 8, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 8, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. ns p > 0.05.(C) Distribution of microglia-PV boutons interactions for the indicated duration during the in vivo imaging experiments in controls (n = 181 PV boutons from 6 mice) and cKOs (Cx3cr1GFP/+;Tmem119CreER/+;GABAB1Rfl/fl, n = 99 PV boutons from 3 mice) cKO mice. The distribution is shown as kernel density estimation of the proportion of PV boutons contacted by microglia for the indicated duration. Control data are the same as in Figure S2D but limited to a total duration of 20 minutes to allow comparison with cKO data (see STAR Methods). **p < 0.01, Kolmogorov–Smirnov test. Note the complex effect of GABAB1R removal on the contact duration, the cKO distribution is bimodal with some contacts having a shorter duration and others being abnormally long.(D) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons at P15 in wild-type control mice (n = 8, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+heterozygous control mice (n = 6, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Holm-Sidak’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4H. Right: Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons at P15 in control (n = 8, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 6, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. *p < 0.05, **p < 0.01.(E) Image, relative frequency histogram and overlaid kernel density estimation of Syt2+Gephyrin+ synapses imaged using STED super-resolution microscopy andmade by PV cells onto excitatory neurons at P15 in wild-type controls (n = 58 cells from 3 Cx3cr1+/+;GABAB1Rfl/fl mice) and GABAB1R cKO mice (n = 66 cells from 3 Cx3cr1Cre/+; GABAB1Rfl/fl mice). ***p < 0.001, Kolmogorov–Smirnov test. The dotted line indicates the soma outline.(F) Left: quantitation of VGlut2+Homer1+ synapses made onto excitatory neurons at P15 in wild-type control mice (n = 6, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 9, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Holm-Sidak’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4I. Right: VGlut2+Homer1+ synapses made onto excitatory neurons at P15 in control (n = 9, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 8, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. ns p > 0.05.(G) Quantitation of VGlut2+Homer1+ synapses onto L4 PV neurons in P15 control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(H) Quantitation of VGlut1+Homer1+ synapses onto L4 excitatory cells (left) or pyramidal cell dendrites (right) in control (n = 7–9, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 6–7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Mann-Whitney test for somatic analysis and Student’s t test for dendritic analysis. (I) and (J) Traces, frequency and amplitude of mIPSCs (n = 26 cells from 3 Tmem119+/+;GABAB1Rfl/fl control mice and n = 28 cells from 4 Tmem119CreER/+; GABAB1Rfl/fl cKO mice) and mEPSCs (n = 26 cells from 3 Tmem119+/+; GABAB1Rfl/fl control mice and n = 31 cells from 4 Tmem119CreER/+;GABAB1Rfl/fl cKO mice) recorded from layer 4 excitatory neurons at P15. ***p < 0.001, ns p > 0.05, Student’s t test except for mEPSC amplitude where Mann-Whitney test was used.(K) mEPSC/mIPSC frequency ratio (n = 26 cells from 3 Tmem119+/+; GABAB1Rfl/fl control mice and n = 28 cells from 4 Tmem119CreER/+;GABAB1Rfl/fl cKO mice). ***p < 0.001, Mann-Whitney test.(L) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto S1 L4 excitatory neurons in control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P30. Right: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto S1 L4 excitatory neurons in control (n = 6, Tmem119+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Tmem119CreER/+;GABAB1Rfl/fl) mice at P30. **p < 0.01, Student’s t test.(M) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto V1 L4 excitatory neurons at P30 in control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. Right: quantitation of PV+ Gephyrin+ synapses made onto NeuN+ cells in the dorsolateral striatum of P30 control (n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. *p < 0.05, **p < 0.01, Student’s t test.(N) Images, 3D reconstruction and quantitation of SST synapses (GAD65+Gephyrin+ synapses with GAD65 presynaptic boutons completely encased within genetically labeled SST axons) in layer 1 of S1 at P15 in control (n = 5; SSTFlp/+; RCE:FRT; Cx3cr1Cre/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 4; SSTFlp/+; RCE:FRT; Cx3cr1+/+; GABAB1Rfl/fl) mice. *p < 0.05, Student’s t test.All data are mean ± SEM, each data point represents one experimental animal except in I-K where they represent cells. Scale bars equal 1 mm. Arrowheads indicate colocalization.
Figure S5. The synaptic phenotype of GABAB1R cKO mice is not due to off-target effects, related to Figure 3 (A) Representative images and quantitation of the density of microglia in control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(B) Representative images and quantitation of the density of PV cells in control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(C) Representative images and quantitation of the density of SST cells (GFP+) in control (n = 8, SSTFlp/+; RCE:FRT; Cx3cr1Cre/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 7, SSTFlp/+; RCE:FRT; Cx3cr1+/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(D) Quantitation of the density of PV and SST cells in control (n = 6, Lhx6GFP/+; Cx3cr1+/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 6, Lhx6GFP/+; Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P10. PV cells were identified as GFP+SST− cells. ns p > 0.05, Student’s t test.(E) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons at P10 in control (n = 3, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(F) Representative images and quantitation of the fraction of L4 PV cells surrounded by perineuronal nets (WFA) in control (n = 4, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15 and P30. ns p > 0.05, One-Way ANOVA and Holm-Sidak multiple comparisons test. Scale bar equal 20 μm.(G) Quantitation of neurons (labeled with NeuN) expressing Gabbr1 mRNA in control (n = 4, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 3, Cx3cr1Cre/+; GABAB1R+/+) mice. Gabbr1 mRNA was detected by single-molecule in situ hybridization specifically for the exons deleted in the cKOs (7 and 8). ns p > 0.05, Student’s t test.(H) Time course of mean Racine scores of seizures induced by intraperitoneal injection of kainic acid (20 mg/kg) into control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. Seizures were scored as described in the STAR Methods. Two-Way ANOVA for repeated-measures with genotype (p > 0.05) and time (p < 0.001) as factors. Interaction between time and genotype p > 0.05.(I) Maximum and cumulative seizure scores over 90 minutes from the time of kainic acid injection. ns p > 0.05, Mann-Whitney test.
(J) Latency to reach the status epilepticus and/or death. ns p > 0.05, Mann-Whitney test.
(K) and (L) Traces, frequency and amplitude of mIPSCs (n = 16 cells from 3 control and n = 22 cells from 3 cKO mice) and mEPSCs (n = 18 cells from 3 control andn = 25 cells from 3 cKO mice) recorded from layer 4 excitatory neurons at P60. **p < 0.01, ns p > 0.05, Mann-Whitney test for mIPSCs and Student’s t test for mEPSCs.(M) Images and quantitation of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons in P60 control (Ctl, n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice as well as cKO (n = 5) and control (n = 3) mice that underwent microglia depletion from P30 to P60. ns p > 0.05, *p < 0.05, One-Way ANOVA followed by Sidak’s multiple comparisons test. Scale bar equal 2 μm.(N) Quantitation of VGlut2+Homer1+ synapses made onto excitatory neurons in P60 control (Ctl, n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(O) Quantitation of PV+ Gephyrin+ synapses made onto NeuN+ cells in the dorsolateral striatum of P60 control (Ctl, n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. *p < 0.05, Student’s t test.All data are mean ± SEM, each data point represents one experimental animal except in K and L where they represent cells. Arrowheads indicate colocalization.
Figure S6. Loss of GABAB1Rs does not fundamentally alter the range of microglial states but causes a downregulation of pruning-related genes, related to Figure 5 (A) UMAP plots showing the integration of P15 wild-type microglia with embryonic, early postnatal and P30 datasets from Hammond et al. (2019).(B) UMAP plot of Cre-Het control (Cx3cr1Cre/+) microglia showing 5 clusters and representative genes enriched in each cluster.(C) UMAP plot of Cre-Het control microglia from cluster 1 further subjected to subclustering. Representative enriched genes are shown.
(D) UMAP plot of GABAB1R cKO microglia from P15 S1 cortex showing 7 main clusters and representative genes enriched in each cluster.(E) Pie charts indicating the contribution of each wild-type or cKO cluster to the clusters of the integrated dataset. The overall filled area is proportional to the total number of cells within each cluster.
(F) UMAP plots of Cre-Het control and cKO integrated scRNaseq dataset. See methods for the different integration of this dataset as compared to WT control andcKO data.(G) Same as (F), showing 10 mixed clusters and representative genes enriched in each cluster.
(H) Mixed cluster contributions to total differentially expressed genes (DEGs) between Cre-Het control and cKO microglia.
(I) Violin plots of normalized log-expression values for representative genes significantly downregulated in cKO microglia from Cre-Het-cKO cluster 3. See methods for the different integration of Cre-Het control and cKO data as compared to WT control and cKO data.(J) Violin plots of normalized log-expression values for representative genes significantly downregulated in cKO microglia from Cre-Het-cKO cluster 5.
(K) Heatmap showing the output of an elastic net regularized classifier trained to learn which genes are predictive of control wild-type and GABAB1R cKO microglia using all genes as features and wild-type or cKO as labels. Genes are ordered by increasing mean expression. The top 5 genes are predictive of wild-type microglia (downregulated in cKO) and the remaining 3 genes are predictive of cKO microglia (upregulated in cKO).(L) UMAP plots of control and cKO integrated scRNaseq dataset after denoising expression data with a weighted affinity kernel and self-supervision method(DEWÄ KSS, see STAR Methods).(M) Same as (L) followed by unsupervised clustering and showing 8 mixed clusters. See also Table S2 for genes enriched in each cluster.(N) Volcano plot showing differentially expressed genes between control and cKO microglia from cluster 4 after denoising. The negative log10-transformed p values are plotted against the log2 fold change. Differentially expressed genes with an absolute log2 fold change higher than 0.5 and -log10 (p value) higher than or equal to 20 are depicted in orange. Top downregulated genes are highlighted. A full list is shown in Table S2.
Figure S7. MoSeq reveals a biphasic behavioral phenotype in GABAB1R cKO mice. Related to Figure 7 (A) Expression probability of syllable usage in P30 wild-type control (n = 11), cKO (n = 15) and Cre-Het control (n = 5) female mice. Syllables are ordered by differential usage (left: cKO-enriched syllables; right: cKO-downregulated syllables). Here and in the rest of the figure, only relevant or significantly differentially used syllables are shown. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM. Here and in the rest of the figure, syllable labels were assigned by a human observer.(B) Expression probability of syllable usage in P30 wild-type control (n = 6), cKO (n = 14) and Cre-Het control (n = 7) male mice. Syllables are ordered by differential usage. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.
(C) Expression probability of syllables enriched in P60 control (n = 19, both sexes) and cKO (n = 14, both sexes) mice. Syllables are ordered by differential usage(left: cKO-enriched syllables; right: cKO-downregulated syllables). *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.
(D) Heatmap depicting the position of P60 control (n = 10 males and 9 females) and cKO (n = 5 males and 9 females) mice during MoSeq.(E) Graph showing the mean syllable speed in control (n = 19) and GABAB1R cKO (n = 14) mice. Syllables are ordered by differential speed (left: syllables having a higher speed in cKOs and right: syllables with a lower speed in the cKOs). *p < 0.05, t test on bootstrapped syllable speed distribution corrected for false discovery rate. Data are mean ± SEM.(F) Expression probability of syllables enriched in P60 Cx3cr1Cre cKOs (n = 9 females and 10 males) and Tmem119CreER cKOs (n = 3 females and 3 males) mice. Only significantly differentially used syllables are shown. In females, almost all syllables were similarly used. Males from both groups used high velocity syllables although Tmem119CreER cKOs appeared even more active than Cx3cr1Cre cKOs. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.(G) Summary of the scalar information for P60 control (n = 19) and GABAB1R cKO (n = 14) mice. Velocity is measured in mm/s; height, width and length are measured in mm. ***p < 0.001, ns p > 0.05, Student’s t test. The inset shows the probability density function (PDF) of the two-dimensional velocity, p < 0.001, Two-sample Kolmogorov–Smirnov test.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||
|---|---|---|---|---|---|
|
| |||||
| Antibodies | |||||
|
| |||||
| mouse anti-Synaptotagmin-2 | ZFIN | Cat #ZDB-ATB-081002–25; RRID:AB_10013783 | |||
| mouse anti-gephyrin | Synaptic Systems | Cat #147 011; RRID:AB_887717 | |||
| rabbit anti-NeuN | Millipore | Cat #ABN78; RRID:AB_10807945 | |||
| guinea-pig anti-NeuN | Millipore | Cat #ABN90P; RRID: AB_2341095 | |||
| guinea-pig anti-VGlut2 | Millipore | Cat #AB2251; RRID: AB_1587626 | |||
| guinea-pig anti-VGlut2 | Synaptic Systems | Cat #135404; RRID: AB_887884 | |||
| guinea-pig anti-VGlutl | Millipore | Cat #AB5905; RRID: AB_2301751 | |||
| rabbit anti-Homer 1b/c | Synaptic Systems | Cat #160023; RRID: AB_2619858 | |||
| mouse anti-parvalbumin | Sigma | Cat#P-3088; RRID:AB_477329 | |||
| rabbit anti-DsRed | Clontech | Cat #632496; RRID:AB_10013483 | |||
| chicken anti-GFP | Aves Lab | Cat #1020; RRID:AB_10000240 | |||
| rabbit anti-Iba1 | Wako Chemicals | Cat #019–19741; RRID: AB_839504 | |||
| guinea-pig anti-Iba1 | Synaptic Systems | Cat #234 004; RRID: AB_2493179 | |||
| guinea-pig anti-parvalbumin | Swant | Cat #GP72; RRID: AB_2665495 | |||
| rabbit anti-C1q | Abcam | Cat #ab182451; RRID: AB_2732849 | |||
| rat anti-CD68 | Bio-Rad | Cat #MCA1957; RRID: AB_322219 | |||
| mouse anti-GABABRI | NeuroMab | Cat #73–183; RRID: AB_10672843 | |||
| guinea-pig anti-GABABR2 | Millipore | Cat #AB2255; RRID: AB_10563515 | |||
| Wisteria Floribundas (WFA) | Sigma | L1516–2mg; RRID: AB_2620171 | |||
| rabbit anti-SST | Peninsula Laboratories | Cat #T4103.0050; RRID:AB_518614 | |||
| mouse anti-GAD65 | Millipore | Cat #MAB351R; RRID: AB_94905 | |||
| Cd11b-PE | BioLegend | Cat #101208; RRID: AB_312791 | |||
| CD45-APC-Cy7 | BioLegend | Cat #103116; RRID: AB_312981 | |||
| Cx3cr1-APC | BioLegend | Cat #149008; RRID: AB_2564492 | |||
|
| |||||
| Bacterial and virus strains | |||||
|
| |||||
| AAV-PHP.eB-PVe(S5E2)-Syp-dTomato | Vormstein-Schneider et al., 2020 | N/A | |||
| AAV-PHP.eB-PVe(S5E2)-Syp-GamiIIus | This paper | N/A | |||
|
| |||||
| Chemicals, peptides, and recombinant proteins | |||||
|
| |||||
| PLX5622 powder | Plexxikon Inc; Spangenberg et al., 2019 | N/A | |||
| PLX5622-formulated AIN-76A diet | Research Diets | N/A, Custom made | |||
| Control AIN-76A diet | Research Diets | Cat D10001i | |||
| Tamoxifen | Sigma-Aldrich | Cat #T5648–1G | |||
| Kainic acid | Sigma-Aldrich | Cat #K0250–10MG | |||
| Percoll® Density Gradient Media | Fisher Scientific | Cat #17–0891-01 | |||
|
| |||||
| Critical commercial assays | |||||
|
| |||||
| Chromium Single Cell 3′ Library and Gel Bead Kit v3.0 | 10x Genomics | Cat# PN-1000075 | |||
| Chromium Single Cell 3′ Library Construction Kit v3.0 | 10x Genomics | Cat# PN-1000078 | |||
| Chromium Chip B Single Cell kit | 10x Genomics | Cat# PN-1000154 | |||
| Chromium i7 Multiplex Kit | 10x Genomics | Cat# PN-120262 | |||
| Imaging Reagent Kit | Vizgen | Cat# IK-18 | |||
|
| |||||
| Deposited data | |||||
|
| |||||
| Raw data files for single-cell RNA-seq | NCBI Gene Expression Omnibus | GSE159947 | |||
|
| |||||
| Experimental models: cell lines | |||||
|
| |||||
| HEK293FT | ThermoScientific | Cat #R70007 | |||
|
| |||||
| Experimental models: organisms/strains | |||||
|
| |||||
| Mouse: B6.129P2(Cg)-Cx3cr1tm1Litt/J | Jackson Laboratories | Cat #005582; RRID: IMSR_JAX:005582 | |||
| Mouse: GABAB1 floxed | A gift from B. Bettler, Haller et al., 2004 | N/A | |||
| Mouse: B6J.B6N(Cg)-Cx3cr1tm11(cre)Jung/J | Jackson Laboratories | Cat #025524; RRID: IMSR_JAX:025524 | |||
| Mouse: C57BL/6-Tmem119em1(cre/ERT2)Gfng/J | Jackson Laboratories Kaiser and Feng, 2019 | Cat #031820; RRID: IMSR_JAX:031820 | |||
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratories | Cat #007914; RRID: IMSR_JAX:007914 | |||
| Mouse: B6.Cg-Gt(ROSA)26Sortm32(CAG-COP4*H134R/EYFP)Hze/J | Jackson Laboratories | Cat #024109; RRID: IMSR_JAX:024109 | |||
| Mouse: B6;129P2-Pvalbtm1(cm)Arbr/J | Jackson Laboratories | Cat #008069; RRID: IMSR_JAX:008069 | |||
| Mouse: Tg(Cx3cr1-cre)MW126Gsat/Mmucd | MMRRC | Cat #036395-UCD; RRID:MMRRC_036395-UCD | |||
| Mouse: B6;129S-Gt(ROSA)26Sortm34’1(CAG-Syp/tdTomato)Hze/j | Jackson Laboratories | Cat #012570; RRID: IMSR_JAX:012570 | |||
| Mouse: C57BL/6-Tmem119em2(EGFP)Gfng/j | Jackson Laboratories; Kaiser and Feng, 2019 | Cat #031823; RRID: IMSR_JAX:031823 | |||
| Mouse: Ssttm3.1(flpo)Zjh/J | Jackson Laboratories | Cat #028579; RRID: IMSR_JAX:028579 | |||
| Mouse: Gt(ROSA)26Sortm12(CAG-EGFP)Fsh/Mmjax | Jackson Laboratories | Cat #032038; RRID: MMRRC_032038-JAX | |||
| Mouse: C1qa−/− | 3otto et al., 1998 | N/A | |||
| Mouse: Ssttm2.1(cre)Zh/J | Jackson Laboratories | Cat #013044; RRID: IMSR_JAX:013044 | |||
| Mouse: B6;129S-Slc 17a7tm1.1(cre)Hze/J | Jackson Laboratories | Cat #023527; RRID: IMSR_JAX:023527 | |||
| Mouse: Gt(ROSA)26Sortm1.1(CAG-EGFP)Fsh/Mmjax | Jackson Laboratories | Cat #032037; RRID: RRID:MMRRC_032037-JAX | |||
| Mouse: Tg(Lhx6-EGFP)BP221Gsat/Mmmh | MMRRC | Cat #000246-MU; RRID:MMRRC_000246-MU | |||
| Mouse: C57BL/6J | Jackson Laboratories | Cat #000664; RRID: IMSR_JAX:000664 | |||
| Mouse: BALB/cJ | Jackson Laboratories | Cat #000651; RRID: IMSR_JAX:000651 | |||
|
| |||||
| Oligonucleotides | |||||
|
| |||||
| RNAscope® Probe-Mm-Gabbr1 | ACDBio | Cat #425181; | |||
| RNAscope® Probe-Mm-Gabbr2 | ACDBio | Cat #317971; | |||
| RNAscope® Probe-Mm-Ex7–8-Gabbr1 | ACDBio | Custom made | |||
| MERFISH 12-bit probe library | Vizgen | Cat #VZG114 | |||
| Primer WPRE sequence Forward: AGC TCC III CCG GGA CTT TC | This paper | N/A | |||
| Primer WPRE sequence Forward: CAC CAC GGA ATT GTC AGT GC | This paper | N/A | |||
|
| |||||
| Recombinant DNA | |||||
|
| |||||
| AAV-PVe(S5E2)-Syp-dTomato | Vormstein-Schneider et al., 2020 | N/A | |||
| AAV-PVe(S5E2)-Syp-Gamillus | This paper | N/A | |||
|
| |||||
| Software and algorithms | |||||
|
| |||||
| ImageJ - Fiji 2.0.0 | Schneider et al., 2012 | https://imagej.nih.gov/ij/ | |||
| MATLAB R2014b/R2017a (Imaris analysis) | MathWorks | https://www.mathworks.com/ | |||
| MATLAB R2019a (9.6.0.1072779) 64-bit (glnxa64) (MERFISH analysis) | MathWorks | https://www.mathworks.com/ | |||
| GraphPad Prism 8/9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | |||
| Zen blue 2.6 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html | |||
| Imaris 8.1.2/9.3.1/9.5.0 | Bitplane | https://imaris.oxinst.com | |||
| Clampfit | Molecular Devices | https://mdc.custhelp.com/app/answers/detail/a_id/18779/~/axon%E2%84%A2pclamp%E2%84%A2–10-electrophysiology-data-acquisition-%26-analysis-software-download | |||
| Minianalysis | Synaptosoft | http://www.synaptosoft.com/MiniAnalysis/ | |||
| ThorlmageLS 3.2 | Thorlabs | https://www.thorlabs.com/newgrouppage9.cfm?objectgroup_id=9072#ad-image-0 | |||
| Cell Ranger v3.0.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation | |||
| Monocle3 v0.2.3.0 | Cao et al., 2019 | https://cole-trapnell-lab.github.io/monocle3/ | |||
| Seurat v4.0.0 | Satija et al., 2015 | https://satijalab.org/seurat/ | |||
| Scanpy 1.6.1.dev78+gd6457902 | Amir et al., 2013; Wolf et al., 2018 | https://scanpy.readthedocs.io/en/stable/ | |||
| R 4.0.0 | The R Foundation | http://www.r-project.org/ | |||
| DEWÄKSS | Tjärnberg et al., 2020 | N/A | |||
| Python 3.8.5 | Python Software Foundation | https://www.python.org/ | |||
| MERlin | Emanuel and Babcock, 2020 | https://github.com/emanuega/MERlin | |||
| Scran 1.18.5 | Lun et al., 2016b | https://rdrr.io/bioc/scran/ | |||
| Scater 1.18.6 | McCarthy et al., 2017 | https://rdrr.io/github/davismcc/scater/ | |||
| SingleCellExperiment 1.12.0 | Amezquita et al., 2020 | https://rdrr.io/bioc/SingleCellExperiment/ | |||
| Igraph 1.2.6 | Csardi and Nepusz, 2006 | https://igraph.org/ | |||
| MoSeq | Wiltschko et al., 2015 | N/A | |||
| Deposited data | |||||
| Raw and analyzed scRNA-seq data | This paper | GEO: GSE159947 | |||
| Raw and analyzed MERFISH data | This paper | https://drive.google.com/drive/folders/1TG4KkAW6–0HBmKnYqHCD4vfbBZXzSXkA | |||
| Code for scRNA-seq analysis | This paper; https://doi.org/10.5281/zenodo.4899812 | https://github.com/emiliafavuzzi/Favuzzi_et_al_2021 | |||
| Code for synaptic analysis | This paper; https://doi.org/10.5281/zenodo.4899956 | https://github.com/emiliafavuzzi/synaptic-analyses | |||
| Code for MERFISH analysis | This paper; https://doi.org/10.5281/zenodo.4899812 | https://github.com/emiliafavuzzi/Favuzzi_et_al_2021 | |||
Highlights.
GABA-receptive microglia interact with inhibitory synapses during development
Removing GABABRs from microglia alters inhibitory but not excitatory connectivity
Synapse pruning genes are altered in GABA-receptive microglia lacking GABABRs
Mice lacking microglial GABAB1Rs exhibit behavioral abnormalities
ACKNOWLEDGMENTS
We thank Plexxikon for PLX5622, B. Bettler for Gabbr1-floxed allele, and G. Feng forTmem119CreER and Tmem119GFP alleles. We thank L. Dissing-Olesen for advice on microglia isolation and suggestions on the project. We thank V. Regulapati for help with IHC, J. Dimidschstein for sharing the S5E2 plasmid, and L. Guo for help with AAV production. We are grateful to members of the Fishell laboratory for stimulating discussions. We thank B. Dejanovic, E. Lebois, and J. Dimidschstein for feedback on the manuscript. E.F. was supported by EMBO (ALTF 444-2018) and FY21 Hearst Fellowships. Work in the G.F. laboratory is supported by the NIH (R01 NS081297, R01 MH071679, UG3 MH120096, and P01 NS074972), the Simons Foundation SFARI (566615), and the Harvard’s Dean Initiative. MERFISH work was supported by RF1MH121289 from NIMH and a Joint Broad-Israel Science Foundation Research Grant. The HMS Neuro Imaging facility is funded by NINDS P30 Core Center (NS072030).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.06.018.
DECLARATION OF INTERESTS
B.S. serves on the SAB and is a minor shareholder of Annexon. G.F. is a founder of Regel. The other authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors ofthis paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
REFERENCES
- American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5) (APA Publishing; ). [Google Scholar]
- Amezquita RA, Lun ATL, Becht E, Carey VJ, Carpp LN, Geistlinger L, Marini F, Rue-Albrecht K, Risso D, Soneson C, et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, and Pe’er D. (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A, Ikegami A, Hwang P, Chan AT, Graves SM, Uweru JO, et al. (2020). Negativefeedback control of neuronal activity by microglia. Nature 586, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Friedman BA, Dejanovic B, and Sheng M. (2019). Microglia in Brain Development, Homeostasis, and Neurodegeneration. Annu. Rev. Genet 53, 263–288. [DOI] [PubMed] [Google Scholar]
- Botto M, Dell’Agnola C, Bygrave AE, Thompson EM, Cook HT, Petry F, Loos M, Pandolfi PP, and Walport MJ (1998). HomozygousC1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet 19, 56–59. [DOI] [PubMed] [Google Scholar]
- Braat S, and Kooy RF (2015).TheGABAA ReceptorasaTherapeuticTarget for Neurodevelopmental Disorders. Neuron 86, 1119–1130. [DOI] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot G, and Cherubini E. (2013). Functional role ofambientGABA in refining neuronal circuits early in postnatal development. Front. Neural Circuits 7,136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Jouannet S, Boucheix C, and Rubinstein E. (2014). Tetraspanins at a glance. J. Cell Sci 127, 3641–3648. [DOI] [PubMed] [Google Scholar]
- Cheadle L, Rivera SA, Phelps JS, Ennis KA, Stevens B, Burkly LC, Lee WA, and Greenberg ME (2020). Sensory Experience Engages Microglia to Shape Neural Connectivity through a Non-Phagocytic Mechanism. Neuron 108, 451–468.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jalabi W, Hu W, Park H-J, Gale JT, Kidd GJ, Bernatowicz R, Gossman ZC, Chen JT, Dutta R, and Trapp BD (2014). Microglial displacement of inhibitory synapses provides neuroprotection in the adult brain. Nat. Commun 5, 4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JA, Peñagarikano O, Belgard TG, Swarup V, and Geschwind DH (2015a). The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol 10, 111–144. [DOI] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, and Zhuang X. (2015b). RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapser JD., Ochaba J, Soni N, Reidling JC, Thompson L,M, and Green KN (2020). Microglial depletion prevents extracellular matrix changes and striatal volume reduction in a model of Huntington’s disease. Brain 143, 266–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, and Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9. [Google Scholar]
- Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, Orsolits B, Molnár G, Heindl S, Schwarcz AD, et al. (2020). Microglia monitorand protect neuronal function through specialized somatic purinergic junctions. Science 367, 528–537. [DOI] [PubMed] [Google Scholar]
- Cubillo A, Halari R, Smith A, Taylor E, and Rubia K. (2012). A review of fronto-striatal and fronto-cortical brain abnormalities in children and adults with Attention Deficit Hyperactivity Disorder (ADHD) and new evidence for dysfunction in adults with ADHD during motivation and attention. Cortex 48, 194–215. [DOI] [PubMed] [Google Scholar]
- Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, and Amit I. (2018). Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Doolen S, Cook J, Riedl M, Kitto K, Kohsaka S, Honda CN, Fairbanks CA, Taylor BK, and Vulchanova L. (2017). Complement 3a receptor in dorsal horn microglia mediates pronociceptive neuropeptide signaling. Glia 65, 1976–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Crocetti D, Zhu H, Gilbert DL, and Mostofsky SH (2012). Reduced GABA concentration in attention-deficit/hyperactivity disorder. Arch. Gen. Psychiatry 69, 750–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel G, and Babcock H. (2020). ZhuangLab/MERlin: MERlin v0.1.6 (Zenodo; ). [Google Scholar]
- Favuzzi E, and Rico B. (2018). Molecular diversity underlying cortical excitatory and inhibitory synapse development. Curr. Opin. Neurobiol 53, 8–15. [DOI] [PubMed] [Google Scholar]
- Favuzzi E, Marques-Smith A, Deogracias R, Winterflood CM, Sánchez-Aguilera A, Mantoan L, Maeso P, Fernandes C, Ewers H, and Rico B. (2017). Activity-Dependent Gating of Parvalbumin Interneuron Function by the Perineuronal Net Protein Brevican. Neuron 95, 639–655.e10. [DOI] [PubMed] [Google Scholar]
- Favuzzi E, Deogracias R, Marques-Smith A, Maeso P, Jezequel J, Exposito-Alonso D, Balia M, Kroon T, Hinojosa AJ, F Maraver E, and Rico B. (2019). Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science 363, 413–417. [DOI] [PubMed] [Google Scholar]
- Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, Erreni M, Markicevic M, Starvaggi-Cucuzza C, Otero K, et al. (2018). The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 48, 979–991.e8. [DOI] [PubMed] [Google Scholar]
- Fontainhas AM, Wang M, Liang KJ, Chen S, Mettu P, Damani M, Fariss RN, Li W, and Wong WT (2011). Microglial morphology and dynamic behavior is regulated by ionotropic glutamatergic and GABAergic neurotransmission. PLoS ONE 6, e15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiarsa J-L, and Porcher C. (2013). Emerging neurotrophic role of GABAB receptors in neuronal circuit development. Front. Cell. Neurosci 7, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin C-H, Shiue L, Ares M Jr., Mody I, and Black DL (2011). The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet 43,706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DL, Isaacs KM, Augusta M, Macneil LK, and Mostofsky SH (2011). Motor cortex inhibition: a marker of ADHD behavior and motor development in children. Neurology 76, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller C, Casanova E, Muller M, Vacher C-M, Vigot R, Doll T, Barbieri S, Gassmann M, and Bettler B. (2004). Floxed allele for conditional inactivation of the GABAB(1) gene. Genesis 40, 125–130. [DOI] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50,253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B,Tamm JA, Craig DA, Durkin MM, Dai M, Yao W-J, Johnson M, Gunwaldsen C, Huang L-Y, et al. (1998). GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature 396, 674–679. [DOI] [PubMed] [Google Scholar]
- Kaiser T, and Feng G. (2019). Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia. eNeuro 6, ENEURO.0448–18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Yamamoto A, Mizushima N, Yoshimori T, and Miyawaki A. (2008). GFP-like proteins stably accumulate in lysosomes. Cell Struct. Funct 33, 1–12. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017).A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 769, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn SA, van Landeghem FKH, Zacharias R, Färber K, Rappert A, Pavlovic S, Hoffmann A, Nolte C, and Kettenmann H. (2004). Microglia express GABA(B) receptors to modulate interleukin release. Mol. Cell. Neurosci 25, 312–322. [DOI] [PubMed] [Google Scholar]
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 707, 207–223.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YU, Ying Y, Li Y, Eyo UB, Chen T, Zheng J, Umpierre AD, Zhu J, Bosco DB, Dong H, and Wu LJ (2019). Neuronal network activity controls microglial process surveillance in awake mice via norepinephrine signaling. Nat. Neurosci 22, 1771–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-J, Spangenberg E, Tang B, Holmes TC, Green KN, and Xu X. (2021). Microglia elimination increases neural circuit connectivity and activity in adult mouse cortex. J. Neurosci 47, 1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang H-Y, Shang Y, Oldham MC, Martens LH, Gao F, et al. (2016). Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell 765, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Bach K, and Marioni JC (2016a). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 77,75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016b). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5,2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu Y-WA, Garcia AJ 3rd, Gu X, Zanella S, et al. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci 75, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, and Attwell D. (2015). Receptors, ion channels, and signaling mechanisms underlying microglial dynamics. J. Biol. Chem 290, 12443–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, Bhagat ND, Peterson RE, Peterson E, Hyun M, Linderman SW, et al. (2018). The Striatum Organizes 3D Behavior via Moment-to-Moment Action Selection. Cell 774, 44–58.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederos S, and Perea G. (2019). GABAergic-astrocyte signaling: A refinement of inhibitory brain networks. Glia 67, 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, and Zhuang X. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Rajbhandari AK, Gangwani MR, Hachisuka A, Coppola G, Masmanidis SC, Fanselow MS, and Khakh BS (2019). Hyperactivity with Disrupted Attention by Activation of an Astrocyte Synaptogenic Cue. Cell 177, 1280–1292.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neniskyte U, and Gross CT (2017). Errant gardeners: glial-cell-dependent synaptic pruning and neurodevelopmental disorders. Nat. Rev. Neurosci 18, 658–670. [DOI] [PubMed] [Google Scholar]
- Nguyen PT, Dorman LC, Pan Vainchtein ID, Han RT, Nakao-Inoue H, Taloma SE, Barron JJ, Molofsky AB, Kheirbek MA., et al. (2020). Microglial Remodeling of the Extracellular Matrix Promotes Synapse Plasticity. Cell 182, 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WC, Lutzu S, Castillo PE, and Kwon H-B (2016). De novo synaptogenesis induced by GABA in the developing mouse cortex. Science 353, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Parslow A, Cardona A, and Bryson-Richardson RJ (2014). Sample Drift Correction Following 4D Confocal Time-lapse Imaging. J. Vis. Exp (86), 51086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JGA, Jones DG, and Murphy KM (2013). Comparing development of synaptic proteins in rat visual, somatosensory, and frontal cortex. Front. Neural Circuits 7, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock JM, and Kettenmann H. (2007). Neurotransmitter receptors on microglia. Trends Neurosci. 30, 527–535. [DOI] [PubMed] [Google Scholar]
- Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park J-E (2020). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine RJ (1972). Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol 32, 281–294. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, and Perry VH (2009). Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol 27, 119–145. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda H, Ma Y, Nakashima R, Sakurai K, Matsuda T, and Nagai T. (2018a). Acid-Tolerant Monomeric GFP from Olindias formosa. Cell Chem. Biol 25, 330–338.e7. [DOI] [PubMed] [Google Scholar]
- Shinoda H, Shannon M, and Nagai T. (2018b). Fluorescent Proteins for Investigating Biological Events in Acidic Environments. Int. J. Mol. Sci 19, 1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, and Rubenstein JLR (2019). Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol. Psychiatry 24, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg E, Severson PL, Hohsfield LA, Crapser J, Zhang J, Burton EA, Zhang Y, Spevak W, Lin J, Phan NY, et al. (2019). Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun 10, 3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, and Garel S. (2014). Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8, 1271–1279. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, and Tyers M. (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Stowell RD, Sipe GO, Dawes RP, Batchelor HN, Lordy KA, Whitelaw BS, Stoessel MB, Bidlack JM, Brown E, Sur M, and Majewska AK (2019). Noradrenergic signaling in the wakeful state inhibits microglial surveillance and synaptic plasticity in the mouse visual cortex. Nat. Neurosci 22, 1782–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 (D1), D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, Ginhoux F, and Garel S. (2018). Microglia and early brain development: An intimate journey. Science 362, 185–189. [DOI] [PubMed] [Google Scholar]
- Thion MS, Mosser C-A, Ferezou I, Grisel P, Baptista S, Low D, Ginhoux F, Garel S, and Audinat E. (2019). Biphasic Impact of Prenatal Inflammation and Macrophage Depletion on the Wiring of Neocortical Inhibitory Circuits. Cell Rep. 28, 1119–1126.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Lee BR, Chong P, Soler-Llavina G, Cobbs C, Koch C, Zeng H, and Lein E. (2018). Preparation of Acute Brain Slices Using an Optimized N-Methyl-D-glucamine Protective Recovery Method. J. Vis. Exp (132), 53825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjärnberg A, Mahmood O, Jackson CA, Saldi G-A, Cho K, Christiaen LA, and Bonneau RA (2020). Optimal tuning of weighted kNN- and diffusion-based methods for denoising single cell genomics data. bioRxiv. 10.1101/2020.02.28.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci 22, 1021–1035. [DOI] [PubMed] [Google Scholar]
- Vogels TP, and Abbott LF (2009). Gating multiple signals through detailed balance of excitation and inhibition in spiking networks. Nat. Neurosci 12, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ehr A, Attaai A, Neidert N, Potru PS, Ruß T, Zöller T, and Spittau B. (2020). Inhibition of Microglial TGFβ Signaling Increases Expression of Mrc1. Front. Cell. Neurosci 14, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, Allaway K, Sakopoulos S, Schneider G, Stevenson O, et al. (2020). Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci 23, 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B, Jaglin XH, Favuzzi E, Quattrocolo G, Nigro MJ, Yusuf N, Khodadadi-Jamayran A, Rudy B, and Fishell G. (2018). Rbfox1 Mediates Cell-type-Specific Splicing in Cortical Interneurons. Neuron 100, 846–859.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD, and Kriegstein AR (2009). Defining the role of GABA in cortical development. J. Physiol 587, 1873–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton DK, Dissing-Olesen L, and Stevens B. (2019). Neuron-Glia Signaling in Synapse Elimination. Annu. Rev. Neurosci 42, 107–127. [DOI] [PubMed] [Google Scholar]
- Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, and Datta SR (2015). Mapping Sub-Second Structure in Mouse Behavior. Neuron 88, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Fu Y, Knott G, Lu J, Di Cristo G, and Huang ZJ (2012). GABA signaling promotes synapse elimination and axon pruning in developing cortical inhibitory interneurons. J. Neurosci 32, 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer H, Riese S, and Regnier-Vigouroux A. (2003). Functional characterization of mannose receptor expressed by immunocompetent mouse microglia. Glia 42, 89–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Consequences of early postnatal microglia depletion for cortical connectivity, related to Figure 1 (A) Representative images and quantitation of the density and layer distribution of PV cells in control (n = 4) and microglia-depleted (n = 4) mice at P15 after P1-P15 microglia depletion. ns p > 0.05, Student’s t test (for density) and One-Way ANOVA followed by Sidak’s multiple comparisons test (for layer distribution). Scale bar equal 100 μm.
(B) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in P10 control (n = 4) or depleted (n = 4) mice after P1-P10 microglia depletion and in P12 control (n = 3) or depleted (n = 5) mice after P1-P12 microglia depletion. ns p > 0.05, One-Way ANOVA followed by Tukey’s multiple comparisons test.
(C) Representative images and quantitation of Iba1+ microglia density in control (n = 4) and microglia-depleted (n = 4) mice at P30 after P15-P30 microglia depletion. ***p < 0.001, Student’s t test. Scale bar equal 100 μm.
(D) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in P30 control (n = 4) and depleted (n = 3) mice after P15-P30 microglia depletion. One brain with incomplete depletion was excluded from the analysis. ns p > 0.05, Student’s t test.
(E) Representative images and quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons in the primary visual cortex (V1) of P15 control (n = 6) and depleted (n = 9) mice after P1-P15 microglia depletion and of P25 control (n = 12) and depleted (n = 10) mice after P1-P25 microglia depletion. ns p > 0.05 and *p < 0.05, One-Way ANOVA followed by Tukey’s multiple comparisons test. Scale bar equal 2 μm.
(F) Representative images and quantitation of synapses made in L1 of S1 by SST+ cells (SSTCre;Ai34 Synaptophysin-tdTomato labeled SST+ presynaptic terminals colocalizing with Gephyrin) in P15 control (n = 8) and depleted (n = 6) mice after P1-P15 microglia depletion. *p < 0.05, Student’s t test. Scale bar equal 2 μm.
(G) Quantitation of the density of SST cells in P15 control (n = 6) and microglia-depleted (n = 5) mice after P1-P15 microglia depletion. ns p > 0.05, Mann-Whitney test.
(H) Traces, frequency and amplitude of sIPSCs (n = 14 cells from 3 control and n = 17 cells from 5 depleted mice). ***p < 0.001, ns p > 0.05, Student’s t test.
(I) Traces, frequency and amplitude of sEPSCs (n = 17 cells from 4 control and n = 18 cells from 5 depleted mice). ***p < 0.001, ns p > 0.05, Student’s t test.
(J) sEPSC/sIPSC ratio (n = 14 cells from 3 control and n = 15 cells from 3 depleted mice). ns p > 0.05, Mann-Whitney test.
(K) Schematic of synapses analyzed in L and M.
(L) Representative images and quantitation of VGlut2+Homer1+ synapses onto excitatory cell dendrites (TdTomato+, labeled by virus injection of pyramidalneurons) in P15 control (n = 6) and depleted (n = 4) mice. **p < 0.01, Student’s t test. Scale bar equal 2 μm.
(M) Quantitation of VGlut1+Homer1+ synapses made onto L4 excitatory cells (NeuN) in P15 control (n = 6) and depleted (n = 4) mice (left), onto PV cells in P15 control (n = 9) or depleted (n = 10) mice (middle) and onto excitatory cell dendrites (TdTomato+, labeled by virus injection of pyramidal neurons) in P15 control (n = 7) or depleted (n = 4) mice. ns p > 0.05, Mann-Whitney test for excitatory neurons and Student’s t test for PV cells and dendrites.
(N) Representative images of Iba1+ microglia in control and P1-P15 microglia-depleted brains at the indicated stages of repopulation and quantitation of microglia density in control and microglia-repopulated brains at P17, P19, P21, P25 and P30 (n = 3–4 mice per condition). ***p < 0.001, ns p > 0.05, One-Way ANOVA followed by Sidak’s multiple comparisons test. Scale bar equal 100 μm.
(O) Quantitation of synapses made in L1 of S1 by SST+ cells (SSTCre;Ai34 labeled presynaptic terminals colocalizing with Gephyrin) in P30 control (n = 4) and microglia-repopulated (n = 5) mice after P1-P15 microglia depletion. *p < 0.05, Student’s t test.
(P) Quantitation of Syt2+Gephyrin+ synapses (left), SSTCre;Ai34+Gephyrin+ synapses (middle) and VGlut2+Homer1+ synapses (right) made by PV, SST and thalamic cells in P60 control (n = 6–8) and microglia-repopulated (n = 4–7) mice after P1-P15 microglia depletion. ExCs: excitatory cell soma. ns p > 0.05, Student’s t test for SST+ and thalamic synapses and Mann-Whitney test for PV synapses.
All data are mean ± SEM, each data point represents one experimental animal except in F, I and J where it represents one cell. Arrowheads indicate colocalization.
Figure S2. Microglia-PV synapse interactions during development, related to Figure 2 (A) Masks showing the criteria for contact identification in the in vivo imaging experiments. Percentages indicate the fraction of bouton area contacted by microglia.
(B) Proportion of PV boutons contacted by one microglia during the in vivo imaging experiments in deep layer 3 (n = 15 cells contacting 68 boutons) or layer 4 (n = 36 cells contacting 154 boutons). Data are the same as in (C) and (I) where they are shown as pooled dataset. ns p > 0.05, Mann-Whitney test. Data are mean ± SEM, each data point represents one cell.
(C) Distribution of Cx3cr1GFP/+microglia contacting the indicated percentages of PV boutons in a 20 μmradius around the microglia cell body over 20 minutes (n = 51 cells from 6 mice) during the in vivo imaging experiments. PV boutons are labeled with PVe-Syp-tdTomato. The distribution is shown as density histogram and kernel density estimation. The first peak corresponds to 11.7% of PV boutons contacted by microglia and the second peak to 60.9%. The local minimum of the bimodal distribution is 26.4%. These data were pooled with data in (I) to generate the graph in Figure 3C.
(D) Distribution of microglia-PV boutons interactions for the indicated duration during the in vivo imaging experiments. The distribution is shown as density histogram and kernel density estimation of the proportion of Syp-tdTomato+ PV synaptic boutons contacted by microglia for the indicated duration (n = 222 PV boutons from 6 mice).
(E) Duration of contacts between microglia interacting with a minority (n = 11 cells) or the majority (n = 25 cells) of local PV boutons during the in vivo imaging experiments. *p < 0.01, Mann-Whitney test. These data represent the subset of data shown in (D) for which it was possible to visualize the microglia cell body and they were pooled with data in (K) to generate the graph shown in Figure 3D. Data were split based on the local minimum of the kernel density estimation in (C). Data are mean ± SEM, each data point represents one cell.
(F) Time-lapse images from Video S2 showing a Syp-tdTomato+ PV bouton being enwrapped by a microglia process (Cx3cr1GFP/+) and contacted by a second process over 10 minutes. Scale bar equal 10 μm.
(G) Time-lapse images from Video S3 showing a Cx3cr1GFP/+ microglial process that terminates into what resembles a phagocytic cup engulfing a Syp-tdTomato+ PV bouton over 20 minutes. Scale bar equal 10 μm.
(H) Time-lapse image from in vivo imaging of Syp-Gamillus+ PV boutons and microglia genetically labeled using Tmem119CreER/+;Ai14 mice. Scale bar equal 10 μm.
(I) Distribution of Tmem119CreER/+;Ai14 microglia contacting the indicated percentages of PV boutons (labeled with PVe-Syp-Gamillus) in a 20 μm radius around the cell body over 20 minutes (n = 37 cells from 6 mice). The distribution is shown as density histogram and kernel density estimation. The first peak corresponds to 18.1% of PV boutons contacted by microglia and the second peak to 57.4%. The local minimum of the bimodal distribution is 34.4%. These data were pooled with data in (C) to generate the graph in Figure 3C.
(J) Distribution of microglia-PV boutons interactions for the indicated duration. The distribution is shown as density histogram and kernel density estimation of the proportion of Syp-Gamillus+ PV synaptic boutons contacted by microglia for the indicated duration (n = 115 PV boutons from 6 mice).
(K) Duration of contacts between microglia interacting with a minority (n = 13 cells) or the majority (n = 23 cells) of local PV boutons. *p < 0.01, Student’s t test. These data represent the subset of data shown in (J) for which it was possible to visualize the microglia cell body and they were pooled with data in E to generate the graph shown in Figure 3D. Data are mean ± SEM, each data point represents one cell.
(L) Representative images of the same 3D Imaris space in two different orientations. Note that, in the visualization of a 3D volume, boutons can appear closer than they are depending on the orientation. In contrast, “real contacts” appear close regardless of the orientation. Scale bar equal 5 μm.
(M) Representative 3D reconstruction of a microglia process and surrounding PV boutons. Contacted PV boutons are reconstructed as red spots whereas PV boutons not contacted by microglia are reconstructed as light blue spots. Full and empty arrowheads indicate positive and negative (i.e., boutons that did not meet the contact criteria) examples, respectively. The yellow empty arrowhead indicates a bouton that is far from the process. In all relevant figures, only contacted boutons (see methods for criteria) have been highlighted as “reconstructed spots.” Scale bar equal 5 μm.
Figure S3. Expression of GABAb receptors in microglia, related to Figure 2 (A) Schematic of analysis and heatmaps showing ligands-receptor pairs that are leading candidates for mediating interneuron-microglia communication. The heatmap on the left is showing the expression (at the indicated developmental time points) of putative ligands enriched in interneurons as compared to pyramidal cells. For P5 and P10 interneurons, the corresponding values in SST and PV cells were averaged. The list is ranked according to specificity and developmental upregulation. For GABA, the expression of the GABA-synthetizing enzyme Gad1 is shown. The heatmap on the right is showing the expression (at the indicated developmental time points) of receptors expressed in microglia that are experimentally validated interactors of the ligands shown on the left. For the P3-P9 time point, values at P3 and P9 were averaged.
(B) Venn diagram depicting the fraction of microglia expressing Gabbr1+ mRNA, Gabbr2+ mRNA as well as double positive for both Gabbr1+ and Gabbr2+ mRNAs at P15 (smFISH: single molecule fluorescent in situ hybridization). The diagram is a visualization of the data shown in Figure 3F.
(C) Validation of the specificity of GABAB1R antibody. Representative images from P15 controls and VGlut1Cre;Gabbr1 cKO mice, lacking GABAB1R in cortical excitatory neurons. Note the absence of GABAB1R signal within NeuN+ excitatory neurons in VGlut1Cre cKO mice. Scale bar equal 10 μm.
(D) Left: quantitation of the percentage of microglia (Iba1+) expressing GABAB1R protein at P0, P10 and P15 in control and Cx3cr1Cre GABAB1R cKO mice (n = 3–6 each). ns p > 0.05, **p < 0.01, ***p < 0.001, One-Way ANOVA followed by Sidak’s multiple comparisons test. Note the higher expression of GABAB1R in controls at P15 compared to P0 or P10. Right: quantitation of the percentage of microglia (Iba1+) expressing GABAB1R protein at P15 in control and Tmem119CreER GABAB1R cKO mice (n = 4 each). *p < 0.05, Student’s t test. Data are mean ± SEM, each data point represents one experimental animal.(E) Representative images and quantitation of microglia (Iba1+) expressing both GABAb-|R and GABAB2R proteins in P15 control mice (n = 3) within layer 4 of S1. Scale bar equal 5 μm. See STAR Methods for details about noise in the IHC experiment versus smFISH. Arrowheads indicate colocalization. Data are mean ± SEM, each data point represents one experimental animal.(F) Gabbr1 and Gabbr2 expression in adult microglia/macrophages from different brain regions. Data and visualization are from Saunders et al. (2018). FC: frontal cortex, TH: thalamus, STR: striatum, PC: posterior cortex, CB: cerebellum, SN: substantia nigra, HC: hippocampus, GP: globus pallidus. The reported confidence intervals reflect statistical sampling noise.(G) Fraction of GABA-receptive microglia (defined as expressing both Gabbr1 and Gabbr2 mRNAs by single molecule fluorescent in situ hybridization) in layer 4 of S1 and the CA1 region of the hippocampus of Cx3cr1GFP/+ mice at P15. S1 data are the same as shown in Figure 3F. Data are mean ± SEM, each data point represents one experimental animal.(H) 3D reconstruction and fraction of PV boutons (PVe-Syp-tdTom) encased by Gabbr2+ and Gabbr2-microglia (Cx3cr1GFP/+) at P15 (n = 58 Gabbr2+ and 59 Gabbr2-cells from 4 mice). ***p < 0.001, Mann-Whitney test. Scale bar equal 1 μm. Data are mean ± SEM, each data point represents one cell.(I) 3D reconstruction and fraction ofVGlut2+ boutons encased by Gabbr2+ and Gabbr2-microglia (Cx3cr1GFP/+) at P15 (n = 23 Gabbr2+ and 36 Gabbr2-cells from 4 mice). *p < 0.05, Student’s t test. Scale bar equal 1 mm. Data are mean ± SEM, each data point represents one cell.
Figure S4. Synaptic phenotype of GABAB1R cKO mice, related to Figure 3 (A) Left: quantitation of PV boutons contacted by microglia at P15 in wild-type control mice (n = 7, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 8, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Tukey’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4A. Right: quantitation of PV boutons contacted by microglia at P15 in control (n = 4, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 6, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. *p < 0.05, **p < 0.01.(B) Left: quantitation of VGlut2 boutons contacted by microglia at P15 in wild-type control mice (n = 7, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 6, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Tukey’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4B. Right: quantitation of VGlut2 boutons contacted by microglia at P15 in control (n = 8, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 8, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. ns p > 0.05.(C) Distribution of microglia-PV boutons interactions for the indicated duration during the in vivo imaging experiments in controls (n = 181 PV boutons from 6 mice) and cKOs (Cx3cr1GFP/+;Tmem119CreER/+;GABAB1Rfl/fl, n = 99 PV boutons from 3 mice) cKO mice. The distribution is shown as kernel density estimation of the proportion of PV boutons contacted by microglia for the indicated duration. Control data are the same as in Figure S2D but limited to a total duration of 20 minutes to allow comparison with cKO data (see STAR Methods). **p < 0.01, Kolmogorov–Smirnov test. Note the complex effect of GABAB1R removal on the contact duration, the cKO distribution is bimodal with some contacts having a shorter duration and others being abnormally long.(D) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons at P15 in wild-type control mice (n = 8, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+heterozygous control mice (n = 6, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Holm-Sidak’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4H. Right: Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons at P15 in control (n = 8, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 6, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. *p < 0.05, **p < 0.01.(E) Image, relative frequency histogram and overlaid kernel density estimation of Syt2+Gephyrin+ synapses imaged using STED super-resolution microscopy andmade by PV cells onto excitatory neurons at P15 in wild-type controls (n = 58 cells from 3 Cx3cr1+/+;GABAB1Rfl/fl mice) and GABAB1R cKO mice (n = 66 cells from 3 Cx3cr1Cre/+; GABAB1Rfl/fl mice). ***p < 0.001, Kolmogorov–Smirnov test. The dotted line indicates the soma outline.(F) Left: quantitation of VGlut2+Homer1+ synapses made onto excitatory neurons at P15 in wild-type control mice (n = 6, Cx3cr1+/+;GABAB1Rfl/fl), GABAB1R cKO mice (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) and Cx3cr1−/+ heterozygous control mice (n = 9, Cx3cr1Cre/+; GABAB1R+/+) mice. One-Way ANOVA followed by Holm-Sidak’s multiple comparisons test. Wild-type control and cKO data are the same as shown in Figure 4I. Right: VGlut2+Homer1+ synapses made onto excitatory neurons at P15 in control (n = 9, Tmem119+/+;GABAB1Rfl/fl) and cKO (n = 8, Tmem119CreER/+;GABAB1Rfl/fl) mice. Student’s t test. ns p > 0.05.(G) Quantitation of VGlut2+Homer1+ synapses onto L4 PV neurons in P15 control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(H) Quantitation of VGlut1+Homer1+ synapses onto L4 excitatory cells (left) or pyramidal cell dendrites (right) in control (n = 7–9, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 6–7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Mann-Whitney test for somatic analysis and Student’s t test for dendritic analysis. (I) and (J) Traces, frequency and amplitude of mIPSCs (n = 26 cells from 3 Tmem119+/+;GABAB1Rfl/fl control mice and n = 28 cells from 4 Tmem119CreER/+; GABAB1Rfl/fl cKO mice) and mEPSCs (n = 26 cells from 3 Tmem119+/+; GABAB1Rfl/fl control mice and n = 31 cells from 4 Tmem119CreER/+;GABAB1Rfl/fl cKO mice) recorded from layer 4 excitatory neurons at P15. ***p < 0.001, ns p > 0.05, Student’s t test except for mEPSC amplitude where Mann-Whitney test was used.(K) mEPSC/mIPSC frequency ratio (n = 26 cells from 3 Tmem119+/+; GABAB1Rfl/fl control mice and n = 28 cells from 4 Tmem119CreER/+;GABAB1Rfl/fl cKO mice). ***p < 0.001, Mann-Whitney test.(L) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto S1 L4 excitatory neurons in control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P30. Right: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto S1 L4 excitatory neurons in control (n = 6, Tmem119+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Tmem119CreER/+;GABAB1Rfl/fl) mice at P30. **p < 0.01, Student’s t test.(M) Left: quantitation of Syt2+Gephyrin+ synapses made by PV cells onto V1 L4 excitatory neurons at P30 in control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. Right: quantitation of PV+ Gephyrin+ synapses made onto NeuN+ cells in the dorsolateral striatum of P30 control (n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. *p < 0.05, **p < 0.01, Student’s t test.(N) Images, 3D reconstruction and quantitation of SST synapses (GAD65+Gephyrin+ synapses with GAD65 presynaptic boutons completely encased within genetically labeled SST axons) in layer 1 of S1 at P15 in control (n = 5; SSTFlp/+; RCE:FRT; Cx3cr1Cre/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 4; SSTFlp/+; RCE:FRT; Cx3cr1+/+; GABAB1Rfl/fl) mice. *p < 0.05, Student’s t test.All data are mean ± SEM, each data point represents one experimental animal except in I-K where they represent cells. Scale bars equal 1 mm. Arrowheads indicate colocalization.
Figure S5. The synaptic phenotype of GABAB1R cKO mice is not due to off-target effects, related to Figure 3 (A) Representative images and quantitation of the density of microglia in control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(B) Representative images and quantitation of the density of PV cells in control (n = 6, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(C) Representative images and quantitation of the density of SST cells (GFP+) in control (n = 8, SSTFlp/+; RCE:FRT; Cx3cr1Cre/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 7, SSTFlp/+; RCE:FRT; Cx3cr1+/+; GABAB1Rfl/fl) mice at P15. ns p > 0.05, Student’s t test. Scale bar equal 100 μm.(D) Quantitation of the density of PV and SST cells in control (n = 6, Lhx6GFP/+; Cx3cr1+/+; GABAB1Rfl/fl) and GABAB1R cKO (n = 6, Lhx6GFP/+; Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P10. PV cells were identified as GFP+SST− cells. ns p > 0.05, Student’s t test.(E) Quantitation of Syt2+Gephyrin+ synapses made by PV cells onto L4 excitatory neurons at P10 in control (n = 3, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 6, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(F) Representative images and quantitation of the fraction of L4 PV cells surrounded by perineuronal nets (WFA) in control (n = 4, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 5, Cx3cr1Cre/+; GABAB1Rfl/fl) mice at P15 and P30. ns p > 0.05, One-Way ANOVA and Holm-Sidak multiple comparisons test. Scale bar equal 20 μm.(G) Quantitation of neurons (labeled with NeuN) expressing Gabbr1 mRNA in control (n = 4, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 3, Cx3cr1Cre/+; GABAB1R+/+) mice. Gabbr1 mRNA was detected by single-molecule in situ hybridization specifically for the exons deleted in the cKOs (7 and 8). ns p > 0.05, Student’s t test.(H) Time course of mean Racine scores of seizures induced by intraperitoneal injection of kainic acid (20 mg/kg) into control (n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and GABAB1R cKO (n = 7, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. Seizures were scored as described in the STAR Methods. Two-Way ANOVA for repeated-measures with genotype (p > 0.05) and time (p < 0.001) as factors. Interaction between time and genotype p > 0.05.(I) Maximum and cumulative seizure scores over 90 minutes from the time of kainic acid injection. ns p > 0.05, Mann-Whitney test.
(J) Latency to reach the status epilepticus and/or death. ns p > 0.05, Mann-Whitney test.
(K) and (L) Traces, frequency and amplitude of mIPSCs (n = 16 cells from 3 control and n = 22 cells from 3 cKO mice) and mEPSCs (n = 18 cells from 3 control andn = 25 cells from 3 cKO mice) recorded from layer 4 excitatory neurons at P60. **p < 0.01, ns p > 0.05, Mann-Whitney test for mIPSCs and Student’s t test for mEPSCs.(M) Images and quantitation of Syt2+Gephyrin+ synapses made by PV cells onto excitatory neurons in P60 control (Ctl, n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice as well as cKO (n = 5) and control (n = 3) mice that underwent microglia depletion from P30 to P60. ns p > 0.05, *p < 0.05, One-Way ANOVA followed by Sidak’s multiple comparisons test. Scale bar equal 2 μm.(N) Quantitation of VGlut2+Homer1+ synapses made onto excitatory neurons in P60 control (Ctl, n = 5, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. ns p > 0.05, Student’s t test.(O) Quantitation of PV+ Gephyrin+ synapses made onto NeuN+ cells in the dorsolateral striatum of P60 control (Ctl, n = 7, Cx3cr1+/+;GABAB1Rfl/fl) and cKO (n = 8, Cx3cr1Cre/+; GABAB1Rfl/fl) mice. *p < 0.05, Student’s t test.All data are mean ± SEM, each data point represents one experimental animal except in K and L where they represent cells. Arrowheads indicate colocalization.
Figure S6. Loss of GABAB1Rs does not fundamentally alter the range of microglial states but causes a downregulation of pruning-related genes, related to Figure 5 (A) UMAP plots showing the integration of P15 wild-type microglia with embryonic, early postnatal and P30 datasets from Hammond et al. (2019).(B) UMAP plot of Cre-Het control (Cx3cr1Cre/+) microglia showing 5 clusters and representative genes enriched in each cluster.(C) UMAP plot of Cre-Het control microglia from cluster 1 further subjected to subclustering. Representative enriched genes are shown.
(D) UMAP plot of GABAB1R cKO microglia from P15 S1 cortex showing 7 main clusters and representative genes enriched in each cluster.(E) Pie charts indicating the contribution of each wild-type or cKO cluster to the clusters of the integrated dataset. The overall filled area is proportional to the total number of cells within each cluster.
(F) UMAP plots of Cre-Het control and cKO integrated scRNaseq dataset. See methods for the different integration of this dataset as compared to WT control andcKO data.(G) Same as (F), showing 10 mixed clusters and representative genes enriched in each cluster.
(H) Mixed cluster contributions to total differentially expressed genes (DEGs) between Cre-Het control and cKO microglia.
(I) Violin plots of normalized log-expression values for representative genes significantly downregulated in cKO microglia from Cre-Het-cKO cluster 3. See methods for the different integration of Cre-Het control and cKO data as compared to WT control and cKO data.(J) Violin plots of normalized log-expression values for representative genes significantly downregulated in cKO microglia from Cre-Het-cKO cluster 5.
(K) Heatmap showing the output of an elastic net regularized classifier trained to learn which genes are predictive of control wild-type and GABAB1R cKO microglia using all genes as features and wild-type or cKO as labels. Genes are ordered by increasing mean expression. The top 5 genes are predictive of wild-type microglia (downregulated in cKO) and the remaining 3 genes are predictive of cKO microglia (upregulated in cKO).(L) UMAP plots of control and cKO integrated scRNaseq dataset after denoising expression data with a weighted affinity kernel and self-supervision method(DEWÄ KSS, see STAR Methods).(M) Same as (L) followed by unsupervised clustering and showing 8 mixed clusters. See also Table S2 for genes enriched in each cluster.(N) Volcano plot showing differentially expressed genes between control and cKO microglia from cluster 4 after denoising. The negative log10-transformed p values are plotted against the log2 fold change. Differentially expressed genes with an absolute log2 fold change higher than 0.5 and -log10 (p value) higher than or equal to 20 are depicted in orange. Top downregulated genes are highlighted. A full list is shown in Table S2.
Figure S7. MoSeq reveals a biphasic behavioral phenotype in GABAB1R cKO mice. Related to Figure 7 (A) Expression probability of syllable usage in P30 wild-type control (n = 11), cKO (n = 15) and Cre-Het control (n = 5) female mice. Syllables are ordered by differential usage (left: cKO-enriched syllables; right: cKO-downregulated syllables). Here and in the rest of the figure, only relevant or significantly differentially used syllables are shown. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM. Here and in the rest of the figure, syllable labels were assigned by a human observer.(B) Expression probability of syllable usage in P30 wild-type control (n = 6), cKO (n = 14) and Cre-Het control (n = 7) male mice. Syllables are ordered by differential usage. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.
(C) Expression probability of syllables enriched in P60 control (n = 19, both sexes) and cKO (n = 14, both sexes) mice. Syllables are ordered by differential usage(left: cKO-enriched syllables; right: cKO-downregulated syllables). *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.
(D) Heatmap depicting the position of P60 control (n = 10 males and 9 females) and cKO (n = 5 males and 9 females) mice during MoSeq.(E) Graph showing the mean syllable speed in control (n = 19) and GABAB1R cKO (n = 14) mice. Syllables are ordered by differential speed (left: syllables having a higher speed in cKOs and right: syllables with a lower speed in the cKOs). *p < 0.05, t test on bootstrapped syllable speed distribution corrected for false discovery rate. Data are mean ± SEM.(F) Expression probability of syllables enriched in P60 Cx3cr1Cre cKOs (n = 9 females and 10 males) and Tmem119CreER cKOs (n = 3 females and 3 males) mice. Only significantly differentially used syllables are shown. In females, almost all syllables were similarly used. Males from both groups used high velocity syllables although Tmem119CreER cKOs appeared even more active than Cx3cr1Cre cKOs. *p < 0.05, z-test on bootstrapped syllable usage distribution corrected for false discovery rate. Data are mean ± SEM.(G) Summary of the scalar information for P60 control (n = 19) and GABAB1R cKO (n = 14) mice. Velocity is measured in mm/s; height, width and length are measured in mm. ***p < 0.001, ns p > 0.05, Student’s t test. The inset shows the probability density function (PDF) of the two-dimensional velocity, p < 0.001, Two-sample Kolmogorov–Smirnov test.
Data Availability Statement
scRNA-seq data have been deposited in NCBI GEO and assigned the following accession number GSE159947.
MERFISH raw and processed data are available at: https://drive.google.com/drive/folders/1TG4KkAW6-0HBmKnYqHCD4vfbBZXzSXkA
Should this link not be accessible, MERFISH processed data are also available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021/tree/main/MERFISH while the raw data will be shared upon request in a manner determined on a case by case basis (e.g., hard drive, cloud server).
Scripts for synaptic analyses are available at: https://github.com/emiliafavuzzi/synaptic-analyses (https://doi.org/10.5281/zenodo.4899956)
Scripts reproducing scRNA-seq and MERFISH analyses are available at: https://github.com/emiliafavuzzi/Favuzzi_et_al_2021 (https://doi.org/10.5281/zenodo.4899812)