Abstract
Biological data support the hypothesis that there are multiple species in the genus Cryptosporidium, but a recent analysis of the available genetic data suggested that there is insufficient evidence for species differentiation. In order to resolve the controversy in the taxonomy of this parasite genus, we characterized the small-subunit rRNA genes of Cryptosporidium parvum, Cryptosporidium baileyi, Cryptosporidium muris, and Cryptosporidium serpentis and performed a phylogenetic analysis of the genus Cryptosporidium. Our study revealed that the genus Cryptosporidium contains the phylogenetically distinct species C. parvum, C. muris, C. baileyi, and C. serpentis, which is consistent with the biological characteristics and host specificity data. The Cryptosporidium species formed two clades, with C. parvum and C. baileyi belonging to one clade and C. muris and C. serpentis belonging to the other clade. Within C. parvum, human genotype isolates and guinea pig isolates (known as Cryptosporidium wrairi) each differed from bovine genotype isolates by the nucleotide sequence in four regions. A C. muris isolate from cattle was also different from parasites isolated from a rock hyrax and a Bactrian camel. Minor differences were also detected between C. serpentis isolates from snakes and lizards. Based on the genetic information, a species- and strain-specific PCR-restriction fragment length polymorphism diagnostic tool was developed.
Cryptosporidiosis is a coccidian infection of humans, domestic animals, and wild vertebrates. In farm animals, Cryptosporidium parasites cause disorders of the digestive and respiratory systems, which lead to poor health of infected animals and significant economic losses. In immunocompetent humans, Cryptosporidium parasites cause acute infections of the digestive system, but in immunocompromised patients they cause a chronic, life-threatening disease. The general consensus is that one species, Cryptosporidium parvum from mammals, is the only Cryptosporidium species responsible for infections in humans (11, 24). In contrast, a recent study suggested that all Cryptosporidium parasites, including those from lower vertebrates, should be considered hazardous to humans (35).
The contrasting views are largely the result of confusion in the taxonomy of Cryptosporidium parasites. Since the discovery of Cryptosporidium muris and C. parvum in rodents at the turn of the century (31–33), more than 20 Cryptosporidium species from various animals have been described (24). Studies conducted in late 1970s and early 1980s, however, indicated that cross-transmission of some isolates among various animal species was possible. Thus, it was suggested that all Cryptosporidium parasites belong to the same species, C. muris (34). Later, it was shown that various Cryptosporidium isolates (at least isolates obtained from members of different classes of vertebrates) exhibit host specificity. Based on these observations, Levine (18, 19) proposed that the parasites from mammals, birds, reptiles, and fish should be classified as C. muris, Cryptosporidium meleagridis, Cryptosporidium serpentis, and Cryptosporidium nasorum, respectively. Subsequently, it was realized that C. parvum from mammals and Cryptosporidium baileyi from birds are biologically and morphologically different from C. muris and C. meleagridis (8, 36). Thus, C. parvum, C. muris, C. baileyi, C. meleagridis, C. serpentis, and C. nasorum have been considered the six valid Cryptosporidium species by many researchers (24). More recently, based on host specificity data, Fayer et al. (11) added Cryptosporidium felis from cats and Cryptosporidium wrairi from guinea pigs to the list of valid species. The taxonomy of the genus Cryptosporidium was recently challenged by a recent study which suggested that genetic data do not support the currently proposed species designations (35).
In this study, we compared Cryptosporidium isolates from humans and various animals at the 18S small-subunit (SSU) rRNA gene locus. The results of our study revealed that the genus Cryptosporidium consists of multiple species, as proposed previously on the basis of biological characteristics and host specificity data. The species analyzed in this study formed two clades; C. parvum and C. baileyi were most closely related to each other in one clade, and C. muris and C. serpentis were most closely related to each other in the other clade. The differences in the rRNA gene at the species and strain levels allowed us to develop specific diagnostic procedures for accurate identification of Cryptosporidium parasites in clinical and environmental samples.
MATERIALS AND METHODS
Cryptosporidium isolates and oocyst and DNA isolation.
The isolates used in this study were obtained from humans, cattle, snakes, lizards, a guinea pig, a camel, a hyrax, and a chicken (Table 1). Oocysts were purified from fecal samples by a combination of discontinuous density sucrose gradient centrifugation and isopycnic Percoll centrifugation or cesium chloride gradient centrifugation. After treatment in a 5.25% sodium hypochlorite solution (100% commercial bleach) at 4°C for 10 min, oocysts were washed five times in sterile water. DNA was extracted from purified oocysts by a previously described technique (14).
TABLE 1.
Isolates of Cryptosporidium parasites used in this studya
| Isolate | Species | Host | Location | Length of SSU ribosomal DNA (bp) | A + T content (%) |
|---|---|---|---|---|---|
| HFL2 | C. parvum | Human | Florida | 1,750 | 60.97 |
| HFL5 | C. parvum | Human | Florida | 1,745 | 60.86 |
| HCNV4 | C. parvum | Human | Nevada | 1,750 | 61.03 |
| DR1 | C. parvum | Deer via calf | Georgia | 1,746 | 60.88 |
| GCH1 | C. parvum | Human via calf | Massachusetts | 1,746 | 60.88 |
| BOH6 | C. parvum | Calf | Ohio | 1,746 | 60.88 |
| GP1 | C. wrairi | Guinea pig | Michigan | 1,746 | 60.71 |
| CSP01 | C. serpentis | Corn snake | Kansas | 1,743 | 58.52 |
| CSP05 | C. serpentis | Amazon tree boas | Washington, D.C. | 1,743 | 58.52 |
| CSP02 | C. serpentis | Savannah monitor | Washington, D.C. | 1,743 | 58.29 |
| CSP04 | C. serpentis | Savannah monitor | Washington, D.C. | 1,743 | 58.52 |
| IDVS-811 | C. muris | Cattle | Idaho | 1,743 | 57.74 |
| IDRH-13 | C. muris | Rock hyrax via mouse | Washington, D.C. | 1,746 | 57.85 |
| CMU03 | C. muris | Bactrian camel via mouse | Washington, D.C. | 1,746 | 57.85 |
| CBA01 | C. baileyi | Chicken | Alabama | 1,733 | 58.80 |
Partial sequences (length, about 790 bp) were also obtained from four additional human genotype isolates, 18 bovine genotype isolates, and one C. baileyi isolate.
PCR, cloning, and DNA sequencing.
The full-length SSU rRNA gene was amplified from each sample by conventional PCR by using forward primer 5′-AACCTGGTTGATCCTGCCAGTAGTC-3′ and reverse primer 5′-TGATCCTTCTGCAGGTTCACCTACG-3′. Each PCR consisted of 35 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and extension at 72°C for 60 s; an initial denaturation step consisting of incubation at 94°C for 5 min and a final extension step consisting of incubation at 72°C for 10 min were also included. After PCR amplification, the PCR fragment was cloned into the CloneAmp pAMP1 system (Gibco BRL, Frederick, Md.). At least two positive clones from each sample were sequenced by using a model ABI377 autosequencer (Perkin Elmer, Foster City, Calif.).
Sequence analyses.
The DNA sequences obtained from two different clones of each isolate were compared with each other first. Any discrepancy between the two sequences was resolved by the sequence obtained from a third clone. The sequences were aligned by using the Clustal W program (30) with manual adjustment (a data matrix used in the analysis is available upon request). We performed two phylogenetic analyses. First, a neighbor-joining (NJ) tree (27) was constructed for all species belonging to the Apicomplexa in the study. The tree was rooted by using sequences from dinoflagellates (Perkinsus, Gymnodinium, and Gyrodinium spp.), which are closely related to apicomplexans (10, 17). In the second analysis, we restricted the analysis to the Cryptosporidium species. The reliability of the NJ tree was assessed by the bootstrap method (12) with 1,000 replications. We used 95% as the statistically significant value (9); however, values greater than 70% are reported below since bootstrap values may be conservative estimates of the reliability of clades (13). NJ and bootstrap analyses were performed by using the program Treecon W (37). The relative distances among different Cryptosporidium spp. were calculated by using the Kimura two-parameter method and the Wisconsin Package, version 9.0 from the Genetics Computer Group. SSU rRNA sequences of Babesia divergens (accession no. U16370), Theileria parva (L02366), Cytauxzoon felis (L19080), Eimeria tenella (AF026388), Eimeria praecox (U67120), Cyclospora sp. (U40261), Isospora suis (U97523), Toxoplasma gondii (U03070), Neospora caninum (U03069), Sarcocystis tenella (L24383), Frenkelia microti (AF009244), Gymnodinium simplex (U41086), Gymnodinium sp. (AF022196), Gyrodinium impudicum (AL022197), and Perkinsus marinus (X75762) were obtained from the GenBank database.
PCR-RFLP analysis.
To develop a PCR-restriction fragment length polymorphism (RFLP) technique for species- and strain-specific diagnosis of Cryptosporidium parasites, sequences unique to all Cryptosporidium species were identified from the multiple alignment and used as primers in a nested PCR protocol. For the primary PCR step, a PCR product that was about 1,325 bp long was amplified by using primers 5′-TTCTAGAGCTAATACATGCG-3′ and 5′-CCCTAATCCTTCGAAACAGGA-3′. Each PCR mixture (total volume, 100 μl) contained 10 μl of Perkin-Elmer 10× PCR buffer, 6 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, each primer at a concentration of 200 nM, 2.5 U of Taq polymerase, and 0.25 to 2 μl of DNA template. A total of 35 cycles, each consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, were performed; an initial hot start at 94°C for 3 min and a final extension step at 72°C for 7 min were also included. For the secondary PCR step, a PCR product that was 819 to 825 bp long (depending on the species) was amplified by using 2 μl of the primary PCR product and primers 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ and 5′-AAGGAGTAAGGAACAACCTCCA-3′. The PCR mixture and cycling conditions were identical to the conditions used for the primary PCR step, except that 3 mM MgCl2 was used in the PCR mixture.
For restriction fragment analysis, 20 μl of the secondary PCR products was digested in a 50-μl reaction mixture containing 20 U of SspI (New England BioLabs, Beverly, Mass.) (for species diagnosis) or 20 U of VspI (Gibco BRL, Grand Island, N.Y.) (for genotyping of C. parvum) and 5 μl of the appropriate restriction buffer at 37°C for 1 h, under conditions recommended by the supplier. The digested products were fractionated on a 2.0% agarose gel and visualized by ethidium bromide staining.
Nucleotide sequence accession numbers.
The nucleotide sequences of the SSU rRNA genes of C. parvum, C. wrairi, C. muris, C. baileyi, and C. serpentis have been deposited in the GenBank database under accession no. AF093489 to AF093502 and AF115377.
RESULTS
Complete sequences of the SSU rRNA gene were obtained for six C. parvum isolates, one C. wrairi isolate, one C. baileyi isolate, three C. muris isolates, and four C. serpentis isolates. The length of the gene varied from 1,733 to 1,750 bp depending on the species and isolate; the gene of the C. parvum human genotype (HFL2 and HCNV4) was the longest, and the C. baileyi gene was the shortest (Table 1). The rRNA genes of the Cryptosporidium parasites were AT rich, with A+T contents of 57.74 to 61.03%. Within each species, however, the A+T contents of different isolates were quite similar. All bovine C. parvum SSU rRNA sequences were from the type A unit (15).
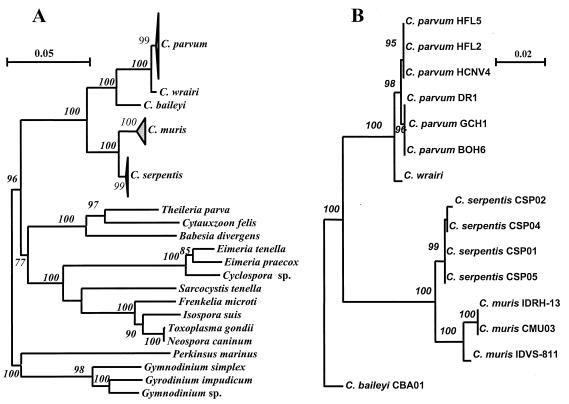
To establish the relationship between Cryptosporidium spp. and other apicomplexans, the SSU rRNA gene sequences of Cryptosporidium spp. were aligned with the previously published sequences of B. divergens (accession no. U16370), T. parva (L02366), C. felis (L19080), E. tenella (AF026388), E. praecox (U67120), Cyclospora sp. (U40261), I. suis (U97523), T. gondii (U03070), N. caninum (U03069), S. tenella (L24383), F. microti (AF009244), Gymnodinium sp. (AF022196), G. simplex (U41086), G. impudicum (AL022197), and P. marinus (X75762) obtained from GenBank. An NJ tree was constructed by using the dinoflagellate parasites as an outgroup (Fig. 1A). The five Cryptosporidium species formed a monophyletic group in the tree, with full statistical reliability (bootstrap value, 100%). Other apicomplexans formed a separate clade, with a bootstrap value of 77%. Within the latter clade the non-Cryptosporidium members of the suborder Eimeriorina (members of the genera Eimeria, Cyclospora, Sarcocystis, Isospora, Toxoplasma, Frenkelia, and Neospora) formed a monophyletic group with full statistical reliability. Among the non-Cryptosporidium coccidians, the parasites once historically placed in the genus Isospora, such as Isospora spp., Sarcocystis spp., Toxoplasma spp., Frenkelia spp., and Neospora spp. (17), formed a monophyletic group distinct from the classical coccidian genus Eimeria and the recently identified genus Cyclospora.
FIG. 1.
Phylogenetic relationships of Cryptosporidium parasites to other apicomplexan parasites (A) and to each other (B).
The five Cryptosporidium species examined in this study formed two clades with full statistical reliability (Fig. 1A). One clade contained C. parvum, C. wrairi, and C. baileyi, and C. parvum and C. wrairi grouped together in this clade. The separation of C. baileyi from C. parvum and C. wrairi and the separation of C. parvum from C. wrairi were statistically reliable (bootstrap values, 99 to 100%). The other clade consisted of C. muris and C. serpentis, which were separated from each other reliably (bootstrap values, 99 to 100%). Most of the interspecies differences in primary sequences occurred in the first half of the SSU rRNA gene.
Intraspecies variations were also found in C. parvum, C. muris, and C. serpentis, and the most noteworthy observations were made with C. parvum (Fig. 1B and Table 2). There were two genotypes of C. parvum, which differed from each other in four areas of the SSU rRNA gene (data not shown). The human genotype included the three isolates from humans, whereas the bovine genotype included the three bovine isolates that originated from cattle, deer, and human sources (Fig. 1B). The genotype of the closely related Cryptosporidium isolate from guinea pigs (designated C. wrairi) was also different from the C. parvum bovine genotype in four areas. All differences among the three genotypes of C. parvum (bovine, human, and guinea pig) occurred in the first half of the SSU rRNA gene. This separation of the three genotypes of C. parvum was also evident on the phylogenetic tree (Fig. 1B).
TABLE 2.
Evolutionary distances among Cryptosporidium spp. and other organismsa
| Organism | No. of nucleotide differences/100 bases
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
C. parvum HFL2 |
C. parvum HFL5 |
C. parvum HCNV4 |
C. parvum DR1 |
C. parvum GCH1 |
C. wrairi | C. baileyi |
C. serpentis CSP01 |
C. serpentis CSP05 |
C. serpentis CSP04 |
C. serpentis CSP02 |
C. muris CMU03 |
C. muris IDRH-13 |
C. muris IDVS-811 |
E. tenella | |
| C. parvum HFL5 | 0.11 | ||||||||||||||
| C. parvum HCNV4 | 0.11 | 0.11 | |||||||||||||
| C. parvum DR1 | 0.23 | 0.17 | 0.23 | ||||||||||||
| C. parvum GCH1 | 0.23 | 0.17 | 0.23 | 0 | |||||||||||
| C. wrairi | 0.63 | 0.58 | 0.63 | 0.46 | 0.46 | ||||||||||
| C. baileyi | 3.93 | 3.93 | 3.93 | 3.69 | 3.69 | 3.69 | |||||||||
| C. serpentis CSP01 | 5.87 | 5.82 | 5.87 | 5.81 | 5.81 | 5.63 | 5.08 | ||||||||
| C. serpentis CSP05 | 5.87 | 5.82 | 5.87 | 5.81 | 5.81 | 5.63 | 5.08 | 0 | |||||||
| C. serpentis CSP04 | 5.99 | 5.94 | 5.99 | 5.94 | 5.94 | 5.75 | 5.20 | 0.11 | 0.11 | ||||||
| C. serpentis CSP02 | 6.24 | 6.13 | 6.24 | 6.19 | 6.19 | 6.00 | 5.45 | 0.34 | 0.34 | 0.23 | |||||
| C. muris CMU03 | 7.00 | 6.95 | 7.00 | 6.89 | 6.89 | 7.01 | 5.71 | 1.98 | 1.98 | 2.10 | 2.33 | ||||
| C. muris IDRH-13 | 7.00 | 6.95 | 7.00 | 6.89 | 6.89 | 7.01 | 5.71 | 1.98 | 1.98 | 2.10 | 2.33 | 0 | |||
| C. muris IDVS-811 | 6.74 | 6.69 | 6.75 | 6.51 | 6.51 | 6.63 | 5.46 | 1.81 | 1.81 | 1.92 | 2.16 | 0.87 | 0.87 | ||
| E. tenella | 20.14 | 19.95 | 20.15 | 20.26 | 20.19 | 20.25 | 19.90 | 18.52 | 18.52 | 18.67 | 18.90 | 19.23 | 19.23 | 19.16 | |
| Cyclospora sp. | 19.86 | 19.67 | 19.87 | 19.84 | 19.76 | 19.91 | 19.60 | 18.06 | 18.06 | 18.21 | 18.44 | 18.77 | 18.77 | 18.84 | 3.24 |
Values were calculated by performing a Kimura two-parameter analysis with the Wisconsin Package, version 9.0, of the Genetics Computer Group.
Two genotypes of C. muris isolates were also distinguished (Fig. 1B). The SSU rRNA gene sequences of the C. muris isolates from a Bactrian camel and a rock hyrax were identical to each other but were different from the SSU rRNA gene sequence of the C. muris bovine isolate at 15 nucleotide positions (data not shown). The evolutionary distance (0.87%) between the two C. muris genotypes was greater than the evolutionary distances (0.17 to 0.63%) among the three C. parvum genotypes (including C. wrairi) (Table 2). Thus, the two genotypes were separated from each other on the phylogenetic tree with full statistical reliability (Fig. 1B). Unlike the differences in the C. parvum genotypes, which occurred in the first half of the SSU rRNA gene, the differences between the two C. muris genotypes were spread over the entire gene.
Minor differences were also detected in the SSU rRNA genes of various C. serpentis isolates. Although the sequences of the two snake isolates (CSP01 and CSP05) were identical, they differed from the sequences of the two isolates from savanna monitors (lizards) at two to four nucleotide positions (nucleotides 371, 560, 692, and 1688).
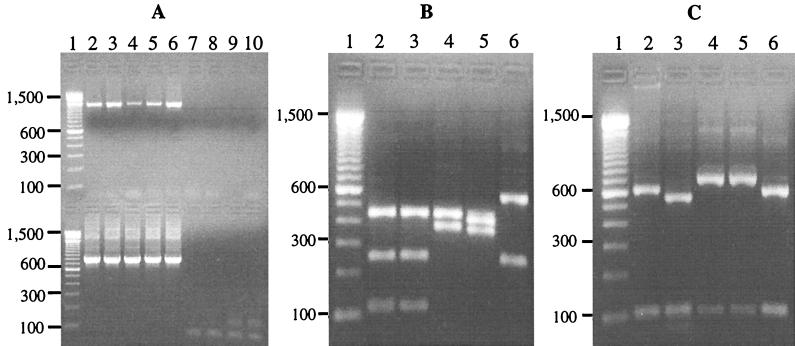
Based on the analysis of the SSU rRNA gene sequences, a PCR-RFLP technique was developed for diagnosis of species and strains of Cryptosporidium parasites. A 819 to 825-bp product was amplified by a nested PCR from Cryptosporidium (C. parvum, C. baileyi, C. muris, and C. serpentis) samples used. The Cryptosporidium specificity of the PCR was confirmed by the failure to amplify samples of Eimeria and Giardia parasites (Fig. 2A). This technique was quite sensitive, as demonstrated by amplification of DNA from a single purified oocyst (data not shown). Species diagnoses were made by digesting the secondary PCR product with SspI (Fig. 2B). C. parvum generated three visible bands of 448, 247, and 106 bp; C. baileyi generated two bands of 572 and 247 bp; C. muris generated two bands of 448 and 377 bp; and C. serpentis generated two visible bands of 414 and 363 bp. To differentiate human and bovine genotypes of C. parvum, the secondary PCR product was digested with VspI (Fig. 2C). The C. parvum bovine genotype produced two visible bands of 628 and 104 bp, whereas the human genotype produced two visible bands of 556 and 104 bp due to the presence of one additional VspI restriction site. In contrast, C. baileyi generated a band pattern similar to that of the C. parvum bovine genotype after VspI digestion, whereas C. muris and C. serpentis both generated two bands of 724 to 730 and 95 to 100 bp.
FIG. 2.
Molecular diagnosis of Cryptosporidium parasites by a nested PCR-RFLP procedure based on SSU rRNA gene sequences. (A) Specific detection of Cryptosporidium spp. by nested PCR. Lane 1, molecular weight markers; lane 2, C. parvum bovine genotype; lane 3, C. parvum human genotype; lane 4, C. muris; lane 5, C. serpentis; lane 6, C. baileyi; lane 7, Eimeria papillata; lane 8, Eimeria nieschulzi; lane 9, Giardia duodenalis; lane 10, negative control. The upper panel contained the primary PCR products, and the lower panel contained the secondary PCR products. (B) Species diagnosis of Cryptosporidium parasites by SspI digestion of the nested PCR products. For lane contents see above. (C) Differentiation of two genotypes of C. parvum by VspI digestion of the nested PCR products. For lane contents see above.
DISCUSSION
Phylogenetic studies of the SSU rRNA gene sequences of Cryptosporidium parasites have been limited, and the few studies that have been done have contributed to the confusion concerning the systematics of Cryptosporidium parasites. An initial SSU rRNA sequence analysis revealed more than 99% identity between C. parvum and C. muris (4). A recent phylogenetic analysis also failed to separate C. parvum, C. muris, and C. baileyi (35). Sample misidentification and sequence inaccuracy, however, may have affected the outcome of these analyses (40).
Phylogenetic analyses of the SSU rRNA genes of various isolates of C. parvum, C. muris, C. baileyi, and C. serpentis in the present study indicated that the Cryptosporidium parasites belong to a multispecies complex that can be divided into two groups. The first group includes various isolates of C. parvum (including C. wrairi) and C. baileyi, whereas the second group includes all isolates of C. muris and C. serpentis. The difference between these two groups is greater than the difference between E. tenella and Cyclospora sp. To minimize the contribution of sequence errors, we used only Cryptosporidium SSU rRNA sequences generated in this study in the phylogenetic analysis.
Importantly, the results of the phylogenetic analyses are in agreement with biologic differences between the two groups of Cryptosporidium parasites. Biologically, the two Cryptosporidium groups have different predilection sites. C. parvum and C. baileyi primarily infect the small intestine and the respiratory tract, respectively, whereas C. muris and C. serpentis primarily infect the stomach. Furthermore, some C. parvum isolates have been shown to infect chicks when large numbers of oocysts are used for inoculation (20, 34). The results of an analysis of an undefined DNA sequence (39) and an antigen reactivity analysis (23) also suggested that C. parvum and C. baileyi are more closely related to each other than they are to C. muris. The results of this study also suggest that Cryptosporidium parasites are not related to other coccidians at the SSU rRNA locus. This observation is in agreement with the results of previous studies (2, 10, 26). Extensive characterization at other genetic loci will be needed to determine the origin of Cryptosporidium parasites.
The results of phylogenetic analyses also indicated that there is significant intraspecies diversity in the genus Cryptosporidium. The three human C. parvum isolates differ from the three bovine isolates in four regions of the SSU rRNA gene. This difference between the two genotypes of C. parvum is in agreement with the presence of two genotypes in other genes observed by us and by other workers (3, 5, 6, 22, 25, 28, 29, 38, 40). Similarly, the Cryptosporidium isolate from guinea pigs, C. wrairi, also differs from the bovine isolates in four regions, two of which are the same polymorphic regions that differ in the human and bovine genotypes. Because the evolutionary distance between C. wrairi and the two C. parvum genotypes is much smaller than the evolutionary distances between other Cryptosporidium spp., C. wrairi might be considered another genotype of C. parvum except for its biological characteristics, such as host specificity. A difference between the human and bovine genotypes of C. parvum at nucleotides 689 to 699 has also been observed recently (21). Using a partial SSU rRNA gene sequence, another group of workers has also identified a new C. parvum genotype (5, 7). The new genotype sequence (ICP), however, is identical to the sequence of our C. muris bovine isolate.
Sequence variation in the SSU rRNA gene was also observed in C. muris and C. serpentis. The SSU rRNA gene sequence of the C. muris bovine isolate is different from the sequences of the C. muris Bactrian camel isolate and the C. muris rock hyrax isolate, whereas the latter two are similar to each other. Interestingly, both the Bactrian camel isolate and the rock hyrax isolate infect young mice, whereas the C. muris bovine isolate does not infect these animals. Similarly, there are small differences in the SSU rRNA gene of C. serpentis. Two snake isolates differed in minor ways from two lizard isolates. It has been suggested previously that C. serpentis is a multispecies complex consisting of at least five species (36). Our results suggest that there may be some intraspecies biological differences among these Cryptosporidium parasites.
In addition to further refining the taxonomic relationships among Cryptosporidium parasites, the results of our sequence analysis allowed us to develop a species- and strain-specific diagnostic tool, which is urgently needed for monitoring the presence of the pathogens in the environment. This PCR-RFLP technique amplifies the sequences of all Cryptosporidium parasites, but species and genotypes can be identified by digesting the secondary PCR product with SspI and VspI, respectively. Previously, two PCR-RFLP techniques based on the SSU rRNA gene were developed to differentiate C. parvum from other Cryptosporidium parasites (1, 16). One technique (16) used conserved sequences for primers and therefore probably amplified the SSU rRNA genes of all eukaryotic organisms. The other technique (1) used an erroneous sequence (4) as the primer, which reduced the efficiency of amplification and made interpretation of data difficult.
In summary, the results of this study confirmed biological observations that the genus Cryptosporidium comprises multiple species. Based on the SSU rRNA gene sequence, the four species used in this study form two clades. Further studies with C. meleagridis, C. felis, and C. nasorum are needed to obtain a more complete phylogenetic profile. The data which we obtained contradict the results of a recent study (35) and are supported by host specificity data and other biological characteristics. In addition to redefining the species structure of the genus, the results of this study facilitated the development of species- and strain-specific diagnostic tools that are needed for monitoring the safety of drinking water.
ACKNOWLEDGMENTS
This work was supported in part by CDC/EPA interagency agreements (DW75937730-01-0 and DW7593784-01-0) and by funds from CDC’s Opportunistic Infectious Diseases Program.
We thank Bruce Anderson of the University of Idaho, Clarence Chrisp of the University of Michigan, and Ann Bratthauer and Donna Fischer of the National Zoological Park (Washington, D.C.) for providing samples. We also thank Mary E. Bartlett for editorial assistance and Daniel G. Colley, Suzanne Binder, Thomas R. Navine, and William Mac Kenzie for helpful comments.
REFERENCES
- 1.Awad-el-Kariem F M, Warhurst D C, McDonald V. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1994;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 2.Barta J R, Martin D S, Liberator P A, Dashkevicz M, Anderson J W, Feighner S D, Elbrecht A, Perkins-Barrow A, Jenkins M C, Danforth H D, Ruff M D, Profous-Juchelka H. Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J Parasitol. 1997;83:262–271. [PubMed] [Google Scholar]
- 3.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil G, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 4.Cai J, Collins M D, McDonald V, Thompson D E. PCR cloning and nucleotide sequence determination of the 18S rRNA genes and internal transcribed spacer 1 of the protozoan parasites Cryptosporidium parvum and Cryptosporidium muris. Biochim Biophys Acta. 1992;1131:317–320. doi: 10.1016/0167-4781(92)90032-u. [DOI] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraway M, Widmer G, Tzipori S. Genetic markers differentiate C. parvum isolates. J Eukaryot Microbiol. 1994;41:26S. [PubMed] [Google Scholar]
- 8.Current W L, Upton S J, Haynes T B. The life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) infecting chickens. J Protozool. 1986;33:289–296. doi: 10.1111/j.1550-7408.1986.tb05608.x. [DOI] [PubMed] [Google Scholar]
- 9.Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci USA. 1996;93:13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escalante A A, Ayala F J. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92:5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayer R, Spear C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 1–41. [Google Scholar]
- 12.Felsenstein J. Confidence limits on phylogenies: an approach using bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 13.Hillis D M, Bull J J. An empirical test of bootstraping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;8:189–191. [Google Scholar]
- 14.Kim K, Gooze L, Petersen C, Gut J, Nelson R G. Isolation, sequence and molecular karyotype analysis of the actin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1992;50:105–113. doi: 10.1016/0166-6851(92)90248-i. [DOI] [PubMed] [Google Scholar]
- 15.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 16.Leng X, Mosier D A, Oberst R D. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18S rRNA gene. Vet Parasitol. 1996;62:1–7. doi: 10.1016/0304-4017(95)00863-2. [DOI] [PubMed] [Google Scholar]
- 17.Levine N D. The protozoan phylum Apicomplexa, vol. I and II. Boca Raton, Fla: CRC Press; 1988. [Google Scholar]
- 18.Levine N D. Taxonomy and review of the coccidian genus Cryptosporidium (protozoa, apicomplexa) J Protozool. 1984;31:94–98. doi: 10.1111/j.1550-7408.1984.tb04296.x. [DOI] [PubMed] [Google Scholar]
- 19.Levine N D. The taxonomy of Sarcocystis (Protozoa, Apicomplexa) species. J Parasitol. 1986;72:372–382. [PubMed] [Google Scholar]
- 20.Lindsay D S, Blagburn B, Ernest J A. Experimental Cryptosporidium parvum infections in chickens. J Parasitol. 1987;73:242–243. [PubMed] [Google Scholar]
- 21.Morgan U M, Constantine C C, Forbes D A, Thompson R C. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 22.Morgan U M, Constantine C C, O’Donoghue P, Meloni B P, Pa O B, Thompson R C. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 23.Nina J M, McDonald V, Dyson D A, Catchpole J, Uni S, Iseki M, Chiodini P L, McAdam K P. Analysis of oocyst wall and sporozoite antigens from three Cryptosporidium species. Infect Immun. 1992;60:1509–1513. doi: 10.1128/iai.60.4.1509-1513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Donoghue O D. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- 25.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S, Mac Kenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Relman D A, Schmidt T M, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis. 1996;173:440–445. doi: 10.1093/infdis/173.2.440. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 29.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyzzer E. Cryptosporidium parvum (sp. nov.), a coccidium found in the small intestine of the common mouse. Arch Protistenkd. 1912;26:394–412. [Google Scholar]
- 32.Tyzzer E. An extracellular coccidium, Cryptosporidium muris (gen. & sp. nov.), of the gastric glands of the common mouse. J Med Res. 1910;18:487–509. [PMC free article] [PubMed] [Google Scholar]
- 33.Tyzzer E. A sporozoon found in the peptic glands of the common mouse. Proc Soc Exp Biol Med. 1907;5:12–13. [Google Scholar]
- 34.Tzipori S, Angus K W, Campbell I, Gray E W. Cryptosporidium: evidence for a single-species genus. Infect Immun. 1980;30:884–886. doi: 10.1128/iai.30.3.884-886.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzipori S, Griffiths J K. Natural history and biology of Cryptosporidium parvum. Adv Parasitol. 1998;40:5–36. doi: 10.1016/s0065-308x(08)60116-5. [DOI] [PubMed] [Google Scholar]
- 36.Upton S J, Current W L. The species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) infecting mammals. J Parasitol. 1985;71:625–629. [PubMed] [Google Scholar]
- 37.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 38.Vasquez J R, Gooze L, Kim K, Gut J, Petersen C, Nelson R G. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–165. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 39.Webster K A, Pow J D E, Giles M, Catchpole J, Woodward M J. Detection of Cryptosporidium parvum using a specific polymerase chain reaction. Vet Parasitol. 1993;50:35–44. doi: 10.1016/0304-4017(93)90005-8. [DOI] [PubMed] [Google Scholar]
- 40.Xiao L, Sulaiman I, Fayer R, Lal A A. Species and strain-specific typing of Cryptosporidium parasites in clinical and environmental samples. Mem Inst Oswaldo Cruz Rio D J. 1998;93:687–691. doi: 10.1590/s0074-02761998000500022. [DOI] [PubMed] [Google Scholar]