Abstract
Serine/one-carbon metabolism provides critical resources for nucleotide biosynthesis and epigenetic maintenance and is thus necessary in cancer cell growth, although the detailed regulatory mechanisms remain unclear. We uncover a critical role of glycogen synthase kinase 3 (GSK3) in regulating the expression of serine/one-carbon metabolic enzymes. Nuclear enrichment of GSK3 significantly suppresses genes that mediate de novo serine synthesis, including PHGDH, PSAT1, PSPH, and one-carbon metabolism, including SHMT2 and MTHFD2. FRAT1 promotes nuclear exclusion of GSK3, enhances serine/one-carbon metabolism, and, as a result, confers cell vulnerability to inhibitors that target this metabolic process such as SHIN1, a specific SHMT1/2 inhibitor. Furthermore, pharmacological or genetic suppression of GSK3 promotes serine/one-carbon metabolism and exhibits a significant synergistic effect in combination with SHIN1 in suppressing cancer cell proliferation in cultured cells and in vivo. Our observations indicate that inhibition of nuclear GSK3 signaling creates a vulnerability, which results in enhanced efficacy of serine/one-carbon metabolism inhibitors for the treatment of cancer.
Inhibition of nuclear GSK3 enhances the anticancer effect of SHMT inhibitor SHIN1 in non–small cell lung cancer (NSCLC) cells.
INTRODUCTION
Glycogen synthase kinase 3 (GSK3), as is implied by the name, was initially found as the protein kinase that phosphorylates and suppresses glycogen synthase (1). This ubiquitously expressed serine/threonine kinase was then soon determined to phosphorylate over a hundred protein substrates and link various extracellular cues to intracellular signals to maintain proper homeostasis. Accordingly, dysregulation of GSK3 has been implicated in a wide variety of disorders including psychiatric diseases, neurological diseases, cardiovascular diseases, diabetes, and certain cancers (2, 3). GSK3 is expressed as two isoforms known as GSK3α (51 kDa) and GSK3β (47 kDa), which are encoded by independent genes present in all mammalian cells. The overall homology of these isoforms is greater than 67%, with the catalytic domain demonstrating greater than 98% sequence similarity (4).
GSK3 is regulated by multiple mechanisms. Within the WNT signaling pathway, for example, GSK3 is recruited by the scaffolding protein AXIN and subsequently forms a multiprotein complex that phosphorylates β-catenin, leading to its inactivation (2). GSK3α/β is also regulated by direct phosphorylation at Ser21/9 in the N-terminal tail, which acts as a pseudosubstrate domain that competes for GSK3 substrate docking to suppress GSK3 signaling (2, 5). GSK3 phosphorylation is directly regulated by several basophilic protein kinases including Akt, protein kinase C (PKC), RSK, and PKA (2, 5, 6). Generally, GSK3 is active in resting cells and is inhibited upon phosphorylation. Recently, a new mechanism for modulating GSK3 signaling through regulation of its nuclear-cytoplasmic location has been observed by our laboratory and others (7–9). Mechanistic target of rapamycin complex 1 (mTORC1) promotes nuclear exclusion of GSK3, whereas inhibition of mTORC1 by either nutrient insufficiency, or deprivation of growth regulators, or treatment with mTORC1 inhibitors results in the nuclear accumulation of GSK3. Furthermore, drug resistance is observed under conditions where rapamycin cannot promote sufficient nuclear accumulation of GSK3 (8). By highlighting the critical role of GSK3 in cellular metabolism, we have demonstrated that GSK3 mediates mTORC1-associated regulation of global metabolic genes that regulate glycolysis, serine/one-carbon metabolism, purine synthesis, and others (9).
One-carbon metabolism is a well-conserved metabolic process supported by the cofactor folate that serves to transfer one-carbon units for critical biosynthetic processes including nucleotide biosynthesis, various methylation reactions, redox status, and others. One-carbon metabolism is linked to serine catabolism as one-carbon units are predominantly derived from this nonessential amino acid. Because of its critical role in the synthesis of building blocks necessary for cell growth and proliferation, one-carbon metabolism has been associated with various biological processes such as stem cell maintenance and cancer development (10). It has been reported that serine and one-carbon metabolism are significantly up-regulated in many types of cancers, and furthermore, multiple enzymes that function within these pathways are well-known targets for the development of anticancer therapeutics (10). For example, the enzyme thymidylate synthase (TYMS), which catalyzes the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP) through methylation reactions, as well as dihydrofolate reductase (DHFR), which reduces dihydrofolic acid to tetrahydrofolic acid, are two important targets within the serine and one-carbon metabolic pathways that have been the focus of efforts to target this pathway in cancer for therapeutic benefit. For instance, the pyrimidine analog 5-fluorouracil acts as a potent inhibitor of TYMS and was approved decades ago for the treatment of multiple cancers such as breast, colorectal, stomach, and skin cancers (10). Another well-known anticancer agent, methotrexate, potently inhibits DHFR and has been shown to be effective in the treatment of a number of cancers including that of the breast, head and neck, and lung, as well as leukemia and lymphoma (10). Recently, a meta-analysis of gene expression in tumor tissues revealed that two mitochondrial enzymes that mediate one-carbon metabolism, serine hydroxymethyltransferase 2 (SHMT2) and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), are consistently overexpressed in cancer compared to corresponding normal tissues (11–13).
SHMT2 catalyzes the conversion of serine and tetrahydrofolate (THF) into glycine and 5,10-methylene, thus linking serine catabolism with one-carbon metabolism. MTHFD2 has both dehydrogenase and cyclohydrolase activity that facilitates the production of 5,10-methylene-THF and 10-formyl-THF in mitochondria, both of which are important carbon donors for subsequent nucleotide synthesis. Therefore, both SHMT2 and MTHFD2 play important roles in regulating cell proliferation in cancer cells and are considered important targets in anticancer drug development. For example, suppression of MTHFD2 in acute myeloid leukemia cells decreased cancer cell growth in vitro and in vivo (14), and Woo and colleagues (15) showed a robust suppression of tumor growth using short hairpin RNAs (shRNAs) against SHMT2 in a hepatocellular carcinoma xenograft model.
Despite the significant contribution of one-carbon metabolism to cancer development and progression, the underlying mechanism that regulates the expression of one-carbon metabolic enzymes in cancer remains unclear. In this study, we report a role for GSK3 in the regulation of genes that mediate mitochondrial serine/one-carbon metabolism in non–small cell lung cancer (NSCLC) cell lines. Our observation indicates a critical role for GSK3 in the regulation of serine/one-carbon metabolism and demonstrates that suppression of nuclear GSK3 signaling improves the anticancer efficacy of inhibitors of enzymes involved in serine/one-carbon metabolism.
RESULTS
Nuclear GSK3 regulates extensive gene expression
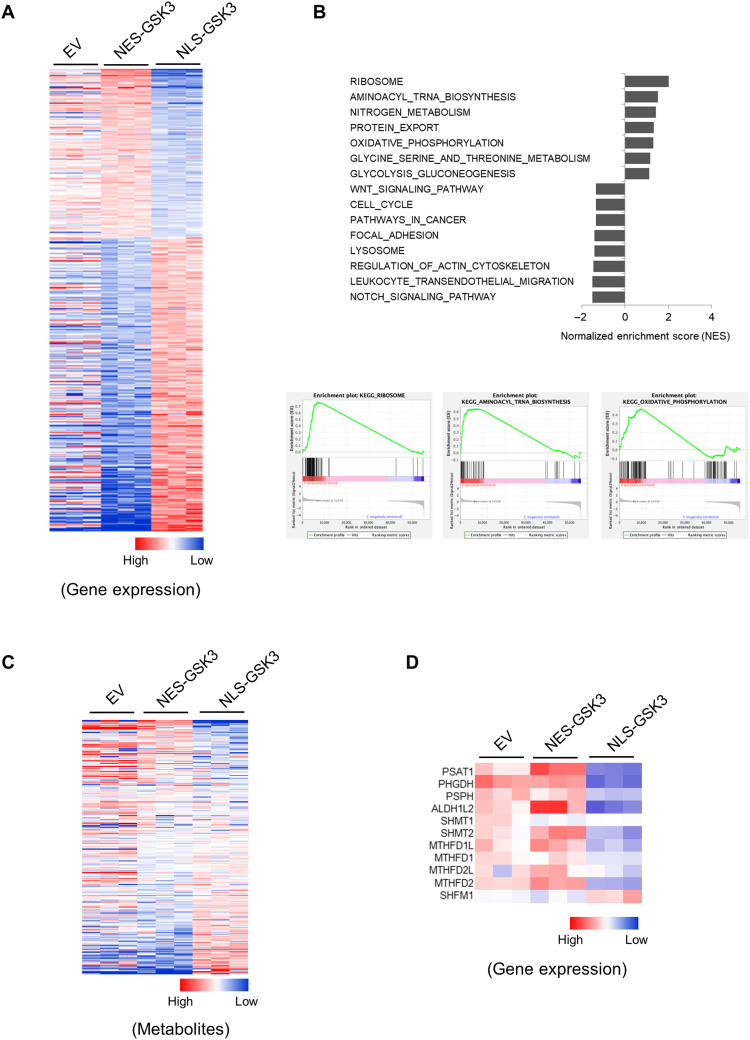
We and others have previously reported a significant nuclear enrichment of GSK3 upon mTORC1 suppression in several breast and lung cancer cell lines (7, 8). To further investigate the role of nuclear GSK3 in cancer, we performed global gene expression profiling in established cell lines with empty vector (EV)–, nuclear export signal (NES)–, and nuclear localization signal (NLS)–tagged GSK3β as previously described (8) (fig. S1, A and B). We found that the expression of about 4000 genes was significantly altered in cells expressing NLS-GSK3β as compared with those expressing NES-GSK3β (P < 0.001) (Fig. 1A), demonstrating that the subcellular distribution of GSK3 plays a critical role in regulating the expression of an extensive number of genes. Gene set enrichment analysis (GSEA) revealed that multiple pathways are significantly altered in NLS-GSK3β–expressing cells, including ribosome biosynthesis, transfer RNA (tRNA) biosynthesis, nitrogen metabolism, oxidative phosphorylation, glycolysis, and others (Fig. 1B). Furthermore, global metabolite profiling confirmed that over 100 metabolites are significantly altered in cells that express NLS-GSK3β as compared with NES-GSK3, revealing extensive metabolic rewiring upon NLS-GSK3 overexpression (Fig. 1C). We also observed that genes associated with serine/one-carbon metabolism are among those most significantly altered (Fig. 1D). Consistent with this observation, levels of intracellular dAMP and dTMP were suppressed, whereas dUMP levels were increased in NLS-GSK3β–expressing cells (fig. S1, C to E).
Fig. 1. Nuclear GSK3 regulates extensive gene expression.
Cell lines were established with pTRIPZ-EV, pTRIPZ-HA-NES-GSK3β, and pTRIPZ-HA-NLS-GSK3β in NCI-H1299 cells. Cells were exposed to 1 μM doxycycline for 24 hours. (A) Heatmap of the most significantly altered genes in NCI-H1299 cells expressing EV, NES-GSK3, or NLS-GSK3. (B) GSEA of NES-GSK3– versus NLS-GSK3–expressing cells. NESs and GSEA of ribosome, tRNA biosynthesis, and oxidative phosphorylation are shown. (C) Heatmap of the most significantly altered metabolites from EV-, NES-GSK–, and NLS-GSK3–expressing cells. (D) Heatmap of the expression of genes that mediate serine/one-carbon metabolism in cells expressing EV, NES-GSK3, or NLS-GSK3.
Nuclear GSK3 suppresses serine/one-carbon metabolism
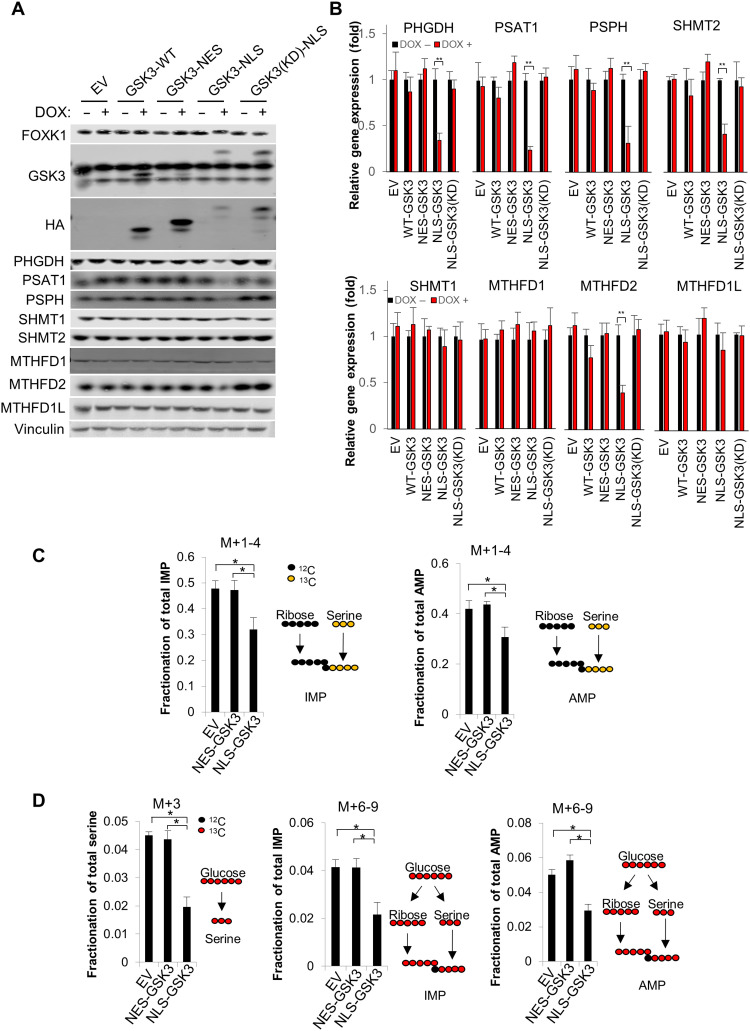
Given the critical role of serine/one-carbon metabolism in nucleoside biosynthesis and cancer progression, we sought to further investigate the role of nuclear GSK3 in the regulation of serine/one-carbon metabolism. Consistent with our gene expression profiling (fig. S2A), we further confirmed that multiple enzymes that mediate de novo serine synthesis (PHGDH, PSAT1, and PSPH) and mitochondrial one-carbon metabolism (SHMT2 and MTHFD2) are significantly suppressed by NLS-GSK3. In addition, overexpression of wild-type (WT), NES-tagged GSK3, or NLS-tagged kinase dead (KD) GSK3 minimally affected the expression of these genes as determined by immunoblot and gene expression analysis, supporting the critical role of nuclear GSK3 in regulating gene expression to mediate serine/one-carbon metabolism (Fig. 2, A and B).
Fig. 2. Nuclear GSK3 suppresses serine/one-carbon metabolism.
Cell lines were established with pTRIPZ-EV, pTRIPZ-HA-GSK3-(WT), pTRIPZ-HA-NES-GSK3β, pTRIPZ-HA-NLS-GSK3β, or pTRIPZ-HA-NLS-GSK3β-(KD) in NCI-H1299 cells. WT, wild type; HA, hemagglutinin. (A and B) Cells were exposed to 1 μM doxycycline for 24 hours, and (A) whole-cell lysate was subjected to immunoblot analysis with the indicated antibodies, or (B) total RNA was extracted and subjected to reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis with indicated primers. (C and D) EV, NES-GSK3, or NLS-GSK3 cells were exposed to 1 μM doxycycline for 24 hours before U-13C-Serine or U-13C-Glucose labeling. Cells were fed with U-13C-Serine or U-13C-Glucose for 4 hours, and metabolites were subsequently extracted and subjected to liquid chromatography–mass spectrometry (LC-MS) analysis. *P < 0.05, **P < 0.01.
To further explore this relationship, we next traced the fate of both U-13C-Serine and U-13C-Glucose following NES-GSK3 or NLS-GSK3 expression. De novo purine synthesis requires two 10-formyl-tetrahydrofolates and one glycine, both of which are products of serine/one-carbon metabolism (fig. S2, B and C). Therefore, to understand the contribution of these pathways, we determined the production of labeled nucleotides and M+3 serine in cells fed U-13C-Serine and U-13C-Glucose, respectively. As expected, production of both 13C incorporated inosine monophosphate (IMP) and adenosine monophosphate (AMP) from both U-13C-Serine– and U-13C-Glucose–fed cells as well as M+3 serine from U-13C-Glucose–fed cells was significantly reduced upon NLS-GSK3 expression (Fig. 2, C and D and table S1), thus supporting the critical role of nuclear GSK3 in suppressing cellular serine/one-carbon metabolism.
Frequently rearranged in advanced “T cell” lymphomas-1 promotes serine/one-carbon metabolism by promoting GSK3 nuclear exclusion.
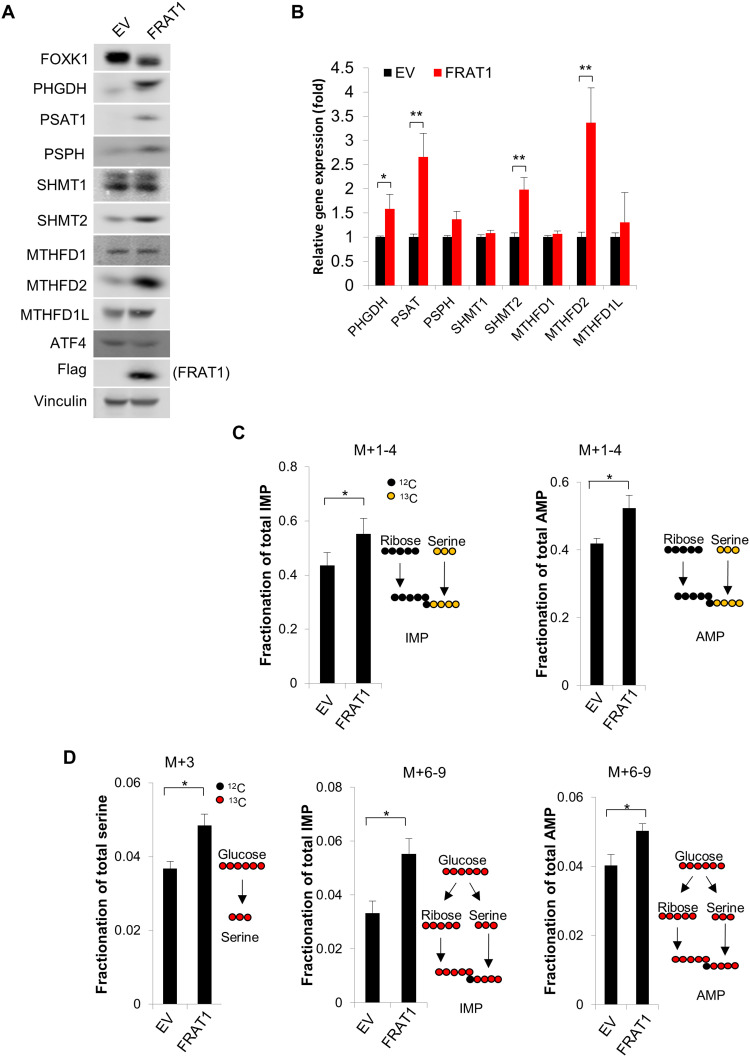
Frequently rearranged in advanced T cell lymphomas-1/2 (FRAT1/2) are oncoproteins that interact with and promote the nuclear exclusion of GSK3 (16, 17). Furthermore, FRAT1/2 overexpression is sufficient to prevent nuclear enrichment of GSK3 (fig. S3A) and thus subsequently reduces GSK3-dependent phosphorylation of several nuclear proteins (8). Consistent with the idea that nuclear GSK3 suppresses serine/one-carbon metabolism, FRAT1/2 expression significantly promotes the expression of genes that mediate de novo serine metabolism (PHGDH, PSAT1, and PSPH) and mitochondrial one-carbon metabolism (SHMT2 and MTHFD2), while genes that mediate cytoplasmic one-carbon metabolism (SHMT1, MTHFD1, and MTHFD1L) were minimally affected (Fig. 3, A and B, and fig. S3B). Supporting this idea, U-13C-Serine and U-13C-Glucose tracing analysis further revealed that production of both 13C-labeled IMP and AMP was significantly increased by ectopically expressing FRAT1 (Fig. 3, C and D, and table S1). The production of M+3 serine from U-13C-Glucose–fed cells was also significantly increased upon FRAT1 expression, further confirming the critical role of FRAT1-driven nuclear exclusion of GSK3 in promoting serine/one-carbon metabolism (Fig. 3D).
Fig. 3. Nuclear GSK3 exclusive protein FRAT1 promotes serine/one-carbon metabolism.
Cell lines were established with EV or FRAT1 overexpression in NCI-H1299 cells. (A) Whole-cell lysate was subjected to immunoblot analysis with the indicated antibodies. (B) Total RNA was extracted and subjected to RT-qPCR analysis with the indicated primers. (C and D) Cells were fed with U-13C-Serine or U-13C-Glucose for 4 hours, and the metabolites were subsequently extracted and subjected to LC-MS analysis. *P < 0.05, **P < 0.01.
FRAT1 expression sensitizes H1299 cells to SHIN1 treatment
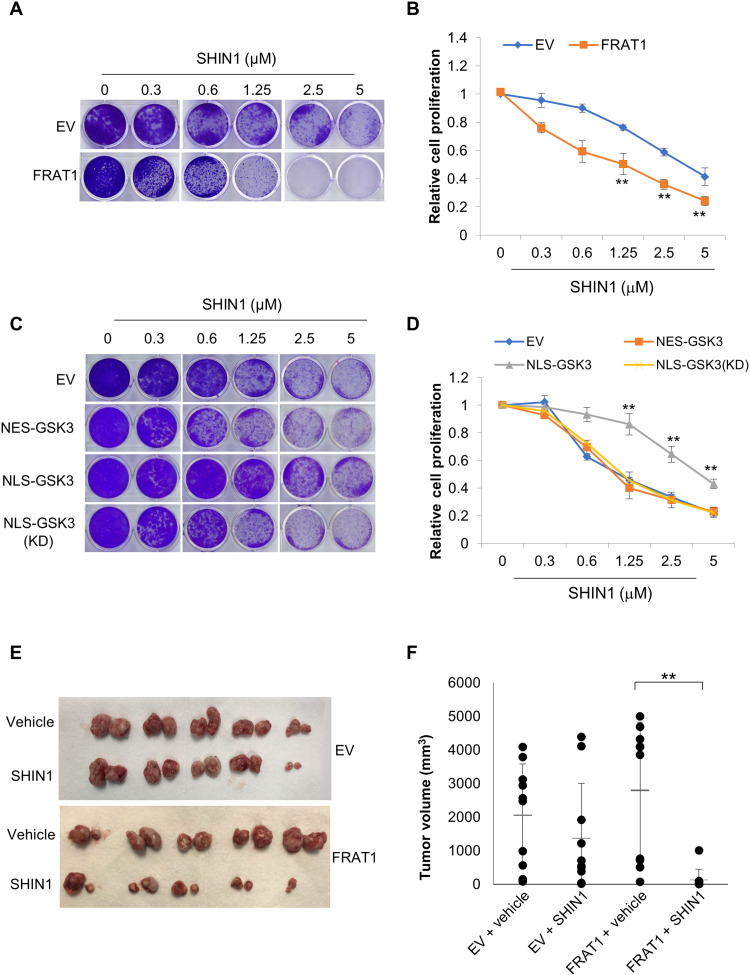
Given the role of the proto-oncogene FRAT1 in the promotion of serine/one-carbon metabolism, we next sought to investigate if FRAT1 overexpression confers dependence on serine/one-carbon metabolism and therefore renders these cells vulnerable to its inhibition. Strikingly, we observed that compared to EV, proliferation of FRAT1-overexpressing cells is relatively more sensitive to the SHMT1/2 inhibitor SHIN1 [with median inhibitory concentration (IC50) of 4.3 and 0.6 μM, respectively] and the MTHFD1/2 inhibitor DS18561882 (with IC50 of 4.8 and 2.6 μM, respectively), with a more significant response to SHIN1 treatment (Fig. 4, A and B, and fig. S4, A and C). Suppression of cell proliferation in both EV- and FRAT1-expressing cells can be nearly completely rescued by supplementation with either formate or nucleosides (fig. S4, A, B, D, and E), indicating that both SHIN1 and DS18561882 suppress cell proliferation via specifically inhibiting serine/one-carbon metabolism–dependent nucleotide synthesis. Furthermore, NLS-GSK3 expression reduced the up-regulation of serine/one-carbon metabolic enzymes observed upon ectopic expression of FRAT1, whereas NES-GSK3 or NLS-GSK3(KD) had little impact (fig. S4, F and G). Consistently, we observed that upon NLS-GSK3 expression, cells are less sensitive to SHIN1 treatment (with IC50 of ~4 μM), whereas cells expressing EV, NES-GSK3, or NLS-GSK3(KD) (with IC50 of ~0.93, 0.78, and 0.97 μM, respectively) retain sensitivity (Fig. 4, C and D). These observations were further supported by in vivo xenograft analysis using NCI-H1299 cells. SHIN1 treatment slightly suppressed tumor growth of H1299-EV cells but significantly decreased the tumor growth of H1299 cells with overexpression of FRAT1 (Fig. 4, E and F). These results support an important link between suppression of nuclear GSK3 signaling and vulnerability of cancer cells to inhibition of serine/one-carbon metabolism.
Fig. 4. FRAT1 expression sensitizes H1299 cells to SHIN1 treatment.
Cell lines were established with either EV or FRAT1 overexpression in NCI-H1299 cells. (A) Clonogenic assay was performed with cells expressing either EV or FRAT1 in the presence of the indicated concentrations of SHIN1, and (B) cell proliferation was estimated by quantification of clonogenic cell growth as described in Materials and Methods. (C and D) Cell lines were established with FRAT1 overexpression and overexpression of pTRIPZ-EV, pTRIPZ-HA-NES-GSK3β, pTRIPZ-HA-NLS-GSK3β, or pTRIPZ-HA-NLS-GSK3β-(KD) in NCI-H1299 cells. (C) Clonogenic assay was performed using the above cell lines in the presence of the indicated concentrations of SHIN1, and (D) cell proliferation was estimated by quantification of clonogenic cell growth as described in Materials and Methods. Values are expressed as means ± SEM. Student’s t test, **P < 0.001, n = 3. (E and F) 1 × 106 of either EV- or FRAT1-expressing NCI-H1299 cells were injected into mice through subcutaneous inoculation as described in Materials and Methods. Mice were treated with either vehicle or SHIN1 (100 mg/kg) three times a week, and tumors were extracted after 3 weeks (n = 5 mice per group). Values are expressed as means ± SD. Two-tailed Student’s t test, **P < 0.01.
Inhibition of GSK3 promotes serine/one-carbon metabolism
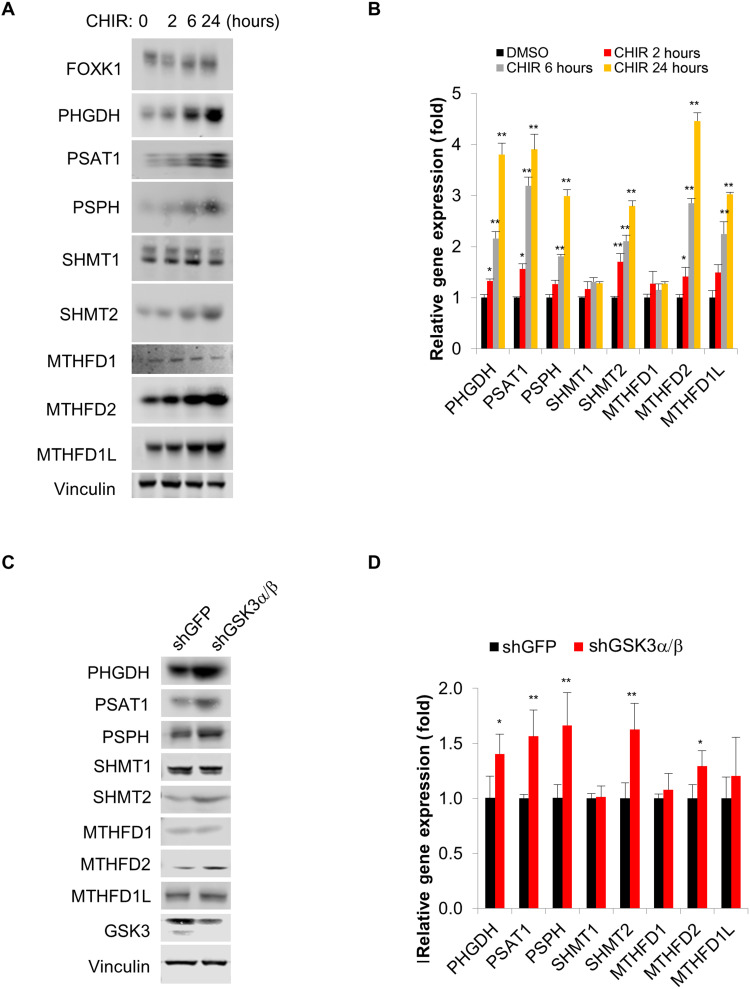
Suppression of GSK3 has been reported to promote numerous metabolic processes such as glucose metabolism and mitochondrial energy metabolism (9, 18, 19). Thus, we next sought to determine whether general inhibition of GSK3 promotes serine/one-carbon metabolism. Upon treatment with the GSK3 inhibitor CHIR99021, expression of multiple genes that mediate serine/one-carbon metabolism increased in a time-dependent manner, which is consistent with our previous observations (Fig. 5, A and B) (9). We further confirmed that two alternate GSK3 inhibitors (LY2090314 and SB216763) with differing chemical structures also have a similar effect in promoting the expression of genes that mediate serine/one-carbon metabolism (fig. S5, A and B). We see a very limited effect on metabolic gene expression in FRAT1 overexpression cells upon treatment with a GSK3 inhibitor (fig. S5C). In further support of this relationship, we generated shRNAs to suppress GSK3α/β expression. While suppression of GSK3α or GSK3β alone had little effect on the expression of associated metabolic genes, a significant increase was observed upon suppression of both GSK3α and GSK3β (Fig. 5, C and D, and fig. S5D). Consistently, upon overexpression of FRAT1 in GSK3α/β-deficient cells, we observed no further elevation of proteins involved in serine/one-carbon metabolism, further supporting our previous observation (fig. S5E).
Fig. 5. Suppression of GSK3 promotes serine/one-carbon metabolism.
(A and B) NCI-H1299 cells were exposed to 5 mM CHIR99021 for the indicated time, and (A) whole-cell lysate was subjected to immunoblot analysis with the indicated antibodies, or (B) total RNA was extracted and subjected to RT-qPCR analysis with the indicated primers. (C and D) NCI-H1299 cell lines were established with either overexpression of short hairpin green fluorescent protein (shGFP) or dual overexpression of shGSK3a#1 and shGSK3b#1. (C) Whole-cell lysate was subjected to immunoblot analysis with the indicated antibodies, and (D) total RNA was extracted and subjected to RT-qPCR analysis with the indicated primers. *P < 0.05, **P < 0.01.
Suppression of GSK3 sensitizes cancer cells to SHIN1 treatment
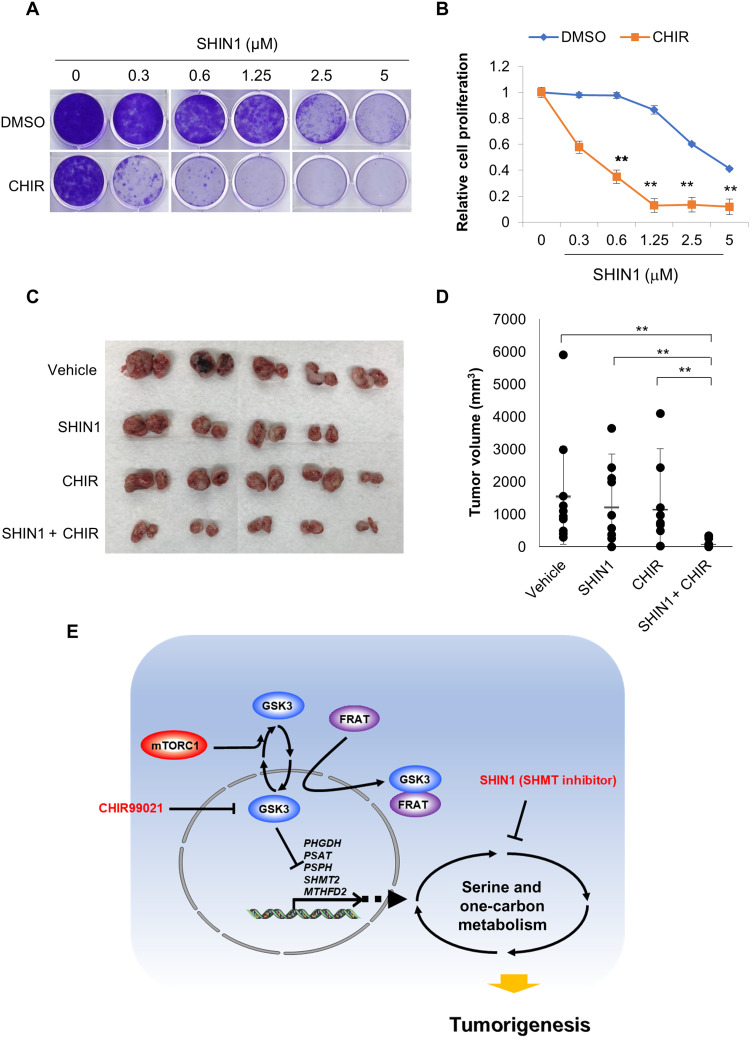
Given that suppression of GSK3 significantly promotes serine/one-carbon metabolism, we next asked whether GSK3 inhibition confers a metabolic vulnerability to potentiate treatment with the SHMT1/2 inhibitor SHIN1 in cancer cells. Strikingly, SHIN1 treatment displayed a synergistic effect when combined with the GSK3 inhibitor CHIR99021 as determined using a clonogenic assay. Alone, H1299 cells had an IC50 of approximately 4.2 μM in response to SHIN1 treatment, which decreased to approximately 0.4 μM when combined with 1 μM CHIR99021 (Fig. 6, A and B). Notably, treatment with 1 μM CHIR99021 alone did not significantly affect H1299 cell proliferation. Furthermore, GSK3α/β double knockdown cells, which have increased serine/one-carbon metabolism, were more sensitive to SHIN1 treatment (IC50 of ~2.4 μM) as compared with the short hairpin green fluorescent protein (shGFP) control cells (IC50 of ~4.3 μM), further supporting this interaction (fig. S6, A and B). Last, in vivo xenograft analysis using H1299 cells further indicated that single treatment of either SHIN1 or CHIR99021 had little effect in suppressing tumor growth, whereas combination treatment significantly suppressed tumor growth, which is consistent with our observations in vitro (Fig. 6, C and D).
Fig. 6. Suppression of GSK3 sensitizes cells to SHIN1 treatment.
(A) Clonogenic assay was performed with cells treated with the indicated concentration of SHIN1 in the presence or absence of 1 μM CHIR99021. (B) Cell proliferation was estimated by quantification of clonogenic cell growth as described in Materials and Methods. (C and D) 1 × 106 of NCI-H1299 cells were injected into mice through subcutaneous inoculation as described in Materials and Methods. Mice were treated with vehicle, SHIN1 (100 mg/kg), CHIR99021 (10 mg/kg), or SHIN1 plus CHIR99021 three times a week, and tumors were extracted after 3 weeks (n = 5 mice per group, and n = 4 in the SHIN1 group). Values are expressed as means ± SD. Two-tailed Student’s t test, **P < 0.01. (E) Model: Nuclear enrichment of GSK3 suppresses the expression of multiple genes including PHGDH, PSAT1, PSPH, SHMT2, and MTHFD2 and subsequently suppresses serine/one-carbon metabolism. Increased expression of FRAT1/2 facilitates nuclear exclusion of GSK3 and promotes serine/one-carbon metabolism, which therefore confers a metabolic vulnerability to inhibition of this pathway. Consequently, FRAT1/2-expressing cells are relatively sensitive to inhibitors targeting serine/one-carbon metabolism such as the SHMT1/2 inhibitor, SHIN1. Thus, inhibition of GSK3 through treatment with the pharmacologic inhibitor CHIR99021 effectively mimics FRAT1/2 expression and synergizes with SHIN1 treatment to enhance its anticancer effect both in cells and in vivo. **P < 0.01.
DISCUSSION
In support of growth and proliferation, most cancer cells require considerable amounts of one-carbon units to ensure sufficient nucleotide biosynthesis. Therefore, alteration of one-carbon metabolism is one of the most frequently observed forms of metabolic reprogramming beyond the Warburg effect in multiple cancer cell types (20). Targeting one-carbon metabolism is one of the most well-known strategies in anticancer drug development. For example, aminopterin and methotrexate are analogs of folic acid that target one-carbon metabolism and have the unique distinction of being the first approved chemotherapeutic agents for certain cancer therapies (10). Numerous groups have reported that multiple genes, which mediate de novo serine synthesis and mitochondrial one-carbon pathways, are up-regulated in several cancers. In addition, extensive profiling of these cancers using genomic and metabolomic approaches has revealed that SHMT2 and MTHFD2 are among the most consistently amplified genes that positively regulate cancer cell growth (12, 13).
In this study, we have found that GSK3 is one of the major regulators of serine/one-carbon metabolism in cancer cells. Notably, nuclear enrichment of GSK3 significantly suppressed the expression of genes that mediate de novo serine synthesis such as PHGDH, PSAT1, and PSPH, as well as mitochondrial one-carbon metabolism such as SHMT2 and MTHFD2. Enzymes that mediate cytosolic one-carbon metabolism such as SHMT1 and MTHFD1, however, are largely unaffected by GSK3 signaling (Fig. 2, A and B). Reduced levels of nuclear GSK3 localization induced by FRAT1 overexpression promote serine/one-carbon metabolism through up-regulation of PHGDH, PSAT1, PSPH, SHMT2, and MTHFD2 and confer a metabolic vulnerability to inhibition of this pathway (Figs. 3, A to D, and 4, A to F). ATF4 is one of a few transcription factors that are known to regulate multiple enzymes that mediate serine/one-carbon metabolism (21, 22). However, we did not observe a significant increase in ATF4 levels upon FRAT1 overexpression (Fig. 3A), nor did we observe an alteration of ATF4 DNA binding affinity upon either overexpression of NLS-GSK3 or inhibition of GSK3 using treatment with CHIR99021 (fig. S7A). We have previously shown that Forkhead Box K1 (FOXK1) plays an important role in regulating several metabolic processes including serine/one-carbon metabolism in mouse embryonic fibroblasts (9). Consistently, multiple enzymes, which mediate serine/one-carbon metabolism, were also significantly decreased upon FOXK1 suppression in H1299 cells (fig. S7B). Given previous studies with ATF4 and other transcriptional regulators (22–24), it appears that in addition to nuclear GSK3-dependent regulation of serine/one-carbon metabolism via FOXK1, additional mechanisms also contribute to the process, underscoring the importance of this metabolic pathway to cell growth and survival.
We have previously reported that rapamycin significantly promotes nuclear enrichment of GSK3 (8), and consistent with this notion, we have found that multiple enzymes that mediate serine/one-carbon metabolism are significantly down-regulated by treatment with rapamycin and induced by serum stimulation (fig. S7, C and D). Overexpression of FRAT1, which decreases nuclear GSK3 signaling, enhances serine/one-carbon metabolism and thus makes cancer cells more sensitive to both SHMT and MTHFD inhibitors (fig. S4, A to D). It is worth noting that FRAT1-expressing cells grow at similar rates in vitro (Fig. 4A), whereas they form tumors faster than the control in xenograft models in vivo (Fig. 4F), suggesting differences in cell culture versus in vivo regulation of tumor growth by FRAT. Conversely, overexpression of nuclear GSK3 in NSCLC cells confers resistance to these inhibitors (Fig. 4, C and D). However, we only observed a partial correlation between SHIN1 sensitivity and FRAT1 expression in vitro, which is consistent with the complexity of serine/one-carbon metabolism regulation (fig. S8).
In addition, overexpression of FRAT1 did not confer increased sensitivity to PHGDH inhibition as demonstrated using the compounds NCT-503 and CBR-5884 (fig. S6E), despite significant alterations in the expression of enzymes that mediate de novo serine synthesis (25, 26). It is possible that a sufficient amount of serine is available in the culture media to support cell growth and proliferation even when de novo serine synthesis is repressed via PHGDH inhibition. In support of this idea, our U-13C-Glucose tracing experiment demonstrated that less than 5% of cellular serine was derived from de novo serine synthesis in our cell culture system. However, it is worth noting that in vivo, serine is unable to pass through the blood-brain barrier, leading to an increased dependence on de novo serine synthesis in most brain tumors. Therefore, although we did not observe an increased sensitivity upon overexpression of FRAT1 in vitro, it is possible that brain tumors with higher expression level of FRAT1 are more sensitive to PHGDH inhibitors in vivo. Expression of serine/one-carbon metabolic genes was refractory to single knockdown of GSK3α or GSK3β but was significantly up-regulated when both GSK3α and GSKβ were suppressed (fig. S5C). In addition, treatment with the GSK3 inhibitor CHIR99021 significantly promoted de novo serine synthesis and mitochondrial one-carbon metabolism, and thus sensitized cells to treatment with the SHMT1/2 inhibitor SHIN1 or the MTHFD2 inhibitor DS18561882 (Fig. 6, A and B, and fig. S6, C and D). In support of this, two structurally distinct GSK3 inhibitors (LY2090314 and SB216763) also promoted the expression of genes involved in serine/one-carbon metabolism (fig. S5, A and B). It is interesting that GSK3 inhibition leads to an up-regulation of mitochondrial MTHFD1L expression, which was minimally altered by NLS-GSK3 or FRAT1/2 expression, indicating a possible role for cytoplasmic GSK3 in the regulation of MTHFD1L expression (Fig. 5, A and B, and fig. S5, A and B). Now, GSK3 inhibitors are undergoing phase 2 clinical trials for the treatment of certain cancers, although the efficacy of single agent treatment remains limited (27). Our observations demonstrate a potential strategy for the use of combination therapy, in which GSK3 inhibitors and inhibitors of serine/one-carbon metabolism are administered concurrently to improve anticancer efficacy and, ultimately, patient outcomes.
MATERIALS AND METHODS
RNA extraction and reverse transcription quantitative polymerase chain reaction analysis
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and subjected to RNA sequencing analysis (performed by the Genomics Resources Core Facility at Weill Cornell Medicine) or used for cDNA synthesis with SuperScript III First-Strand Synthesis Supermix Kit (Life Technologies) according to the manufacturers’ instructions. Quantitative polymerase chain reaction (qPCR) was performed using the QuantiTect SYBR Green qPCR Kit on Roche LightCycler 480. Primers were purchased from IDT, and melting curve analysis was performed at the end of the PCR analysis.
Immunoblot analysis
Whole-cell lysate was extracted with 1X SDS sample buffer [50 mM tris-HCl (pH 6.8), 10% glycerol, 2% SDS, 10% 2-mercaptoethanol, and 0.1% bromophenol blue]. Samples were boiled and separated using electrophoresis by SDS–polyacrylamide gel electrophoresis. Proteins were then transferred to nitrocellulose membranes. The membranes were blocked for 1 hour before probing with primary antibodies overnight. The membranes were subsequently incubated with secondary antibodies for 1 hour and developed with LI-COR Odyssey. All immunoblots in this study are representative of at least three independent experiments.
Total and 13C metabolite flux and analysis in cells
Cells were plated the day before labeling at a concentration of 5 × 105 cells per 6-cm dish. To perform the 13C metabolite profiling, the medium was changed to 5 mM U-13C-Glucose or 0.5 mM U-13C-Serine–containing medium for the indicated time points. Cells were collected, and the intracellular metabolites were extracted using 80% (v/v) aqueous methanol. Targeted liquid chromatography tandem mass spectrometry was performed using a 5500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC System (Shimadzu) with Amide HILIC chromatography (Waters). Data were acquired in selected reaction monitoring (SRM) mode using positive/negative ion polarity switching for steady-state polar profiling of greater than 260 molecules. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/SCIEX). Informatics analysis was carried out using MetaboAnalyst.ca free online software.
Clonogenic assay
Cells (2000) were seeded in 12-well plates and incubated for 1 to 2 weeks. After colonies were clearly observed, they were fixed with 4% formaldehyde and stained with crystal violet (0.5%, w/v). After rinsing four times with phosphate-buffered saline (PBS) buffer, the images of the wells were scanned. For quantification after imaging, methanol was added to each 12-well plate, and optical density was measured at 570 nm as described (28).
Immunofluorescence staining
Immunofluorescence was performed as described previously with some modifications. Cells were plated on cover glass and, the following day, were fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were then rinsed with PBS three times and incubated with a blocking solution containing 5% bovine serum albumin in PBS for 15 min. The cells were next incubated with indicated antibodies in blocking solution for 3.5 hours, followed by four washes with PBS. Secondary antibodies conjugated to a fluorochrome (Alexa Fluor, Thermo Fisher Scientific) diluted in blocking buffer were then added to the cover glasses and incubated for another 1.5 hours at room temperature. Cells were rinsed with PBS four times and incubated with Hoechst 33258 solution (DNA staining) for 15 min. After washing with PBS four times, cells were then mounted with mounting buffer, and images were observed by fluorescence microscopy.
Animal studies
For our mouse xenograft studies, we followed the Institutional Animal Care and Use Committee–approved protocols and guidelines. Indicated cell lines (1 × 106 cells) were injected subcutaneously into 5- to 6-week-old female nude mice (Envigo or Taconic). After subcutaneous tumors were formed, mice were randomly divided into two to four groups for intraperitoneal injection 3 days/week with vehicle, SHIN1 (100 mg/kg) (29), CHIR99021 (10 mg/kg), or SHIN1 plus CHIR99021, and tumors were allowed to grow for an additional 3 to 4 weeks.
Establishment of stable cell lines
To generate lentiviruses, shRNA plasmids or overexpression plasmids were transfected into 293T cells together with the packaging and envelope plasmids, and the medium was changed the next day. After 24 hours, viral supernatants were harvested. Cells were infected with viral supernatants in the presence of a serum-containing medium supplemented with polybrene (4 μg/ml). After 16 hours, viral-containing medium was removed, and cells were grown in serum-containing medium for 24 hours. Cells were treated with puromycin (2 μg/ml) or blasticidin (10 μM) for selection. The knockdown or overexpression of target protein was confirmed by immunoblot analysis.
Statistics
Data were expressed as average ± SEM of at least three independent experiments performed in triplicate. One-way analysis of variance (ANOVA) or two-tailed Student’s t test was used to determine differences between each group, followed by Dunnett’s or Tukey’s posttest or pairwise comparisons as appropriate.
Acknowledgments
We thank M. Nagiec, J. Park, and B. Parang for reading the manuscript and providing valuable feedback. We also thank all other members of the Blenis’ laboratory for critical discussions and technical assistance.
Funding: The work was supported by NIH grant RO1 GM051405 to J.B.
Author contributions: L.H. and J.B. designed the study. L.H. conducted all the experiments unless otherwise indicated. S.C., Z.L., and Y.Z. helped with data analysis and discussion. J.M.A. performed metabolomics analysis. L.H., J.E., and J.B. wrote the manuscript. All authors read and commented on the manuscript appropriately.
Competing interests: The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.Embi N., Rylatt D. B., Cohen P., Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur. J. Biochem. 107, 519–527 (1980). [PubMed] [Google Scholar]
- 2.Beurel E., Grieco S. F., Jope R. S., Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad F., Woodgett J. R., Emerging roles of GSK-3α in pathophysiology: Emphasis on cardio-metabolic disorders. Biochim. Biophys. Acta. Mol. Cell Res. 1867, 118616 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Doble B. W., Woodgett J. R., GSK-3: Tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning B. D., Toker A., AKT/PKB signaling: Navigating the network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anjum R., Blenis J., The RSK family of kinases: Emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9, 747–758 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Bautista S. J., Boras I., Vissa A., Mecica N., Yip C. M., Kim P. K., Antonescu C. N., mTOR complex 1 controls the nuclear localization and function of glycogen synthase kinase 3β. J. Biol. Chem. 293, 14723–14739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L., Fei D. L., Nagiec M. J., Mutvei A. P., Lamprakis A., Kim B. Y., Blenis J., Regulation of GSK3 cellular location by FRAT modulates mTORC1-dependent cell growth and sensitivity to rapamycin. Proc. Natl. Acad. Sci. U.S.A. 116, 19523–19529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L., Gomes A. P., Wang X., Yoon S. O., Lee G., Nagiec M. J., Cho S., Chavez A., Islam T., Yu Y., Asara J. M., Kim B. Y., Blenis J., mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol. Cell 70, 949–960.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducker G. S., Rabinowitz J. D., One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehrmohamadi M., Liu X., Shestov A. A., Locasale J. W., Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 9, 1507–1519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee G. Y., Haverty P. M., Li L., Kljavin N. M., Bourgon R., Lee J., Stern H., Modrusan Z., Seshagiri S., Zhang Z., Davis D., Stokoe D., Settleman J., de Sauvage F. J., Neve R. M., Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 74, 3114–3126 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Nilsson R., Jain M., Madhusudhan N., Sheppard N. G., Strittmatter L., Kampf C., Huang J., Asplund A., Mootha V. K., Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5, 3128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pikman Y., Puissant A., Alexe G., Furman A., Chen L. M., Frumm S. M., Ross L., Fenouille N., Bassil C. F., Lewis C. A., Ramos A., Gould J., Stone R. M., DeAngelo D. J., Galinsky I., Clish C. B., Kung A. L., Hemann M. T., Vander Heiden M. G., Banerji V., Stegmaier K., Targeting MTHFD2 in acute myeloid leukemia. J. Exp. Med. 213, 1285–1306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo C. C., Chen W. C., Teo X. Q., Radda G. K., Lee P. T., Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 7, 53005–53017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franca-Koh J., Yeo M., Fraser E., Young N., Dale T. C., The regulation of glycogen synthase kinase-3 nuclear export by Frat/GBP. J. Biol. Chem. 277, 43844–43848 (2002). [DOI] [PubMed] [Google Scholar]
- 17.van Amerongen R., Nawijn M. C., Lambooij J.-P., Proost N., Jonkers J., Berns A., Frat oncoproteins act at the crossroad of canonical and noncanonical Wnt-signaling pathways. Oncogene 29, 93–104 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Jellusova J., Cato M. H., Apgar J. R., Ramezani-Rad P., Leung C. R., Chen C., Richardson A. D., Conner E. M., Benschop R. J., Woodgett J. R., Rickert R. C., Gsk3 is a metabolic checkpoint regulator in B cells. Nat. Immunol. 18, 303–312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin S. A., Souder D. C., Miller K. N., Clark J. P., Sagar A. K., Eliceiri K. W., Puglielli L., Beasley T. M., Anderson R. M., GSK3β regulates brain energy metabolism. Cell Rep. 23, 1922–1931.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenzweig A., Blenis J., Gomes A. P., Beyond the Warburg effect: How do cancer cells regulate one-carbon metabolism? Front. Cell Dev. Biol. 6, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Sahra I., Hoxhaj G., Ricoult S. J., Asara J. M., Manning B. D., mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeNicola G. M., Chen P. H., Mullarky E., Sudderth J. A., Hu Z., Wu D., Tang H., Xie Y., Asara J. M., Huffman K. E., Wistuba I. I., Minna J. D., DeBerardinis R. J., Cantley L. C., NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Song L., Wan Q., Wu G., Li X., Wang Y., Wang J., Liu Z., Zhong X., He X., Shen S., Pan X., Li A., Wang Y., Gao P., Tang H., Zhang H., cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 25, 429–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye J., Fan J., Venneti S., Wan Y. W., Pawel B. R., Zhang J., Finley L. W. S., Lu C., Lindsten T., Cross J. R., Qing G., Liu Z., Simon M. C., Rabinowitz J. D., Thompson C. B., Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4, 1406–1417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullarky E., Lucki N. C., Beheshti Zavareh R., Anglin J. L., Gomes A. P., Nicolay B. N., Wong J. C. Y., Christen S., Takahashi H., Singh P. K., Blenis J., Warren J. D., Fendt S. M., Asara J. M., DeNicola G. M., Lyssiotis C. A., Lairson L. L., Cantley L. C., Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc. Natl. Acad. Sci. U.S.A. 113, 1778–1783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pacold M. E., Brimacombe K. R., Chan S. H., Rohde J. M., Lewis C. A., Swier L. J. Y. M., Possemato R., Chen W. W., Sullivan L. B., Fiske B. P., Cho S., Freinkman E., Birsoy K., Abu-Remaileh M., Shaul Y. D., Liu C. M., Zhou M., Koh M. J., Chung H., Davidson S. M., Luengo A., Wang A. Q., Xu X., Yasgar A., Liu L., Rai G., Westover K. D., Vander Heiden M. G., Shen M., Gray N. S., Boxer M. B., Sabatini D. M., A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12, 452–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzieri D. A., Cooley S., Odenike O., Moonan L., Chow K. H., Jackson K., Wang X., Brail L., Borthakur G., An open-label phase 2 study of glycogen synthase kinase-3 inhibitor LY2090314 in patients with acute leukemia. Leuk. Lymphoma 57, 1800–1806 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Feoktistova M., Geserick P., Leverkus M., Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, pdb.prot087379 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Pikman Y., O.-Martinez N., Alexe G., Dimitrov B., Kitara S., Diehl F. F., Robichaud A. L., Conway A. S., Ross L., Su A., Ling F., Qi J., Roti G., Lewis C. A., Puissant A., Heiden M. G. V., Stegmaier K., Targeting serine hydroxymethyltransferases 1 and 2 for T-cell acute lymphoblastic leukemia therapy. Leukemia 36, 348–360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1