Abstract
Anti-HER2 therapies have markedly improved prognosis of HER2-positive breast cancer. However, different mechanisms play a role in treatment resistance. Here, we identified AXL overexpression as an essential mechanism of trastuzumab resistance. AXL orchestrates epithelial-to-mesenchymal transition and heterodimerizes with HER2, leading to activation of PI3K/AKT and MAPK pathways in a ligand-independent manner. Genetic depletion and pharmacological inhibition of AXL restored trastuzumab response in vitro and in vivo. AXL inhibitor plus trastuzumab achieved complete regression in trastuzumab-resistant patient-derived xenograft models. Moreover, AXL expression in HER2-positive primary tumors was able to predict prognosis. Data from the PAMELA trial showed a change in AXL expression during neoadjuvant dual HER2 blockade, supporting its role in resistance. Therefore, our study highlights the importance of targeting AXL in combination with anti-HER2 drugs across HER2-amplified breast cancer patients with high AXL expression. Furthermore, it unveils the potential value of AXL as a druggable prognostic biomarker in HER2-positive breast cancer.
AXL is a prognostic biomarker and a potential therapeutic target to restore trastuzumab response in HER2+ breast cancer.
INTRODUCTION
HER2 is a member of the human epidermal growth factor transmembrane receptor family and is overexpressed in about 20% of breast cancer (BC) (1). The HER2-positive (HER2+) BC subtype correlates with an aggressive phenotype and poor prognosis (2). Several anti-HER2 drugs have been developed and are currently being used following positive clinical outcomes in both localized and metastatic HER2+ BC (3). HER2 is thus a well-established therapeutic target, and anti-HER2 treatment has become the standard of care for patients with HER2-amplified BC. Despite a large fraction of HER2+ BC patients benefiting from anti-HER2 therapy, a subset of these patients develop resistance and ultimately experience disease progression (4). Hence, furthering our understanding and defining the molecular mechanisms involved in treatment response and the metastatic process may reveal novel personalized and more efficient therapeutic strategies.
Metastasis is a complex multistep process that involves intratumoral cell invasion followed by circulatory spread and is completed once cancer cells invade and survive in distant tissues to grow as macrometastatic lesions in permissive niches (5). Epithelial-to-mesenchymal transition (EMT) plays an important role in this malignant progression, allowing cells to metastasize and promoting acquired resistance to several treatment strategies (6, 7).
AXL is a member of the TAM (TYRO3, AXL, MER) receptor tyrosine kinase family that has been linked to EMT. AXL signaling promotes cell survival, resistance to anoikis, drug resistance, invasion, and metastasis across different cancers (8). AXL is activated through multiple mechanisms, including binding to its ligand growth arrest–specific protein 6 (GAS6) precursor and extracellular domain–mediated dimerization or cross-talk with other transmembrane receptor tyrosine kinase (9). Furthermore, AXL up-regulation causes both targeted agents and chemotherapy resistance in several types of cancer (10–19). In BC, AXL expression correlates with metastasis and worse survival (20–22). In addition, AXL inhibition has been proposed as a therapeutic strategy in triple-negative BC (23–25) and other cancers (26). However, there is still a lack of clinical information on the role of AXL in HER2+ BC.
In this study, we demonstrate that AXL high expression independently predicts poor prognosis in HER2+ BC patients. We present our findings, which delineate in a systematic fashion how AXL activation modulates trastuzumab response in in vitro and in vivo patient-derived xenograft (PDX) models. Concordant with this observation, neoadjuvant treatment with lapatinib and trastuzumab led to an increase in AXL mRNA levels at week 2 in HER2+ BC patients from the PAMELA trial. The rapid increase in AXL expression suggests its potential role in acquired resistance to anti-HER2 treatment in BC patients.
We propose AXL overexpression as a new mechanism of resistance to anti-HER2 therapy and as a new potential druggable prognostic biomarker. AXL inhibition plus anti-HER2 could be a new therapeutic approach in HER2+ BC.
RESULTS
Characterization of trastuzumab-resistant HER2+ BC cell lines
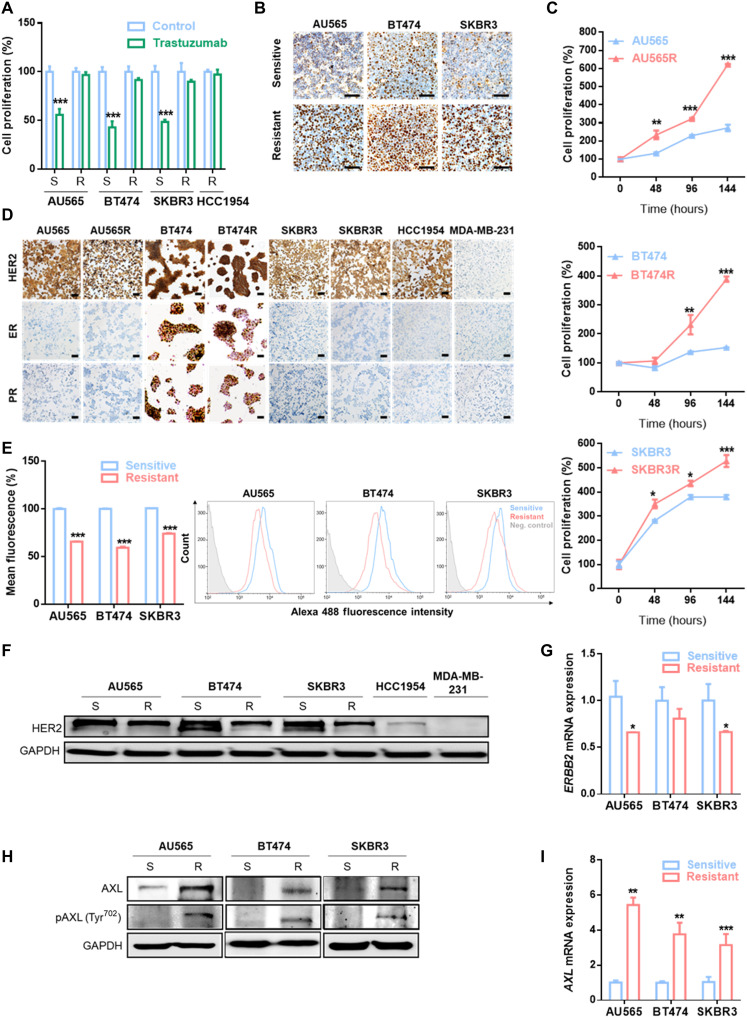
To identify novel mechanisms of trastuzumab resistance, we used three in vitro BC cell lines with acquired resistance to trastuzumab: AU565R, BT474R, and SKBR3R. In resistant models, trastuzumab treatment decreased the growth rate by less than 20%, in contrast with the approximately 50% reduction observed in parental cell lines (Fig. 1A). Given that standard of care for HER2+ BC consists of dual HER2 blockade with trastuzumab plus pertuzumab, we checked response to pertuzumab and combination of both agents in trastuzumab-resistant cell lines to give clinical value to our study. Results showed a significantly lower response of AU565R, BT474R, and SKBR3R to these treatments in comparison to parental cells (fig. S1A). In addition, the three acquired trastuzumab-resistant cell lines were significantly more proliferative at basal level than parental cell lines, as demonstrated by Ki67+ staining and cell proliferation (Fig. 1, B and C).
Fig. 1. Trastuzumab-resistant HER2+ BC cell line characterization.
(A) Cell proliferation analysis by WST-1 in trastuzumab-sensitive and trastuzumab-resistant cell lines with and without trastuzumab treatment at 15 μg/ml for 7 days. (B) IHC staining of Ki67 in the indicated cell lines. (C) Comparison of basal cell proliferation by WST-1 in trastuzumab-sensitive and trastuzumab-resistant cell lines over 144 hours. (D) IHC staining ER, PR, and HER2 in the indicated cell lines. MDA-MB-231 was used as a negative control for HER2, ER, and PR staining. Flow cytometry analysis of HER2 expression in cell membrane (E), Western blot analysis for total HER2 expression (F) and RT-qPCR analysis of ERBB2 mRNA expression (G) in trastuzumab-sensitive and trastuzumab-resistant cell lines. Western blot analysis of AXL and phospho-AXL (H) and RT-qPCR analysis of AXL mRNA expression (I) and in trastuzumab-sensitive and trastuzumab-resistant cell lines. Scale bars, 100 μm; magnifications, ×20 (B) and ×10 (D). GAPDH was used as an endogenous control in (F) to (I). Flow cytometry negative control was stained with isotype primary antibody in (E). *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed Student’s t test in (A), (C), (E), (G), and (I).
Immunohistochemistry (IHC) analysis revealed no substantial changes in pathological biomarkers between resistant and parental cell lines. BT474 and BT474R cell lines express HER2, estrogen receptor (ER), and progesterone receptor (PR). In contrast, AU565, AU565R, SKBR3, SKBR3R, and an innate trastuzumab-resistant cell line (HCC1954) were HER2+, while ER and PR were not expressed (Fig. 1D). All cell lines were HER2 amplified, yet trastuzumab-resistant cell lines expressed lower HER2 total expression and cell membrane expression (Fig. 1, E and F). Notably, ERBB2 mRNA remains lower in our acquired resistant cell lines compared to the parental sensitive cell lines (Fig. 1G). The above data suggest that our resistant models have an increased proliferation rate and lower ERBB2 expression despite maintaining HER2 amplification and the same molecular subtype.
AXL promotes trastuzumab resistance in HER2+ BC cell lines
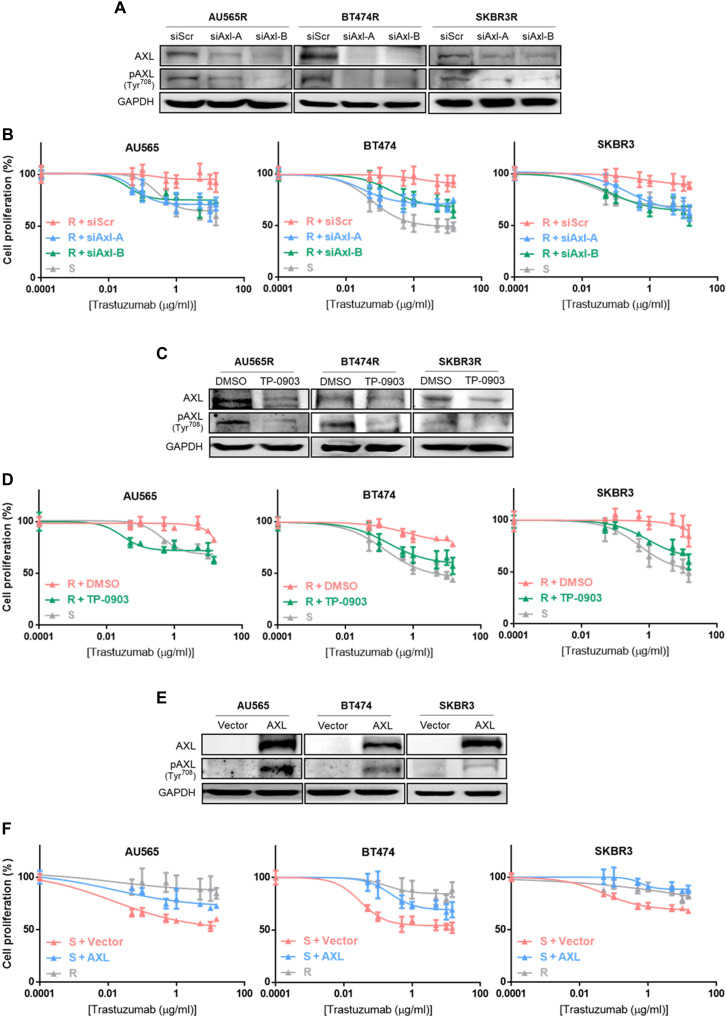
Given that AXL has been previously associated with resistance to therapies (27), we compared AXL expression between trastuzumab-sensitive and trastuzumab-resistant cell lines, finding that both AXL mRNA and protein expression were up-regulated in the three resistant cell lines compared to their respective parental cell lines. In addition, AXL phosphorylation was also higher in resistant cell lines (Fig. 1, H and I). These data suggest that AXL expression could mediate trastuzumab resistance in our models. To address this question, AXL was inhibited using both a genetic and a pharmacological approach. First, we depleted AXL expression using two independent small interfering RNAs (siRNAs): In both cases, AXL down-regulation restored the sensitivity of resistant cells to trastuzumab, and we observed over 20% growth inhibition in AXL-depleted acquired resistant cell lines after trastuzumab treatment (Fig. 2A and fig. S2A). Trastuzumab IC50 (half-maximal inhibitory concentration) values in the three acquired resistant cell lines with AXL knockdown were similar to IC50 of their parental cell lines (Fig. 2B and table S1). However, AXL down-regulation in sensitive cell lines and in the innate resistant cell line HCC1954 did not enhance trastuzumab growth rate inhibition, underscoring its role as mechanism of acquired resistance (fig. S2, A and B).
Fig. 2. AXL has a role in trastuzumab response in HER2+ BC cell lines.
(A) Western blot analysis of AXL and phospho-AXL protein expression after 72 hours of siRNA-mediated AXL knockdown. (B) Cell proliferation analysis by WST-1 in sensitive cell lines and siRNA-mediated AXL knockdown acquired resistant cell lines treated with trastuzumab for 7 days. (C) Western blot analysis of AXL and phospho-AXL protein expression after 72 hours of TP-0903 treatment. (D) Cell proliferation analysis by WST-1 in sensitive cell lines and TP-0903–treated resistant cell lines with trastuzumab for 7 days. (E) Western blot analysis of AXL and phospho-AXL protein expression after 72 hours of AXL overexpression. (F) Cell proliferation analysis by WST-1 in resistant cell lines and AXL-overexpressed sensitive cell lines with trastuzumab treatment for 7 days. GAPDH was used as an endogenous control in (A), (C), and (E).
These results were confirmed using a selective AXL tyrosine kinase inhibitor (TP-0903) (Fig. 2C). Reduced proliferation caused by the combination of trastuzumab with TP-0903 in acquired resistant cell lines reached growth inhibition levels observed in parental cell lines with trastuzumab (fig. S2C). Subsequently, TP-0903 combined with increasing trastuzumab concentrations led to trastuzumab IC50 values in acquired resistant cell lines similar to those in sensitive cells (Fig. 2D and table S1). However, growth inhibition and IC50 values for trastuzumab were not modified in parental cell lines or HCC1954 upon addition of AXL inhibitor (fig. S2, C and D).
Conversely, AXL gain of function in parental cell lines significantly reduced growth rate inhibition after trastuzumab treatment, thus leading to trastuzumab resistance (Fig. 2E and fig. S2E). Moreover, treatment with a range of trastuzumab concentrations showed an increased IC50 value in AXL-overexpressed cell lines compared to controls (AU565, P = 0.0012; BT474, P < 0.0001; SKBR3, P = 0.0001) (Fig. 2F and table S1). On the other hand, AXL overexpression had no effect on trastuzumab response in resistant cell lines (fig. S2, E and F). Proliferation was not affected by AXL levels (fig. S3). Collectively, these results suggest that AXL promotes acquired resistance to trastuzumab, and AXL inhibition restores sensitivity in in vitro acquired trastuzumab-resistant models.
AXL-mediated trastuzumab resistance is associated with EMT-like phenotype
Acquisition of mesenchymal characteristics by epithelial cells, in particular, the ability to migrate and invade the extracellular matrix, is a well-defined mechanism of drug resistance (28). Given that the mesenchymal phenotype is related to worse response to trastuzumab and that AXL has been described as a component of the acquired mesenchymal phenotype (29), we next analyzed the association between AXL expression and EMT markers.
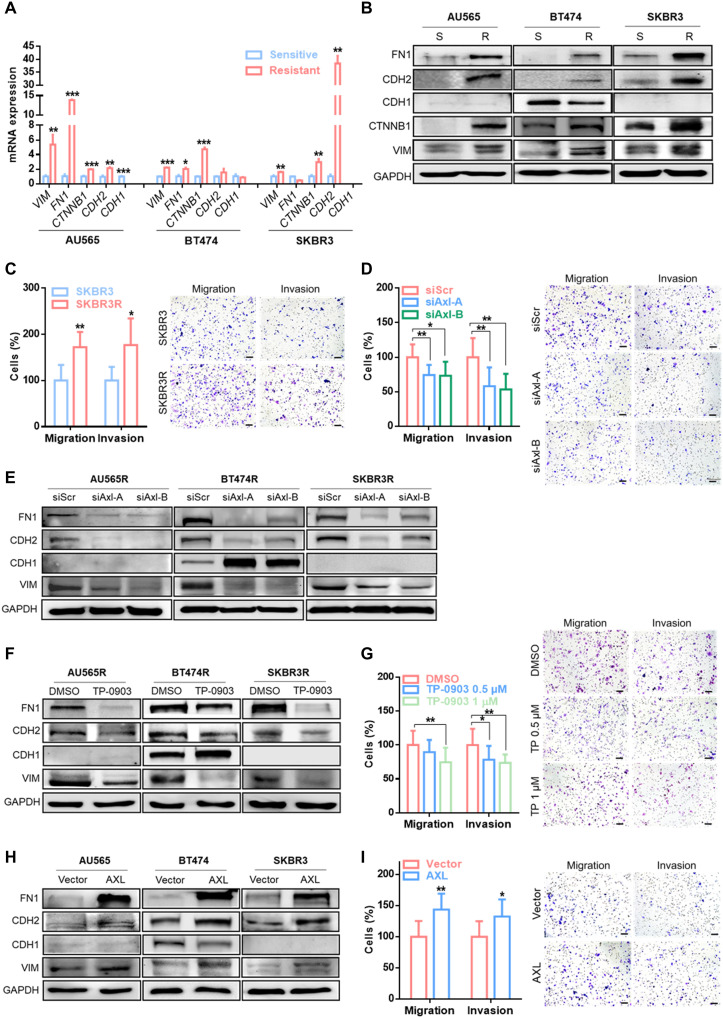
First, we observed that acquired trastuzumab-resistant cell lines, which overexpressed AXL, also showed an EMT-like phenotype by up-regulation of vimentin (VIM), fibronectin (FN1), β-catenin (CTNNB1), and N-cadherin (CDH2) and a decrease in E-cadherin (CDH1) at mRNA and protein levels compared with their respective parental cell lines (Fig. 3, A and B). As expected, the SKBR3R cell line presented an enhanced cancer cell invasion and migration capability through AXL signaling (Fig. 3C and fig. S4A).
Fig. 3. AXL expression is associated with EMT-like phenotype.
RT-qPCR analysis of mRNA expression (A) and Western blot analysis of protein expression (B) of EMT markers: VIM, FN1, CTNNB1, CDH2, and CDH1 in sensitive versus trastuzumab-resistant cell lines. Transwell migration and invasion assay in SKBR3 versus SKBR3R (C), siRNA-mediated AXL knockdown (D), and pharmacological AXL inhibition by TP-0903 (G) in the SKBR3R cell line. Western blot analysis of EMT markers in siRNA-mediated AXL knockdown (E) and pharmacological AXL inhibition by TP-0903 (F) in SKBR3R and in AXL-overexpressed SKBR3 (H). (I) Transwell migration and invasion assay in AXL-overexpressed SKBR3. GAPDH was used as an endogenous control in (A), (B), (E), (F), and (H). Images show representative migrating/invading cells, and bars show percentage of the mean (±SD, n = 8) of migrating/invading cells. Scale bar, 100 μm; magnification, ×10 (C, D, G, and I). *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed Student’s t test in (A), (C), (D), (G), and (I).
Second, we determined whether AXL expression was sufficient to modulate a mesenchymal-like phenotype. The results showed that AXL inhibition by two different siRNAs or by TP-0903 was sufficient to reverse the mesenchymal-like phenotype (Fig. 3, E and F, and fig. S4B). AXL down-regulation by siRNA and inhibition by TP-0903 reduced cell migration and invasion in the SKBR3R cell line (Fig. 3, D and G, and fig. S4, C and D).
Consistent with the above results, AXL overexpression increased both EMT markers expression (Fig. 3H) and migration and invasion capability in the SKBR3 cell line (Fig. 3I and fig. S4E). Moreover, modulation of migration and invasion ability by AXL was also validated in BT474 and BT474R cells (fig. S5). These results suggest that AXL orchestrates a mesenchymal-like phenotype in acquired resistance to trastuzumab in HER2+ BC cell lines.
Acquired trastuzumab resistance is mediated by ligand-independent heterodimerization of AXL and HER2
Several studies have demonstrated that both ligand-dependent and ligand-independent AXL activation initiates downstream signaling in several cancer types (30, 31). We undertook to determine the biological mechanism underlying the resistance to trastuzumab mediated by AXL overexpression. To do so, we evaluated the role of its single ligand GAS6 without detecting consistent overexpression of GAS6 mRNA or protein in acquired resistant versus parental cell lines (fig. S6, A and B). Neither GAS6 protein expression nor the percentage of GAS6-positive cells changed after treatment with protein transport inhibitor, thus confirming that GAS6 ligand production did not vary between resistant and sensitive cell lines (fig. S6, B to E).
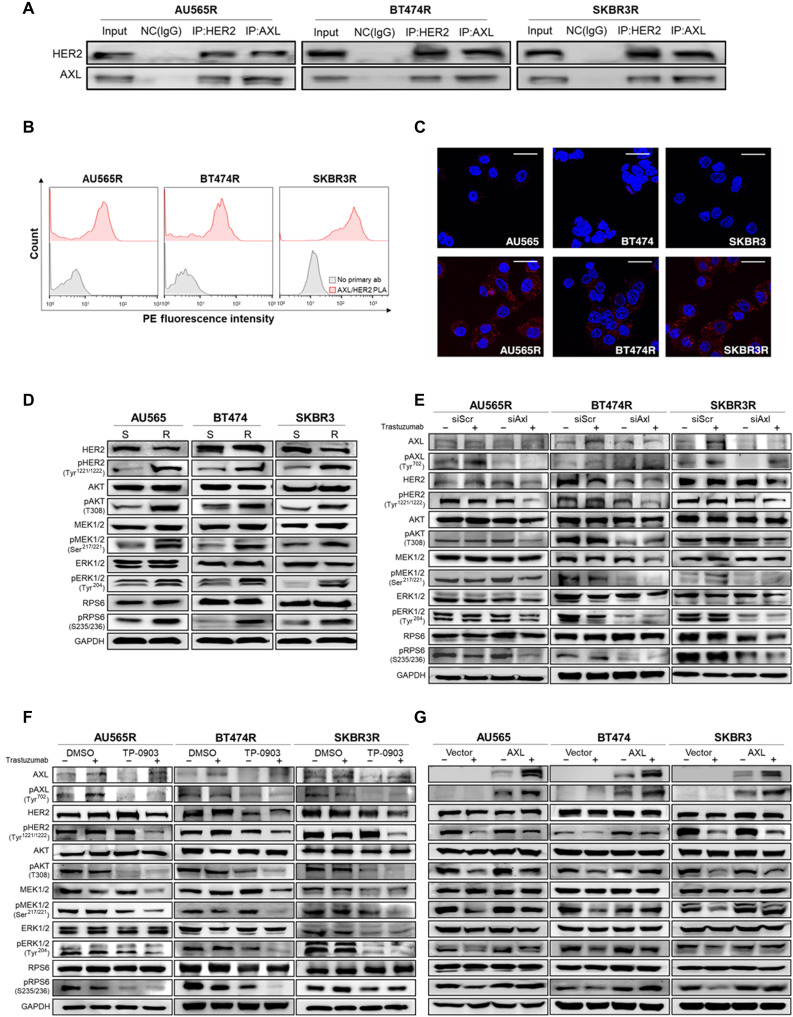
On the basis of these data, we hypothesized that AXL activation is ligand independent and proposed the heterodimerization between AXL and HER2 as the trigger in our acquired trastuzumab-resistant models. To test this hypothesis, we co-immunoprecipitated AXL and HER2 demonstrating a direct interaction between these two proteins (Fig. 4A). In agreement with this result, AXL-HER2 interaction was also detectable by proximity ligation assay (PLA). Flow cytometry and immunofluorescence confirmed the presence in the plasma membrane of AXL-HER2 heterodimer in our acquired trastuzumab-resistant models (Fig. 4, B and C, and fig. S7A). These results suggest a ligand-independent interaction between AXL and HER2.
Fig. 4. AXL-HER2 heterodimer regulates PI3K/AKT and MAPK/ERK pathways.
(A) Co-immunoprecipitation (IP) of AXL, HER2, and IgG as a negative control (NC) followed by immunoblotting of AXL and HER2 in acquired trastuzumab-resistant cell lines. (B) PLA of AXL and HER2 (red) in acquired resistant cell lines by flow cytometry. Negative control: primary antibodies were omitted (gray). (C) PLA of AXL and HER2 in trastuzumab-sensitive and acquired resistant cell lines by confocal microscopy. Red spots indicate AXL-HER2 interaction. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (D) Western blot analysis of AXL-HER2 downstream proteins in trastuzumab-sensitive versus trastuzumab-resistant cell lines at basal level. Western blot analysis of AXL-HER2 downstream proteins with and without trastuzumab for 72 hours in siRNA-mediated AXL knockdown (E) after pharmacological AXL inhibition by TP-0903 (F) in acquired trastuzumab-resistant cell lines and in AXL-overexpressed sensitive cell lines (G). GAPDH was used as an endogenous control in (D) to (G). Scale bar, 25 μm; magnification, ×40 (C).
AXL-HER2 dimerization induces phosphorylation of PI3K/AKT and MAPK/ERK pathways
We next studied signaling on HER2-AXL downstream pathways. AXL-HER2 heterodimer downstream adapter proteins, such as AKT, RPS6, extracellular signal–regulated kinase (ERK), and mitogen-activated protein kinase (MAPK), were found to be hyperphosphorylated in trastuzumab-resistant models compared to sensitive parental cell lines (Fig. 4D). Furthermore, genetic knockdown or pharmacological inhibition of AXL plus trastuzumab achieved the greatest inactivation of HER2 downstream pathways (Fig. 4, E and F, and fig. S7, B and C). Although AXL overexpression alone was not sufficient for promoting the phosphorylation of phosphoinositide 3-kinase (PI3K)/AKT and MAPK/ERK, pathway inhibition by trastuzumab was considerably less effective in AXL-overexpressed cell lines than in control cell lines (Fig. 4G). Together, our data indicate that AXL-HER2 dimerization activates PI3K/AKT and MAPK/ERK signaling pathways, thus leading to a reduced trastuzumab treatment efficacy.
AXL inhibition overcomes trastuzumab resistance in PDX models
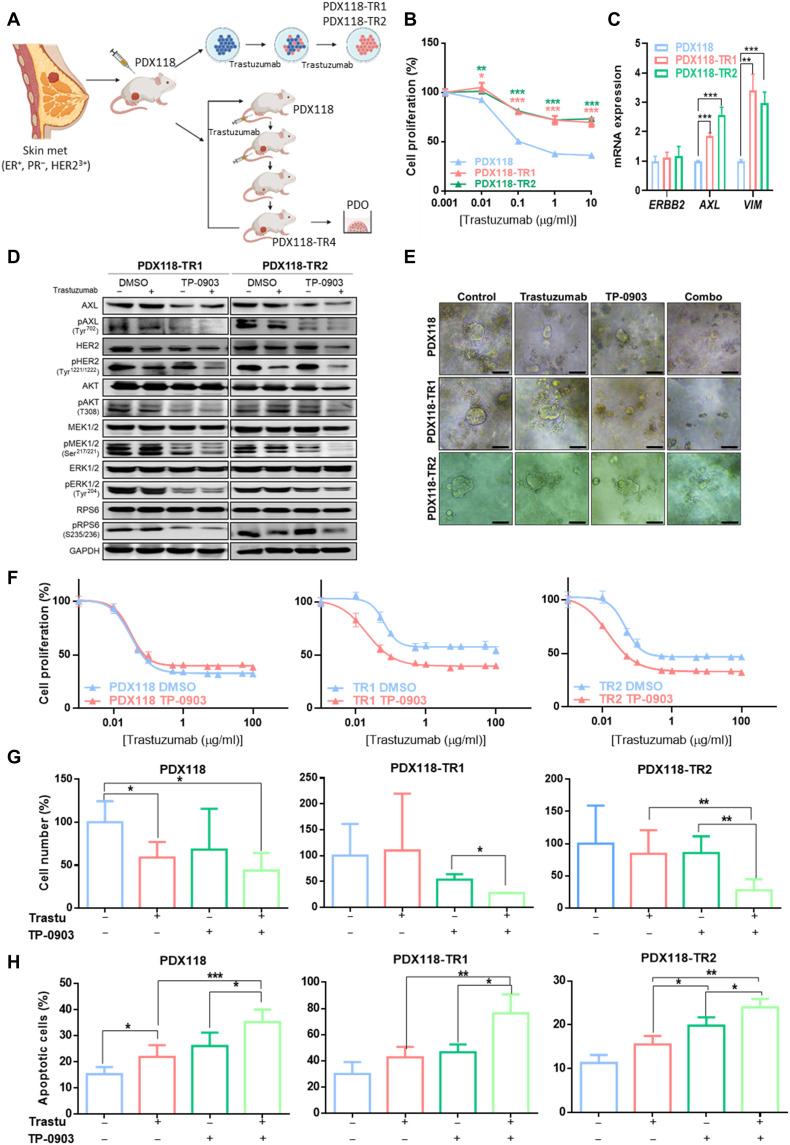
On the basis of the previous results demonstrating that AXL leads to trastuzumab resistance, we tested whether the combination of trastuzumab and AXL inhibition could be used to blunt tumor growth in different in vitro and in vivo acquired trastuzumab-resistant PDX models from a HER2+ BC patient to obtain more translational results. First, two in vitro PDX-derived resistant models were established by chronic treatment for 6 months with trastuzumab (Fig. 5, A and B, and table S1). The two obtained PDX-derived trastuzumab-resistant cell lines (PDX118-TR1 and PDX118-TR2) also presented decreased response to pertuzumab and dual HER2 blockade (fig. S1, B and C) and maintained HER2 overexpression but exhibited up-regulation of AXL and VIM in comparison to the parental cells (PDX118) (Fig. 5C). Co-immunoprecipitation assay confirmed AXL-HER2 dimerization in these PDX-derived trastuzumab-resistant models (fig. S8A). These results reproduced those previously obtained in our panel of acquired trastuzumab-resistant cell lines. In addition, the in vitro combination of TP-0903 plus trastuzumab resensitized resistant cells by significantly decreasing trastuzumab IC50 (PDX118-TR1 and PDX118-TR2 P < 0.0001) and inhibiting phosphorylation of PI3K/AKT and MAPK/ERK pathways. Furthermore, growth inhibition caused by trastuzumab in PDX118 was independent of AXL blockade (Fig. 5, D and F, and table S1). These results were next validated in three-dimensional (3D) in vitro models from PDX118, TR1, and TR2 cell lines. In resistant organoids, trastuzumab or TP-0903 alone did not induce significant growth inhibition, while the combination achieved a decrease to 27% in both models compared to control. In PDX118 organoids, trastuzumab significantly decreased cell numbers with no greater effect of combination with TP-0903 (Fig. 5, E and G). Besides, apoptosis assay revealed a significant increase in the percentage of apoptotic cells achieved by the combination of trastuzumab plus TP-0903 compared to any single agent in both parental and resistant organoids (Fig. 5H).
Fig. 5. AXL inhibition overcomes trastuzumab resistance in PDX-derived HER2+ BC cell lines.
(A) Schematic representation of in vitro trastuzumab–resistant PDX cell lines (PDX118-TR1 and TR2) and in vivo trastuzumab–resistant PDX model (PDX118-TR4) generation. (B) Cell proliferation assay of PDX-derived cell lines treated with trastuzumab for 6 days. Cell numbers were estimated with the crystal violet staining assay. (C) RT-qPCR analysis of ERBB2, AXL, and VIM mRNA expression of PDX-derived cell lines. (D) Western blot analysis of HER2 downstream pathways with and without trastuzumab and TP-0903 treatment for 72 hours in acquired trastuzumab-resistant PDX-derived cell lines. (E) Representative images of organoids treated with trastuzumab, TP-0903, and combination. (F) Cell proliferation analysis of PDX-derived cell lines treated with trastuzumab combined with pharmacological AXL inhibition by TP-0903. Cell number (G) and apoptosis assay (H) of PDX118, TR1, and TR2 organoids treated with trastuzumab (100 μg/ml), TP-0903 (1 μM in PDX118 and TR1, 0.15 μM in TR2), or combination. Viable and apoptotic cells were quantified by flow cytometry using propidium iodide and annexin V staining. GAPDH was used as an endogenous control in (C) and (D). Scale bar, 100 μm; magnification, ×4 (E). *P < 0.05, **P < 0.01, ***P < 0.001, by two-tailed Student’s t test in (B), (C), (G), and (H).
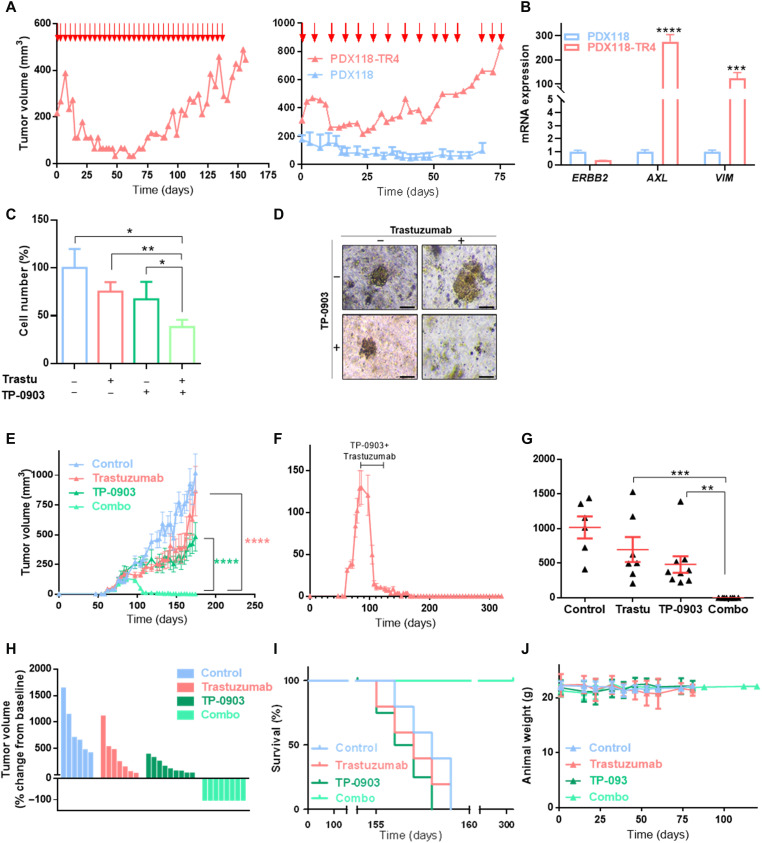
Second, an in vivo PDX-resistant model (PDX118-TR4) was established by chronic treatment with trastuzumab for 4 months and two serial passages in mice (Figs. 5A and 6A). The level of HER2 amplification was similar in parental PDX118 and PDX118-TR4, but ERBB2 expression was down-regulated (32, 33). Consistent with the phenotype observed in vitro, these resistant PDX tumors up-regulated AXL and VIM (Fig. 6B) and presented AXL-HER2 heterodimers (fig. S8B). Notably, GAS6 was undetectable in our PDX models, which supports the ligand-independent mechanism. In 3D organoid models from PDX118-TR4, trastuzumab or TP-0903 alone did not significantly reduced cell numbers, whereas in combination they decreased cell numbers to 38.2% compared to control (Fig. 6, C and D).
Fig. 6. AXL inhibition overcomes trastuzumab resistance in HER2+ BC PDX models.
(A) Generation of an in vivo trastuzumab–resistant model (PDX118-TR4). Parental PDX118 was implanted in an immunocompromised mouse, and when tumors reached ~200 mm3, animals were treated intraperitoneally with trastuzumab (left). Next, resistant tumors were reimplanted in immunocompromised mice and treated with trastuzumab (right). Red arrows represent days of trastuzumab treatment. (B) RT-qPCR analysis of ERBB2, AXL, and VIM mRNA expression of in vivo PDX–resistant model PDX118-TR4. GAPDH was used as an endogenous control. (C) Cell number analysis of trastuzumab-resistant patient-derived organoids treated with trastuzumab at 100 μg/ml, TP-0903 at 150 nM, and a combination of these two drugs. Viable cells were quantified by flow cytometry using EpCAM as a marker. (D) Representative images of organoids treated with trastuzumab, TP-0903, and combination of these two drugs. Tumor growth of PDX118-TR4 in vivo model injected in NOD-SCID mice treated with vehicle, trastuzumab alone (10 mg/kg), TP-0903 alone (50 mg/kg), or a combination of these two drugs represented in time-course line (E), endpoint dot plot (G), and percentage change from baseline of each individual (H). (F) Tumor growth of PDX118-TR4 in vivo model in NOD-SCID mice treated with trastuzumab and TP-0903 combination. Black line represents treatment time period. (I) OS Kaplan-Meier curve of PDX mice divided by treatment. (J) Animals’ weight in grams during treatment and follow-up period. Treatment was performed between days 89 and 110 after injection. N = 30 mammary fat pad tumors; data represent means ± SEM in (E) to (G). Scale bar, 100 μm; magnification, ×4 (D). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by two-tailed Student’s t test in (B) and (C). ****P < 0.0001 by two-way ANOVA with Bonferroni correction posttest in (E). **P < 0.01 and ***P < 0.001 by Mann-Whitney test in (G).
In a PDX118-TR4 in vivo model experiment, mice were treated with vehicle, trastuzumab, TP-0903, or a combination of these two drugs. The results showed that trastuzumab and the AXL inhibitor alone had no significant effect on tumor growth; however, the two drugs in combination abrogated tumor growth and achieved a complete regression after 21 days of treatment (Fig. 6, E to H, and fig. S8, C to E). Notably, animals from the combined treatment group remained alive with no evidence of tumor growth after therapy during a total of 321 days of follow-up (Fig. 6I); likewise, treatment did not affect animal weight, showing that AXL inhibition did not lead to a significant toxic effect in preclinical models (Fig. 6J).
With these results, we conclude that the combination of TP-0903 with trastuzumab completely abrogates tumor growth in acquired trastuzumab-resistant PDX models and achieves complete tumor regression safely. Therefore, this is a promising therapeutic strategy to treat HER2+ BC resistant to standard of care.
AXL expression in HER2+ BC patients correlates with a strong negative prognostic factor
On the basis of these preclinical data, we hypothesized that up-regulation of AXL might promote acquired resistance to trastuzumab treatment in HER2+ BC patients. To test this hypothesis and assess the clinical relevance of our preclinical findings, we analyzed AXL expression in a retrospective cohort of HER2+ BC patients (n = 50) (table S2). The median age at BC diagnosis was 51 years (range, 33 to 73); median follow-up was 7 years. Median disease-free survival (DFS) was 65.5 months (range, 5 to 177), and median overall survival (OS) was 80.5 months (range, 7 to 177).
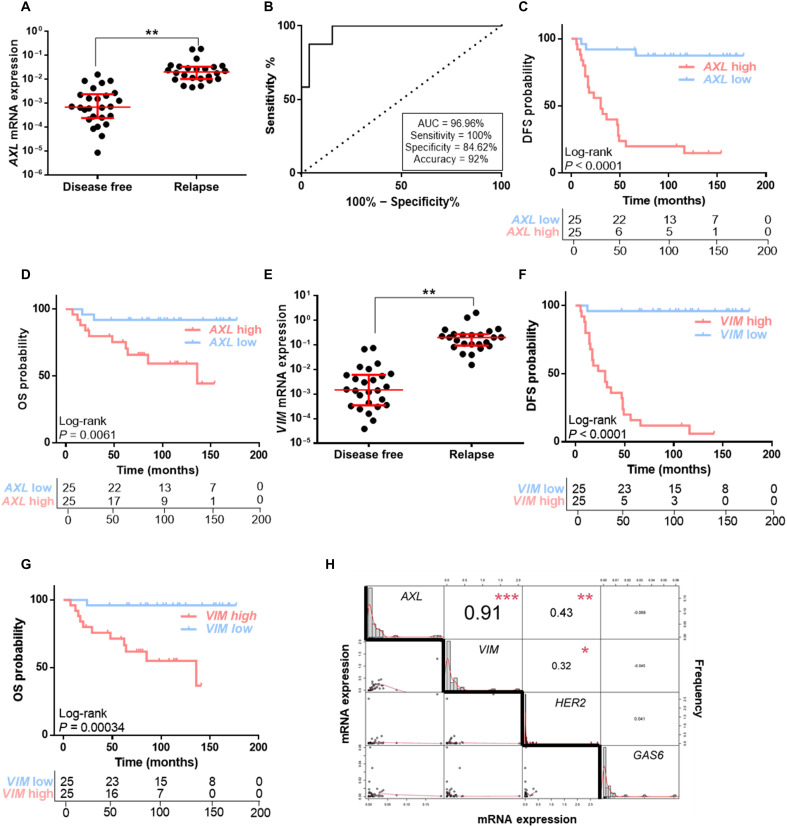
AXL expression was significantly up-regulated at the time of diagnosis in primary tumor tissue from patients who later experienced disease relapse (P = 0.003) (Fig. 7A). Receiver operating curve (ROC) analysis also revealed that AXL expression may discriminate between HER2+ BC patients who will relapse from those who will be free of disease with area under the curve (AUC) of 96.96% [95% confidence interval (CI), 0.9292 to 1.00; P < 0.0001]. Applying the best cutoff value, biomarker performance parameters showed 100% sensitivity, 84.62% specificity, and 92% accuracy (Fig. 7B). We found that patients with high levels of AXL had significantly shorter DFS and OS (for DFS: log-rank P < 0.001; hazard ratio, 12.56; 95% CI, 3.70 to 42.66; for OS: log-rank P = 0.006; hazard ratio, 6.91; 95% CI, 1.50 to 31.78). Median DFS and OS were 30 and 136 months, respectively, in patients with high AXL expression, whereas in patients with low AXL expression these were not reached (Fig. 7, C and D).
Fig. 7. AXL expression correlates with a strong negative prognostic factor in HER2+ BC patients.
(A) RT-qPCR analysis of AXL mRNA expression in HER2+ BC patients’ FFPE samples. (B) ROC curve analysis for relapse prediction based on AXL mRNA expression in HER2+ BC patients. DFS (C) and OS (D) Kaplan-Meier curves of HER2+ BC patients stratified by median AXL expression levels. (E) RT-qPCR analysis of VIM mRNA expression in HER2+ BC patients’ FFPE samples. DFS (F) and OS (G) Kaplan-Meier curves of HER2+ BC patients stratified by median VIM expression levels. (H) Correlation matrix of AXL, VIM, ERRB2, and GAS6 mRNA expression in HER2+ BC patients. N = 50. Red lines represent median and interquartile range. GAPDH was used as an endogenous control in (A) and (E). *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed Student’s t test with Welch’s correction in (A), (E), and (H).
The prognostic value of AXL was also confirmed in silico in an independent public early HER2+ BC patients’ cohort (34). Patients with high AXL levels had significantly shorter DFS (log-rank P = 0.0018; hazard ratio, 2.03; 95% CI, 1.29 to 3.19) and distant metastasis–free survival (log-rank P = 0.0043; hazard ratio, 2.62; 95% CI, 1.32 to 5.19) (fig. S9, A and B).
As AXL expression correlated with VIM in our resistant models, we sought to validate this correlation in our patients’ cohort. We found a strong positive correlation between VIM and AXL expression [correlation (cor) = 0.91, P < 0.001] (Fig. 7H), and VIM expression was associated with worse prognosis. The results revealed that higher VIM expression in primary tumor was related to relapse (P = 0.0033), worse DFS (log-rank P < 0.001; hazard ratio, 50.63; 95% CI, 6.74 to 380.5), and OS (log-rank P < 0.001; hazard ratio, 16.48; 95% CI, 2.11 to 128.80) compared to low VIM HER2+ BC patients (Fig. 7, E to G).
Next, we analyzed GAS6 mRNA expression in our patients’ cohort. No association between GAS6 and AXL expression was found (cor = −0.069, P = 0.647) (Fig. 7H), in accordance with our preclinical data. Furthermore, GAS6 was not a negative prognostic factor; there was no significant relationship between GAS6 levels and DFS or OS (fig. S9, C to F). A similar result was observed with ERBB2, whose expression was not strongly correlated with either AXL (cor = 0.43, P = 0.003) or VIM (cor = 0.32, P = 0.031) (Fig. 7H).
In the multivariable model, AXL was selected the best model for both DFS (hazard ratio, 22.95; 95% CI, 3.07 to 171.87; P = 0.002) and OS (hazard ratio, 15.92; 95% CI, 0.849 to 298.32; P = 0.064) according to Akaike’s information criterion (AIC). Collectively, these findings reveal a ligand-independent association between high AXL expression and worse prognosis in patients with HER2+ BC treated with trastuzumab and taxanes plus anthracycline-based chemotherapy.
Early in vivo biological changes during dual HER2 blockade in BC patients
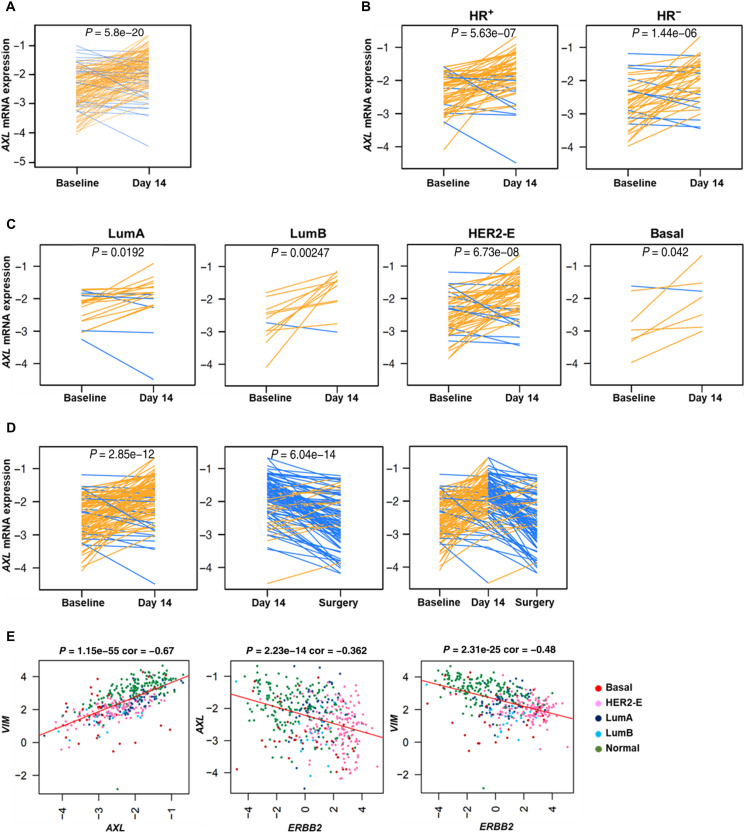
To identify early molecular changes induced by dual HER2 blockade in patients with HER2+ BC, gene expression profiling was performed in 139 tumor samples obtained before treatment and at day 14 with lapatinib and trastuzumab [and endocrine therapy in hormonal receptor–positive (HR+) tumors] in the PAMELA phase 2 clinical trial (35). AXL gene expression was explored at both time points. Of particular interest was that AXL significantly increased at day 14 compared to baseline (P = 5.8 × 10−20) (Fig. 8A). Overall, this effect was observed in HER2+/HR+ (P = 5.63 × 10−7), in HER2+/HR− (P = 1.446 × 10−6), and across all PAM50 subtypes (luminal A, P = 0.0192; luminal B, P = 0.00247; HER2-enriched, P = 6.73 × 10−8; basal-like, P = 0.042) (Fig. 8, B and C). This rapid response suggests that AXL is involved in determining resistance to HER2 inhibition.
Fig. 8. AXL mRNA biological changes after dual HER2 blockade.
AXL mRNA expression changes between baseline and day 14 of treatment in all 139 tumor samples from PAMELA clinical trial (A) and across histological subtypes (B) and PAM50 subtypes (C). (D) AXL mRNA expression changes between baseline, day 14, and surgery in 97 residual tumors at surgery. (E) Correlation between AXL, VIM, and ERBB2 in 96 tumor samples from the PAMELA clinical trial. Each line represents a tumor sample. Increases are represented in orange, and decreases in blue. P values were determined by two-tailed paired t tests.
We completed the analysis of AXL in residual tumors from the surgery in those patients failing to achieve pathological complete response (n = 97). Of note, the median time from the last treatment dose to surgery, during which the tumor samples were obtained, was 29.5 days (range, 7 to 76 days). AXL was found to be differentially expressed in residual tumors at surgery compared to day 14 (P = 6.04 × 10−14) (Fig. 8D). In agreement with previous gene expression analysis in the PAMELA trial (36), we found a rebound effect between day 14 and surgery samples (P = 6.04 × 10−14) potentially due to that discontinuation of dual HER2 inhibition, which reverses its biological effects. According to our previous preclinical data, VIM expression presented similar biological changes than AXL, whereas ERBB2 expression decreased after dual HER2 blockade (fig. S10, A and B).
Other interesting findings were that AXL and VIM expression were strongly correlated (cor = 0.67, P = 1.15 × 10−55), and ERBB2 had a negative correlation with both AXL and VIM (cor = −0.362, P = 2.23 × 10−14; cor = −0.48, P = 2.31 × 10−25, respectively) (Fig. 8E). These observations were particularly significant as the biological changes observed in our preclinical models recapitulated the biological changes observed in patients from a phase 2 clinical trial.
DISCUSSION
Trastuzumab is an essential and effective targeted anticancer agent for HER2+ BC patients. Unfortunately, despite the efficacy of this anti-HER2 treatment, a significant number of patients develop progressive disease (2), requiring additional therapeutic strategies. Several molecular mechanisms such as alternative signaling pathways, activation, and/or down-regulation of ERBB2 could contribute to development of resistance to anti-HER2 agents (37, 38). This scenario highlights an urgent need to develop new therapeutic approaches to effectively treat these patients. The current study was conducted to investigate the potential role of AXL activation in acquired resistance to trastuzumab with the aim of exploring AXL inhibition as a new potential therapeutic strategy for HER2+ BC patients. Several tyrosine kinase inhibitors, monoclonal antibodies, and antibody-drug conjugates against AXL are currently under development in different clinical trials as effective targeted therapy in solid tumors (39).
We demonstrated that acquired trastuzumab-resistant HER2+ BC cells have significantly higher AXL expression than sensitive cells. AXL up-regulation, together with a mesenchymal phenotype, has been associated with acquired resistance to several drugs in cancer, including triple-negative BC (17, 19, 40). However, the role of AXL in the EMT process remains unclear (14). We found that AXL up-regulation occurred in the context of an EMT-associated transcriptional program. The data obtained are consistent with previous studies in which VIM up-regulation was associated with AXL overexpression in BC cells (41). Trastuzumab-resistant cell lines showed increased migration and invasion capability and an EMT-like phenotype, which are related to metastatic behavior. AXL gain and loss of function experiments across our in vitro models corroborated that AXL activity is associated with trastuzumab response and induces a mesenchymal-like phenotype. AXL genetic or therapeutic inhibition is sufficient to reduce migration and invasion capacity, EMT marker expression, and acquired trastuzumab resistance in our models. Together, these results highlight the role of AXL in metastatic cascade in HER2+ BC, in agreement with previous findings by Goyette et al. (20).
Previous studies show evidence that AXL could be activated via both ligand-dependent and ligand-independent mechanisms (42). Recent data suggest that AXL heterodimerization with other transmembrane receptors occurs in several solid tumors (42–44). Our results provide evidence that AXL activation arises through AXL-HER2 heterodimerization followed by the trigger of PI3K and MAPK cascades. This is consistent with previous work showing that AXL can drive oncogenic signaling of these pathways in other cancer types (45). In addition, we demonstrated in our models that AXL activation occurs in a ligand-independent manner, and no correlation was observed between GAS6 and AXL expression in either in vitro or in vivo models and patients’ samples.
We further elucidated the contribution of AXL in acquired resistance to trastuzumab in a PDX model derived from a HER2+ BC patient, and interestingly, AXL overexpression was confirmed in acquired trastuzumab-resistant PDX in vitro and in vivo models. Moreover, we reported that simultaneous therapeutical ablation of AXL and HER2 activity results in complete regression of trastuzumab-resistant HER2+ tumors. The potential effect of TP-0903 has been studied in vivo in other cancers such as leukemia (46, 47) and lung cancer (48). To our knowledge, this is the first report to demonstrate the in vivo efficacy and tolerability of TP-0903 combined with trastuzumab in the treatment of HER2+ BC. All PDX animals treated with this combination remained tumor free for over 211 days after therapy discontinuation. This observation also suggests that the combination of HER2 and AXL inhibition might avoid treatment escape by tumor cells with acquired trastuzumab resistance, leading to a potential long-term benefit for HER2+ BC patients.
In agreement with previous research, our data further current understanding of the role of AXL as a prognostic biomarker in human cancer (21). Analysis of primary tumor HER2+ BC samples showed that high AXL expression at the moment of diagnosis was strongly related with poorer prognosis in terms of DFS and OS. This observation demonstrates a direct association between AXL activation and poor response to anti-HER2 therapies, leading to metastatic disease development. In our patients’ cohort, AXL was also found to be strongly linked to VIM overexpression, which has been described as a poor prognosis biomarker in triple-negative BC (49). However, little is known about its relationship with prognosis in HER2+ BC. Here, we demonstrated that VIM is also related to poor prognosis in HER2+ BC patients.
Another marked observation across tumor sample studies deriving from the PAMELA clinical trial is the increase of AXL expression in residual disease during dual anti-HER2 therapy. This biological acquisition was relatively rapid in the first 2 weeks of treatment, suggesting the importance of AXL overexpression in development of anti-HER2 resistance and the consequent high risk of metastases. This underlines that the selective pressures of therapy could shape cancer evolution, leading to selection of tumor subclones enriched with aberrations causing drug resistance.
The increase in AXL mRNA expression upon trastuzumab resistance observed in our in vitro and in vivo models and also in patients’ samples from the PAMELA trial indicates enhanced transcription activity. On that basis, further investigation exploring potential transcription factors regulating AXL in the context of trastuzumab resistance would be of special interest.
In conclusion, this study demonstrates the substantial biological significance of AXL activation in acquired trastuzumab resistance in HER2+ BC. AXL plays a critical role in EMT and in development of metastatic disease. Our findings demonstrate cross-talk between AXL and HER2 receptors. This heterodimerization induces resistance through PI3K and MAPK pathways activation. The combination of trastuzumab with AXL inhibitor overcomes resistance to trastuzumab both in vitro and in vivo. This dual inhibition induced complete tumor regression in in vivo PDX models without tumor regrowth after 321 days of total follow-up. The strong independent correlation of AXL expression with reduced DFS and OS in HER2+ BC patients suggests that patients with high AXL expression could benefit from AXL-targeted therapy. Moreover, we proved that high AXL expression is not genetically driven (neither amplified nor activating mutations) (50); thus, this key drug target will be missed when using DNA-based tests.
Given the retrospective nature of this study, and to determine the potential heterogeneity of treatment effects associated with AXL functions, our results warrant further validation in a prospective clinical trial. Despite this limitation, our findings provide a strong rationale for developing and testing AXL inhibitors for clinical use in AXL up-regulated HER2+ BC patients to either prevent or overcome resistance to trastuzumab.
MATERIALS AND METHODS
Cell lines and reagents
The BC cell lines BT474, SKBR3, AU565, HCC1954, and MDA-MB-231 were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained as recommended by the suppliers. TP-0903 was purchased from Selleckchem (#S7846, Houston, TX, USA). Trastuzumab was obtained from Roche Pharma (Basel, Switzerland). Trastuzumab-resistant cell lines (AU565R, BT474R, and SKBR3R) were obtained from F.R.’s group at the Fundación Jiménez Díaz Hospital. Cells were established by treating with trastuzumab for 6 months, starting at 10 μg/ml trastuzumab for the first 30 days and increasing the dose at each passage until resistant clones emerged. The final trastuzumab concentration was 15 μg/ml (51). In vitro trastuzumab treatment was maintained at 15 μg/ml. TP-0903 was given at IC50 for each cell line for 72 hours.
In vitro cell proliferation assays
Cell proliferation was measured using WST-1 Assay Reagent Cell Proliferation (#155902, Abcam, Cambridge, UK). A total of 5000 cells per well were seeded in a 96-well plate with at least four technical replicates for each condition. After the treatment, WST-1 was added to each well under sterile conditions (to a final concentration of 7% of the total volume) and the plates were incubated for 4 hours at 37°C. Absorbance was measured at 450 nm in a microplate reader with background correction at 650 nm.
Immunohistochemistry
IHC of cell lines was performed in a μ-slide VI plate (#80826, ibidi GmbH, Martinsried, Germany) for HR and HER2 staining and in Cytospin cytocentrifuge for Ki67 staining. Cells were fixed with methanol/acetone before antibody staining. For expression of HR and HER2, Benchmark XT instrument in a fully automated system (Ventana System, Roche) was used with the UltraView Universal DAB Detection Kit (#0526980600, Roche) and corresponding antibodies (see table S3). For IHC analysis of tumor samples from in vivo experiment, tissue specimens were embedded in paraffin and sectioned at 4-μm thickness. Ki67 staining was performed by automated Ventana System, and for phospho-Axl and cleaved caspase 3 staining, tissue specimens were pretreated in the DAKO PT Link Pre-treatment module (#PT100/PT101, Dako, Glostrup, Denmark) and epitope retrieval was performed with Dako Low Retrieval Solution (citrate buffer, pH 6) for 30 min. After quenching endogenous peroxidase with EnVision FLEX Peroxidase-Blocking Reagent (#SM801, Dako), slides were incubated for 1 hour with primary antibody (table S4) diluted in EnVision FLEX antibody diluent (#K8006, Dako). Slides were incubated with Real EnVision HRP Rabbit (#K5007, Dako) for 30 min following the manufacturer’s protocol. Detection of peroxidase was revealed with 3,3′-diaminobenzidine EnVision FLEX DAB+ Chromogen (#DM827, Dako), and samples were counterstained with Harris hematoxylin and mounted for visualization in Dako Coverstainer.
RNA isolation and RT-qPCR
mRNA expression was measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) on RNA isolated from BC cells and patients’ samples. RNA from eight to ten 10-μm formalin-fixed paraffin-embedded (FFPE) slides was extracted using the High Pure FFPET RNA Isolation Kit (#06650775001, Roche) following the manufacturer’s protocol. TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract RNA from cell lines according to the manufacturer’s protocol. RNA was quantified with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA (1000 ng) was retro-transcribed to complementary DNA (cDNA) using a High-Capacity cDNA Reverse Transcription kit (#34368814, Applied Biosystems, Waltham, MA, USA). Gene expression was quantified with TaqMan Universal Master Mix (#0123456, Applied Biosystems) using TaqMan probes (see table S3). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as an internal reference to normalize input cDNA. The threshold cycle value (CT) was determined for each measurement, and mRNA expression was calculated relative to the control using the comparative critical threshold (2−ΔCT) method, where ΔCT = CTmRNA – CThousekeeping control. Triplicates were performed for each sample.
Protein extraction, Western immunoblotting, and co-immunoprecipitation
Protein expression was measured through Western blot. Cells were lysed with radioimmunoprecipitation assay buffer (#89900, Thermo Fisher Scientific) and sonicated for five cycles of 10 s at 50% pulse and 50 s of pause. Protein quantity was measured with the Pierce BCA Protein Assay Kit (#23227, Thermo Fisher Scientific). For immunoprecipitation, whole-cell lysates were precleared with the appropriate control immunoglobulin G (IgG) together with agarose conjugate (Protein G PLUS-Agarose; sc-2002, Santa Cruz Biotechnology, Dallas, TX, USA) at 4°C for 30 min. After bead pelleting, 500 μg of total cellular protein was incubated rotating head over tail for 2 hours at 4°C with anti-HER2 or anti-AXL antibodies and then reacted overnight with Protein G PLUS-Agarose beads (#2002, Santa Cruz Biotechnology). These beads were then washed three times with lysis buffer, and immunoblotting was performed with anti-HER2 and anti-AXL antibodies. Cell lysates or immunoprecipitates were resolved by SDS–polyacrylamide gel electrophoresis gel, transferred to nitrocellulose membrane, and blocked with 5% bovine serum albumin on tris-buffered saline–Tween. Membranes were incubated with the primary antibody overnight at 4°C and then washed and incubated with secondary antibody. Blots were developed with a chemiluminescence system. GAPDH expression was used as an internal endogenous control. The antibodies used are listed in table S3.
Flow cytometry
For intracellular staining, fixation and permeabilization were performed with the Cytofix/Cytoperm Plus Fixation/Permeabilization Solution Kit (#555028, Becton Dickinson, Franklin Lake, NJ, USA) before staining. For GAS6 staining, cells were previously treated with GolgiStop (#554724, Becton Dickinson). For antibody staining, cells were incubated for 30 min in fluorescence-activated cell sorting (FACS) buffer at the preferred antibody dilution (table S3). Samples were analyzed using a BD LSRFortessa flow cytometer (Becton Dickinson), and data were analyzed by FlowJo V10 (FlowJo LLC).
Annexin V assay
To assay the number of apoptotic cells, cells were stained with allophycocyanin–annexin V (#550475, BD Pharmingen) for 15 min and then washed and resuspended in 1:500 propidium iodide (#81845, Sigma-Aldrich) viability marker and acquired on LSRFortessa. Data were analyzed with FlowJo software. Annexin V+ cells were considered as apoptotic cells.
siRNA and plasmid transfection
Cells were transfected with Lipofectamine 2000 (#11668019 Invitrogen) according to the manufacturer’s instructions. We used pcDNA AXL construct for overexpression donated by R. M. Melillo [#105932, Addgene (52)] and AXL siRNA (#AM16708 Id1313 and Id1218, Invitrogen) or scramble siRNA (#AM4635, Invitrogen). Knockdown and overexpression efficiency was assessed at 72 hours after transfection by RT-qPCR or Western blotting as previously described.
Wound-healing assay
Cells were seeded at 100% confluence on a 24-well plate. For AXL gain or loss of function, cells were seeded 72 hours after transfection, and for pharmacological inhibition of AXL, the drug was added to media during the assay. The wound was induced by scratching the monolayer with a micropipette tip, and cells were allowed to move into the gap. Migration rate was calculated as the proportion of initial scratch distance and the mean distance between the two borders remaining cell free after migration. Triplicates were performed for each condition.
In vitro cell migration and invasion assay
Invasion and migration assay was performed using an 8-μm-pore Boyden chamber (24-well, #353097, Corning, NY, USA). For invasion assay, the upper chamber was coated with Matrigel (#356235, Corning). For pharmacological inhibition of AXL, cells were pretreated with dimethyl sulfoxide (DMSO) or TP-0903 for 24 hours before seeding. For AXL down-regulation or overexpression, cells were seeded 72 hours after transfection. A total of 50,000 cells were seeded in serum-free medium in the top chamber and allowed to migrate for 48 hours toward the bottom chamber containing 10% fetal bovine serum medium. Cells were fixed and permeabilized with cold 70% ethanol and methanol and stained with 0.4% crystal violet. The average number of migrating cells was evaluated from at least four independent microscope fields.
Proximity ligation assay
Cells were fixed and permeabilized with the Cytofix/Cytoperm Plus Fixation/Permeabilization Solution Kit. Primary antibodies (see table S3) were incubated overnight at 4°C. Hybridization with the PLA probes and negative control conditions, ligation, and amplification were performed with Duolink reagents according to the manufacturer’s recommendations (#DUO92007, Sigma-Aldrich, St. Louis, MO, USA). Technical negative controls were included by omission of primary antibodies. PLA results were obtained with a Leica TCS SP8 confocal microscope and by flow cytometry with BD FACSAria III instrument (Becton Dickinson) and analyzed by FlowJo V10.
In vitro PDX–resistant model
PDX118 (ER+/PR−/HER23+) was derived from skin metastasis collected by core needle biopsy. PDXs were established at Vall d’Hebron Institute of Oncology (VHIO) following institutional guidelines. The Institutional Review Board at Vall d’Hebron Hospital provided approval for this study in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients who provided tissue samples. Trastuzumab in vitro PDX–resistant models (PDX118-TR1 and PDX118-TR2) were established by culturing cells in the presence of increasing concentration of trastuzumab (starting from 5 to 1000 ng/ml). In both models, resistance was obtained after 4 months of treatment. For proliferation experiments, PDX118-TR1 and PDX118-TR2 cell lines were treated with trastuzumab for 7 days combined with TP-0903 at a concentration of 150 nM during the last 72 hours of treatment.
Patient-derived organoid generation and treatment
Tumor cells were seeded in 48-well plates (5 × 104 cells per well) in a drop of 20 μl of Matrigel. Each drop was dispensed in the center of the well and incubated for 15 min at room temperature. After the Matrigel solidified, 250 μl of 3D breast tumor organoid medium was added to each well (53). Medium was replaced twice weekly, and 3D formation was assessed after 15 days. Organoids per well were counted and each one assumed to contain approximately 50 cells. Organoid medium was removed, and 3D structures were cultured with trastuzumab and/or TP-0903. At the end point, organoids were disaggregated by adding 500 μl of trypsin for 30 min at 37°C, after which cells were collected and incubated for 30 min on ice to liquify the Matrigel. Fully disaggregated organoids were washed and stained as previously described in full (53). The number of live cells was quantified with FlowJo software by means of EpCAM counts. Viability marker Zombie Aqua was used at 1:1000 (#423101, BioLegend, San Diego, CA, USA).
In vivo PDX–resistant model
To generate the PDX118 model resistant to trastuzumab, nonobese diabetic (NOD)–severe combined immunodeficient (SCID) mice were transplanted with fragments of PDX118 tumors. Once tumor size reached 300 mm3, animals were intraperitoneally injected with trastuzumab (10 mg/kg) twice per week. After two passages, resistant tumors (termed PDX118-TR4) were obtained. Samples for flow cytometry and reinjection were digested in collagenase IA (300 U/ml; #C2674, Sigma-Aldrich) and hyaluronidase IS (100 U/ml; #H3506, Sigma-Aldrich) in Dulbecco’s modified Eagle’s medium/F-12. After 1 hour of incubation at 37°C with shaking at 80 rpm, the mixture was filtered through 100-μm strainers. Red blood cells were lysed with 1× RBC lysis buffer for 5 min at room temperature. After a wash with 1× phosphate-buffered saline, samples were counted and either reinjected or stained for HER2 and EpCAM as previously described and acquired on LSRFortessa. Data were analyzed with FlowJo software. For the in vivo experiment, mice were injected with either trastuzumab (10 mg/kg, intraperitoneally) biweekly, TP-0903 orally (50 mg/kg) daily 5/7, or a combination of both across 21 days (from days 89 to 110). A follow-up of 321 days was recorded. Animal experiments received ethical approval from Generalitat de Catalunya/Comissió d’Experimentació Animal Òrgan Habilitat (Authorised Establishment B9900062, ethical protocol CEA-OH/10303).
Clinical samples
Samples were obtained from patients with early-stage HER2+ BC treated at the Hospital Clínico de Valencia Department of Medical Oncology from 2004 to 2016 by standard guidelines. Patients received (neo)adjuvant chemotherapy and anti-HER2 therapy (trastuzumab ± pertuzumab) for 6 months, followed by surgery. Adjuvant treatment was completed with trastuzumab for up to 1 year, and a minimum of 5 years of hormonal therapy for patients with HR+ tumors. Radiation therapy was administered according to local guidelines. Patients provided signed informed consent for experimental analysis of samples. The study is compliant with all relevant ethical regulations regarding research involving human participants and received ethical approval from the Hospital Clínico Research Ethics Committee. All samples were FFPE and had been analyzed by an expert pathologist to ensure >30% tumor infiltration. HR status was evaluated by IHC (ER+ and PR+ were defined as ≥1% positively stained nuclei), and HER2 was assessed by IHC and/or fluorescence in situ hybridization (FISH) (IHC: complete 3+ membrane staining ≥10% invasive cells; FISH:HER2:CEP17 ratio ≥ 2.0). Clinicopathological data of this cohort are detailed in table S2.
Analysis of public human data
HER2+ BC cohort from Kaplan-Meier plotter software was analyzed. The prognostic value of AXL and GAS6 mRNA expression was analyzed in 252 HER2+ by IHC BC patients with available data. The median value was used to identify the high and low groups. The hazard ratio with 95% CI and P value were calculated, and Kaplan-Meier curves were plotted (34).
PAMELA study
The main results of the PAMELA neoadjuvant phase 2 study have been previously reported (35). This completed study is registered with ClinicalTrials.gov (NCT01973660). The study protocol was approved by independent ethics committees at each center. In this trial, 151 patients with early HER2+ BC were treated with neoadjuvant lapatinib (1000 mg daily) and trastuzumab (8 mg kg−1 intravenous loading dose followed by 6 mg kg−1) for 18 weeks. Patients with HR+ BC received letrozole or tamoxifen according to menopausal status. Tumor samples were collected at baseline, day 14, and surgery and then FFPE. HER2+ was defined as 3+ or 2+ overexpression by IHC and positive for chromogenic in situ hybridization. RNA samples of the PAMELA trial at baseline, at day 14 of treatment, and in residual tumors were previously analyzed using a panel of 560 genes (including AXL, VIM, and ERBB2) at the nCounter platform (NanoString Technologies, Seattle, WA, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 6.01; GraphPad Software Inc., La Jolla, CA, USA) and R (version 3.6.2). Results were expressed as means ± SD. Normality was checked using Shapiro-Wilks’s test. Mean comparison was carried out using two-tailed Student’s t test. All experiments were reproduced at least three times.
GAS6, ERBB2, AXL, and VIM gene expression in the retrospective cohort was dichotomized by the median. Prognostic potential was evaluated by ROC analysis, and optimal cutoff was calculated with Youden index (54). DFS and OS were described graphically using Kaplan-Meier curves, and differences were evaluated using the log-rank test. Cox regression was conducted to calculate hazard ratio and 95% CI. Cox multivariable model included dichotomized GAS6, ERBB2, AXL, VIM, tumor size, and affected nodes. Selection of the best model was done according to AIC using a stepwise forward-backward approach.
To identify AXL gene expression changes in clinical samples of the PAMELA trial across time points (baseline versus day 14 or day 14 versus surgery), we used two-tailed paired t tests. Two-way analysis of variance (ANOVA) with Bonferroni correction post hoc was used for in vivo statistical analysis. The cutoff for statistical significance in all tests was 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Acknowledgments
We thank the INCLIVA Biobank (PT17/0015/0049; B.000768 ISCIII) and the Valencian Biobanking Network integrated into the Spanish National Biobanks Network for its collaboration. We are also grateful to SOLTI. We are especially grateful to the patients who agreed to participate in this research and to Associations Amunt Contra el Cáncer, Asociación Cultural Falla Palleter, and Asociación en tu Seno.
Funding: A.A.-A., E.J.A., and F.B.-M. were supported by Asociación Española contra el Cáncer AECC (PRDVA18013LLUC to A.A.-A., POSTD211413AREN to E.J.A., and AECC_Postdoctoral17-1062 to F.B.-M.). A.M.-S. was funded by the Spanish Government (PFIS FI20/00188). J.Ar. is supported by Breast Cancer Research Foundation (BCRF-20-08), Instituto de Salud Carlos III Project reference number AC15/00062, and the EC under the framework of the ERA-NET TRANSCAN-2 initiative cofinanced by FEDER, Instituto de Salud Carlos III (CB16/12/00449 and PI19/01181), and Asociación Española Contra el Cáncer (AECC). A.P. was supported by Instituto de Salud Carlos III—PI19/01846, Breast Cancer Now—2018NOVPCC1294. P.E. and A.L. were funded by Instituto de Salud Carlos III and cofinanced by FEDER (PI18/01219 to P.E. and CB16/12/00481 to A.L.). J.M.C. was funded by Sociedad Española de Oncología Médica (Rio Hortega-SEOM) and Compromiso ADAMED.
Author contributions: A.A.-A., P.E., and J.M.C. conceptualized the study. A.A.-A., E.J.A., F.B.-M., A.M.-S., J.Ar., A.P., and J.M.C. designed the study methodology. A.A.-A., E.J.A., F.B.-M., A.M.-S., J.C.-A., O.B., and J.M.C. interpreted the data and/or performed statistical analysis. A.A.-A., E.J.A., R.C., F.B.-M., A.M.-S., D.M., and E.T. conducted experiments. A.A.-A., E.J.A., F.B.-M., A.M.-S., C.H., M.T.M., S.S., J.P., S.M., S.Z., A.R., O.B., F.R., J.Al., B.B., A.L., A.P., J.Ar., P.E., and J.M.C. interpreted data and/or wrote the manuscript. A.A.-A., E.J.A., and J.M.C. were responsible for data curation and supervised the project. A.L., B.B., J.Ar., A.P., and P.E. provided resources. J.M.C. supervised the study.
Competing interests: A.P. reports advisory and consulting fees from Roche, Pfizer, Novartis, Amgen, BMS, Puma, Oncolytics Biotech, MSD, Guardan Health, Peptomyc, and Lilly; lecture fees from Roche, Pfizer, Novartis, Amgen, BMS, NanoString Technologies, and Daiichi Sankyo; institutional financial interests from Boehringer, Novartis, Roche, NanoString, Sysmex Europa GmbH, Medica Scientia inno. Research, SL, Celgene, Astellas, and Pfizer; and a leadership role in Reveal Genomics, SL; A.P. is an inventor on a patent related to this work filed by SOLTI Breast Cancer Research Group, Hospital Clinic de Barcelona, Vall d’Hebron Institute of Oncology (VHIO) University and FISABIO (no. PCT/EP2016/080056, filed on 7 December 2016, published on 14 June 2018). J.Al. has received consulting or advisory role fees from Roche, Pfizer, Amgen, MSD, Lilly, AstraZeneca, and Daiichi-Sankyo; research funding or grant support trials by Roche, Pfizer, Amgen, MSD, Lilly, and Daiichi-Sankyo; and travel and accommodation support from Roche, Pfizer, Amgen, MSD, Lilly, and Daiichi-Sankyo. A.L. reports advisory and consulting fees from Novartis, Pfizer, Roche/Genentech, Eisai, and Celgene. J.M.C. has received honoraria as Speaker from Pfizer and Novartis, and travel expenses from Roche. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The patient-derived xenografts (PDXs) can be provided by J.Ar. (VHIO) pending scientific review and a completed material transfer agreement. Requests for the PDX models should be submitted to the corresponding author (jmcejalvo@incliva.es) and to J.Ar. (jarribas@vhio.net).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S3
REFERENCES AND NOTES
- 1.Moasser M. M., The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26, 6469–6487 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez E. A., Romond E. H., Suman V. J., Jeong J. H., Davidson N. E., Geyer C. E., Martino S., Mamounas E. P., Kaufman P. A., Wolmark N., Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: Joint analysis of data from NCCTG N9831 and NSABP B-31. J. Clin. Oncol. 29, 3366–3373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Xu B., Targeted therapeutic options and future perspectives for her2-positive breast cancer. Signal Transduct. Target. Ther. 4, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukohara T., Mechanisms of resistance to anti-human epidermal growth factor receptor 2 agents in breast cancer. Cancer Sci. 102, 1–8 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kaplan R. N., Psaila B., Lyden D., Bone marrow cells in the “pre-metastatic niche”: Within bone and beyond. Cancer Metastasis Rev. 25, 521–529 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Nieto M. A., Epithelial-mesenchymal transitions in development and disease: Old views and new perspectives. Int. J. Dev. Biol. 53, 1541–1547 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Antony J., Huang R. Y. J., AXL-driven EMT state as a targetable conduit in cancer. Cancer Res. 77, 3725–3732 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Asiedu M., Beauchamp-Perez F., Ingle J., Radisky D., Knutson K., AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene 33, 1316–1324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Greger J., Shi H., Liu Y., Greshock J., Annan R., Halsey W., Sathe G. M., Martin A. M., Gilmer T. M., Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: Activation of AXL. Cancer Res. 69, 6871–6878 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Miller M. A., Oudin M. J., Sullivan R. J., Wang S. J., Meyer A. S., Im H., Frederick D. T., Tadros J., Griffith L. G., Lee H., Weissleder R., Flaherty K. T., Gertler F. B., Lauffenburger D. A., Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov. 6, 383–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller J., Krijgsman O., Tsoi J., Robert L., Hugo W., Song C., Kong X., Possik P. A., Cornelissen-Steijger P. D. M., Foppen M. H. G., Kemper K., Goding C. R., McDermott U., Blank C., Haanen J., Graeber T. G., Ribas A., Lo R. S., Peeper D. S., Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 5, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byers L. A., Diao L., Wang J., Saintigny P., Girard L., Peyton M., Shen L., Fan Y., Giri U., Tumula P. K., Nilsson M. B., Gudikote J., Tran H., Cardnell R. J. G., Bearss D. J., Warner S. L., Foulks J. M., Kanner S. B., Gandhi V., Krett N., Rosen S. T., Kim E. S., Herbst R. S., Blumenschein G. R., Lee J. J., Lippman S. M., Ang K. K., Mills G. B., Hong W. K., Weinstein J. N., Wistuba I. I., Coombes K. R., Minna J. D., Heymach J. V., An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19, 279–290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi H., Yamada T., Wang R., Tanimura K., Adachi Y., Nishiyama A., Tanimoto A., Takeuchi S., Araujo L. H., Boroni M., Yoshimura A., Shiotsu S., Matsumoto I., Watanabe S., Kikuchi T., Miura S., Tanaka H., Kitazaki T., Yamaguchi H., Mukae H., Uchino J., Uehara H., Takayama K., Yano S., AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat. Commun. 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terry S., Abdou A., Engelsen A. S. T., Buart S., Dessen P., Corgnac S., Collares D., Meurice G., Gausdal G., Baud V., Saintigny P., Lorens J. B., Thiery J. P., Mami-Chouaib F., Chouaib S., AXL targeting overcomes human lung cancer cell resistance to NK- And CTL-mediated cytotoxicity. Cancer Immunol. Res. 7, 1789–1802 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Boshuizen J., Koopman L. A., Krijgsman O., Shahrabi A., Van Den Heuvel E. G., Ligtenberg M. A., Vredevoogd D. W., Kemper K., Kuilman T., Song J. Y., Pencheva N., Mortensen J. T., Foppen M. G., Rozeman E. A., Blank C. U., Janmaat M. L., Satijn D., Breij E. C. W., Peeper D. S., Parren P. W. H. I., Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med. 24, 203–212 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z., Lee J. C., Lin L., Olivas V., Au V., Laframboise T., Abdel-Rahman M., Wang X., Levine A. D., Rho J. K., Choi Y. J., Choi C. M., Kim S. W., Jang S. J., Park Y. S., Kim W. S., Lee D. H., Lee J. S., Miller V. A., Arcila M., Ladanyi M., Moonsamy P., Sawyers C., Boggon T. J., Ma P. C., Costa C., Taron M., Rosell R., Halmos B., Bivona T. G., Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 44, 852–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okura N., Nishioka N., Yamada T., Taniguchi H., Tanimura K., Katayama Y., Yoshimura A., Watanabe S., Kikuchi T., Shiotsu S., Kitazaki T., Nishiyama A., Iwasaku M., Kaneko Y., Uchino J., Uehara H., Horinaka M., Sakai T., Tanaka K., Kozaki R., Yano S., Takayama K., ONO-7475, a Novel AXLInhibitor, suppresses the adaptive resistance to initial EGFR-TKI treatment in EGFR-mutated non-small cell lung cancer. Clin. Cancer Res. 26, 2244–2256 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Elkabets M., Pazarentzos E., Juric D., Sheng Q., Pelossof R. A., Brook S., Benzaken A. O., Rodon J., Morse N., Yan J. J., Liu M., Das R., Chen Y., Tam A., Wang H., Liang J., Gurski J. M., Kerr D. A., Rosell R., Teixidó C., Huang A., Ghossein R. A., Rosen N., Bivona T. G., Scaltriti M., Baselga J., AXL mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell 27, 533–546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyette M. A., Duhamel S., Aubert L., Pelletier A., Savage P., Thibault M. P., Johnson R. M., Carmeliet P., Basik M., Gaboury L., Muller W. J., Park M., Roux P. P., Gratton J. P., Côté J. F., The receptor tyrosine kinase AXL is required at multiple steps of the metastatic cascade during HER2-positive breast cancer progression. Cell Rep. 23, 1476–1490 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Gjerdrum C., Tiron C., Høiby T., Stefansson I., Haugen H., Sandal T., Collett K., Li S., McCormack E., Gjertsen B. T., Micklem D. R., Akslen L. A., Glackin C., Lorens J. B., Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc. Natl. Acad. Sci. U.S.A. 107, 1124–1129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davra V., Kumar S., Geng K., Calianese D., Mehta D., Gadiyar V., Kasikara C., Lahey K. C., Chang Y. J., Wichroski M., Gao C., de Lorenzo M. S., Kotenko S. V., Bergsbaken T., Mishra P. K., Gause W. C., Quigley M., Spires T. E., Birge R. B., Axl and mertk receptors cooperate to promote breast cancer progression by combined oncogenic signaling and evasion of host antitumor immunity. Cancer Res. 81, 698–712 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leconet W., Chentouf M., Du Manoir S., Chevalier C., Sirvent A., Aït-Arsa I., Busson M., Jarlier M., Radosevic-Robin N., Theillet C., Chalbos D., Pasquet J. M., Pèlegrin A., Larbouret C., Robert B., Therapeutic activity of anti-AXL antibody against triple-negative breast cancer patient-derived xenografts and metastasis. Clin. Cancer Res. 23, 2806–2816 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Goyette M. A., Cusseddu R., Elkholi I., Abu-Thuraia A., El-Hachem N., Haibe-Kains B., Gratton J. P., Côté J. F., AXL knockdown gene signature reveals a drug repurposing opportunity for a class of antipsychotics to reduce growth and metastasis of triple-negative breast cancer. Oncotarget 10, 2055–2067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei J., Sun H., Zhang A., Wu X., Li Y., Liu J., Duan Y., Xiao F., Wang H., Lv M., Wang L., Wu C., A novel AXL chimeric antigen receptor endows T cells with anti-tumor effects against triple negative breast cancers. Cell. Immunol. 331, 49–58 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Schoumacher M., Burbridge M., Key roles of AXL and MER receptor tyrosine kinases in resistance to multiple anticancer therapies. Curr. Oncol. Rep. 19, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D., Bach D. H., Fan Y. H., Luu T. T. T., Hong J. Y., Park H. J., Lee S. K., AXL degradation in combination with EGFR-TKI can delay and overcome acquired resistance in human non-small cell lung cancer cells. Cell Death Dis. 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brabletz T., Kalluri R., Nieto M. A., Weinberg R. A., EMT in cancer. Nat. Rev. Cancer 18, 128–134 (2018). [DOI] [PubMed] [Google Scholar]
- 29.del Pozo Martin Y., Park D., Ramachandran A., Ombrato L., Calvo F., Chakravarty P., Spencer-Dene B., Derzsi S., Hill C. S., Sahai E., Malanchi I., Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Rep. 13, 2456–2469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antony J., Tan T. Z., Kelly Z., Low J., Choolani M., Recchi C., Gabra H., Thiery J. P., Huang R. Y. J., The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci. Signal. 9, ra97 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Scaltriti M., Elkabets M., Baselga J., Molecular pathways: AXL, a membrane receptor mediator of resistance to therapy. Clin. Cancer Res. 22, 1313–1317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vicario R., Peg V., Morancho B., Zacarias-Fluck M., Zhang J., Martínez-Barriocanal Á., Navarro Jiménez A., Aura C., Burgues O., Lluch A., Cortés J., Nuciforo P., Rubio I. T., Marangoni E., Deeds J., Boehm M., Schlegel R., Tabernero J., Mosher R., Arribas J., Patterns of HER2 gene amplification and response to anti-HER2 therapies. PLOS ONE 10, e0129876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang S. A., Guan J. S., Tan H. J., Chu T., Thike A. A., Bernado C., Arribas J., Wong C. Y., Tan P. H., Gudi M., Putti T. C., Sohn J., Lim S. H., Lee S. C., Lim Y. P., Elevated WBP2 expression in HER2-positive breast cancers correlates with sensitivity to trastuzumab-based neoadjuvant therapy: A retrospective and multicentric study. Clin. Cancer Res. 25, 2588–2600 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z., An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Llombart-Cussac A., Cortés J., Paré L., Galván P., Bermejo B., Martínez N., Vidal M., Pernas S., López R., Muñoz M., Nuciforo P., Morales S., Oliveira M., de la Peña L., Peláez A., Prat A., HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 18, 545–554 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Brasó-Maristany F., Griguolo G., Pascual T., Paré L., Nuciforo P., Llombart-Cussac A., Bermejo B., Oliveira M., Morales S., Martínez N., Vidal M., Adamo B., Martínez O., Pernas S., López R., Muñoz M., Chic N., Galván P., Garau I., Manso L., Alarcón J., Martínez E., Gregorio S., Gomis R. R., Villagrasa P., Cortés J., Ciruelos E., Prat A., Phenotypic changes of HER2-positive breast cancer during and after dual HER2 blockade. Nat. Commun. 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griguolo G., Brasó-Maristany F., González-Farré B., Pascual T., Chic N., Saurí T., Kates R., Gluz O., Martínez D., Paré L., Tsvetkova V., Pesantez D., Vidal M., Adamo B., Muñoz M., Galván P., Barberá L., Cuatrecasas M., Christgen M., Kreipe H., Monge-Escartín I., Villagrasa P., Soy D., Giarratano T., Dieci M. V., Conte P., Harbeck N., Guarneri V., Prat A., ERBB2 mRNA expression and response to ado-trastuzumab emtansine (T-DM1) in HER2-positive breast cancer. Cancers 12, 1902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prat A., Pascual T. A., De Angelis C., Gutierrez C., Llombart-Cussac A., Wang T., Cort J., Rexer B., Par L., Forero A., Wolff A. C., In Morales S., Adamo B., O-Maristany F. B., Vidal M., Veeraraghavan J., Krop I., Galv P., Pavlick A. C., Bermejo N., Izquierdo M., Rodrik-Outmezguine V., Reis-Filho J. S., Hilsenbeck S. G., Oliveira M., Dieci M. V., Griguolo G., Fasani R., Nuciforo P., Parker J. S., Conte P., Schiff R., Guarneri V., Osborne C. K., Rimawi M. F., HER2-enriched subtype and ERBB2 expression in HER2-positive breast cancer treated with dual HER2 blockade. J. Natl. Cancer Inst. 112, 46–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boshuizen J., Pencheva N., Krijgsman O., Altimari D. D., Castro P. G., de Bruijn B., Ligtenberg M. A., Gresnigt-Van den Heuvel E., Vredevoogd D. W., Song J.-Y., Visser N., Apriamashvili G., Janmaat M. L., Plantinga T. S., Franken P., Houtkamp M., Lingnau A., Jure-Kunkel M., Peeper D. S., Cooperative targeting of immunotherapy-resistant melanoma and lung cancer by an AXL-targeting antibody–drug conjugate and immune checkpoint blockade. Cancer Res. 81, 1775–1787 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Wang C., Jin H., Wang N., Fan S., Wang Y., Zhang Y., Wei L., Tao X., Gu D., Zhao F., Fang J., Yao M., Qin W., Gas6/Axl axis contributes to chemoresistance and metastasis in breast cancer through Akt/GSK-3β/β- catenin signaling. Theranostics 6, 1205–1219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuoriluoto K., Haugen H., Kiviluoto S., Mpindi J. P., Nevo J., Gjerdrum C., Tiron C., Lorens J. B., Ivaska J., Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 30, 1436–1448 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Meyer A. S., Miller M. A., Gertler F. B., Lauffenburger D. A., The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci. Signal. 6, ra66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan G.-X., Kazlauskas A., Axl is essential for VEGF-A-dependent activation of PI3K/Akt. EMBO J. 31, 1692–1703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salian-Mehta S., Xu M., Wierman M. E., AXL and MET crosstalk to promote gonadotropin releasing hormone (GnRH) neuronal cell migration and survival. Mol. Cell. Endocrinol. 374, 92–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keating A. K., Kim G. K., Jones A. E., Donson A. M., Ware K., Mulcahy J. M., Salzberg D. B., Foreman N. K., Liang X., Thorburn A., Graham D. K., Inhibition of Mer and Axl receptor tyrosine kinases in astrocytoma cells leads to increased apoptosis and improved chemosensitivity. Mol. Cancer Ther. 9, 1298–1307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park I. K., Mundy-Bosse B., Whitman S. P., Zhang X., Warner S. L., Bearss D. J., Blum W., Marcucci G., Caligiuri M. A., Receptor tyrosine kinase Axl is required for resistance of leukemic cells to FLT3-targeted therapy in acute myeloid leukemia. Leukemia 29, 2382–2389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeon J. Y., Buelow D. R., Garrison D. A., Niu M., Eisenmann E. D., Huang K. M., Zavorka Thomas M. E., Weber R. H., Whatcott C. J., Warner S. L., Orwick S. J., Carmichael B., Stahl E., Brinton L. T., Lapalombella R., Blachly J. S., Hertlein E., Byrd J. C., Bhatnagar B., Baker S. D., TP-0903 is active in models of drug-resistant acute myeloid leukemia. JCI Insight 5, e140169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen T., Tong P., Diao L., Li L., Fan Y., Hoff J., Heymach J. V., Wang J., Byers L. A., Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin. Cancer Res. 23, 6239–6253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita N., Tokunaga E., Kitao H., Hisamatsu Y., Taketani K., Akiyoshi S., Okada S., Aishima S., Morita M., Maehara Y., Vimentin as a poor prognostic factor for triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 139, 739–746 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Recio S., Thennavan A., East M. P., Parker J. S., Cejalvo J. M., Garay J. P., Hollern D. P., He X., Mott K. R., Galván P., Fan C., Selitsky S. R., Coffey A. R., Marron D., Brasó-Maristany F., Burgués O., Albanell J., Rojo F., Lluch A., De Dueñas E. M., Rosen J. M., Johnson G. L., Carey L. A., Prat A., Perou C. M., FGFR4 regulates tumor subtype differentiation in luminal breast cancer and metastatic disease. J. Clin. Invest. 130, 4871–4887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zazo S., González-Alonso P., Martín-Aparicio E., Chamizo C., Cristóbal I., Arpí O., Rovira A., Albanell J., Eroles P., Lluch A., Madoz-Gúrpide J., Rojo F., Generation, characterization, and maintenance of trastuzumab-resistant HER2+ breast cancer cell lines. Am. J. Cancer Res. 6, 2661–2678 (2016). [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnamoorthy G. P., Guida T., Alfano L., Avilla E., Santoro M., Carlomagno F., Melillo R. M., Molecular mechanism of 17-allylamino-17-demethoxygeldanamycin (17-AAG)-induced AXL receptor tyrosine kinase degradation. J. Biol. Chem. 288, 17481–17494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arenas E. J., Martínez-Sabadell A., Rius Ruiz I., Román Alonso M., Escorihuela M., Luque A., Fajardo C. A., Gros A., Klein C., Arribas J., Acquired cancer cell resistance to T cell bispecific antibodies and CAR T targeting HER2 through JAK2 down-modulation. Nat. Commun. 12, 1237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youden W. J., Index for rating diagnostic tests. Cancer 3, 32–35 (1950). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S3