Abstract
Determining the viability of waterborne Cryptosporidium parvum oocysts remains a technical challenge. rRNA and mRNA were evaluated in a reverse transcription (RT)-PCR assay as potential markers of oocyst viability. The rationale for this approach is the rapid turnover and postmortem decay of cellular RNA. The β-tubulin mRNA and an anonymous mRNA transcript were chosen as potential markers because they are the only mRNA species in C. parvum known to possess introns. This feature facilitated the distinction between genuine RT-PCR products and PCR products originating from copurifying DNA. Prolonged incubation at room temperature of initially viable oocysts resulted in a gradual decrease in mRNA levels, which correlated with the loss of oocyst infectivity to neonatal mice. In contrast, oocysts stored at 4°C for over 39 weeks maintained their infectivity and displayed no decrease in the level of β-tubulin RT-PCR product. The postmortem decay of two mRNA species demonstrates that RT-PCR analysis can provide information on the viability of C. parvum oocysts. The methodological similarity between PCR detection and RT-PCR viability analysis could facilitate the development of a combined detection and viability assay.
Cryptosporidium parvum is an enteric apicomplexan parasite infecting several mammalian species. In immunocompromised individuals, C. parvum can establish persistent infections leading to chronic diarrhea and wasting.
Oocysts of C. parvum are commonly found in surface water and can contaminate public water supplies (15, 17, 23). Information on the viability of waterborne oocysts is critical for assessing the risk of waterborne transmission. Since standard, antibody-based detection methods do not discriminate between viable and dead oocysts, it is difficult to assess the risk posed by the presence of oocysts in drinking water. Although some authors distinguish between viable and infectious oocysts, for the purpose of this report the terms “viable” and “infectious” are considered synonyms, as are “nonviable,” “inactivated,” and “dead.”
In the laboratory, the benchmark for viability assessment of C. parvum oocysts is animal infectivity. Commonly used animal surrogates are neonatal or immunosuppressed rodents (28). Since animal infectivity assays do not meet the requirements of the water industry for fast and cost-effective tests, a number of in vitro methods which differentiate between viable and dead oocysts have been developed. These methods take advantage of several changes associated with oocyst death, such as increased oocyst wall permeability to vital dyes (4, 11), loss of ability to excyst (8, 30), loss of infectivity to tissue culture cells (22, 24), and absence of transcriptional activity (26, 29). The fact that waterborne oocysts are typically recovered among a heterogeneous mixture of other organisms and organic matter limits the use of viability assays requiring microscopic examination of water filtrates such as vital dyes. These commonly used methods are also affected by exposure of oocysts to disinfectants (1, 7, 14) and will not indicate inactivation by exposure to low doses of UV light (2a). Cell culture methods are sensitive indicators of viability, particularly when combined with PCR detection (21), but are relatively slow and labor-intensive. A 24- to 48-h incubation time limits their use in emergency situations. In contrast, techniques involving PCR do not depend on microscopic examination and can take advantage of the specificity of PCR amplification to distinguish among different species or genotypes (31). Results can be obtained within a day, and the use of newer PCR analysis tools can significantly shorten the procedure by eliminating the need for electrophoretic analysis (9).
On the basis of the observed rapid postmortem RNA decay in certain mammalian tissues (16, 20), rRNA and mRNA transcripts from C. parvum oocysts were examined with the aim of identifying suitable markers of oocyst viability. In this report we show that rRNA and mRNA decay at different rates. β-Tubulin mRNA was a suitable target for this assay because it decays quickly and because the presence of an intron (3) facilitates the differentiation between PCR products originating from mRNA and from copurifying genomic DNA.
MATERIALS AND METHODS
Oocyst preparation.
C. parvum oocysts of isolate GCH1 (27), propagated in neonatal calves, were isolated from fecal material by flotation on 2 volumes of saturated NaCl followed by sedimentation on a 15% to 25% (wt/vol) Nycodenz (Sigma, St. Louis, Mo.) step gradient in phosphate-buffered saline for 1.5 h at 100,000 × g (32). Recovered oocysts were surface sterilized in 10% bleach (0.5% sodium hypochloride) on ice for 10 min and resuspended in 1% penicillin-streptomycin in sterile phosphate-buffered saline. The ratio of excystation was estimated by counting excysted and unexcysted oocysts after a 45-min incubation in 0.75% taurocholic acid at 37°C.
Purified oocysts were divided into two groups. One group was stored at 4°C, and the other was stored at room temperature. At 5-week intervals, two aliquots of 107 oocysts were removed from the sample stored at room temperature. One aliquot was tested for the ability to infect mice, and the remaining aliquot was stored at −80°C for later RNA extraction and reverse transcription (RT)-PCR analysis. Oocysts stored at 4°C were processed in the same manner except that samples were collected at 10-week intervals.
Animal inoculation.
Litters of ICR mice (Taconic, Germantown, N.Y.), 6 to 7 days of age, were inoculated with 106 purified oocysts per mouse. Typically, eight mice were infected per sample. Modified acid-fast staining was used to monitor inoculated mice three times a week for oocyst shedding. Fecal oocyst counts were estimated on acid-fast stained smears as described and expressed on a scale from 0 to 5 (27).
RNA extraction, RT-PCR, and gel analysis.
Oocysts were lysed by three cycles of freeze-thawing, and RNA was extracted with Trireagent (Sigma). Contaminating DNA was digested with RQ1 DNase (Promega, Madison, Wis.). The DNase was inactivated before RT by heat treatment at 65°C for 15 min.
RNA from approximately 2 × 105 oocysts were used in each RT reaction. The RNA was denatured in diethylpyrocarbonate-treated water by incubation at 80°C in 10 mM EDTA–2 μM reverse primer for 2 min. The reaction mixtures were cooled slowly to 37°C, and 100 U of Moloney murine leukemia virus reverse transcriptase, 20 U of RNase inhibitor, 1 mM (each) deoxynucleoside triphosphates, and 1× Moloney murine leukemia virus RT buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol) were added. The reaction mixtures were incubated at 37°C for 1 h. RT primers included cry20 (positions 1091 to 1072; GenBank accession no. L16996) and btub2 (32) (positions 708 to 689; GenBank accession no. Y12615).
A fragment of the C. parvum β-tubulin cDNA was amplified from RT reaction mixtures following a nested-PCR protocol. In the first-round RT-PCR, a 454-bp fragment of the β-tubulin cDNA spanning the exon1-exon2 splice site was amplified by using sense primer btub5 (positions 165 to 184; accession no. Y12615) and antisense primer btub2 (positions 708 to 689) (32). It is important to note that a putative artifactual 6-bp duplication in the sequence deposited under accession no. Y12615 causes a discrepancy between calculated and expected amplicon sizes. In the second round of amplification, an internal 282-bp fragment was amplified by using sense primer btub3 (5′ GTCATTTCTGATGAGCACGG 3′; positions 187 to 206) and antisense primer btub6 (5′ ACAGCATCTAAGAGTTCAGCTCC 3′; positions 536 to 558) also located in exon 1 and exon 2, respectively. First-round amplifications were performed by using a hot-start protocol with Taq DNA polymerase, 1/10 volume of RT reaction mixtures as a template, and 30 cycles of 95°C for 50 s, 52°C for 50 s, and 72°C for 1 min followed by a final extension at 72°C for 3 min. The same PCR protocol was used with the internal primers except that the primer annealing temperature was raised to 57°C and 1 μl of first-round amplicon was used as a template. Oocyst RNA and sterile water were used as negative RT-PCR controls.
The ribosomal PCR products were amplified from 1/10 volume of the RT reaction mixtures primed with cry20 (5′ AAGTTTCAGCCTTGCGACC 3′) by using primers cry4 and cry2 as described by Carraway et al. (5), except that the annealing temperature was raised to 55°C. A single 35-cycle PCR amplification was performed with the ribosomal primers. A 408-bp fragment of the mRNA deposited under GenBank accession no. AA224676 was amplified by RT-PCR using reverse primer AA2241 (5′ GATACTTTCGAAGGGCAAGG 3′; positions 527 to 507) and forward primer AA2242 (5′ TGGTTCGAGTAAAATCCAAGG 3′; positions 120 to 139). The reverse primer was also used to prime the RT.
PCR products were visualized on 1.5% agarose stained with ethidium bromide. Gel images were digitized with MCID-M4 software (Imaging Research Inc., St. Catharines, Ontario, Canada), and band intensities were measured as integrated optical density with the Direct Band Analysis program.
RESULTS
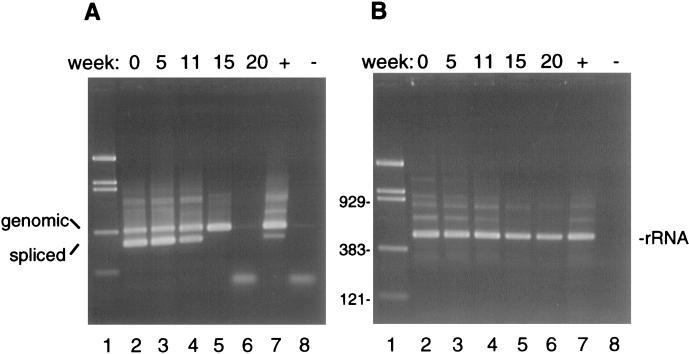
To assess the rate of RNA decay following oocyst inactivation, viable C. parvum oocysts were inactivated at 65°C for 15 min (6) and stored at room temperature. RT-PCR analysis of the β-tubulin mRNA and small-subunit (SSU) rRNA revealed different rates of decay between these transcripts. Whereas the SSU rRNA was still detectable for at least 11 weeks after oocyst inactivation when a single 35-cycle PCR was used, the β-tubulin transcript was not detected, even when a nested-PCR protocol was used, in RNA extracted within 1 h of oocyst inactivation (data not shown).
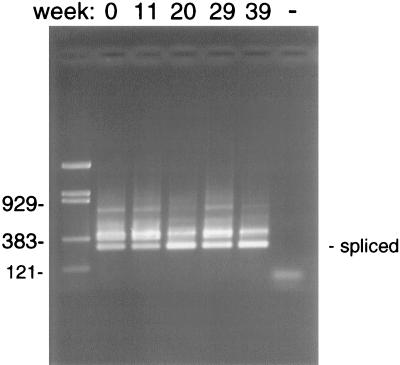
The rapid decay of the β-tubulin mRNA in inactivated oocysts suggested that this transcript might be an adequate marker of oocyst viability. RNA stability was examined in live oocysts stored for prolonged periods of time at room temperature or at 4°C. Oocysts of isolate GCH1 were purified from the stool of an experimentally infected calf within 3 days of excretion. The viability of the oocysts at the onset of the experiment was estimated microscopically by determining the rate of excysted oocysts in an in vitro excystation assay and determining their infectivity to neonatal mice. An excystation rate of 90%, and 100% infectivity (eight mice infected out of eight inoculated), confirmed that this sample was fully viable. RT-PCR analysis was performed on RNA extracted from oocysts aged over periods of 20 and 39 weeks at room temperature and 4°C, respectively. To minimize sample-to-sample variation in the analysis, oocyst samples removed at specific time points were stored at −70°C and RNA was extracted at all time points simultaneously. At each time point, the infectivity of the oocysts was determined in neonatal mice (Table 1). As expected, the infectivity decreased faster at room temperature. Loss of infectivity occurred by week 20. In the 4°C sample, oocysts were still infectious after 39 weeks, although some reduction in mouse fecal oocyst scores was found. Confirming the initial observations with inactivated oocysts, RT-PCR analysis of the room temperature samples revealed a relative rapid decay in the RT-PCR signal. By week 15, no β-tubulin mRNA was detected (Fig. 1A). Except for one sample (Fig. 1A, lane 6), a PCR product was also amplified from the unspliced, genomic sequence. The disappearance of the DNA signal in the 20-week sample suggests that DNA degradation might also occur at later time points. In contrast to the β-tubulin signal, the SSU rRNA showed little sign of decay after 20 weeks of storage (Fig. 1B). Due to the relative abundance of this transcript, a single 35-cycle amplification was sufficient for its detection. Control RT-PCRs with the rRNA primers performed in the absence of reverse transcriptase were negative (data not shown), confirming that the persistence of the RT-PCR signal was not due to DNA contamination. Attempts at identifying less stable regions of the SSU rRNA by using RT-PCR primers closer to the 3′ end did not reveal differences in the rate of decay within this transcript. In oocysts stored at 4°C, the spliced β-tubulin PCR signal was still visible after 39 weeks of storage. In fact, there was no apparent decrease in signal intensity (Fig. 2).
TABLE 1.
Infectivity of C. parvum oocysts stored at room temperature and 4°C
| Length of time (wk) at temp | Mouse infectivity (% infected) | Fecal oocyst scorea |
|---|---|---|
| Room temp | ||
| 0 | 100 | 2–3 |
| 5 | 80 | 2 |
| 11 | 60 | 2 |
| 15 | 20 | 1 |
| 20 | 0 | 0 |
| 4°C | ||
| 0 | 100 | 2–3 |
| 11 | 100 | 1–3 |
| 20 | 100 | 1–3 |
| 29 | 100 | 1–3 |
| 39 | 100 | 1–2 |
For explanation of oocyst score, see reference 27.
FIG. 1.
RT-PCR detection of RNA in oocysts aged at room temperature for 20 weeks. (A) mRNA and genomic DNA amplification using the β-tubulin primers btub3 and btub6. The size difference between the genomic (366-bp) and the spliced (mRNA; 282-bp) product is due to the presence of an 84-bp intron. Notice the decay of the mRNA signal starting at week 11 and the disappearance of the DNA signal at week 20 (lane 6). A trace of PCR product is visible in the negative control (lane 8). (B) RT-PCR amplification of SSU rRNA showing no time-dependent decay of this transcript. Lanes 1, pBR/Bst/NI DNA ladder, with sizes of relevant bands (in base pairs) shown left of panel B; lanes 7, positive PCR control; lanes 8, negative PCR control.
FIG. 2.
Stability of β-tubulin RT-PCR signal in oocysts stored at 4°C for 39 weeks. Notice the persistence of the RT-PCR signal in oocysts maintained at this temperature. Leftmost lane, pBR/Bst/NI DNA markers, with sizes of relevant bands (in base pairs) shown on the left. The negative PCR control is shown in the rightmost lane (−).
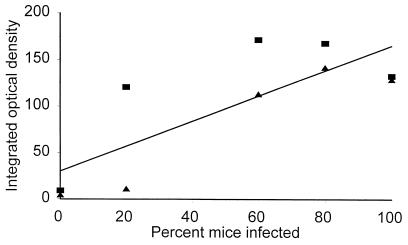
The infectivity of aged oocysts was monitored in neonatal mice with the aim of examining the correlation between the decay of two mRNA species and infectivity. Although the correlation between β-tubulin mRNA and infectivity was not very high (correlation coefficient [r] = 0.64; n = 10), two independent experiments confirmed the absence of β-tubulin mRNA in oocysts that were no longer infective (Fig. 3). In contrast, all infectious oocyst samples showed some level of β-tubulin mRNA. Samples displaying a high RT-PCR signal infected 60 to 100% of the mice. The decrease in infectivity was paralleled by the fecal oocyst concentration (Table 1); fecal scores ranged from 2 to 3 for fresh oocysts and gradually decreased to 0 for aged samples. In agreement with the persistence of the RT-PCR signal, all mice inoculated with the 4°C oocysts developed an infection.
FIG. 3.
The β-tubulin RT-PCR signal as a marker of oocyst viability. The integrated optical density of the spliced RT-PCR signal was plotted against oocyst infectivity in mice (Table 1) for oocysts aged at room temperature. Different symbols represent two independent series of RT-PCR amplifications.
Because of the advantage of targeting the viability assay to transcripts bearing introns, we monitored the rate of decay of a second mRNA transcript reported to contain an intron (GenBank accession no. AA224676) (18). This transcript, of unknown coding function, also decayed in a time-dependent fashion (r = 0.92). In contrast to β-tubulin mRNA, a PCR signal was still detectable in nonviable oocysts (data not shown), suggesting a higher stability of this transcript than of β-tubulin mRNA. It also appears that this transcript is present at a higher copy number in oocysts, because a single round of PCR amplification was sufficient to generate visible PCR products.
Since RT-PCR was performed with oocyst numbers unlikely to be encountered in environmental samples, a serial dilution experiment was performed with the aim of identifying the detection limit of the β-tubulin RT-PCR assay. A nested-PCR amplification of 10-fold serial dilutions of an RT reaction detected spliced and unspliced β-tubulin signal in a 10-oocyst equivalent (data not shown).
DISCUSSION
Much attention has focused on the use of vital dyes for C. parvum viability assessment (1, 2, 7, 11, 14). The effect of disinfectants on oocyst dye uptake has generated some controversy regarding the use of dye-based methods. In spite of such assays being technically simple and cost-effective, it is conceivable that in the future the need for microscopic examination of individual oocysts will limit the application of vital dyes to small operations. As nucleic acid-based diagnostic technologies amenable to automation are being developed (9), such methods will become attractive for environmental testing. Detection and viability methods based on PCR may facilitate automation for water testing.
PCR has been evaluated for the detection of waterborne C. parvum oocysts in many laboratories (12, 21, 25, 31). Because loss of viability is not thought to immediately affect DNA integrity, few viability methods relying on nucleic acid amplification have been described. Originally, Filkorn et al. (8) and Wagner-Wiening and Kimmig (30) proposed a PCR viability method based on the detection of excysted sporozoites by PCR. In our hands, exposure of intact oocysts to high temperatures during thermal cycling resulted in the release of some DNA from oocysts, suggesting that a PCR test relying solely on the capacity of viable oocysts to excyst could be prone to generating false positives. A different approach was described by Stinear et al. (26). The heat shock protein 70 (hsp70) transcript was chosen as a target for RT-PCR amplification because it is known to be induced in some experimental organisms in response to heat shock. Although the hsp70 RT-PCR proved to be a sensitive detection method (13), the hsp70 mRNA was not induced in oocysts exposed to heat. In addition, the fate of this RNA species in aged oocysts was not investigated (26). Using fluorescent in situ hybridization (FISH) directed at the SSU rRNA 5′ region, Vesey et al. (29) demonstrated a good correlation between oocyst fluorescence and excystation. At first sight, this observation is in conflict with the stability of rRNA described here. Several differences in the RT-PCR and FISH protocols could explain the discrepancy, namely, (i) differential sensitivities of the RT-PCR and FISH and the exponential amplification (PCR) versus the nonamplified (FISH) signal, (ii) the different regions of the SSU targeted in the assays, and (iii) the use of excystation versus animal infectivity as the second variable.
The sensitivity of any PCR is dependent on the initial concentration of target molecules. Because visualization of the mRNA deposited under accession no. AA224676 required a single round of amplification, we assume that this transcript is present in oocysts at a higher copy number than β-tubulin mRNA. We initially focused on the β-tubulin transcript because it was the only transcript in C. parvum known to contain an intron. This feature allowed us to eliminate the interference from DNA contaminating our RNA extracts. For the purpose of a viability assay, the advantage of a single-step amplification with this transcript (accession no. AA224676) is offset by its lower rate of decay. An extrapolation of the RT-PCR data from this transcript shows that, assuming a constant and linear rate of decay, mRNA would still be detected more than 20 weeks postmortem. This property makes this mRNA (accession no. AA224676) unsuitable as a marker of viability. The rates of decay of rRNA and of two mRNA species investigated in this study demonstrate that transcripts in oocysts vary considerably in their abundance and postmortem stability. An ideal RNA-based viability assay requires the identification of an mRNA species of higher abundance than β-tubulin and with a higher decay rate than that of the mRNA deposited under accession no. AA224676.
Because of the effects of sodium hypochloride on the oocyst wall (7, 19), it would have been preferable to omit the bleach treatment in the preparation of aged oocyst stocks. Initial trials showed that without sterilization even highly purified oocyst samples would rapidly become contaminated with fungi and bacteria. This made it difficult to obtain precise oocyst counts. Therefore, the bleach treatment was retained as a means of removing contaminants. Although this resulted in an artificially homogeneous oocyst suspension, it was considered adequate for the purpose of this study, namely, establishing the correlation between RNA levels and viability. Assessing the performance of the RNA-based viability assay in environmental samples is a separate goal, which is currently being addressed.
In conclusion, although the development of an optimal viability test for waterborne oocysts remains a technical challenge, RNA analysis, whether by RT-PCR or hybridization, compares favorably with other viability methods. Since RNA decay was examined in heat-inactivated and in untreated, aged oocysts only, the effect of disinfectants on the performance of this assay remains to be investigated. A significant advantage of this approach is the potential for the development of a combined detection-viability assay. In addition, this method can be tailored to species- or genus-specific sequences depending on the requirements of the test.
ACKNOWLEDGMENT
This work was supported by U.S. Department of Agriculture grant 9604051.
REFERENCES
- 1.Black E K, Finch G R, Taghi-Kilani R, Belosevic M. Comparison of assays for Cryptosporidium parvum oocyst viability after chemical disinfection. FEMS Microbiol Lett. 1996;135:187–189. doi: 10.1111/j.1574-6968.1996.tb07987.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown M A, McDonald V, Denton H, Coombs G H. The use of a new viability assay to determine the susceptibility of Cryptosporidium and Eimeria sporozoites to respiratory inhibitors and extremes of pH. FEMS Microbiol Lett. 1996;142:203–208. doi: 10.1111/j.1574-6968.1996.tb08431.x. [DOI] [PubMed] [Google Scholar]
- 2a.Bukhari, Z. (Clancy Environmental Consultants). Personal communication.
- 3.Cacciò S, La Rosa G, Pozio E. The beta-tubulin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1997;89:307–311. doi: 10.1016/s0166-6851(97)00122-9. [DOI] [PubMed] [Google Scholar]
- 4.Campbell A T, Robertson L J, Smith H V. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl Environ Microbiol. 1992;58:3488–3493. doi: 10.1128/aem.58.11.3488-3493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fayer R. Effect of high temperature on infectivity of Cryptosporidium parvum oocysts in water. Appl Environ Microbiol. 1994;60:2732–2735. doi: 10.1128/aem.60.8.2732-2735.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fayer R. Effect of sodium hypochloride exposure on infectivity of Cryptosporidium parvum oocysts for neonatal BALB/c mice. Appl Environ Microbiol. 1995;61:844–846. doi: 10.1128/aem.61.2.844-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filkorn R, Wiedenmann A, Botzenhart K. Selective detection of viable Cryptosporidium oocysts by PCR. Zentbl Hyg. 1994;195:489–494. [PubMed] [Google Scholar]
- 9.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Hodgson J. Shrinking DNA diagnostics to fill the markets of the future. Nat Biotechnol. 1998;16:725–727. doi: 10.1038/nbt0898-725. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins M B, Anguish L J, Bowman D D, Walker M J, Ghiorse W C. Assessment of dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl Environ Microbiol. 1997;63:3844–3850. doi: 10.1128/aem.63.10.3844-3850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaucner C, Stinear T. Sensitive and rapid detection of viable Giardia cysts and Cryptosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription-PCR. Appl Environ Microbiol. 1998;64:1743–1749. doi: 10.1128/aem.64.5.1743-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korich D G, Mead J R, Madore M S, Sinclair N A, Sterling C R. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl Environ Microbiol. 1990;56:1423–1428. doi: 10.1128/aem.56.5.1423-1428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeChevallier M W, Norton W D. Giardia and Cryptosporidium in raw and finished water. J Am Water Works Assoc. 1995;87:54–68. [Google Scholar]
- 16.Noguchi I, Arai H, Iizuka R. A study on postmortem stability of vasopressinmessenger RNA in rat brain compared with those in total RNA and ribosomal RNA. J Neural Transm. 1991;83:171–178. doi: 10.1007/BF01253387. [DOI] [PubMed] [Google Scholar]
- 17.Ongerth J E, Stibbs H H. Identification of Cryptosporidium oocysts in river water. Appl Environ Microbiol. 1987;53:672–676. doi: 10.1128/aem.53.4.672-676.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piper M B, Bankier A T, Dear P H. A HAPPY map of Cryptosporidium parvum. Genome Res. 1998;8:1299–1307. doi: 10.1101/gr.8.12.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reduker D W, Speer C A, Blixt J A. Ultrastructural changes in the oocyst wall during excystation of Cryptosporidium parvum (Apicomplexa; Eucoccidiorida) Can J Zool. 1985;63:1892–1896. [Google Scholar]
- 20.Reichert G H, Issinger O G. In vitro study of the biological activity of RNAs after incubation of hog liver, heart and brain at room temperature. Biochimie. 1985;67:657–661. doi: 10.1016/s0300-9084(85)80208-x. [DOI] [PubMed] [Google Scholar]
- 21.Rochelle P A, De Leon R, Stewart M H, Wolfe R L. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl Environ Microbiol. 1997;63:106–114. doi: 10.1128/aem.63.1.106-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochelle P A, Ferguson D M, Handojo T J, De Leon R, Stewart M H, Wolfe R L. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl Environ Microbiol. 1997;63:2029–2037. doi: 10.1128/aem.63.5.2029-2037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose J B, Gerba C P, Jakubowski W. Survey of potable water supplies for Cryptosporidium and Giardia. Environ Sci Technol. 1991;25:1393–1400. [Google Scholar]
- 24.Slifko T R, Friedman D, Rose J B, Jakubowski W. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl Environ Microbiol. 1997;63:3669–3675. doi: 10.1128/aem.63.9.3669-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluter S D, Tzipori S, Widmer G. Parameters affecting polymerase chain reaction detection of waterborne Cryptosporidium parvum oocysts. Appl Microbiol Biotechnol. 1997;48:325–330. doi: 10.1007/s002530051057. [DOI] [PubMed] [Google Scholar]
- 26.Stinear T, Matusan A, Hines K, Sandery M. Detection of a single viable Cryptosporidium parvum oocysts in environmental water concentrates by reverse transcription-PCR. Appl Environ Microbiol. 1996;62:3385–3390. doi: 10.1128/aem.62.9.3385-3390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. Evaluation of an animal model system for Cryptosporidiosis: the therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol. 1994;1:450–463. doi: 10.1128/cdli.1.4.450-463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzipori S. Cryptosporidiosis: laboratory investigations and chemotherapy. Adv Parasitol. 1998;40:188–221. doi: 10.1016/s0065-308x(08)60121-9. [DOI] [PubMed] [Google Scholar]
- 29.Vesey G, Ashbolt N, Fricker E J, Deere D, Williams K L, Veal D A, Dorsch M. The use of a ribosomal RNA targeted oligonucleotide probe for fluorescent labelling of viable Cryptosporidium parvum oocysts. J Appl Microbiol. 1998;86:429–440. doi: 10.1046/j.1365-2672.1998.853496.x. [DOI] [PubMed] [Google Scholar]
- 30.Wagner-Wiening C, Kimmig P. Detection of viable Cryptosporidium parvum oocysts by PCR. Appl Environ Microbiol. 1995;61:4514–4516. doi: 10.1128/aem.61.12.4514-4516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:224–241. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 32.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the β-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]