Abstract
The genes encoding the six polypeptide components of the alkene monooxygenase from Xanthobacter strain Py2 (Xamo) have been located on a 4.9-kb fragment of chromosomal DNA previously cloned in cosmid pNY2. Sequencing and analysis of the predicted amino acid sequences indicate that the components of Xamo are homologous to those of the aromatic monooxygenases, toluene 2-, 3-, and 4-monooxygenase and benzene monooxygenase, and that the gene order is identical. The genes and predicted polypeptides are aamA, encoding the 497-residue oxygenase α-subunit (XamoA); aamB, encoding the 88-residue oxygenase γ-subunit (XamoB); aamC, encoding the 122-residue ferredoxin (XamoC); aamD, encoding the 101-residue coupling or effector protein (XamoD); aamE, encoding the 341-residue oxygenase β-subunit (XamoE); and aamF, encoding the 327-residue reductase (XamoF). A sequence with >60% concurrence with the consensus sequence of ς54 (RpoN)-dependent promoters was identified upstream of the aamA gene. Detailed comparison of XamoA with the oxygenase α-subunits from aromatic monooxygenases, phenol hydroxylases, methane monooxygenase, and the alkene monooxygenase from Rhodococcus rhodochrous B276 showed that, despite the overall similarity to the aromatic monooxygenases, XamoA has some distinctive characteristics of the oxygenases which oxidize aliphatic, and particularly alkene, substrates. On the basis of the similarity between Xamo and the aromatic monooxygenases, Xanthobacter strain Py2 was tested and shown to oxidize benzene, toluene, and phenol, while the alkene monooxygenase-negative mutants NZ1 and NZ2 did not. Benzene was oxidized to phenol, which accumulated transiently before being further oxidized. Toluene was oxidized to a mixture of o-, m-, and p-cresols (39.8, 18, and 41.7%, respectively) and a small amount (0.5%) of benzyl alcohol, none of which were further oxidized. In growth studies Xanthobacter strain Py2 was found to grow on phenol and catechol but not on benzene or toluene; growth on phenol required a functional alkene monooxygenase. However, there is no evidence of genes encoding steps in the metabolism of catechol in the vicinity of the aam gene cluster. This suggests that the inducer specificity of the alkene monooxygenase may have evolved to benefit from the naturally broad substrate specificity of this class of monooxygenase and the ability of the host strain to grow on catechol.
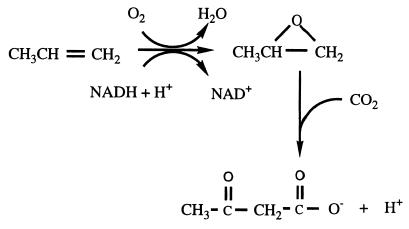
Xanthobacter strain Py2 is a gram-negative bacterial strain which was isolated on propene as a sole carbon and energy source (34). The metabolism of propene involves an alkene-specific monooxygenase which converts propene to epoxypropane. Further metabolism involves isomerization and carboxylation of the epoxide, ultimately yielding acetoacetate (Fig. 1), which feeds into the central metabolism (1, 6, 36).
FIG. 1.
Initial steps in the metabolism of propene in Xanthobacter strain Py2.
The Xanthobacter strain Py2 alkene monooxygenase (Xamo) will catalyze the epoxidation of a range of alkenes, some with a high degree of stereospecificity, but will not hydroxylate the homologous alkanes (35). This contrasts with alkane monooxygenases such as those based on cytochrome P-450 (28) or nonheme iron (e.g., ω-hydroxylase [27] and methane monooxygenase [MMO] [11]), which appear to generate a highly reactive iron-oxygen intermediate which can attack both unactivated C-H bonds and carbon-carbon double bonds. This reaction specificity and stereospecificity make Xamo an enzyme of considerable interest as a biocatalyst for the production of chiral 1,2-epoxides. Additionally, Xamo has been shown to be responsible for catalyzing the initial step in the cometabolic degradation of a number of chlorinated alkenes of environmental concern, including vinyl chloride, trichloroethene, and 1,3-dichloropropene (9), and it can even be induced by the presence of these chlorinated alkenes, although there is no evidence for growth on these substrates (10). Xamo has been resolved into four components: an NADH-dependent reductase, a Rieske-type ferredoxin, an oxygenase, and a small protein which may be a coupling or effector protein (29). The oxygenase is an α2β2γ2 hexamer which was reported to contain approximately four atoms of nonheme iron per hexamer on the basis of colorimetric iron analysis and the lack of a significant UV- or visible-light chromophore. Sequence (25) and electron paramagnetic resonance evidence (12) has demonstrated that an alkene-specific monooxygenase derived from the gram-positive bacterium Rhodococcus rhodochrous (formerly Nocardia corallina) B276 has a binuclear nonheme iron center of the type found in soluble MMO. However, the Xanthobacter enzyme is more complex than that from Rhodococcus, having a two-component (reductase and ferredoxin) redox system typical of aromatic dioxygenases (3) and a more complex oxygenase structure.
We have recently reported the sequencing of the first open reading frame (ORF) in the Xamo gene cluster (42). The predicted polypeptide sequence showed strong homology to the nonheme iron binding subunit in aromatic monooxygenases and MMO, particularly around the iron binding domain. Modelling of the predicted Xanthobacter polypeptide on the coordinates of the α-subunit in the MMO crystal structure (24) allowed us to conclude that Xamo is also a nonheme iron monooxygenase. In this paper, we report the entire sequence of the six ORFs which encode Xamo. The predicted amino acid sequences of all six polypeptides encoded by these ORFs show strong homology with those of aromatic monooxygenases, including benzene monooxygenase and toluene 4-monooxygenase. This led us to investigate whether Xamo could catalyze the monohydroxylation of aromatic hydrocarbons or grow on them as sole sources of carbon and energy.
MATERIALS AND METHODS
Bacterial strains.
The following strains were used: Escherichia coli DH 10B F− mcrA Δ(mrr hsdRMS mcrBC) φ80d lacZΔM15 ΔlacX74 deoR reclA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG (GIBCO BRL) and Xanthobacter strains Py2 (34), NZ1, and NZ2. The latter two are independently isolated alkene monooxygenase-negative (Amo−) mutants of Xanthobacter strain Py2 (41).
Materials.
Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were obtained from Promega. Restriction enzymes were supplied by Boehringer and used as recommended by the manufacturer. DNA ligase was purchased from New England Biolabs.
Bacterial growth.
E. coli cells were routinely grown in liquid Luria-Bertani medium (10 g of casein peptone/liter, 5 g of yeast extract/liter, 5 g of NaCl/liter [pH 7.2]) or on Luria-Bertani plates at 37°C. For biotransformation assays, Xanthobacter strain Py2 was grown on an ammonia mineral salts medium (AMS) supplemented with 10% (vol/vol) propene or 0.5% fructose, and the mutants NZ1 and NZ2 were grown on AMS supplemented with 10% (vol/vol) propene and 0.01% (vol/vol) propene oxide, as previously described (41).
For testing growth on aromatic substrates, Xanthobacter strain Py2 and its mutants were grown on AMS supplemented with 2 and 4 mM benzene, toluene, or phenol. Where growth was not seen, the toxicity of aromatic substrates was tested by comparing the growth of Xanthobacter strain Py2 on 0.5% fructose in AMS with and without aromatic substrate supplementation.
Sequencing strategy and sequence analysis.
An 11.2-kb EcoRI fragment from the cosmid clone pNY2, previously subcloned as pNY2C and shown to complement Amo− mutants of Xanthobacter strain Py2 (41), was used to prepare nested deletions by using the Deletion Factory system, version 2 (GIBCO BRL), as described previously (42). These were sequenced in both directions by using BigDye Terminator Cycle Sequencing (Perkin-Elmer) with AmpliTaq FS DNA polymerase and were analyzed on an ABI 377 automated sequencer (Applied Biosystems). DNA and deduced amino acid sequences were analyzed by using the MacVector (version 4.5.3) and AssemblyLIGN (version 1.07) software packages (Eastman Kodak Co.). Homologous protein searches were carried out with FASTA 3 (22) and PSI-BLAST (2). Pairwise alignments were made with the GAP program in the Wisconsin package (7). Multiple alignments were run under Clustal W (32). The search for ς54-dependent promoter sequences was done remotely with the SEQSCAN program (26a).
Biotransformation assays with alkene monooxygenase (AMO).
Assays were carried out on resting cell suspensions of Xanthobacter strain Py2 and the mutants NZ1 and NZ2. Cells were harvested in late-exponential phase, washed once, and resuspended in 25 mM phosphate buffer, pH 7.5, to an optical density at 600 nm of 10. Cell suspensions (1 ml) were preincubated for 2 min at 30°C in 7-ml Suba-sealed (W. H. Freeman, Barnsley, United Kingdom) flasks before addition of either benzene, toluene, or phenol to a final concentration of 2 mM. The rate of product formation was assayed by gas chromatography (Philips PU4500) by taking 5-μl liquid samples at suitable time intervals, separating them on a Tenax TA (60/80 mesh) column (Phase Separations, Deeside, United Kingdom) at 210°C with nitrogen as the carrier gas at 40 ml · min−1, and carrying out flame ionization detection. Product concentrations were determined by reference to external standards, by using a Shimadzu CR3A recording integrator. The identities of the reaction products were confirmed by coretention with authentic standards on both the Tenax column and (for phenol) 5% (wt/vol) OV-17 on a Chromosorb W.HP (80/100 mesh) column at 130°C with nitrogen as the carrier gas at 40 ml · min−1. To separate and identify o-, m-, and p-cresol and benzyl alcohol, the samples were analyzed by capillary gas chromatography (model 436; United Technologies Packard) on a 50-m by 0.25-mm Lipodex C column (Macharey-Nagel) at 120°C with helium as the carrier gas at 0.77 ml · min−1. The split ratio was 1:100. For analysis on the latter two columns, the reaction mixtures were extracted with 0.5 ml of ethyl acetate and the extract was dried over sodium sulfate.
Propene oxide formation and degradation were measured as previously described (41).
Nucleotide sequence accession number.
The sequences described in this report have been deposited with the EMBL data bank and are available under accession no. AJ012090.
RESULTS AND DISCUSSION
Preliminary sequence analysis of Xanthobacter aam genes.
The genes encoding Xamo have previously been cloned as a 25.7-kb insert in the broad-host-range cosmid vector pLAFR5 and shown to express propene-inducible AMO when transferred to Xanthobacter autotrophicus JW33, although it was not expressed in E. coli (41). Mapping of the cosmid showed that it overlapped with a region sequenced by Swaving et al. (31) which contained the genes encoding components of the epoxide isomerase-carboxylase complex. Additionally, deletions from one end of the cosmid resulted in loss of expression of the AMO and complementation of Amo− mutants, suggesting that the AMO was encoded in a fragment of 11.2 kb flanked on one side by the cosmid junction and on the other by the isomerase-carboxylase genes. By using a nested deletion strategy, a large part of this fragment has now been sequenced, revealing the presence of six closely spaced ORFs in a 4.9-kb DNA fragment, consistent in size and predicted amino acid sequence with the reported components of Xamo (29). These ORFs have been designated aamA through aamF (Table 1); the designations refer to “alkene and aromatic monooxygenase,” for reasons that will become clear in this manuscript. We have already reported (42) that the predicted amino acid sequence of the aamA gene product (497 residues; predicted mass, 58,037 Da; gene previously referred to as xamoA [42]) has a high degree of sequence similarity to the α-subunit of nonheme iron monooxygenases, particularly the aromatic monooxygenases, allowing us to model part of the sequence of this polypeptide on the coordinates of the MMO α-subunit, obtained from the crystal structure.
TABLE 1.
Genes with predicted products which are most homologous to those of aamA through aamF
| Gene | Size (aa)a | % Identity | % Similarity | Organism | Accession no.b | Reference |
|---|---|---|---|---|---|---|
| aamA | 497 | |||||
| bmoA | 500 | 47.69 | 67.0 | Pseudomonas aeruginosa JI104 | D83068 | 16 |
| tbhA | 501 | 46.23 | 64.39 | Burkholderia cepacia AA1 | AF001356 | |
| tmoA | 500 | 45.07 | 63.98 | Pseudomonas mendocina KR1 | M65106 | 39 |
| tbuA1 | 501 | 44.06 | 64.39 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlK | 501 | 43.86 | 64.59 | Ralstonia eutropha JMP134 | AF06589 | |
| phhN | 516 | 27.16 | 46.91 | Pseudomonas putida P35X | X79063 | 19 |
| memA | 527 | 22.86 | 45.16 | Methylococcus capsulatus (Bath) | M90050 | 30 |
| memA | 525 | 22.82 | 46.47 | Methylosinus trichosporium OB3b | X55394 | 5 |
| amoC | 501 | 22.08 | 48.33 | Rhodococcus rhodochrous B-276 | D37875 | 25 |
| aamB | 88 | |||||
| tbuU | 86 | 36.47 | 60.00 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlL | 86 | 35.29 | 60.00 | Ralstonia eutropha JMP134 | AF06589 | |
| tmoB | 84 | 34.94 | 53.00 | Pseudomonas mendocina KR1 | M65106 | 39 |
| bmoB | 88 | 31.76 | 60.00 | Pseudomonas aeruginosa JI104 | D83068 | 16 |
| aamC | 122 | |||||
| bmoC | 111 | 44.95 | 71.56 | Pseudomonas aeruginosa JI104 | D83068 | 16 |
| tbhC | 99 | 42.86 | 68.37 | Burkholderia cepacia AA1 | AF001356 | |
| tmoC | 112 | 39.45 | 66.05 | Pseudomonas mendocina KR1 | M65106 | 39 |
| tbuB | 111 | 38.53 | 68.81 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlM | 111 | 37.61 | 68.81 | Ralstonia eutropha JMP134 | AF06589 | |
| bphA3 | 107 | 34.91 | 51.89 | Rhodococcus sp. strain RHA1 | D32142 | 17 |
| hcaA3 | 106 | 30.48 | 54.29 | Escherichia coli | Y11070 | |
| msmC | 123 | 23.64 | 48.18 | α-Proteobacterium M2 | U59152 | 13 |
| aamD | 101 | |||||
| bmoD1 | 147 | 45.00 | 66.00 | Pseudomonas aeruginosa JI104 | D83068 | 16 |
| tbuV | 104 | 43.00 | 66.00 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlN | 105 | 43.00 | 65.00 | Ralstonia eutropha JMP134 | AF065891 | |
| tbhD | 104 | 43.00 | 63.00 | Burkholderia cepacia AA1 | AF001356 | |
| tmoD | 103 | 39.39 | 62.63 | Pseudomonas mendocina KR1 | M65106 | 39 |
| dmpM | 90 | 23.60 | 48.31 | Pseudomonas sp. strain CF600 | M60276 | 20 |
| aamE | 341 | |||||
| tbuA2 | 329 | 38.34 | 56.44 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlO | 332 | 37.08 | 56.53 | Ralstonia eutropha JMP134 | AF065891 | |
| tbhE | 332 | 34.36 | 53.37 | Burkholderia cepacia AA1 | AF001356 | |
| tmoE | 327 | 34.26 | 55.56 | Pseudomonas mendocina KR1 | M65106 | 39 |
| dsoB | 333 | 24.84 | 50.97 | Acinetobacter sp. strain 2OB | D85083 | 15 |
| amoA | 343 | 24.77 | 43.50 | Rhodococcus rhodochrous B-276 | D37875 | 25 |
| 333 | 22.96 | 49.06 | Acinetobacter calcoaceticus | Z36090 | 8 | |
| dmpL | 331 | 22.93 | 46.18 | Pseudomonas sp. strain CF600 | M60276 | 20 |
| phhL | 331 | 22.29 | 45.22 | Pseudomonas putida P35X | X79063 | 19 |
| aamF | 327 | |||||
| tbuC | 334 | 41.92 | 63.04 | Ralstonia pickettii PKO1 | U04052 | 4 |
| phlP | 332 | 41.87 | 64.69 | Ralstonia eutropha JMP134 | AF065891 | |
| xylA | 346 | 37.3 | 57.68 | Sphingomonas aromaticivorans F199 | AF079317 | |
| caraD | 329 | 37.11 | 59.43 | Pseudomonas sp. strain CA10 | D89064 | 26 |
| 329 | 37.1 | 59.75 | Pseudomonas stutzeri OM1 | AB001723 | 21 | |
| tmoF | 326 | 33.02 | 57.00 | Pseudomonas mendocina KR1 | M65106 | 40 |
| amoD | 342 | 32.6 | 57.99 | Rhodococcus rhodochrous B-276 | D37875 | 25 |
aa, amino acids.
Refers to NCBI nucleotide accession no.
aamB encodes a polypeptide of 88 amino acids (9,740 Da) with a high degree of overall similarity to the γ-subunit of the α2β2γ2 hexameric nonheme iron monooxygenases; aamC encodes a polypeptide of 122 amino acids (13,359 Da) with a high degree of similarity to the ferredoxins of four-component nonheme iron monooxygenases and aromatic ring-hydroxylating bacterial dioxygenases; aamD encodes a polypeptide of 101 amino acids (11,193 Da) which is most similar to the small “coupling” proteins of four-component nonheme iron monooxygenases and also the dmpM gene product (P2) of the three-component phenol hydroxylase from Pseudomonas sp. strain CF600 (19); aamE encodes a polypeptide of 341 amino acids (38,188 Da) which is homologous to the oxygenase β-subunits of α2β2γ2 hexameric monooxygenases and also the β-subunit of the R. rhodochrous B276 AMO, which is probably tetrameric (18, 25). As is the case with most other multicomponent mono- and dioxygenases, the final ORF (aamF) encodes a reductase homolog (327 amino acids [34,171 Da]).
Gene order.
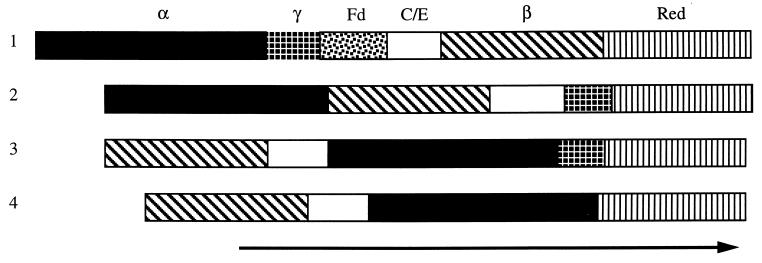
On the basis of the comparisons above, it is reasonable to conclude that aamA through aamF encode the oxygenase α-subunit, oxygenase γ-subunit, ferredoxin, coupling or effector protein, oxygenase β-subunit, and reductase, respectively (Fig. 2). This gene order is identical to that of the four-component benzene and toluene monooxygenases (4, 39, 40) and the Ralstonia eutropha JMP134 phenol hydroxylase, to which the individual subunits show the highest homology. The extent of sequence identity or similarity between the component polypeptides of the hexameric monooxygenase subunits of these enzymes also shows a clear grouping, separating them from MMO (5, 30) and the almost identical pair of phenol hydroxylases from Pseudomonas sp. strain CF600 (20) and Pseudomonas putida P35X (19), all of which have hexameric oxygenases, and the tetrameric R. rhodochrous B276 AMO (25). All of the latter appear to be three-component systems, lacking the separate ferredoxin component, and have a gene order different from that of the Xamo–aromatic ring monooxygenase family. In MMO the genes encoding the β- and γ-subunits are in reverse order with respect to that for the “coupling” protein, while in the R. rhodochrous B276 AMO and the pseudomonad phenol hydroxylases, the first structural gene encodes the oxygenase β-subunit, and the α-subunit is actually the third structural gene in the sequence.
FIG. 2.
Gene orders of the Xamo and related gene clusters. The monooxygenase components encoded are indicated at the top, as follows: α, β, and γ, oxygenase α-, β-, and γ-subunits; Fd, ferredoxin; C/E, coupling or effector protein; Red, reductase. The arrow indicates the direction of transcription. Diagram 1, Xamo and the benzene (bmo), toluene 3- (tbh), toluene 4- (tmo), toluene/benzene (tbu), and phenol (phl) monooxygenases; diagram 2, MMO (an additional ORF [not shown] lies between the genes encoding the γ-subunit and the reductase); diagram 3, phenol hydroxylase (dmp and phh); diagram 4, R. rhodochrous B276 AMO.
Regulatory features and codon usage.
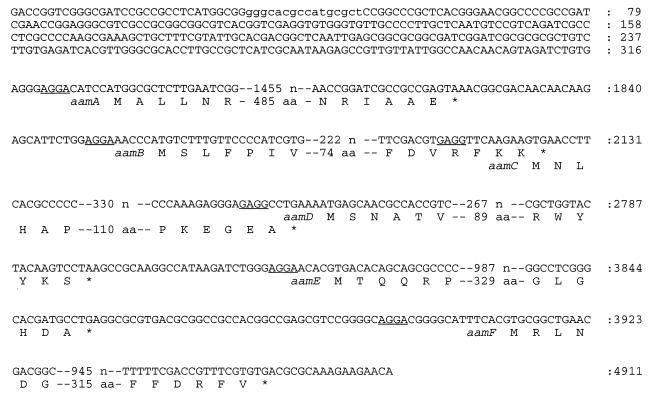
Potential ribosome binding sites precede the ATG or GTG translational initiation sites by 4 to 13 bp in all six ORFs, namely AGGA for aamA, aamB, aamE, and aamF and GAGG for aamC and aamD (Fig. 3). The translational stop codon is TAA for aamA and aamD and TGA for the other genes. No homolog of the −35 and −10 consensus sequence of ς70-dependent promoters was found immediately upstream of the aamA gene, which might explain the lack of expression of the cosmid pNY2 in E. coli. However, a possible ς54 (RpoN)-dependent promoter sequence (GGGCACGCCATGCGCT) is present approximately 300 bp upstream of the translational start site of aamA.
FIG. 3.
Regulatory features of the aam gene cluster, showing the putative ς54 (RpoN)-dependent promoter sequence (lowercase letters in the top line) and ribosome binding sites (underlined).
Xanthobacter spp. have genomes which are typically composed of 65 to 70% G+C (38). While this feature is evident when the combined frequencies of all six ORFs (67% GC) are considered, there is significant variation, from 62.4% (aamC) to 71.9% (aamF, among the different coding regions. A high GC content implies that codons in which the final base in the triplet is G or C should be more frequently used, and this is also evident (90.5% G/C). The codon usage is similar to that described for other genes in Xanthobacter spp. (31, 33).
Intriguingly, the codons UUA (Leu), CUA (Leu), GUA (Val), UCA (Ser), and AGA (Arg) are not used once either in the 1,476 codons characterized here or in the 1,469 codons described by Swaving et al. (31) from the same organism.
Further analysis of aamC and aamD. (i) aamC.
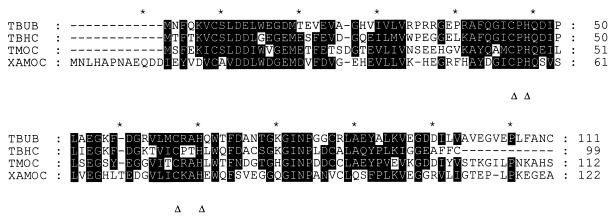
Alignment of the amino acid sequence of the aamC gene product with that of the homologous monooxygenase and dioxygenase ferredoxins (Fig. 4) clearly shows that a number of key residues are absolutely conserved, including the pairs of cysteine and histidine residues which coordinate the 2Fe–2S cluster. The latter are diagnostic of Rieske-type iron-sulfur proteins and confirm the spectroscopic assignments (14, 29). Although it is clear that the aamC gene product is most similar to the ferredoxins present in the four-component aromatic ring monooxygenases, XamoC contains an additional 12 amino acids at the N terminus which are not seen in any of the near homologs. This enzyme contains a high proportion of acidic residues and could well be important in protein-protein interactions.
FIG. 4.
Homology between the putative ferredoxin of Xamo (XAMOC), encoded by aamC, and ferredoxins from four-component aromatic ring monooxygenases. Dark highlighting indicates residues which are identical or functionally conserved in at least three sequences. The two cysteines and two histidines which form the metal binding sites are shown. TBUB, TBHC, and TMOC, ferredoxins from the Ralstonia pickettii PKO1 toluene 3-monooxygenase (4), the Burkholderia cepacia AA1 toluene 3-monooxygenase, and the Pseudomonas mendocina KR1 toluene 4-monooxygenase (39), respectively.
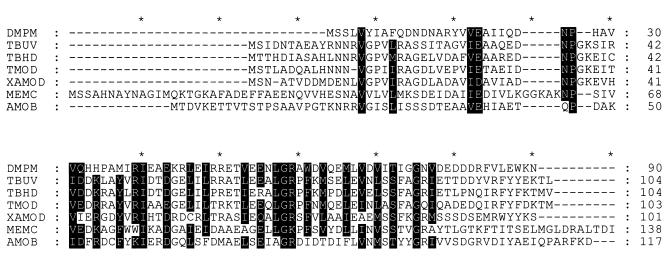
(ii) aamD.
Alignments of the predicted amino acid sequence of the aamD gene product (Fig. 5) show that it clearly falls into the aromatic ring monooxygenase family and is more distantly related to the coupling protein in MMO (not shown). The role of this small protein, which does not appear to contain any cofactors, in the catalytic cycle of monooxygenases is still unclear. Small and Ensign (29) reported that this small protein was obligately required for steady-state alkene epoxidation. In MMO the coupling protein is necessary to couple electron transfer to substrate oxidation, and it is also known to affect the regioselectivity of substrate oxidation and the redox potential of the oxygenase binuclear nonheme iron center. dmpM encodes the homologous coupling protein (P2) in the phenol hydroxylase from Pseudomonas sp. strain CF600. A solution structure for this protein has recently been determined by nuclear magnetic resonance (23), and although the structure was poorly defined in places, it is evident that this small protein has a hydrophobic cavity which could well act to bind substrate, either to deliver it to the oxygenase, or to act as a substrate-dependent regulator of electron transport. The conserved residues indicated in Fig. 5 are all either within, or at the ends of, regions of secondary structure evident from the P2 structure (23).
FIG. 5.
Homology between the putative small coupling or effector protein of Xamo (XAMOD), encoded by aamD, and the coupling or effector proteins of aromatic ring monooxygenases, MMO, and the R. rhodochrous B276 AMO. Dark highlighting indicates residues which are identical or functionally conserved in at least six sequences. DMPM, TBUV, TBHD, TMOD, MEMC, and AMOB, small coupling or effector proteins from the Pseudomonas sp. strain CF600 phenol hydroxylase (20), the R. pickettii PKO1 toluene 3-monooxygenase (4), the B. cepacia AA1 toluene 3-monooxygenase, the P. mendocina KR1 toluene 4-monooxygenase (39), the Methylosinus trichosporium OB3b MMO (5), and the R. rhodochrous B276 AMO (25), respectively.
Does Xamo function as an aromatic hydrocarbon monooxygenase?
Despite the similarity of the primary sequence of the Xamo oxygenase α-subunit to that of the aromatic monooxygenases, there are certain features which clearly link it to the R. rhodochrous B276 AMO and also to MMO. In particular, it was noted previously that at the position equivalent to Cys151 in the memA gene product, the AMO enzymes have an acid residue, whereas the aromatic hydroxylases have a Gln residue (Fig. 6). Additionally, residues 206 and 208 in the memA protein sequence are small aliphatic amino acids, whereas in all of the aromatic ring monooxygenases they are Phe’s. This is particularly significant because Glu209 in the memA-encoded gene product is one of the acidic residues which coordinate the binuclear nonheme iron center, and it is probable that the residue at position 208 (and its equivalent in homologous polypeptides) has a significant effect on the approach and orientation of the substrate. Aromatic residues at positions equivalent to positions 206 and 208 in the memA-encoded protein sequence should therefore facilitate the approach of aromatic substrates.
FIG. 6.
Homology between the putative oxygenase iron binding subunit of Xamo (XAMOA), encoded by aamA, and the iron binding subunits of aromatic ring monooxygenases, MMO, and the R. rhodochrous B276 AMO. Only the sequence around the iron binding sites is shown. Dark highlighting indicates residues which are identical or functionally conserved in at least four sequences. The glutamate and histidine ligands that coordinate the binuclear iron center (24) in MMO (and are completely conserved in the other homologs) are shown as subscripts in capital letters, and the residues referred to in the text are indicated by triangles. TMOA, BMOA, AMOB, and MEMA, iron binding subunits from the P. mendocina KR1 toluene 4-monooxygenase (39), the Pseudomonas aeruginosa JI 104 benzene monooxygenase (16), the R. rhodochrous B276 AMO (25), and the M. trichosporium OB3b MMO (5), respectively.
Despite these differences, the evident homology between all of the Xamo components and those of aromatic ring monooxygenases suggested that Xamo might hydroxylate aromatic substrates. By using propene-grown resting cells, Xanthobacter strain Py2 was shown to convert benzene to phenol and to convert toluene to a mixture of cresols and a trace of benzyl alcohol (Table 2). It should be noted that, although the rates of aromatic hydrocarbon oxidation are similar to those cited for propene oxidation, the latter was carried out at pH 9 to limit the further metabolism of epoxypropane. Propene oxidation assays measured by substrate disappearance at pH 7.2 give rates at least 5 times higher (10) than those observed here for aromatic hydrocarbon oxidation at a similar pH. Confirmation that oxidation was due to the AMO was obtained by using the Amo− mutants NZ1 and NZ2, which cannot oxidize propene but retain the ability to grow on propene oxide. Additionally, resting cells of noninduced control cultures of the wild type grown on AMS fructose were also unable to oxidize propene or the aromatic substrates. In the biotransformation of benzene, it was noticed that phenol accumulation was transient and that phenol was metabolized as soon as the benzene had been converted. By repetition of the same assays with phenol as a substrate, it was evident that phenol was also oxidized by the AMO, and thus, benzene was being oxidized consecutively by the same enzyme. However, tests with pure cresols indicated that these were not good substrates for Xamo. Therefore, in the biotransformation of toluene, the resulting cresols remained at their final concentrations once the toluene had been completely oxidized.
TABLE 2.
Whole cell propene, benzene, and toluene monooxygenase activities, and phenol hydroxylase and propene oxide degradation activities, of Xanthobacter strain Py2 and the Amo− mutants NZ1 and NZ2a
| Strain | Activityb on:
|
||||
|---|---|---|---|---|---|
| Benzene | Toluene | Phenol | Propene | Propene oxide | |
| Xanthobacter strain Py2 | 13.8 | 19.5 | 8.4 | 20.1 | 50.5 |
| NZ1 | 0 | 0 | 0 | 0 | 51.2 |
| NZ2 | 0 | 0 | 0 | 0 | 49.8 |
No activity was detected with any of the substrates with Xanthobacter strain Py2 cells grown with 0.5% (wt/vol) fructose as the sole carbon source.
Activities, in nanomoles per minute per milligram of protein, refer to the rate of product formation, except for the activities on phenol and propene oxide, which were measured as rates of substrate disappearance.
Biotransformation of a range of substrates is a common feature of iron-containing monooxygenases, but it is generally observed that the range of substrates used for growth is much more restricted, usually because of the greater inducer specificity or inability to further metabolize the hydroxylated product. Given the evidence for metabolism of propene oxide via isomerization and carboxylation, it seemed unlikely that aromatic hydrocarbons would act as growth substrates. Thus we were surprised to find that although benzene and toluene were not growth substrates (even at nontoxic concentrations), growth of Py2 was observed on 2 and 4 mM phenol, and this required a functional Xamo, as seen by the fact that the Amo− mutants failed to grow at any concentration. Cells grown on phenol were able to oxidize propene and metabolize propene oxide in resting cell assays, but at rates lower than those obtained with propene-grown cells, confirming that phenol induces both Xamo and the epoxide carboxylase. Growth on phenol presumably occurred as a result of oxidation to catechol and subsequent ring cleavage reactions; certainly Xanthobacter strain Py2 (and the Amo− mutants) can grow on catechol as a sole carbon and energy source. However, although the sequencing is not complete, we have found no evidence for genes encoding ring cleavage enzymes in the vicinity of the aamA-through-aamF cluster. At this stage we can only speculate that the broad range of inducers previously described for Xamo also includes phenol and that the resulting catechol can induce the necessary ring cleavage pathway. The ability to grow on phenol via the hybrid route may well have provided selective pressure for the evolution of inducer specificity to include phenol. Interestingly, we have been unable to detect induction with the nongrowth substrates, benzene and toluene (2 mM), in cells grown on either glucose or isopropanol, although at this stage we cannot exclude the possibility that they are weak inducers.
ACKNOWLEDGMENT
We are grateful to BBSRC for financial support to C.K.N.C.K.C. (grant 28/T07300).
REFERENCES
- 1.Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 4.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 5.Cardy D L, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 6.Chan Kwo Chion C K N, Leak D J. Purification and characterization of 2 components of epoxypropane isomerase/carboxylase from Xanthobacter Py2. Biochem J. 1996;319:499–506. doi: 10.1042/bj3190499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programmes for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrt S, Schirmer F, Hillene W. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIM8250. Mol Microbiol. 1995;18:13–20. doi: 10.1111/j.1365-2958.1995.mmi_18010013.x. [DOI] [PubMed] [Google Scholar]
- 9.Ensign S A, Hyman M R, Arp D J. Cometabolic degradation of chlorinated alkenes by alkene monooxygenase in a propylene-grown Xanthobacter strain. Appl Environ Microbiol. 1992;58:3038–3046. doi: 10.1128/aem.58.9.3038-3046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensign S A. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl Environ Microbiol. 1996;62:61–66. doi: 10.1128/aem.62.1.61-66.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feig A L, Lippard S J. Reactions of nonheme iron (II) centers with dioxygen in biology and chemistry. Chem Rev. 1994;94:759–805. [Google Scholar]
- 12.Gallagher S C, Cammack R, Dalton H. Alkene monooxygenase from Nocardia corallina B-276 is a member of the class of dinuclear iron proteins capable of stereospecific epoxygenation reactions. Eur J Biochem. 1997;247:635–641. doi: 10.1111/j.1432-1033.1997.00635.x. [DOI] [PubMed] [Google Scholar]
- 13.Higgins T P, DeMarco P, Murrell J C. Purification and molecular characterization of the electron transfer protein on methanesulfonic acid monooxygenase. J Bacteriol. 1997;179:1974–1979. doi: 10.1128/jb.179.6.1974-1979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holz R C, Small F J, Ensign S A. Proton nuclear magnetic resonance investigation on the [2Fe-2S](1-)-containing “Rieske-type” protein from Xanthobacter strain Py2. Biochemistry. 1997;36:14690–14696. doi: 10.1021/bi971831t. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol Lett. 1997;155:99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x. [DOI] [PubMed] [Google Scholar]
- 16.Kitayama A, Suzuki E, Kawakami Y, Nagamune T. Gene organization and low regiospecificity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104 J. Ferment Bioeng. 1996;82:421–425. [Google Scholar]
- 17.Masai E, Yamada A, Healy J M, Hatta T, Kimbara K, Fukuda M, Yano K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:2079–2085. doi: 10.1128/aem.61.6.2079-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura A, Dalton H. Purification and characterization of the alkene monooxygenase from Nocardia corallina B276. Biosci Biotech Biochem. 1995;59:853–859. [Google Scholar]
- 19.Ng L C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first 8 genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 20.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouchiyama N, Miyachi S, Omori T. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol. 1998;44:57–63. doi: 10.2323/jgam.44.57. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian H, Edlund U, Powlowski J, Shingler V, Sethson I. Solution structure of phenol hydroxylase protein component P2 determined by NMR spectroscopy. Biochemistry. 1997;36:495–504. doi: 10.1021/bi9619233. [DOI] [PubMed] [Google Scholar]
- 24.Rosenzweig A C, Frederick C A, Lippard S J, Nordlund P. Crystal structure of a bacterial nonheme iron hydroxylase that catalyzes the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 25.Saeki H, Furuhashi K. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J Ferment Bioeng. 1994;78:399–406. [Google Scholar]
- 26.Sato S I, Nam J-W, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.SEQSCAN Website. 26 August 1998, analysis date. [Online.] http://www.bmb.psu.edu/seqscan/matrixes/sig54pro.htm. [19 February 1999, last date accessed.]
- 27.Shanklin J, Whittle E, Fox B G. 8 Histidine-residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 28.Sligar S G, Murray R I. Cytochrome P-450cam and other bacterial P-450 enzymes. In: Ortiz de Montellano P R, editor. Cytochrome P450. New York, N.Y: Plenum Press; 1986. pp. 429–503. [Google Scholar]
- 29.Small F J, Ensign S A. Alkene monooxygenase from Xanthobacter strain Py2—purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J Biol Chem. 1997;272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 30.Stainthorpe A C, Lees V, Salmond G P C, Dalton H, Murrell J C. The methane monooxygenase gene cluster from Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 31.Swaving J, Weijers C A G M, Van Ooyen J J, de Bont J A M. Complementation of Xanthobacter Py2 mutants defective in epoxyalkane degradation; expression and nucleotide sequence of the complementing DNA fragment. Microbiology. 1995;141:477–484. doi: 10.1099/13500872-141-2-477. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Ploeg J, Janssen D B. Sequence analysis of the upstream region of dhlB, the gene encoding haloalkanoic acid dehalogenase of Xanthobacter autotrophicus GJ10. Biodegradation. 1995;6:257–263. doi: 10.1007/BF00700465. [DOI] [PubMed] [Google Scholar]
- 34.van Ginkel C G, de Bont J A M. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch Microbiol. 1986;145:403–407. [Google Scholar]
- 35.van Ginkel C G, Welten H G J, de Bont J A M. Oxidation of gaseous and volatile hydrocarbons by selected alkene-utilizing bacteria. Appl Environ Microbiol. 1987;53:2903–2907. doi: 10.1128/aem.53.12.2903-2907.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weijers C A G M, Jongejan H, Franssen M C R, de Groot A, de Bont J A M. Dithiol and NAD-dependent degradation of epoxyalkanes by Xanthobacter Py2. Appl Microbiol Biotechnol. 1995;43:775–781. [Google Scholar]
- 37.Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation and some properties of methane utilising bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 38.Wiegel J W, Schlegel H G. Genus Xanthobacter. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams and Wilkins Co.; 1984. pp. 325–333. [Google Scholar]
- 39.Yen K-M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen K-M, Karl M R. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1992;174:7253–7261. doi: 10.1128/jb.174.22.7253-7261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou N-Y, Chan Kwo Chion C K N, Leak D J. Cloning and expression of the genes encoding the propene monooxygenase from Xanthobacter Py2. Appl Microbiol Biotechnol. 1996;44:582–588. [Google Scholar]
- 42.Zhou N-Y, Jenkins A, Chan Kwo Chion C K N, Leak D J. The alkene monooxygenase from Xanthobacter Py2 is a binuclear non-haem iron protein closely related to toluene 4-monooxygenase. FEBS Lett. 1998;430:181–185. doi: 10.1016/s0014-5793(98)00653-x. [DOI] [PubMed] [Google Scholar]