Abstract
During routine genome duplication, many potential replication origins remain inactive or ‘dormant’. Such origin dormancy is achieved, in part, by an interaction with the metabolic sensor SIRT1 deacetylase. We report here that dormant origins are a group of consistent, pre-determined genomic sequences that are distinguished from baseline (i.e. ordinarily active) origins by their preferential association with two phospho-isoforms of the helicase component MCM2. During normal unperturbed cell growth, baseline origins, but not dormant origins, associate with a form of MCM2 that is phosphorylated by DBF4-dependent kinase (DDK) on serine 139 (pS139-MCM2). This association facilitates the initiation of DNA replication from baseline origins. Concomitantly, SIRT1 inhibits Ataxia Telangiectasia and Rad3-related (ATR)-kinase-mediated phosphorylation of MCM2 on serine 108 (pS108-MCM2) by deacetylating the ATR-interacting protein DNA topoisomerase II binding protein 1 (TOPBP1), thereby preventing ATR recruitment to chromatin. In cells devoid of SIRT1 activity, or challenged by replication stress, this inhibition is circumvented, enabling ATR-mediated S108-MCM2 phosphorylation. In turn, pS108-MCM2 enables DDK-mediated phosphorylation on S139-MCM2 and facilitates replication initiation at dormant origins. These observations suggest that replication origin dormancy and activation are regulated by distinct post-translational MCM modifications that reflect a balance between SIRT1 activity and ATR signaling.
INTRODUCTION
Mammalian cells initiate the duplication of their genomes at a series of pre-licensed replication origins (1–3). Origin licensing starts as soon as cells emerge from mitosis and is completed in the early G1 phase of the cell cycle. Licensing entails the assembly of an ORC1-6 protein complex on replication origins, which in turn engages the CDC6 and CDT1 clamps to recruit two anti-parallel MCM2-7 hexamers, forming a pre-replication complex (pre-RC) (1–3). The MCM2-7 complex is initially recruited to replication origins in an inactive configuration, which can be activated to form a complete CMG (MCM-CDC45-GINS) helicase that initiates DNA replication upon entry into S-phase. During unperturbed cell cycle progression, only about 10% of all potential, licensed replication origins initiate DNA replication (we henceforth refer to origins that are activated during unperturbed growth as baseline origins). Baseline origins can be activated each cell cycle (constitutive) or intermittently in a proliferating cell population (flexible), whereas the rest of the potential origins remain inactive (dormant origins). Dormant origins are activated only under specific growth conditions such as during differentiation or in response to endogenous or exogenous stress (2,4–7). A role for the excess licensing of dormant origins is evident when cells encounter events that stall or slow the progression of DNA synthesis (‘replication stress’), which can be triggered by DNA lesions, secondary DNA structures, depletion of dNTP pools or DNA polymerase inhibition. When replication forks stall or their progression is delayed, adjacent dormant origins can initiate replication, facilitating complete genome duplication (6,8–12).
Although dormant origins are crucial for genome stability under conditions of replicative stress, activation of dormant origins during unperturbed growth is deleterious, and specific signaling pathways are in place to maintain origin dormancy (13). The tight regulation of origin activity is evident from single-fiber analyses of DNA replication, which demonstrate that dormant replication origins are activated when DNA replication forks slow, suggesting that decreases in fork speed are compensated by an increase in the number of active origins (4,6,11). The stochasticity and inherent inefficiency of baseline replication initiation events (14,15) could support a model for a passive activation of dormant origins, whereby slow replication would increase the probability of initiation from infrequent origins that are normally passively replicated by adjacent origins. However, recent observations suggest that dormant origins are also modulated by active mechanisms (9,16,17).
We have previously shown that dormant origins are activated in cells in which the NAD+-dependent deacetylase SIRT1 is depleted and in cells containing a form of SIRT1 harboring a mutation preventing its phosphorylation on threonine residue 530, thus inhibiting its protein deacetylase activity (18). These observations suggest that the phosphorylated, active form of SIRT1 prevents excess initiation at dormant origins. SIRT1 is responsible for the deacetylation of a myriad of histone and nonhistone proteins and is widely recognized as a key factor involved in many cellular and biological processes, including but not restricted to aging, metabolism, genome stability, reprogramming, tumorigenesis, transcription and DNA replication (reviewed in (18,19)). As it is responsible for the deacetylation of histones H1, H3 and H4 and for increasing the prevalence of repressed chromatin marks H3K9me3 and H4K20me3, SIRT1 is key to modulating chromatin structure (20). Our previous work demonstrated that acetyl H4K16, an important target of SIRT1, binds both active and dormant replication origins (18). However, it is reasonable to think that SIRT1 may have broader effects on replication since several chromatin marks, some of which are directly or indirectly linked to SIRT1 activity, were found to associate with different subsets of origins (i.e. early versus late origins) (21).
Sir2, the yeast SIRT1 ortholog, is involved in the distribution of the MCM2-7 helicase on ribosomal DNA sequences, with Sir2 activity on MCM2 reducing the frequency of replication initiation (22). The MCM2-7 helicase may play a role in dormant origin activation, as reduced levels of individual MCM2-7 subunits decrease the activation of dormant origins (23–25). Other key factors involved in dormant origin activation are the DNA damage response kinases ATR and CHK1 (10). ATR is activated upon replication stress and phosphorylates CHK1 to inhibit replication initiation at distal origins while allowing dormant origins to initiate DNA synthesis and resume replication at loci that are experiencing replicative stress (16,17), presumably within the vicinity of stalled forks (9). The underlying mechanism for this phenomenon is not yet known, but one possibility is that ATR signaling converges with SIRT1 signaling, resulting in differential activation of baseline versus dormant origins.
Here, we mapped replication initiation events at baseline and dormant origins in SIRT1-proficient and depleted cells and investigated the interdependence between SIRT1, ATR and MCM helicase subunits in regulating replication dynamics. Under unperturbed conditions, we observed that both baseline and dormant replication origins exhibit DBF4-dependent kinase (DDK)-mediated phosphorylation of serine residue 139 on MCM2 (S139-MCM2), but unlike baseline origins, which can be directly activated by DDK, dormant origins require an additional, ATR-mediated phosphorylation of S108-MCM2 for initiation. ATR recruitment to chromatin and the subsequent dormant origin activation is normally inhibited by SIRT1-mediated deacetylation of the ATR-interacting protein TOPBP1. However, when the inhibition is relieved and ATR is recruited to chromatin (e.g. in cells undergoing replication stress or in cells with mutated SIRT1 that cannot deacetylate TOPBP1), ATR-mediated phosphorylation on S108-MCM2 at dormant origins enhances S139-MCM2 phosphorylation and enables initiation. This interplay between the SIRT1 and ATR pathways modulates replication initiation levels by maintaining origin dormancy during normal growth and facilitating dormant origin activation when replication is perturbed, thus maintaining genomic stability.
MATERIALS AND METHODS
Cell culture, chemicals and establishment of stable cell lines
Human HCT116, HCT116 with MCM2-mAID (26) (a gift from Dr Andre Nussenzweig and Dr Masato Kanemaki; mAID refers to mini auxin-inducible degron), HEK293 and U2OS cells were grown at 37°C in 5% CO2 atmosphere in RPMI medium (Themo Fisher, 11875–119), supplemented with 10% FBS. All original cancer cell lines were obtained from ATCC (www.atcc.org), and all cell lines were tested as negative for mycoplasmas (Lonza, LT07–418). To evaluate SIRT1 activity, cells were maintained in Trichostatin A (10 nM, SIGMA T8552) for 24 h prior to harvesting in order to block HDAC class I and II. For details see Supplementary Data.
Flow cytometry
Cells were pulse-labeled with 10 μM EdU for 20 min before harvest. EdU staining was performed using the Click-iT EdU kit (ThermoFisher, C10634 (for AL647) or C10633 (AL488)) according to the manufacturer’s protocol. For immunodetection of chromatin-bound phospho-MCM2 (pS139) (Cell Signaling, 12958) and phospho-MCM2 (pS108) (Bethyl Labs A300-094A) were performed using pre-extracted cells as reported earlier (27). A BD LSR Fortessa cell analyzer with FACSDiva software and/or FlowJo10.6 was used for cell cycle analysis.
Chromatin fractionation, co-immunoprecipitation, mass spectrometry (MS) and immunoblotting
For measurements of chromatin binding, 5 × 106 harvested cells were incubated in cytosolic extraction buffer (10 μM HEPES, pH 7.9; 10 μM KCl; 1 μM EGTA; 0.25% NP40; 1X protease inhibitor cocktail and phosphatase inhibitor cocktail) for 10 min on ice. Nuclei were collected using centrifugation at 2700 × g for 5 min at 4°C and washed twice with a cytosolic extraction buffer (without NP40) and resuspended in cold nuclear extraction buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 20% [v/v] glycerol, 1 mM EDTA, 1 μg/ml RNase A and 20 U/ml DNase, 1× proteinase inhibitor cocktail and phosphatase inhibitor cocktail) and incubated on ice for 5 min and chromatin fractions were collected by centrifugation at 5000 × g for 5 min at 4°C. Chromatin-bound fractions were immunoprecipitated (co-IP) using specific antibodies. For evaluating protein interactors of SIRT1 and MCM2, chromatin-bound fraction was co-IPed using antibody against pT530-SIRT1 (18) or MCM2 (Cell Signaling, cat#12079), and mass spectrometry (MS) was performed as reported earlier (28) (see Supplementary Methods). Obtained hits were grouped based on gene-ontology molecular function using GSEA. For immunoblots, chromatin-bound fraction lysates were mixed with 200 μl 1 × SDS loading buffer per 1 million cells. Samples were heated at 100°C for 5 min, centrifuged and the supernatants used for immunoblots. For details on antibodies used, see Supplementary Methods.
DNA replication analysis by molecular combing
Analysis of DNA replication by molecular combing was performed as previously described (29), for details please see supplementary methods. Briefly, asynchronous cells were sequentially labelled with 20 μM CIdU for 30 min and 20 μM ldU for 30 min, then chased with 100 μM thymidine for 60 min. DNA fibers were stretched onto salinized coverslips (Genomic Vision, cov-002-RUO) as previously described (29) using an in-house DNA combing machine. Slides were scanned with a FiberVision Automated Scanner (Genomic Vision). Replication signals on single DNA fibers were analyzed using FiberStudio (Genomic Vision) and ImageJ. Median fiber lengths were measured and recorded for all slides to control for equal fiber stretching. Only replication signals from high-quality ssDNA (signals from long fibers, omitting replication signals from DNA bundles or signals located at the ends of DNA strands) were selected for analyses. Experiments were performed at least in duplicate using independent biological isolations of DNA fibers for each experimental condition. The statistical analyses were performed using Prism 8 (GraphPad software) and the non-parametric Mann–Whitney rank sum test.
Nascent strand DNA sequencing (NS-seq)
HCT116 cells (harboring WT, KO, or H363Y SIRT1 or WT-MCM2, KO-MCM2 or mutant-MCM2) without or with (aphidicolin or IAA) treatment were harvested. Genomic DNA was purified, and nascent strands were isolated as described previously (27,30,31). Briefly, DNA was denatured by boiling for 10 min, immediately cooled on ice and fractionated on a neutral sucrose gradient. Fragments of 0.5–2 kb (containing nascent strand DNA and broken genomic DNA) were collected and treated with lambda exonuclease (λexo) to remove non-RNA-primed broken genomic DNA. Because short DNA fragments were enriched for origin-proximal nascent DNA due to size fractionation (Supplementary Figure S2A; see Supplementary Methods for details), we isolated high molecular weight genomic DNA from the same gradient as a control for peak calling. Sonicated, RNase treated and λexo digested high molecular DNA served as a standard control for λexo biases as described (7,21,32). Single-stranded nascent DNA was converted to double-stranded DNA using the BioPrime DNA Labeling System (ThermoFisher, 18094011). Double-stranded nascent DNA (1 μg) was sequenced using the Illumina genome analyzer II (Solexa). Peak calling was performed as described (27) (for details, see Supplementary Methods).
Chromatin immunoprecipitation and sequencing (ChIP-seq)
Chromatin immunoprecipitations were performed as described in Supplementary Methods. For phospho-SIRT1 (T530) ChIP-seq, we used asynchronous WT-, KO- and H363Y- HCT116 cells as described earlier (18). For antibodies targeted against specific phospho-MCM2 targets, we used HCT116 cells that were synchronized at the G1/S boundary (using a double-thymidine block: 2.5 mM thymidine for 18 h followed by a release into fresh media for 9 h and 2.5 mM thymidine for 16 h). Each antibody was tested for target recognition in formaldehyde-fixed and native chromatin extracts using co-immunoprecipitation followed by immunoblotting, and then subjected to ChIP-qPCR validation using origin proximal and distal primer-probe combinations. For ChIP-Seq, sequencing libraries (each from 10 ng of ChIP samples) were prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7805S) and NEBNext Multiplex Oligos for Illumina (Index Primers Set 1 and Set 2) (NEB, E7335S and E7500S). Sequencing was done by Illumina NextSeq 75 cycle High Output kit. Peak calling was performed as described (27) (for details, see Supplementary Methods).
RESULTS
Cells with inactive SIRT1 constitutively activate dormant origins
Our previous studies have demonstrated that a form of SIRT1 with DYRK2-dependent phosphorylation at threonine residue 530 (pT530) binds replication origins and prevents excessive initiation during unperturbed cell cycle progression (18). These results align with the observation that a phosphorylation of the murine ortholog of T530 (T522) facilitates genomic stability in mice (33). To determine directly whether SIRT1 enzymatic activity is a prerequisite for preventing dormant origin initiation, we characterized the phosphorylation status, cell cycle progression and replication initiation frequency in cells harboring a point mutant of SIRT1 (H363Y) that lacks deacetylase activity without significant changes in its structure (34,35). H363Y mutant SIRT1 could be phosphorylated at T530 and showed similar chromatin binding to that of wild-type (WT) SIRT1 (Supplementary Figure S1A). SIRT1-knockout cells (KO cells) or KO cells complemented with the inactive SIRT1 mutant (H363Y cells) showed similar overall growth rates (Supplementary Figure S1B–D). Because dormant origins are activated in the absence of SIRT1, these cell lines provide a unique isogenic model to study the characteristics of dormant origins and molecular cues for origin dormancy.
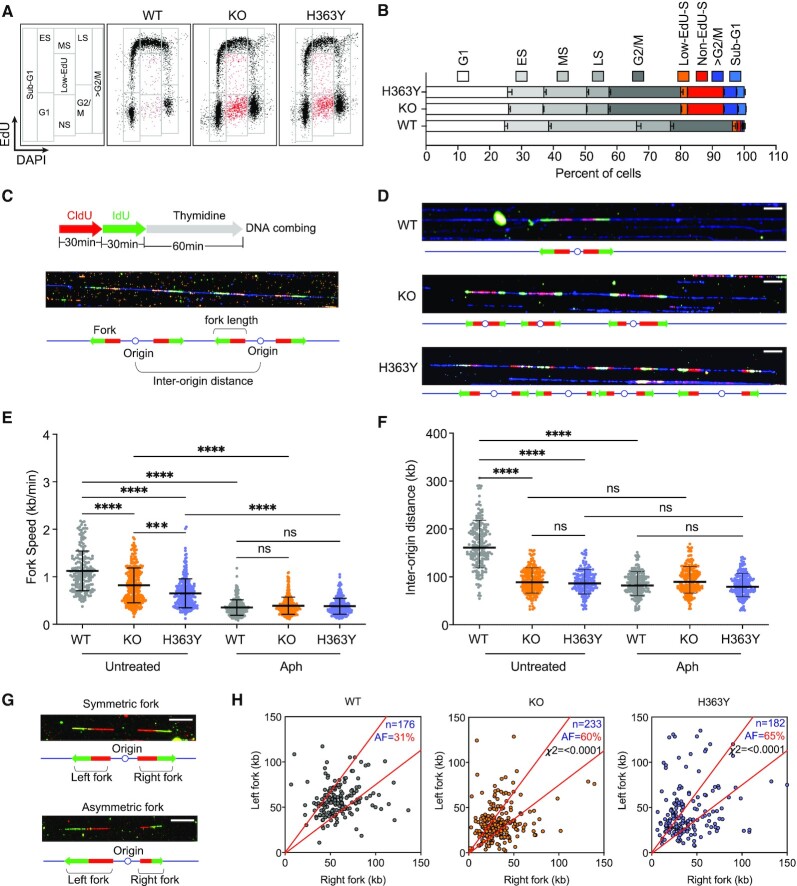
As shown in Figure 1A,B, in KO and H363Y cells, we observed a cell population with skewed S-phase progression, presented as low- or no-EdU incorporating cells with S-phase DNA content (Low-EdU-S and no-EdU-S, hence termed Low-EdU-S, labeled as red dots in Figure 1A). WT, KO and H363Y cell populations showed similar G1 or G2/M fractions (Figure 1B), but cell populations devoid of SIRT1 activity exhibited a high prevalence of low-EdU-S (Supplementary Figure S1E: KO and H363Y cells exhibited 24% and 25%, respectively, compared to 7.2% in WT). The low-EdU-S populations showed increased levels of γ-H2Ax (Supplementary Figure S1F) indicating elevated replication stress and DNA breakage in the absence of functional SIRT1.
Figure 1.
SIRT1 deacetylase activity facilitates normal S-phase progression and origin dormancy. (A) SIRT1 knock-out (KO) HCT116 cells were reconstituted with either wild-type (WT) or mutant (H363Y, lacks deacetylase activity) SIRT1. S-phase progression in HCT116 cells harboring WT, KO or H363Y SIRT1 was measured by flow cytometry using a standard EdU incorporation/DAPI assay. Cell cycle phases were gated as shown in the far-left panel and representative cell cycle profiles from each cell line are shown (A). ES-early S, MS-mid S, LS-late S and NS- non-EdU + low EdU (red dots). (B) A bar plot showing the percentage of WT, KO and H363Y cells in G1, S (ES, MS, LS) and G2/M phases, along with cells exhibiting skewed replication (low-EdU, non-EdU), >G2/M and sub-G1 phases. The data were obtained from four replicates. (C) Schematic for the experimental strategy measuring replication profile changes using DNA combing. (D) Representative images shown DNA combing images of fiber samples from WT, KO and H363Y cells without any treatment (white scale bar = 50 kb). (E and F) Replication fork progression rates (E) and replication inter-origin distances (F) in untreated WT, KO and H363Y cells in the presence or absence of Aphidicolin (Aph). Aph was added (0.8 μM) 24 h prior to CldU. The Mann–Whitney test was used to determine statistical significance. ns = non-significant, *** represents P< 0.001 and **** represents P< 0.0001. (G) Representative DNA-combing image of symmetric and asymmetric replication forks on a single DNA fiber (white scale bar = 50 kb). (H) Inactive SIRT1 (KO or H363Y) increases the frequency of asymmetric replication forks in HCT116 cells. DNA-combing analysis was performed as in Figure 1C. Lengths of replicating signals from left and right forks emanating from the same origins were compared, and replication forks were classified as asymmetric if the difference in length between the right and left fork was >30%. n = total number of forks, AF = the percentage of asymmetric forks. Chi-square test was used to determine the statistical significance between results for WT versus KO and WT versus H363Y.
In WT cells, SIRT1-specific deacetylase activity on chromatin decreased during S-phase whereas, as expected, we did not detect any deacetylase activity in both KO cells and H363Y cells (Supplementary Figure S1G,H). Because the levels of free NAD+ (a co-factor/substrate for SIRT1 activity) also decreased during S-phase, the reduced SIRT1 activity on chromatin might reflect the cellular metabolic state (Supplementary Figure S1I). The skewed replication dynamics in cells with inactive SIRT1 (KO and H363Y cells) observed in Figure 1A and B was detected despite the presence of higher deacetylase activities of other, zinc-dependent HDACs (Supplementary Figure S1J).
Since phosphorylation of T530 on SIRT1 is required for SIRT1 activity, whereas loss of SIRT1 activity does not affect phosphorylation, we asked if SIRT1 deacetylase activity was required to maintain unperturbed DNA synthesis during normal cell cycle progression. To that end, we sequentially incorporated CldU followed by IdU (each analog for 30 min) in WT, KO, and H363Y cells and analyzed replication dynamics using single fiber DNA combing (Figure 1C) with or without exposure to aphidicolin. Aphidicolin is an inhibitor of DNA polymerase that slows replication and activates stress-induced replication origins (18,36,37). We detected CldU (red) and IdU (green) on combed, isolated DNA and used the CldU and IdU tracks to measure replication fork speed and inter-origin distances for replication initiation frequencies (29). In concordance with the EdU incorporation profiles observed in Figure 1A, in untreated cells both KO and H363Y cells demonstrated slower median replication speeds compared to WT cells (Figure 1D–E). Although DNA fibers from the KO and H363Y cells exhibited similar length distributions as DNA fibers isolated from WT cells (Supplementary Figure S1K and L), fibers from KO and H363Y cells exhibited significantly decreased inter-origin distances (Figure 1D and F), indicating dormant origin activation. Initiation from dormant origins in the absence of SIRT1 activity was also observed in the other cell lines HEK293 and U2OS (Supplementary Figure S1M).
Interestingly, the distances between origins further decreased upon aphidicolin treatment in WT cells but not in KO and H363Y cells (Figure 1F), suggesting that aphidicolin-induced dormant origins were persistently activated in cells that lacked SIRT1 activity. Additionally, we observed an escalation in replication fork asymmetry rates in KO (60%) and H363Y (65%) cells compared to WT (31%) (Figure 1G and H), indicating unequal bidirectional fork progression and fork stalling. Combined with our flow cytometry analyses (Figure 1A), which measured normal EdU incorporation in the majority of S-phase cells in KO and H363Y cells, these results imply that in most cells devoid of SIRT1 activity the increased frequency of replication initiation could balance slow replication fork progression to facilitate the timely completion of S-phase. However, the increased prevalence of slowed or stalled replication fork progression in KO and H363Y cells (Figure 1H) also coincided with a marked increase in the fraction of low-EdU-S cells in those cells (Figure 1A). To test if cells devoid of SIRT1 activity with skewed replication patterns were likelier to sustain DNA damage, we have performed flow cytometry analysis with cells concomitantly labeled for EdU and γ-H2AX. As shown in Supplementary Figure S1F, low-EdU-S cell populations in KO and H363Y cells were predominantly represented in the population that exhibited elevated levels of γ-H2AX. These results were consistent with the marked increase in the fraction of sub-G1 cell populations in KO and H363Y (Figure 1B), suggesting that cells with skewed progression during S-phase can incur DNA damage and subsequently be eliminated from the proliferating cell population. These observations are in line with our previously reported finding that active SIRT1 promotes replication origin dormancy, suppresses replication stress and safeguards genome stability (18), and further imply that origin dormancy requires the activity of SIRT1 and not merely its presence on chromatin.
Dormant replication origins are located at consistent chromosomal sites
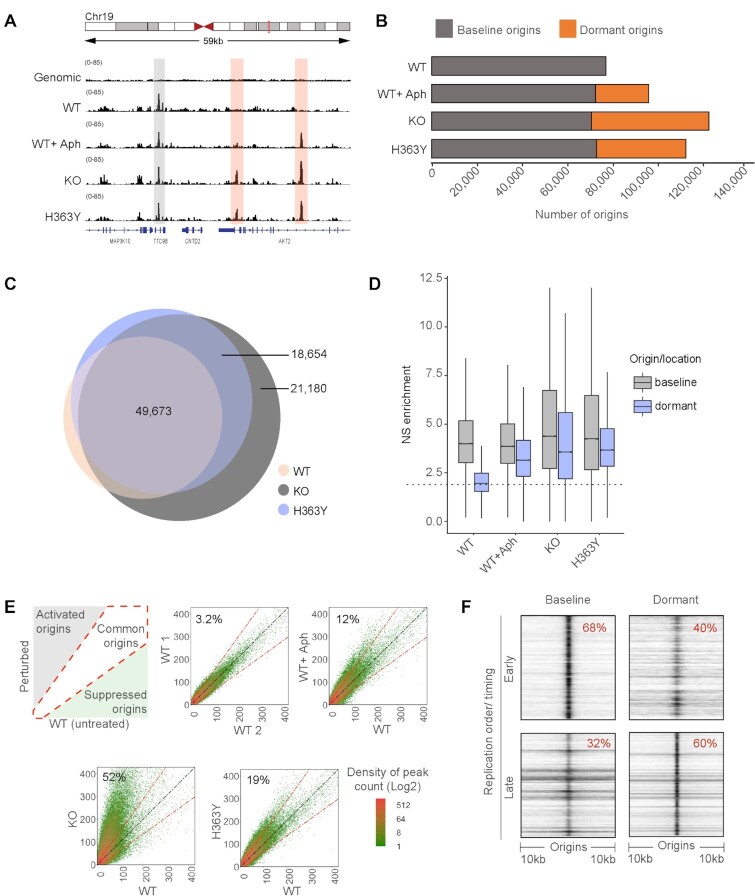
We next asked if both SIRT1-repressed dormant origins and stress-induced origins are derived from the same pool of potential initiation sites or represent two distinct genomic entities. We mapped replication initiation sites in cells that differed in SIRT1 status using nascent strand (NS-seq) sequencing (30,31). For replication origin peak calling, we used both sonicated genomic DNA and RNase and λexo digested high molecular weight DNA (Supplementary Figure S2A–C) to control for potential exonuclease biases (7,21,32). As shown in Figure 2A and Supplementary Figure S2A, KO and H363Y cells contained replication origins that were not detected in WT cells (‘dormant origins’, red shaded) in addition to origins that were active in all cells (‘baseline origins’, gray-shaded). These additional peaks were detected when peaks were called either against sonicated genomic DNA or against RNase and λexo digested high molecular weight DNA (Figure 2B,C and Supplementary Figure S2C). These dynamic changes suggest that the peak distribution we observed reflected the activation of replication origins and was not simply due to the in vitro resistance of purified DNA to λexo digestion. As shown in Figure 2B,C, 49,673 (90%) of origins detected in WT cells also initiated replication in KO and H363Y cells, but KO, H363Y and aphidicolin-treated WT cells also activated an additional group of dormant origins (Figure 2B,C; Supplementary Figure S2C).
Figure 2.
A similar pool of dormant origins is activated in SIRT1 deficient cells and in cells undergoing replication stress. (A) IGV (Integrative genomics viewer) (67) screenshot of active origins mapped by NS-seq in WT, KO and H363Y HCT116 cells. Origins activated in WT are referred to as baseline origins whereas origins that are additionally activated in KO and H363Y but not in WT are referred to as dormant origins. Gray and red highlights show the genomic locations of baseline and dormant origins, respectively. (B) Bar plot representing the comparison between (active) baseline and dormant origins obtained in the HCT116 SIRT1 model system (WT, WT + Aph, KO and H363Y, and replicates of each sample). Here ‘baseline’ origins refer to origins initiated in untreated WT and ‘dormant’ origins were those with auxiliary origin initiation relative to that of baseline origins in respective cells/conditions. (C) Overlaps among replication origins in WT, KO and H363Y cells. Overlaps were calculated using ‘Intervene Venn 3-way’ comparison module and plotted using ‘Eulerr’. Numbers in the Venn diagram represent the number of origins in each group. (D) Box-whisker plot comparing replication initiation activity among baseline and dormant origins. Nascent strand (NS) enrichment was calculated using BAMScale as a function of library size reads per coordinates (origins) for respective NS-seq/ genomic control. The dotted line represents median NS enrichment on random genomic intervals (generated using ‘bedtools random’ module). (E) The left upper panel is a schematic of density plots comparing origins usage in treated/perturbed WT cells compared to unperturbed WT cells. The diagonal white area represents origins that initiate equally in both treated and untreated WT cells. The gray-shaded area indicates origins activated upon perturbation (1.5-fold higher origin initiation activity compared to untreated WT), whereas the green-shaded area represents origins that are suppressed upon perturbation. From left to right, top to bottom: density plot illustrations for origin initiation between two replicates of untreated WT; WT without versus WT with aphidicolin treatment (WT + Aph, 0.8 μM for 24 h); WT versus KO cells; and WT versus H363Y cells. Percentages represent dormant origin initiation in respective (Y-axis) samples. (F) A heatmap generated using ColoWeb (42) representing the association between genomic locations of early and late-replicating regions with baseline (WT) or dormant (from KO) origins in HCT116 cells.
Because the increase in replication initiation frequency was evident using two orthogonal methods (single-fiber analyses and SNS-seq), thus lending confidence in the sequencing approach, we next asked if baseline and dormant origins represent consistent, mutually exclusive groups of DNA sequences. We measured the fraction of overlap between total and dormant origins from each NS-seq replicate and two sets of random locations (Supplementary Figure S2D, black dotted box) and compared replication activity among baseline and dormant origins in WT, KO and H363Y cells (Figure 2D). In untreated cells, we observed similar median baseline origin activities in WT, KO, and H363Y with higher variance in KO and H363Y cells. Aphidicolin treatment partially reduced baseline origin activity but the absence of SIRT1 activity did not affect initiation at baseline origins. As expected, dormant origin locations showed minimal activity in WT and higher median activity in WT + Aph, KO and H363Y cells.
These observations suggest that KO or H363Y-specific dormant origins mapped to the same genomic sites as the ‘backup’ dormant origins known to be activated by replication stress induced by Aph (red highlighted peaks in Figure 2A and Supplementary Figure S2D, red solid squares). Reciprocal comparisons among dormant origins from KO and WT + Aph or H363Y showed a partial overlap (Supplementary Figure S2D, black solid line squares). A direct comparison between dormant origins activated in KO cells with Aph-treated WT cells (WT + Aph) or H363Y demonstrated that origins activated in both WT + Aph and H363Y consisted of a subset of the dormant origins that initiated replication in KO cells (Supplementary Figure S2E, top and middle panel). Origins activated in H363Y and WT + Aph cells showed partial overlap (Supplementary Figure S2E, lower panel).
To determine whether the excess initiation at KO and H363Y cells reflected a higher frequency of initiation or indicated infrequent initiation at multiple ‘weak’ origins, we used the BAMScale tool (38), which compares sequencing read density among next-generation sequencing (NGS) peaks. We measured the extent of initiation at each origin sequence by calculating the ratio of normalized sequencing reads per peak at each origin (Figure 2E). As represented in the upper left panel of Figure 2E, these peak-density plots compare the abundance of origins commonly activated in two samples (present on the diagonal slope, white shade) and sample-specific origins that are preferentially active in one of the samples (>1.5-fold deviation from the slope, gray and green shades). As a validation, we first compared two sets of replication origin peaks in two untreated replicates derived from WT cells, detecting only a 3.2% variation between the two replicates (Figure 2E, upper middle panel). In contrast, exposure to aphidicolin activated 12% additional origins in the Aph treated sample, corresponding to dormant origins (Figure 2E, upper right panel). Similarly, when we compared origin activity in WT cells with origin activity in KO and H363Y cells, we observed 52% and 19% dormant origin activation in samples derived from KO and H363Y cells, respectively (Figure 2E, lower panels).
NS-seq data also showed a reduction in median inter-origin distances (Supplementary Figure S2F), in concordance with the DNA-combing observations. We measured median inter-origin distances of 60 kb in WT cells; this was reduced to 47, 33 and 45 kb in WT + Aph, KO and H363Y cells, respectively. This observation supports the hypothesis that dormant origins are present within clusters of baseline origins and can recover stalled replication. It should be noted that, because NS-seq is a population-based technique that captures all initiation sites utilized in a cell population (2,5,27) whereas DNA combing measures distances in single fibers, we observed that NS-seq inter-origin distances were shorter (∼27–65 kb) than the distances measured by DNA-combing (∼135–170 kb, as shown in Figure 1F and Supplementary Figure S1M). These observations are consistent with the patterns of replication initiation events in mammalian chromosomes, which can each start replication stochastically from one of several licensed origins (2,5).
We noticed that baseline and dormant origins also differed in their replication timing. Only 32% of baseline origins were associated with late-replicating regions whereas 60% of dormant origins were associated with late-replicating regions (Figure 2F). This observation suggests that dormant origins might be associated preferentially with heterochromatin. Hence, it is clear that the dormant origins, which are maintained by SIRT1 and triggered to initiate replication either by SIRT1 inhibition or by replication stress, require a distinct molecular cue for their activation.
SIRT1 interacts with MCM complexes
To uncover the differential molecular cue(s) that distinguish(es) dormant origins from baseline origins, we first compared the reported binding sites of ORC1 (39), MCM7 (40) and CDT1 (27) (components of pre-RC) with baseline and dormant origins. We observed that baseline origins colocalized with 20%, 40% and 54% of the binding sites for ORC1, MCM7 and CDT1 respectively (Supplementary Figure S3A), and likewise, dormant origins also showed partial overlaps, with 25%, 50% and 30% colocalizations with these sites. This indicates that ORC1, MCM7 and CDT1 are all likely associated with potential origin sites; however, variations in the binding of pre-RC components do not sufficiently differentiate between baseline versus dormant origins to account for the observed differences in initiation patterns.
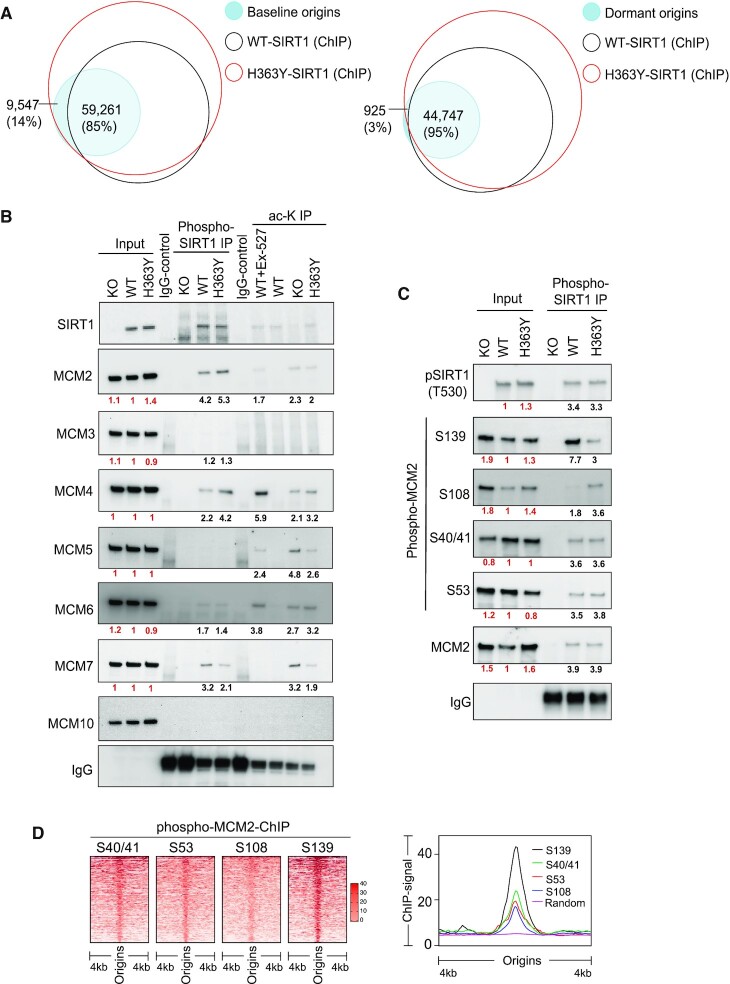
Next, we mapped the DNA-binding sites (Figure 3A, empty circles) of SIRT1 phosphorylated at T530 (pT530-SIRT1), in cells harboring both wild-type (WT) and mutant (H363Y) SIRT1. We then determined the extent of overlap between the pT530-SIRT1 binding sites and baseline and dormant origins (Figure 3A, filled circles). For WT-SIRT1, there was an 85% overlap between active SIRT1 binding sites and baseline origins, whereas mutant SIRT1 (H363Y) was associated with all baseline origins (Figure 3A, left panel). Both WT and H363Y SIRT1 were also associated with 95% of dormant origins (Figure 3A, right panel), consistent with our previous finding (18). The association of WT- or H363Y- SIRT1 with baseline or dormant origins was not a random occurrence (Supplementary Figure S3B).
Figure 3.
SIRT1 activity modulates chromatin binding of MCM helicases. (A) SIRT1-binding sites (empty circles), obtained from WT and H363Y HCT116 cells, aligned with baseline (all WT origins, left panel) or dormant (KO, right panel) origins (filled circles). Dormant origins were defined as origins that did not initiate replication in untreated WT cells but initiated replication in SIRT1 KO cells. Dormant origins from KO cells were used for the analysis this group overlaps with dormant origins from both Aph treated WT and H363Y cells. Three-way comparisons were calculated and represented as in Figure 2C. Numbers and percentages represent the overlap between origins and chromosomal sites shared with ChIP-seq peaks (pSIRT1). (B) Validation of pT530 SIRT1 immunoprecipitation/mass spectrometry observations. pT530 SIRT1 was immunoprecipitated from chromatin extracts of WT, KO and H363Y cells, and immunoblots were probed with the indicated antibodies targeting components of the MCM helicase. Extracts from KO served as negative controls. About 5% of input was added as a control. The four right lanes evaluate the extent of SIRT1-specific deacetylation by measuring the acetylation (ac-K-IP) of MCM-helicase components from WT, KO and H363Y cells. Cells were pretreated with 5 nM trichostatin A (24 h) to block zinc-dependent HDACs, and chromatin immunoprecipitation was performed using an antibody against acetyl-lysine. Immunoprecipitated protein blots were probed for components of MCM helicases. Ex-527 treatment was used as SIRT1 inhibitor (positive control) in WT cells. The numbers under each blot represent intensity ratios for each protein or IP, normalized to WT (red font) or against the immunoblot background in the IgG control lane (black font). For quantification of replicate blots see supplementary Figure 4C,D. (C) Immunoblot analysis of pT530 SIRT1 immunoprecipitates from the chromatin extracts of WT, KO and H363Y cells. Immunoprecipitates were probed for specific serine phosphorylation sites of MCM2. About 5% of chromatin extract was used as input and chromatin extracts from SIRT1 KO were used as negative controls. The numbers under each blot represent intensity ratios for each protein or IP normalized to WT (red font) and blot background from IgG controls (black font). For quantification of replicate blots see Supplementary Figure S5A. (D) A heatmap (left) and a line-plot (right) representing the occupancies of four phosphoisoforms of MCM2 (S40/41, S53, S108 and S139, mapped by ChIP-Seq in WT SIRT1 cells synchronized at G1/S phase) at replication origin sites. Using an R-script (heatmapper2 (38), see Materials and methods), coverage for each MCM2 phosphoisoform was calculated and 5000 randomly selected genomic coordinates for origins were represented as in the heatmap.
Because SIRT1 binds most replication origins and is required for origin dormancy, we hypothesized that substrate(s) of SIRT1 could facilitate the differential initiation of DNA replication. To identify SIRT1 substrates involved in the differential regulation of baseline versus dormant origins, we first performed a GIGGLE (41) search and a colocalization analysis using ColoWeb (42) to identify histone marks that differentially colocalize with baseline and dormant origins. We found that open chromatin marks tended to associate preferentially with baseline origins, whereas H3K9me3, a marker for heterochromatin, showed a limited colocalization with dormant origins (Supplementary Figure S3C–F). This is in line with our previous observation (Figure 2F) that dormant origins are preferentially present in late replicating regions of the genome.
Next, we performed SIRT1 chromatin interactome analyses using mass-spectrometry (co-IP-MS) in WT and H363Y cells. Co-IP-MS of pT530 SIRT1 from chromatin extracts obtained from both WT and H363Y cells identified 208 protein targets, which were further classified based on gene function ontology using GSEA (https://www.gsea-msigdb.org/gsea/index.jsp) (Supplementary Figure S4A). This analysis identified MCM proteins (GO cell cycle processes), which are key components of pre-replication complexes, as binding partners for SIRT1 on chromatin. Interestingly, MCMs chromatin binding is modulated by the yeast SIRT1 homolog Sir2 (22,43). Mass spectrometry of MCM2 immunoprecipitates from chromatin extracts of WT, KO an H363Y cells validated the interactions between MCM2 proteins and SIRT1 in both WT and H363Y cells (Supplementary Figure S4B). Co-immunoprecipitation of pT530-SIRT1 from chromatin extracts probed for MCM proteins detected interactions of pT530-SIRT1 with MCM2, MCM4, MCM6 and MCM7 (Figure 3B and Supplementary Figure S4C). To further identify enzymatic substrates of SIRT1 among components of MCM helicases, we performed immunoprecipitations using an antibody directed against acetylated lysines. Such immunoprecipitates from HDAC I and II inhibitor-treated chromatin extracts of cells with WT or mutant SIRT1 were probed for components of MCM helicase (Figure 3B and Supplementary Figure S4D). Consistent with reports in the literature (44–47), MCM2, MCM4, MCM5, MCM6 and MCM7 could be acetylated in KO or H363Y cells but not in WT cells. We further focused on MCM2 as transgenic mice expressing hypomorphic mutants of this protein showed a reduction in the number of dormant origins (48,49). Since phosphorylation of MCM2 was proposed to be a key activation step in the activation of pre-replication complexes and for the modulation of initiation (50), we next investigated the relationship between four different MCM2 phosphorylation sites and SIRT1.
Phosphorylation of MCM2 on S40/41, S53, S139 and S108 were associated with DNA replication in human fibroblasts (51) and were proposed to play key roles in the activation of pre-replication complexes (50). We asked if the various forms of phospho-MCM2 were associated with phospho-SIRT1. We probed pT530-SIRT1 immunoprecipitants with antibodies directed against different MCM2 phosphoisoforms (Figure 3C; see Supplementary Figure S4E and 4F for validation of antibody specificity). We observed that MCM2 molecules harboring phosphorylations at S40/41 and S53 interacted with pT530-SIRT1 in both WT and H363Y cells (Supplementary Figure S5A). MCM2 phosphorylated at S139 (pS139-MCM2) interacted more strongly with pT530-SIRT1 in WT cells than in H363Y mutant cells, whereas more MCM2 molecules phosphorylated at S108 (pS108-MCM2) interacted with pT530-SIRT1 in SIRT1 H363Y cells. We next assessed the cell cycle distribution of chromatin-bound pS139-MCM2 and pS108-MCM2 in WT, KO and H363Y cells using flow cytometry (Supplementary Figure S5B). Both pS139-MCM2 and pS108-MCM2 were detected at higher levels in early S-phase (Supplementary Figure S5B).
S108-MCM2 phosphorylation associates with dormant origins
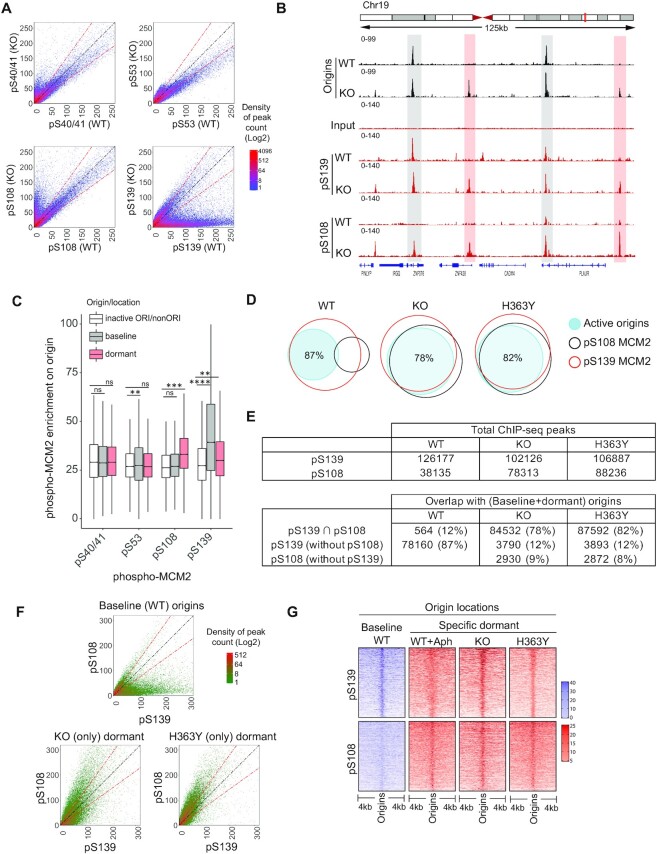
To ask if SIRT1 activity affected the association of MCM2 post-translational modifications with replication origins, we mapped the genome-wide locations of pS40/41-MCM2, pS53-MCM2, pS108-MCM2 and pS139-MCM2 in G1/S synchronized cells with active and inactive SIRT1. Most forms of phosphorylated MCM2 were associated with replication origins (Figure 3D). Binding sites for phosphorylated pS40/41- and pS53-MCM2 in WT and KO cells were located at similar chromosomal locations, whereas a distinct group of pS139-MCM2 binding sites was associated with chromatin in WT cells but not in KO cells (Figure 4A). As phospho-SIRT1 binding sites from cells harboring WT and mutant (H363Y) SIRT1 were associated with both baseline and dormant origin locations (Supplementary Figure S5D,E), dormant origin suppression was not merely due to binding of phospho-SIRT1.
Figure 4.
Phosphorylation of MCM2 on S108 is differentially associated with activation of dormant origins in SIRT1-deficient cells. (A) Density plots comparing the binding of four MCM2 phosphoisoforms (phosphorylated S40/41, S53, S108 and S139) on chromatin from WT and KO HCT116 cells. (B) IGV screenshot of overlaps between origin sites mapped by NS-seq in WT and KO cells and binding sites for phospho-MCM2 (pS139 and pS108) mapped by ChIP-seq. Examples of interactions at baseline and dormant origins are highlighted in gray and red highlights, respectively. (C) Box-whisker plot illustrating pMCM2 enrichment on origins as a function of the relative occupancy of four MCM2 phosphoisoforms on baseline origins (KO), dormant (KO) or inactive/non-Ori (remaining binding sites of phospho-MCM2 from KO) (see Supplementary Figure S5C for phospho-MCM2 enrichment in WT, KO and H363Y cells). Serine phospho-MCM2 ChIP-seq enrichment was calculated from ChIP-seq library normalized signals on each origin category using BAMScale and represented as a ratio between ChIP/Input. pMCM2 enrichment in KO cells was used as these cells initiate replication at both baseline and dormant origins. Kruskal–Wallis test was used to determine statistical significance. Ns = non-significant, ** represents P< 0.01, *** represents P< 0.001 and **** represents P< 0.0001, respectively. (D) Venn diagrams comparing the binding occupancy pS139-MCM2 (red) and pS108-MCM2 (black) on total (baseline + dormant) active origins (blue shaded) in WT, KO and H363Y cells. The numbers of common and exclusive peaks (binding sites or origins) were calculated by ‘intersection’ module of bedtools. Active origin peaks obtained from NS-Seq were compared with pS139-MCM2 or pS108-MCM2 binding sites obtained from ChIP-Seq in the indicated cells. Percentages indicate fractions of origins overlapping with phosphor-MCM2. See (E) for additional comparisons. (E) The number of total (top) and overlapping (bottom) chromosomal sites for ChIP-seq peaks (pS139 and pS108-MCM2) and origins within a comparison group. ‘∩’ represents shared sites between comparative groups. (F) Density plots showing comparative coverages of pS139-MCM2 and pS108-MCM2 on baseline origins (total origins from WT cells) or dormant origins in KO or H363Y cells. (G) A heatmap representing the binding of pS108-MCM2 and pS139-MCM2 (as determined by ChIP-seq) to baseline and dormant origins in the indicated cells, with or without Aph treatment. Baseline origins in WT cells are shown in purple; specific dormant origins are in red. Using an R-script (heatmapper2(38), see Methods), coverage for each MCM2 phosphoisoform was calculated and 5000 randomly selected peaks were represented as heatmap.
To explore which phosphorylated forms of MCM2 affect baseline and/or dormant origin activation, we compared the abundance of binding sites for all four MCM2 phosphoisoforms on baseline and dormant (from SIRT1 KO cells) origins. As shown in Figure 4B,C, pS139-MCM2 binding was highest on baseline origins (gray highlighted in Figure 4B) and did not associate with dormant origins in WT cells, whereas pS108-MCM2 showed a very low association with origins in WT cells yet associated with both baseline and dormant origins in KO cells (red highlighted in Figure 4B). S40/41- and S53-MCM2 were associated with a similar extent with both baseline and dormant origins (Figure 4C). We also observed an overall comparable enrichment of pS139-MCM2 in samples derived from all three cells with a slight enrichment of pS139-MCM2 in WT compared to KO and H363Y cells (Supplementary Figure S5C). In contrast, pS108-MCM2 showed increased chromatin enrichment in KO and H363Y cells compared to WT (Figure 4A,B; Supplementary Figure S5C). The other two serine phosphorylation sites (40/41 and 53) on MCM2 did not show changes in chromatin binding patterns (Figure 4C and Supplementary Figure S5C). Hence, pS108-MCM2 and pS139-MCM2 differentially associate with baseline and dormant origins.
We next compared the extent of pS139-MCM2 and pS108-MCM2 binding on baseline and dormant origins. In WT cells, active (baseline) origins associated with 87% of pS139-MCM2 binding sites and only 12% of pS108-MCM2 binding sites (Figure 4D,E). In contrast, active origins in KO and mutant cells (including both baseline and activated dormant origins) were highly associated (∼78–82%) with both pS139- and pS108-MCM2 binding sites (Figure 4D,E; Supplementary Figure S5F and G). This would imply that the baseline origins, which did not associate with pS108-MCM2 in WT cells, associated with pS108-MCM2 in the SIRT1 mutant/KO cells. Sites that could bind both pS139-MCM2 and pS108-MCM2 were associated with 85% and 81% of (active) dormant origins site from KO and H363Y cells, respectively (Supplementary Figure S5G). In addition, we observed that 83% of stress-induced (Aph) dormant origins were associated pS108-MCM2 and pS139-MCM2 binding sites (Supplementary Figure S5G).
As shown in Figure 4F, pS139-MCM2 showed a higher enrichment on baseline origins (from WT cells) compared to pS108-MCM2 (top panel). In contrast, pS108-MCM2 showed a higher degree of binding on dormant origins from KO and H363Y compared to pS139-MCM2 (Figure 4F, lower panels), indicating a preferential association between pS108-MCM2 and dormant origins. Further, as shown in Figure 4G, Aph treatment significantly increased the association of pS108-MCM2 with dormant origin sites.
Next, we analyzed the distribution of pS139-MCM2 and pS108-MCM2 binding sites at distinct replication timing domains. We observed that pS139-MCM2 and pS108-MCM2 were enriched in early and late replicating regions, respectively (Supplementary Figure S5H). Our observations are consistent with a model suggesting that phosphorylation of MCM2 at S108 associated with dormant origins.
ATR-dependent phosphorylation of S108-MCM2 affects replication in cells with inactive SIRT1
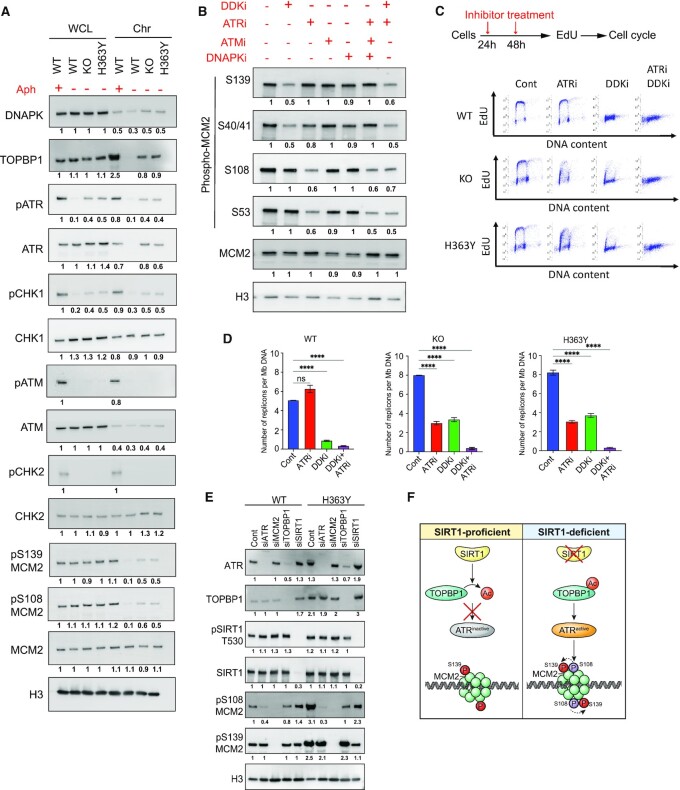
We next sought to identify the kinase(s) responsible for differential MCM2 phosphorylation. Since we observed replication stress in SIRT1-deficient cells, we first determined the levels of PI3-kinase related kinases (PIKKs) and downstream effector signaling in KO and H363Y cells. As shown in Figure 5A, increased chromatin binding of phospho-ATR and TOPBP1, as well as the ATR downstream target, phospho-CHK1, was observed in cells with inactive SIRT1. Chromatin bound phosphor-ATR and TOPBP1 were also observed in Aph treated, but not untreated, cells harboring WT SIRT1.
Figure 5.
TOPBP1-mediated ATR activation phosphorylates MCM2 on S108 in SIRT1-deficient cells. (A) Whole cell lysate (WCL) and chromatin extracts (Chr) from HCT116 cells (WT, KO and H363Y) were used to estimate the levels of stress-responding kinases (DNA-PK, ATR, ATM, CHK1, CHK2) and their activated forms (pATM, pATR, pCHK1, pCHK2) as well as TOPBP1 and MCM2. Aphidicolin-treated WT cells (0.8 μM, 24 h) were used as positive controls. This panel shows representative blots among three replicates. The number under each blot represents intensity ratios for each protein normalized to the levels measured in WCL in WT cells treated with Aph. Histone 3 (H3) was used as a loading control. (B) HCT116 cells were treated without or with specific inhibitors as indicated for 48 h and total cell extracts were used to estimate phosphorylation levels of four MCM2 serine residues (S108, S53, S40/41 and S139). Shown is a representative immunoblot from three replicates (see Supplementary Figure S6A for quantification). Histone 3 (H3) was used as a loading control. Protein intensity ratios were normalized with intensity levels of untreated WT. (C) Top panel, scheme for inhibition of ATR or/and DDK using specific inhibitor to study their effect on replication initiation. Cells were treated with inhibitors at 24 h intervals (as indicated by red arrows). Bottom panel, representative EdU/DAPI cell cycle analyses without or with ATR or DDK inhibitors (48 h) as shown in the top panel. (D) The number of replication signals detected by DNA combing per Mb of total DNA fibers was calculated as a measure for active replication from WT, KO and H363Y cells with or without ATR or DDK inhibitors (48 h). The Mann–Whitney test was used to determine the statistical significance. ns = non-significant and **** represents P< 0.0001. (E) Endogenous ATR, MCM2, TOPBP1 and SIRT1 were depleted using siRNA (72 h) in WT and H363Y HCT116 cells and chromatin-bound levels of ATR, TOPBP1, phospho-SIRT1 (T530) and phospho-MCM2 (S139 and S108) were quantified using Immunoblotting and densitometry. The intensity of each signal was normalized with WT control (see Supplementary Figure S6F for quantification). Histone 3 (H3) was used as a loading control. (F) Illustration of the role of TOPBP1/ATR on S108-MCM2 phosphorylation in SIRT1-deficient cells. SIRT1-mediated deacetylation of TOPBP1 coordinates its activity and chromatin binding. In the absence of SIRT1, acetylated TOPBP1 is recruited to chromatin in excess, which increases ATR kinase activity. Active ATR further phosphorylates S108-MCM2, which in turn modulates S139-MCM2 phosphorylation (dotted arrow).
As MCM2 is also a reported DDK substrate (50–53), we treated cells with a specific inhibitor of DDK and probed with specific phospho-MCM2 antibodies. DDK inhibition reduced the phosphorylation of S40/41 and S139 on MCM2 but did not alter the phosphorylation of the S108 and S53 residues (Figure 5B and Supplementary Figure S6A). These results suggested that DDK was a major kinase involved in the phosphorylation of S139 and S40/41 on MCM2, but that it does not catalyze the phosphorylation of S108-MCM2.
S108-MCM2 is a part of a ‘SQ’ motif, a specialized amino-acid sequence which may be phosphorylated in a PIKK-dependent manner (54,55), and previous reports suggested that S108-MCM2 was an ATR substrate (56). We also identified ATR and DNAPK as binding partners of MCM2 (Supplementary Figure S4B). The use of specific inhibitors of ATR, ATM or DNA-PK revealed that only ATR inhibition was able to reduce phosphorylation of S108 and S53 on MCM2, with no effect observed on S40/41 MCM2 and S139-MCM2. Inhibition of ATR + ATM + DNAPK or of ATR + DDK did not cause a synergistic decrease in MCM2 phosphorylation (Figure 5B). These findings indicate that DDK and ATR are the major kinases catalyzing MCM2 phosphorylation on S139 and S108, respectively.
Because pS108-MCM2 was associated with dormant origins, we asked if ATR-mediated phosphorylation of S108-MCM2 would modulate replication in cells with inactive SIRT1. Indeed, ATR inhibition drastically and differentially affected the progression of DNA replication in KO and H363Y cells (Figure 5C and Supplementary Figure S6B–D). By contrast, DDK inhibition completely blocked DNA replication initiation in WT cells while some degree of replication was still observed in KO and H363Y cells. Indeed, DDK inhibition completely blocked DNA synthesis in KO and H363Y cells only when used in combination with ATR inhibitors. In addition, both flow and DNA combing data showed that inhibition of ATR did not affect ongoing replication in WT cells but reduced the abundance of active replicons in KO and H363Y cells by 50% (Figure 5C,D; Supplementary Figure S6B–E). The combination of DDK and ATR inhibition synergistically stopped all ongoing replication in all cell types.
To further explore the link between SIRT1 and MCM2 phosphorylation, we depleted ATR, TOPBP1, MCM2 and SIRT1 in cells harboring WT or H363Y SIRT1 (Figure 5E and Supplementary Figure S6F). In concordance with previous reports showing that SIRT1 deacetylates and regulates the ATR–TOPBP1 interaction (57–59), we observed that TOPBP1 depletion reduced ATR chromatin binding and pS108-MCM2 phosphorylation. This suggests that SIRT1-mediated TOPBP1 deacetylation would inhibit ATR chromatin recruitment and the subsequent S108-MCM2 phosphorylation, thus preventing dormant origin activation (Figure 5F). Altogether, these observations suggest that SIRT1 activity prevents excess initiation by restraining ATR-mediated pS108-MCM2 to block the activation of pre-RC on dormant origins.
S108-MCM2 phosphorylation is required for activation of dormant origins
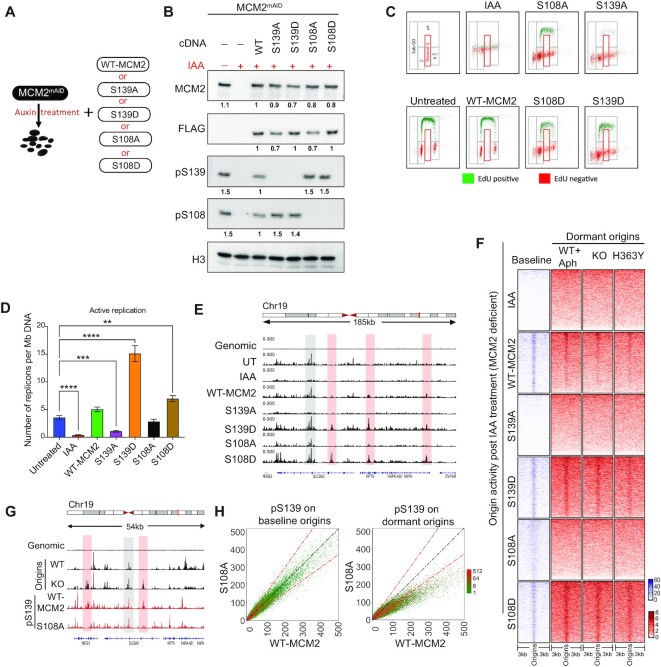
To examine the direct effects of ATR-mediated MCM2 S108 phosphorylation on dormant origins, we mapped the distribution of replication initiation sites upon blocking either MCM2 S108 or S139 phosphorylation. Since MCM2 depletion is lethal, we used a mini-auxin inducible degron (mAID) (60)-dependent degradation strategy as represented in Figure 6A and complemented these cells with mutant variants of MCM2. Immunoblot analyses using specific antibodies against total MCM2 (FLAG-tag), as well as pS139-MCM2 and pS108-MCM2, demonstrated comparable levels of expression of MCM mutants and WT MCM2 (Figure 6B).
Figure 6.
Stress-dependent pS108-MCM2 is also required for dormant origin activation. (A) Scheme for the depletion of endogenous MCM2 and compensatory expression of WT or mutant MCM2 in HCT116 cells. Five cell lines were generated by exogenously expressing cDNA for WT-MCM2 or mutant MCM2 at serine residues S139A, S139D, S108A and S108D in cells expressing the endogenous MCM2mAID. MCM2mAID is degraded upon treatment with auxin (IAA) leading to cells with MCM deficient/null phenotype. (B) HCT116-MCM2mAID cells were treated with IAA (500 μM) for 48 h and expression levels of total MCM2, pS108-MCM2 and pS139-MCM2 were measured in total cell extracts using immunoblotting. FLAG tag present in the MCM2-cDNA serves as a control for comparative expression of all MCM2 cDNA. Untreated MCM2-mAID cells were used as a positive control. The number under each signal represents the ratio of the intensity of each protein normalized to the intensity of the same protein in WT-MCM2 transfected cells. Histone 3 (H3) was used as a loading control. (C) EdU/DAPI flow analyses for mutants described in A,B after 48 h IAA treatment. Top left panel represents the gating strategy used for quantification and other panels represent EdU incorporation profiles for each mutant MCM2. IAA treatment depleted the endogenous MCM2 protein. (D) The number of active replication forks calculated using DNA-combing in cells harboring either WT-MCM2 or mutant MCM2s as described in the legend to Figure 5D. The Mann–Whitney test was used to determine the statistical significance. **represents P< 0.01 and **** represents P< 0.0001. (E) IGV screenshot of active origins mapped by NS-seq in cells harboring WT-MCM2 or phospho-mutant MCM2. Selected initiation of baseline and dormant origins in mutant MCM2-expressing cells are shown in gray and red highlights, respectively. (F) Heatmap comparing coverage of active origins between cells with WT- or mutant MCM2 and cells with SIRT1 WT, KO and H363Y mutant, as well as stress-induced dormant origins. (G) IGV screenshot of coverage of NS-seq in WT and KO (SIRT1), (as represented in Figure 2A) and ChIP-seq for pS139-MCM2 obtained from HCT116 cells harboring either WT or S108A MCM2. Red and gray highlights represent examples of baseline and dormant origin sites. See Supplementary Figure SS7E for total number and percent overlap between baseline and dormant origins. H: Comparative dot plot for occupancy of pS139-MCM2 at chromosomal sites for baseline (from WT-SIRT1 cells, left) and dormant (from KO-SIRT1 cells, right) origins in cells harboring WT MCM2 and S108A MCM2.
Flow cytometry experiments demonstrated that Auxin (indol-acetic acid: IAA) treatment, which triggered the inactivation of mAID-MCM2, could completely abolish ongoing replication, which was reversed by WT-MCM2 expression (Figure 6B,C; Supplementary Figure S7A). Although both S108A (phospho-deficient) and S108D (phospho-mimicking) mutants rescued the loss of MCM2-function, they did not fully restore cell cycle progression, showing a lower fraction of replicating cells compared to WT-MCM2 (Figure 6C; Supplementary Figure S7A,B). Phospho-mutants of S139 (S139A and S139D) show strong effects on cell cycle progression, with S139A completely unable to restore replication in the MCM2-depleted cells and S139D complementing the MCM2 deficiency and exhibiting faster cell cycle progression (Figure 6C; Supplementary Figure S7A,B).
In order to elucidate the effects of S108-MCM2 and S139-MCM2 phosphorylations on origin activation, we next used DNA combing to quantify the frequency of replication initiation and replication fork progression dynamics in the cells expressing the mutant MCM2 variants. While MCM2 depletion (IAA treatment) abrogated replication activity and increased inter-origin distances (Figure 6D; Supplementary Figure S7C,D), this phenotype was rescued by complementing these cells with exogenously expressed WT-MCM2. IAA-treated cells complemented with an MCM2 mutant that cannot be phosphorylated on S139 (S139A mutant) were unable to rescue replication initiation (Figure 6D and Supplementary Figure S7C), whereas the frequency of replication initiation events increased in these IAA-treated cells with the phospho-mimicking mutant S139D MCM2, indicating a vital role of MCM2 phosphorylation on S139 for pre-replication complex activation. The phosphorylation-deficient S108A MCM2 mutant was able to rescue IAA-mediated abrogation of replication initiation, and the phosphorylation-mimic S108D MCM2 mutant increased the frequency of initiation (Figure 6D). In addition, IAA-treated cells carrying the phosphorylation-deficient MCM2 mutants S108A and S108D showed increased and reduced inter-origin distances, respectively (Supplementary Figure S7D). Nascent strand abundance analyses (Figure 6E,F) revealed that S139-MCM2 phosphorylation was necessary for initiation at all origins, whereas S108-MCM2 phosphorylation was required for dormant origins initiation, as the S108A MCM2 mutant did not activate dormant origins. The S108D MCM2 mutant exhibited higher levels of dormant origin activation (Supplementary Figure S7E).
To observe the effects of MCM2 phospho-mutants in cells harboring SIRT1 variants, we depleted the endogenous MCM2 in WT-, KO- or H363Y-SIRT1 cells using specific 5′-UTR siRNA and complemented these cells with WT- or mutant MCM2 (Supplementary Figure S8A). Expression of WT or mutated MCM2, with the exception of S139A, complemented the MCM2 depletion and facilitated DNA synthesis in all three SIRT1 variants (WT, KO and H363Y). Replication fork speeds ranged within 0.8–1.5 kb/min in all SIRT1 variants, with the parental cell lines exhibiting replication fork speeds between 0.8 and 1.2 kb/min (Supplementary Figure S8B). Despite these similarities in replication speeds, we have observed a marked effect on inter-origin distances, as the parental cell lines, but not cells harboring WT-SIRT1, exhibited longer inter-origin distances when carrying the MCM2 S108A mutation. In accordance, SIRT1 WT cells, but not SIRT1 deficient or cells harboring the mutated SIRT1, displayed shorter inter-origin distances when carrying the MCM2 S108D mutation (Supplementary Figure S8C).
We also tested directly if SIRT1 catalytic activity could modulate the S108-MCM2 phosphorylation-dependent initiation of dormant origins by blocking SIRT1 activity with the specific inhibitor Ex-527. For this, we analyzed replication profiles in cells harboring WT- or mutant-MCM2 with and without Ex-527. No significant changes in inter-origin distances were observed when comparing replication patterns between cells expressing WT and S108D MCM2, while longer inter-origin distances were present in cells harboring S108A MCM2 (Supplementary Figure S8D). These observations are consistent with the hypothesis that dormant origin activation observed in SIRT1-deficient cells is dependent on ATR-mediated phosphorylation of S108-MCM2.
Finally, we asked if phosphorylation of S108-MCM2 was a prerequisite for phosphorylation of S139-MCM2 on baseline and dormant origins. For this, we mapped binding sites of pS139-MCM2 on baseline and dormant origins in cells carrying the wild-type MCM2 or the non-phosphorylable S108A MCM2 mutant. Indeed, although pS139-MCM2 occupancy on baseline origins was similar in cells expressing either WT or S108A MCM2, there was a significantly reduced level of pS139-MCM2 binding on dormant origins in the S108A MCM2 mutant compared to cells harboring WT-MCM2 (Figure 6G,H). Altogether, these observations indicate that S108-MCM2 phosphorylation facilitates the phosphorylation of S139-MCM2 on dormant origins and was essential for replication initiation at dormant origins.
DISCUSSION
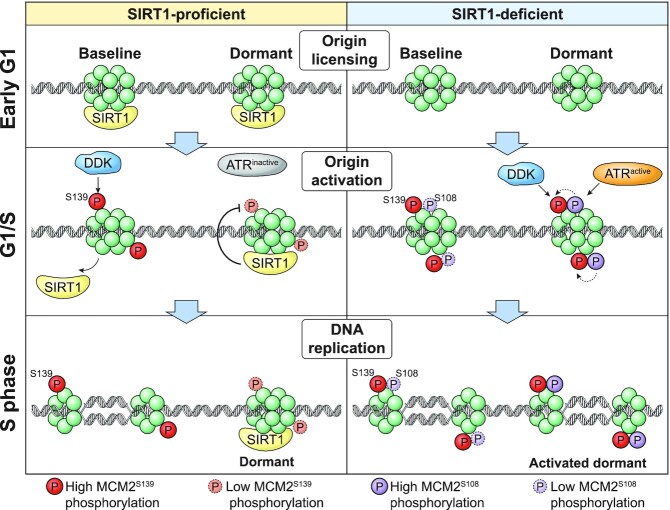
The separation of the assembly and activation stages during the initiation of DNA replication (1,2,61,62) facilitates genomic stability by limiting DNA replication to the S-phase of the cell cycle and preventing re-replication of already-duplicated genomic regions. However, this safety mechanism involves the assembly of excess pre-replication complexes on dormant origins that normally do not initiate DNA replication, necessitating a process that prioritizes replication initiation from some pre-replication complexes. The studies reported here suggest that dormant and baseline origins differ in post-translational MCM modifications that reflect an interplay between the SIRT1 and ATR signaling pathways. Initiation of DNA replication from dormant origins involves actively removing, or circumventing, a SIRT1-mediated barrier that normally maintains origin dormancy. We propose a model (Figure 7) suggesting that SIRT1 activity maintains genome-wide origin dormancy during unperturbed growth by preventing ATR-mediated phosphorylation on S108-MCM2. When SIRT1 activity is low, or if cells are exposed to replication stress and ATR activity increases, ATR-mediated phosphorylation events at S108-MCM2 overcome SIRT1-mediated inhibition and can activate dormant origins. When ATR activity increases further, for example, when cells are exposed to higher levels of replication stress, ATR can phosphorylate additional substrates regardless of SIRT1 status and globally suppress origin activity (10,63).
Figure 7.
Model: SIRT1 and ATR converge to control origin dormancy. Dormant origin activation is determined by post-translational MCM modifications that reflect an interplay between SIRT1 and ATR. Left panel, in SIRT1 proficient cells, SIRT1 binds origins and prevents the activation and recruitment of ATR to chromatin during normal unperturbed replication (Figure 5F). In such cells, baseline origins undergo DDK-mediated phosphorylation of S139-MCM2, which is required to initiate DNA replication, but dormant origins show no or low levels of S139-MCM2 phosphorylation and therefore cannot initiate replication. Right panel, in SIRT1-deficient cells, ATR is activated and recruited to chromatin (Figure 5F), catalyzing S108-MCM2 phosphorylation at dormant origins. S108-MCM2 phosphorylation, in turn, facilitates DDK-mediated S139-MCM2 phosphorylation on dormant origins, rendering both baseline and dormant origins capable of initiation of DNA replication.
Our observations are in concordance with studies in Xenopus, Caenorhabditis elegans, mice models and human cell lines demonstrating that reduction in chromatin-bound MCM2-7 levels does not affect overall replication, but that these ‘minimally licensed’ cells fail to complete replication under replicative stress (4,8,9,11,64). In addition, simulations of 250 kb origin clusters showed that replication initiates stochastically from particular origins within a replication cluster (25) and mirrors the levels of chromatin-bound pre-RC complexes, which include MCM2-7 (27,40). These studies, combined with our observations and the association between pre-RC abundance, replication timing and initiation frequency (15,65,66) suggest that post-translational modifications, in particular the phosphorylation of MCM2, define an activation threshold for origins. Baseline origins, which exhibit a high level of phosphorylation of S139-MCM2, can initiate replication regardless of the phosphorylation status of S108-MCM2, whereas the activation of dormant origins requires the phosphorylation of the S108-MCM2 residue. The structural basis for this distinction between baseline and dormant origins has yet to be investigated but given the tissue-specific nature of a large cohort of potential replication origins, it is likely to be modulated by (an) epigenetic chromatin modification(s).
Because many chromatin-associated proteins, including multiple lysines on MCM2, can be acetylated (45–47), it is attractive to hypothesize that SIRT1-mediated deacetylation of MCM2 or other chromatin-associated substrates can modulate the accessibility of the MCM helicase to activating phosphorylation to maintain origin dormancy. We observed a higher association of the active site mutant of SIRT1 with baseline origins, which could indicate that SIRT1 can normally leave chromatin after substrate deacetylation, triggering initiation. Our findings further suggest that during normal unperturbed mitotic growth, baseline origins exhibit an above-threshold phosphorylation of S139-MCM2, which is sufficient to activate the CMG helicase. By contrast, dormant origins exhibit a lower level of S139-MCM2 phosphorylation, which is not sufficient to activate the CMG helicase.
Our studies suggest that phosphorylation of S108-MCM2 by ATR could enhance S139 phosphorylation and thus activate dormant origins. However, origin dormancy is maintained by the metabolic sensory protein SIRT1, which prevents the interaction of origins with the ATR kinase via deacetylation of TOPBP1. Under these circumstances, replication preferentially initiates from baseline origins, which are preferred sites of replication initiation in both SIRT1-proficient and deficient cells. In the absence of SIRT1, hyper-acetylation of the TOPBP1 docking protein recruits and activates the ATR kinase and promotes ATR-mediated phosphorylation of MCM2 on S108. The presence of pS108-MCM2 can counter SIRT1-mediated inhibition of replication initiation, facilitating the phosphorylation of S139 on MCM2, thus activating dormant origins. Excess initiation of DNA replication, including initiation at dormant origins, can increase replication stress due to a massive load of progressing replication forks. SIRT1 activity, therefore, is essential to maintain the stability of replicating fork progression and ensures a low frequency of replication initiation, thereby maintaining genome stability.
Our model (Figure 7) predicts that mammalian cells with active SIRT1 exhibit an above-threshold level of pS139-MCM2 in early replicating regions during the early S-phase of the cell cycle. In concordance, we observed that SIRT1-proficient cells exhibit a low frequency of pS139-MCM2 sites in late replicating regions. In contrast, pS108-MCM2 showed preferential binding to late replicating regions, enriched sites for dormant origins. In SIRT1-deficient cells, pS108-MCM2 also showed binding sites preferentially associated with early-replicating regions in addition to the late replication-associated binding sites. This dynamic agrees with a recent study in yeast (65), which demonstrates the influence of SIRT1 activity on the occupancy of MCM2-7 helicases on chromatin. These observations strongly suggest that pSIRT1 activity maintains the low levels of pS108-MCM2 on dormant origins under unperturbed conditions.
Origin dormancy is an integral part of maintaining genome stability through timely activation of replication forks restarting halted DNA synthesis and facilitating DNA damage repair. This process is evident from studies with fibroblasts from MCM2 and MCM4 hypomorphic mice which showed increased fork stalling and DNA damage (48,49). DNA damage kinases ATR, ATM, CHK1 and CHK2 activate cell cycle checkpoints upon replication stress and fork stalling, but inhibition of these kinases in the absence of stress increases the frequency of replication initiation events, indicating that when replication elongation stalls, these kinases play a role in suppressing dormant origins (10,63). Our study shows that replication dormancy can be circumvented either by attenuation of SIRT1 activity, resulting in an increased abundance of TOPBP1 on replication origins, or by stalling of replication forks, leading to the direct activation of ATR. In both cases, ATR-mediated phosphorylation of S108-MCM2 triggers initiation events at dormant origins. The interplay between SIRT1 and ATR, which respond to distinct stress conditions, integrates signaling to chromatin to modulate origin activity and maintain genomic stability.
DATA AVAILABILITY
We have deposited the MS data in the Proteome Xchange repository (accession numbers: PXD028487 and MSV000088134). Both NS-seq and ChIP-seq data is deposited in GEO, accession number GSE184353.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the CCR core sequencing facility headed by Bao Tran and Jyoti Shetty for expert help with DNA sequencing. We thank Dr Andre Nussenzweig and Dr Masato Kanemaki for the gift of HCT116-mAID cell line, Thorkell Andresson (NIH mass-spectrometry facility) for expert help with mass-spectrometry and protein identification, and Dr Michael Kruhlak, Dr Langston Lim and Dr Andy Tran (Confocal Microscopy Core Facility, CCR, NCI, NIH) and Ms. Kiran Parwani (Emory University) for expert technical assistance in the initial characterization of SIRT1 cell lines. We thank the CCR Genomics Core led by Liz Conner for the expert help with ChIP-sequencing, especially Madeline Wong for her quick and excellent work. We thank Dr. Urbain Weyemi (University of Texas at Austin) for critical reading of the manuscript and helpful suggestions and Ms. Iris Helen Indig for the ‘Baseline Origin’ suggestion.
Notes
Present address: Adrian M. Baris, Program in Cancer Biology, Oregon Health and Science University, Portland, OR, USA.
Present address: Sang-Min Jang, Department of Biochemistry, Chungbuk National University, Chungdae-ro, Seowon-gu, Cheongju, Korea, 28644.
Present address: Koichi Utani, Department of Microbiology, Kanazawa Medical University, Uchinada Ishikawa, Japan 920-0293.
Contributor Information
Bhushan L Thakur, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Adrian M Baris, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Haiqing Fu, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Christophe E Redon, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Lorinc S Pongor, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Sara Mosavarpour, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Jacob M Gross, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Sang-Min Jang, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Robin Sebastian, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Koichi Utani, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Lisa M Jenkins, Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
Fred E Indig, Confocal Imaging Facility, National Institute on Aging, NIH, Baltimore, MD 21224, USA.
Mirit I Aladjem, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892-4255, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [ZIA BC010411 to M.I.A.]; National Institute on Aging [ZIC AG000615-14 to F.E.I.]. Funding for open access charge: National Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Marks A.B., Fu H., Aladjem M.I.. Regulation of replication origins. Adv. Exp. Med. Biol. 2017; 1042:43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aladjem M.I., Redon C.E.. Order from clutter: selective interactions at mammalian replication origins. Nat. Rev. Genet. 2017; 18:101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ekundayo B., Bleichert F.. Origins of DNA replication. PLos Genet. 2019; 15:e1008320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alver R.C., Chadha G.S., Blow J.J.. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst.). 2014; 19:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ganier O., Prorok P., Akerman I., Mechali M.. Metazoan DNA replication origins. Curr. Opin. Cell Biol. 2019; 58:134–141. [DOI] [PubMed] [Google Scholar]
- 6. Techer H., Koundrioukoff S., Nicolas A., Debatisse M.. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 2017; 18:535–550. [DOI] [PubMed] [Google Scholar]
- 7. Akerman I., Kasaai B., Bazarova A., Sang P.B., Peiffer I., Artufel M., Derelle R., Smith G., Rodriguez-Martinez M., Romano M.et al.. A predictable conserved DNA base composition signature defines human core DNA replication origins. Nat. Commun. 2020; 11:4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Branzei D., Foiani M.. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005; 17:568–575. [DOI] [PubMed] [Google Scholar]
- 9. Courtot L., Hoffmann J.S., Bergoglio V.. The protective role of dormant origins in response to replicative stress. Int. J. Mol. Sci. 2018; 19:3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moiseeva T.N., Yin Y., Calderon M.J., Qian C., Schamus-Haynes S., Sugitani N., Osmanbeyoglu H.U., Rothenberg E., Watkins S.C., Bakkenist C.J.. An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:13374–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woodward A.M., Gohler T., Luciani M.G., Oehlmann M., Ge X., Gartner A., Jackson D.A., Blow J.J.. Excess mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006; 173:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ibarra A., Schwob E., Mendez J.. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8956–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blow J.J., Ge X.Q., Jackson D.A.. How dormant origins promote complete genome replication. Trends Biochem. Sci. 2011; 36:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lob D., Lengert N., Chagin V.O., Reinhart M., Casas-Delucchi C.S., Cardoso M.C., Drossel B.. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun. 2016; 7:11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S.P., Borrman T., Liu V.W., Yang S.C., Bechhoefer J., Rhind N.. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015; 25:1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge X.Q., Blow J.J.. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 2010; 191:1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ge X.Q., Jackson D.A., Blow J.J.. Dormant origins licensed by excess mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007; 21:3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Utani K., Fu H., Jang S.M., Marks A.B., Smith O.K., Zhang Y., Redon C.E., Shimizu N., Aladjem M.I.. Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability. Nucleic Acids Res. 2017; 45:7807–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alves-Fernandes D.K., Jasiulionis M.G.. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int. J. Mol. Sci. 2019; 20:3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007; 450:440–444. [DOI] [PubMed] [Google Scholar]
- 21. Smith O.K., Kim R., Fu H., Martin M.M., Lin C.M., Utani K., Zhang Y., Marks A.B., Lalande M., Chamberlain S.et al.. Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin. 2016; 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foss E.J., Gatbonton-Schwager T., Thiesen A.H., Taylor E., Soriano R., Lao U., MacAlpine D.M., Bedalov A.. Sir2 suppresses transcription-mediated displacement of mcm2-7 replicative helicases at the ribosomal DNA repeats. PLos Genet. 2019; 15:e1008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawabata T., Yamaguchi S., Buske T., Luebben S.W., Wallace M., Matise I., Schimenti J.C., Shima N.. A reduction of licensed origins reveals strain-specific replication dynamics in mice. Mamm. Genome. 2011; 22:506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shima N., Pederson K.D.. Dormant origins as a built-in safeguard in eukaryotic DNA replication against genome instability and disease development. DNA Repair (Amst.). 2017; 56:166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blow J.J., Ge X.Q.. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009; 10:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Natsume T., Kiyomitsu T., Saga Y., Kanemaki M.T.. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 2016; 15:210–218. [DOI] [PubMed] [Google Scholar]
- 27. Fu H., Redon C.E., Thakur B.L., Utani K., Sebastian R., Jang S.M., Gross J.M., Mosavarpour S., Marks A.B., Zhuang S.Z.et al.. Dynamics of replication origin over-activation. Nat. Commun. 2021; 12:3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jang S.M., Nathans J.F., Fu H., Redon C.E., Jenkins L.M., Thakur B.L., Pongor L.S., Baris A.M., Gross J.M., O’Neill M.J.et al.. The repid-CRL4 ubiquitin ligase complex regulates metaphase to anaphase transition via BUB3 degradation. Nat. Commun. 2020; 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu H., Martin M.M., Regairaz M., Huang L., You Y., Lin C.M., Ryan M., Kim R., Shimura T., Pommier Y.et al.. The DNA repair endonuclease mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat. Commun. 2015; 6:6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu H., Besnard E., Desprat R., Ryan M., Kahli M., Lemaitre J.M., Aladjem M.I.. Mapping replication origin sequences in eukaryotic chromosomes. Curr. Protoc. Cell Biol. 2014; 65:22.20.1–22.20.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin M.M., Ryan M., Kim R., Zakas A.L., Fu H., Lin C.M., Reinhold W.C., Davis S.R., Bilke S., Liu H.et al.. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011; 21:1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foulk M.S., Urban J.M., Casella C., Gerbi S.A.. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015; 25:725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J., Xu Q., Ji M., Guo X., Xu X., Fargo D.C., Li X.. The phosphorylation status of T522 modulates tissue-specific functions of SIRT1 in energy metabolism in mice. EMBO Rep. 2017; 18:841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y.et al.. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004; 303:2011–2015. [DOI] [PubMed] [Google Scholar]
- 35. Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A.. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001; 107:149–159. [DOI] [PubMed] [Google Scholar]
- 36. Marheineke K., Hyrien O.. Aphidicolin triggers a block to replication origin firing in xenopus egg extracts. J. Biol. Chem. 2001; 276:17092–17100. [DOI] [PubMed] [Google Scholar]
- 37. Ozeri-Galai E., Lebofsky R., Rahat A., Bester A.C., Bensimon A., Kerem B.. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell. 2011; 43:122–131. [DOI] [PubMed] [Google Scholar]
- 38. Pongor L.S., Gross J.M., Vera Alvarez R., Murai J., Jang S.M., Zhang H., Redon C., Fu H., Huang S.Y., Thakur B.et al.. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenetics Chromatin. 2020; 13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G.. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013; 23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugimoto N., Maehara K., Yoshida K., Ohkawa Y., Fujita M.. Genome-wide analysis of the spatiotemporal regulation of firing and dormant replication origins in human cells. Nucleic Acids Res. 2018; 46:6683–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Layer R.M., Pedersen B.S., DiSera T., Marth G.T., Gertz J., Quinlan A.R.. GIGGLE: a search engine for large-scale integrated genome analysis. Nat. Methods. 2018; 15:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim R., Smith O.K., Wong W.C., Ryan A.M., Ryan M.C., Aladjem M.I.. ColoWeb: a resource for analysis of colocalization of genomic features. BMC Genomics. 2015; 16:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoggard T.A., Chang F., Perry K.R., Subramanian S., Kenworthy J., Chueng J., Shor E., Hyland E.M., Boeke J.D., Weinreich M.et al.. Yeast heterochromatin regulators sir2 and sir3 act directly at euchromatic DNA replication origins. PLos Genet. 2018; 14:e1007418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 45. Bienvenut W.V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C.. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol. Cell. Proteomics. 2012; 11:M111 015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., Kim D.S., De Juan-Pardo E., Demeyer K., Hole K., Larrea E.et al.. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase natB. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Z., Xu X.. Post-translational modifications of the mini-chromosome maintenance proteins in DNA replication. Genes (Basel). 2019; 10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pruitt S.C., Bailey K.J., Freeland A.. Reduced mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007; 25:3121–3132. [DOI] [PubMed] [Google Scholar]
- 49. Kunnev D., Rusiniak M.E., Kudla A., Freeland A., Cady G.K., Pruitt S.C.. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010; 29:3630–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsuji T., Ficarro S.B., Jiang W.. Essential role of phosphorylation of MCM2 by cdc7/dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell. 2006; 17:4459–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C.. Identification of mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 2006; 281:10281–10290. [DOI] [PubMed] [Google Scholar]
- 52. Mahbubani H.M., Chong J.P., Chevalier S., Thommes P., Blow J.J.. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol. 1997; 136:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oehlmann M., Score A.J., Blow J.J.. The role of cdc6 in ensuring complete genome licensing and s phase checkpoint activation. J. Cell Biol. 2004; 165:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y.et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 55. Cortez D., Glick G., Elledge S.J.. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang J., Zhang J., Bellani M.A., Pokharel D., Gichimu J., James R.C., Gali H., Ling C., Yan Z., Xu D.et al.. Remodeling of interstrand crosslink proximal replisomes is dependent on ATR, FANCM, and FANCD2. Cell Rep. 2019; 27:1794–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumagai A., Lee J., Yoo H.Y., Dunphy W.G.. TopBP1 activates the ATR-ATRIP complex. Cell. 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- 58. Wang R.H., Lahusen T.J., Chen Q., Xu X., Jenkins L.M., Leo E., Fu H., Aladjem M., Pommier Y., Appella E.et al.. SIRT1 deacetylates topbp1 and modulates intra-S-phase checkpoint and DNA replication origin firing. Int. J. Biol. Sci. 2014; 10:1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu T., Lin Y.H., Leng W., Jung S.Y., Zhang H., Deng M., Evans D., Li Y., Luo K., Qin B.et al.. A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint. Mol. Cell. 2014; 56:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Natsume T., Nishimura K., Minocherhomji S., Bhowmick R., Hickson I.D., Kanemaki M.T.. Acute inactivation of the replicative helicase in human cells triggers MCM8-9-dependent DNA synthesis. Genes Dev. 2017; 31:816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prioleau M.N., MacAlpine D.M.. DNA replication origins-where do we begin?. Genes Dev. 2016; 30:1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ilves I., Petojevic T., Pesavento J.J., Botchan M.R.. Activation of the MCM2-7 helicase by association with cdc45 and GINS proteins. Mol. Cell. 2010; 37:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moiseeva T.N., Bakkenist C.J.. Dormant origin signaling during unperturbed replication. DNA Repair (Amst.). 2019; 81:102655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Remus D., Beuron F., Tolun G., Griffith J.D., Morris E.P., Diffley J.F.. Concerted loading of mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009; 139:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hoggard T., Muller C.A., Nieduszynski C.A., Weinreich M., Fox C.A.. Sir2 mitigates an intrinsic imbalance in origin licensing efficiency between early- and late-replicating euchromatin. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:14314–14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mei L., Kedziora K.M., Song E.A., Purvis J.E., Cook J.G.. The consequences of differential origin licensing dynamics in distinct chromatin environments. Nucleic Acids Res. 2022; 10.1093/nar/gkac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have deposited the MS data in the Proteome Xchange repository (accession numbers: PXD028487 and MSV000088134). Both NS-seq and ChIP-seq data is deposited in GEO, accession number GSE184353.