Abstract
Argonaute (Ago) proteins are programmable nucleases found in eukaryotes and prokaryotes. Prokaryotic Agos (pAgos) share a high degree of structural homology with eukaryotic Agos (eAgos), and eAgos originate from pAgos. Although eAgos exclusively cleave RNA targets, most characterized pAgos cleave DNA targets. This study characterized a novel pAgo, MbpAgo, from the psychrotolerant bacterium Mucilaginibacter paludis which prefers to cleave RNA targets rather than DNA targets. Compared to previously studied Agos, MbpAgo can utilize both 5′phosphorylated(5′P) and 5′hydroxylated(5′OH) DNA guides (gDNAs) to efficiently cleave RNA targets at the canonical cleavage site if the guide is between 15 and 17 nt long. Furthermore, MbpAgo is active at a wide range of temperatures (4–65°C) and displays no obvious preference for the 5′-nucleotide of a guide. Single-nucleotide and most dinucleotide mismatches have no or little effects on cleavage efficiency, except for dinucleotide mismatches at positions 11–13 that dramatically reduce target cleavage. MbpAgo can efficiently cleave highly structured RNA targets using both 5′P and 5′OH gDNAs in the presence of Mg2+ or Mn2+. The biochemical characterization of MbpAgo paves the way for its use in RNA manipulations such as nucleic acid detection and clearance of RNA viruses.
INTRODUCTION
Eukaryotic Argonautes (eAgos) are key components of the RNA-induced silencing complex (RISC) and participate in posttranscriptional gene regulation and antivirus defense (1). Argonautes (Agos) are also found in many bacterial and archaeal organisms (2). Currently, only a few prokaryotic Agos (pAgos) have been characterized, and biochemical studies showed that they could function as programmable endonucleases in vitro and protect cells from foreign genetic elements in vivo (3–6). A recent study showed that pAgos may function in DNA replication (7), suggesting that the cellular functions of pAgos are still obscure and remain to be explored.
Genomic studies revealed that pAgos are much more diverse than eAgos (8), and pAgos can be classified into long pAgos, short pAgos and PIWI-RE (9). Despite these divergent cellular functions and diversity, eAgos and most long pAgos adopt a highly conserved six-domain architecture, including the N-terminal (N), linker 1 (L1), middle (MID), linker 2 (L2), P-element induced wimpy testis (PIWI), and PIWI-Ago-Zwille (PAZ) domains (5). Structural studies of Agos showed that the 5′ end and the 3′ end of nucleic acid guides are anchored in the MID and PAZ domains, respectively (10,11). Agos with endonuclease activity have a catalytic tetrad DEDX (X is D, H, N, or K) in the PIWI domain, which is essential in binding divalent metal ions and responsible for catalysis (5,12,13). After binding small nucleic acid guides, active Agos can precisely cleave the complementary targets between the 10′ and 11′ nucleotides of the guide nucleic acid, which is the canonical cleavage pattern (14).
Although eAgos exclusively cleave RNA targets, most previously characterized pAgos cleave DNA targets. Initially, pAgos have been characterized from thermophilic prokaryotes, including TtAgo (Thermus thermophilus) (3), PfAgo (Pyrococcus furiosus) (4), MpAgo (Marinitoga piezophila) (13), and MjAgo (Methanocaldococcus jannaschii) (15,16), which can be programmed with DNA guides (gDNAs) or RNA guides (gRNAs) to cleave DNA targets effectively at elevated temperatures but not moderate temperatures. Recently, active pAgos from mesophilic prokaryotes including CbAgo (Clostridium butyricum) (17,18), LrAgo (Limnothrix rosea) (17), SeAgo (Synechococcus elongatus) (19), KmAgo (Kurthia massiliensis) (20,21), CpAgo (Clostridium perfringens) and IbAgo (Intestinibacter bartlettii) (22) were characterized to search for a pAgo that can cleave both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) at moderate temperatures and be used for genome editing. Some of these also have RNA target cleavage activity, including TtAgo, MpAgo, CbAgo, CpAgo and KmAgo, but they all cleave DNA targets better than RNA targets (13,17,20–23).
The clustered regularly interspaced short palindromic repeats-associated (CRISPR-Cas) system is the most widely used enzymatic tool for programmable nucleic acid cleavage. As another programmable nuclease with diverse binding and cleavage activities, pAgos are being exploited in genome editing applications (9), molecular cloning (24), and nucleic acid detection (25–28). To date, catalytically active pAgos characterized have been isolated from thermophilic and mesophilic species (29). Interestingly, pAgos from mesophilic species analyzed so far are active in a wide temperature range from 37°C to 50°C (17,22), suggesting the possibility of finding pAgos cleaving targets with a high efficiency at 37°C also in psychrotolerant or psychrophilic species. In addition, the properties of pAgos from psychrotolerant or psychrophilic species remain unknown.
In this work, we characterized a novel pAgo, MbpAgo, from the psychrotolerant organism Mucilaginibacter paludis (30), which is distantly related to other characterized pAgos, and contains the canonical catalytic tetrad in the PIWI domain (residues D566, E601, D635, and D768). This study demonstrated that, different from other pAgos, MbpAgo binds gDNAs to cleave RNA with high efficiency but DNA with very low efficiency at physiological temperatures. Furthermore, we demonstrated that MbpAgo could utilize both 5′phosphorylated(5′P) and 5′hydroxylated(5′OH) gDNAs to cleave unstructured and highly-structured RNA, suggesting that it can expand the toolkit for RNA manipulations.
MATERIALS AND METHODS
Protein expression and purification
Considering possible applications in human cells, the nucleotide sequence of the MbpAgo gene (WP_008504757.1; M. paludis) and MbpAgo double mutant (MpbAgo_DM: D566A, D635A) gene were codon-optimized for expression in Homo sapiens. The codon-optimized genes were synthesized by Wuhan Genecreate Biotechnology Co., Ltd and cloned into pET28a expression vectors in frame with the N-terminal His6 tag (Supplementary Figure S1B; Supplementary Table S4). MbpAgo variants H482Y/K486 (YK), H482R/K486 (RK), H482A/K486 (AK), H482/K486A (HA) and H482A/K486A (AA) that were used for in vitro activity assays were constructed by polymerase chain reaction (PCR) mediated site-directed mutagenesis (31); the primers, template, and purpose are listed in Supplementary Table S1. All cloned constructs were verified by DNA sequencing. Escherichia coli Rosetta (DE3) (Novagen), containing eukaryotic tRNAs rarely used in E. coli, was used to express MbpAgo or MbpAgo variant proteins. Cultures were grown at 37°C in Luria-Bertani (LB) medium containing 50 μg/ml kanamycin and induced by adding isopropyl-β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM until OD600 reached 0.8. Cells were incubated at 18°C for 16 h with continuous shaking for expression. Centrifugally collected cells were stored in a −80°C refrigerator for further protein purification.
The cell pellet was resuspended in Buffer A [20 mM Tris–HCl (pH 7.4), 500 mM NaCl, and 20 mM imidazole] supplemented with 1 mM phenylmethylsulfonyl fluoride and disrupted by sonication (SCIENTZ-IID: 400 W, 2 s on/4 s off for 20 min). The lysate was clarified by centrifugation and the supernatant was loaded onto Ni-NTA agarose resin for 1 h with rotation. The beads were washed with Buffer A, then with the same buffer containing 50 mM imidazole and eluted with Buffer A containing 150 mM imidazole. Fractions containing MbpAgo or variants were concentrated by ultrafiltration using an Amicon 50K filter unit (Millipore) and purified on a Superdex 200 16/600 column (GE Healthcare) equilibrated with Buffer B [20 mM Tris-HCl (pH 7.4), and 500 mM NaCl]. Fractions containing MbpAgo were concentrated with an Amicon 50K filter unit (Millipore), placed in Buffer B, aliquoted and flash-frozen in liquid nitrogen.
Single-stranded nucleic acid cleavage assays
Cleavage assays were performed using the synthetic guides and targets (see Supplementary Table S2 for oligonucleotide sequences) under conditions described previously study (20). 5′-FAM-labeled targets and 5′-P guides were synthesized for some experiments. 800 nM MbpAgo was mixed with 400 nM gDNA or gRNA, and incubated for 10 min at 37°C for guide loading in buffer RB [10 mM HEPES–NaOH (pH 7.5), 100 mM NaCl, and 5% glycerol] with 5 mM MnCl2. Target nucleic acids were added to the final concentration of 200 nM. The cleavage reactions were performed in PCR tubes at 37°C, stopped after the indicated time intervals by mixing the samples with equal volumes of 2 × RNA loading dye [95% formamide, 18 mM EDTA, and 0.025% sodium dodecyl sulfate (SDS), and 0.025% bromophenol blue], and heated for 5 min at 95°C. Kinetic analyses of RNA cleavage were performed under single- or multiple-turnover conditions, and the detailed methods were described in the literature (17,18). Briefly, in single-turnover reactions (the [MbpAgo–gDNA] /RNA target ratio was > 1.0), the data were fitted to the following single-exponential equation: C = Cmax × [1 – exp(–kobs × t)], where C is the cleavage efficiency at a given time point, Cmax is the maximum cleavage, and kobs is the observed rate constant. In multiple-turnover reactions (the [MbpAgo–gDNA] /RNA target ratio was < 1.0), the data were fitted to the following two-phase equation: C = B × [1 – exp(–kburst × t)] + V × t, where B is the burst fraction, kburst is the observed cleavage rate in the burst fraction, and V is the steady-state velocity of RNA cleavage. To analyze the effect of various divalent cations, 5 mM Mn2+, Mg2+, Ni2+, Co2+, Cu2+, Fe2+, Ca2+, or Zn2+ was used in the reactions, respectively. To analyze the temperature dependence of RNA cleavage, MbpAgo was loaded with gDNAs for 10 min at 37°C; the samples were transferred to indicated temperatures in a PCR thermocycler (T100, Bio-Rad), RNA targets were added and the samples were incubated for 15 min. All reactions were carried out at 37°C if not indicated. For reactions not containing fluorescent labels, the gels were resolved by 20% denaturing PAGE, stained with SYBR Gold (Invitrogen), and visualized with Gel DocTM XR+ (Bio-Rad). For reactions containing FAM labels, the gels were visualized with Gel DocTM XR+ (Bio-Rad) and analyzed by the NIH program ImageJ and Prism 8 (GraphPad).
Highly-structured RNA cleavage assays
The HIV-1 ΔDIS 5′-untranslated region (UTR) RNA (32) was in vitro transcribed using T7 RNA polymerase (Thermo Fisher Scientific) and synthetic DNA templates carrying a T7 promoter sequence. The transcripts were treated with DNase I, gel-purified, and ethanol precipitated. The gDNAs used for cleavage (Supplementary Table S3) were 5′-phosphorylated using T4 PNK (New England Biolabs) except for cleavage with 5′-OH guides. 800 nM MbpAgo was mixed with 400 nM gDNA and incubated for 10 min at 37°C for guide loading. The RNA target was added to the final concentration of 200 nM and incubated for 30 min at 37°C for cleavage. Reactions were stopped with 2 × RNA loading dye and heated for 5 min at 95°C. The cleavage products were resolved by 8% denaturing PAGE and stained with SYBR Gold.
Electrophoretic mobility shift assay (EMSA)
To examine the loading of the guide onto MbpAgo, MbpAgo and the guide were incubated in buffer RB with 5 mM Mn2+ or 1 mM EDTA for 10 min at 37°C. The concentration of the internally FAM-labeled fluorescent guide was fixed as 2.5 nM, whereas the concentration of MbpAgo varied. The samples were mixed with 1 μl of 10 × loading buffer [250 mM Tris–HCl (pH 7.5), 40% glycerol] and resolved by 10% native PAGE with 0.5 × Tris–borate–EDTA (TBE) buffer. The gels were visualized with the ChemiDoc MP Imaging System (Bio-Rad). To analyze the loading of the target onto the MbpAgo-gDNA complex, MbpAgo_DM and label-free guide (1:1) with varying concentrations were incubated for 10 min at 37°C, and the 100 nM 5′-FAM-labeled target was added to the reaction for 30 min at 37°C. The samples were mixed with 1 μl of 10 × loading buffer and resolved by 10% native PAGE with 0.5 × TBE. For reactions containing FAM-labeled targets, the gels were visualized with the Gel DocTM XR+ (Bio-Rad). To determine the apparent dissociation constants (Kd) for guide binding by MbpAgo and target binding by the MbpAgo–gDNA complex, the gel images obtained were analyzed with the NIH program ImageJ and Prism 8 (GraphPad). Data were fitted with the Hill equation with a Hill coefficient of 2–2.5. All nucleic acids used in this study are listed in Supplementary Table S2.
CD measurements for thermal denaturation
CD spectra and melting curves were acquired with a Chirascan V100 CD spectrometer (Applied Photophysics) equipped with a thermostatted cell holder at 25°C under a constant nitrogen flow, holding the protein solutions in quartz cuvettes of 0.1 mm optical path, scanning from 290–195 nm at 1 nm/s with a slit width of 1 nm against an air background obtained with the same settings without any cuvette. The protein concentration was 0.32 mg/ml (3.5 μM) in all experiments with addition of 3.5 μM gDNA or without gDNA, and the buffer contained 7.5 mM Tris–HCl (pH 7.4), and 150 mM NaCl with 5 mM Mn2+ or 1 mM EDTA. A reference spectrum of the corresponding buffer showed only noise within ± 0.5 mdeg. Melting curves were acquired with 2.5 °C stepwise increments and 30 s intervals from 25°C to 75 °C. Measurements were performed in duplicate. Data were normalized to the fraction of unfolded protein and analyzed using the software GraphPad Prism 8 in nonlinear regression using the Boltzmann sigmoidal function for melting temperature (Tm) determination.
Co-purification nucleic acids
Isolation of co-purification nucleic acids was carried out as described in Hegge et al.’s (18) method, with minor modifications. Briefly, to 2 mg of purified MbpAgo in Buffer B, CaCl2 and proteinase K (Zomanbio) were added to final concentrations of 5 mM CaCl2 and 1 mg/ml proteinase K. The sample was incubated for 50 min at 55°C. The nucleic acids were separated from the organic fraction by adding Roti-phenol/chloroform/isoamyl alcohol (pH 7.5–8.0) at a 25:24:1 ratio. The top layer was isolated and nucleic acids were precipitated using ethanol precipitation by adding 99% ethanol at a 1:2 ratio supplied with 0.5% linear polymerized acrylamide as a co-precipitating agent. This mixture was incubated for 16 h at −20°C and centrifuged at 13,000 rpm for 30 min. Next, the nucleic acid pellet was washed with 500 μl of 70% ethanol and solved in 50 μl nuclease-free water. The purified nucleic acids were treated with either 100 μg/ml RNase A (Thermo Fisher Scientific), 2 units DNase I (NEB), or both for 1 h at 37°C and resolved on 20% denaturing PAGE and stained with SYBR Gold.
RESULTS
MbpAgo strongly prefers to cleave RNA rather than DNA with small gDNAs at a physiological temperature
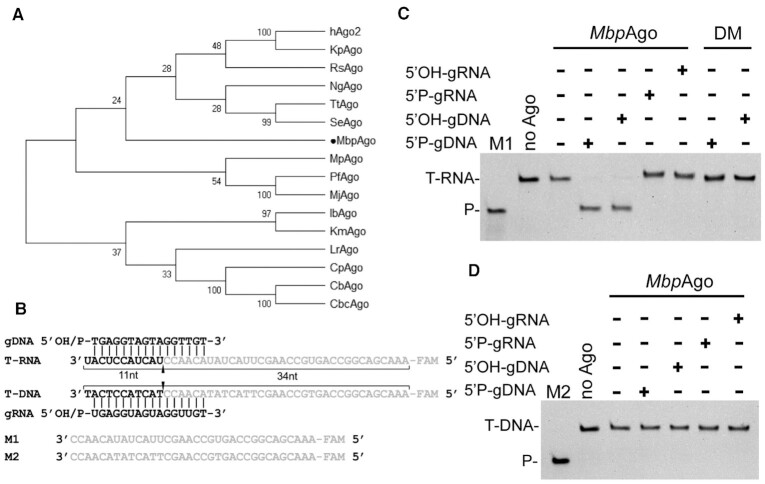
MbpAgo (accession no. WP_008504757.1 in the National Center for Biotechnology Information protein database) is distantly related to other characterized eAgos and pAgos (sequence identity < 20%; Figure 1A; Supplementary Figure S1A) and contains the canonical catalytic tetrad in the PIWI domain (residues D566, E601, D635, and D768; Supplementary Figure S1A). To study its biochemical properties and in vivo functions, MbpAgo was expressed and purified. The codon-optimized gene encoding MbpAgo was chemically synthesized and cloned into the pET28a plasmid (Supplementary Figure S1B). In addition to the wild-type protein, its catalytically inactive variant (MbpAgo_DM) was obtained by substituting two of four catalytic tetrad residues (D566A/D635A) (Supplementary Figure S1A). The protein was expressed in E. coli and purified using Ni-NTA-affinity and size-exclusion chromatography (see Supplementary Figure S1 and Materials and Methods for details). Examination of purified MbpAgo showed high purity of the samples (Supplementary Figure S1).
Figure 1.
MbpAgo exhibits DNA-guided RNA endonuclease activity at 37°C. (A) Maximum likelihood phylogenetic tree of characterized Ago proteins. (B) Guide and target oligonucleotides. gDNAs and RNA targets (T-RNA) were used in most experiments. The black triangle indicates the cleavage site. (C)MbpAgo exhibits DNA-guided RNA endonuclease activity. (D)MbpAgo exhibits no DNA cleavage activity. Positions of the cleavage products (P) are indicated on the left of the gels. MbpAgo, guide and target were mixed at a 4:2:1 molar ratio (800 nM MbpAgo preloaded with 400 nM guide, plus 200 nM target) and incubated for 30 min at 37°C. Catalytic dead mutant MbpAgo_DM (DM) was used as a control. Lanes M1 and M2 contain chemically synthesized 34-nt RNA and DNA corresponding to the cleavage products of T-RNA and the DNA target (T-DNA), respectively.
The nucleic acid specificity of MbpAgo was next studied by in vitro cleavage assay using synthetic fluorescently labeled oligonucleotide targets (Figure 1B). MbpAgo was loaded with 16 nt gDNAs or gRNAs containing a 5′P or 5′OH group at 37°C for 10 min followed by the addition of complementary 5′-end FAM-labeled 45-nt-long ssDNA or RNA targets (Figure 1B, Supplementary Table S2). After incubation for 30 min at 37°C, the cleavage products were resolved by 20% denaturing gel (Figure 1C and D). Unexpectedly, although most studied pAgos strongly prefer to cleave DNA targets (5,17,20), MbpAgo can use both 5′P-gDNA and 5′OH-gDNA to cleave almost all RNA targets, and no DNA target cleavage was observed in 30 min (Figure 1C and D). In addition, only very weak DNA target cleavage was observed even after incubation for 6 h (Supplementary Figure S1E). Interestingly, eAgos also can use gDNA to cleave RNA targets with decreased activity relative to the use of gRNA (32,33). However, for the gRNAs, no MbpAgo-mediated cleavage was observed, neither of DNA nor RNA targets, even after incubation for 12 h (Figure 1C and D; Supplementary Figure S1E). This DNA-guided RNA target preference has not been observed in other eAgos or pAgos homologs, as characterized eAgos use gRNAs to cleave RNA targets and characterized pAgos always prefer to cleave DNA targets. Cleavage of the target strand by MbpAgo occurs at a single site after the 10th nucleotide counting from the 5′-end of the 5′P-gDNA, consistent with previously characterized Ago homologs (Supplementary Figure S1F and 1G) (14). Surprisingly, when guided with 5′OH-gDNA, target cleavage also only occurred between target position 10′-11′ relative to the 5′-end of the guide (Supplementary Figure S1F and 1G), different from that catalyzed by LrAgo (17) and KmAgo (20), as they cleave beyond the canonical cleavage site when guided with 5′OH-gDNA. No cleavage activity was detected for a catalytically dead MbpAgo variant with substitutions of two of the tetrad residues (D566A/D635A; Figure 1C). Moreover, the nucleic acids that co-purify with MbpAgo after expression in E. coli were analyzed (Supplementary Figure S1H). Although no small DNAs were observed, DNAs longer than 45 nt and RNAs of undefined length were found in association with MbpAgo. Previous studies showed that no small DNAs were found in association with the catalytically dead version of TtAgo and SeAgo, demonstrating that their catalytic activity is essential for small DNA biogenesis (4,19). MbpAgo might not acquire short DNA as guides in E. coli for its weak DNA catalytic activity. Future studies are necessary to explore the in vivo function of MbpAgo.
In summary, in contrast to previously characterized pAgos that prefer to cleave DNA targets, MbpAgo prefers to cleave RNA targets rather than DNA targets and can potentially be exploited for RNA manipulation.
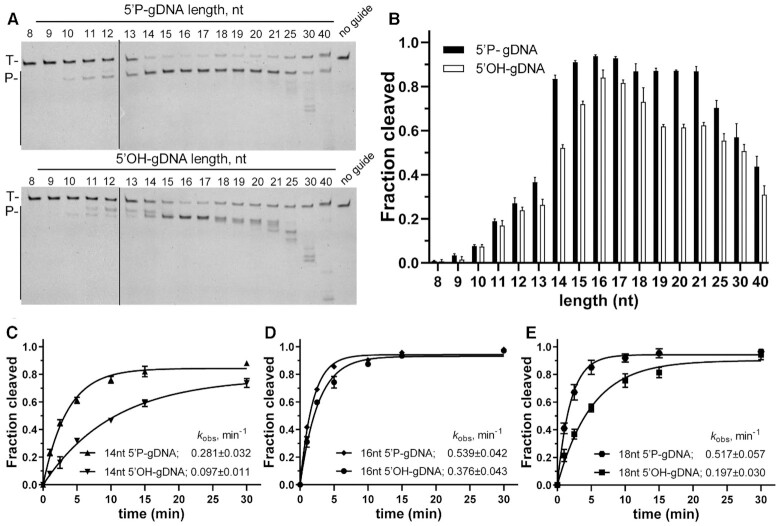
Effects of guide length and presence of 5′P on RNA cleavage by MbpAgo
Previous studies indicated that the presence or absence of 5′P may affect cleavage efficiency and cause the cleavage site to shift relative to the canonical cleavage site (17,22), but MbpAgo can cleave RNA targets with no obvious difference between 5′P-gDNA and 5′OH-gDNA (Figure 1C and Supplementary Figure S1G). Furthermore, previous studies showed that the guide length could also cause the same effect on cleavage efficiency and precision (21). Thus, MbpAgo cleavage efficiency was first investigated using 5′P-gDNA and 5′OH-gDNA of different lengths. MbpAgo was most active with 14–21 nt for 5′P -gDNA and 15–18 nt for 5′OH-gDNA, with a lower efficiency observed with longer or shorter guides (Figure 2A and B). Similar to KmAgo, the cleavage positions were shifted if shorter (11–13 nt) and longer (21–40 nt) 5′P-gDNAs were used (21). Interestingly, although the cleavage position was shifted if shorter (10–14 nt) or longer (18–30 nt) 5′OH -gDNA were used, the cleavage occurred only between the 10th and 11th guide positions when using 15 to 17 nt long 5′OH-gDNA, and the same occurred with 14 to 21 nt long 5′P-gDNA (Figure 2A).
Figure 2.
Both the length and the 5′P of the guide affect cleavage efficiency and precision. (A) Cleavage assays with 5′P-gDNA (Upper panel) and 5′OH-gDNA (Lower panel) of varying lengths. The positions of the targets (T) and cleavage products (P) are indicated on the left of the gels. Representative gels from three independent measurements are shown. (B) Quantification of cleavage efficiencies (the percentage of target cleavage). The fraction of the cleaved target for each guide length is shown. Experiments in (A) and (B) were carried out for 10 min at 37°C. (C–E) Kinetics analyses of RNA cleavage by MbpAgo with 14, 16 and 18 nt gDNAs, respectively. The kobsvalues were determined from the single-exponential fits of the data. Data are represented as the mean ± standard deviation (SD) from three independent experiments. In all experiments, MbpAgo, guide and target were mixed at a 4:2:1 molar ratio (800 nM MbpAgo preloaded with 400 nM guide, plus 200 nM target) and incubated at 37°C.
In this experiment, a change in the guide length and 5′-group (5′P or 5′OH) can affect the target cleavage efficiency and even the precision of the cleavage site, also found in some other pAgos, such as LrAgo and KmAgo (17,20). To further explore the catalytic properties, the kinetics of RNA cleavage by MbpAgo were analyzed with 14 and 18 nt gDNA, which had a markedly different cleavage efficiency or precision compared to 16 nt gDNA (Figure 2A). Under single-turnover conditions, the observed rates (kobs) of RNA cleavage with 5′P-gDNA were faster than those with 5′OH-gDNA, and this distinction was bigger in the 14 and 18 nt guides than in the 16 nt guides (Figure 2C-E; Supplementary Figure S2A-D). For 5′P-gDNA, the kobs value of the 16 nt (0.539 ± 0.042 min–1) was almost identical to that of the 18 nt (0.517 ± 0.057 min–1), but almost two times faster than that of 14 nt (0.281 ± 0.032 min–1). Besides, 18 nt gDNA had an adverse effect on cleavage activity under multiple-round conditions, which is discussed in the next section. However, for 5′OH-gDNA, the kobs value of the 16 nt (0.376 ± 0.043 min–1) was markedly faster than those of 18 nt (0.197 ± 0.030 min–1) and 14 nt (0.097 ± 0.011 min–1). Overall, the most appropriate length of the guides is 16 nt for MbpAgo, and both 16 nt 5′P-gDNA and 5′OH-gDNA direct effective and accurate cleavage of RNA targets. Therefore, most experiments were performed with 16 nt guides.
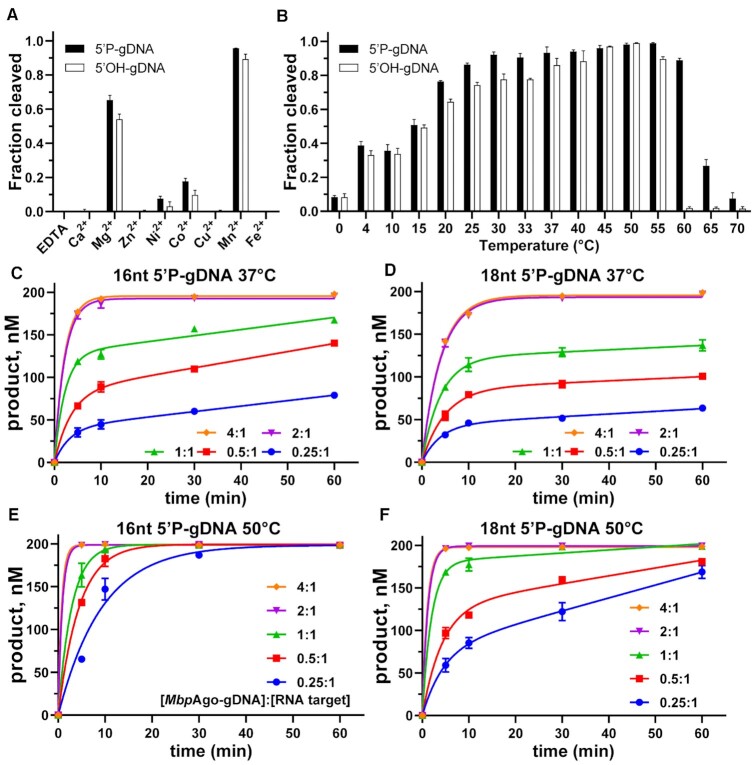
MbpAgo is active under a wide range of reaction conditions and is a multiple-turnover enzyme
To further determine the prerequisites for MbpAgo-mediated target cleavage, the influence of divalent cation type, temperature, and [MbpAgo–gDNA]/RNA target ratios was tested. As divalent metal ions are crucial for Ago protein activity, we test which divalent cations MbpAgo can utilize to mediate DNA-guided RNA target cleavage. MbpAgo was active with Mn2+, Mg2+, Ni2+, and Co2+ as cations, with Mn2+ and Mg2+ giving higher activity than the others (Figure 3A; Supplementary Figure S3A). Titration of Mn2+ and Mg2+ ions showed that, whether the guide is 5′P-gDNA or 5′OH-gDNA, MbpAgo was active at Mn2+ concentrations ≥ 0.01 mM and showed increased cleavage activity at Mn2+ concentrations > 1.0 mM (Supplementary Figure S3B). However, MbpAgo was active at Mg2+ concentrations ≥ 0.2 mM and showed increased cleavage activity at Mg2+ concentrations > 5.0 mM (Supplementary Figure S3C). Thus, MbpAgo-mediated cleavage was more efficient the in presence of Mn2+.
Figure 3.
Characteristics of nuclease activity of MbpAgo. (A) DNA-guided RNA cleavage by MbpAgo with various divalent cations. (B) Temperature dependence of RNA cleavage by MbpAgo. Experiments in (A) and (B) were performed for 15 min at 37°C. (C and D) Quantified data of the MbpAgo-mediated 16 and 18 nt 5′P DNA-guided RNA cleavage turnover experiments using 200 nM RNA target and increasing concentrations of MbpAgo–gDNA (50–800 nM) at 37°C, respectively. (E and F) Quantified data of MbpAgo-mediated 16 and 18 nt 5′P DNA-guided RNA cleavage turnover experiments using 200 nM RNA target and increasing concentrations of MbpAgo–gDNA (50–800 nM) at 50°C, respectively. The MbpAgo–gDNA complex was prepared by mixing MbpAgo with a 1:1 molar ratio of 5′P-gDNA and incubating for 10 min at 37°C. Data were fitted using single-exponential functions if the [MbpAgo–gDNA]/RNA target ratio was > 1. Data were fitted using two-phase functions if the [MbpAgo–gDNA]/RNA target ratio was ≤ 1. Data are the mean ± SD from three independent measurements.
Analyses of the temperature-dependent RNA cleavage activity revealed that MbpAgo bound to 5′P-gDNA displayed comparable levels of RNA cleavage activity between 25°C and 60°C and retained good target RNA cleavage activity at 4–20°C (Figure 3B). The observed absence of differences in the efficiencies of RNA cleavage between 25°C and 60°C is most likely explained by the fact that almost all target is already cleaved at 25°C, so a further increase in the cleavage rate with increasing temperature could not be detected. In the case of 5′OH-gDNA, MbpAgo cleavage activity increased from 0 to 50°C and decreased at >50°C (Figure 3B; Supplementary Figure S3D), suggesting that interactions with the 5′P may be essential in stabilizing the binary MbpAgo–guide complex at elevated temperatures (17). Therefore, although MbpAgo originates from a psychrotolerant bacterium, it is an active DNA-guided RNA nuclease at physiological temperatures and even sufficiently stable to cleave RNA at elevated temperatures.
To investigate the substrate turnover kinetics of MbpAgo, the cleavage in a time course was monitored using variable [MbpAgo–gDNA]/RNA target ratios at 37°C (Figure 3C). Two-phase kinetics with a rapid burst in the initial phase was followed by a slower linear steady state at 37°C under multiple-turnover conditions (when the [MbpAgo–gDNA]/RNA target ratio was < 1.0), which were noticeably slower than the rate of RNA cleavage under single-turnover conditions (when the [MbpAgo–gDNA]/RNA target ratio was > 1.0). According to previously studied Ago proteins (17,18,21,34), the initial burst phase corresponds to the first round of catalysis, following the formation of the ternary pAgo/gDNA/RNA target complex. In contrast, the slow linear phase corresponds to the subsequent rounds of catalysis, after the pAgo–gDNA complex dissociates from the first substrate and rebinds another. From the burst amplitude in the reaction in which the molar ratio of the MbpAgo–gDNA complex to target is 0.25:1, the concentration of active binary MbpAgo/gDNA complexes was estimated as 40 nM (of 50 nM MbpAgo taken in the reaction), corresponding to ∼80% of active MbpAgo in the preparations. The steady-state velocity of RNA cleavage by MbpAgo under these conditions was 0.6436 nM × min−1 [95% confidence interval (95% CI): 0.4593–0.7967 nM × min−1], corresponding to the kobs value of 0.0161 min−1 (based on the 40 nM effective concentration of the binary complex). Then the substrate turnover kinetics experiment was performed with 18 nt gDNA at 37°C (Figure 3D). From the burst amplitude in the reaction in which the molar ratio of the MbpAgo-gDNA complex to target is 0.25:1, the concentration of active binary MbpAgo–gDNA complexes (44 nM) was comparable with the concentration for 16 nt gDNA (40 nM). Interestingly, the steady-state velocity of MbpAgo–mediated target RNA cleavage directed by 18-nt gDNA (0.3121 nM × min−1; 95% CI: 0.2166 – 0.4008 nM × min−1), corresponding to the kobs value of 0.0071 min−1 (based on the 44 nM effective concentration of the binary complex), was two times slower than the velocity measured in the same setup using a 16 nt gDNA. This suggested that the velocity of the binary complex dissociation from the target substrate is decreased in the case of longer gDNAs. In addition, almost all RNA target was cleaved within 60 min when the reaction was performed at 50°C, indicative of a multiple-round reaction (Figure 3E and F). In conclusion, although MbpAgo functions as a multiple-turnover enzyme, its steady-state rate may be limited by product release and exchange of target molecules, such as CbAgo and KmAgo at 37°C (18,21).
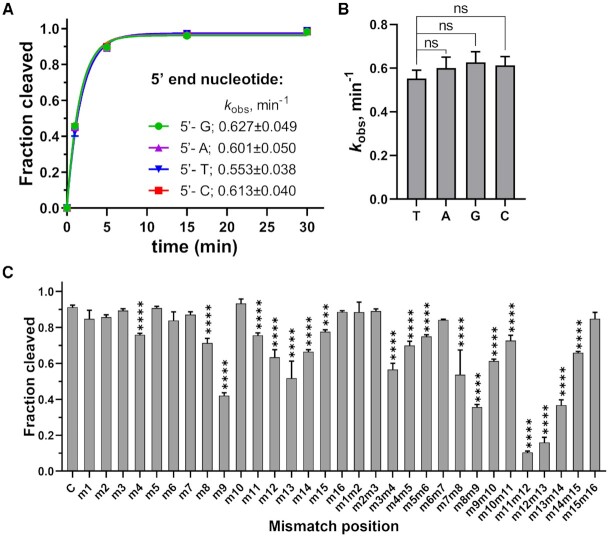
Effects of the 5′-nucleotide of the guide and guide-target mismatches on target cleavage
Previous studies demonstrated that the 5′-nucleotide of the guide strand is bound in the MID pocket of Ago, and many studied eAgos and pAgos have a certain bias for it (5). To determine whether MbpAgo has specificity for the 5′-nucleotide of gDNAs, four guide variants were tested with different 5′-nucleotides but otherwise identical sequences. Like in case of CbAgo and KmAgo, there were no obvious changes in cleavage efficiency and rate when MbpAgo was loaded with gDNAs with different 5′-nucleotides (Figure 4A and B, Supplementary Figure S4A) (17,21).
Figure 4.
Effects of the 5′-nucleotide of the guide and guide-target mismatches on target cleavage. (A) Preferences for the 5′-nucleotide of the guide. The kobsvalues were determined from the single-exponential fits of the data. Data are the mean ± SD from three independent experiments. (B) P-values for all comparisons of kobs values from (A). nsP > 0.05, compared to the 5′-T guide using Student's t-test. (C) Effects of guide–target mismatches on RNA cleavage by MbpAgo. Data are the mean ± SD from three independent measurements. The reaction was performed with 16 nt 5′P-gDNA at 37°C for 5 min. ***P < 0.001 and ****P < 0.0001, compared to C, the control reactions with guide containing no mismatches, using Student′s t-test. In all experiments, MbpAgo, guide and target were mixed at a 4:2:1 molar ratio (800 nM MbpAgo preloaded with 400 nM guide, plus 200 nM target) and incubated at 37°C.
Mismatches between the guide and target affect the cleavage activities of Ago proteins (17,20,32). To check the mismatch tolerance of MbpAgo, the effects of mismatches between the guide and target strands on its RNA cleavage activity were analyzed. A set of gDNAs was designed, each containing a single-nucleotide or dinucleotide mismatches at a certain position (Supplementary Table S2), and tested them in the RNA cleavage reaction with MbpAgo (Figure 4C; Supplementary Figure S4B). When a single-nucleotide mismatch was introduced, mismatches at positions 8 and 9 and 11–15 affected the cleavage efficiency. However, a dramatic decrease in cleavage efficiency was not observed, and only mismatches at positions 9 and 13 reduced cleavage efficiency to ∼50%. Furthermore, although most dinucleotide mismatches affected cleavage efficiency, dinucleotide mismatches at positions 11–13 dramatically reduced target cleavage. Thus, MbpAgo has a high tolerance for mismatches between the guide and target strands.
Binding analyses of guides and targets by MbpAgo
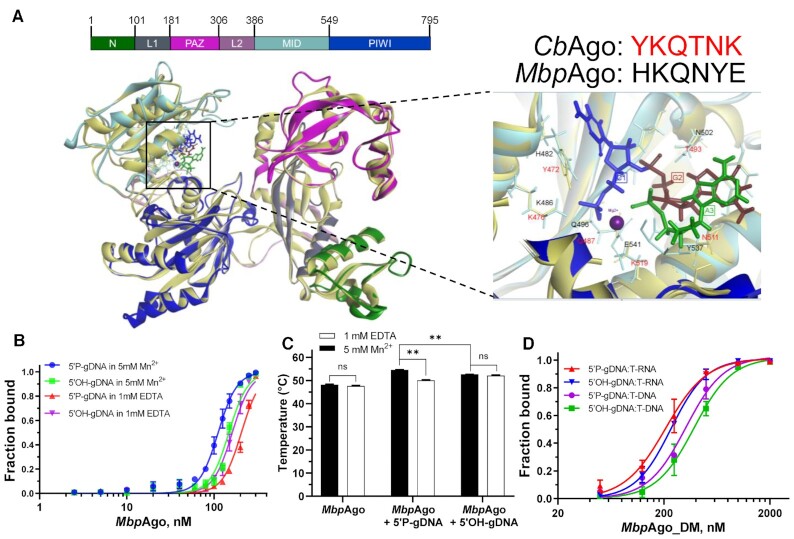
Previous structural and bioinformatic studies showed that the vast majority of pAgos contain the MID and PIWI domains to anchor the 5′-end of a guide by a set of conserved amino acid residues, which constitute a conserved six-amino-acid motif (8). The first two residues of this motif are the most conserved, they might be important for the function of different groups of pAgos, by which pAgos can be classified into several subtypes, including YK, HK, RK, and MID-OH (8). The multiple sequence alignment of the 5′-end guide binding pocket of the MID domain from MbpAgo with several other characterized Ago proteins showed that the first two residues of most characterized Ago proteins are YK, whereas the first two residues of MbpAgo are HK (H482 and K486; Supplementary Figure S5A). Furthermore, the three-dimensional (3D) model built using SWISS-MODEL revealed that the first two amino acid residues in the specific motif involved in the interaction with the guide 5′-end consistent with the multi-sequence alignment results and interactions of the guide 5′end with the MID pocket are similar overall for MbpAgo and CbAgo (Figure 5A and Supplementary Figure S5A). Analysis of the 5′-binding pocket in the MID domain of MbpAgo revealed H482 and K486 in the specific motif involved in interactions with the Me2+ ion and the guide 5′P similar to Y472 and K476 in CbAgo, respectively (Figure 5A) (17).
Figure 5.
Binding analyses of guides and targets by MbpAgo. (A) (Left panel) A 3D model of the MbpAgo aligned to the structure of CbAgo in complex with a gDNA and a DNA target (with bound Mg2+ ions; PDB: 6QZK). MbpAgo domains are colored according to the colored domain architecture of MbpAgo with numbered residues and CbAgo is light yellow. The model was built using the SWISS-MODEL portal. (Right panel) Amino acid residues of the conserved MID-domain motif (shown for MbpAgo and CbAgo above the structure) and Mg2+ ions (purple) involved in interactions with the first nucleotide (blue) and the second nucleotide (deep red) of the guide are highlighted. Elements of the secondary structure and amino residues specific to MbpAgo and CbAgo are shown in cyan and light yellow, respectively. (B) Binding of 16 nt guides by MbpAgo with 5 mM Mn2+ or 1 mM EDTA. The fraction of bound guides was plotted against the protein concentration and fitted using the model of specific binding with the Hill slope. Data are represented as the mean ± SD from three independent experiments. (C) Thermostability of the MbpAgo and MbpAgo–gDNA complex with 5 mM Mn2+ or 1 mM EDTA. The melting temperature of MbpAgo and the MbpAgo-gDNA complex was measured by circular dichroism. Data are the mean ± SD from three independent experiments. P-values for all comparisons of the melting temperature were calculated using Student's t-test. nsP > 0.05 and **P < 0.01. (D) Binding of the target binding by the MbpAgo–gDNA complex with 5 mM Mn2+. The fraction of the bound target was plotted against the MbpAgo–gDNA complex concentration and fitted using the model of specific binding with the Hill slope. Data are represented as the mean ± SD from three independent experiments.
These results indicated that MbpAgo could use 5′OH-gDNAs to cleave RNA targets, with only slightly lower efficiency than 5′P-gDNAs of identical sequence (Figure 2D; Supplementary Figure S2D). The Kd value was further measured for guide binding by MbpAgo using an EMSA (Figure 5B; Supplementary Figure S5B and C). In the presence of Mn2+, the Kd value of MbpAgo associated with a 5′P-gDNA (111.2 ± 3 nM) was lower than that for complexes composed of MbpAgo and 5′OH-gDNA (147.7 ± 5.8 nM), which may be the reason why MbpAgo prefers to use 5′P-gDNAs for efficient target RNA cleavage. Interestingly, the Kd value for binding 5′OH-gDNA in the absence of Mn2+ (161.5 ± 4.8 nM) was slightly higher than the Kd value measured in the presence of Mn2+. In contrast, the Kd value for binding 5′P-gDNA in the absence of Mn2+ (205.4 ± 7.6 nM) was relatively higher than the Kd value measured in the presence of Mn2+. Meanwhile, the thermostability of MbpAgo was also measured using circular dichroism (Figure 5C; Supplementary Figure S5D–F). gDNA binding remarkably improves the thermostability of MbpAgo, similar to hAgo2 and KmAgo (10,20). MbpAgo associated with 5′P-gDNAs has higher thermostability than 5′OH-gDNAs in the presence of Mn2+, which may explain why MbpAgo guided with 5′P-gDNAs can cleave RNA at higher temperatures (Figure 3B). Moreover, the addition of Mn2+ can significantly improve the thermostability of MbpAgo associated with 5′P-gDNAs but has no significant effect on the thermostability of MbpAgo associated with 5′OH-gDNAs. Both the gDNA-binding kinetics and thermal denaturation analyses suggested that divalent metal ions are important for MbpAgo to bind the 5′P group of the guide. The Kd value for target binding was also measured by the MbpAgo–gDNA complex using an EMSA (Figure 5D; Supplementary Figure S5G and H). The Kd value of MbpAgo–gDNA complex binding RNA targets is 207.22 ± 22 nM, lower than the Kd value of MbpAgo–gDNA complex binding DNA targets (322.1 ± 35.5 nM). The higher affinity to RNA targets might be one of the reasons why MbpAgo prefers RNA targets. The real Kd values for guide binding by MbpAgo and target binding by the MbpAgo–gDNA complex are probably lower, given that the effective concentration of MbpAgo is < 100%, owing to the detection limits of measurements.
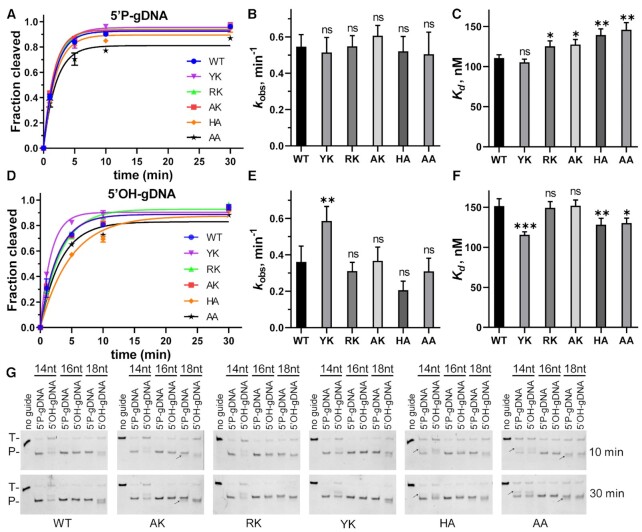
To determine the role of HK in the specific motif, HK was substituted with YK, RK, AK, HA or AA and the resulting MbpAgo variants were tested in cleavage assays. When guided with 5′P-gDNA, the cleavage efficiency and kobs of the variants were comparable to the wild-type (WT) MbpAgo (Figure 6A and B; Supplementary Figure S6A–F). WT MbpAgo had a slightly higher binding affinity for 5′P-gDNA than other variants, except for the YK variant (Figure 6C; Supplementary Figure S7A). However, when guided with 5′OH-gDNA, although the cleavage efficiency and kobs of RK, AK, HA and AA variants were comparable to the WT, the kobs value of the YK variant increased compared with that of the WT (Figure 6D and E; Supplementary Figure S6A–6F). In addition, for the YK variant, the cleavage efficiencies with 5′P-gDNA and 5′OH-gDNA as well as the kobs were similar, which is not the case for all other variants. The binding affinity of the YK variant to 5′OH-gDNA was ∼25% higher than that of the WT for the same 5′OH-gDNA (Figure 6F; Supplementary Figure S7B), which might be one of the reasons why the YK variant has a higher cleavage activity with 5′OH-gDNA. Although previously studied pAgos with YK or RK in the motif have good activity for DNA targets, the DNA target cleavage efficiency of these MbpAgo variants did not change significantly compared to WT (Supplementary Figure S6G). This suggested that the preference of MbpAgo for RNA targets may not be related to the motif. Moreover, the cleavage sites of WT, YK and RK variants guided with 14 or 18 nt 5′P-gDNAs occurred only between target position 10′-11′ relative to the guide 5′-end. Still, the cleavage sites of HA, AK and AA variants guided with 14 or 18 nt 5′P-gDNAs were shifted (Figure 6G). Overall, these results suggested that the HK in the motif indeed was involved in interactions with the 5′-end of a guide.
Figure 6.
Cleavage analyses of MbpAgo variants. (A) Comparison of the kinetic analysis of RNA target cleavage by MbpAgo variants guided by 16 nt 5′P-gDNA. Data were fitted using single exponential functions. In all experiments, MbpAgo variants, guide and target were mixed at a 4:2:1 molar ratio (800 nM MbpAgo preloaded with 400 nM guide, plus 200 nM target) and incubated at 37°C. (B) Comparisons of the kobs values from (A). (C)Kd values of the MbpAgo variants for binding of 16 nt 5′P-gDNA. Experiments in (D–F) were performed with 16 nt 5′OH-gDNA. Data are represented as the mean ± SD from three independent experiments. nsP > 0.05, *P < 0.05, **P < 0.01 and ***P < 0.001 compared to the WT using Student's t-test. (G) Cleavage analyses of different MbpAgo variants with 14, 16 and 18 nt gDNAs, respectively. Positions of the targets (T) and cleavage products (P) are indicated on the left of the gels. Reaction time is indicated on the right of the gels. The black arrow indicates the shifted cleavage products of AK, HA and AA variants. All experiments were performed at 37°C.
MbpAgo can use 5′P-gDNAs and 5′OH-gDNAs to cleave highly-structured RNA
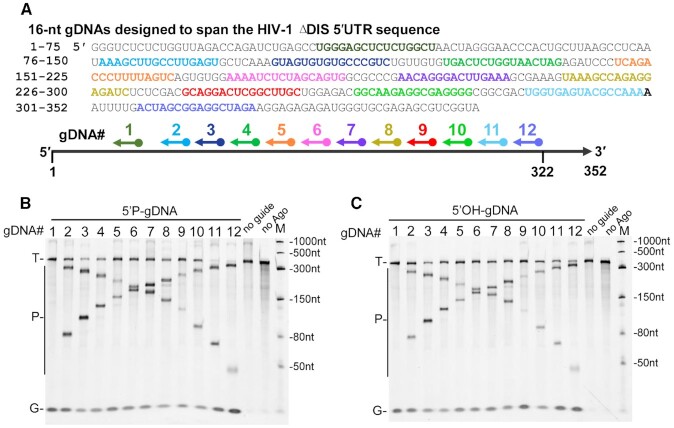
Considering that MbpAgo guided with both 5′P-gDNAs and 5′OH-gDNAs could cleave RNA with high efficiency, this study next examined whether the MbpAgo–guide complex can cleave target sequences in a highly-structured RNA that contains a diverse set of conformational features, such as varying lengths of helices, bulges, hairpin loops, and single-stranded regions. Dayeh et al. predicted the secondary structure of HIV-1 ΔDIS 5-′UTR with SHAPE and designed a set of gDNAs to span the HIV-1 ΔDIS 5-′UTR sequence in 23-nt increments (Supplementary Figure S8A) (32). As 45-nt unstructured RNA targets are cleaved best when gDNAs are 16 nt long (Figure 2A and B), 12 gDNAs (16 nt long) were designed, with target regions (TR) 2 and 4–14 in Dayeh et al.’s research to examine whether MbpAgo could cleave at the same corresponding sites (Figure 7A; Supplementary Figure S8A). Cleavage products were detected with most 5′P-gDNAs and 5′OH-gDNAs, except gDNA-1 located within the highly-stable stems of the transactivation response (TAR; nt 1–57) element when reactions were performed with 5 mM Mn2+ (Figure 7B and C). Then, we tested whether MbpAgo can also cleave highly-structured RNAs under Mg2+, because Mg2+ is the recommended divalent metal ion in the reaction system of some RNA manipulation tool enzymes (such as reverse transcriptase RNA polymerase, and RNA ligase). The results showed that MbpAgo can cleave the highly-structured RNA at the expected position with most gDNA except gDNA-1 with 5 mM Mg2+, albeit a little less efficiently (Supplementary Figure S8B and C). This suggested the possibility of the combined use of MbpAgo and these RNA manipulation tool enzymes. Besides, in vitro cleavage of a double-stranded RNA target could not be detected (Supplementary Figure S8D, lanes 5 and 6). Thus, similar to KpAgo (Kluyveromyces polysporus) and KmAgo (21,32), the MbpAgo–gDNA complex can cleave the target sequences in highly-structured RNAs, and the cleavage efficiency is modulated by the secondary RNA structure.
Figure 7.
Cleavage of highly-structured HIV-1 ΔDIS 5′-UTR RNA by the MbpAgo–gDNA complex. (A) Schematic overview of the MbpAgo-gDNA complex-mediated cleavage assay. Twelve regions (shown in different colors) were selected from the RNA target sequence, with each region targeted by a different 16 nt gDNA. (B) Analyses of the cleavage products obtained after incubation of 5′P-gDNA-MbpAgo complex with HIV-1 ΔDIS 5′-UTR RNA. (C) Analyses of the cleavage products obtained after incubation of the 5′OH-gDNA-MbpAgo complex with HIV-1 ΔDIS 5-′UTR RNA. Experiments in (B) and (C) were carried out with 5 mM Mn2+ at 37°C for 30 min. The positions of the targets (T), gDNAs (G) and cleavage products (P) are indicated on the left of the gels. M, RNA marker.
DISCUSSION
This study characterized a novel pAgo from the psychrotolerant bacterium M. paludis. Compared to known Agos, MbpAgo has an unusual preference for cleaving RNA targets with high efficiency at moderate temperatures but has very weak activity in cleaving DNA targets. Previously, most characterized mesophilic pAgo proteins strongly prefer DNA targets, and have only very weak or undetectable RNA target cleavage activity, including CbAgo, LrAgo, SeAgo, CpAgo and IbAgo (17–19,22). KmAgo can efficiently and precisely cleave RNA targets, but KmAgo also prefers to cleave DNA targets (20). Therefore, MbpAgo is the first example of a pAgo protein that prefers to cleave RNA targets at moderate temperatures. Furthermore, MbpAgo can target RNA under a wide range of reaction conditions. The efficiency and accuracy of cleavage are modulated by temperature, divalent ions, and the phosphorylation and length of gDNAs and their complementarity to the RNA targets.
MbpAgo can utilize both 16 nt 5′OH guides and 5′P guides for efficient RNA target cleavage. MbpAgo is most active with 14–21 nt lengths for 5′P-gDNA and 15–18 nt lengths for 5′OH-gDNA, with a lower cleavage efficiency observed with shorter or longer guides, which is similar to that of other studied pAgos. Although some mesophilic pAgos, including CbAgo and KmAgo, were reported to can use 5′P guides for RNA target cleavage, almost no 5′OH guide-mediated RNA target cleavage activity was detected (17,20). Moreover, target cleavage by most eAgos and pAgos, including hAgo2, CpAgo, IbAgo, LrAgo and KmAgo, with 5′OH guides resulted in a shift of the cleavage site compared to cleavage using 5′P-gDNAs (17,20,22,35). For MbpAgo, although the cleavage site is shifted if shorter or longer guides are used, the cleavage position is no longer shifted with 15 to 17 nt long 5′OH guides. Thus, except for the 5′P that can help to determine the correct register of the guide–target duplex relative to the active site of pAgo, the length of the guide can also modulate the cleavage efficiency and position.
MbpAgo utilizes Mn2+ and Mg2+ as cations, and MbpAgo-mediated cleavage is more efficient in the presence of Mn2+. MbpAgo can cleave RNA target at a wide range of temperatures and still has good RNA cleavage activity at 4–20°C, probably because MbpAgo is from a psychrotolerant bacterium. This suggested that pAgos with good activity at mesophilic temperatures could also be found from psychrotolerant and psychrophilic bacteria. Furthermore, MbpAgo cannot use 5′OH guides to cut targets at 60°C. This indicated that MbpAgo loaded with 5′P-gDNAs may be more stable than that with 5′OH-gDNAs, as demonstrated by measuring the thermostability of MbpAgo using circular dichroism. Several Agos have strong sequence preferences for the 5′-nucleotide of a guide, including KpAgo and MjAgo (32,36). However, MbpAgo has no obvious preference for the 5′-nucleotide of a guide, which is similar to several other pAgos, including KmAgo and CbAgo (17,20). Previous studies on eAgos and pAgos demonstrated the importance of complementarity between the guide and the target for efficient repression. MbpAgo has a high tolerance for mismatches between the guide and target strands. However, only activity comparisons were performed at a single time point. The real effects of mismatches at other positions may be higher in different conditions (e.g. shorter incubation times). Given the weak effects of mismatches on MbpAgo dependent cleavage, MbpAgo can potentially be used to detect or clear the RNA virus, because the virus will not be able to escape easily by mutating single bases.
Except for several pAgos, including MpAgo, which exclusively bind 5′OH guides (13), most eAgos and pAgos were shown to use 5′P guides for RNA cleavage, and multiple interactions between the 5′P group and the MID domain are observed in the structure of some Ago–guide complexes (5,18). A bioinformatic study revealed several subtypes of the MID domain, with substitutions of key residues involved in interactions with the 5′-end of guide molecule (8). The MID domain of most Agos, including CbAgo, RsAgo, and KmAgo, contains the YK subtype, whereas MbpAgo contains the HK subtype (Supplementary Figure S5A). As far as we know, MbpAgo might be the first studied pAgo with the HK subtype in the motif. Although substituting HK with YK, RK, AK, HA or AA has no obvious effect on 16 nt 5′P-gDNA mediated RNA cleavage activity, a single alanine mutation in the HK motif affects the 5′P-gDNA binding affinity and the 14 and 18 nt 5′P-gDNA mediated cleavage precision. Thus, H482 and K486 in the motif contribute to anchoring the 5′phosphate of 5′P-gDNA to ensure the cleavage precision. Furthermore, substituting HK with YK can increase the 5′OH-gDNA mediated RNA cleavage activity, and substituting HK with RK, AK, HA or AA has little effect on 16-nt 5′OH-gDNA mediated RNA cleavage activity. In addition, the affinity of MbpAgo for 5′P-gDNA is 30% higher than the affinity of MbpAgo for 5′-OH-gDNA. The actual differences may be much higher because of the limitations of the method. Interactions with other parts of the guide may be important for stabilizing the 5′OH guide in MbpAgo to ensure that MbpAgo can use 5′OH-gDNA for effective cleavage. Furthermore, the affinity of the MbpAgo–gDNA complex for RNA targets is higher than that for DNA targets, which may be one of the reasons why MbpAgo prefers to cleave RNA targets. Thus, structural studies are necessary to determine the structural basis of the apparent preference for RNA targets.
Finally, we have demonstrated that MbpAgo can efficiently cleave highly-structured RNA targets using both 5′P-gDNAs and 5′OH-gDNAs in the presence of Mg2+ or Mn2+. Previously, eAgo from the budding yeast K. polysporus and pAgo from the mesophilic bacterium K. massiliensis were used for highly-structured RNA cleavage (20,21,32). Similar to these findings, the cleavage efficiency of MbpAgo is modulated by the secondary structure at 37°C. However, using MbpAgo to cleave RNA is more convenient and cost-effective, as the synthesis of 5′OH-gDNAs is easier and more inexpensive than 5′P-gDNAs. Thus, MbpAgo can potentially be applied in RNA-centric in vivo and in vitro methods, such as RNA targeting, antiviral and nucleic acid detection (37,38). In conclusion, MbpAgo is a unique programmable nuclease with a strong preference for RNA targets and can potentially be used in RNA manipulations.
Supplementary Material
Contributor Information
Wenqiang Li, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Yang Liu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Ruyi He, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Longyu Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Yaping Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Wanting Zeng, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Zhiwei Zhang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Fei Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Lixin Ma, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by China National Key Research and Development (R&D) Program (2021YFC2100100); and the Open Funding Project of the State Key Laboratory of Biocatalysis and Enzyme Engineering [SKLBEE2018003]. Funding for open access charge: Ministry of Science and Technology of the People's Republic of China.
Conflict of interest statement. Hubei University has applied for a patent (application no. 202110581929.8) for MbpAgo with M.L., L.W., W.F., H.R. and L.Y. listed as co-inventors.
REFERENCES
- 1. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.J.et al.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lisitskaya L., Aravin A.A., Kulbachinskiy A.. RNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018; 9:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A., Maslova O., Ninova M., Ryazansky S., Leach D.et al.. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020; 587:632–637. [DOI] [PubMed] [Google Scholar]
- 7. Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D.. Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell. 2020; 182:1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryazansky S., Kulbachinskiy A., Aravin A.A.. The expanded universe of prokaryotic Argonaute proteins. Mbio. 2018; 9:e01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools. Nat. Rev. Microbiol. 2017; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 10. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. PNAS. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. PNAS. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu J., Yang J., Cho W.C., Zheng Y.. Argonaute proteins: structural features, functions and emerging roles. J. Adv. Res. 2020; 24:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stockl S., Tinnefeld P., Schneider S., Klingl A.et al.. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 17. Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A.. Programmable DNA cleavage by ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019; 47:5822–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J.. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019; 47:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olina A., Kuzmenko A., Ninova M., Aravin A.A., Kulbachinskiy A., Esyunina D.. Genome-wide DNA sampling by ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 2020; 17:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Li W., Jiang X., Wang Y., Zhang Z., Liu Q., He R., Chen Q., Yang J., Wang L.et al.. A programmable omnipotent argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021; 49:1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kropocheva E., Kuzmenko A., Aravin A.A., Esyunina D., Kulbachinskiy A.. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 2021; 49:4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T., Shi W., Li C., Wang Y., Zhao F.et al.. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 °C. Cell Discov. 2019; 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Enghiad B., Zhao H.. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017; 6:752–757. [DOI] [PubMed] [Google Scholar]
- 25. Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J.et al.. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2020; 48:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He R., Wang L., Wang F., Li W., Liu Y., Li A., Wang Y., Mao W., Zhai C., Ma L.. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. (Camb). 2019; 55:13219–13222. [DOI] [PubMed] [Google Scholar]
- 27. Wang F., Yang J., He R., Yu X., Chen S., Liu Y., Wang L., Li A., Liu L., Zhai C.et al.. PfAgo-based detection of SARS-CoV-2. Biosens. Bioelectron. 2021; 177:112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L., He R., Lv B., Yu X., Liu Y., Yang J., Li W., Wang Y., Zhang H., Yan G.et al.. Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction for detection of SARS-CoV-2 and HPV. Talanta. 2021; 227:122154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin S., Zhan J., Zhou Y.. Argonaute proteins: structures and their endonuclease activity. Mol. Biol. Rep. 2021; 48:4837–4849. [DOI] [PubMed] [Google Scholar]
- 30. Pankratov T.A., Tindall B.J., Liesack W., Dedysh S.N.. Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 2007; 57:2349–2354. [DOI] [PubMed] [Google Scholar]
- 31. Carey M.F., Peterson C.L., Smale S.T.. PCR-mediated site-directed mutagenesis. Cold Spring Harb. Protoc. 2013; 2013:738–742. [DOI] [PubMed] [Google Scholar]
- 32. Dayeh D.M., Cantara W.A., Kitzrow J.P., Musier-Forsyth K., Nakanishi K.. Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA. Nucleic Acids Res. 2018; 46:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lima W.F., Wu H., Nichols J.G., Sun H., Murray H.M., Crooke S.T.. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009; 284:26017–26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haley B., Zamore P.D.. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004; 11:599–606. [DOI] [PubMed] [Google Scholar]
- 35. Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L.. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005; 12:340–349. [DOI] [PubMed] [Google Scholar]
- 36. Willkomm S., Zander A., Grohmann D., Restle T.. Mechanistic insights into archaeal and human argonaute substrate binding and cleavage properties. PLoS One. 2016; 11:e0164695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D., Kellner M.J., Regev A.et al.. RNA targeting with CRISPR-Cas13. Nature. 2017; 550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R.et al.. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020; 181:865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.