Abstract
Botrytis cinerea is a plant-pathogenic fungus infecting over 200 different plant species. We use a molecular genetic approach to study the process of pectin degradation by the fungus. Recently, we described the cloning and characterization of an endopolygalacturonase (endoPG) gene from B. cinerea (Bcpg1) which is required for full virulence. Here we describe the cloning and characterization of five additional endoPG-encoding genes from B. cinerea SAS56. The identity at the amino acid level between the six endoPGs of B. cinerea varied from 34 to 73%. Phylogenetic analysis, by using a group of 35 related fungal endoPGs and as an outgroup one plant PG, resulted in the identification of five monophyletic groups of closely related proteins. The endoPG proteins from B. cinerea SAS56 could be assigned to three different monophyletic groups. DNA blot analysis revealed the presence of the complete endoPG gene family in other strains of B. cinerea, as well as in other Botrytis species. Differential gene expression of the gene family members was found in mycelium grown in liquid culture with either glucose or polygalacturonic acid as the carbon source.
Botrytis cinerea Pers.:Fr. Botryotinia fuckeliana (de Bary) Wetz., also known as the gray mold fungus, is a plant pathogen infecting more than 200 different plant species, including many economically important crops (18). Primary infection often involves invasion of weak, damaged, or senescent tissues. After the initial establishment in the host, the fungus spreads throughout the plant causing severe damage by tissue maceration. During all stages of infection the fungus produces a spectrum of cell-wall-degrading enzymes (CWDEs), among which are several pectin-degrading enzymes such as pectin methyl esterase, pectin lyase, and a number of different polygalacturonases (PGs) (23, 26, 30). Although much biochemical research has been performed, the importance of these enzymes for pathogenesis of B. cinerea was not well understood until recently (13, 22, 34). We set out a molecular genetic approach to study this process.
Recently we described the cloning and characterization of an endopolygalacturonase (endoPG) from B. cinerea (Bcpg1) (38). Elimination of this gene resulted in a mutant with reduced virulence on different hosts, indicating that CWDEs can be involved in pathogenesis. Here we describe the cloning of five additional endoPG genes from B. cinerea and the partial characterization of the gene family.
MATERIALS AND METHODS
Fungal strains and culturing methods.
B. cinerea strains used in this study are indicated in Table 1. Isolates of four other Botrytis species were also used as indicated in Table 1. Fungal strains were grown on malt extract agar (Oxoid, Basingstoke, United Kingdom) at 20°C. For liquid cultures conidia were harvested from 10-day-old plates and used to inoculate Gamborg’s B5 medium (Duchefa Biochemie BV, Haarlem, The Netherlands) supplemented with 1% (wt/vol) glucose and 10 mM (NH4)H2PO4. Cultures were incubated in a rotary shaker at 180 rpm and 20°C. Depending on the growth rate of the different strains and isolates used, cultures were grown for between 16 and 48 h postinoculation prior to harvesting the mycelium.
TABLE 1.
Strains of B. cinerea and other Botrytis species used
| Strain or isolate | Type | Mating type | Origin |
|---|---|---|---|
| Strains | |||
| SAS56 | Monoascospore isolate | MAT1-1 | Italy (of parental strains) |
| SAS405 | Monoascospore isolate | MAT1-2 | Italy (of parental strains) |
| B05.10 | Haploid derivative SAS56 | NDc | Italy (of parental strains) |
| Isolatesa | |||
| Bc7 (tomato, 1970) | The Netherlands | ||
| Bc12 (gerbera, 1986) | The Netherlands | ||
| Bc21 (rose, 1990) | The Netherlands | ||
| Bc29 (gerbera, 1991) | The Netherlands | ||
| J13 (Vitis vinifera, before 1985) | Jena, Germany | ||
| M15 (Vitis vinifera, 1993) | Freiburg, Germany | ||
| Other speciesb | |||
| B. aclada (onion) | |||
| B. gladiorum (gladiolus) | |||
| B. paeoniae (peony) | |||
| B. squamosa (onion) |
The host from which the isolate was obtained and the year of isolation are indicated in parentheses.
The known host is indicated in parentheses.
ND, not determined.
DNA recombinant techniques.
Standard DNA recombinant protocols were used as described before (32). Host strains used were Escherichia coli LE392 for λEMBL3 phages and E. coli DH5α for plasmid propagation. The plasmid vectors pBluescript II SK/KS (Stratagene, La Jolla, Calif.) and pGEM-T-Easy (Promega, Madison, Wis.) were used for DNA fragment cloning.
DNA blot analysis.
DNA was isolated as described previously (25), digested with EcoRI or HindIII (1 μg), separated on a 0.7% (wt/vol) agarose gel, and subsequently alkali blotted onto Hybond N+ membranes according to the manufacturer’s instructions (Amersham). Membranes were hybridized as described earlier (41) at 65 and 55°C for high and low stringency, respectively. High-stringency hybridizations were followed by washing in 0.3 M NaCl plus 0.03 M sodium citrate (pH 7.0) (2× SSC)–0.1% (wt/vol) sodium dodecyl sulfate (SDS), 0.5× SSC–0.1% (wt/vol) SDS, and 0.2× SSC–0.1% (wt/vol) SDS at 65°C for 30 min each. Low-stringency hybridizations were followed by washing in 2× SSC–0.1% (wt/vol) SDS and then 0.5× SSC–0.1% (wt/vol) SDS at 55°C for 15 min each. Autoradiographs were made by 96-h exposure of Kodak-LS/Kodak-AR films at −70°C with one intensifying screen. The following fragments were used for probe preparation for the different genes (numbers indicate the distances of restriction sites from the translation start site): Bcpg1, PstI-BamHI (+161, +862); Bcpg2, NcoI-EcoRI (+858, +1413); Bcpg3, KpnI-KpnI (+766, +1313); Bcpg4, BamHI-BamHI (+384, +891); Bcpg5, BglII-HindIII (+92, +1072); and Bcpg6, ClaI-ClaI (+308, +980).
Medium shift and RNA blot analysis.
The expression of the endoPG gene family was analyzed on two different carbon sources. Gamborg’s B5 medium supplemented with 1% (wt/vol) glucose, 0.05% yeast extract, and 10 mM NaH2PO4-Na2HPO4 (pH 6.0) was inoculated with 106 conidia ml−1 as described above. After 16 h of growth in a rotary shaker at 180 rpm and 20°C, the mycelium was harvested by using Miracloth (Calbiochem, La Jolla, Calif.) and washed thoroughly with Gamborg’s B5 medium supplemented with 10 mM NaH2PO4-Na2HPO4 (pH 6.0). Wet mycelium was transferred to fresh Gamborg’s B5 medium with 10 mM NaH2PO4-Na2HPO4 (pH 6.0) and supplemented with either 1.0% (wt/vol) glucose or 1.0% polygalacturonic acid (U.S. Biochemical Corp., Cleveland, Ohio). After transfer, the fungus was grown for 6, 12, 24, and 30 h prior to harvest of the mycelium by using Miracloth. The harvested mycelium was blotted dry on filter paper, quickly frozen in liquid nitrogen, and stored at −80°C prior to extraction of the RNA. RNA was extracted from frozen mycelium by using the Trizol reagent (Life Technologies, Inc., Gaithersburg, Md.). Then, 10 μg of total RNA was denatured by using glyoxal as described before (32) separated on 1.2% (wt/vol) agarose gel and blotted onto Hybond N membranes with 10× SSC according to the manufacturer’s instructions (Amersham). Membranes were hybridized as described previously (41) at 65°C and washed with 2× SSC–0.1% (wt/vol) SDS (two times for 30 min) and 0.5× SSC–0.1% (wt/vol) SDS (30 min). Autoradiographs were made by exposure of Kodak-LS/Kodak-AR films at −70°C with two intensifying screens. DNA fragments used for probe preparation were similar to those described above for DNA blot analysis. A B. cinerea 27S ribosomal fragment (kindly provided by Theo Prins, Laboratory of Phytopathology, Wageningen Agricultural University, The Netherlands) was used to demonstrate equal loading of the gels.
Screening of genomic library.
A genomic library (λEMBL3) of B. cinerea SAS56 (kindly provided by Theo Prins) was screened (105 phages) with an internal PstI/BamHI fragment (0.7 kb) of Bcpg1 as a probe. Hybridizations and washings were performed as described earlier (41) at 60°C and resulted in the isolation of positive phages. Hybridizing fragments of these phages were subcloned into pBluescript II SK/KS plasmids and further characterized by restriction analysis and Southern hybridizations. This resulted in the identification of different classes of hybridizing clones. Within each class, DNA fragments were further characterized by sequence analysis.
Nucleotide sequence analyses.
Sequencing reactions were performed by using the ThermoSequenase fluorescent-labelled primer cycle sequencing kit (Amersham) with universal sequencing primers and the Cy5 Autoread Sequencing kit (Pharmacia Biotech, Uppsala, Sweden) with gene-specific oligonucleotides. The sequencing reactions were analyzed on an ALF Express sequencer (Pharmacia Biotech). Nucleotide sequence data were analyzed by using the Lasergene Biocomputing Software for Windows (DNASTAR, Inc., Madison, Wis.). BLAST database searches were performed by using the National Center for Biotechnology Information BLAST WWW server. Phylogenetic analyses were performed with PAUP 3.1 (35).
Nucleotide sequence accession numbers.
The nucleotide sequences for the endoPG-encoding genes of B. cinerea are in the GenBank database under accession numbers U68715 (Bcpg1), U68716 (Bcpg2), U68717 (Bcpg3), U68719 (Bcpg4), U68721 (Bcpg5), and U68722 (Bcpg6).
RESULTS
Cloning of the Bcpg gene family members encoding endoPGs.
The isolation and characterization of the Bcpg1 gene has been described (38). Southern analysis of genomic DNA of B. cinerea SAS56, with the Bcpg1 gene as a probe at low-stringency conditions, revealed the presence of at least three additional genes that are homologous to the Bcpg1 gene (38). Screening of the genomic library of B. cinerea (SAS56) with the Bcpg1 gene as a probe resulted in the identification of numerous positive phages, of which 100 hybridizing phages were chosen randomly. A total of 21 clones appeared to contain the Bcpg1 gene as determined by PCR analysis. Eighteen hybridizing phage clones (not containing Bcpg1) were further characterized by using restriction, Southern, and nucleotide sequence analyses. The phages were assigned to five groups of (overlapping) clones, each covering a separate region of the B. cinerea genome. This resulted in the identification of five additional Bcpg genes (Bcpg2 to -6). The complete nucleotide sequences of these genes have been determined and deposited in GenBank.
Genomic organization of the endoPG gene family.
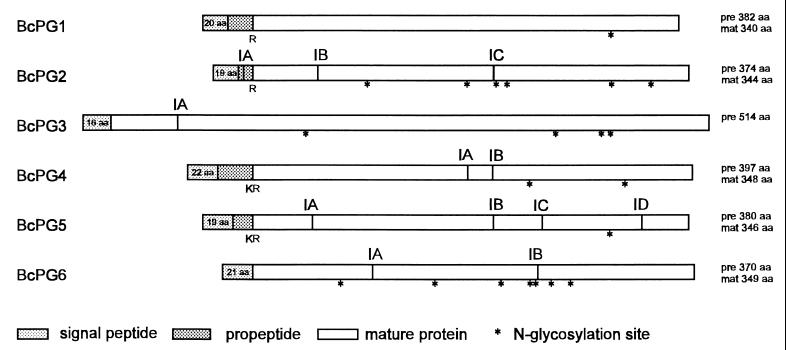
Intron positions in the nucleotide sequences were predicted based on codon usage by using the program Testcode (14) and amino acid sequence alignment with homologous fungal endoPGs from other species. Between 1 and 4 introns are present in the different Bcpg genes, exception for Bcpg1, which is intron-less (Fig. 1). The position of intron A of Bcpg3 was confirmed by sequencing of the reverse transcriptase PCR (RT-PCR) products (37). Indirect evidence for the presence of introns was provided for Bcpg2 (intron C), Bcpg4 (introns A and B), Bcpg5 (introns B and C), and Bcpg6 (intron B) by RT-PCR with primers specifically annealing to regions flanking either side of the intron (37). The border sequences of the introns in the Bcpg genes and the internal consensus for lariat formation corresponded with previously reported 5′ and 3′ splice sites in the fungal genes (39). The introns in the Bcpg genes varied in size between 46 and 60 nucleotides. Conservation of intron positions was only observed for Bcpg2 (intron C), Bcpg4 (intron B), and Bcpg5 (intron B) (Fig. 1).
FIG. 1.
Genomic organization of the endopolygalacturonase gene family of B. cinerea. Indicated are the positions of the introns in the original DNA sequence (IA, IB, IC, or ID), the presence of a putative monobasic (R) or dibasic (KR) cleavage sites, and the presence of N-glycosylation signals (*). Also depicted in the figure are the derived lengths of unprocessed proteins (pre) and mature processed proteins (mat). The lengths of predicted signal peptides for each of the proteins are indicated in the respective boxes.
Analysis of the deduced amino acid sequences.
The amino acid sequences were deduced from the predicted open reading frames present in the genomic sequences of each of the five endoPG genes. The predicted endoPG proteins ranged in length from 371 to 515 amino acids (Fig. 1). All protein sequences contain a predicted signal sequence as determined according to the method of Nielsen et al. (27). Analogous to Aspergillus niger endoPGs, monobasic (Arg) and dibasic (Lys-Arg) cleavage sites (3) were present in most of the Botrytis endoPGs (Arg for BcPG1 and BcPG2; Lys-Arg for BcPG4 and BcPG5). The functionality of the cleavage sites remains to be confirmed by sequencing of the N terminus of the processed proteins. For BcPG6 no apparent propeptide cleavage site could be predicted. The BcPG3 structure is different from the other five genes. The protein is enlarged by the predicted presence of an N-terminal extension of approximately 150 amino acids. BcPG3 contains a predicted signal peptide of 16 amino acids (27) but no putative mono- or dibasic cleavage sites. Sequence identity between the unprocessed endoPGs varied between 34 and 73% (Table 2). Nine amino acid residues which are strictly conserved in all PGs (3) are present in each of the Botrytis endoPGs (also mentioned in reference 38). The presence of N-linked glycosylation signals in all of the endoPGs of B. cinerea (Fig. 1) indicates that they might be excreted as glycosylated enzymes, as is the case for A. niger (43).
TABLE 2.
Sequence pair distance of the endoPG family of B. cinerea as determined by the CLUSTAL method
| endoPG | Sequence pair distance (% identity) of endoPG:
|
||||
|---|---|---|---|---|---|
| BcPG2 | BcPG3 | BcPG4 | BcPG5 | BcPG6 | |
| BcPG1 | 72.0 | 38.7 | 65.8 | 72.7 | 55.0 |
| BcPG2 | 34.8 | 59.2 | 63.7 | 54.3 | |
| BcPG3 | 33.6 | 36.7 | 48.3 | ||
| BcPG4 | 67.7 | 48.5 | |||
| BcPG5 | 55.2 | ||||
Protein sequence alignment of fungal endoPGs.
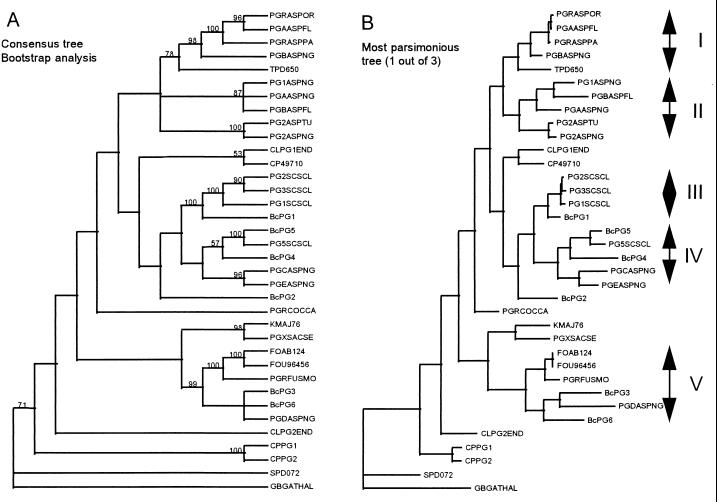
BLAST protein sequence similarity searches were performed by using deduced amino acid sequences of the six endoPG-encoding genes of B. cinerea. The striking homology between amino acid sequences of the Botrytis endoPGs and those from other filamentous fungi prompted us to perform a detailed phylogenetic analysis with a large number of fungal endoPGs. Related fungal endoPGs (Table 3) were used to generate a protein sequence alignment which was used for phylogenetic analysis. A general heuristic search was performed by using PAUP 3.1, and clade stability was assessed by bootstrap replications. Gaps in the alignment were treated as missing values. The Arabidopsis thaliana PG (GBGATHAL) was used as an outgroup (Fig. 2). Figure 2A shows the consensus tree generated from the three most parsimonious trees found in the analysis. Figure 2B shows one of the three most parsimonious trees. The phylogenetic analysis indicated the presence of several groups of related PGs as predicted from the BLAST protein sequence similarity searches. Five different monophyletic groups were distinguished, each containing a minimum of three endoPGs originating from more than one fungal species. Among the fungal species represented in the tree, several possessed PGs belonging to more than one group: for example, A. niger (groups I, II, IV, and V), Sclerotinia sclerotiorum (groups III and IV), Aspergillus flavus (groups I and II) and B. cinerea (groups III, IV, and V). With respect to the endoPGs of B. cinerea, BcPG1 belongs to group III together with three endoPGs of S. sclerotiorum, with sequence identities of around 90%. BcPG3 and BcPG6 cluster with PGD (A. niger) and several endoPGs isolated from different Fusarium species (group V). PGD (28) and BcPG3 are distinct from most endoPGs because of the presence of an N-terminal extension of approximately 150 amino acids. BcPG4 and BcPG5 were assigned to group IV together with PG5 of S. sclerotiorum and PGC and PGE of A. niger. BcPG5 and PG5 of S. sclerotiorum are 89.5% identical at the amino acid level. BcPG2 was related to BcPG1 and BcPG5; however, it was assigned to neither group III nor group IV but was assigned to a separate branch of the tree.
TABLE 3.
EndoPGs used for the construction of the protein sequence alignments
| Organism | Protein | Accession number | Length (aa)a | Source or reference |
|---|---|---|---|---|
| Arabidopsis thaliana | GBGATHAL | S34266 | 445 | Unpublished |
| Aspergillus flavus | PGAASPFL | P41749 | 363 | 42 |
| PGBASPFL | P41750 | 366 | 42 | |
| Aspergillus niger | PG1ASPNG | P26213 | 368 | 4 |
| PG2ASPNG | P26214 | 362 | 7 | |
| PGAASPNG | Y18804 | 370 | 28 | |
| PGBASPNG | Y18805 | 362 | 28 | |
| PGCASPNG | X64356 | 383 | 5 | |
| PGDASPNG | Y18806 | 495 | 28 | |
| PGEASPNG | Y14386 | 378 | 29 | |
| Aspergillus oryzae | PGRASPOR | P35335 | 363 | 21 |
| Aspergillus parasiticus | PGRASPPA | P49575 | 363 | 9 |
| Aspergillus tubingensis | PG2ASPTU | P19805 | 362 | 6 |
| Botrytis cinerea | BcPG1 | U68715 | 382 | 38 |
| BcPG2 | U68716 | 374 | This study | |
| BcPG3 | U68717 | 514 | This study | |
| BcPG4 | U68719 | 397 | This study | |
| BcPG5 | U68721 | 380 | This study | |
| BcPG6 | U68722 | 370 | This study | |
| Claviceps purpurea | CPPG1 | Y10165 | 369 | 36 |
| CPPG2 | Y10165 | 369 | 36 | |
| Cochliobolus carbonum | PGRCOCCA | P26215 | 364 | 33 |
| Colletotrichum lindemuthianum | CLPG1END | X89370 | 363 | 10 |
| CLPG2END | X95475 | 366 | 11 | |
| Cryphonectria parasitica | CP47910 | U49710 | 369 | 16 |
| Fusarium moniliforme | PGRFUSMO | Q07181 | 373 | 8 |
| Fusarium oxysporum | FOAB124 | AB000124 | 371 | Unpublished |
| FOU96456 | U96456 | 370 | 12 | |
| Kluyveromyces marxianus | KMAJ76 | AJ000076 | 361 | Unpublished |
| Saccharomyces cerevisiae | PGXSACSE | P47180 | 361 | Unpublished |
| Sclerotinia sclerotiorum | PG1SCSCL | L12023 | 380 | 31 |
| PG2SCSCL | S62742 | 380 | 15 | |
| PG3SCSCL | S63743 | 380 | 15 | |
| PG5SCSCL | Y13669 | 387 | Unpublished | |
| Stereum purpureum | SPD072 | D45072 | 404 | 24 |
| Trichosporon penicillatum | TPD650 | D89650 | 367 | 17 |
aa, amino acids.
FIG. 2.
Phylogenetic analysis of fungal endoPGs. The analysis was performed by using an optimal alignment generated from the PGs depicted in Table 3. Panel A shows the consensus tree derived from three most parsimonious trees calculated by using PAUP 3.1. The different values represent the percentage of occurrence obtained after bootstrap analysis (1,000 iterations) of the phylogenetic analysis. Panel B shows the one most parsimonious tree and identifies the different monophyletic groups that we defined as a result of the analysis. The abbreviations of protein names are indicated in Table 3.
Presence of the endoPG gene family in different B. cinerea strains and Botrytis species.
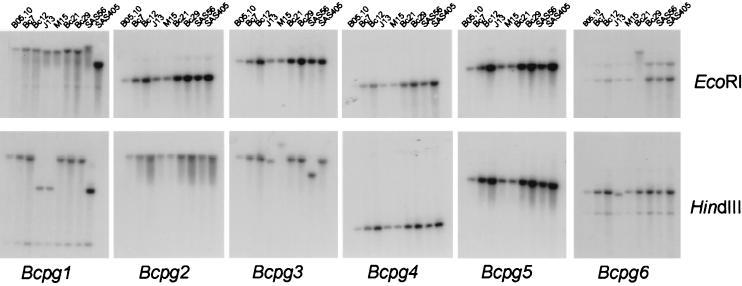
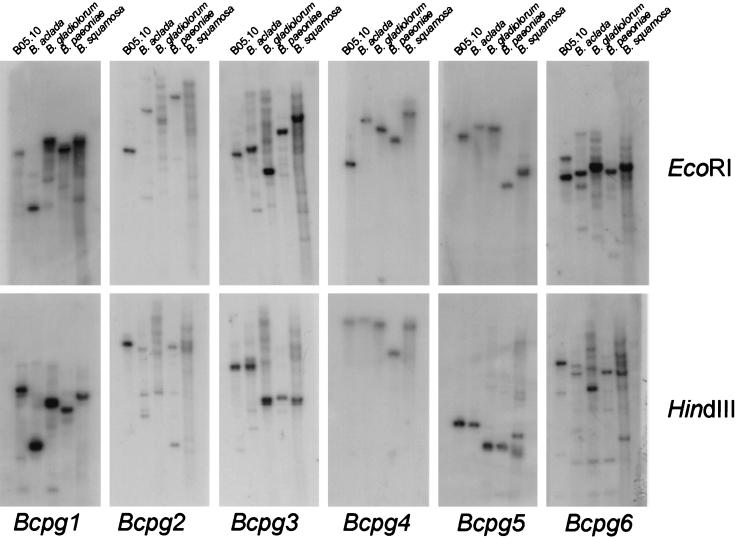
All genes have been cloned from a genomic library of strain SAS56 and might thus be present in only this particular strain. In order to examine the presence of the genes throughout the species B. cinerea, a high-stringency DNA blot analysis was performed with DNA obtained from nine different strains (Table 1). All strains tested showed a hybridizing fragment; however, some restriction fragment length polymorphisms were observed (Fig. 3). In addition, we performed a low-stringency DNA blot analysis with DNA isolated from Botrytis aclada, Botrytis gladiorum, Botrytis paeoniae, and Botrytis squamosa. All Botrytis species tested displayed at least one hybridizing fragment specific for each of the probes used (Fig. 4). For some probes (for example, Bcpg2) the signal was not strong but the observed hybridization pattern was distinct for each probe used. This excludes the occurrence of possible cross-hybridization between the different genes in this experiment, since blots used for the different hybridizations were identical. Apparently, homologues of the entire endoPG gene family found in B. cinerea are also present in other Botrytis species.
FIG. 3.
Southern blot analysis of different strains of B. cinerea with the Bcpg genes as a probe. Fungal DNA isolated from the different strains of B. cinerea was digested with EcoRI (top panel) or HindIII (lower panel) and subjected to Southern hybridization with gene-specific probes (Bcpg1 to -6) under high-stringency conditions as described in Materials and Methods.
FIG. 4.
Southern blot analysis of different species of Botrytis with the Bcpg genes as a probe. Fungal DNA was isolated from the different Botrytis species, digested with EcoRI (top panel) or HindIII (lower panel), and subjected to Southern hybridization with gene-specific probes (Bcpg1 to -6) under low-stringency conditions as described in Materials and Methods.
Expression of the Bcpg gene family on glucose and polygalacturonic acid.
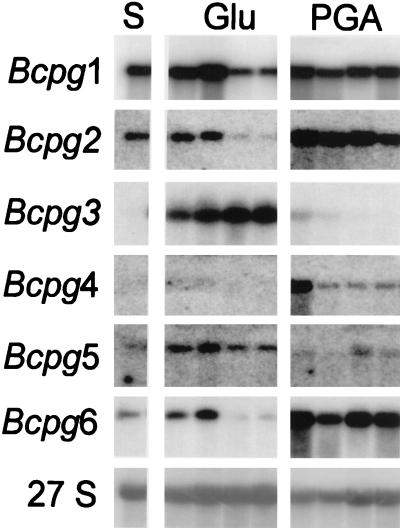
The expression of the Bcpg gene family members in B. cinerea was analyzed in a medium shift experiment in which the fungus was precultured on glucose-containing medium and transferred to medium supplemented with glucose or polygalacturonic acid as sole carbon sources (Fig. 5). RNA blot analysis revealed that each member of the gene family was expressed in liquid culture. High expression of the Bcpg1 gene was observed both on glucose and polygalacturonic acid; however, expression on glucose decreased after a longer incubation period (24 and 30 h posttransfer). The pattern of expression of the Bcpg2 gene was comparable to the Bcpg1 gene expression. Expression of the Bcpg3 gene was low at the time of mycelium transfer (lane S) but increased after prolonged periods of growth on glucose. On polygalacturonic acid hardly any expression of the Bcpg3 gene was observed. For Bcpg4, the highest expression was observed early after transfer to polygalacturonic acid. Expression of the Bcpg4 gene was low or not detectable on glucose. Expression of the Bcpg5 gene was observed on glucose and only at later time points on polygalacturonic acid. Bcpg6 expression was relatively low on glucose and an increased expression was observed after transfer to polygalacturonic acid. Three independent medium shift experiments resulted in the same expression pattern for each of the genes as shown in Fig. 5.
FIG. 5.
Northern blot analysis of B. cinerea B05.10 in a medium shift experiment with the Bcpg genes as a probe. Fungal RNA was isolated from mycelium grown in liquid culture on glucose (Glu) and polygalacturonic acid (PGA) harvested 6, 12, 24, and 30 h after transfer from glucose (S). RNA was subjected to Northern blot hybridization with gene-specific probes (Bcpg1 to -6) under high-stringency conditions as described in Materials and Methods. As loading control, RNA was hybridized with a ribosomal probe (27S) from B. cinerea. Incubation times for autoradiography after hybridization with the different probes were adjusted to obtain equally exposed films.
DISCUSSION
Over the last three decades, plant pathology research has focused on the identification of extracellular enzymes involved in fungal pathogenicity (1, 34). Among them were CWDEs, which include enzymes involved in pectin degradation. It was shown before that the broad-host-range pathogen B. cinerea produces a range of pectinolytic enzymes (19, 23, 26, 30), including up to 13 different PG isoforms (40). The complex pectinolytic system of B. cinerea prompted us to initiate a molecular genetic analysis to unravel the functional role of individual enzymes.
Six different endoPG genes were isolated from B. cinerea SAS56. The sequence identity at the amino acid level within the endoPG family of B. cinerea varied between 34 and 73%. Highly homologous proteins were found in other fungal species by using BLAST protein sequence similarity searches. The phylogenetic analysis performed with a group of 35 related endoPGs clearly resulted in the identification of distinct monophyletic groups of endoPGs originating from different species. This suggests that ancestor genes for these clusters existed prior to the divergence of these fungal species. It is also interesting to note that several species have endoPGs belonging to different monophyletic groups, while others produce only a single known PG. It is possible that other endoPG genes are present in some of these species but that they have not been found yet. The presence of three very closely related PGs (>98%) originating from a single species (S. sclerotiorum) in one group was explained as a recent gene duplication (15). The biological significance for the presence of more than one endoPG in B. cinerea is not known. Enzymes produced by the saprophytic fungus A. niger display considerable differences with respect to substrate specificity, cleavage rate, and optimal pH for activity (2, 20, 29). Information on endoPGs of other fungal species is required to show whether enzymes belonging to the same monophyletic group share biochemical properties which could also be related with a biological function.
We analyzed different B. cinerea strains and other Botrytis species for the presence of DNA homologous to the endoPG-encoding genes of B. cinerea SAS56. Without exception, each member of the gene family was present in all of the strains and species tested, although for some of the genes the hybridizing signal was not strong. Therefore, the presence of the endoPG gene family in B. cinerea is presumably not the sole explanation for its broad host range. The other Botrytis species tested can only infect a single host plant species, yet they contain the homologues of the complete gene family. Whether these genes are functional in these species remains to be determined. Moreover, the saprophytic fungus A. niger produces a spectrum of endoPGs which, nevertheless, do not enable the fungus to infect living plant tissue.
EndoPG genes of B. cinerea appear to be differentially regulated in liquid culture since most of the isoforms produced are only found when the fungus is cultivated on pectin-related carbon sources (19, 40). Those factors affecting the expression pattern in liquid culture might also influence the expression of the endoPG-encoding genes during infection of plants. Previously, we reported that the Bcpg1 gene is expressed during the infection of tomato leaves (38). We analyzed here the expression of the Bcpg gene family when the fungus is grown on two different carbon sources: glucose and polygalacturonic acid. Clear differences in gene expression levels could be observed between members of the Bcpg gene family. Expression of some of the gene family members was observed on both carbon sources (Bcpg1, -2, and -6), while others were predominantly expressed on either glucose (Bcpg3 and -5) or polygalacturonic acid (Bcpg4). We are currently studying the expression of the Bcpg gene family on a range of other carbon sources to identify inducers that may affect gene expression during growth on different plant species. It can be envisaged that a coordinated regulation of gene expression occurs during infection of plants. EndoPGs from constitutively expressed genes might release pectin degradation products which can induce the expression of other endoPG-encoding genes.
Our aim is to unravel the role of the induced and concerted action of endoPGs of B. cinerea during infection of plants. We have already reported that gene replacement of Bcpg1 yielded a fungal mutant with reduced virulence (38). Similar experiments are in progress with other Bcpg gene family members. It will be interesting to investigate whether each individual endoPG of B. cinerea has a specific function during the course of infection. Differential gene expression in combination with specific enzymatic properties of the endoPGs would justify the need for several enzymes in order to optimally degrade pectin polymers under different environmental conditions.
ACKNOWLEDGMENTS
This research was supported by the Dutch Technology Foundation (STW), grant no. WBI 33.3046.
We thank M. A. Kusters-van Someren for her advice at the start of the project, D. K. Aanen for his assistance and advise on the phylogenetic analysis, T. Prins for construction of the genomic library, and M. Hulst who contributed to the gene cloning and sequencing. We also thank J. A. E. Benen and L. Pařenicová for critical reading of the manuscript.
REFERENCES
- 1.Annis S L, Goodwin P H. Recent advances in the molecular genetics of plant cell wall-degrading enzymes produced by plant pathogenic fungi. Eur J Plant Pathol. 1997;103:1–14. [Google Scholar]
- 2.Benen J A E, Kester H C M, Visser J. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur J Biochem. 1999;259:577–585. doi: 10.1046/j.1432-1327.1999.00080.x. [DOI] [PubMed] [Google Scholar]
- 3.Benen J A E, Pařenicová L, Kusters van Someren M, Kester H C M, Visser J. Molecular genetic and biochemical aspects of pectin degradation in Aspergillus. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 331–346. [Google Scholar]
- 4.Bussink H J D, Brouwer K B, De Graaff L H, Kester H C M, Visser J. Identification and characterization of a second polygalacturonase gene of Aspergillus niger. Curr Genet. 1991;20:301–308. doi: 10.1007/BF00318519. [DOI] [PubMed] [Google Scholar]
- 5.Bussink H J D, Buxton F P, Fraaye B A, De Graaff L H, Visser J. The polygalacturonases of Aspergillus niger are encoded by a family of diverged genes. Eur J Biochem. 1992;208:83–90. doi: 10.1111/j.1432-1033.1992.tb17161.x. [DOI] [PubMed] [Google Scholar]
- 6.Bussink H J D, Buxton F P, Visser J. Expression and sequence comparison of the Aspergillus niger and Aspergillus tubingensis genes encoding polygalacturonase II. Curr Genet. 1991;19:467–474. doi: 10.1007/BF00312738. [DOI] [PubMed] [Google Scholar]
- 7.Bussink H J D, Kester H C M, Visser J. Molecular cloning, nucleotide sequence and expression of the gene encoding prepro-polygalacturonase II of Aspergillus niger. FEBS lett. 1991;273:127–130. doi: 10.1016/0014-5793(90)81066-w. [DOI] [PubMed] [Google Scholar]
- 8.Caprari C, Richter A, Bergmann C, Lo Cicero S, Salvi G, Cervone F, De Lorenzo G. Cloning and characterization of a gene encoding the endopolygalacturonase of Fusarium moniliforme. Mycol Res. 1993;97:497–505. [Google Scholar]
- 9.Cary J W, Brown R, Cleveland T E, Whitehead M P, Dean R A. Cloning and characterization of a novel polygalacturonase-encoding gene from Aspergillus parasiticus. Gene. 1995;153:129–133. doi: 10.1016/0378-1119(94)00749-i. [DOI] [PubMed] [Google Scholar]
- 10.Centis S, Dumas B, Fournier J, Marolda M, Esquerré Tugayé M T. Isolation and sequence analysis of clpg1, a gene coding for an endopolygalacturonase of the phytopathogenic fungus Colletotrichum lindemuthianum. Gene. 1996;170:125–129. doi: 10.1016/0378-1119(95)00867-5. [DOI] [PubMed] [Google Scholar]
- 11.Centis S, Guillas I, Sejalon N, Esquerré Tugayé M T, Dumas B. Endopolygalacturonase genes from Colletotrichum lindemuthianum: cloning of clpg2 and comparison of its expression to that of clpg1 during saprophytic and parasitic growth of the fungus. Mol Plant-Microbe Interact. 1997;10:769–775. doi: 10.1094/MPMI.1997.10.6.769. [DOI] [PubMed] [Google Scholar]
- 12.Di Pietro A, Roncero M I G. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol Plant-Microbe Interact. 1998;11:91–98. doi: 10.1094/MPMI.1998.11.2.91. [DOI] [PubMed] [Google Scholar]
- 13.Edlich W, Lorenz G, Lyr H, Nega E, Pommer E-H. New aspects on the infection mechanism of Botrytis cinerea Pers. Neth J Plant Pathol. 1989;95:53–62. [Google Scholar]
- 14.Fickett J W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraissinet Tachet L, Reymond Cotton P, Fèvre M. Characterization of a multigene family encoding an endopolygalacturonase in Sclerotinia sclerotiorum. Curr Genet. 1995;29:96–99. doi: 10.1007/BF00313199. [DOI] [PubMed] [Google Scholar]
- 16.Gao S, Choi G H, Shain L, Nuss D L. Cloning and targeted disruption of enpg-1, encoding the major in vitro extracellular endopolygalacturonase of the chestnut blight fungus, Cryphonectria parasitica. Appl Environ Microbiol. 1996;62:1984–1990. doi: 10.1128/aem.62.6.1984-1990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iguchi K I, Hirano H, Kishida M, Kawasaki H, Sakai T. Cloning of a protopectinase gene of Trichosporon penicillatum and its expression in Saccharomyces cerevisiae. Microbiology. 1997;143:1657–1664. doi: 10.1099/00221287-143-5-1657. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis W R. Botryotinia and Botrytis species: taxonomy, physiology, and pathogenicity: a guide to the literature. Ottawa, Ontario, Canada: Canada Department of Agriculture; 1977. [Google Scholar]
- 19.Johnston D J, Williamson B. An immunological study of the induction of polygalacturonases in Botrytis cinerea. FEMS Microbiol Lett. 1992;97:19–24. [Google Scholar]
- 20.Kester H C M, Visser J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem. 1990;12:150–160. [PubMed] [Google Scholar]
- 21.Kitamoto N, Kimura T, Kito Y, Ohmiya K, Tsukagoshi N. Structural features of a polygalacturonase gene cloned from Aspergillus oryzae KBN616. FEMS Microbiol Lett. 1993;111:37–41. doi: 10.1111/j.1574-6968.1993.tb06358.x. [DOI] [PubMed] [Google Scholar]
- 22.Leone G. Significance of polygalacturonase production by Botrytis cinerea in pathogenesis. In: Verhoeff K, Malathrakis N E, Williamson B, editors. Recent advances in Botrytis research. Wageningen, The Netherlands: Pudoc Scientific Publishers; 1992. pp. 63–68. [Google Scholar]
- 23.Leone G, Schoffelmeer E A M, Van den Heuvel J. Purification and characterization of a constitutive polygalacturonase associated with the infection process of French bean leaves by Botrytis cinerea. Can J Bot. 1990;68:1921–1930. [Google Scholar]
- 24.Miyairi K, Senda M, Watanabe M, Hasui Y, Okuno T. Cloning and sequence analysis of cDNA encoding endopolygalacturonase I from Stereum purpureum. Biosci Biotechnol Biochem. 1997;61:655–659. doi: 10.1271/bbb.61.655. [DOI] [PubMed] [Google Scholar]
- 25.Möller E M, Bahnweg G, Sandermann H, Geiger H H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20:6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Movahedi S, Heale J B. The roles of aspartic proteinase and endo-pectin lyase enzymes in the primary stages of infection and pathogenesis of various host tissues by different isolates of Botrytis cinerea Pers ex. Pers Physiol Mol Plant Pathol. 1990;36:303–324. [Google Scholar]
- 27.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Pařenicová, L. 1998. Unpublished results.
- 29.Pařenicová L, Benen J A E, Kester H C M, Visser J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur J Biochem. 1998;251:72–80. doi: 10.1046/j.1432-1327.1998.2510072.x. [DOI] [PubMed] [Google Scholar]
- 30.Reignault P, Mercier M, Bompeix G, Boccara M. Pectin methylesterase from Botrytis cinerea: physiological, biochemical and immunochemical studies. Microbiology. 1994;140:3249–3255. [Google Scholar]
- 31.Reymond P, Deléage G, Rascle C, Fèvre M. Cloning and sequence analysis of a polygalacturonase-encoding gene from the phytopathogenic fungus Sclerotinia sclerotiorum. Gene. 1994;146:233–237. doi: 10.1016/0378-1119(94)90298-4. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Scott-Craig J S, Panaccione D G, Cervone F, Walton J D. Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell. 1990;2:1191–1200. doi: 10.1105/tpc.2.12.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staples R C, Mayer A M. Putative virulence factors of Botrytis cinerea acting as a wound pathogen. FEMS Microbiol Lett. 1995;134:1–7. [Google Scholar]
- 35.Swofford D L. PAUP: phylogenetic analysis using parsimony version 3.1.1. Champaign, Ill: Illinois Natural History Survey; 1993. [Google Scholar]
- 36.Tenberge K B, Homann V, Oeser B, Tudzynski P. Structure and expression of two polygalacturonase genes of Claviceps purpurea oriented in tandem and cytological evidence for pectinolytic enzyme activity during infection of rye. Phytopathology. 1996;86:1084–1097. [Google Scholar]
- 37.Ten Have, A. 1998. Unpublished results.
- 38.Ten Have A, Mulder W, Visser J, Van Kan J A L. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant-Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 39.Unkles S E. Gene organization in filamentous fungi. In: Kinghorn J R, Turner G, editors. Applied molecular genetics of filamentous fungi. Glasgow, Scotland: Blackie Academic and Professional; 1992. pp. 28–53. [Google Scholar]
- 40.Van der Cruyssen G, De Meester E, Kamoen O. Expression of polygalacturonases of Botrytis cinerea in vitro and in vivo. Meded Fac Landbouwkd Toegep Biol Wet Univ Gent. 1994;59:895–905. [Google Scholar]
- 41.Van der Vlugt Bergmans C J B, Wagemakers C A M, Van Kan J A L. Cloning and expression of the cutinase A gene of Botrytis cinerea. Mol Plant-Microbe Interact. 1997;10:21–29. doi: 10.1094/MPMI.1997.10.1.21. [DOI] [PubMed] [Google Scholar]
- 42.Whitehead M P, Shieh M T, Cleveland T E, Cary J W, Dean R A. Isolation and characterization of polygalacturonase genes (pecA and pecB) from Aspergillus flavus. Appl Environ Microbiol. 1995;61:3316–3322. doi: 10.1128/aem.61.9.3316-3322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Bergmann C, Benen J, Orlando R. Identification of the glycosylation site and glycan structures of recombinant endopolygalacturonase II by mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1257–1262. doi: 10.1002/(SICI)1097-0231(199708)11:12<1257::AID-RCM19>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]