Abstract
Although previous studies identified numerous single nucleotide polymorphisms (SNPs) and their target genes predisposed to prostate cancer (PrCa) risks, SNP-related splicing associations are rarely reported. In this study, we applied distance-based sQTL analysis (sQTLseekeR) using RNA-seq and SNP genotype data from benign prostate tissue (n = 467) and identified significant associations in 3344 SNP-transcript pairs (P ≤ 0.05) at PrCa risk loci. We characterized a common SNP (rs7247241) and its target gene (PPP1R14A) located in chr19q13, an sQTL with risk allele T associated with upregulation of long isoform (P = 9.99E−7). We confirmed the associations in both TCGA (P = 2.42E−24) and GTEX prostate cohorts (P = 9.08E−78). To functionally characterize this SNP, we performed chromatin immunoprecipitation qPCR and confirmed stronger CTCF and PLAGL2 binding in rs7247241 C than T allele. We found that CTCF binding enrichment was negatively associated with methylation level at the SNP site in human cell lines (r = −0.58). Bisulfite sequencing showed consistent association of rs7247241-T allele with nearby sequence CpG hypermethylation in prostate cell lines and tissues. Moreover, the methylation level at CpG sites nearest to the CTCF binding and first exon splice-in (ψ) of PPP1R14A was significantly associated with aggressive phenotype in the TCGA PrCa cohort. Meanwhile, the long isoform of the gene also promoted cell proliferation. Taken together, with the most updated gene annotations, we reported a set of sQTL associated with multiple traits related to human prostate diseases and revealed a unique role of PrCa risk SNP rs7247241 on PPP1R14A isoform transition.
Introduction
Since 2005, approximately 4000 GWASs have been published, identifying ~136 287 SNPs associated with susceptibility to over 1000 unique traits and common diseases. Among these findings, over 700 SNPs were recognized to be associated with an increased risk to prostate cancer (PrCa) (1,2). Thus far, as most of these risk SNPs have been found in non-coding regions of the genome, with many residing some distance from nearby annotated genes, it is believed that many of these (or their closely linked causal SNPs) will be located in regulatory domains of the genome that control gene expression rather than in coding regions that directly affect protein function (3,4). Additionally, a small fraction of these SNPs is located in the exon–intron junction region that determines allele-specific RNA splicing, which may serve as an important driving force for tumorigenesis (5,6). However, due to the weak effect of each SNP, there has been a significant knowledge gap between GWAS findings and their underlying biological mechanisms. This issue has been challenging both population and basic scientists since the first GWAS report.
In the past decades, expression quantitative trait loci (eQTL) analysis has been widely applied to identify numerous PrCa risk SNP gene associations by integrating gene expression and genotype information. However, only a few of these associations have been validated by low-throughput function assays (7,8). Despite providing insights into the biological significance of risk SNPs, eQTL analysis showed several constraints in determining the causal variants. Because of linkage disequilibrium (LD), eQTL analysis can identify significant associations between one certain gene and many highly correlated SNPs but is not able to determine causal SNPs. Additionally, allele-dependent isoform change may only reflect subtle fluctuations at the gene level, thus resulting in weak eQTL signals. Therefore, there is a need to expand eQTL analysis from the whole gene to transcription levels, leading to splicing quantitative trait loci (sQTL) (9–11). This approach utilizes isoform percentages within each gene to dissect genotypes that can exactly categorize different isoform distributions, thereby linking variants with isoform transitions (9).
Previously, we had performed RNA sequencing and germline genotyping on a large number of normal prostate tissues (12). In this study, we performed isoform percentage-based sQTL analysis (9) to identify allelic transcript alterations on risk SNPs related to PrCa risk. This analysis identified a set of novel sQTL revealing transcript transitions between different germline variants. Furthermore, we characterized a PrCa risk variant rs7247241 for its allelic transcription factor (TF) binding and differential methylation, as well as its target gene protein phosphatase 1 regulatory subunit 14A (PPP1R14A), which encodes a small protein with phosphatase activities, and has been reported to be relevant to pancreatic cancer (13), melanoma (14), colorectal cancer and prostate cancer (15). Additionally, we further characterized the unique role of CCCTC-binding factor (CTCF) in regulating DNA CpG methylation. The data from this study will provide insight about cross talk between TF binding and epigenetic changes and help fill the knowledge gap between phenotypical associations and PrCa etiology.
Results
sQTL analysis identified risk SNP-related isoform alterations
To identify the PrCa-related sQTL, we first performed a two-phase population-based imputation analysis using our PrCa cohort including 467 germline genotype data from Infinium Omni2.5–8 BeadChip array. After removing SNPs by Hardy–Weinberg Equilibrium P-value (≤10E−6) and missing rate (≥5%), we converted Illumina kgp identifiers into rsID according to Illumina manifest file. After imputation against the 1000G European population reference, we received genotype data for a total of 17 076 866 SNPs. By applying filtering criteria of MAF ≥ 1% and imputation quality (R2 > 0.3), a total of 9 256 807 high-quality SNPs were kept as the final genotype source.
To identify PrCa-related SNPs, we queried GWAS-catalog and retrieved 766 PrCa risk SNPs by keywords ‘prostate carcinoma’ (EFO_0001663), 68 PSA risk SNPs by ‘prostate-specific antigen measurement’ (EFO_0004264) and 30 BPH risk SNPs by ‘benign prostatic hyperplasia’ (EFO_0000284). These SNPs were further batch queried in LDproxy to generate 21 986 for PrCa, 2252 for PSA and 1497 for BPH risk-associated SNPs (based on R2 ≥ 0.4) in the European population. Genotypes were obtained from imputation results to run the downstream analysis.
To quantify isoform expression from RNA-seq data, we used the RSEM program and ENSEMBL gene annotations. To find cis sQTL, we applied the sQTLseekeR package to index the genotype table, transform isoform expression and perform association analysis within each chromosome. The sQTLseekeR implemented a unique svQTL P-value in the output, which identifies false-positive sQTL through testing variance heterogeneity of transcript relative expression between genotypes (9). For PrCa sQTL, we were able to identify a total of 3344 associations between risk-associated SNPs and isoform transitions with a P-value below 0.05 or 1549 associations with an svQTL P-value below 0.1. For PSA sQTL, we found a total of 645 associations with a P-value below 0.05 or 78 associations with an svQTL P-value below 0.1. For BPH sQTL, we found a total of 170 associations with a P-value below 0.05 or 34 associations with an svQTL P-value below 0.1. Three representative PrCa-related sQTL were shown with sashimi plots for merged sample reads from each genotype, including BABAM1-rs10424178 (Supplementary Material, Fig. S1A and B, P = 9.99E−7, svQTL P-value = 0.070), LILRB2-rs443874 (Supplementary Material, Fig. S1C and D, P = 3E−5, svQTL P-value = 0.046) and FAM118A-rs1569414 (Supplementary Material, Fig. S1E and F, P = 9.99E−7, svQTL P-value = 9.9E−5). For PSA and BPH sQTL, only a small fraction was intersected with PrCa sQTL (Supplementary Material, Fig. S2A). We also showed two additional sQTL, including WDR11-rs35980300 (Supplementary Material, Fig. S2B and C, P = 2.9E−5, svQTL P-value = 0.077) and RNASEH2BAS1-rs1870839 (Supplementary Material, Fig. S2D and E, P = 9.99E−7, svQTL P-value = 9.9E−5). The detailed list and boxplot for each association can be found in Supplementary Material, Table S1 and Materials and Methods.
rs7247241 T allele is associated with PPP1R14A long isoform
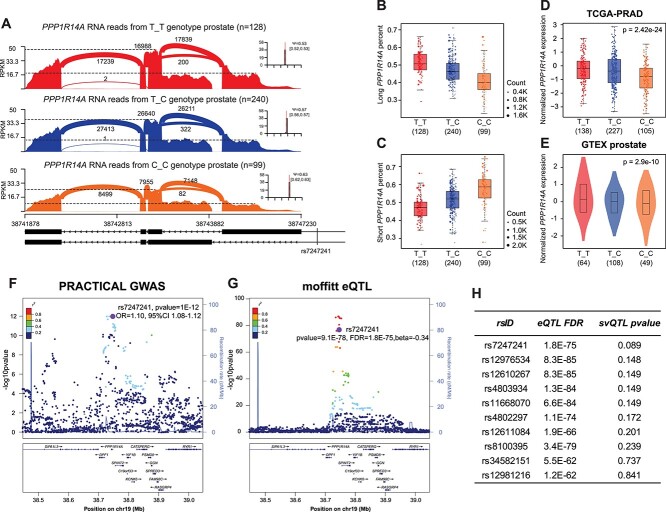
Following the sQTL analysis pipeline (Fig. 1A), we found a significant association of SNP rs7247241 with long isoforms of PPP1R14A gene (Fig. 1B, P = 9.99E−7, svQTL P-value = 0.089). rs7247241 is a common SNP located in PPP1R14A 5UTR region, with 60.78% of T and 39.22% of C allele in the European population. Sashimi plot in PPP1R14A region showed clear difference between T_T and C_C genotype (Fig. 2A). The percentage of splice-in (PSI, ψ) demonstrated more PPP1R14A first exon usage in the T_T genotype (Fig. 2B and C). In TCGA and GTEX eQTL data, T_T genotype samples also exhibited higher PPP1R14A gene expression (Fig. 2D and E). In GWAS meta-analysis, rs7247241 T allele conferred a 10% higher PrCa risk in the European population (Fig. 2F). It’s worth mentioning that rs7247241 showed a statistical advantage in the sQTL analysis. Among its LD SNPs, the gene-level eQTL analysis (Fig. 2G) at the PPP1R14A locus demonstrated a significant but not the most significant signal for rs7247241. However, sQTL analysis showed the smallest svQTL P-value for rs7247241 (Fig. 2H).
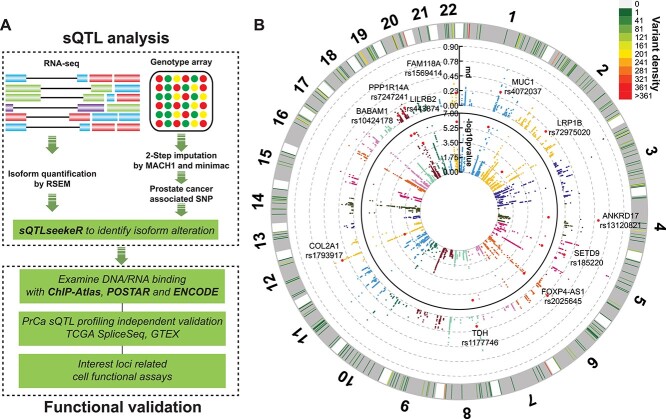
Figure 1.
Study design and representative sQTL associated with PrCa risk. (A) The current study consists of two major parts: sQTL analysis (raw data analysis) and functional validation (experimental). (B) The circos plot demonstrated main findings from sQTL analysis with PrCa risk SNPs. Inside circle denotes negative log base 10 of sQTL P-value, while outside circle denotes the maximum difference in splicing ratios between genotype groups (md).
Figure 2.
Associations between rs7247241 T allele and PPP1R14A long isoform. (A) Sashimi plot of PPP1R14A gene coverages between rs7247241 genotypes in benign prostate tissues. (B, C) Relative expressions of long (ENST00000301242) (B) and short (ENST00000591585) (C) PPP1R14A isoform in three rs7247241 genotypes. (D, E) Association between PPP1R14A gene expressions and rs7247241 genotypes in TCGA prostate cancer (D) and GTEX prostate (E) cohorts. (F, G) Prostate cancer GWAS signals (F) and PPP1R14A eQTL signals (G) at rs7247241 locus. (H) Top-ranked svQTL P-value for PPP1R14A gene eQTL and isoform sQTL at rs7247241 locus.
Transcription factor binding at rs7247241 locus is allele-specific
To characterize biological function around the rs7247241 locus, we inspected the SNP region within the ChIP-Atlas database (16). As expected, we found variable CTCF occupancies in multiple prostate cell lines (Fig. 3A) near the SNP, centered on a highly confident CTCF binding motif according to JASPAR TF prediction (Fig. 3B, score = 16.4). Additionally, rs7247241 was also residing in a PLAGL2 factor binding site (Fig. 3B). To confirm their regulatory role, we knocked down CTCF and PLAGL2 and observed increased PPP1R14A expression in primary prostate epithelial cells (PrEC, Fig. 3C). We further performed correlation analysis using primary tumor tissue RNA profiling datasets and found a negative correlation between CTCF/PLAGL2 and PPP1R14A expression in ICGC (Fig. 3D), TCGA (Fig. 3E) and Broad Cornell (Fig. 3F) prostate cancer cohorts. To ascertain the TF binding at rs7247241, we performed chromatin immunoprecipitation (ChIP) reactions with antibodies against CTCF and PLAGL2, which could bind around rs7247241 (rs7247241-TFBS) in 22Rv1 and RWPE-1 cell lines. We found that this genomic locus was significantly enriched by both antibodies used to capture 22Rv1 chromatins (Fig. 3G), although only enriched CTCF binding in RWPE-1 cells (Fig. 3H). Compared with negative control, the enrichment at rs7247241-containing region was 15.7-fold higher in 22Rv1 and 6-fold higher in RWPE-1 for CTCF binding.
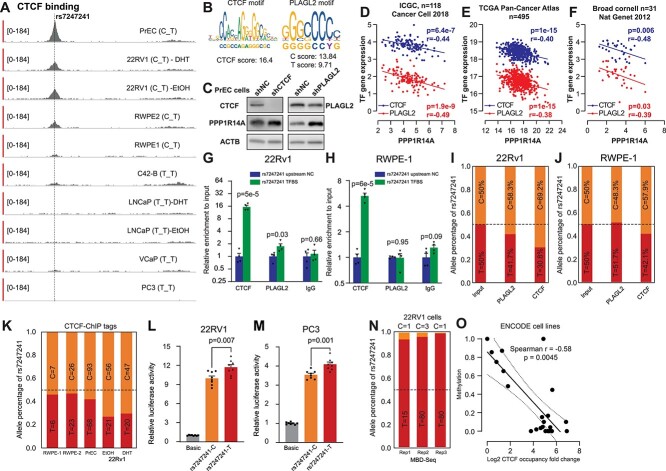
Figure 3.
Allele-specific transcription factor binding at rs7247241. (A) rs7247241 is located in the PPP1R14A promoter region with CTCF binding across human prostate cell lines. (B) JASPAR prediction of rs7247241 flanking sequences with allele difference. (C) Western blot analysis of CTCF, PLAGL2 and PPP1R14A protein expressions after stably knocking down CTCF and PLAGL2 in PrEC cells. (D–F) mRNA correlation between a putative regulatory transcription factor and PPP1R14A in ICGC (D), TCGA (E) and Broad Cornell (F) prostate primary tumor samples. (G, H) Relative enrichment measured by CTCF and PLAGL2 ChIP-qPCR at rs7247241 locus in 22Rv1 (G) and RWPE-1 cells (H). Columns represent the means of a total of four independent experiments. Error bars represent the standard error of the mean. (I, J) rs7247241 allele dosages in ChIP-qPCR amplicons from 22Rv1 (I) and RWPE-1 cells (J). (K) rs7247241 allele dosages in CTCF ChIP-seq tags from rs7247241 heterozygote prostate cells. (L, M) Quantification of promoter activities between rs7247241 T and C alleles by reporter assays in 22Rv1 (L) and PC3 (M) cells. Columns represent the means of a total of eight independent experiments. Error bars represent the standard error of the mean. (N) rs7247241 allele dosages in MBD-seq reads in 22Rv1 cells. (O) Correlations between CTCF occupancies and methylation status at rs7247241 locus in 22 human cell lines from ENCODE database. Each dot represents a cell line, and the solid line indicates the linear regression coefficient, and the dashed lines depict 95% confidence interval.
To confirm the allele-specific TF binding, we sequenced this region from each ChIP-enriched chromatin and input control DNA. We found that rs7247241 C allele was significantly enriched than T allele in both antibody-captured chromatins in 22Rv1 (Fig. 3I). For RWPE-1 cells, rs7247241 C allele count was also higher than T allele in CTCF-captured chromatins (Fig. 3J). To independently validate our finding, we retrieved ChIP-seq (Supplementary Material, Table S3) reads covering rs7247241 from heterozygote cells and summarized allele percentages for both alleles. As expected, the rs7247241 C allele showed higher read counts than T alleles, especially in those cell lines with higher ChIP-seq coverages (Fig. 3K). Additionally, similar tendencies were also present in immortalized lymphocyte cell lines (Supplementary Material, Fig. S3A and B). Since the variant resided in the promoter region, we performed luciferase reporter assay and confirmed higher transcription potential of T allele than C allele in both 22Rv1 (Fig. 3L) and PC3 (Fig. 3M) cells. Since CTCF binding was reported to increase nearby genome CpG methylation viability (17,18), we further examined methyl-CpG-binding domain (MBD) sequencing datasets (PRJNA300503, Supplementary Material, Table S3) and observed a significant hypermethylation for rs7247241 T compared with C allele in 22Rv1 cells (Fig. 3N). Interestingly, the methylation status around the rs7247241 locus was negatively correlated with CTCF occupancy (Fig. 3O) in ENCODE cell lines.
CpG methylation at rs7247241 locus is allele-specific
Due to distinct cytosine methylation between rs7247241 alleles in 22Rv1 MBD-seq, we further investigated the effect of CTCF binding on CpG methylation. To confirm this allele-specific methylation at single-base resolution, we designed a primer pair that specifically amplified a 241 bp crick strand. The base content of rs7247241 comprises A and G in the Crick strand, allowing tracing allele-specific CpG methylation after bisulfite conversion (Fig. 4A). High-throughput sequencing analysis showed an overall reduction of methylation near CTCF and PLAGL2 binding sites in prostate cell lines (Fig. 4B) and tissues (Fig. 4C). To determine the allelic effect on the methylation, we first counted the total number of methylated CpGs per sequence read and normalized the methylated read counts to the total read count of T and C alleles. We then calculated the methylated CpG count ratios between normalized T-containing and C-containing reads. This analysis showed that T allele–containing sequences tended to be hypermethylated in heterozygote prostate cell lines (Fig. 4D) and tissues (Fig. 4E). The lollipop plot showed an example of the allele-specific methylation pattern in 22Rv1 cells (Fig. 4F).
Figure 4.
Allele-specific methylation at rs7247241. (A) Bisulfite PCR design for retaining original allele in the amplicon. (B, C) Overall methylation status in sequencing amplicon from prostate cells (B) and tissues (C). (D, E) Normalized count ratios between rs7247241 T and C fragments within different cumulative methylated CpG count in prostate cell lines (D) and tissues (E). (F) Lollipop plot showing the top five abundant methylation pattern surrounding rs7247241 locus in 22Rv1 cells. Each circle denotes a CpG site. Dashed boxes indicated the two CpG sites located in the CTCF motif, and dashed lines indicated rs7247241 location. (G) Strategy of dissecting DNA reads with CpG methylation in CTCF binding motif. (H, I) CpG methylation based on nearby CTCF motif methylation status in prostate cells (H) and tissues (I). (J) Strategy of disrupting crucial adenine (A) in CTCF binding motif with the adenine-base editor. (K, L) CpG methylation calculated from DNA molecules with crucial A or disruptive guanine (G) according to the above strategy in PrEC (K) and 22Rv1 (L) cells. NTC, non-transfected control.
CTCF binding is crucial for CpG methylation near rs7247241 site
Since CTCF loss may cause localized DNA hypermethylation in human cancers (18), we thus explored the relationships between rs7247241 locus methylation and CTCF motif integrities. As CTCF binding is sensitive to the methylation of two CpG sites within its motif (CpG1 and CpG2), we first analyzed read pairs with designated patterns in bisulfite-converted DNA (Fig. 4G) and calculated the methylation ratio for each subgroup. In primary prostate epithelial PrEC cells, compared with non-methylated reads, methylated CpG1 (mCpG1) fragment showed higher methylation level in a nearby CpG cluster (Fig. 4H left). In prostate cancer 22Rv1 cells, mCpG1 and mCpG2 led to a 10% methylation gain at the same nearby CpG cluster (Fig. 4H right). In benign prostate tissue pools, DNA fragments with mCpG1 or mCpG2 tended to be hypermethylated in the same CpG cluster region (Fig. 4I).
To further investigate the causal effect of CTCF binding on DNA methylation at this risk locus, we designed base editing guide RNA with a crucial A in the editing window, aiming to change it into a disruptive G using the xCas9-ABE system (Fig. 4J). Intriguingly, the base switch from A to G increased methylation level at the two CpG sites in the same cluster region in PrEC and 22Rv1 cells (Fig. 4K and L). To rule out potential sequencing error, we also examined methylation results from non-transfection control A (NTCA) and G (NTCG) and did not observe methylation changes.
Long isoform of PPP1R14A implicates aggressive phenotype and promoted prostate cell proliferation
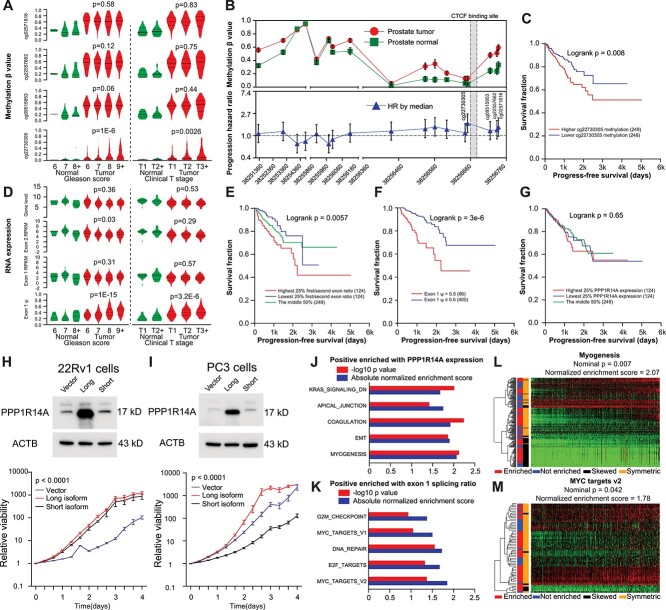
To determine if the methylation status near CTCF binding sites can be validated by other techniques measuring CpG methylation, we retrieved methylation data of TCGA prostate cohorts from the UCSC Xena data browser (https://xena.ucsc.edu/). We plotted methylation percentages associated with PPP1R14A gene along with common clinicopathological features related to prostate tumors (Fig. 5A). We found that only the CpG site nearest to CTCF binding motif (cg22730305) showed a clear trend of hypermethylation as Gleason score and clinical T stages increased in primary tumors. Expectedly, higher methylation of cg22730305 in primary tumor tissues was associated with worse progression-free survival (Fig. 5B and C). For RNA expression, we analyzed PPP1R14A abundance from UCSC Xena, PPP1R14A first/second exon RPKM from the Broad GDAC Firehose repository (https://gdac.broadinstitute.org) and PPP1R14A first exon ψ from TCGA SpliceSeq database (https://bioinformatics.mdanderson.org/TCGASpliceSeq/). We found that first exon ψ showed positive associations with Gleason score and clinical T stages (Fig. 5D). We also found that the read count ratio between exon 1 and exon 2 (Fig. 5E) and the first exon ψ (Fig. 5F) was both negatively associated with progression-free survival. At the same time, gene-level or exonic expression didn’t show any association (Fig. 5G and Supplementary Material, Fig. S4E, F).
Figure 5.
Functional characterization of PPP1R14A in prostate cancer. (A) Methylation status of probes near CTCF binding site in TCGA prostate cancer cohorts. ‘Normal’ indicated normal prostate tissues. P-value indicates testing for linear trend between means and subgroups in primary cancer tissues. (B) Probe-based methylation status in TCGA prostate cancer cohorts (Upper, error bars represent standard error of the mean of beta value) and progression hazard ratio for each probe (Lower, bars represent 95% confidence interval of HR). (C) Kaplan–Meier curve of progression-free survival stratified by median cg22730305 percentages. (D) RNA expression of PPP1R14A gene expression, exon quantification and first exon ψ in TCGA prostate cancer cohorts. Normal indicated normal prostate tissues. P-value indicates testing for linear trend between means and subgroups in primary cancer tissues. (E, G) Kaplan–Meier curve of progression-free survival stratified by first/second exon ratio (E), first exon ψ (F) and PPP1R14A gene expression (G). (H, I) Western blotting for stably overexpressing PPP1R14A isoform constructs, followed by proliferation assays in 22Rv1 (H) and PC3 (I) cells. (J, K) Top five HALLMARK gene sets positively enriched with PPP1R14A expression (J) and exon one ψ ratio (K) in TCGA prostate cancer cohort. (L) Heatmap showing myogenesis pathway gene expressions in TCGA prostate cancer column-wise ordered by ascending PPP1R14A expressions. (M) Heatmap showing MYC target gene profiling in TCGA prostate cancer column-wise ordered by ascending first exon ψ.
To evaluate the role of the splicing variants, we subsequently overexpressed the PPP1R14A long and short isoforms in 22Rv1 and PC3 cells. We found that only long isoform produced an expected protein product. Cell proliferation analysis in these stable cell lines showed an accelerated cell growth by PPP1R14A long isoform upregulation (Fig. 5H and I). To identify pathways that were associated with PPP1R14A gene expression and its first exon ψ, we performed GSEA analysis in whole transcriptome data of TCGA prostate cancer with HALLMARK gene set collection. Distinct pathways such as MYC targets, DNA repair and G2M checkpoint were enriched with PPP1R14A first exon ψ. In contrast, the myogenesis pathway, a well-known function of PPP1R14A, was enriched with its gene expression in prostate cancer. Top five pathways significantly enriched with either PPP1R14A gene expression or first exon ψ were listed in Figure 5J and K, respectively. When ranking samples according to respective phenotypes (PPP1R14A gene expression or first exon ψ), heatmap analysis showed a consistent enriched pattern of myogenesis (Fig. 5L) and MYC target (Fig. 5M) genes in TCGA RNA data.
Discussion
Given matched genotyping and transcriptome information, large-scale eQTL analysis has provided a valuable resource for investigating genetic regulation and deciphering disease etiology (19). Despite providing insights into the biological significance of risk SNPs, allele-dependent isoform change may only reflect subtle fluctuation at the gene level, thus resulting in weak eQTL signals. To address this issue, sQTL analysis has been used to directly correlate the normalized gene isoform expressions with genotypes (11). In this study, we applied a novel approach (9) that associates genotypes with distinct relative isoform expression distributions. With the most updated gene annotations, we reported a set of sQTL associated with multiple traits related to human prostate diseases. We also reported functional characterization of a PrCa risk SNP rs7247241 and its regulatory role on the target gene PPP1R14A.
The association between rs7247241 and PrCa was newly reported in a European cohort (20) (n = 421 142). Another variant rs8102476 at chr19q13, highly linked with rs7247241 (EUR population: R2 = 0.7992, D′ = 0.9956), is also associated with prostate cancer risk in large cohorts (21,22). Based on sashimi plots, risk allele (T)-driven PPP1R14A expression, mainly from the long isoform, is attributable to increased first exon expressions. ChIP-seq data showed allele-dependent CTCF binding at the rs7247241 site, supporting a critical role of CTCF in regulating PPP1R14A. In fact, previous studies have demonstrated that CTCF may function as a ‘splicing’ factor at the DNA level (23–25), further supporting our observation. Meanwhile, we also examined the JASPAR database and found the potential effect of this SNP on another TF PLAGL2. The anti-correlation between CTCF/PLAGL2 and PPP1R14A in primary PrCa tissues suggests the presence of negative regulations, which is supported by an elevated PPP1R14A expression followed by CTCF/PLAGL2 knockdown in primary prostate cell lines. However, this observation is not consistent in metastatic PrCa (Supplementary Material, Fig. S4A and B) and in another two immortalized cell lines 22Rv1 and RWPE-1 (Supplementary Material, Fig. S4C and D). These results suggested the complexity of disrupted gene regulation under malignant context and raised a similar challenge by using transformed cell lines for functionally validating germline variants (19,26). Additionally, we only used one shRNA to knock down each transcription factor (CTCF and PLAGL2) in multiple cell lines. Further proteome investigations regarding the rs7247241 locus will be warranted in our future experiments.
The core CTCF motif includes two CpG sites (27). The pre-existed methylation at these CpG sites has been reported to attenuate CTCF binding (28–30). In a previous work, by fluorescence-based DNA binding assay (31), Hideharu et al. revealed that CTCF binding was more sensitive to CpG methylation at core motif position 2 (corresponding to CpG site1) than the one at core motif position 12 (corresponding to CpG site2). This is consistent with our observation that CTCF motif methylation is associated with CpG methylation in the nearby sequence in prostate cells and normal tissues. Recent work also demonstrated that inducible CTCF factor silencing conferred localized DNA hypermethylation (18). In another study, the CTCF core motif position 6 adenine (A) was identified to be more affinitive with the CTCF ZN4–7 domain than guanine (G) or thymidine (T) (31). To confirm the effect of CTCF binding on nearby DNA methylation, we disrupted the binding site by CRISPR base editing and validated the relations between the CTCF binding site integrities and nearby genome methylation. Although disruptive G at this crucial position was not enough to increase overall methylation in the DNA sequence, two consistent CpG sites were highly methylated. We thus concluded that CTCF binding played an important role in maintaining its nearby genome hypomethylation status.
The important role of CTCF in DNA methylation was also supported by our analysis using TCGA prostate datasets, a decreased methylation level near the CTCF binding site. Our data showed a significant positive association of methylation level in the most adjacent probeset (cg22730305) and PPP1R14A first exon ψ with Gleason score, clinical T stage and progression-free survival in primary tumor tissue. Our data also showed that PPP1R14A first exon ψ was positively correlated with a series of oncogenic HALLMARK pathways. These results suggest the potential use of genetic and epigenetic changes at this locus as a biomarker for PrCa risk assessment and outcome prediction. Our results also suggest the potential use of sQTL analysis to uncover the most biologically relevant splicing event to the phenotypes of interest. PPP1R14A encodes a 17 kDa protein with phosphatase activities reported in smooth muscle and cancer cells (32,33). Studies have demonstrated that PPP1R14A is crucial to the oncogenic potential of pancreatic cancer (13) and melanoma cells (14). Furthermore, a large-scale transcriptome-wide association study (15) showed that PPP1R14A gene expression was associated with PrCa risk in trans-ethnic meta-analysis. Our study further confirmed the pro-proliferation effect of PPP1R14A and determined the critical role of its long isoform. Taken together, our data strongly support that the long isoform of PPP1R14A plays an important role in driving prostate initiation and progression.
The svQTL nominal P-value implemented in the sQTLseekeR package was originally provided as an additional approach to exclude false-positive sQTL based on transcript relative expression variance heterogeneity between genotypes. Notably, the variance heterogeneity may originate from multiple sources, including inaccurate isoform ratios calculated from inadequate RNA sequencing depth, biased variance estimation from unbalanced genotype groups and biological differences reflecting causal variants splicing effect. In our case, we did observe suspicious svQTLs such as rs12486932-POU1F1 (Supplementary Material, Fig. S5A) and rs72646967-TBX1 (Supplementary Material, Fig. S5B), exhibiting random isoform ratio changes between genotypes. However, for isoforms with reliable quantification, svQTL testing within linked SNPs could determine the variants with the greatest splicing fluctuation and thus help prioritize causal candidates. For example, we confirmed previously reported variants (rs10424178-BABAM1 and rs4072037-MUC1, see Supplementary Material, Figs S1A, B and S5C) involved in splicing sites disruption (6,34). A recent study also demonstrated the same benefit of using the svQTL P-value in finding causal variants in breast cancer (35). Based on these facts, we believe that the svQTL P-value has great potential in refining causal variants.
In summary, we performed sQTL analysis in our prostate cohort and identified the association between risk SNP rs7247241 and its target gene PPP1R14A. We further characterized the novel regulatory role of rs7247241 on PPP1R14A expression through allelic TF binding and the nearby genome methylation and revealed the functional potential of PPP1R14A isoform to the aggressive phenotype of prostate cancer. This study demonstrates the feasibility of sQTL analysis in identifying critical risk SNP-splicing transcript associations, which may be missed in eQTL analysis, and further broadens our knowledge on the complicated regulation caused by cancer risk SNPs. Understanding the biological effect of these sQTLs will eventually be translated into clinical practice and further benefit patients with prostate cancer.
Materials and Methods
Population-based genotype imputation and PrCa risk SNP genotype preparation
The genotyping data used for this study have been deposited in the dbGaP database under accession phs000985.v1.p1, which originates from a total of 467 prostate normal tissue genomic DNA. To perform imputation, we first input per chromosomal genotype data to MACH1 (36) to generate estimated phased haplotypes. The estimated haplotype data were further used by minimac to impute SNP based on 1000G phase 1 European population reference (n = 379). To identify high-quality SNPs, we applied minor allele frequency (MAF ≥ 1%) and imputation quality (R2 > 0.3) as a filter to determine the final genotype source.
Isoform expression quantification
From the same prostate normal tissue cohort (phs000985.v1.p1), we analyzed the raw sequence reads using the RSEM package (37) to quantify gene and isoform expression from ENSEMBL GRCh38 human gene annotation, which catalogued 58 884 genes and 208 527 transcripts. The raw RNA-seq reads were trimmed with universal illumina adaptor sequence before being sent to RSEM package for quantification. RSEM outputs were merged into sample-gene matrix for downstream sQTL analysis.
sQTL analysis and visualization
To perform sQTL analysis, we used the sQTLseekeR package (9) to integrate risk-associated SNP genotype and transcript percentage. After removing transcripts with low expression (relative transcript expression >1%, the relative transcription expression was calculated by dividing the transcript FPKM by entire gene-level FPKM) and genes with only one expressed transcript, we identified a total of 12 627 genes, including 114 670 transcripts, in the sQTL analysis. For candidate sQTL, we merged alignments in the gene region of interest in each homozygote genotype and visualized coverages in sashimi plots with the miso package (38). In addition, boxplots representing isoform transitions were generated with an in-house R script and can be found in Supplementary Information (Boxplot of significant sQTLs).
ChIP analysis
For candidate SNP rs7247241, we queried ChIP-Atlas and ENCODE for possible peak signals at the loci of interest. Independent ChIP-seq experiments and available controls were downloaded from source repositories (Supplementary Material, Table S3). Raw reads were aligned to the hg38 human genome using bowtie2. Unique alignments were sorted and indexed. Integrative Genomics Viewer was used to visualize ChIP peaks. To further verify transcription factor binding and histone modification in prostate cells, we performed ChIP-qPCR in RWPE-1 and 22Rv1 cells. Chromatin was prepared from these cell lines with ChIP-IT High Sensitivity kit (Active Motif, 53 040) and used for three independent ChIP reactions. To assess TF occupancies around the rs7247241 locus, we designed primers to amplify the SNP containing (rs7247241-TFBS, TF binding site) and an upstream negative control (rs7247241-upstream-NC) region. We performed quantification reactions with input or ChIPed DNA by PowerUp SYBR Green Master Mix on CFX96 qPCR instrument (Bio-Rad). Enrichment at each region (primer) was calculated by normalizing to the respective input control. The primers used are listed in Supplementary Material, Table S2.
Allele-specific methylation quantification
To determine allele-dependent methylation surrounding rs7247241 locus, we designed bisulfite PCR primers according to SNP strand with A or G alleles, which will retain the original genotypes. We chose heterozygous cell lines, RWPE-1, RWPE-2, 22Rv1 and PrEC to extract RNA-free genomic DNA. We used EZ DNA Methylation-Lightning Kits (D5030, Zymo Research) to prepare bisulfite PCR template from 500 ng genomic DNA and EpiMark® Hot Start Taq DNA Polymerase (M0490S, New England Biolabs) to amplify rs7247241 regions. We individually amplified the target region and combined the amplicons from genomic DNA sample from 192 prostate tissue into two separate pools. The pooled amplicons were further ligated with adapters and submitted for sequencing. We used the Bismark bisulfite mapper (39) to align these reads to the human genome and separate aligned reads by allele contents at the rs7247241 loci. We counted methylated (non-converted) CpG sites from reads for both alleles and plotted the read count according to the CpG site number.
Plasmid construction and lentivirus production
For investigation of PPP1R14A gene overexpression, we retrieved the open reading frame and corresponding five prime UTR sequences from ENSEMBL GRCh38 build for both isoforms (Long: ENST00000301242; Short: ENST00000587515). These sequences were cloned into pLV vector with C-terminal 3xFLAG fusion. The transfer plasmids were co-transfected with pMD2G and pxPAX2 into 293FT cells to generate lentiviral particles. When stably overexpressing PPP1R14A isoforms, we aimed for a low multiplicity of infection (MOI ≤ 0.5) to ensure the majority of the cells only received one copy of corresponding ORF, which mimicked the gradual gene expression changes related to QTL changes. For knocking down CTCF and PLAGL2, we cloned one shRNA sequence for each TF (see Supplementary Material, Table S2) into pLV vector with hU6 promoter and generated lentiviral particles with the same packaging plasmids. For the luciferase reporter assay, we cloned a 223 bp promoter surrounding rs7247241 locus from 22Rv1 cells (C_T genotype) into pGL3-basic vectors for the luciferase reporter assay between NheI and XhoI restriction sites. Promoters with C or T alleles were selected according to Sanger sequencing results from subclones.
Reagents and cell culture
The antibodies against PPP1R14A (ab32213) and PLAGL2 (ab139509) were purchased from Abcam. Antibodies against β-actin (4970) were purchased from Cell Signaling Technology. Antibodies against CTCF (61311) were purchased from Active Motif. PC3 (RRID: CVCL_0035), 22Rv1 (RRID: CVCL_1045), RWPE-1 (RRID: CVCL_3791), RWPE-2 (RRID: CVCL_3792) and PrEC (RRID: CVCL_0061) cells were obtained from the ATCC. 293FT cells (R70007) were purchased from Thermo Fisher Scientific. All cell lines were verified with short tandem repeat (STR) profiling before use. Cell lines were disposed of and replaced with low passage aliquots after being subcultured 15 times. Unless specified otherwise, all cell culture reagents were obtained from Thermo Fisher Scientific. PC3 and 22Rv1 cells were grown in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS). 293FT cells were grown in DMEM medium supplemented with 10% FBS and 500 ug/ml Geneticin selective antibiotics. RWPE-1 and RWPE-2 cells were grown in Keratinocyte Serum-Free Medium. PrEC cells were grown in Prostate Epithelial Cell Growth Kit (PCS-440-040, ATCC). All cell lines were examined for mycoplasma contamination with Venor GeM Mycoplasma Detection Kit (Sigma-Aldrich) and were confirmed to be mycoplasma-free before experiments.
Western blot analysis
Total protein was extracted and electrophoresed as described previously (40), with minor modification of using Tricine Sample Buffer and Mini-PROTEAN® Tris/Tricine Precast Gels (BioRad). SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) produced luminescent signals on the LICOR imaging system. Captured images were aligned in Photoshop and assembled in Illustrator.
Luciferase reporter assay
The cells were seeded into a 24-well plate one day before 500 ng of pGL3 reporter plasmids were transfected per-well using Lipofectamine 3000. The media were replaced 24 h after transfection. After 48 h of transfection, the cells were lysed for the luciferase assay according to the Dual-Luciferase® Reporter Assay (E1960, Promega) protocol. The GlowMax plate reader measured luminescence. After normalization to Renilla luciferase readout, relative firefly luciferase activities driven by corresponding promoters were represented by luminescence unit fold changes.
CRISPR base editing
To change the CTCF motif A allele to G allele, we created a GFP labeled xCas9(3.7)-ABE(7.10) plasmid based on the backbone from David Liu’s lab (41). Guide RNA template was synthesized in a gblock fragment with an hU6 promoter and a scaffold (https://benchling.com/rrm38/f/h4fdYFOi-protocols/prt-10T3UWFo-detailed-gblocks-based-crispr-protocol/edit) (Guide RNA sequence can be found in Supplementary Material, Table S2). We co-transfected 2.5 ug of xCas9(3.7)-(ABE7.10) plasmid and 1.2 ug of guide RNA gblock into cells 80% confluent in each well of a 6-well plate. After 48 h, GFP-positive cells were sorted by flow cytometry and collected for DNA extraction and downstream bisulfite PCR analysis.
Clinical association analysis
The PPP1R14A promoter CpG methylation microarray data were retrieved from UCSC Xena data browser (https://xena.ucsc.edu/). The PPP1R14A exon RPKM data were retrieved from Broad GDAC Firehose repository (https://gdac.broadinstitute.org). The PPP1R14A first exon ψ, i.e. the percentage-splice-in of alternative promoter was downloaded from TCGA SpliceSeq database (https://bioinformatics.mdanderson.org/TCGASpliceSeq/).
Supplementary Material
Acknowledgements
The funders had no role in study design, data collection and analysis, decision to publish or manuscript preparation. We thank Flow Cytometry and Microscopy Core Facility at the Moffitt Cancer Center, an NCI designated Comprehensive Cancer Center (P30CA076292).
Conflict of Interest statement: No potential conflicts of interest were disclosed.
Contributor Information
Yijun Tian, Department of Tumor Biology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Alex Soupir, Department of Tumor Biology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Qian Liu, Department of Tumor Biology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Lang Wu, Division of Cancer Epidemiology, Population Sciences in the Pacific Program, University of Hawaii Cancer Center, University of Hawaii at Manoa, Hawaii, HI 96822, USA.
Chiang-Ching Huang, Joseph J. Zilber School of Public Health, University of Wisconsin, Milwaukee, WI 53226, USA.
Jong Y Park, Department of Cancer Epidemiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Liang Wang, Department of Tumor Biology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA.
Funding
National Institutes of Health (R01CA250018 and R01CA212097 to L.W.).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability
All data generated or analyzed in this study are included in this article or the Supplemental Information files. In addition, codes for bisulfite sequencing and sQTL analysis are available upon request.
Authors’ contributions
Conception and design: Y. Tian.
Development of methodology: Y. Tian and L. Wang.
Acquisition of data: Y. Tian and L. Wang.
Analysis and interpretation of data: Y. Tian and L. Wu.
Writing, review and revision of the manuscript: Y. Tian, L. Wang, J. Park, C-C. Huang, A.C. Soupir and Q. Liu.
Study supervision: L. Wang.
References
- 1. Buniello, A., MacArthur, J.A.L., Cerezo, M., Harris, L.W., Hayhurst, J., Malangone, C., McMahon, A., Morales, J., Mountjoy, E., Sollis, E. et al. (2019) The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res., 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farashi, S., Kryza, T., Clements, J. and Batra, J. (2019) Post-GWAS in prostate cancer: from genetic association to biological contribution. Nat. Rev. Cancer, 19, 46–59. [DOI] [PubMed] [Google Scholar]
- 3. Nicolae, D.L., Gamazon, E., Zhang, W., Duan, S., Dolan, M.E. and Cox, N.J. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet., 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen, H., Yu, H., Wang, J., Zhang, Z., Gao, Z., Chen, Z., Lu, Y., Liu, W., Jiang, D., Zheng, S.L. et al. (2015) Systematic enrichment analysis of potentially functional regions for 103 prostate cancer risk-associated loci. Prostate, 75, 1264–1276. [DOI] [PubMed] [Google Scholar]
- 5. Amin Al Olama, A., Dadaev, T., Hazelett, D.J., Li, Q., Leongamornlert, D., Saunders, E.J., Stephens, S., Cieza-Borrella, C., Whitmore, I., Benlloch Garcia, S. et al. (2015) Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among Europeans. Hum. Mol. Genet., 24, 5589–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng, L., Zhu, C., Gu, J., Xi, P., Du, J. and Jin, G. (2013) Functional polymorphism rs4072037 in MUC1 gene contributes to the susceptibility to gastric cancer: evidence from pooled 6,580 cases and 10,324 controls. Mol. Biol. Rep., 40, 5791–5796. [DOI] [PubMed] [Google Scholar]
- 7. Xiao, F., Zhang, P., Wang, Y., Tian, Y., James, M., Huang, C.C., Wang, L. and Wang, L. (2020) Single-nucleotide polymorphism rs13426236 contributes to an increased prostate cancer risk via regulating MLPH splicing variant 4. Mol. Carcinog., 59, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao, P., Xia, J.H., Sipeky, C., Dong, X.M., Zhang, Q., Yang, Y., Zhang, P., Cruz, S.P., Zhang, K., Zhu, J. et al. (2018) Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell, 174, 576–589 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monlong, J., Calvo, M., Ferreira, P.G. and Guigo, R. (2014) Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat. Commun., 5, 4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qu, W., Gurdziel, K., Pique-Regi, R. and Ruden, D.M. (2017) Identification of splicing quantitative trait loci (sQTL) in Drosophila melanogaster with developmental lead (Pb(2+)) exposure. Front. Genet., 8, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Battle, A., Mostafavi, S., Zhu, X., Potash, J.B., Weissman, M.M., McCormick, C., Haudenschild, C.D., Beckman, K.B., Shi, J., Mei, R. et al. (2014) Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res., 24, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larson, N.B., McDonnell, S., French, A.J., Fogarty, Z., Cheville, J., Middha, S., Riska, S., Baheti, S., Nair, A.A., Wang, L. et al. (2015) Comprehensively evaluating cis-regulatory variation in the human prostate transcriptome by using gene-level allele-specific expression. Am. J. Hum. Genet., 96, 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eto, M., Kirkbride, J.A., Chugh, R., Karikari, N.K. and Kim, J.I. (2013) Nuclear localization of CPI-17, a protein phosphatase-1 inhibitor protein, affects histone H3 phosphorylation and corresponds to proliferation of cancer and smooth muscle cells. Biochem. Biophys. Res. Commun., 434, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riecken, L.B., Zoch, A., Wiehl, U., Reichert, S., Scholl, I., Cui, Y., Ziemer, M., Anderegg, U., Hagel, C. and Morrison, H. (2016) CPI-17 drives oncogenic Ras signaling in human melanomas via Ezrin-Radixin-Moesin family proteins. Oncotarget, 7, 78242–78254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emami, N.C., Kachuri, L., Meyers, T.J., Das, R., Hoffman, J.D., Hoffmann, T.J., Hu, D., Shan, J., Feng, F.Y., Ziv, E. et al. (2019) Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nat. Commun., 10, 3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oki, S., Ohta, T., Shioi, G., Hatanaka, H., Ogasawara, O., Okuda, Y., Kawaji, H., Nakaki, R., Sese, J. and Meno, C. (2018) ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep., 19, e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kemp, C.J., Moore, J.M., Moser, R., Bernard, B., Teater, M., Smith, L.E., Rabaia, N.A., Gurley, K.E., Guinney, J., Busch, S.E. et al. (2014) CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep., 7, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Damaschke, N.A., Gawdzik, J., Avilla, M., Yang, B., Svaren, J., Roopra, A., Luo, J.H., Yu, Y.P., Keles, S. and Jarrard, D.F. (2020) CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin. Epigenetics, 12, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cano-Gamez, E. and Trynka, G. (2020) From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet., 11, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rashkin, S.R., Graff, R.E., Kachuri, L., Thai, K.K., Alexeeff, S.E., Blatchins, M.A., Cavazos, T.B., Corley, D.A., Emami, N.C., Hoffman, J.D. et al. (2020) Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun., 11, 4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schumacher, F.R., Al Olama, A.A., Berndt, S.I., Benlloch, S., Ahmed, M., Saunders, E.J., Dadaev, T., Leongamornlert, D., Anokian, E., Cieza-Borrella, C. et al. (2018) Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet., 50, 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gudmundsson, J., Sulem, P., Gudbjartsson, D.F., Blondal, T., Gylfason, A., Agnarsson, B.A., Benediktsdottir, K.R., Magnusdottir, D.N., Orlygsdottir, G., Jakobsdottir, M. et al. (2009) Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet., 41, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz-Velasco, M., Kumar, M., Lai, M.C., Bhat, P., Solis-Pinson, A.B., Reyes, A., Kleinsorg, S., Noh, K.M., Gibson, T.J. and Zaugg, J.B. (2017) CTCF-mediated chromatin loops between promoter and gene body regulate alternative splicing across individuals. Cell Syst., 5, 628–637 e626. [DOI] [PubMed] [Google Scholar]
- 24. Kornblihtt, A.R. (2012) CTCF: from insulators to alternative splicing regulation. Cell Res., 22, 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shukla, S., Kavak, E., Gregory, M., Imashimizu, M., Shutinoski, B., Kashlev, M., Oberdoerffer, P., Sandberg, R. and Oberdoerffer, S. (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature, 479, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang, J., Li, Y., Chan, L., Tsai, Y.T., Wu, W.H., Nguyen, H.V., Hsu, C.W., Li, X., Brown, L.M., Egli, D. et al. (2014) Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Hum. Mol. Genet., 23, 3445–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Schones, D.E., Wang, Z., Wei, G., Chepelev, I. and Zhao, K. (2007) High-resolution profiling of histone methylations in the human genome. Cell, 129, 823–837. [DOI] [PubMed] [Google Scholar]
- 28. Kanduri, C., Pant, V., Loukinov, D., Pugacheva, E., Qi, C.F., Wolffe, A., Ohlsson, R. and Lobanenkov, V.V. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol., 10, 853–856. [DOI] [PubMed] [Google Scholar]
- 29. Hark, A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M. and Tilghman, S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature, 405, 486–489. [DOI] [PubMed] [Google Scholar]
- 30. Bell, A.C. and Felsenfeld, G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature, 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 31. Hashimoto, H., Wang, D., Horton, J.R., Zhang, X., Corces, V.G. and Cheng, X. (2017) Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell, 66, 711–720 e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao, G., Zhong, Y., Su, W., Liu, S., Song, X., Hou, T., Mu, X., Gong, M.C. and Guo, Z. (2019) Transcriptional suppression of CPI-17 gene expression in vascular smooth muscle cells by tumor necrosis factor, kruppel-like factor 4, and Sp1 is associated with lipopolysaccharide-induced vascular hypocontractility, hypotension, and mortality. Mol. Cell. Biol., 39, e00070–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eto, M. (2009) Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. J. Biol. Chem., 284, 35273–35277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caswell, J.L., Camarda, R., Zhou, A.Y., Huntsman, S., Hu, D., Brenner, S.E., Zaitlen, N., Goga, A. and Ziv, E. (2015) Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum. Mol. Genet., 24, 7421–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Machado, J., Magno, R., Xavier, J.M. and Maia, A.-T. (2019) Alternative splicing regulation by GWAS risk loci for breast cancer. BioRxiv, 766394 in press. [Google Scholar]
- 36. Li, Y., Willer, C.J., Ding, J., Scheet, P. and Abecasis, G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li, B. and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katz, Y., Wang, E.T., Airoldi, E.M. and Burge, C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krueger, F. and Andrews, S.R. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian, Y., Liu, Q., Yu, S., Chu, Q., Chen, Y., Wu, K. and Wang, L. (2020) NRF2-driven KEAP1 transcription in human lung cancer. Mol. Cancer Res., 18, 1465–1476. [DOI] [PubMed] [Google Scholar]
- 41. Hu, J.H., Miller, S.M., Geurts, M.H., Tang, W., Chen, L., Sun, N., Zeina, C.M., Gao, X., Rees, H.A., Lin, Z. et al. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature, 556, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this study are included in this article or the Supplemental Information files. In addition, codes for bisulfite sequencing and sQTL analysis are available upon request.