Abstract
Objectives
To describe the propensity of carbapenem-resistant Pseudomonas aeruginosa to spread within a hospital critical care setting.
Methods
The study was conducted in a 700-bed tertiary centre in Cologne, Germany. P. aeruginosa resistant to piperacillin, ceftazidime, cefepime, imipenem, meropenem and ciprofloxacin, isolated from clinical and screening specimens from four critical care units from 2015 to 2020 were analysed. Genotyping was carried out by WGS (Illumina and MinION). MLST, core genome MLST (cgMLST) and resistome analysis was performed and merged with epidemiological data.
Results
Fifty-five out of 79 non-duplicate P. aeruginosa isolates were available, of which 20 were carbapenemase producers as follows: blaVIM-1 (n = 1), blaVIM-2 (n = 17), blaVIM-4 (n = 1), and blaNDM-1/blaGES-5 (n = 1). Forty-two of 55 isolates were hospital-acquired. cgMLST revealed three clusters: Cluster 1 (n = 15, ST111, blaVIM-2, recovered between 2015 and 2020); Cluster 2 (n = 4, ST970, carbapenemase negative); and Cluster 3 (n = 2, ST357, carbapenemase negative). The blaVIM-2 gene of Cluster 1 was integrated on the chromosome in a class 1 integron (type In59). Using conventional epidemiology, we were only able to confirm two patient-to-patient transmissions and one room-to-patient transmission on three different ICUs within Cluster 1. Isolates from Cluster 2 represented an outbreak occurring in 2019.
Conclusions
These data give insight into the epidemiology of carbapenem-resistant P. aeruginosa. Transmission dynamics differed between carbapenemase- and non-carbapenemase-producing isolates. A continuous acquisition of clonally related ST111 VIM-2 P. aeruginosa, being the main carbapenemase-producing strain, was observed over the whole study period, as well as an overall higher genomic diversity among non-carbapenemase-producing P. aeruginosa.
Introduction
Pseudomonas aeruginosa is an environmental bacterium that can colonize the human body. As a leading nosocomial pathogen P. aeruginosa may cause surgical site infections, ventilator-associated pneumonia, catheter-associated urinary tract infections or central-line-associated bloodstream infections in healthcare settings.1 The organism is especially problematic for immunocompromised patients within special units (ICU, haematology-oncology ward or burn unit).2 Infections can be difficult to treat because of intrinsic resistance to many antimicrobial agents as well as rapid development of antimicrobial resistance to nearly all available antimicrobials through chromosomal mutations and acquisition of transferable resistance genes.3 Of particular interest is carbapenem resistance mediated by intrinsic resistance mechanisms (a combination of efflux pumps, AmpC overexpression and porin loss) or acquisition of a carbapenemase, especially an MBL.3 Carbapenemase production is linked to globally distributed and emerging MDR or even XDR high-risk clones.4,5 While the emergence of carbapenem-resistant P. aeruginosa is well described, less is known about the different propensity of carbapenemase-producing and carbapenemase-non-producing P. aeruginosa to spread within the hospital setting. This is important, as a relevant part of hospital-acquired infections caused by P. aeruginosa is transmission-associated, either patient-to-patient (mostly via the hands of healthcare workers) or environment-to-patient.6,7 The hospital environmental is a known reservoir, especially in moist sites. Reports show that contaminated tap water as well as washbasins are linked to transmission events.8–11
A previous study from our research group performed in three hospitals of different levels of healthcare has shown a prevalence of approximately 20% carbapenemase producers, mostly VIM-2, amongst MDR/XDR P. aeruginosa over a 3 year period.12 Using PFGE nearly all VIM-2-producing isolates were clonally related. However, only a few epidemiologically proven transmission events were confirmed, exclusively on several ICUs of the tertiary care centre.12 The present investigation aims to define the local epidemiology and transmission dynamics of MDR/XDR P. aeruginosa irrespective of carbapenemase production in these ICUs of the tertiary care centre over a period of 5 years using a genomics-based approach.
Materials and methods
Study setting
The study was conducted in a 700-bed tertiary care centre in Cologne, Germany. Based on data of the implemented active surveillance system following the protocol of the German healthcare-associated infection surveillance on intensive care units (ITS-KISS)13 and previous studies,12 four out of six available critical care units with a frequent detection and/or possible transmission events of carbapenem-resistant and MDR/XDR P. aeruginosa were chosen: three ICUs (ICU 1–3) and one intermediate care unit (ImCU1). ICU1 and ICU3 provide care for surgical patients including burn patients (max. of 32 and 14 beds, respectively), whereas ICU2 and ImCU1 are primarily reserved for medical patients (max. of 18 and 16 beds, respectively). Overall, there were approximately 3500 admissions (20 000 patient days) per year on these units. The number of patients colonized/infected with P. aeruginosa was assessed using the laboratory surveillance information system (Hybase v.6, epiNET AG, Germany).
Identification and susceptibility testing
All isolates were identified with standard microbiological procedures using the Vitek 2 system (Vitek GN-ID, bioMérieux, Marcy l’Étoile, France) or MALDI-TOF (Bruker Daltonics, Bremen, Germany). First susceptibility testing was performed with automated systems (Vitek 2 system from bioMérieux or the BD-Phoenix system from BD Diagnostics, Heidelberg, Germany) or disc diffusion (BD Sensi-Disc, BD Diagnostics) and later confirmed by broth microdilution using Micronaut-S Pseudomonas MIC panels (Merlin Diagnostika, Bornheim, Germany) according to the manufacturers’ instructions. MICs were determined for piperacillin, piperacillin/tazobactam, ceftazidime, cefepime, ceftazidime/avibactam, ceftolozane/tazobactam, imipenem, meropenem, aztreonam, gentamicin, tobramycin, amikacin, ciprofloxacin, levofloxacin, fosfomycin and colistin. EUCAST breakpoints (v11.0, 2021) were used for interpretation. P. aeruginosa resistant to piperacillin, ceftazidime, cefepime, imipenem, meropenem and ciprofloxacin isolated on the designated wards from clinical and screening specimens from January 2015 to June 2020 were included. This basically corresponds to an MDR/XDR phenotype according to the ECDC/CDC classification.5 Two VIM-2-producing P. aeruginosa isolates detected on other wards of the same hospital analysed in a previous study (PSA-2016-03 and PSA-2017-02) were also included in this study.12
WGS
To prepare short-read sequencing libraries, fresh cultures were grown overnight on Mueller–Hinton agar and DNA was isolated using the DNeasy Ultra Clean Microbial Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Sequencing libraries were prepared with the purified DNA using the Ultra II FS DNA Library Prep Kit (New England Biolabs, Frankfurt, Germany) for a 250 bp paired-end sequencing run on an Illumina MiSeq sequencer. De novo assembly was performed using Velvet (version 1.1.04).14 The raw sequencing short reads generated in this project were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) under the Project Accession number PRJEB43695.
To understand the genetic location of the blaVIM-2 gene, three strains belonging to the same core genome MLST (cgMLST) cluster (PSA-2015-07, PSA-2017-03 and PSA-2020-04) were selected for long-read sequencing. DNA was extracted from bacteria grown overnight in Luria broth using the Genomic-Tips 100/G kit and Genomic DNA Buffers kit (Qiagen). Libraries were prepared using the Ligation Sequencing Kit (SQK-LSK109) combined with Native Barcoding Kit (EXP-NBD114) and were loaded onto a R9.4 flow cell (Oxford Nanopore Technologies, Oxford, UK) for a MinION sequencing run. Finally, a hybrid assembly of the long- and short-reads was performed using Unicycler.15 The long-read raw data have been deposited to the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the BioProject Accession number PRJNA771632.
Genotyping and resistome analysis
Relatedness of all isolates was assessed by a cgMLST genotyping approach. Assembled genomes were analysed by SeqSphere+ software (v.7.2.3, Ridom, Germany) using a validated cgMLST scheme recently proposed by Tönnies et al.16 and based on 3867 target genes. During comparison of the allelic profile the ‘pairwise ignoring missing values’ option was turned on. Genomes containing at least 95% of the defined cgMLST targets were included. Isolates with less than 12 different alleles in the cgMLST target gene set were considered as highly related (and termed a cluster). Additionally, based on the assembled genomes, the conventional 7-loci MLST scheme was retrieved from the MLST database.17 Furthermore, acquired resistance genes on assembled genomes were identified by the ResFinder Bacterial Analysis Pipeline v. 2.1.18
The genetic environment of blaVIM-2 was annotated and curated manually and visualized using the SnapGene® software (Insightful Science, GSL Biotech, San Diego, CA, USA) based on the hybrid assemblies. Insertion sequence elements were investigated using the ISfinder database (http://www-is.biotoul.fr).19
Infection prevention and control (IPC) management
A general rectal admission screening for MDR Gram-negative organisms was in place on all units. Additionally, weekly tracheal secretions were taken from intubated patients, and wound swabs from burn patients (surveillance cultures). Weekly rectal screenings were performed on ICU1 only. Standard and contact precautions were applied for every patient found colonized or infected with MRD/XDR P. aeruginosa (single room and use of gowns and gloves). Relevant clinical and epidemiological data was collected from patients’ clinical records or the attending physician. If the collection of the specimen occurred on or before the second day of admission, and there was no prior contact to the healthcare system within the previous 30 days, bacterial isolates were considered community-acquired. If prior contact with the healthcare system (other than our hospital) was observed within the previous 30 days and collection occurred on or before the second day of admission, isolates were considered healthcare-associated. If the collection of the specimen occurred after the second day, or if the patient stayed at our hospital within the last 30 days, bacterial isolates were defined as hospital-acquired. Transmission analysis was based on epidemiological data (direct room or ward contact, and/or documented care by the same staff) and genetic data. Proven transmission events were defined as isolation of genetically related isolates (cluster) in two or more patients who were hospitalized during overlapping periods on the same ward (at least 24 h, patient-to-patient transmission) or in the same room with a maximum time interval of 6 months (room-to-patient transmission).12 An interval of 6 months was chosen because transmission of P. aeruginosa from environmental sources can continue over longer periods and can be sporadic.8,12 Hospital-acquired infections were classified according to the CDC/NHSN definitions.20 Patients without related signs of infection were considered to be colonized.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Faculty of Health of the Witten/Herdecke University (study number S-33/2021).
Results
Isolate and patient characteristics
Seventy-nine first MDR/XDR P. aeruginosa clinical and screening isolates were detected from the designated wards during the time period and 55 non-duplicate isolates were available for further analysis.
Susceptibility testing by broth microdilution showed that all bacterial isolates displayed an MDR or XDR phenotype as defined by the inclusion criteria; no isolate was pandrug resistant. One isolate was not cultivable for broth microdilution. The remaining 54 isolates displayed a susceptibility rate of 50%, 61.1% and 100% for tobramycin, amikacin and colistin, respectively. In all MBL-producing isolates (n = 20) the susceptibility rate for aztreonam was 85% and was much lower in non-MBL-producing isolates (n = 34), being 26.4%. All MBL-producing isolates were resistant to ceftolozane/tazobactam and ceftazidime/avibactam, whereas in all non-MBL-producing isolates, susceptibility rates to ceftolozane/tazobactam and ceftazidime/avibactam were both 72.7%. MICs are shown in Table S1 (available as Supplementary data at JAC-AMR Online).
Sixteen of these isolates were previously analysed by conventional genotyping methods.12 Sequence analysis confirmed 20 carbapenemase-producing P. aeruginosa isolates as follows: blaVIM-1 (n = 1), blaVIM-2 (n = 17), blaVIM-4 (n = 1) and blaNDM-1/blaGES-5 (n = 1). Other relevant acquired resistance genes are shown in Table S1.
As all patients were from critical care units, devices, surgical and nonsurgical interventions and antibiotic therapy were common (Table 1). The mode of acquisition was mostly either hospital-acquired or healthcare-associated, and only one isolate was considered as community-acquired.
Table 1.
Characteristics of 55 patients with the analysed MDR/XDR P. aeruginosa
| Patient characteristics (n = 55) | Value |
|---|---|
| Age, years, median (Q1–Q3) | 59 (46–70) |
| Sex, male | 40 (72.7) |
| Medical departments | |
| surgery | 32 (58.2) |
| internal medicine | 23 (41.8) |
| Mode of acquisition | |
| hospital-acquired | 42 (76.4) |
| healthcare-associated | 12 (21.8) |
| community-acquired | 1 (1.8) |
| Day of acquisition during hospital stay (hospital-acquired only; n = 42), median (IQR) | 29 (30.25) |
| Hospital-acquired infection (CDC/NHSN) | |
| pneumonia | 11 (20) |
| surgical site | 5 (9.1) |
| urinary tract | 2 (3.6) |
| skin | 2 (3.6) |
| CLABSI | 1 (1.8) |
| Antipseudomonal antibiotic treatmenta | 38 (69.1) |
| Surgerya | 40 (72.7) |
| Nonsurgical interventiona | 51 (92.7) |
| Dialysisa | 18 (32.7) |
| Mechanical ventilationa | 48 (87.3) |
| Central linea | 47 (85.5) |
| Urinary cathetera | 50 (90.9) |
Values are shown as n (%) unless specified otherwise.
Q1, lower quartile; Q3, upper quartile; CLABSI, central line-associated bloodstream infection.
Within a maximal interval of 7 days before first isolation of MDR/XDR P. aeruginosa.
Genotyping and transmission analysis
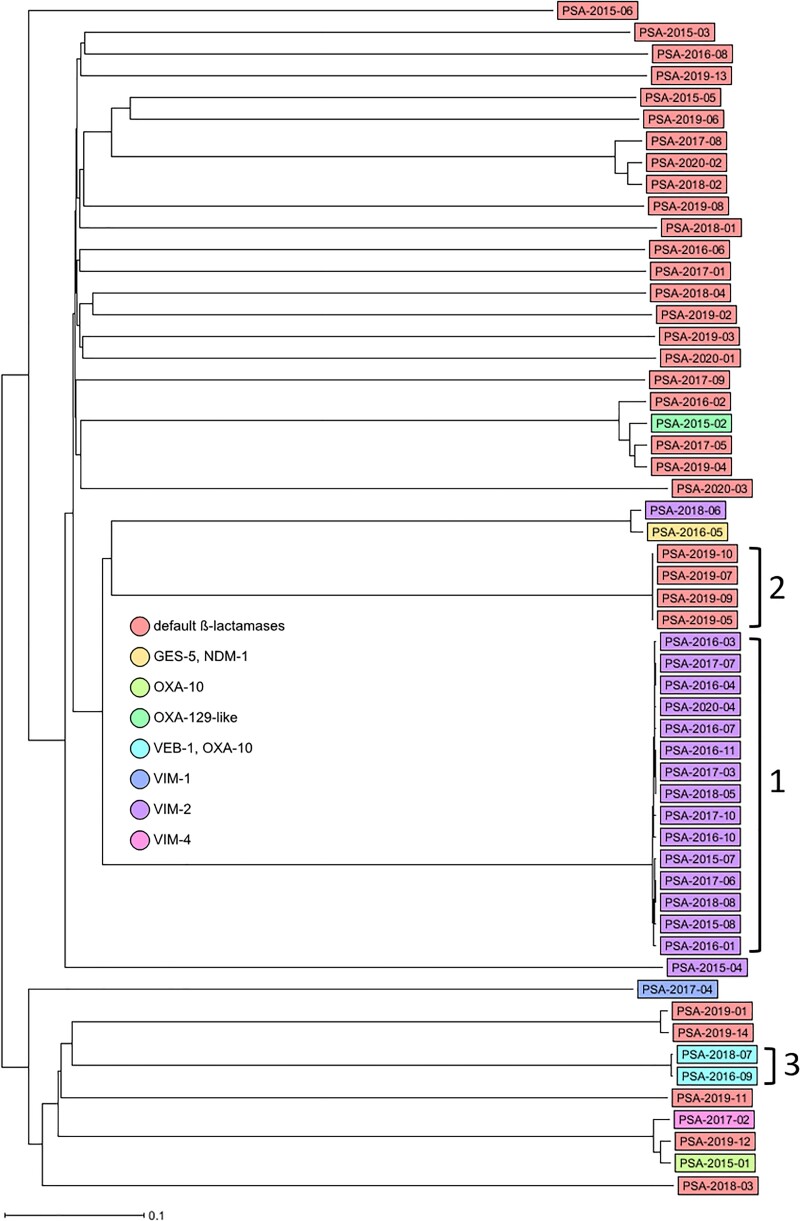
Carbapenemase-producing P. aeruginosa isolates (n = 20) were assigned to five STs, predominantly ST111, but also ST273, ST654, ST235 and ST3618, the latter being a newly assigned ST. A high diversity was detected within the remaining 35 non-carbapenemase-producing P. aeruginosa isolates, which comprised 24 different STs. cgMLST revealed three clusters that were represented by different STs: Cluster 1 (n = 15, ST111), Cluster 2 (n = 4, ST970) and Cluster 3 (n = 2, ST357). Within Clusters 1, 2 and 3, a maximum difference of 21, 1 and 5 alleles, respectively, was observed. Carbapenemase production in P. aeruginosa was significantly associated with belonging to a clonal cluster (P < 0.001, Fisher’s exact test). Table 2 summarizes isolate characteristics, genotyping results and presence of ESBL/carbapenemase genes, and Figure 1 shows the relatedness of the isolates. All Cluster 1 isolates carried the blaVIM-2 gene and the aacA29-like gene. Moreover, all but two isolates from Cluster 1 were hospital-acquired; the other two were healthcare-associated and had contact with two different institutions. Cluster 1 isolates were detected sporadically on all four wards and during the whole study period from 2015 to 2020. Using epidemiological data, we were only able to confirm one room-to-patient and two patient-to-patient transmissions (on three different ICUs and during three different time periods; Figure 2).
Table 2.
Characteristics of MDR/XDR P. aeruginosa ranked by cgMLST cluster type and date of isolation
| Isolate no. | Date (month-year) | Specimen type | Epidemiological link to ward | ST | cgMLST cluster | Acquired β-lactamase genes |
|---|---|---|---|---|---|---|
| PSA-2015-07 | Dec-15 | wound | ICU1 | ST111 | 1 | bla VIM-2 |
| PSA-2015-08 | Dec-15 | respiratory tract | ICU1 | |||
| PSA-2016-01 | Feb-16 | urine | ICU3 | |||
| PSA-2016-03 | Apr-16 | respiratory tract | other | |||
| PSA-2016-04 | Apr-16 | respiratory tract | ICU3 | |||
| PSA-2016-07 | Oct-16 | respiratory tract | ImCU1 | |||
| PSA-2016-10 | Dec-16 | respiratory tract | ICU3 | |||
| PSA-2016-11 | Dec-16 | unknown | ICU2 | |||
| PSA-2017-03 | Jul-17 | screening (rectal) | ICU2 | |||
| PSA-2017-06 | Oct-17 | wound | ICU1 | |||
| PSA-2017-07 | Oct-17 | screening (nose/throat) | ICU3 | |||
| PSA-2017-10 | Dec-17 | respiratory tract | ICU3 | |||
| PSA-2018-05 | Jun-18 | urine | ICU2 | |||
| PSA-2018-08 | Nov-18 | wound | ICU1 | |||
| PSA-2020-04 | Apr-20 | screening (rectal) | ICU3 | |||
| PSA-2019-05 | Aug-19 | respiratory tract | ICU1 | ST970 | 2 | — |
| PSA-2019-07 | Sep-19 | screening (nose/throat) | ICU1 | |||
| PSA-2019-09 | Sep-19 | wound | ICU1 | |||
| PSA-2019-10 | Sep-19 | screening (rectal) | ICU1 | |||
| PSA-2016-09 | Nov-16 | respiratory tract | ICU2 | ST357 | 3 | bla OXA-10-like, blaVEB-1 |
| PSA-2018-07 | Nov-18 | respiratory tract | ICU3 | |||
| PSA-2015-01 | Jan-15 | screening (rectal) | ICU1, ICU3 | ST235 | singleton | bla OXA-10 |
| PSA-2015-02 | Feb-15 | wound | ImCU1 | ST395 | singleton | bla OXA-129-like |
| PSA-2015-03 | Apr-15 | screening (nose/throat) | ICU1 | ST1233 | singleton | — |
| PSA-2015-04 | Jun-15 | screening (rectal) | ICU2 | ST273 | singleton | bla VIM-2, blaACT-5-like |
| PSA-2015-05 | Oct-15 | respiratory tract | ICU2 | ST980 | singleton | — |
| PSA-2015-06 | Oct-15 | wound | ICU3 | ST17 | singleton | — |
| PSA-2016-02 | Mar-16 | respiratory tract | ImCU1 | ST395 | singleton | — |
| PSA-2016-05 | Sep-16 | urine | ICU3 | ST654 | singleton | bla NDM-1, blaGES-5 |
| PSA-2016-06 | Oct-16 | respiratory tract | ImCU1 | ST918 | singleton | — |
| PSA-2016-08 | Nov-16 | wound | ICU3 | ST1743 | singleton | — |
| PSA-2017-01 | Jan-17 | respiratory tract | ICU2 | ST1044 | singleton | — |
| PSA-2017-02 | Feb-17 | screening (rectal) | other | ST235 | singleton | bla VIM-4 |
| PSA-2017-04 | Aug-17 | urine | ICU2, ImCU1 | ST3618 | singleton | bla VIM-1 |
| PSA-2017-05 | Oct-17 | screening (nose/throat) | ICU1 | ST395 | singleton | — |
| PSA-2017-08 | Oct-17 | respiratory tract | ICU3 | ST274 | singleton | — |
| PSA-2017-09 | Oct-17 | wound | ICU1 | ST2069 | singleton | — |
| PSA-2018-01 | Jan-18 | wound | ICU1 | ST2167 | singleton | — |
| PSA-2018-02 | Feb-18 | respiratory tract | ICU1 | ST274 | singleton | — |
| PSA-2018-03 | Mar-18 | respiratory tract | ICU2 | ST701 | singleton | — |
| PSA-2018-04 | May-18 | wound | ICU1 | ST291 | singleton | — |
| PSA-2018-06 | Nov-18 | urine | ICU2, ICU3 | ST654 | singleton | bla VIM-2 |
| PSA-2019-01 | Feb-19 | respiratory tract | ICU2, ImCU1 | ST207 | singleton | — |
| PSA-2019-02 | Mar-19 | screening (nose/throat) | ICU1, ICU3 | ST3480 | singleton | — |
| PSA-2019-03 | Jul-19 | screening (rectal) | ImCU1 | ST1320 | singleton | — |
| PSA-2019-04 | Jul-19 | respiratory tract | ImCU1 | ST395 | singleton | — |
| PSA-2019-06 | Aug-19 | screening (rectal) | ICU1 | ST508 | singleton | — |
| PSA-2019-08 | Sep-19 | respiratory tract | ICU2 | ST2332 | singleton | — |
| PSA-2019-11 | Oct-19 | respiratory tract | ICU2 | ST309 | singleton | — |
| PSA-2019-12 | Oct-19 | screening (rectal) | ImCU1 | ST235 | singleton | — |
| PSA-2019-13 | Nov-19 | screening (rectal) | ICU3 | ST27 | singleton | — |
| PSA-2019-14 | Nov-19 | screening (rectal) | ICU3 | ST207 | singleton | — |
| PSA-2020-01 | Jan-20 | respiratory tract | ICU2 | ST399 | singleton | — |
| PSA-2020-02 | Feb-20 | screening (nose/throat) | ICU1 | ST274 | singleton | — |
| PSA-2020-03 | Apr-20 | respiratory tract | ICU2, ImCU1 | ST871 | singleton | — |
PSA, P. aeruginosa; ST, sequence type (conventional 7-loci MLST).
Figure 1.
Ridom SeqSphere+ neighbour joining tree for 55 samples based on 3867 columns, pairwise ignoring missing values, percentage columns difference. Isolates are coloured based on their carbapenem-resistance mechanism. ‘Default β-lactamases’ is defined as those where only the blaOXA-50-like and blaPAO-like genes were detected. Clonal Clusters 1 to 3 are indicated by brackets and numbers.
Figure 2.
Epidemiological timeline and transmission route of Cluster 1 VIM-2-producing P. aeruginosa. Each circle represents one patient at time of first isolation. An arrow indicates genetically and epidemiologically confirmed transmission events (dashed line, room-to-patient; continuous line, patient-to-patient). Wards of transmission are indicated in different colours. Circle colour indicates mode of transmission (grey, healthcare-associated, white, hospital-acquired). Isolate numbers can be inferred by combining year and number in the node. Isolate no. PSA-2016-03 was isolated on another ward than the four critical care units.
Clusters 2 and 3 represented non-carbapenemase-producing isolates. Cluster 2 isolates were obtained from four patients, all from the same ward, with three proven patient-to-patient transmissions. We were not able to identify the transmission route of the index patient. This single outbreak among burn patients was actively identified by the IPC team at the time of detection and immediately terminated. The Cluster 3 isolates both harboured the ESBL genes blaVEB-1 and blaOXA-10, and differed by five alleles suggesting a transmission event, however the two patients had no known epidemiological link.
Singletons were generally separated from all other isolates by over 2785 alleles. Exceptions to this were the two ST207 isolates, the two ST654 isolates, the three ST235 isolates, the three 274 isolates and the four ST395 isolates, which differed by 39, 59, 96, 151 and 169 alleles, respectively.
We compared our Cluster 1 blaVIM-2-harbouring ST111 isolates with those from previous studies in the UK and the Netherlands.21,22 Raw reads were downloaded from the European Nucleotide Archive with the study number ERP010395 and PRJEB39528, and assembled as described in the materials and methods. The resulting minimum spanning tree is shown in Figure S1. The isolates from Germany clustered closely with the isolates from the UK and the Netherlands, with 19–58 allelic differences.
Genetic environment of the MBL blaVIM-2
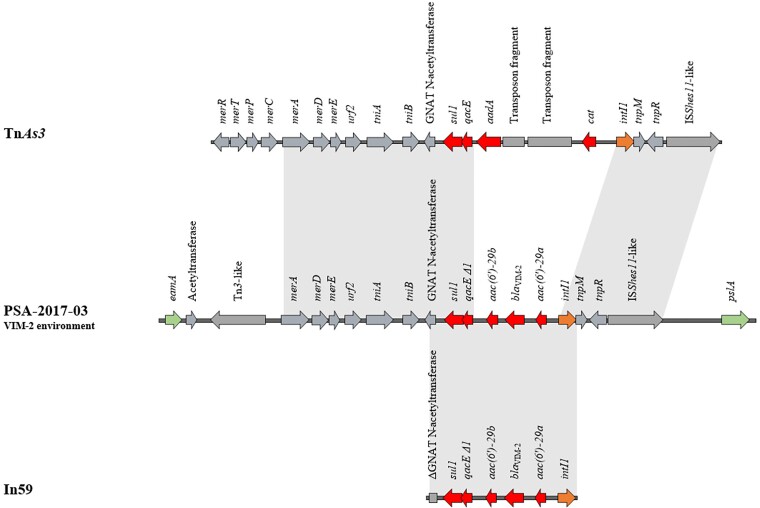
Hybrid genome assemblies (short and long reads) of three ST111 VIM-2-producing isolates from the cgMLST Cluster 1 revealed that the genetic environment of the blaVIM-2 gene was identical in the three isolates: a class 1 integron located in the chromosome. The blaVIM-2 gene was flanked by aminoglycoside resistance genes aacA(6′)-29a and aacA(6′)-29b, differing in four amino acid substitutions, as well as the integrase gene intI1. Further upstream the antiseptic resistance cassette qacE was detected, followed by sul1 conferring resistance to sulphonamides. The genetic environment of the MBL was highly similar (blastn coverage 100% and identity 99.5%, accession number AF263519.1) to the integron In59.23 Finally, the integron was inserted into a transposon TnAs3-like structure (blastn coverage 72% and identity 99%, accession number CP000645) (Figure 3). TnAs3 belongs to the Tn3 family and the Tn21 subgroup.24
Figure 3.
Schematic diagram of the genetic environment of blaVIM-2 in the isolate PSA-2017-03 compared with the transposon TnAs3 (accession number CP000645) and the integron In59 (accession number AF263519.1). Arrows indicate the deduced open reading frames (ORFs) and their orientations. Homologous regions are shaded in grey. Antimicrobial resistance genes are shown in red and the intI1 gene is shaded orange. The ORFs EamA family transporter (putative membrane protein) and PslA (biofilm formation protein) coloured in green represent chromosomal markers. The remaining genes are shown in grey.
Discussion
P. aeruginosa is an important hospital-acquired pathogen causing infections and outbreaks in ICUs.1 Over the last few years we have seen a dramatic increase of MDR isolates worldwide.2,3,25 In this study, from a collection of MDR/XDR P. aeruginosa isolates from four critical care units of a tertiary care centre transmission was nearly exclusively observed among ST111 VIM-2-producing isolates over a period of 5 years. Another short-term outbreak caused by ST970 non-carbapenemase-producing P. aeruginosa was observed in vulnerable patients in the burn unit. We observed a higher diversity in non-carbapenemase-producing isolates, most of the isolates being genetically unrelated to each other. Overall, the study reveals a remarkable clonal diversity, with most isolates represented by sporadic single genotypes, and a few epidemic strains.
Studies comparing clonal diversity among susceptible and MDR/XDR resistant isolates have shown a lower diversity among MDR and especially XDR strains and that XDR/MDR P. aeruginosa infections are disproportionally caused by a small subset of globally distributed ‘high-risk clones’, linked to mutational resistance determinants but also transferable resistance genes.4,26–28 Traditionally, these clones are classified according to the MLST scheme developed by Curran et al.29 over 15 years ago. Four out of the worldwide top 10 P. aeruginosa high-risk clones (ST235, ST111, ST357 and ST654) were found in this study.4
In our study the appearance of high-risk clones was also linked to carbapenemase production, mostly ST111 having acquired VIM-2, a combination that has also been described previously.4,30 Other surveillance studies from the UK and the Netherlands reported that ST111 P. aeruginosa was linked mostly to VIM-2 production.21,22 Epidemiological data and high resolution genotyping by WGS showed evidence for spread within and between hospitals in different regions of the UK.21 It is important to note that in our study most isolates were hospital-acquired and a limited number of isolates were also present on admission and healthcare-associated. As there is no continuous molecular surveillance on a regional or national level in Germany, we are unable to determine the extent to which ST111 carrying carbapenemases has spread throughout the region. However, VIM-2 is the leading carbapenemase in P. aeruginosa based on data from the German national reference centre.31 Moreover, previous studies have shown ST111 VIM-2-producing P. aeruginosa in Hamburg in 2001,32 and Wendel et al.8 reported the spread of ST111 GIM-1-producing P. aeruginosa in a hospital located close to Cologne. Furthermore, we also demonstrated that the ST111 isolates from the UK and the Netherlands clustered closely, although not overlapping, with those from the current study, highlighting that these high-risk clones are not confined to small geographical regions but have in fact spread to other countries. The genetic environment of the blaVIM-2 gene of the ST111 clone was highly similar to the integron In59 described in a VIM-2-positive P. aeruginosa isolate recovered in 1997 in France and since then in different European countries, and mostly in ST111 isolates.23,33–35 As high-risk clones tend to harbour several resistance traits they are also linked to aminoglycoside resistance, with the aacA29a gene being the most common determinant in ST111, also confirmed in our study.30
Microbiological and infection control monitoring of carbapenem resistance in P. aeruginosa is of utmost importance with regard to the clonal structure and mobile genetic elements such as carbapenemases. Both SNP-based and cgMLST-based typing have been successfully applied in various studies.9,10,36–39 Recently, several validated cgMLST schemes were published enabling a standardized approach and a consistent nomenclature.16,40,41 The cgMLST scheme proposed by Tönnies et al.16 (3867 targets) used in this study was comparable to an ad-hoc cgMLST scheme previously established by one of the authors (4547 targets).39
Depending on the mode of transmission, different infection and control approaches are needed. Individual nosocomial acquisition of P. aeruginosa is either endo- or exogenous and can subsequently lead to transmission chains. Sporadic or low-frequency transmissions from the environment (mostly moist sites) to the patient are difficult to trace back epidemiologically as shown in several studies.8,10,11 However, we were unable to confirm many transmission events within the hospital despite sporadic appearance of the clone. A hidden environmental reservoir (especially sinks) or complex epidemiological links might be an explanation. There is growing evidence supporting a water-free patient environment and removing sinks from the patient’s room to eliminate patient-side biofilm reservoirs.9,42,43 This is possible on the ICU where the patient generally does not need a bathroom. Nevertheless, we did not find a pattern of room transmissions as patients from the biggest cluster were found on all four wards.
There are a few limitations in this study. Unfortunately, we were not able to provide full prevalence data, as only two-thirds of the non-duplicate isolates detected during this period were available for further study. Moreover, transmissions by unidentified colonized patients might have been overlooked as there was no periodic rectal screening in place on all units. However, we are confident that the study gives a good overview of the epidemiological pattern. Secondly, the inclusion criteria were probably not sensitive enough to detect carbapenemases, as we chose to include isolates based on the German classification guideline for Gram-negative MDR organisms (Gram-negative MDR organisms with resistance to four out of four major antibiotic classes, 4MRGN).44 However, carbapenemase production is often linked to MDR/XDR phenotypes.45
Thirdly, the mutational resistome was not analysed and we only performed analysis of acquired/intrinsic resistance genes in silico using a web-based tool. This is more complex and not completely validated yet and out of the scope of our investigation, which is basically epidemiological.46 Fourthly, we did not conduct environmental sampling in this study to detect an inanimate reservoir; however we want to point out that on two out of four of the ICUs, sinks in most of the patient rooms were removed during the study period.
In conclusion, to ensure surveillance of P. aeruginosa high-risk clones and carbapenemase genes, it is necessary to implement diagnostic tools at local level for their early detection and the combination with epidemiological data, in order to guide IPC strategies. This is especially the case among carbapenemase-producing high-risk clones that were associated with ongoing acquisitions. Therefore, the timely detection of carbapenemases can potentially lead to strategies to halt transmission.
Supplementary Material
Acknowledgements
We thank Ingo Winterfeld and Alfons Schön from the Institute of Hygiene Cologne for technical help. We also want to thank the team of the private microbiology laboratory MVZ synlab Leverkusen for routine microbiological analysis.
Funding
This study was supported by internal funding.
Transparency declarations
None to declare.
Supplementary data
Table S1 and Figure S1 are available as Supplementary data at JAC-AMR Online.
References
- 1. Maraolo AE, Cascella M, Corcione Set al. Management of multidrug-resistant Pseudomonas aeruginosa in the intensive care unit: state of the art. Expert Rev Anti Infect Ther 2017; 15: 861–71. [DOI] [PubMed] [Google Scholar]
- 2. Buhl M, Peter S, Willmann M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant Pseudomonas aeruginosa: a systematic review. Expert Rev Anti Infect Ther 2015; 13: 1159–70. [DOI] [PubMed] [Google Scholar]
- 3. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011; 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Del Barrio-Tofino E, Lopez-Causape C, Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents 2020; 56: 106196. [DOI] [PubMed] [Google Scholar]
- 5. Magiorakos AP, Srinivasan A, Carey RBet al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 6. Grundmann H, Barwolff S, Tami Aet al. How many infections are caused by patient-to-patient transmission in intensive care units? Crit Care Med 2005; 33: 946–51. [DOI] [PubMed] [Google Scholar]
- 7. Otter JA, Yezli S, French GL. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol 2011; 32: 687–99. [DOI] [PubMed] [Google Scholar]
- 8. Wendel AF, Kolbe-Busch S, Ressina Set al. Detection and termination of an extended low-frequency hospital outbreak of GIM-1-producing Pseudomonas aeruginosa ST111 in Germany. Am J Infect Control 2015; 43: 635–9. [DOI] [PubMed] [Google Scholar]
- 9. Wendel AF, Mattner F. Contamination of the water system with Pseudomonas aeruginosa after implementation of antiseptic bathing with a leave-on product. J Hosp Infect 2020; 104: 81–2. [DOI] [PubMed] [Google Scholar]
- 10. De Geyter D, Vanstokstraeten R, Crombe Fet al. Sink drains as reservoirs of VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa in a Belgian intensive care unit: relation to patients investigated by whole genome sequencing. J Hosp Infect 2021; 115: 75–82. [DOI] [PubMed] [Google Scholar]
- 11. Hopman J, Meijer C, Kenters Net al. Risk assessment after a severe hospital-acquired infection associated with carbapenemase-producing Pseudomonas aeruginosa. JAMA Netw Open 2019; 2: e187665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schafer E, Malecki M, Tellez-Castillo CJet al. Molecular surveillance of carbapenemase-producing Pseudomonas aeruginosa at three medical centres in Cologne, Germany. Antimicrob Resist Infect Control 2019; 8: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schroder C, Schwab F, Behnke Met al. Epidemiology of healthcare associated infections in Germany: nearly 20 years of surveillance. Int J Med Microbiol 2015; 305: 799–806. [DOI] [PubMed] [Google Scholar]
- 14. Junemann S, Sedlazeck FJ, Prior Ket al. Updating benchtop sequencing performance comparison. Nat Biotechnol 2013; 31: 294–6. [DOI] [PubMed] [Google Scholar]
- 15. Wick RR, Judd LM, Gorrie CLet al. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tönnies H, Prior K, Harmsen Det al. Establishment and evaluation of a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Pseudomonas aeruginosa. J Clin Microbiol 2021; 59: e01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zankari E, Hasman H, Cosentino Set al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siguier P, Perochon J, Lestrade Let al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34: D32–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008; 36: 309–32. [DOI] [PubMed] [Google Scholar]
- 21. Turton JF, Wright L, Underwood Aet al. High-resolution analysis by whole-genome sequencing of an international lineage (sequence type 111) of Pseudomonas aeruginosa associated with metallo-carbapenemases in the United Kingdom. J Clin Microbiol 2015; 53: 2622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pirzadian J, Persoon MC, Severin JAet al. National surveillance pilot study unveils a multicenter, clonal outbreak of VIM-2-producing Pseudomonas aeruginosa ST111 in the Netherlands between 2015 and 2017. Sci Rep 2021; 11: 21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poirel L, Lambert T, Turkoglu Set al. Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla(VIM-2) carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother 2001; 45: 546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ross K, Varani AM, Snesrud Eet al. TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. mBio 2021; 12: e0206021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oliver A, Mulet X, Lopez-Causape Cet al. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 2015; 21-22: 41–59. [DOI] [PubMed] [Google Scholar]
- 26. Cabot G, Ocampo-Sosa AA, Dominguez MAet al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 2012; 56: 6349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pincus NB, Bachta KER, Ozer EAet al. Long-term persistence of an extensively drug-resistant subclade of globally distributed Pseudomonas aeruginosa clonal complex 446 in an academic medical center. Clin Infect Dis 2020; 71: 1524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawa T, Momiyama K, Mihara Tet al. Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: practical overview. Microbiol Immunol 2020; 64: 331–44. [DOI] [PubMed] [Google Scholar]
- 29. Curran B, Jonas D, Grundmann Het al. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 2004; 42: 5644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kocsis B, Gulyas D, Szabo D. Diversity and distribution of resistance markers in Pseudomonas aeruginosa international high-risk clones. Microorganisms 2021; 9: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfennigwerth N, Schauer J. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger – 2020. Epidemiologisches Bulletin 2020; 36: 4–11. [Google Scholar]
- 32. Hentschke M, Goritzka V, Campos CBet al. Emergence of carbapenemases in Gram-negative bacteria in Hamburg, Germany. Diagn Microbiol Infect Dis 2011; 71: 312–5. [DOI] [PubMed] [Google Scholar]
- 33. Samuelsen O, Toleman MA, Sundsfjord Aet al. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother 2010; 54: 346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liakopoulos A, Mavroidi A, Katsifas EAet al. Carbapenemase-producing Pseudomonas aeruginosa from central Greece: molecular epidemiology and genetic analysis of class I integrons. BMC Infect Dis 2013; 13: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duljasz W, Gniadkowski M, Sitter Set al. First organisms with acquired metallo-β-lactamases (IMP-13, IMP-22, and VIM-2) reported in Austria. Antimicrob Agents Chemother 2009; 53: 2221–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kossow A, Kampmeier S, Willems Set al. Control of multidrug-resistant Pseudomonas aeruginosa in allogeneic hematopoietic stem cell transplant recipients by a novel bundle including remodeling of sanitary and water supply systems. Clin Infect Dis 2017; 65: 935–42. [DOI] [PubMed] [Google Scholar]
- 37. Parcell BJ, Oravcova K, Pinheiro Met al. Pseudomonas aeruginosa intensive care unit outbreak: winnowing of transmissions with molecular and genomic typing. J Hosp Infect 2018; 98: 282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willmann M, Bezdan D, Zapata Let al. Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: a molecular epidemiological study. J Antimicrob Chemother 2015; 70: 1322–30. [DOI] [PubMed] [Google Scholar]
- 39. Paul G, Meissner A, Neuneier Jet al. Outbreak of Pseudomonas aeruginosa infections after CT-guided spinal injections. J Hosp Infect 2021; 116: 1–9. [DOI] [PubMed] [Google Scholar]
- 40. de Sales RO, Migliorini LB, Puga Ret al. A core genome multilocus sequence typing scheme for Pseudomonas aeruginosa. Front Microbiol 2020; 11: 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stanton RA, McAllister G, Daniels JBet al. Development and application of a core genome multilocus sequence typing scheme for the health care-associated pathogen Pseudomonas aeruginosa. J Clin Microbiol 2020; 58: e00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hopman J, Tostmann A, Wertheim Het al. Reduced rate of intensive care unit acquired Gram-negative bacilli after removal of sinks and introduction of ‘water-free’ patient care. Antimicrob Resist Infect Control 2017; 6: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weinbren MJ. The handwash station: friend or fiend? J Hosp Infect 2018; 100: 159–64. [DOI] [PubMed] [Google Scholar]
- 44. Muller J, Voss A, Kock Ret al. Cross-border comparison of the Dutch and German guidelines on multidrug-resistant Gram-negative microorganisms. Antimicrob Resist Infect Control 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz-Garbajosa P, Canton R. Epidemiology of antibiotic resistance in Pseudomonas aeruginosa. Implications for empiric and definitive therapy. Rev Esp Quimioter 2017; 30Suppl 1: 8–12. [PubMed] [Google Scholar]
- 46. Cortes-Lara S, Barrio-Tofino ED, Lopez-Causape Cet al. Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin Microbiol Infect 2021; 27: 1631–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.