Abstract
COVID-19 quickly became a global pandemic after only four months of its first detection. It is crucial to detect this disease as soon as possible to decrease its spread. The use of chest X-ray (CXR) images became an effective screening strategy, complementary to the reverse transcription-polymerase chain reaction (RT-PCR). Convolutional neural networks (CNNs) are often used for automatic image classification and they can be very useful in CXR diagnostics. In this paper, 21 different CNN architectures are tested and compared in the task of identifying COVID-19 in CXR images. They were applied to the COVIDx8B dataset, a large COVID-19 dataset with 16,352 CXR images coming from patients of at least 51 countries. Ensembles of CNNs were also employed and they showed better efficacy than individual instances. The best individual CNN instance results were achieved by DenseNet169, with an accuracy of 98.15% and an F1 score of 98.12%. These were further increased to 99.25% and 99.24%, respectively, through an ensemble with five instances of DenseNet169. These results are higher than those obtained in recent works using the same dataset.
Keywords: Convolutional neural networks, Transfer learning, Chest X-ray images
1. Introduction
COVID-19 is an infectious disease caused by the Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) (Khan et al., 2021). It quickly became a global pandemic in less than four months after its first detection in December 2019 in Wuhan, China (Monshi et al., 2021). As of February 2022, it has over 434 million confirmed cases and almost 6 million deaths reported to World Health Organization (World Health Organization, 2022). Early detection of positive COVID-19 cases is critical for avoiding the virus’s spread.
The most common technique for diagnosing COVID-19 is known as transcriptase-polymerase chain reaction (RT-PCR). It detects SARS-CoV-2 through collected respiratory specimens of nasopharyngeal or oropharyngeal swabs. However, RT-PCR testing is expensive, time-consuming, and shows poor sensitivity (Monshi et al., 2021, Mostafiz et al., 2020), especially in the first days of exposure to the virus (Long et al., 2020). Up to 54% of COVID-19 patients may have an initial negative RT-PCR result (Arevalo-Rodriguez et al., 2020).
Patients that receive a false negative diagnosis may contact and infect other people before they are tested again. Therefore, it is important to have alternative methods to detect the disease, such as Chest X-ray (CXR) images. CXR equipment is widely available in hospitals and CXR images are cheap and fast to acquire. They can be inspected by radiologists to find visual indicators of the virus (Feng et al., 2020).
In the past decade, the rise of deep learning methods (Goodfellow et al., 2016, LeCun et al., 2015, Schmidhuber, 2015), especially the convolutional neural networks (CNNs), were responsible for many advances in automatic image classification (Krizhevsky et al., 2012). CXR diagnostic using deep learning methods is a mechanism that can be explored to surpass the limitations of RT-PCR insufficient test kits, waiting time of test results, and test costs (Mostafiz et al., 2020).
Many studies concerning the application of CNNs to COVID-19 diagnostic on CXR images were published since the last year (Abbas et al., 2021, Alawad et al., 2021, Chhikara et al., 2021, Heidari et al., 2020, Hira et al., 2021, Ismael and Şengür, 2021, Jia et al., 2021, Karthik et al., 2021, Khan et al., 2021, Mohammad Shorfuzzaman, 2020, Monshi et al., 2021, Mostafiz et al., 2020, Narin et al., 2021, Nigam et al., 2021). However, most of them used relatively small and more homogeneous datasets. In this paper, the COVIDx8B dataset1 (Zhao et al., 2021) is used. It has 16,352 CXR images, from which 2,358 are COVID-19 positive and the remaining are from both healthy and pneumonia patients. Released in March 2021, this dataset is composed of images from six other open-source chest radiography datasets. Therefore it is larger and more heterogeneous than earlier available datasets. However, there are only a few works that used this dataset so far (Dominik, 2021, Pavlova et al., 2021, Zhao et al., 2021). A recent survey on applications of artificial intelligence in the COVID-19 pandemic (Khan et al., 2021) reviewed dozens of papers, including papers on CNNs applied to CXR images and all of them used earlier available datasets which are smaller than COVIDx8B.
In this paper, a comparison of different CNN models applied to the COVIDx8B dataset is presented, including popular architectures such as VGG (Simonyan & Zisserman, 2015), ResNet (He et al., 2016a), DenseNet (Huang et al., 2017), and EfficientNet (Tan & Le, 2019). They were all trained in the same conditions with the training and test subsets defined by the dataset authors. The initial weights of all methods were defined to those trained on the ImageNet dataset (Russakovsky et al., 2015), which is commonly used in transfer learning scenarios (Oquab et al., 2014). The accuracy, sensitivity (TPR), precision (PPV), and F1 score were evaluated using the test subset. Later, some models’ continuous output (before the classification layer) were combined (ensembles) to overcome individual limitations and provide better classification results.
The remainder of this paper is organized as follows. Section 2 shows related work, in which CNNs were used to detect COVID-19 on CXR images. Section 3 presents the COVIDx8B dataset. Section 4 shows the CNN architectures employed in this paper. Section 5 shows the computer simulations comparing the proposed models and other recent approaches from the literature for COVID-19 classification on CXR images using the same dataset. Section 6 shows the computer simulations with CNN ensembles, improving the classification performance of individual models. Finally, the conclusions are drawn in Section 7.
2. Related work
Many studies have investigated the use of machine learning techniques to detect COVID-19. Many of the researchers used CNN techniques and CXR images and faced challenges due to the lack of available datasets (Alawad et al., 2021). While many authors provided tables comparing results achieved in different works, the comparisons are not fair, since the used datasets are frequently different and pose different levels of challenge. Therefore, here the related works are described focusing on what architectures have been used to handle the problem of COVID-19 detection on CXR images and the size of the evaluated datasets.
Nigam et al. (2021) used VGG16, DenseNet121, Xception, NASNet, and EfficientNet in a dataset with 16,634 images. Though this dataset is slightly larger than COVIDx8B, unfortunately, the authors did not make it publicly available. The highest accuracy was 93.48% obtained with EfficientNetB7.
Ismael and Şengür (2021) used ResNet18, ResNet50, ResNet101, VGG16, and VGG19 for deep feature extraction and support vector machines (SVM) for CXR images classification. The highest accuracy was 94.7% obtained with ResNet50. However, they used a small dataset with only CXR images.
Abbas et al. (2021) validated a deep CNN called Decompose, Transfer, and Compose (DeTraC) for COVID-19 CXR images classification with 93.1% accuracy. They used a combination of two small datasets, totaling images.
Hira et al. (2021) used the AlexNet, GoogleNet, ResNet-50, Se-ResNet-50, DenseNet121, Inception V4, Inception ResNet V2, ResNeXt-50, and Se-ResNeXt-50 architectures. Se-ResNeXt-50 achieved the highest classification accuracy of 99.32%. They used a combination of four datasets, totaling 8,830 CXR images.
Alawad et al. (2021) used VGG16 both as a stand-alone classifier and as a feature extractor for SVM, Random-Forests (RF), and Extreme-Gradient-Boosting (XGBoost) classifiers. VGG-16 and VGG16+SVM models provide the best performance with 99.82% accuracy. They used a combination of five datasets, totaling 7,329 CXR images.
Narin et al. (2021) used ResNet50, ResNet101, ResNet152, InceptionV3, and Inception-ResNetV2. ResNet50 achieved the highest classification performance with 96.1%, 99.5%, and 99.7% accuracy on three different datasets, totaling 7,406 CXR images.
Monshi et al. (2021) focused on data augmentation and CNN hyperparameters optimization, increasing VGG19 and ResNet50 accuracy. They also proposed CovidXrayNet, a model based on EfficientNet-B0, which achieved an accuracy of 95.82% on an earlier version of the COVIDx dataset with 15,496 CXR images.
Heidari et al. (2020) focused on preprocessing algorithms to improve the performance of VGG16. They used a dataset with 8,474 CXR images and reached 94.5% accuracy.
Jia et al. (2021) proposed a modified MobileNet to classify CXR and CT images. They applied their method to a CXR dataset with 7,592 CXR images and achieved 99.3% accuracy. They also applied it to an earlier version of COVIDx with 13,975 CXR images, achieving 95.0% accuracy.
Karthik et al. (2021) proposed a custom CNN architecture which they called Channel-Shuffled Dual-Branched (CSDB). They achieved an accuracy of 99.80% on a combination of seven datasets, totaling 15,265 images.
Mostafiz et al. (2020) used a hybridization of CNN (ResNet50) and discrete wavelet transform (DWT) features. The random forest-based bagging approach was used for classification. They combined different datasets and used data augmentation techniques to produce a total of CXR images and achieved 98.5% accuracy.
Mohammad Shorfuzzaman (2020) used VGG16, ResNet50V2, Xception, MobileNet, and DenseNet121 in a transfer learning scenario. They collected CXR images from different sources to compose a dataset with images. The best accuracy (98.15%) was achieved with ResNet50V2. They also made an ensemble of the four best models (ResNet50V2, Xception, MobileNet, and DenseNet121) with the final output obtained by majority voting, raising the accuracy to 99.26%.
Chhikara et al. (2021) proposed a InceptionV3 based-model and applied it to three different datasets with 11,244, 8,246, and 14,486 CXR images, respectively. The model has reached an accuracy of 97.7%, 84.95%, and 97.03% on the mentioned datasets, respectively.
Pavlova et al. (2021) proposed the COVIDx8B dataset, which they claim is the largest and most diverse COVID-19 CXR dataset in open access form, and the COVID-Net CXR-2 model, a CNN specially tailored for COVID-19 detection on CXR images using machine-driven design, which achieved an accuracy of 95.5%.
Zhao et al. (2021) used ResNet50V2 to classify the COVIDx8B dataset with an accuracy of 96.5% in the best scenario.
Dominik (2021) proposed a lightweight architecture called BaseNet and achieved an accuracy of 95.50% on COVIDx8B. He also used an ensemble composed of BaseNet, VGG16, VGG19, ResNet50, DenseNet121, and Xception to achieve 97.75% accuracy. It was further increased to 99.25% using an optimal classification threshold.
3. Dataset
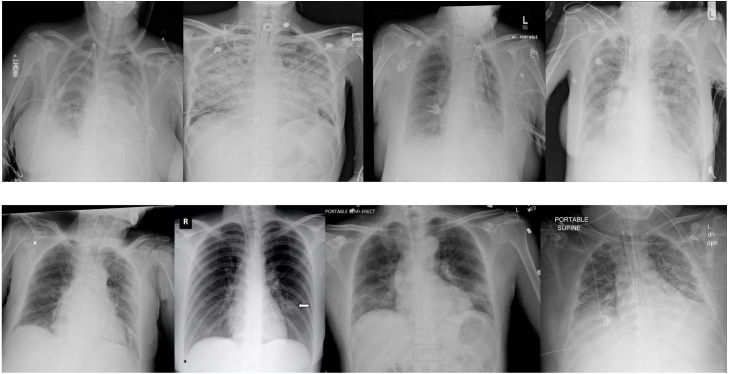
Most of the early research regarding COVID-19 detection on CXR images suffered from the lack of available datasets (Alawad et al., 2021). The authors would frequently combine different smaller datasets, so fairly comparing the results was impossible. COVIDx8B is a large and heterogeneous COVID-19 CXR benchmark dataset with 16,352 CXR images coming from patients of at least countries (Pavlova et al., 2021). It is constructed with images extracted from six open-source chest radiography datasets, which are shown in Table 1. Notice that the sum of the images in the source datasets is much larger than the size of COVIDx8B since not all of them were selected by the authors. Example images from the COVIDx8B dataset are shown in Fig. 1.
Table 1.
List of datasets that compose the COVIDx8B benchmark dataset.
| Source dataset | Size | Reference |
|---|---|---|
| Covid-chestxray-dataset | 950 | Cohen et al. (2020) |
| COVID-19 Chest X-ray Dataset Initiative | 55 | Chung (2020b) |
| Actualmed COVID-19 Chest X-ray Dataset Initiative | 238 | Chung (2020a) |
| COVID-19 Radiography Database-Version 3 | 21,165 | Chowdhury et al., 2020, Rahman et al., 2021 |
| RSNA Pneumonia Detection Challenge | 29,684 | Wang et al. (2017) |
| RSNA International COVID-19 Open Radiology Database (RICORD) | 1,257 | Tsai et al. (2021) |
Fig. 1.
Examples of CXR images from the COVIDx8B dataset. The first row shows COVID-19 negative patient cases, and the second row shows COVID-19 positive patient cases.
Though COVIDx8B does not include information on patients’ demographics, half of their source datasets do. The covid-chestxray-dataset has registers from male patients and registers from female patients. The average age is years old. The COVID-19 positive registers are from male and female patients, with an average age of years old. The Fig. 1 COVID-19 Chest X-ray Dataset Initiative has only registers, most of them do not indicate sex. Among the remaining, there are male patients and female patients. Only patients have their exact age registered and the average is years old. All patients with the exact age described are COVID-19 positive or unlabeled. The RSNA International COVID-19 Open Radiology Database (RICORD) only has COVID-19 positive cases. They come from male and female patients, with an average age of years old.
Four of the source datasets have both COVID-19 positive and negative cases. The RSNA Pneumonia Detection Challenge has only COVID-19 negative cases (non-COVID pneumonia, normal, etc.) and The RSNA International COVID-19 Open Radiology Database (RICORD) has only COVID-19 positive cases.
COVID8xB training subset is composed of 15,952 images, from which 2,158 are COVID-19 positive and 13,794 are COVID-19 negative. The negative group also includes images of patients with non-COVID-19 pneumonia, which poses a major challenge as it is usually difficult to distinguish between COVID-19 and non-COVID19 pneumonia. The test subset has COVID-19 positive images from different patients and COVID-19 negative images. In the negative group, images are from healthy patients. The other images are from non-COVID pneumonia patients. The test images were randomly selected from international patient groups curated by the Radiological Society of North America (RSNA) (Tsai et al., 2021, Wang et al., 2017). The images were annotated by an international group of scientists and radiologists from different institutes around the world. The test set was selected in such a way to ensure no patient overlap between training and test sets (Pavlova et al., 2021).
4. CNN architectures
This section presents the CNN architectures explored in this work. It also describes the layers added to complete the models and perform the CXR images classification. Table 2 shows the tested architectures, some of their characteristics, and their respective literature references.
Table 2.
CNN architectures, some of their characteristics, and their references.
| Model | Input Image Resolution | Output of Last Conv. Layer | Trainable Parameters | Reference |
|---|---|---|---|---|
| DenseNet121 | 224 × 224 | Huang et al. (2017) | ||
| DenseNet169 | 224 × 224 | Huang et al. (2017) | ||
| DenseNet201 | 224 × 224 | Huang et al. (2017) | ||
| EfficientNetB0 | 224 × 224 | Tan and Le (2019) | ||
| EfficientNetB1 | 240 × 240 | Tan and Le (2019) | ||
| EfficientNetB2 | 260 × 260 | Tan and Le (2019) | ||
| EfficientNetB3 | 300 × 300 | Tan and Le (2019) | ||
| InceptionResNetV2 | 299 × 299 | Szegedy et al. (2017) | ||
| InceptionV3 | 299 × 299 | Szegedy et al. (2016) | ||
| MobileNet | 224 × 224 | Howard et al. (2017) | ||
| MobileNetV2 | 224 × 224 | Sandler et al. (2018) | ||
| NASNetMobile | 224 × 224 | Zoph et al. (2018) | ||
| ResNet101 | 224 × 224 | He et al. (2016a) | ||
| ResNet101V2 | 224 × 224 | He et al. (2016b) | ||
| ResNet152 | 224 × 224 | He et al. (2016a) | ||
| ResNet152V2 | 224 × 224 | He et al. (2016b) | ||
| ResNet50 | 224 × 224 | He et al. (2016a) | ||
| ResNet50V2 | 224 × 224 | He et al. (2016b) | ||
| VGG16 | 224 × 224 | Simonyan and Zisserman (2015) | ||
| VGG19 | 224 × 224 | Simonyan and Zisserman (2015) | ||
| Xception | 299 × 299 | Chollet (2017) |
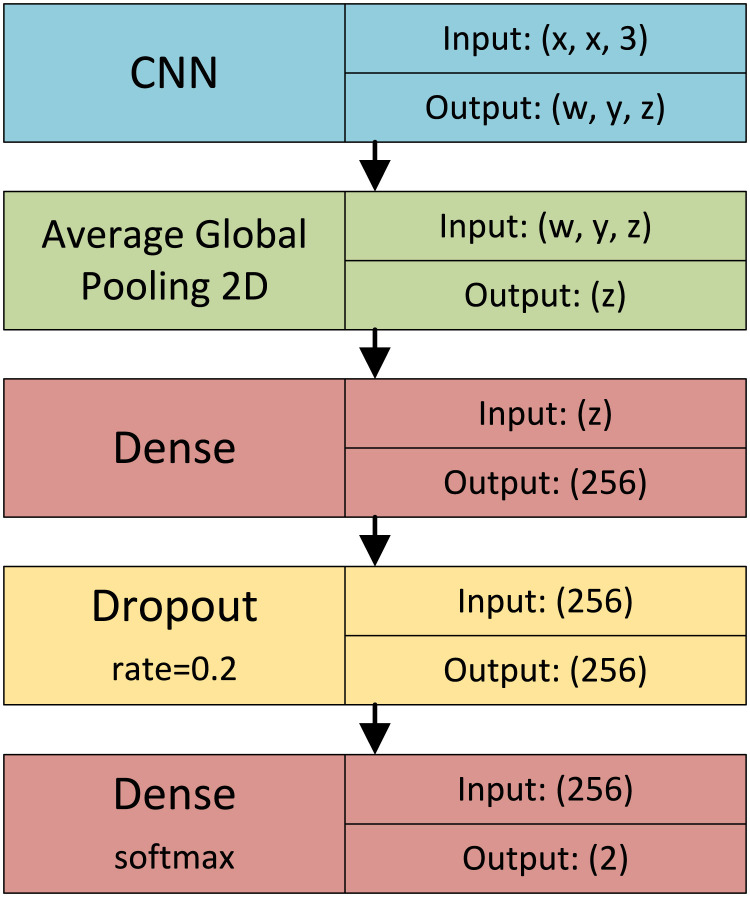
The output of the last convolutional layer of the original CNN is fed to a global average pooling layer. Following, there is a dense layer with neurons using ReLU (Rectified Linear Unit) activation function, a dropout layer with a 20% rate, and a softmax classification layer. This proposed architecture is illustrated in Fig. 2, where indicates the horizontal and vertical input size of the CNN (image size), while , , and indicate the size of the CNN output in its last convolutional layer. These values depend on the original CNN architecture and they are indicated in Table 2. The table also shows the number of trainable parameters in each CNN architecture, including both their original layers and the dense layers added for COVID-19 classification.
Fig. 2.
The proposed CNN Transfer Learning architecture.
5. CNN comparison
In this section, the computer simulations comparing the CNN models applied to the COVIDx8B dataset are presented. All simulations were performed using Python and TensorFlow in three desktop computers with NVIDIA GeForce GPU boards: GTX 970, GTX 1080, and RTX 2060 SUPER, respectively.2
No pre-processing was applied, except for those steps pre-defined by each CNN architecture, which is basically resizing the image to the CNN input size and normalizing the input range. In all tested scenarios, each CNN had its weights initially set to those pre-trained on the Imagenet dataset (Russakovsky et al., 2015), which has millions of images and hundreds of classes. This dataset is frequently used in transfer learning scenarios.
The training phase was conducted using the Adam optimizer (Kingma & Ba, 2014). The learning rate was set to 10−5 for the original CNN layers and 10−3 for the dense layers proposed in this work. The idea is to allow bigger weight changes in the classification layers, which need to be trained from scratch, while only fine-tuning the CNN layers, taking advantage of the weights previously learned from the Imagenet dataset.
From the training subset, 20% of the images are randomly taken to compose the validation subset, using stratification to keep the same classes proportion. Since the training subset is unbalanced, different class weights were defined for each class: 0.5782 and 3.6960 for negative and positive classes, respectively. These values were calculated based on TensorFlow documentation3 :
| (1) |
where is the class weight, is the amount of examples belonging to class , and is the total amount of examples.
All models were trained for up to epochs. An early stopping criterion was set to interrupt the training phase if the loss on the validation set did not decrease during the last epochs. The final weights are always restored to those adjusted in the epoch that achieved the lowest validation loss.
For each CNN model, the training phase was performed five times with different training/validation splits, generating five instances with different adjusted weights. The same five training/validation splits were used for all models. Each instance was evaluated on the test subset and the following measures were obtained: accuracy (ACC), sensitivity (TPR), precision (PPV), and F1 score. The results are shown in Table 3. Each value is the average of the measures obtained on the five different instances of each model. The standard deviation is also presented. Results of the same evaluation applied to the training and validation subsets are available in Appendix.
Table 3.
Comparison of 21 different CNN models applied to the COVIDx8B dataset. Each model is executed five times. The highest values for each measure are highlighted in bold.
| Model | ACC |
TPR |
PPV |
F1 |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | |
| DenseNet169 | 0.9815 | 0.0056 | 0.9700 | 0.0138 | 0.9930 | 0.0075 | 0.9812 | 0.0058 |
| EfficientNetB2 | 0.9760 | 0.0049 | 0.9600 | 0.0141 | 0.9918 | 0.0051 | 0.9756 | 0.0052 |
| InceptionResNetV2 | 0.9755 | 0.0099 | 0.9590 | 0.0246 | 0.9919 | 0.0051 | 0.9749 | 0.0106 |
| InceptionV3 | 0.9750 | 0.0065 | 0.9520 | 0.0144 | 0.9979 | 0.0041 | 0.9744 | 0.0069 |
| MobileNet | 0.9710 | 0.0060 | 0.9430 | 0.0136 | 0.9990 | 0.0021 | 0.9701 | 0.0064 |
| EfficientNetB0 | 0.9705 | 0.0051 | 0.9510 | 0.0086 | 0.9896 | 0.0033 | 0.9699 | 0.0053 |
| EfficientNetB3 | 0.9700 | 0.0163 | 0.9470 | 0.0337 | 0.9927 | 0.0051 | 0.9690 | 0.0177 |
| DenseNet201 | 0.9695 | 0.0176 | 0.9400 | 0.0342 | 0.9989 | 0.0022 | 0.9683 | 0.0186 |
| ResNet152V2 | 0.9695 | 0.0244 | 0.9420 | 0.0510 | 0.9970 | 0.0040 | 0.9679 | 0.0268 |
| ResNet152 | 0.9660 | 0.0223 | 0.9370 | 0.0443 | 0.9947 | 0.0033 | 0.9644 | 0.0243 |
| DenseNet121 | 0.9630 | 0.0053 | 0.9270 | 0.0103 | 0.9989 | 0.0022 | 0.9616 | 0.0057 |
| Xception | 0.9615 | 0.0077 | 0.9230 | 0.0154 | 1.0000 | 0.0000 | 0.9599 | 0.0083 |
| VGG19 | 0.9580 | 0.0198 | 0.9170 | 0.0385 | 0.9989 | 0.0023 | 0.9558 | 0.0216 |
| EfficientNetB1 | 0.9570 | 0.0224 | 0.9240 | 0.0413 | 0.9892 | 0.0075 | 0.9551 | 0.0242 |
| ResNet50 | 0.9545 | 0.0172 | 0.9090 | 0.0344 | 1.0000 | 0.0000 | 0.9520 | 0.0192 |
| VGG16 | 0.9525 | 0.0123 | 0.9090 | 0.0282 | 0.9958 | 0.0052 | 0.9501 | 0.0138 |
| ResNet101V2 | 0.9530 | 0.0302 | 0.9100 | 0.0643 | 0.9959 | 0.0050 | 0.9497 | 0.0342 |
| MobileNetV2 | 0.9485 | 0.0172 | 0.9030 | 0.0359 | 0.9935 | 0.0019 | 0.9457 | 0.0190 |
| ResNet101 | 0.9410 | 0.0170 | 0.8830 | 0.0333 | 0.9988 | 0.0023 | 0.9370 | 0.0190 |
| ResNet50V2 | 0.9280 | 0.0075 | 0.8590 | 0.0153 | 0.9966 | 0.0046 | 0.9226 | 0.0087 |
| NASNetMobile | 0.8530 | 0.0653 | 0.7090 | 0.1317 | 0.9960 | 0.0034 | 0.8212 | 0.0918 |
| Average | 0.9569 | 0.0162 | 0.9178 | 0.0334 | 0.9957 | 0.0036 | 0.9536 | 0.0187 |
It is worth noticing that most related work only shows the results of a single execution on each tested CNN architecture. This may lead to wrong conclusions as there is always some expected variance on multiple executions of neural networks, which are stochastic by nature.
DenseNet169 achieved the highest accuracy (98.15%), TPR (97. 00%), and F1 score (98.12%) among all the tested models. The highest PPV (100%) was achieved by Xception and ResNet50 models. EfficientNetB2 achieved the second-best accuracy, PPV, and F1 score.
Compared to other recent approaches applied to the same dataset, DenseNet169, EfficientNetB2, and InceptionResNetV2 achieved the best accuracy, TPR, and F1 score, as shown in Table 4. It is worth noticing that EfficientNetB2 has less trainable parameters (8.06 million) than all the other architectures in this comparison, including the Covid-Net CXR-2 (9.2 million), which was specially tailored for the COVIDx8B dataset.
Table 4.
Comparison of the best four models tested in this paper (in italic) with other recently proposed models applied to the COVIDx8B dataset. The highest values for each measure are highlighted in bold. The results obtained by other authors were compiled from the respective cited references.
| Model | ACC | TPR | PPV | F1 | Source |
|---|---|---|---|---|---|
| DenseNet169 | 0.9815 | 0.9700 | 0.9930 | 0.9812 | this paper |
| EfficientNetB2 | 0.9760 | 0.9600 | 0.9918 | 0.9756 | this paper |
| InceptionResNetV2 | 0.9755 | 0.9590 | 0.9919 | 0.9749 | this paper |
| InceptionV3 | 0.9750 | 0.9520 | 0.9979 | 0.9744 | this paper |
| VGG16 (ImageNet) | 0.9750 | 0.9500 | 1.0000 | 0.9744 | Dominik (2021) |
| Covid-Net | 0.9400 | 0.9350 | 1.0000 | 0.9664 | Pavlova et al. (2021) |
| DenseNet121 (ChestXray) | 0.9650 | 0.9350 | 0.9947 | 0.9639 | Dominik (2021) |
| ResNet50V2 (Bit-M) | 0.9650 | 0.9300 | 1.0000 | 0.9637 | Zhao et al. (2021) |
| Covid-Net CXR-2 | 0.9630 | 0.9550 | 0.9700 | 0.9624 | Pavlova et al. (2021) |
| VGG19 (ImageNet) | 0.9625 | 0.9250 | 1.0000 | 0.9610 | Dominik (2021) |
| ResNet-50 (ImageNet) | 0.9575 | 0.9200 | 0.9946 | 0.9558 | Dominik (2021) |
| DenseNet121 (ImageNet) | 0.9575 | 0.9150 | 1.0000 | 0.9556 | Dominik (2021) |
| Xception (ImageNet) | 0.9550 | 0.9100 | 1.0000 | 0.9529 | Dominik (2021) |
| ResNet50V2 (Bit-S) | 0.9480 | 0.8950 | 1.0000 | 0.9446 | Zhao et al. (2021) |
| ResNet50V2 (Random) | 0.9280 | 0.8550 | 1.0000 | 0.9218 | Zhao et al. (2021) |
| ResNet50 | 0.9050 | 0.8850 | 0.9220 | 0.9031 | Pavlova et al. (2021) |
There are some common characteristics among the two best- performing CNN architectures. DenseNet and EfficientNet are newer approaches (2017 and 2019, respectively) than VGG (2015) and ResNet (2016). DenseNet and EfficientNet also focus on architecture efficiency to use less trainable parameters than the earlier approaches. In this case, the strategy used in these newer models was more suitable for these types of CXR images. Unfortunately, many related works compared fewer and/or earlier models only. Therefore, future studies should consider a wider variety of models to verify if this tendency confirms. In particular, from the related work section, only Nigam et al. (2021) and Monshi et al. (2021) explored EfficientNet, but they also reported good results with it, showing this is a promising architecture for CXR images.
Table 5 compares results reported in individual papers described in Section 2, where the authors are motivated to use a setup such their algorithm is the best performing, with the best result found in this paper for an individual CNN architecture, in which there is no motivation to implement optimizations to boost any particular architecture. Despite that, the best result from this paper is still in the top half of the best accuracy ranking. For each paper, the CNN architecture used and the dataset size are provided for reference.
Table 5.
Comparison of different CNN-based models applied to different COVID-19 datasets found in individual papers and the best result by an individual CNN model applied to the COVIDx8B dataset in this paper.
| Reference | Architecture | Dataset Size | Accuracy |
|---|---|---|---|
| Alawad et al. (2021) | VGG16 | 7,329 | 99.82% |
| Karthik et al. (2021) | CSDB | 15,265 | 99.80% |
| Hira et al. (2021) | Se-ResNeXt-50 | 8,830 | 99.32% |
| Jia et al. (2021) | MobileNet | 7,592 | 99.30% |
| Mostafiz et al. (2020) | ResNet50 | 4,809 | 98.50% |
| Narin et al. (2021) | ResNet50 | 7.406 | 98.43% |
| this paper | DenseNet169 | 16,352 | 98.15% |
| Chhikara et al. (2021) | InceptionV3 | 11,244 | 97.70% |
| Chhikara et al. (2021) | InceptionV3 | 14,486 | 97.03% |
| Monshi et al. (2021) | EfficientNetB0 | 15,496 | 95.82% |
| Jia et al. (2021) | MobileNet | 13.975 | 95,00% |
| Ismael and Şengür (2021) | ResNet50 | 380 | 94.70% |
| Heidari et al. (2020) | VGG16 | 8,474 | 94.50% |
| Nigam et al. (2021) | EfficientNetB7 | 16,634 | 93.48% |
| Abbas et al. (2021) | DeTrac | 196 | 93.10% |
| Chhikara et al. (2021) | InceptionV3 | 8,246 | 84.95% |
6. CNN ensembles
This section presents the computer simulations with ensembles of different CNN models and ensembles of multiple instances of the same model. All the ensembles experiments used the output of the last dense layer, just before the softmax activation function. Therefore, for each image, each model will output two continuous values, which can be interpreted as the probability of each class. Then, the output of the ensemble will be the average of its members’ output. The same weights trained for the experiments in Section 5 were used for the experiments in this section.
In the first ensemble experiment, the two models that achieved the best individual F1 score (DenseNet169 and EfficientNetB2) were combined in the first ensemble configuration. The second ensemble configuration adds the third-best model (InceptionResNetV2). The third ensemble configuration adds the fourth-best model (InceptionV3) and so on, with up to seven models. Then, in the last ensemble configuration, all the models are combined. In this first experiment, only one instance of each model composes each ensemble, thus there are five ensembles for each configuration. Table 6 shows the average and standard deviation of the measures obtained for each ensemble configuration.
Table 6.
Ensembles of CNN models applied to the COVIDx8B dataset. Each ensemble configuration is executed five times with different instances of the models. The highest values for each measure are highlighted in bold.
| Models | ACC |
TPR |
PPV |
F1 |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | |
| Top 2 models | 0.9855 | 0.0024 | 0.9730 | 0.0040 | 0.9980 | 0.0025 | 0.9853 | 0.0025 |
| Top 3 models | 0.9885 | 0.0034 | 0.9770 | 0.0068 | 1.0000 | 0.0000 | 0.9884 | 0.0035 |
| Top 4 models | 0.9870 | 0.0019 | 0.9740 | 0.0037 | 1.0000 | 0.0000 | 0.9868 | 0.0019 |
| Top 5 models | 0.9865 | 0.0020 | 0.9730 | 0.0040 | 1.0000 | 0.0000 | 0.9863 | 0.0021 |
| Top 6 models | 0.9880 | 0.0010 | 0.9760 | 0.0020 | 1.0000 | 0.0000 | 0.9879 | 0.0010 |
| Top 7 models | 0.9865 | 0.0025 | 0.9730 | 0.0051 | 1.0000 | 0.0000 | 0.9863 | 0.0026 |
| All models | 0.9775 | 0.0032 | 0.9550 | 0.0063 | 1.0000 | 0.0000 | 0.9770 | 0.0033 |
The best accuracy, TPR, and F1 score were achieved when the three best models were combined (DenseNet169, EfficientNetB2, and InceptionResNetV2). All the ensembles, except for the one with the best two models, achieved a PPV of 100%. Except for the ensemble of all models, all the other ensembles achieved higher accuracy, TPR, and F1 scores than the best individual model.
For the second ensembles experiment, the five instances of each model are combined to form an ensemble. It is expected that five instances, even if they are from the same model, will improve the measures by alleviating the randomness effects of the training.
Table 7 shows the measures obtained with these ensembles for each model. It also shows the gain obtained by the ensemble when compared to the average of the single instances. All the models had gained with the ensembles. The highest measures were obtained by DenseNet169, with an F1 score of 99.24% and an accuracy of 99.25%. This is the same accuracy obtained by Dominik (2021) using an ensemble of multiple models and an optimized threshold. To the best of my knowledge, this is the highest accuracy achieved in this dataset at the time this paper is being written.
Table 7.
Ensembles of CNN models applied to the COVIDx8B dataset. Each ensemble is composed of five instances of the same model, with different training/validation splits. The highest values for each measure and the highest gains in comparison to single instances of each model are highlighted in bold.
| Models | ACC |
TPR |
PPV |
F1 |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | Gain | Mean | Gain | Mean | Gain | Mean | Gain | |
| DenseNet169 | 0.9925 | 1.12% | 0.9850 | 1.55% | 1.0000 | 0.70% | 0.9924 | 1.14% |
| EfficientNetB2 | 0.9850 | 0.92% | 0.9750 | 1.56% | 0.9949 | 0.31% | 0.9848 | 0.94% |
| InceptionResNetV2 | 0.9875 | 1.23% | 0.9750 | 1.67% | 1.0000 | 0.82% | 0.9873 | 1.27% |
| InceptionV3 | 0.9800 | 0.51% | 0.9600 | 0.84% | 1.0000 | 0.21% | 0.9796 | 0.53% |
| MobileNet | 0.9825 | 1.18% | 0.9650 | 2.33% | 1.0000 | 0.10% | 0.9822 | 1.25% |
| EfficientNetB0 | 0.9750 | 0.46% | 0.9600 | 0.95% | 0.9897 | 0.01% | 0.9746 | 0.48% |
| EfficientNetB3 | 0.9850 | 1.55% | 0.9750 | 2.96% | 0.9949 | 0.22% | 0.9848 | 1.63% |
| DenseNet201 | 0.9825 | 1.34% | 0.9650 | 2.66% | 1.0000 | 0.11% | 0.9822 | 1.44% |
| ResNet152V2 | 0.9900 | 2.11% | 0.9800 | 4.03% | 1.0000 | 0.30% | 0.9899 | 2.27% |
| ResNet152 | 0.9800 | 1.45% | 0.9650 | 2.99% | 0.9948 | 0.01% | 0.9797 | 1.59% |
| DenseNet121 | 0.9725 | 0.99% | 0.9450 | 1.94% | 1.0000 | 0.11% | 0.9717 | 1.05% |
| Xception | 0.9625 | 0.10% | 0.9250 | 0.22% | 1.0000 | 0.00% | 0.9610 | 0.11% |
| VGG19 | 0.9700 | 1.25% | 0.9400 | 2.51% | 1.0000 | 0.11% | 0.9691 | 1.39% |
| EfficientNetB1 | 0.9725 | 1.62% | 0.9500 | 2.81% | 0.9948 | 0.57% | 0.9719 | 1.76% |
| ResNet50 | 0.9650 | 1.10% | 0.9300 | 2.31% | 1.0000 | 0.00% | 0.9637 | 1.23% |
| VGG16 | 0.9550 | 0.26% | 0.9100 | 0.11% | 1.0000 | 0.42% | 0.9529 | 0.29% |
| ResNet101V2 | 0.9650 | 1.26% | 0.9300 | 2.20% | 1.0000 | 0.41% | 0.9637 | 1.47% |
| MobileNetV2 | 0.9650 | 1.74% | 0.9350 | 3.54% | 0.9947 | 0.12% | 0.9639 | 1.92% |
| ResNet101 | 0.9575 | 1.75% | 0.9150 | 3.62% | 1.0000 | 0.12% | 0.9556 | 1.99% |
| ResNet50V2 | 0.9350 | 0.75% | 0.8700 | 1.28% | 1.0000 | 0.34% | 0.9305 | 0.86% |
| NASNetMobile | 0.8750 | 2.58% | 0.7500 | 5.78% | 1.0000 | 0.40% | 0.8571 | 4.37% |
| Average | 0.9683 | 1.20% | 0.9383 | 2.28% | 0.9983 | 0.26% | 0.9666 | 1.38% |
For the third and last ensembles experiment, the first experiment is repeated, but now using all the five instances of each model in the ensemble. Table 8 shows the measures obtained with each ensemble and the gain obtained by these ensembles when compared to the ensembles which used only a single instance of each model. In this case, there were only small differences and some of them were negative. Therefore, the best ensemble overall is still the one with multiple instances of DenseNet169.
Table 8.
Ensembles of CNN models applied to the COVIDx8B dataset. Each ensemble configuration has five instances of each participant model. The highest values for each measure and the highest gains in comparison with the ensembles of single instances for each model are highlighted in bold.
| Models | ACC |
TPR |
PPV |
F1 |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | Gain | Mean | Gain | Mean | Gain | Mean | Gain | |
| Top 2 models | 0.9850 | −0.05% | 0.9700 | −0.31% | 1.0000 | 0.20% | 0.9848 | −0.05% |
| Top 3 models | 0.9875 | −0.10% | 0.9750 | −0.20% | 1.0000 | 0.00% | 0.9873 | −0.11% |
| Top 4 models | 0.9875 | 0.05% | 0.9750 | 0.10% | 1.0000 | 0.00% | 0.9873 | 0.05% |
| Top 5 models | 0.9875 | 0.10% | 0.9750 | 0.21% | 1.0000 | 0.00% | 0.9873 | 0.10% |
| Top 6 models | 0.9875 | −0.05% | 0.9750 | −0.10% | 1.0000 | 0.00% | 0.9873 | −0.06% |
| Top 7 models | 0.9875 | 0.10% | 0.9750 | 0.21% | 1.0000 | 0.00% | 0.9873 | 0.10% |
| All models | 0.9775 | 0.00% | 0.9550 | 0.00% | 1.0000 | 0.00% | 0.9770 | 0.00% |
| Average | 0.9857 | 0.01% | 0.9714 | −0.01% | 1.0000 | 0.03% | 0.9855 | 0.00% |
7. Conclusions
In this paper, different CNN architectures are applied to the detection of COVID-19 on CXR images. The comparison was performed using the COVIDx8B, a large and heterogeneous COVID-19 CXR images dataset, which is composed of six open-source CXR datasets. The training was repeated five times for each model, with different training and validation splits to get more reliable results, while most related works tested fewer models and performed only a single execution for each one.
CNN ensembles were also explored in this work, combining both different models and multiple instances of the same model. DenseNet169 achieved the best results regarding the accuracy and the F1 score, both as a single instance and with an ensemble of five instances. The classification accuracies were 98.15% and 99.25% for the single instance and the ensemble, respectively, while the F1 scores were 98.12% and 99.24%, also respectively. These results are better than those achieved in recent works where the same dataset was used.
The simulations performed for this paper add more evidence of the efficacy of CNNs in the detection of COVID-19 on CXR images, which is very important to assist in quick diagnostics and to avoid the spread of the disease. Moreover, these experiments may also guide future research as they tested a large amount of CNN architectures and identified which of them produces the best results for this particular task.
CRediT authorship contribution statement
Fabricio Aparecido Breve: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
The code (and data) in this article has been certified as Reproducible by Code Ocean: (https://codeocean.com/). More information on the Reproducibility Badge Initiative is available at https://www.elsevier.com/physical-sciences-and-engineering/computer-science/journals.
Scripts to build the COVIDx8B dataset are available at https://github.com/lindawangg/COVID-Net/blob/master/docs/COVIDx.md.
The source code is available at https://github.com/fbreve/covid-cnn.
TensorFlow documentation on class weights is available at https://www.tensorflow.org/tutorials/structured_data/imbalanced_data.
Appendix. Classification results in training, validation, and test subsets
This appendix shows the classification results obtained by the individual CNN classifiers, with the same weights learned in Section 5 experiment when applied to the training, validation, and test subsets individually. All results are the average of the five executions, with different training/validation splits. Tables A.9, A.10, A.11, A.12 show the results of accuracy (ACC), sensitivity (TPR), precision (PPV), and F1 score, respectively.
These results show that accuracy and sensitivity are a little higher in the training and validation subsets than in the test subset for all the architectures. On the other hand, precision is higher on the test subset for all the architectures. Finally, the F1 score shows the closest results in the training and test subsets. Some architectures achieved a higher F1 score in the training subset, while others achieved a higher F1 score in the test subset. This behavior may be related to the class imbalance, even though class weights were used to minimize its effects. Future work may focus on other techniques to handle class imbalance, like data augmentation. Moreover, future benchmark datasets will probably minimize imbalance as more COVID-19 CXR images become available.
Table A.9.
Classification accuracy (ACC) achieved by the CNN architectures when applied to the train, validation, and test subsets individually.
| Dataset/Subset | Train | Validation | Test |
|---|---|---|---|
| DenseNet169 | 0.9951 | 0.9794 | 0.9815 |
| EfficientNetB2 | 0.9936 | 0.9793 | 0.9760 |
| InceptionResNetV2 | 0.9835 | 0.9681 | 0.9755 |
| InceptionV3 | 0.9960 | 0.9784 | 0.9750 |
| MobileNet | 0.9936 | 0.9788 | 0.9710 |
| EfficientNetB0 | 0.9894 | 0.9761 | 0.9705 |
| EfficientNetB3 | 0.9948 | 0.9803 | 0.9700 |
| ResNet152V2 | 0.9945 | 0.9757 | 0.9695 |
| DenseNet201 | 0.9971 | 0.9816 | 0.9695 |
| ResNet152 | 0.9923 | 0.9783 | 0.9660 |
| DenseNet121 | 0.9962 | 0.9806 | 0.9630 |
| Xception | 0.9909 | 0.9777 | 0.9615 |
| VGG19 | 0.9922 | 0.9804 | 0.9580 |
| EfficientNetB1 | 0.9802 | 0.9697 | 0.9570 |
| ResNet50 | 0.9955 | 0.9806 | 0.9545 |
| ResNet101V2 | 0.9909 | 0.9707 | 0.9530 |
| VGG16 | 0.9913 | 0.9772 | 0.9525 |
| MobileNetV2 | 0.9987 | 0.9808 | 0.9485 |
| ResNet101 | 0.9923 | 0.9803 | 0.9410 |
| ResNet50V2 | 0.9859 | 0.9662 | 0.9280 |
| NASNetMobile | 0.9798 | 0.9660 | 0.8530 |
Table A.10.
Classification sensitivity (TPR) achieved by the CNN architectures when applied to the train, validation, and test subsets individually.
| Dataset/Subset | Train | Validation | Test |
|---|---|---|---|
| DenseNet169 | 0.9987 | 0.9611 | 0.9700 |
| EfficientNetB2 | 0.9936 | 0.9662 | 0.9600 |
| InceptionResNetV2 | 0.9830 | 0.9491 | 0.9590 |
| InceptionV3 | 0.9965 | 0.9458 | 0.9520 |
| EfficientNetB0 | 0.9760 | 0.9361 | 0.9510 |
| EfficientNetB3 | 0.9935 | 0.9662 | 0.9470 |
| MobileNet | 0.9957 | 0.9440 | 0.9430 |
| ResNet152V2 | 0.9928 | 0.9375 | 0.9420 |
| DenseNet201 | 0.9964 | 0.9454 | 0.9400 |
| ResNet152 | 0.9854 | 0.9509 | 0.9370 |
| DenseNet121 | 0.9973 | 0.9421 | 0.9270 |
| EfficientNetB1 | 0.9750 | 0.9505 | 0.9240 |
| Xception | 0.9847 | 0.9333 | 0.9230 |
| VGG19 | 0.9874 | 0.9338 | 0.9170 |
| ResNet101V2 | 0.9882 | 0.9278 | 0.9100 |
| VGG16 | 0.9915 | 0.9324 | 0.9090 |
| ResNet50 | 0.9918 | 0.9338 | 0.9090 |
| MobileNetV2 | 0.9933 | 0.9241 | 0.9030 |
| ResNet101 | 0.9701 | 0.9162 | 0.8830 |
| ResNet50V2 | 0.9758 | 0.8921 | 0.8590 |
| NASNetMobile | 0.8874 | 0.8255 | 0.7090 |
Table A.11.
Classification precision (PPV) achieved by the CNN architectures when applied to the train, validation, and test subsets individually.
| Dataset/Subset | Train | Validation | Test |
|---|---|---|---|
| Xception | 0.9502 | 0.9057 | 1.0000 |
| ResNet50 | 0.9759 | 0.9242 | 1.0000 |
| MobileNet | 0.9587 | 0.9040 | 0.9990 |
| VGG19 | 0.9566 | 0.9228 | 0.9989 |
| DenseNet121 | 0.9754 | 0.9169 | 0.9989 |
| DenseNet201 | 0.9822 | 0.9215 | 0.9989 |
| ResNet101 | 0.9727 | 0.9366 | 0.9988 |
| InceptionV3 | 0.9748 | 0.8998 | 0.9979 |
| ResNet152V2 | 0.9682 | 0.8904 | 0.9970 |
| ResNet50V2 | 0.9244 | 0.8630 | 0.9966 |
| NASNetMobile | 0.9602 | 0.9159 | 0.9960 |
| ResNet101V2 | 0.9501 | 0.8701 | 0.9959 |
| VGG16 | 0.9474 | 0.9044 | 0.9958 |
| ResNet152 | 0.9598 | 0.8964 | 0.9947 |
| MobileNetV2 | 0.9973 | 0.9337 | 0.9935 |
| DenseNet169 | 0.9672 | 0.8946 | 0.9930 |
| EfficientNetB3 | 0.9702 | 0.8977 | 0.9927 |
| InceptionResNetV2 | 0.9077 | 0.8408 | 0.9919 |
| EfficientNetB2 | 0.9631 | 0.8925 | 0.9918 |
| EfficientNetB0 | 0.9486 | 0.8928 | 0.9896 |
| EfficientNetB1 | 0.8924 | 0.8470 | 0.9892 |
Table A.12.
Classification F1 score achieved by the CNN architectures when applied to the train, validation, and test subsets individually.
| Dataset/Subset | Train | Validation | Test |
|---|---|---|---|
| DenseNet169 | 0.9825 | 0.9266 | 0.9812 |
| EfficientNetB2 | 0.9774 | 0.9273 | 0.9756 |
| InceptionResNetV2 | 0.9428 | 0.8905 | 0.9749 |
| InceptionV3 | 0.9855 | 0.9221 | 0.9744 |
| MobileNet | 0.9768 | 0.9233 | 0.9701 |
| EfficientNetB0 | 0.9619 | 0.9139 | 0.9699 |
| EfficientNetB3 | 0.9815 | 0.9304 | 0.9690 |
| DenseNet201 | 0.9892 | 0.9331 | 0.9683 |
| ResNet152V2 | 0.9801 | 0.9128 | 0.9679 |
| ResNet152 | 0.9722 | 0.9226 | 0.9644 |
| DenseNet121 | 0.9862 | 0.9292 | 0.9616 |
| Xception | 0.9670 | 0.9190 | 0.9599 |
| VGG19 | 0.9716 | 0.9281 | 0.9558 |
| EfficientNetB1 | 0.9312 | 0.8954 | 0.9551 |
| ResNet50 | 0.9837 | 0.9287 | 0.9520 |
| VGG16 | 0.9688 | 0.9173 | 0.9501 |
| ResNet101V2 | 0.9679 | 0.8966 | 0.9497 |
| MobileNetV2 | 0.9953 | 0.9287 | 0.9457 |
| ResNet101 | 0.9714 | 0.9262 | 0.9370 |
| ResNet50V2 | 0.9493 | 0.8773 | 0.9226 |
| NASNetMobile | 0.9209 | 0.8675 | 0.8212 |
References
- Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawad W., Alburaidi B., Alzahrani A., Alflaj F. A comparative study of stand-alone and hybrid CNN models for COVID-19 detection. International Journal of Advanced Computer Science and Applications. 2021;12(6) doi: 10.14569/IJACSA.2021.01206102. doi: 10.14569/IJACSA.2021.01206102. URL: [DOI] [Google Scholar]
- Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., Del Campo R., Ciapponi A., et al. False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhikara P., Gupta P., Singh P., Bhatia T. A deep transfer learning based model for automatic detection of COVID-19 from chest X-rays. Turkish Journal Electrical Engineering and Computer Sciences. 2021;29(SI-1):2663–2679. [Google Scholar]
- Chollet F. 2017 IEEE conference on computer vision and pattern recognition. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1800–1807. [DOI] [Google Scholar]
- Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., et al. Can AI help in screening viral and COVID-19 Pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- Chung A. 2020. Actualmed COVID-19 chest X-ray dataset initiative. GitHub Repository, GitHub, https://github.com/agchung/Actualmed-COVID-chestxray-dataset. [Google Scholar]
- Chung A. 2020. Figure1-COVID-chestxray-dataset. GitHub Repository, GitHub, https://github.com/agchung/Figure1-COVID-chestxray-dataset. [Google Scholar]
- Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. COVID-19 image data collection: Prospective predictions are the future. arXiv:2006.11988, URL: https://github.com/ieee8023/covid-chestxray-dataset. [Google Scholar]
- Dominik C. 2021. Detection of COVID-19 in X-ray images using neural networks. Czech Technical University in Prague, Faculty of Information Technology. [Google Scholar]
- Feng H., Liu Y., Lv M., Zhong J. A case report of COVID-19 with false negative RT-PCR test: Necessity of chest CT. Japanese Journal of Radiology. 2020;38(5):409–410. doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Bengio Y., Courville A. MIT Press; 2016. Deep learning. [Google Scholar]
- He, K., Zhang, X., Ren, S., & Sun, J. (2016a). Deep residual learning for image recognition. In The IEEE conference on computer vision and pattern recognition (pp. 770–778).
- He K., Zhang X., Ren S., Sun J. In: Computer vision – ECCV 2016. Leibe B., Matas J., Sebe N., Welling M., editors. Springer International Publishing; Cham: 2016. Identity mappings in deep residual networks; pp. 630–645. [Google Scholar]
- Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. International Journal of Medical Informatics. 2020;144 doi: 10.1016/j.ijmedinf.2020.104284. URL: https://www.sciencedirect.com/science/article/pii/S138650562030959X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira S., Bai A., Hira S. An automatic approach based on CNN architecture to detect Covid-19 disease from chest X-ray images. Applied Intelligence: The International Journal of Artificial Intelligence, Neural Networks, and Complex Problem-Solving Technologies. 2021;51(5):2864–2889. doi: 10.1007/s10489-020-02010-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., et al. 2017. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861. [Google Scholar]
- Huang, G., Liu, Z., van der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition.
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. URL: https://www.sciencedirect.com/science/article/pii/S0957417420308198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Lam H.-K., Xu Y. Classification of COVID-19 chest X-Ray and CT images using a type of dynamic CNN modification method. Computers in Biology and Medicine. 2021;134 doi: 10.1016/j.compbiomed.2021.104425. URL: https://www.sciencedirect.com/science/article/pii/S0010482521002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik R., Menaka R., M. H. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Applied Soft Computing. 2021;99 doi: 10.1016/j.asoc.2020.106744. URL: https://www.sciencedirect.com/science/article/pii/S1568494620306827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Mehran M.T., Haq Z.U., Ullah Z., Naqvi S.R., Ihsan M., et al. Applications of artificial intelligence in COVID-19 pandemic: A comprehensive review. Expert Systems with Applications. 2021;185 doi: 10.1016/j.eswa.2021.115695. URL: https://www.sciencedirect.com/science/article/pii/S0957417421010794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. In: Advances in neural information processing systems, vol. 25. Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. Curran Associates, Inc.; 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. URL: http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Long D.R., Gombar S., Hogan C.A., Greninger A.L., O’Reilly-Shah V., Bryson-Cahn C., et al. Occurrence and timing of subsequent severe acute respiratory syndrome coronavirus 2 reverse-transcription polymerase chain reaction positivity among initially negative patients. Clinical Infectious Diseases. 2020;72(2):323–326. doi: 10.1093/cid/ciaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad Shorfuzzaman M.M. On the detection of COVID-19 from chest X-Ray images using CNN-based transfer learning. Computers, Materials & Continua. 2020;64(3):1359–1381. doi: 10.32604/cmc.2020.011326. URL: http://www.techscience.com/cmc/v64n3/39434. [DOI] [Google Scholar]
- Monshi M.M.A., Poon J., Chung V., Monshi F.M. CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Computers in Biology and Medicine. 2021;133 doi: 10.1016/j.compbiomed.2021.104375. URL: https://www.sciencedirect.com/science/article/pii/S0010482521001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafiz R., Uddin M.S., Reza M.M., Rahman M.M., et al. COVID-19 detection in chest X-ray through random forest classifier using a hybridization of deep CNN and DWT optimized features. Journal of King Saud University-Computer and Information Sciences. 2020 doi: 10.1016/j.jksuci.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Analysis and Applications. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam B., Nigam A., Jain R., Dodia S., Arora N., Annappa B. COVID-19: Automatic detection from X-ray images by utilizing deep learning methods. Expert Systems with Applications. 2021;176 doi: 10.1016/j.eswa.2021.114883. URL: https://www.sciencedirect.com/science/article/pii/S0957417421003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquab, M., Bottou, L., Laptev, I., & Sivic, J. (2014). Learning and transferring mid-level image representations using convolutional neural networks. In The IEEE conference on computer vision and pattern recognition.
- Pavlova M., Terhljan N., Chung A.G., Zhao A., Surana S., Aboutalebi H., et al. 2021. COVID-net CXR-2: An enhanced deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv:2105.06640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Computers in Biology and Medicine. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. URL: https://www.sciencedirect.com/science/article/pii/S001048252100113X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., et al. ImageNet Large scale visual recognition challenge. International Journal of Computer Vision (IJCV) 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., & Chen, L.-C. (2018). MobileNetV2: Inverted residuals and linear bottlenecks. In The IEEE conference on computer vision and pattern recognition (pp. 4510–4520).
- Schmidhuber J. Deep learning in neural networks: An overview. Neural Networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. Computational and Biological Learning Society; 2015. Very deep convolutional networks for large-scale image recognition; pp. 1–14. [Google Scholar]
- Szegedy, C., Ioffe, S., Vanhoucke, V., & Alemi, A. A. (2017). Inception-v4, inception-resnet and the impact of residual connections on learning. In Thirty-First AAAI conference on artificial intelligence.
- Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2016). Rethinking the inception architecture for computer vision. In The IEEE conference on computer vision and pattern recognition (pp. 2818–2826).
- Tan M., Le Q. In: Proceedings of the 36th international conference on machine learning. Chaudhuri K., Salakhutdinov R., editors. vol. 97. PMLR; 2019. EfficientNet: Rethinking model scaling for convolutional neural networks; pp. 6105–6114. (Proceedings of machine learning research). URL: https://proceedings.mlr.press/v97/tan19a.html. [Google Scholar]
- Tsai E., Simpson S., Lungren M., Hershman M., Roshkovan L., Colak E., et al. 2021. ‘Data from medical imaging data resource center (MIDRC)-rsna international covid radiology database (RICORD) release 1C—Chest X-ray, covid+(MIDRC-RICORD-1C) The Cancer Imaging Archive. [DOI] [Google Scholar]
- Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., & Summers, R. M. (2017). Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2097–2106).
- World Health Organization X. 2022. WHO coronavirus (COVID-19) dashboard. World Health Organization, URL: https://covid19.who.int/. (Accessed: 28 February 2022) [Google Scholar]
- Zhao W., Jiang W., Qiu X. Fine-tuning convolutional neural networks for COVID-19 detection from chest X-ray images. Diagnostics. 2021;11(10):1887. doi: 10.3390/diagnostics11101887. doi: 10.3390/diagnostics11101887. URL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoph, B., Vasudevan, V., Shlens, J., & Le, Q. V. (2018). Learning transferable architectures for scalable image recognition. In The IEEE conference on computer vision and pattern recognition (pp. 8697–8710).