Abstract
This paper highlights the extraordinarily rapid spread of SARS-CoV-2 loads in wastewater that during the Omicron wave in December 2021–February 2022, compared with the profiles acquired in 2020–21 with 410 samples from two wastewater treatment plants (Trento+suburbs, 132,500 inhabitants).
Monitoring of SARS-CoV-2 in wastewater focused on: (i) 3 samplings/week and analysis, (ii) normalization to calculate genomic units (GU) inh−1 d−1; (iii) calculation of a 7-day moving average to smooth daily fluctuations; (iv) comparison with the ‘current active cases’/100,000 inh progressively affected by the mass vaccination.
The time profiles of SARS-CoV-2 in wastewater matched the waves of active cases. In February–April 2021, a viral load of 1.0E+07 GU inh−1 d corresponded to 700 active cases/100,000 inh. In July–September 2021, although the low current active cases, sewage revealed an appreciable SARS-CoV-2 circulation (in this period 2.2E+07 GU inh−1 d−1 corresponded to 90 active cases/100,000 inh). Omicron was not detected in wastewater until mid-December 2021. The Omicron spread caused a 5–6 fold increase of the viral load in two weeks, reaching the highest peak (2.0–2.2E+08 GU inh−1 d−1 and 4500 active cases/100,000 inh) during the pandemic. In this period, wastewater surveillance anticipated epidemiological data by about 6 days. In winter 2021–22, despite the 4–7 times higher viral loads in wastewater, hospitalizations were 4 times lower than in winter 2020–21 due to the vaccination coverage >80%.
The Omicron wave demonstrated that SARS-CoV-2 monitoring of wastewater anticipated epidemiological data, confirming its importance in long-term surveillance.
Keywords: COVID-19, SARS-CoV-2, Wastewater-based epidemiology, Omicron variant, Sewerage, Viral concentration
Graphical abstract
1. Introduction
SARS-CoV-2, responsible for the COVID-19 pandemic, had caused 513 million infections as of April 30, 2022 (Worldometer, 2022). This pandemic has surged in successive waves, but up to now, no mid-term strategy has been found worldwide to prevent recurrent waves of infections due to variants (Cacciapaglia et al., 2021). Many actions have been taken to prevent the spread and severity of the COVID-19 disease, such as lockdowns, face masks, smart-working, isolation and quarantine, and vaccination. However, these measures have been partially undermined by emerging variants. In December 2021, a remarkable increase of infections occurred all around the world: values in the world increased from 240,000 new cases/week on December 13, 2021, to 5,800,000 cases/week on January 3, 2022 (WHO, 2022). This increase occurred with an unprecedented rate of infection (Soriano et al., 2022).
A new SARS-CoV-2 variant (B.1.1.529), named Omicron, was reported for the first time in Botswana and South Africa on November 24, 2021 (Tong et al., 2021) and designated as the fifth Variant of Concern (VoC) on November 26, 2021 (WHO).
The Omicron variant has over 30 mutations in the spike protein (Lupala et al., 2022, Ferré et al., 2022, Lee et al., 2021) that enhanced its transmissibility, infectivity, and immune evasion (Tong et al., 2021, Khan et al., 2022). The Omicron variant has spread rapidly across the globe and has been detected in more than 100 countries in just a month, giving rise to a new wave of COVID-19 with the number of daily cases soaring to all-time records. Then, other new sub-lineages of the Omicron VoC have appeared, including Omicron 2 (lineage BA.2), that in February 2022 became the predominant variant in South Africa, other African countries, and Denmark (Statens Serum Institut, 2022). The SARS-CoV-2 Omicron variant was able to replace the Delta variant in most of the European Union/European Economic Area (EU/EEA) countries (ECDC, 2022).
In December 2021, the Omicron variant was confirmed in sewage samples in various countries (USA, Canada, UK, and Italy, among others). Some retrospective investigations in Nova Scotia in Canada (Mundie, 2022), France (Ferré et al., 2022), and the Netherlands (BBC News, 2021) detected the Omicron variant in wastewater samples collected in mid-November 2021, that is, before the variant was first reported in South Africa. This suggests that the new variant was circulating earlier than thought.
The symptoms of the Omicron variant appear less severe than those of the previous variants, especially in vaccinated persons. Some asymptomatic people may not be aware of their infection, or they may have decided not to get tested officially. Therefore, the COVID-19 positive people in a community could be far more numerous than those reported. Conversely, the virus is discharged into wastewater through the faeces or other fluids of infected individuals, symptomatic or not. In this context, wastewater-based epidemiology (WBE) is a valuable means with which to acquire epidemiological information and to understand how the virus is spreading in a population as a whole, i.e. including the less symptomatic or asymptomatic cases (Bivins et al., 2020, Bar-Or et al., 2021, Bonanno Ferraro et al., 2021).
The faeces of COVID-19 infected individuals may contain the genetic materials of the virus already after the first few days of infection and even before the onset of the disease (Foladori et al., 2020). Stool samples can remain positive for weeks after the initial infection. The concentration of SARS-CoV-2 in stool can vary widely depending on the course of the disease: the range of 3.7–7.6 log10 of genomic units (GU) per gram of stool was referred to by Foladori et al. (2020); a range of 0.24–6.5 log10 copies/g faeces (median of 3.4 log) was reported by Miura et al. (2021); while Schmitz et al. (2021) calculated the mean shedding rates averaged from wastewater samples (included asymptomatic cases) and found 7.30 ± 0.67 log10 GU/g faeces.
Since infected people shed the virus differently, it is difficult to convert the wastewater data into a corresponding number of positive cases, and complex mathematical models are needed (McMahan et al., 2021). Moreover, viral concentrations acquired for WBE can be influenced by many environmental and climatic factors (season, temperature of water or air, stormwater, etc.), configuration of the sewer network (combined or separate, length, hydraulic retention time, etc.), operational practices (sampling frequency, sample volume and storage, etc.) or population characteristics (density, habits, sanitation level, fluctuations, etc.). These factors are not yet fully considered in WBE, but they can significantly affect the estimation of positive cases in a sewershed, as well documented by Kumar et al. (2022).
Despite this limitation, the numerous applications of WBE have evidenced the benefit of highlighting SARS-CoV-2 trends and peaks with an anticipation of 1–2 weeks at city scale (Kumar et al., 2021), which helps to provide early warnings at low costs (Kumar et al., 2022).
As documented during the previous waves (Alpha, Beta, Gamma and Delta variants), wastewater signals may appear 12–16 days before COVID-19 cases (Randazzo et al., 2020) and 19–21 days earlier than an increase in the number of newly hospitalized patients (Saguti et al., 2021), but the length of this anticipation has not yet been evaluated with the Omicron variant.
With regard to variant surveillance, genomic sequencing is very expensive and time-consuming, thus limiting the number of clinical specimens (nasal or salivary swabs) that can be analysed. Also in this context, wastewater provides an overview of the variants circulating in the entire community, especially when a new variant emerges, considering that a large population can be investigated as a whole with a few samples. Wastewater surveillance of SARS-CoV-2 and its variants in large urban centres has been strongly endorsed in the EU by the EU Commission Recommendation 2021/472 of 17 March 2021 (EU, 2021), which suggests a sampling frequency of at least twice a week.
This paper highlights the evolution of SARS-CoV-2 loads in the wastewater of a medium-sized municipality (Trento, North-East Italy) during the extraordinarily rapid spread of the Omicron variant, which caused the highest concentration of SARS-CoV-2 in wastewater ever seen during the pandemic. In particular, the research reported in the paper focused on: (i) a frequency of wastewater sampling of three times per week, and the need for smoothing; (ii) a long-term monitoring of 17 months, from December 2020 to April 2022, which included 410 wastewater samples; (iii) comparison of the Omicron wave in wastewater with the previous waves from December 2020 onwards and association with diagnostic and clinical data recorded by health authorities; (iv) evaluation of the anticipation of signals in wastewater during the Omicron wave; (v) SARS-CoV-2 loads in wastewater in periods before and after mass vaccinations in the population.
This work adds insights into the SARS-CoV-2 loads during the Omicron wave, given that data on the Omicron variant in municipal wastewater and its correlation with epidemiological data are still scarce.
2. Materials and methods
2.1. Wastewater treatment plants and sampling plan
The wastewater surveillance of SARS-CoV-2 was applied to a medium-sized municipality (Trento, Italy) with 118,100 inhabitants (henceforth ‘inh.’) and served by two WWTPs (WWTP and WWTP; Figure S1 in Supplementary Material). To be noted is that the boundaries of the administrative municipality may not exactly match the WWTPs sewershed. In the present case, the WWTPs are also connected with the sewers of some small peripheral municipalities with 14,400 inh. In total, the population served is 132,500 inh. divided into two WWTPs: (1) WWTP, which comprises 85,200 inh. and has a design capacity of 120,000 population equivalent (PE); (2) WWTP, which comprises 47,300 inh. and has a design capacity of 110,000 PE. The difference between the number of inhabitants and PE used in the design of the plant is due to the need to treat other inflows such as industrial wastewater, tourism fluctuations, stormwater, or hauled wastewater. The design and operational parameters of WWTP and WWTP are set out in Table S1 in Supplementary Material.
Raw wastewater samples were collected and analysed for 17 months, from December 1, 2020, to April 15, 2022 (sampling is still ongoing), for a total of 208 samples for WWTP and 202 for WWTP. The first two months, December 2020 and January 2021, were a run-in period with changes in the analytical methods, and wastewater was collected once or twice a week. From February 2021 to April 2022 (more than one year), three samplings were performed per week, resulting in more data, a more accurate calculation of the 7-day moving average, and a better understanding of trends.
2.2. Raw wastewater sampling and storage
The points of collection of the raw wastewater samples were the inlet of the two WWTPs immediately after sieving and degritting (where coarse materials and sand are removed), but before the primary settling to avoid the removal of settleable solids. Sampling was performed using refrigerated (4 °C) autosamplers and forming 24-h composite samples obtained by combining 96 aliquots of equal volume per day (aliquots collected every 15 min). The travel time of wastewater from the furthest area in the sewershed along the collection system was approximately 2–3 h so that: (i) the day of origin of wastewater was the same as that of the formation of the 24-h composite sample; (ii) the natural decay of the viral genetic material in the sewer could only take place for a few hours, i.e. a time that did not significantly reduce the viral loads at the entrance of the plants (Ahmed et al., 2020, Foladori et al., 2022).
Then, each composite sample was carefully mixed and collected in 250 mL aliquots before its refrigerated transport to the lab and storage at 4 °C until further analysis. Samples were not frozen at any time, except for 7 samples (6 from WWTP and 1 from WWTP) due to unforeseen logistical problems. From October 1, 2021, in accordance with the European Recommendation (EU, 2021), the samples were analysed within 48 h of sampling. In particular, in that period, the analysis of 66% of the samples began within three hours from the time of delivery to the laboratory.
For each sampling day, the daily flow rate (expressed as m3/d) was recorded.
2.3. SARS-CoV-2 detection and quantification by RT-qPCR
Initially, from December 2020 to January 2021, the analysis of SARS-CoV-2 was based on the biphasic separation (PEG–dextran) recommended by WHO for the environmental surveillance of poliovirus (WHO, 2003) according to La Rosa et al. (2020). From February 2021 to April 2022, another analytical protocol, using PEG precipitation combined with centrifugation, was adopted in agreement with the National Institute of Health (Italy) and shared with the Italian network involved in the national programme for wastewater surveillance (SARI project; La Rosa et al., 2021a). This latter method enables detection of SARS-CoV-2 even at low concentrations (102–103 genomic unit (GU)/L) in a complex matrix like raw wastewater. The procedure applied is summarized in the following paragraphs.
Enrichment phase and nucleic acid extraction - A volume of 45 mL of sample underwent the enrichment phase according to Wu et al. (2020), with the minor changes listed below. Firstly, samples were pre-treated in a 56 °C water bath for 30 min to inactivate the virus and guarantee the safety of the operators. After the sample was cooled at room temperature, 100 L of process control virus (Murine Norovirus, MNV-1, viral stock provided by the Experimental Zooprophylactic Institute of Sicily, Italy) were added. Solids were removed by centrifugation at 4500g. According to this protocol, there is no need to incubate in the precipitation step since 40 mL of supernatant are transferred to new tubes with the addition of 4 g of PEG8000 and 0.9 g of NaCl, mixed at a cold temperature for 15 min and, once the PEG is dissolved, centrifuged again at 12,000g for 2 h. Other authors have successfully used the same protocol for SARS-CoV-2 concentration (Zdenkova et al., 2022, Pellegrinelli et al., 2022). After centrifugation, the supernatant was separated, and the pellet was resuspended for the extraction of nucleic acids with an aliquot of 2 mL of lysis buffer containing guanidine thiocyanate (bioMerieux), followed by 20 min incubation for viral lysis. Then 50 L of magnetic silica beads were added to the sample and incubated for 10 min at room temperature to allow the adhesion of RNA to the beads. A semi-automatic extraction platform (eGeneUp, bioMerieux) was used in accordance with the manufacturer’s instructions, to elute nucleic acids in a final volume of 100 L. The extracted RNA was then cleansed of matrix inhibitors with the commercial OneStep PCR Inhibitor Removal Kit (Zymo Research, CA, USA).
Real-time RT-qPCR - Amplification was carried out by a real-time one-step qPCR reaction for the detection of a target gene of the virus, the orf1b (nsp14) with an assay described in La Rosa et al. (2021b). The instrumental platform used was the Applied Biosystems™ 7500 (ThermoFisher Scientific), while used to prepare the mixture for RT-qPCR were AgPath-ID One-Step RT-PCR (Life Technologies), primer 2297-CoV-2-F, primer 2298-CoV-2-R and probe 2299-CoV-2-P. The amplification conditions were: reverse transcription for 30 min at 50 °C, inactivation for 5 min at 95 °C, and 45 cycles of 15 s at 95 °C and 30 s at 60 °C. Finally, the SARS-CoV-2 concentration, expressed in GU/mL, was obtained from the intersection of the threshold cycle value (Ct) with the calibration curve produced by the serial dilutions of the dsDNA standard containing the sequence target of the virus and provided by the National Institute of Health (Italy).
All samples were analysed in duplicate in qPCR. Analyses were considered acceptable if the standard curves for the quantification of SARS-CoV-2 provided a slope close to −3.32 (minimum −3.1, maximum −3.6) and a regression coefficient equal to or above 0.98 (R2 0.98), and if the other quality controls were within acceptability limits (recovery 1% and PCR inhibition 50%).
In the absence of a general agreement on the acceptable recovery rate for SARS-CoV-2 in sewage, we used the criterion of recovery rate 1% according to ISO 15216-1:2017, which concerns the quantification of viruses in complex food categories. Other protocols indicate that a recovery of 1% or higher may be considered acceptable; i.e., the recovery process will not be repeated or the data will not be rejected (inter alia Omura et al., 2022). Instead, when the recovery rate is below 1% the results are discarded and the samples can be re-extracted and analysed, thus providing an overall improvement in data quality. The recovery of the procedure is indeed higher on average. Here, recovery rate of the process control virus was not used to convert the viral gene concentration of the target as suggested by other authors (Kantor et al., 2021).
For the trend analysis, negative samples (i.e. samples with undetermined Ct or Ct 40), were imputed to a quantitative value corresponding to the one expected, in the run in which the sample was processed, for a Ct 40.
2.4. Calculation of normalized SARS-CoV-2 load in wastewater
In order to normalize the natural fluctuations of SARS-CoV-2 concentration ( in GU/mL) in wastewater, which depends on various environmental factors, such as influent flow rates, the normalized SARS-CoV-2 RNA load (indicated with the symbol L) was calculated according to the following expression:
where is the daily volume of wastewater entering the plant (daily flow rate, in m3/d) and P is the population (inhabitants) in the served sewershed.
Due to the characteristics and habits of the Trento municipality, the influence of tourist fluctuations is moderate compared to the number of residents. Hence the use for normalization of biomarkers (e.g. caffeine or Pepper Mild Mottle Virus or others) – which could instead be very useful in the presence of high fluctuations of non-resident population – was not considered.
2.5. Detection of SARS-CoV-2 variants in wastewater samples
In accordance with the EU Commission Recommendations 2021/472 (EU, 2021), systematic analysis of SARS-CoV-2 variants in wastewater has been established in Italy. Since October 2021 (with a pilot study in July), the Italian National Institute of Health (ISS) has carried out monthly monitoring campaigns (named ‘flash surveys’) in collaboration with the laboratories of the SARI network of the national program for SARS-CoV-2 surveillance in wastewater and thereby produce a ‘snapshot’ of the variants present in Italy. For this purpose, RNA from wastewater samples collected and processed by all the local laboratories in the country is shipped periodically to ISS, where amplification and NGS long-amplicon sequencing (Nanopore technologies) are performed. The Omicron variant in wastewater was demonstrated using two real-time PCR assays: a newly designed assay, named ISS assay (La Rosa et al., 2022b) and an assay developed by the European Commission, Joint Research Centre, named JRC assay (Petrillo et al., 2021). Monthly reports detailing the number of samples tested, the analytical approach, and the results of variants analysis are published on the ISS website (ISS, 2022a, La Rosa et al., 2021c, La Rosa et al., 2021d). In addition to the monthly surveys, an ‘ad hoc’ study was organized for the investigation of the increase in the Omicron variant between December 5 and December 25, 2021 (La Rosa et al., 2022a, La Rosa et al., 2022b).
2.6. Epidemiological data
Epidemiological data related to the population of the WWTP’s catchment area derive from the daily report of the province (Napolitano, 2022). The database details the ‘daily new cases’, which are the COVID-19 cases newly detected using molecular swabs or antigen tests. The database registers also the number of hospitalizations, the recoveries, and the deaths for each municipality in the province.
To correlate epidemiological data with wastewater signals, the ‘current active cases’, also known as ‘current infected cases’, have to be considered because SARS-CoV-2 may remain in the faeces of positive individuals during the entire period of positivity or even longer. Active cases (A) can be estimated by subtracting the deaths (D) and recovered cases (R) from the total positive cases (T) (Solanki et al., 2021, Worldometer, 2022), according to the following balance:
where indicates a difference over a certain period of time. If the officially registered data based on swabs are used, some asymptomatic people may be excluded, so that may be an underestimation of the actual number of COVID-19 positive cases.
The calculation of is performed for ‘n’ municipalities or suburbs served by the sewer, obtaining , , …, . If a municipality is served by two WWTPs (as in our case), is divided into two parts proportional to the population served. Finally, the ‘current active cases’ that discharge their wastewater (and positive faeces) in a WWTP can be calculated by summing all the of the municipalities and suburbs served by the WWTP:
In particular, in the present case n 7, which includes two parts of Trento city and 7 suburbs as indicated in Figure S1 in Supplementary Material.
2.7. Statistics
All the analyses were conducted using MS Excel. In Section 3.4, the coefficient of determination, R2, has been calculated to evaluate the best linear regression of wastewater RNA-signal and the COVID-19 positive cases.
3. Results and discussion
3.1. Normalized SARS-CoV-2 loads in wastewater and the need for smoothing
Data on the SARS-CoV-2 concentration in wastewater were collected continuously for 17 months, from December 1, 2020, to April 15, 2022, using composite not-frozen samples. Composite samples are preferable for WBE because they are less affected by the natural fluctuation of wastewater quality during the day (Ahmed et al., 2021a). Avoiding freezing helps to preserve the viral load and viability (Medema et al., 2020). In fact, frozen samples have shown a significant loss in the number of viral copies (Ahmed et al., 2022), with a reduction of 0.5 log GU/L after 10 days of storage at −20 °C (Cutrupi et al., 2021).
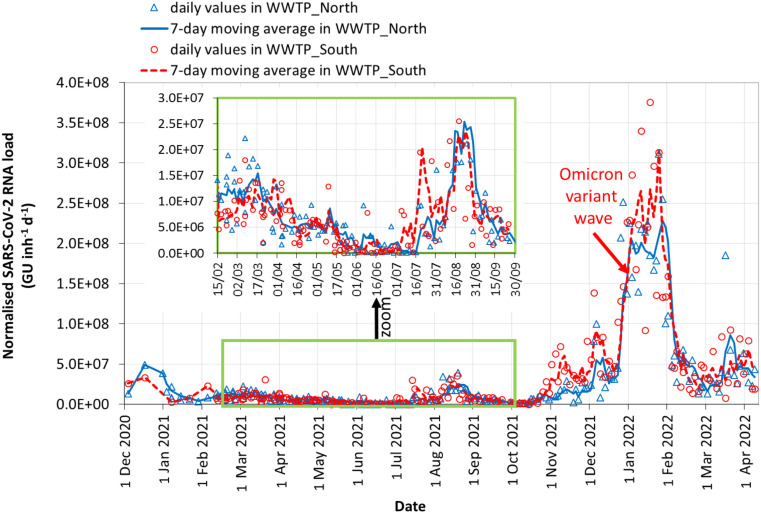
The profiles of the normalized SARS-CoV-2 RNA loads over time (calculated according to the procedure described in Section 2.4) obtained in WWTP and WWTP are shown in Fig. 1. The original daily values acquired by RT-qPCR are represented as discrete points in Fig. 1; data are affected by a ‘natural’ noise that can be associated with the ‘nugget effect’ (Wurtzer et al., 2021), i.e. the problems introduced by the sampling of wastewater but also during the analytical procedures, when coarse, heterogeneous, scarcely mixed particles or microbial clusters (‘nuggets’) can be encountered. Selecting or missing the ‘nugget’ may lead to overestimation or underestimation of the results, respectively. Furthermore, irregular or unpredictable factors such as weather conditions, infiltrations, leakages, the influence of inhibitory substances discharged by – among others – industries, may cause ‘natural’ fluctuations in the daily data.
Fig. 1.
Time-profiles of normalized SARS-CoV-2 RNA loads in wastewater. Open symbols are daily values, lines are the 7-day moving averages.
For these reasons, the use of a smoothing method may help obtain an immediate visualization and a faster interpretation of the underlying trends of the evolution of the outbreak. Similar observations about the need for smoothing have been made by Wurtzer et al. (2021). A simple smoothing technique is the ‘moving average’, which smoothes a series by consolidating the daily data points into a longer unit of time, i.e. a week (or a month). To smooth our data, the 7-day moving average () of the daily normalized SARS-CoV-2 loads (, , …, ) was used. Therefore at day ‘t’ was calculated considering the ‘n’ data acquired within the previous last 7 days, according to the following expression:
where ‘n’ is the number of samples in a week. For example, adopting a frequency of monitoring of 3 times per week, the 7-day average was calculated using 4 data points, i.e comprising the extremes of the week.
As shown in Fig. 1 for the two WWTPs, the use of the 7-day moving average considerably smoothed the daily fluctuations of the viral load. In Fig. 1, the wastewater profiles in the two WWTPs, serving parts of the same municipality and some suburbs, appear very similar and the same waves are apparent.
3.2. Time profile of SARS-CoV-2 in wastewater compared to epidemiological trends
In order to correctly correlate the viral load in wastewater with the epidemiological data, it is very important to conduct detailed calculation of the population and positive cases connected to the sewer and the WWTP (see Figure S1 in Supplementary Material).
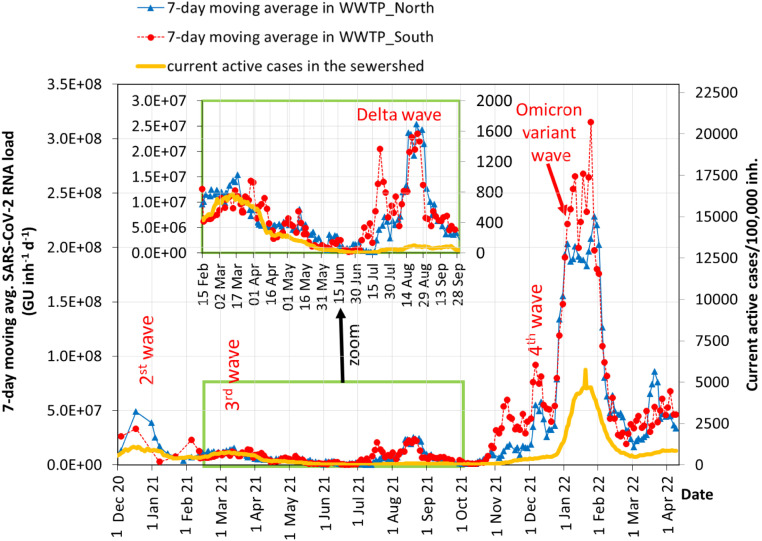
Fig. 2 compares the 7-day moving averages of SARS-CoV-2 loads in wastewater with the ‘current active cases’ in the sewershed: that is, the number of people who may actively discharge SARS-CoV-2 in their faeces and thus into wastewater. The number of ‘current active cases’ is an item of information more relevant to comparison with wastewater signals than ‘new daily cases’; and it is less affected by daily fluctuations because COVID tests increase before weekends, holidays or events, and decrease over weekends or national/local holidays when the labs work on a reduced schedule. Moreover, the number of the ‘current active cases’ is expressed per 100,000 inh.; this normalization allows better comparison between communities of different sizes.
Fig. 2.
Comparison between 7-day moving averages of SARS-CoV-2 RNA loads in wastewater and current active cases.
The methods used to quantify SARS-CoV-2 in wastewater underwent only minor adjustments during the entire monitoring period (17 months). Hence, the concentrations can be easily compared over time. Conversely, the number of active cases over time depends on the vaccination campaign that began on December 27, 2020, with the first doses and moved towards mass vaccination during 2021–22: in particular, as of April 15, 2022, about 83% of the population had received at least one vaccine dose and 61% had received a booster (3rd vaccination dose). As a result of the progressively increased vaccine coverage, the number of active cases was expected to decrease gradually over time.
In Fig. 2, both wastewater profiles appear to be in agreement with the profile of the ‘current active cases’ in the sewershed; and they exhibit the same waves.
In the first months (December 2020–January 2021), the effects of the 2nd wave are evident for both active cases and wastewater signals in WWTP and WWTP. After the lockdown in December 2020–January 2021, the virus load in wastewater gradually decreased, but in February 2021 the 3rd wave began and a new lockdown was imposed.
The 3rd wave, from February to April 2021, can be used as a reference period for the best comparison between wastewater and infections because: (i) the sampling frequency of wastewater was the highest; (ii) the method of analysis was improved; (iii) it was a period with various lockdowns; (iv) efforts were made for contact-tracing; (v) vaccination was in the early phases for sanitary personnel and vulnerable people only. For these reasons, in that period the underestimation of the number of COVID-19 positive cases in the whole population of the sewershed may have been the lowest. On average, in that period, an amount of about 700 active cases/100,000 inh corresponded to a viral load of 1.0E07 GU inh−1 d−1.
Only at the beginning of the summer (June 2021) did we record the absence of detectable SARS-CoV-2 genetic material in some samples, probably due to the more favourable conditions of the summer period (open spaces, closed schools, etc.). However, in July–September 2021, a significant increase in viral circulation was observed in wastewater (with a notable peak from the middle of August), although the number of current active cases remained low thanks to the high number of vaccinations. This wave (July–September 2021) was clearly apparent in wastewater (Fig. 2), and it was due to the Delta variant (lineage B.1.617.2) which was more than 2× as contagious as previous variants (CDC, 2021). The Delta variant was first detected in India in October 2020, but it became predominant in Europe from March 2021 onwards. The presence of the Delta variant in July in WWTP wastewater was confirmed by the analysis carried out by the National Institute of Health (La Rosa et al., 2021c). For comparison, in July 2021, the Delta variant among the population in Italy became dominant with a prevalence of over 90% (ISS, 2021).
The Delta wave during the summer of 2021 was not perceived as a major wave in the population that was mostly covered by vaccinations. In fact, testing is often requested by people with symptoms (even mild), so that, in the presence of scarce or no symptoms due to vaccination, the circulation of the virus may emerge only partially. Therefore, during this Delta wave, there was no epidemiological evidence of the high circulation of the virus, and only wastewater was able to provide a key signature about the spreading of SARS-CoV-2 in the population (Fig. 2). On average, during the Delta wave, an amount of about 90 active cases/100,000 inh corresponded to a viral load of 2.2E07 GU inh−1 d−1. This result indicates once again that wastewater can emit a very sensitive and meaningful signal.
At the end of September 2021, the wave of COVID-19 infections declined significantly, but at the end of October 2021, the SARS-CoV-2 load in wastewater increased progressively with some fluctuations (Fig. 2). In those months the sequencing of the samples in the two WWTPs evidenced the circulation of the Delta variant and Delta sublineages; its presence being confirmed after July in WWTP and, later, in December, also in WWTP (La Rosa et al., 2021d).
At the end of December 2021, a marked increase in the viral signal was detected (Fig. 2). The slope of the curve, i.e. the rate of change of the viral load, explains visually how quickly the viral load increased. From December 23, the 7-day moving average of the viral load (3.2E07 and 4.0E07 GU inh−1 d−1 in WWTP and WWTP, respectively) jumped to 2.0E08 GU inh−1 d−1 in WWTP and 2.2E08 GU inh−1 d−1 in WWTP on January 06, 2022, thus increasing 5–6 fold (500%–600%) in a two-week period. The cause was the spread of the Omicron variant (see Section 3.3), which, because it replicates at a significantly higher speed than SARS-CoV-2 Wilde Type or Delta, is markedly faster in its diffusion rate, with an increased doubling time (Hui et al., 2022).
For comparison among different waves, the peak concentration observed in wastewater during the 3rd wave was about 1.0E07 GU inh−1 d−1, which was only 5% of the viral load circulating in wastewater during the Omicron wave. On average, during the Omicron wave, an amount of about 4500 active cases/100,000 inh corresponded to a viral load of 2.0–2.2E08 GU inh−1 d−1. In short, the viral loads observed between December 2021 and January 2022 were the highest during the period of the study, and the slope was the steepest ever seen since monitoring began in the middle of 2020.
At the end of January 2022, the viral loads in wastewater began to decline rapidly, along with COVID-19 active cases (Fig. 2). On February 15, 2022, the viral load dropped by more than 77% from the peak of the Omicron wave observed in January 2022.
The relationship between SARS-CoV-2 loads in wastewater and current active cases cannot be theoretically or uniquely determined. It depends on viral shedding in faeces, and this value – despite the new data constantly published in the literature (Schmitz et al., 2021, Miura et al., 2021) – is extremely variable for several reasons. First, the viral load in the stool of infected people depends on the category of COVID-19 patients and the severity of the disease; for example, patients with complications shed higher viral loads than those with mild symptoms (see e.g. Lavania et al., 2022). Second, an infected person may shed SARS-CoV-2 RNA for a period of 2–3 weeks or more (Lavania et al., 2022, Kumar et al., 2022) even after a negative swab which excludes that person from the count of active cases. Third, different VOCs could cause different viral loads in infected people; however, preliminary observations suggest viral titres not statistically different in people infected with Omicron compared to Delta (Puhach et al., 2022).
3.3. Confirmation of the presence of the Omicron variant in wastewater in December 2021
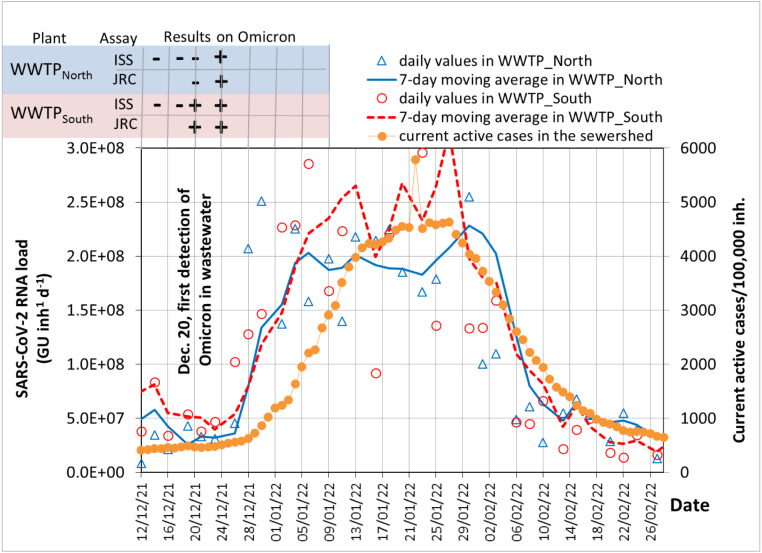
The rapid increase of the SARS-CoV-2 load in wastewater during the Omicron wave and the subsequent rise in the active cases are detailed in the profiles of Fig. 3. The circulation of the Omicron variant in wastewater was first demonstrated around the third week of December 2021 using two real-time PCR assays specific for Omicron (ISS and JRC, indicated at the top of Fig. 3) and Sanger Sequencing for positive results confirmation (La Rosa et al., 2022b).
Fig. 3.
Profiles during the Omicron wave in December 2021–February 2022. 7-day moving averages of SARS-CoV-2 RNA loads in wastewater compared to the current active cases. The presence () or absence () of Omicron variant found by two real-time PCR assays (JRC and ISS) are indicated.
Notably, the Omicron mutations were not detected in the wastewater samples collected on December 13 and 15 in either of the WWTPs. The first positive result of the Omicron variant was found in wastewater on December 20 in WWTP and December 22 in WWTP. The profile of SARS-CoV-2 load in Fig. 3 shows that the rapid rise of the viral load in wastewater began on December 26 in WWTP and on December 28 in WWTP, about one week after the first detection in wastewater by RT-PCR. This indicates that detecting SARS-CoV-2 variants in wastewater can provide valuable information even before the onset of a new wave becomes apparent.
In the week following December 20, 2021 (when the Omicron variant was first detected in wastewater), according to diagnostic tests the active cases in the municipality were stable and only started growing about 10 days later. The rapid growth of active cases then followed, with the Omicron variant rapidly taking over in the first week of January 2022. It is evident from Fig. 3 that wastewater surveillance of SARS-CoV-2 furnished evidence of the increased circulation of the virus even before it became significant according to epidemiological data.
Considering the clinical data from the integrated surveillance system of COVID-19 in Italy, 98.3% of 6.511 sequences submitted to the platform for the genomic surveillance of the SARS-CoV-2 (I-Co-Gen) variants in our country in the period from December 28, 2021 to April 26, 2022 were associated to the Omicron variant (ISS, 2022a). Data for the province of Trento are also available, obtained through clinical “flash surveys” showing that on January 03, 2022, of 25 sequence, 19 were Omicron and 6 Delta (ISS, 2022b). Later on, only Omicron sequences were detected in Trento on the surveys performed on January 31, March 07 and April 04, 2022 with 20, 12, and 14 samples, respectively (ISS, 2022c, ISS, 2022d, ISS, 2022e)
Conversely, the Alpha variant took about 12 weeks to become dominant in the population (Carcereny et al., 2022), while Delta was first detected in England in March 2021 and reached more than 50% of sequenced isolates by May 25, 2021 (Twohig et al., 2022). As evidenced by Fig. 3, just two weeks were enough for the newly emerged Omicron variant to significantly increase active cases and wastewater signals. This rapid diffusion demonstrated the important role of vaccinations in mitigating the consequences of COVID-19 and the number of hospitalizations.
In South Africa, where the Omicron variant first appeared in November 2021, it began to decrease at the end of December 2021 (South Africa Government, 2021). Also in our case, as shown by the profiles in Fig. 3, the decrease in the viral loads from the end of January 2022 was just as rapid as the increase.
AS far as we know, this is one of the first studies where the Omicron wave was completely monitored at high frequency. In particular, once the Omicron variant emerged in the two WWTPs, the time profiles were acquired until the peak was reached and the descending trend was completed. The entire evolution of the Omicron wave lasted about two months. Fig. 2, Fig. 3 clearly show that the dynamics of the viral loads in wastewater follow well the number of COVID-19 cases per 100,000 inh, in agreement with observations during the spread of previous VOCs (Carcereny et al., 2022, Chavarria-Miró et al., 2021).
Since multiple SARS-CoV-2 variants may be present simultaneously in wastewater samples (Agrawal et al., 2022, Novoa et al., 2022), confirmation of the prevalence of the Omicron variant in the community was derived here from clinical data. In this regard, Novoa et al. (2022), in the analysis of variants, demonstrated a significant concordance between the most frequent variants detected in wastewater and positive persons from the same geographical area.
The results also highlighted that wastewater surveillance allows to appreciate the circulation of the virus even with an underestimation of positive cases due to mass vaccination. Understanding the evolution of the diffusion of emerging VOCs is essential for adopting adequate mitigation actions, especially towards the Omicron variant which still has only few reports in wastewater.
3.4. Anticipation of epidemiological data
The Omicron wave (Fig. 3) exhibited a certain temporal shift between the signal in wastewater and the current active cases, confirming that wastewater can anticipate the epidemiological data. Thus, wastewater can provide an early warning signal of the onset of a wave, as maintained by various studies in the literature (Bonanno Ferraro et al., 2021, Ahmed et al., 2021b, La Rosa et al., 2020, Li et al., 2022).
To calculate the time shifting (anticipation) during the Omicron wave, the profile of the positive cases was dragged with a shift t, variable from 0 to 7 days, to overlap as much as possible with the SARS-CoV-2 loads and thus find the best correlation between the two series, analogously to the approach adopted by Wurtzer et al. (2021). The relationship appears quite similar between the two WWTPs: the shift that permits the best correlation is the one with the highest coefficient of determination, R2, and corresponds to the time lag (anticipation) of 6 days. For comparison, an average lag of 3 days was observed by Wurtzer et al. (2021).
Consideration of the entire 17-month monitoring period shows that the anticipation became more evident when the peak was very pronounced during the Omicron wave in Dec. 2021–Jan. 2022 compared to previous waves with lower peaks. Whilst the wastewater data showed an increase in the community about a week before traditional nasal or salivary swabs, by contrast, the anticipation of the declining trend does not seem appreciable (Fig. 3). Instead, the time profile of the viral loads during the decline of the Omicron wave appears to coincide with the time course of the current active cases.
The anticipative capacity of the sewage signal is also highlighted by Novoa et al. (2022) in 11 WWTPs in Spain. In particular, their results indicated that wastewater surveillance and modelling were able to predict the evolution of the pandemics in the municipalities within a time horizon of seven days.
3.5. Comparison of viral loads in winters 2020–21 and 2021–22 due to extended vaccinations
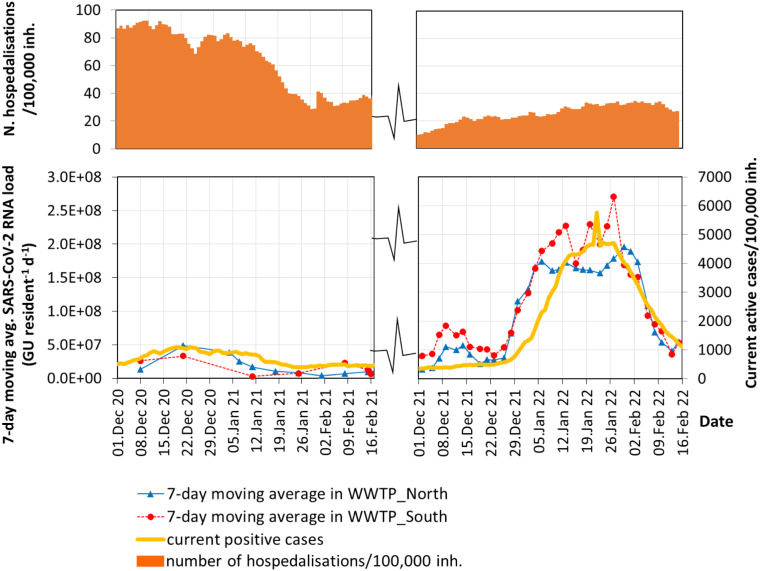
Fig. 4 shows the comparison of the SARS-CoV-2 loads and the current active cases during the months of December, January and February for two consecutive years, i.e. 2020–21 and 2021–22. Immediately apparent are the huge differences in the viral loads and active cases in the two successive winters (Fig. 4).
Fig. 4.
Comparison of two successive winters (December–February 2020–21 and 2021–22). 7-day moving averages of SARS-CoV-2 RNA loads in wastewater, current active cases and hospitalizations are compared.
During the 2nd wave in winter 2020–21, lockdown periods and various restrictions were imposed in the municipality; the SARS-CoV-2 load reached maximum values of 5.0E07 and 3.3E07 GU inh−1 d−1 in WWTP and WWTP, respectively. In that period, the corresponding infections were around 1000 cases/100,000 inh.
Conversely, during winter 2021–22, when the Omicron variant spread rapidly in the community, minor restrictions were applied because of the mass vaccination campaign against COVID-19, which covered more than 80% of the population. The SARS-CoV-2 loads (Fig. 4) reached a maximum plateau with average values of 2.0E08 and 2.5E08 GU inh−1 d in WWTP and WWTP, respectively. These maximum loads were 4 to 7 times higher than those in winter 2020–21. During the Omicron wave, the number of active cases at the peak was 4700 cases/100,000 inh that is 4.7 times higher than the previous winter.
Despite more infections in winter 2021/22, the Omicron variant caused fewer hospital admissions than in the previous winter 2020/21, due to the mostly less severe nature of disease and the mass vaccination campaign, although the Omicron variant may partially evade immunity from vaccines and previous infections. In the first week of December, hospitalizations in the province increased to 22/100,000 inh on December 15, 2021, when the Omicron variant was not yet present in wastewater, and was associated with the Delta variant. Then hospitalizations increased slowly and progressively for the entire month of January 2022, surpassing 30/100,000 inh. during the long maximum plateau (peak on January 27, 2022). By comparison, hospitalizations amounted to more than 90/100,000 inh. at the beginning of winter 2020/21.
The steep spike of positive cases has raised concerns that the healthcare system will once again come under pressure, due to the extremely high rate of the spread of the Omicron variant. Indeed, the wide circulation of the virus among the population, as evidenced by wastewater surveillance (Fig. 4), may have had an impact on the healthcare system and on the number of hospitalizations. However, as shown in Fig. 4, the fast-spreading Omicron variant registered a record number of positive cases but a limited hospitalization increase in January–February 2022, compared with the increase with the Delta variant in December 2021. This is in line with the findings of other studies (Maisa et al., 2022), which confirm mild symptoms in the cases of Omicron detected in France, but underline the great importance of the surveillance system for rapidly detecting new variants and adapting to emergencies.
The comparison among high concentrations of viral RNA in wastewater despite a moderate amount of COVID-19 cases was highlighted by Novoa et al. (2022), confirming once again that infected individuals with less symptoms may go unnoticed (Novoa et al., 2022). In Milan, high viral loads were found in wastewater in combination with high vaccination coverage, suggesting significant circulation of the virus among vaccinated individuals (Nattino et al., 2022) and this could make variants more competitive. However, the authors point out that in these cases standard surveillance metrics cannot accurately describe the spread of the virus, while wastewater surveillance can give early warning of virus circulation. In a different study, Bivins and Bibby (2021) instead measured lower viral concentrations in wastewater after massive vaccination in the closed community living in a college campus, due to the decreased viral shedding.
4. Conclusions
To the best of our knowledge, this is one of the first monitoring of the entire Omicron wave through wastewater surveillance, within a 17-month long-term monitoring (December 2020–April 2022). The main results show that:
-
–
during the Delta wave in summer 2021, a viral load peak emerged in wastewater, not perceived in positive cases due to mass vaccination;
-
–
no Omicron was found in wastewater samples until the mid-December 2021;
-
–
after the first detection, then Omicron variant spread very quickly becoming predominant in a couple of weeks;
-
–
the viral loads in wastewater during the Omicron wave were 4–20 times higher than in the Alpha and Delta waves;
-
–
the Omicron wave lasted two months from December 2021 to February 2022.
-
–
a marked concordance was observed between viral loads in wastewater and ‘current active cases’;
-
–
wastewater is able to show the increase in viral loads with an anticipation of 6 days.
In the rapidly changing and challenging context of emerging VOCs, wastewater surveillance has proven to be a valuable tool, independent of diagnostic testing and important in the long-term monitoring of the pandemic.
CRediT authorship contribution statement
Francesca Cutrupi: Investigation, Writing – original draft, Interpretation of results. Maria Cadonna: Conceiving and design of the study, Investigation, Interpretation of results, Analysis of data, Writing – review. Serena Manara: Conceiving and design of analytical methods, Revision of original draft, Writing – review. Mattia Postinghel: Investigation, Analysis of data. Giuseppina La Rosa: Conceiving and design of analytical methods, Interpretation of results, Writing – review. Elisabetta Suffredini: Conceiving and design of analytical methods, Interpretation of results, Writing – review. Paola Foladori: Conceiving and design of the study, Interpretation of results, Analysis of data, Writing – original draft, Writing – editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The Authors wish to thank the staff at the wastewater treatment plants of Trento Nord and Trento Sud for their support in the sampling and transport of samples and the Agenzia Provinciale per la Protezione dell’Ambiente (Province of Trento) for the availability of the laboratories where the authors carried out the analyses safely. Alejandro R Cinalli is acknowledged for the cooperation in some lab work. The authors thank Giusy Bonanno Ferraro, Pamela Mancini, Carolina Veneri (ISS, Rome) for SARS-CoV-2 sequencing. Financial support for the monitoring from October 1, 2021 was provided by the Ministry of Health and the Ministry of Economy, Italy (decree 30.10.2021). From October 1, 2021, the project benefited also from the financial support allocated by the Directorate General for the Environment of the European Commission for the collaboration agreement EC G.A. NO. 060701/2021/864481/ SUB/ ENV.C2 - “Support to Member States for the creation of systems, local collection points and digital infrastructures for monitoring COVID 19 and its variants in wastewater - Italy”. This research was partially supported by the Internal Call 2020 “Covid 19”, “Surveillance of COVID-19 Pandemic with a Wastewater-Based-Epidemiology approach (SCOPE)” project, awarded by the University of Trento, Italy and by the VRT Foundation with the project “PILLAR”. The authors also acknowledge funding from the Italian Ministry of Education, University and Research (MIUR) in the frame of the “Departments of Excellence” grant L. 232/2016.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.eti.2022.102667.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Figure S1. Map of the municipality served by the two WWTPs. Indication of the parts of the main municipality and suburbs connected to the two sewersheds. Table S1. Capacity of the two WWTPs and average annual characteristics.
References
- Agrawal S., Orschler L., Schubert S., et al. Prevalence and circulation patterns of SARS-CoV-2 variants in European sewage mirror clinical data of 54 European cities. Water Res. 2022;214 doi: 10.1016/j.watres.2022.118162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: Implications for wastewater-based epidemiology. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Weil M., Indenbaum V., Bucris E., Bar-Ilan D., Elul M., Levi N., Aguvaev I., Cohen Z., Shirazi R., Erster O., Sela-Brown A., Sofer D., Mor O., Mendelson E., Zuckerman N.S. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BBC News . BBC; UK: 2021. Covid: Omicron Variant in NetherlandS Earlier than Thought. (accessed 2.15.22) [Google Scholar]
- Bivins, Bibby Wastewater surveillance during mass COVID-19 vaccination on a college campus. Environ. Sci. Technol. Lett. 2021;8(9):792–798. doi: 10.1021/acs.estlett.1c00519. [DOI] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A. Wastewater-based epidemiology: Global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bonanno Ferraro G., Veneri C., Mancini P., Iaconelli M., Suffredini E., Bonadonna L., Lucentini L., Bowo-Ngandji A., Kengne-Nde C., Mbaga D.S., Mahamat G., Tazokong H.R., Ebogo-Belobo J.T., Njouom R., Kenmoe S., Rosa G.La. A state-of-the-art scoping review on SARS-CoV-2 in sewage focusing on the potential of wastewater surveillance for the monitoring of the COVID-19 pandemic. Food Environ. Virol. 2021 doi: 10.1007/s12560-021-09498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciapaglia G., Cot C., Sannino F. Multiwave pandemic dynamics explained: how to tame the next wave of infectious diseases. Sci. Rep. 2021;11:6638. doi: 10.1038/s41598-021-85875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcereny A., Garcia-Pedemonte D., Martínez-Velázquez A., Quer J., Garcia-Cehic D., Gregori J., Antón A., Andrés C., Pumarola T., Chacón-Villanueva C., Borrego C.M., Bosch A., Guix S., Pintó .R.M. Dynamics of SARS-CoV-2 alpha (b.1.1.7) variant spread: The wastewater surveillance approach. Environ. Res. 2022;208 doi: 10.1016/j.envres.2022.112720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2021. Centers for disease control and prevention, USA. URL https://webcache.googleusercontent.com/search?q=cache:xoK8ybpliFUJ: https://www.cdc.gov/coronavirus/2019-ncov/variants/delta-variant.html+&cd=1&hl=it&ct=clnk&gl=it (accessed 2.22.22) [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 2021:87. doi: 10.1128/AEM.02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutrupi F., Cadonna M., Manara S., Foladori P. Surveillance of SARS-CoV-2 in extensive monitoring of municipal wastewater: key issues to yield reliable results. Water Sci. Technol. 2021;84:3508–3514. doi: 10.2166/wst.2021.469. [DOI] [PubMed] [Google Scholar]
- ECDC . European Centre for Disease Prevention and Control; Stockholm: 2022. Assessment of the Further Spread and Potential Impact of the SARS-CoV-2 Omicron Variant of Concern in the EU/EEA, 19th Update - 27 2022. [Google Scholar]
- E.U. 2021. (n.d.) Commission recommendation (EU) 2021/472 of 17 2021 on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variants in wastewaters in the EU [WWW Document] URL http://data.europa.eu/eli/reco/2021/472/oj (accessed 2.15.22) [Google Scholar]
- Ferré V.M., Peiffer-Smadja N., Visseaux B., Descamps D., Ghosn J., Charpentier C. Omicron SARS-CoV-2 variant: What we know and what we don’t. Anaesthesia Crit. Care Pain Med. 2022;41 doi: 10.1016/j.accpm.2021.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Cadonna M., Manara S. Coronaviruses and SARS-CoV-2 in sewerage and their removal: Step by step in wastewater treatment plants. Environ. Res. 2022;207:11220401–11220413. doi: 10.1016/j.envres.2021.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. Sars-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Ho J.C.W., Cheung M.-C., Ng K.-C., Ching R.H.H., Lai K.-L., Kam T.T., Gu H., Sit K.-Y., Hsin M.K.Y., Au T.W.K., Poon L.L.M., Peiris M., Nicholls J.M., Chan M.C.W. SARS-CoV-2 omicron variant replication in human bronchus and lung ex vivo. Nature. 2022 doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- Istituto Superiore di Sanità, Fondazione Bruno Kessler, Ministero della Salute . 2021. Stima della prevalenza delle varianti VOC (variants of concern) in Italia: B.1.1.7, b.1.351, p.1 e b.1.617.2, e altre varianti di SARS-CoV-2 (indagine del 20/7/2021) (accessed 2.17.22) [Google Scholar]
- Istituto Superiore di sanità . ISS, Rome, Italy; 2022. Prevalenza E Distribuzione Delle Varianti Del Virus SARS-CoV-2 Di Interesse Per la Sanità Pubblica in Italia (Iss.It): Rapporto n. 19 del 29 aprile 2022. (accessed 5.12.22) [Google Scholar]
- Istituto Superiore di Sanità . 2022. Comunicato stampa n03/2022 - Covid-19: flash survey iss, il 3 gennaio l’81% dei campioni positivi a omicron - ISS. (accessed 5.12.22) [Google Scholar]
- Istituto Superiore di Sanità . 2022. Comunicato STAMPA n12/2022 - Covid-19, flash survey iss: il 31 gennaio il 99, 1% dei campioni positivi a omicron - ISS. (accessed 5.12.22) [Google Scholar]
- Istituto Superiore di Sanità . 2022. Comunicato stampa n22/2022 - in Italia al 7 marzo variante omicron al 99, 9%, ba.2 al 44, 1% - ISS. (accessed 5.12.22) [Google Scholar]
- Istituto Superiore di Sanità . 2022. Comunicato stampa n30/2022 - covid19: flash survey iss, il 4 aprile omicron al 100%, sottovariante ba.2 predominante - ISS. (accessed 5.12.22) [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Khan N.A., Al-Thani H., El-Menyar A. The emergence of new SARS-CoV-2 variant (omicron) and increasing calls for COVID-19 vaccine boosters-The debate continues. Travel Med. Infect. Dis. 2022;45 doi: 10.1016/j.tmaid.2021.102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Jiang G., Kumar Thakur A., Chatterjee S., Bhattacharya T., Mohapatra S., Chaminda T., et al. Lead time of early warning by wastewater surveillance for COVID-19: Geographical variations and impacting factors. Chem. Eng. J. 2022;441 doi: 10.1016/j.cej.2022.135936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi A.K., Patel M. Unravelling the early warning capability of wastewater surveillance for COVID-19: A temporal study on sarsCoV-2 RNA detection and need for the escalation. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Suffredini E. 2021. Protocollo della sorveglianza di SARS-CoV-2 in reflui urbani (SARI) - rev. 3. [DOI] [Google Scholar]
- La Rosa G., Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brandtner D., Grigioni M., Rossi M., Suffredini E. 2021. Flash survey on SARS-CoV-2 variants in urban wastewater in Italy 4th Report (study period: 30 november – 03 2021) [DOI] [Google Scholar]
- La Rosa G., Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brandtner D., Grigioni M., Rossi M., Suffredini E. 2022. Ad hoc survey on b.1.1.159 (omicron) variant on SARS-CoV-2 in urban wastewater in Italy (study period: 05 december – 25 2021) [DOI] [Google Scholar]
- La Rosa G., Bonanno Ferraro G., Mancini P., Veneri C., Lucentini L., Bonadonna L., Brandtner D., Grigioni M., Rossi M., Suffredini E. 2021. Flash survey on SARS-CoV-2 variants in urban wastewater in Italy 1st report (investigation period: 04–12 2021) [DOI] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Veneri C., Mancini P., Ferraro G.B., Brandtner D., Lucentini L., Bonadonna L., Rossi M., Grigioni M., SARI network ., Suffredini E. The rapid spread of SARS-COV-2 omicron variant in Italy reflected early through wastewater surveillance. Sci. Total Environ. 2022 doi: 10.1016/j.scitotenv.2022.155767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Ferraro G.Bonanno., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavania M., Joshi Madhuri S., Ranshin Sujata S., Potdar Varsha A., Manohar Shinde, Nutan Chavan, Jadhav Santosh M., Prasad Sarkale, Sreelekshmy Mohandas, Sawant Pradeep M., Sanjaykumar Tikute, Vikram Padbidri, Sampada Patwardhan, Rohan Kate. Prolonged shedding of SARS-CoV-2 in Feces of COVID-19 positive patients: Trends in genomic variation in first and second wave. Front. Med. 2022:9. doi: 10.3389/fmed.2022.835168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Gu X., Armas F., Wu F., Chandra F., Chen H., Xiao A., Leifels M., Chua D.F.J., Kwok G.W.C., Tay J.Y.R., Lim C.Y.J., Thompson J., Alm E.J. Quantitative detection of SARS-CoV-2 omicron variant in wastewater through allele-specific RT-qPCR. BioRxiv. 2021 doi: 10.1101/2021.12.21.21268077. [DOI] [Google Scholar]
- Li L., Mazurowski L., Dewan A., Carine M., Haak L., Guarin T.C., Dastjerdi N.G., Gerrity D., Mentzer C., Pagilla K.R. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Sci. Total Environ. 2022;817 doi: 10.1016/j.scitotenv.2022.152958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupala C.S., Ye Y., Chen H., Su X.-D., Liu H. Mutations on RBD of SARS-CoV-2 omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022;590:34–41. doi: 10.1016/j.bbrc.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisa A., Spaccaferri G., Schaeffer J., Fournier L., Deniau J., Rolland P., Coignard B. First cases of omicron in France are exhibiting mild symptoms, 2021–2022. Infect. Dis. now. 2022 doi: 10.1016/j.idnow.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan C.S., Self S., Rennert L., Kalbaugh C., Kriebel D., Graves D., Colby C., Deaver J.A., Popat S.C., Karanfil T., Freedman D.L. Covid-19 wastewater epidemiology: a model to estimate infected populations. Lancet Planet Health. 2021;5:e874–e881. doi: 10.1016/S2542-5196(21)00230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Miura F., Kitajima M., Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021;769(2021) doi: 10.1016/j.scitotenv.2020.144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundie J. 2022. Omicron was in nova scotia wastewater before it was identified in South Africa. Natl. post. (accessed 2.15.22) [Google Scholar]
- Napolitano M. 2022. Situazione sanitaria Covid-19 in provincia autonoma di trento [WWW Document] n.d. Situazione sanitaria Covid-19 in Provincia Autonoma di Trento. URL https://covid19trentino.fbk.eu/ (accessed 4.15.22) [Google Scholar]
- Nattino G., Castiglioni S., Cereda D., et al. Association between SARS-CoV-2 viral load in wastewater and reported cases, hospitalizations, and vaccinations in milan, 2020 to 2021. JAMA. 2022 doi: 10.1001/jama.2022.4908. Published online April 01, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa B., Ríos-Castro R., et al. Wastewater and marine bioindicators surveillance to anticipate COVID-19 prevalence and to explore SARS-CoV-2 diversity by next generation sequencing: One-year study. Sci. Total Environ. 2022;833 doi: 10.1016/j.scitotenv.2022.155140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura ., et al. Japan Society on Water Environment; Japan: 2022. Manual for Detection of SARS-CoV-2 RNA in Wastewater. Edited By COVID-19 Taskforce. February, 2022. [Google Scholar]
- Pellegrinelli L., Castiglioni S., Cocuzza C.E., Bertasi B., Primache V., Schiarea S., Salmoiraghi G., Franzetti A., Musumeci R., Tilola M., Galuppini E., Bertanza G., Callegari M., Stefani F., Turolla A., Ammoni E., Cereda D., Pariani E., Binda S., the WBE Study Group . Evaluation of pre-analytical and analytical methods for detecting SARS-CoV-2 in municipal wastewater samples in Northern Italy. Water. 2022;14(833) doi: 10.3390/w14050833. [DOI] [Google Scholar]
- Petrillo M., Querci M., Corbisier P., Marchini A., Buttinger G., Van den Eede G. 2021. In silico design of specific primer sets for the detection of b.1.1.529 SARS-CoV-2 variant of concern (Omicron) [DOI] [Google Scholar]
- Puhach O., Adea K., Hulo N., Sattonnet-Roche P., Genecand C., Iten A., et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, delta and omicron. MedRxiv. 2022 doi: 10.1101/2022.01.10.22269010. [Preprint] Available at: https://www.medrxiv.org/content/10.1101/2022.01.10.22269010v2. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. Sars-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., Davidsson F., Dotevall L., Mattsson A., Trybala E., Lagging M., Lindh M., Gisslén M., Brezicka T., Nyström K., Norder H. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021;189 doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz BW., Innes GK., Prasek SM., Betancourt WQ., Stark ER., Foster AR., Abraham AG., Gerba CP., Pepper IL. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. 2021 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki R., Varshney A., Gourishetty R., Minase S., Sivadas N., Mahajan A. Correction in active cases data of COVID-19 for the US states by analytical study. Disaster Med. Public Health Prep. 2021:1–4. doi: 10.1017/dmp.2021.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano J.B., Gerli A.G., Centanni S., Ancochea J. Forecasting COVID-19 infection trends and new hospital admissions in Spain due to SARS-CoV-2 variant of concern omicron. Arch. Bronconeumol. 2022 doi: 10.1016/j.arbres.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2021. South Africa Government cabinet approves several changes to the adjusted alert level 1 COVID-19 regulations [WWW Document] URL https://www.gov.za/speeches/cabinet-approves-several-changes-adjusted-alert-level-1-covid-19-regulations-30-dec-2021 (accessed 2.16.22) [Google Scholar]
- Statens Serum Institute . 2022. Now, an omicron variant, BA.2, accounts for almost half of all Danish Omicron-cases (ssi.dk) (accessed 2.02.22) [Google Scholar]
- Tong C., Shi W., Zhang A., Shi Z. Tracking and controlling the spatiotemporal spread of SARS-CoV-2 Omicron variant in South Africa. Travel Med. Infect. Dis. 2021;46 doi: 10.1016/j.tmaid.2021.102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twohig K.A, Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., Seaman S.R., Harris R.J., Hope R., Lopez-Bernal J., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (b.1.617.2) compared with alpha (b.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 2022;22(1):35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation (No. WHO/V & B/03.03) [Google Scholar]
- WHO . 2022. [WWW Document], n.d.. WHO coronavirus (COVID-19) dashboard. URL https://covid19.who.int/ (accessed 2.15.22) [Google Scholar]
- Worldometer . 2022. [WWW Document], n.d.. COVID live - coronavirus statistics - worldometer. URL https://www.worldometers.info/coronavirus/ (accessed 12.05.22) [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MSystems. 2020:5. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Levert M., Cluzel N., Almayrac J.L., Charpentier C., Masnada S., Gillon-Ritz M., Mouchel J.M., Maday Y., Boni M., OBEPINE Consortium, AP-HP Virologist Group, Marechal V., Moulin L. SARS-CoV-2 genome quantification in wastewaters at regional and city scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci. Total Environ. 2021;810 doi: 10.1016/j.scitotenv.2021.152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdenkova K., Bartackova J., Cermakova E., Demnerova K., Dostalkova A., Janda V., Jarkovsky J., Lopez Marin M.A., Novakova Z., Rumlova M., Ambrozova J.R., Skodakova K., Swierczkova I., Sykora P., Vejmelkova D., Wanner J., Bartacek J. Monitoring COVID-19 spread in prague local neighborhoods based on the presence of SARS-CoV-2 RNA in wastewater collected throughout the sewer network. Water Res. 2022;216 doi: 10.1016/j.watres.2022.118343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Map of the municipality served by the two WWTPs. Indication of the parts of the main municipality and suburbs connected to the two sewersheds. Table S1. Capacity of the two WWTPs and average annual characteristics.