Summary
Rapid advances in artificial intelligence (AI) and availability of biological, medical, and healthcare data have enabled the development of a wide variety of models. Significant success has been achieved in a wide range of fields, such as genomics, protein folding, disease diagnosis, imaging, and clinical tasks. Although widely used, the inherent opacity of deep AI models has brought criticism from the research field and little adoption in clinical practice. Concurrently, there has been a significant amount of research focused on making such methods more interpretable, reviewed here, but inherent critiques of such explainability in AI (XAI), its requirements, and concerns with fairness/robustness have hampered their real-world adoption. We here discuss how user-driven XAI can be made more useful for different healthcare stakeholders through the definition of three key personas—data scientists, clinical researchers, and clinicians—and present an overview of how different XAI approaches can address their needs. For illustration, we also walk through several research and clinical examples that take advantage of XAI open-source tools, including those that help enhance the explanation of the results through visualization. This perspective thus aims to provide a guidance tool for developing explainability solutions for healthcare by empowering both subject matter experts, providing them with a survey of available tools, and explainability developers, by providing examples of how such methods can influence in practice adoption of solutions.
Keywords: explainability, artificial intelligence, machine learning, life sciences, clinical research, healthcare
The bigger picture
Adoption of AI tools in practical settings, such as for research/clinical tasks, has been hampered by a lack of transparency/interpretability of the models. After performing a review of different types of AI explainability (XAI) methods developed to better understand the predictions made by a model, we also develop a taxonomy to better classify the different approaches. We think that these XAI techniques are not sufficient to enhance practical implementations and illustrate via an example how user-driven XAI can be useful for different stakeholders in the healthcare domain. We identify and define three key personas involved in healthcare—data scientists, clinical researchers, and clinicians—and present an overview of the different approaches that can address their needs. The ultimate goal of adopting AI in medical practice and patient care goes beyond explainability and will need the development of extra layers of security and confidence, in particular regarding AI trustworthiness, as XAI transparent systems become prone to attacks that may reveal confidential information, and AI fairness, as systems developed and tested in diverse environments need to be expanded to real-world situations.
A significant amount of research has focused on making artificial intelligence (AI) techniques more interpretable and post hoc explanations more accessible. This has been achieved through the development of a wide variety of so-called explainability methods. We here propose a taxonomy and review the most popular AI explainability (XAI) methods, illustrated with examples from healthcare and life sciences. We also propose that user-centered explainability is necessary for further practical adoption of AI approaches and illustrate this using healthcare-centered user examples.
Introduction
With the growing availability of machine learning algorithms and data, there is a rising interest in adopting artificial intelligence (AI) in order to advance not only biological and clinical research but also medical practice and patient care. Machine learning algorithms have been applied to a diversity of biological and medical problems1 like protein folding,2 genomics,3 drug discovery,4 medical imaging,5 and clinical research in chronic diseases, such as AIDS.6 Furthermore, large accessible databases, including genomic data7,8 and electronic health records, such as in Medical Information Mart for Intensive Care (MIMIC)-III,9 or that contain both genomic and clinical data, such as in UK Biobank,10 have opened avenues for active collaboration between researchers in AI, medicine, life sciences, and healthcare applications, as shown by initiatives like Machine Learning for Healthcare (https://www.mlforhc.org/).
Despite its potential benefits, AI and particularly deep learning lack the necessary transparency to generate trust and knowledge. In medicine, although machine learning algorithms can outperform human doctors in making diagnoses,3,11,12 it is difficult to understand how their decision is made,13 a critical question for preventing harm in practical use. For example, a machine learning model made diagnostic predictions of pneumonia by learning the association between a type of X-ray machine and the disease occurrence.14 Moreover, when using gene expression for patient diagnosis, the sample metadata can sometimes predict the outcome perfectly. For high-stakes fields, such as clinical practice, usage of AI without addressing the shortcomings related to trust can often hinder the adoption of such models in practice. Another cause for concern is the danger of amplifying biases in patient sub-populations, given insufficient training data for the group. For instance, one of the most predictive variables for length of stay in a hospital is the postal code, in which longer stays were correlated with relatively poor and predominantly African American neighborhoods.15 Also, participants in genomic studies or clinical trials are often not representative of populations who need treatments in terms of race, gender, and ethnicity groups.16 Finally, explanations and a deeper understanding of the nature of the predictions are necessary because models often make decisions based on unreliable signals from datasets. To mitigate these issues, it is necessary to either continuously inspect and improve models, build methods that can be directly interpreted, or both.
Although some models are inherently “transparent” and provide users with the most important features relevant for a model output, the display of such feature importance is not inherent to deep neural networks. Furthermore, even when model interpretation via feature importance is available at a global level, estimating how the model behaves for an individual example is non-trivial. For example, knowing the feature importance for a random forest model is not sufficient for users to grasp what-if scenarios at a particular example level, such as increasing/decreasing feature values by a certain amount. Recently, a significant amount of research has been focused on postulating many so-called explainability methods for experts to probe global and local explanations and understand why certain predictions are made by trained machine learning models. Providing the tools for interpretation of AI models and potentially answering the “why” question by filling the explainability gap have been identified to be of paramount importance.17
There has been significant research on alleviating these shortcomings by providing various methods to explain AI models, the breadth of which enables choosing the one best fitting the needs.18 However, despite such advances, many critiques stand relative to the methods employed and, more crucially, the explainable problem in AI they are trying to solve. For example, doubts have been raised relative to using attention maps as proxies for feature importance.19,20 Even methods that may otherwise appear grounded in theory and widely adopted, such as SHAP,21 have also been criticized.22 Indeed, counter-arguments exist that outright question the need for explainability, such as when the methods are accurate enough.23 Also, there have been concerns about the generation of misplaced trust when using explanation techniques.24 One of the core challenges in addressing such critiques derives from a lack of a universally accepted definition of explainability. For example, Hind25 argued that the term, explainability, itself is not well-defined unless it is contextualized in a communication between two parties, A and B, where party A provides justification for an action or decision to party B. The more sufficient justifications are provided, the more iterations happen between the two parties, and the more trust and acceptance of the model are built. Defining formally a valid and reliable human-to-human explanation is in itself challenging, so defining a concrete system-to-human explanation is a challenge beyond the realm of AI expertise. Also, the explanations of decisions taken by humans are neither uniform nor consistent26 but derive from different phenomena and contexts of a particular domain. Hence, Hind25 argued in favor of generating explanations for the end user in context and advocated for a persona-driven design. Recent literature27, 28, 29 has further looked into the problem of approaching explainability from an end user perspective, recognizing the fact that explainability also involves a “for whom” in addition to a “why” question. In particular, such end users are usually classified into different personas, and tools for explainability in artificial intelligence (XAI) are developed to adapt the workflow of such personas toward more confident usage of underlying AI models.
From this perspective, we first provide a general overview of the dimensions of explainability and, following,25 identify the personas for healthcare and their unique needs. We employ concrete real-world examples of XAI and connect how such applications were driven by the persona of interest. In the process, we also provide a brief overview of potential pitfalls. Overall, we aim to provide a guidance tool for developing explainability solutions for healthcare by empowering both subject matter experts with a survey of available tools and explainability developers with examples of how such methods can influence adoption of solutions in practice. This approach can readily be extended to the life sciences and medical informatics, where such divisions also exist.
Results
The general dimensions of explainability
The biggest challenge to designing an XAI model is to understand how a human might expect an explanation to be.29 Although there has been research on general metrics that measure some aspect of explainability,18 from a user point of view, universally applicable metrics are difficult to obtain. One can argue there are three major challenges: (1) the definition of explainability is not universal, pointing to the need for user-driven XAI; (2) even when domain experts are involved, they often may not agree, making it hard to define a formal metric; and (3) given the previous two aspects, defining a global metric may not always be feasible. Therefore, wide ranges of XAI techniques have been developed to cater to the diverse needs of explanations in a variety of AI applications. Many generic XAI tools are open-sourced and contain not only the best machine learning tools but also the most used ones (see Table 1).
Table 1.
Summary of available open-source XAI tools
| Toolkit | Data Explanations | Directly Interpretable | Self-explaining | Local Post Hoc Explanation | Global Post Hoc Explanation | Explaina- bility Metrics | URL Links |
|---|---|---|---|---|---|---|---|
| AIX 360 | X | X | X | X | X | X | http://aix360.mybluemix.net |
| Alibi | X | https://github.com/SeldonIO/alibi | |||||
| Skater | X | X | X | https://oracle.github.io/Skater/ | |||
| H2O | X | X | X | https://github.com/h2oai/mli-resources | |||
| InterpretML | X | X | X | https://github.com/interpretml/interpret | |||
| EthicalML-XAI | X | https://github.com/EthicalML/xai | |||||
| DALEX | X | X | https://modeloriented.github.io/DALEX/ | ||||
| tf-explain | X | X | https://github.com/sicara/tf-explain | ||||
| iNNvestigate | X | https://github.com/albermax/innvestigate | |||||
| modelStudio | X | X | X | X | https://bit.ly/3uOnU5y | ||
| ELI5 | X | X | X | https://github.com/TeamHG-Memex/eli5 | |||
| Iml | X | X | X | https://bit.ly/3iBv8Vx |
Coverage is shown along several explainability dimensions: (1) data explanations are provided through data distributions and enable case-based reasoning; (2) directly interpretable refers to a model that inherently provides information about features driving predictions at both global and local levels; (3) in contrast, self-explaining models provide local explanations but may not be globally interpretable; (4) local post hoc explainers can provide explanations around particular data points for black-box models in a post hoc manner; (5) whereas global post hoc explainers provide the same at a global/model level; and (6) explainability metrics cover several state of the art metrics to quantify the explainers/models around several dimensions of explainability.
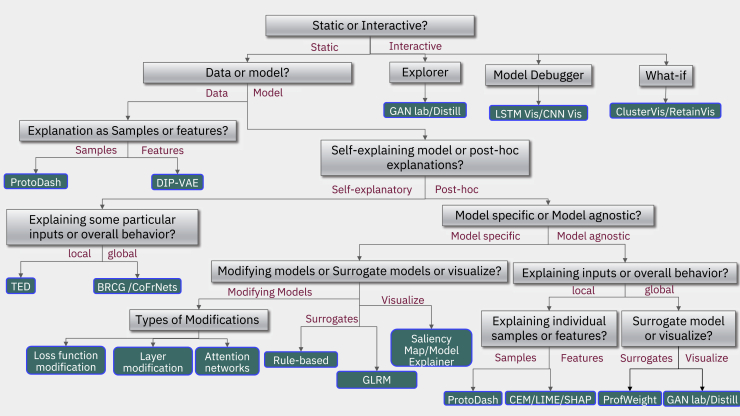
XAI techniques can be categorized broadly into four major classifications: data/model, self-explanatory/post hoc, local/global, and static/interactive (see Figure 1). For a detailed discussion of these different XAI techniques, see Arya et al.,18,30,31 Arrieta et al.,18,30,31 and Linardatos et al.18,30,31 The first division separates whether the data or the model needs to be explained, although different personas might require both. Indeed, to understand data, it is important to have a case-based reasoning approach to compare and place a given sample with respect to other examples in the dataset. Thus, depending on the needs and use case, model developers can only choose to focus on features that can be explained and understood to model the data distributions, potentially at the cost of performance. One possibility is to apply a transformation to the features to make them more explainable.
Figure 1.
Taxonomy tree for explainability in AI models
To figure out the most appropriate explanation method, we propose a taxonomy of questions represented as a decision tree to help navigate the process. The green leaf nodes represent algorithms that are in the current release of AI Explainability 360. Considering the data, different choices are possible relative to its representation and understanding: data understanding based on features, in which case theory can yield disentangled representations, such as in Disentangled Inferred Prior Variational AutoEncoders (DIP-VAEs)32; otherwise, a sample-based approach using ProtoDash33 is possible, which provides a way to do case-based reasoning. If the goal is to explain models instead of data, then the next question is whether a local explanation for individual samples or a global explanation for the entire model is needed. Following the global path, the next question is, Should it be a post hoc method or a self-explaining one? On the self-explaining branch, TED (teaching explanations for decision making)34 is one option, or a global method, such as BRCG (Boolean rule sets with column generation).35 On the model agnostic post hoc branch, again, explaining in terms of samples or features comes up. On the sample side, prototypes come up again, as on the feature side choices among the contrast of explanations methods (CEMs),36 as well as popular algorithms, such as LIME37 or SHAP,21 are available. Finally on the post hoc global side, surrogate models, such as ProfWeight, are available. On the model-specific branch, one has to choose between modifying models, surrogate models, or simply visualizations. Going back up, aiming for global explanations for the entire model, then the question again is whether something post hoc is needed or a directly interpretable model? A directly interpretable model could be a Boolean rule set, such as BRCG or GLRMs (generalized linear rule models),38 can yield the answer.
The second dichotomy is whether the goal is to build a directly interpretable/self-explanatory model (see Table 1 for definitions), something that is simple and easy to understand by itself, or if we are looking for a post hoc explanation of a difficult to explain black-box model.30 Classic approaches in machine learning and AI, such as decision tree and rule-based models, can directly develop interpretable models.31 They are generally learned in a heuristic or greedy manner, but recent advances in discrete optimization have led to new approaches applied to large-scale interpretable models.39 In cases of models that are not interpretable, such as deep neural networks or very large ensembles, a post hoc sort of explanation needs to be performed, and there are various methods for doing so.31 Depending on the problem at hand, directly interpretable approaches can often outperform black-box models.40 Although feature engineering and significant domain knowledge are often needed to make such approaches practical, significant research has expanded such methods to a wide range of problems.41, 42, 43, 44 Nevertheless, black-box models often perform better45 and/or are more broadly applicable.46 The post hoc explanations can be designed by modifying each individual underlying AI algorithm, building surrogate models,38 or visualizing a model’s behavior in a meaningful way.47 Alternatively, generic post hoc XAI models have been developed to provide explanations for any type of AI techniques.
The third division in the explainability taxonomy entails whether we are looking for local or global explanations.18 A local explanation happens at the individual sample level, whereas a global explanation encompasses the entire model. A medical professional society might want a description of the behavior of the entire model, a global explanation, in order to inform best practices for their membership. This can be performed post hoc or with directly interpretable global explanations, or possibly both. Conversely, a local explanation is desired in the case of a patient who wants to know why the model has predicted that he/she has/may have cancer. Note that in the case of a linear and locally consistent global model, the explanations may be the same for both the global and local model; however, for more complex local models, a local neighborhood may have different features with different effects.
Finally, the fourth division type is static versus interactive explanations, where, in the former case, the explanation is just printed out and presented to the user, and, in the latter, the output is interactive and lets the user query the results in some sort of visual or conversational manner.30 Most of the existing technology and way of doing things has led to the static form, but a slew of new ways to think about explanation has produced software that, as people would do in a conversation, explain in an interactive fashion by asking each other questions and are able to dive into details in a conversational way or following visual analytics.48,49
Most of the XAI techniques described so far are generic in nature and typically aimed at a variety of datasets, such as gene expression, text, tabular, images, and their applications. As such, several tools have emerged with open-source codes covering the different categories of the XAI taxonomy we just presented. A brief comparative summary of such freely available XAI tools is provided in Table 1. For more in-depth analysis on this topic please refer to Molnar.50
Ensuring accountability and transparency of XAI models
One critical question that is often asked about any AI model in life sciences or in healthcare before being deployed relates to the robustness, accountability, and transparency of its predictions.51 The XAI models described so far mainly focus on making AI models explainable17,52; although this could be satisfactory for a research question when coupled to a meaningful interpretation, it is just one component of the larger pipelines and life cycles of the healthcare system. Therefore, model explainability alone is not sufficient for ensuring overall accountability and transparency53,54 nor does it entail overall interpretability of the model results.54,55 In the healthcare domain, other tools like datasheets, model cards, factsheets, and documentations in addition to explanations generated from XAI models often become useful for ensuring such overall system transparency.56
Toward user-centric explainability in health
Although XAI models aim to derive generic explanations for complex AI techniques, there exist several challenges for the extension of their use.13,51,53 First, the term explanation itself has been debated in the AI community recently to be more subjective and vague, especially in healthcare and life sciences, where more rational decision making/understanding is desired due to the high-sensitivity/costs associated with each decision made. Second, there is significant debate on the definitions of explainability, the methods employed, and how such methods can be of practical use. As shown in Doshi-Velez and Kim,57 there is no benchmarking of the objective function of explainability; rather, evaluation can only be performed by the end user as, “you’ll know it when you see it.” Third, the level of explanation needed for a complex AI model also depends on the expertise and ability of understanding the explanations by the users who receive and finally interpret them.25 What may be interpretation for a specialist physician is very different for a general physician, biologist, or computational biologist. Past research on this topic has led to attempts at formalizing the requirements in machine readable formats.29 In this paper, we augment such approaches by identifying distinct personas in the healthcare life cycle and further define explanations for addressing the particular requirements and expertise that are specific for different users. Following the perspective of Hind,25 centered on healthcare personas, we further identified three different cases, namely, data scientists, clinical researchers, and clinicians taking ultimate decisions.58 Although patients are excluded from the types of personas of XAI models, data generated from sensors and wearable devices might soon change this. It is to be noted that personas can broadly relate to the role a person plays for a particular use case. Thus, a single person can assume the identity of different personas, e.g., a doctor moving from the clinical researcher persona to the clinician persona when they move to the patient bedside. Furthermore, multiple personas may require deep collaboration to achieve both unique and overlapping tasks performed by them, e.g., several departments of a multi-facility hospitals working together by providing feedback to each other. Another alternative view of a persona can be correlated with the mental model59 of the person of interest in the healthcare life cycle. To understand this categorization, let us explore explainability as a human factor—an answer to a “why” question. It can be argued that this viewpoint then requires an explainer and an explainee. In the case of medical sciences and in the context of AI, the explainee thus usually falls into the mental models or roles described in this manuscript. We conducted interviews with a clinician with clinical research background and, from their perspective, clinical professionals are, in general, constantly interacting with technology that allows them to gain insights into a patient’s condition and thus to the condition of a population. It is not necessary for the patient-facing clinician to understand the inner workings of a particular technology to be able to extract the value represented by the insight provided. There are several examples where technology-driven insights, specifically, software-aided technology, do not require detailed understanding of the inner workings to extract value. Among those, computer-aided imaging techniques are a good example. In general, it is not necessary for a clinician to get an explanation when studying an MRI or tomography60, 61, 62; it is sufficient to understand the basics of the technology, particularly its limitations, sensitivity, and specificity, to be able to extract the necessary value. This could also be true of analog technologies, where understanding of the physics of electricity, sounds, or pressure is not necessary to interpret an electrocardiogram, heart sounds using a stethoscope, or simple measurement of blood pressure. This is not to say that a deeper understanding of the technology does not work in favor of the clinician, but more in the sense of understanding the general inner workings of a machine each time that a technology is used. In the case of AI, it could be argued that a better understanding of the underlying technology by the user could, at least partially, leads to better understanding of the “reasons” behind a specific score or AI-derived insight. The framework presented here also argues that different personas or mental models will require different types and levels of explanation, just like different mental models (radiologist, technician, and medical biophysicist) would require different levels of explanation out of a digital imaging device; even if these mental models existed within the same individual, they are in the end context-driven. Thus, we can argue that the explanation is driven by the value provided by the technology and, as such, the tasks and roles of clinician, clinical researcher, and data scientist while overlapping require different levels of understanding. Ultimately, the human-in-the-loop approach to technology in the aid of medical history is a long thread of successes, and there should be no reason to think that AI as an extension of these technologies would be any different.
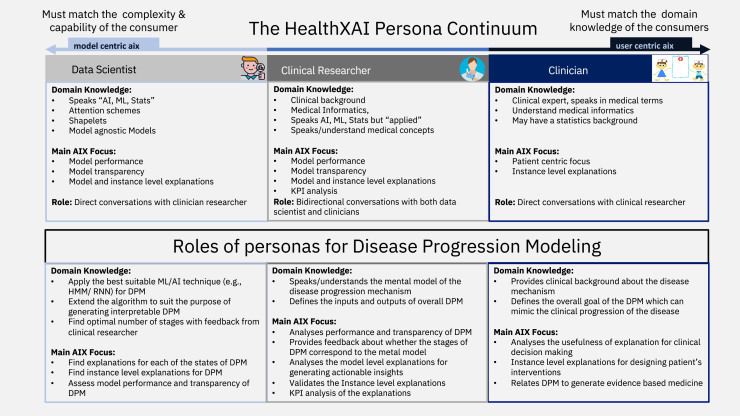
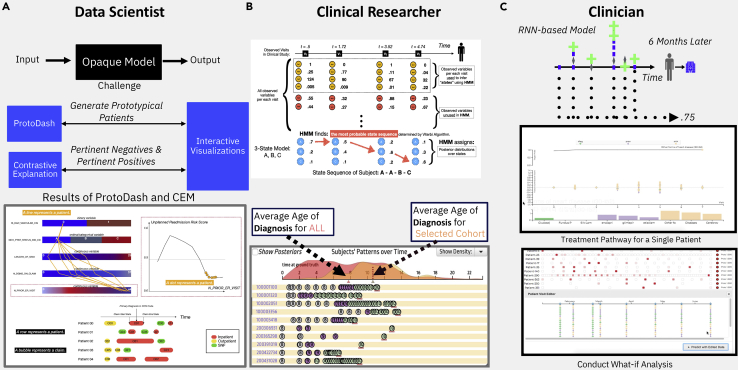
To provide a real-world example, let us take disease progression modeling as a use case to describe what the potential roles each healthcare persona can play (see Figure 2). The goal of disease progression modeling (DPM)63 is to model the natural continuous progression of chronic diseases identifying multiple irreversible stages, each having diverse disease symptoms. Clinicians are responsible for interacting with the end users, the patients, and as such they typically use their medical expertise to define the overall goal of the DPM, mimicking the clinical progression of the disease through the generation of clinical hypothesis. Most of the time, they focus on the clinical impact of such models and how to generate actionable insights at different stages of the disease. Clinical researchers design the overall input/output of the DPM, where the inputs are typically patient prior medical history, electronic health records of patients with the same disease, and any potential clinical hypothesis. The output of the DPM should mimic the mental model of actual disease progression mechanisms. Also, higher-level knowledge of medical informatics and AI both are needed to translate, what clinicians want, to the data scientists who ultimately build the model. The data scientists take these overall input/output features of DPM and make the architecture design of the model based on the clinical goals as defined by the clinical researchers. Then, they build the actual DPM model using the most viable machine learning tools, which can best cater the needs of clinical research. In particular, they are responsible for designing model details, such as number of stages of the model, finding discriminative features to define disease stages, using AI explanations both at the global and instance level. The clinical researcher then validates these explanations, performs key performance indicator (KPI) analysis, and assesses their usefulness for generating clinical insights. After multiple iterations of communications among these three personas, ultimately, clinicians would like to use the explanations generated by DPM for clinical decision making to enhance evidence-based medicine.
Figure 2.
Health XAI Persona continuum and roles
(Top) Different personas relevant for user-centric XAI models and their domain knowledge and XAI roles. (Bottom) Example of specific roles of three personas for a real-world problem of designing an explainable progression model for chronic diseases.
From this motivating example, and a general view of health XAI, the desired level of explanations can vary among these three personas of interest. Researchers/data scientists evaluate explanations as measured by the model performance and AI model transparencies, whereas clinical researchers aim at further interpreting the model-generated explanations by assessing their fitness to a few prototypical instances of particular healthcare applications. In addition, they can perform a KPI analysis of the XAI models to increase their trust in the considered clinical practice. Finally, the explanation should fulfill the requirement of the ultimate end users, i.e., the clinicians who have mainly a patient-centric focus of explainability. Hence, the explanations should mimic the mental model of clinicians comprising diverse information, such as their background training, existing medical knowledge, their own expertise, patients’ prior history, medical norms, patients’ behavioral aspects, etc. Having defined the different explainability-related personas, we will now describe in detail the most common XAI methods, illustrated with examples. As shown in Figure 2, the three different personas using AI models have different requirements, and the term, explanation, has different meanings, depending on the role they play. Specifically, the researcher/data scientist main role is to build core AI or machine learning models, whereas the clinician mostly interacts with medical knowledge specific to a particular disease. On the other hand, clinical researchers act as a bridge between AI experts/researchers and clinicians with brief exposure to both medical terminology and healthcare informatics.
Applying different XAI methods for extracting explanations from biomedical data
We will now describe the application of a few of the XAI models, as described in Figure 1, on specific problems to demonstrate their usefulness in the clinical and life sciences domains. Note that the choice of the XAI methods is not exhaustive; rather, they were chosen based on their overall popularity in the domain of interest and their availability as user-friendly open-source tools for better usability. Similarly, we try to cover a wide range of biomedical applications, including data sources ranging from electronic health records, genomics, clinical images, etc.
The first explainability method we will describe in more detail is LIME (local interpretable model-agnostic explanation), widely used irrespective of domains and data types.64 LIME is a local method that generates locally trusted explanations, mimicking the original predictive model in the neighborhood of a particular sample that is being predicted. LIME falls into the category of a static model-based approach as it tries to provide an explanation using its own optimization framework (Figure S1A). It is also a local model, because it provides an explanation for each individual sample but tries to summarize the local explanations generated from different samples into a global one. Another important feature of LIME is that it is a post hoc model-agnostic method that can extract explanations from any complex model. In contrast to other explanation approaches trying to modify the black-box models themselves to generate a simpler surrogate model that can be interpreted, LIME’s final output is the most important feature used as a best possible explanation of the black-box model. LIME learns a simple linear function, which has similar predictive power to the original complex model within the local neighborhood of a given sample. LIME also provides the flexibility of choosing features that are easier to interpret, either from the original raw features or any other representation of the input features, giving a succinct and short explanation so that anyone can interpret them.
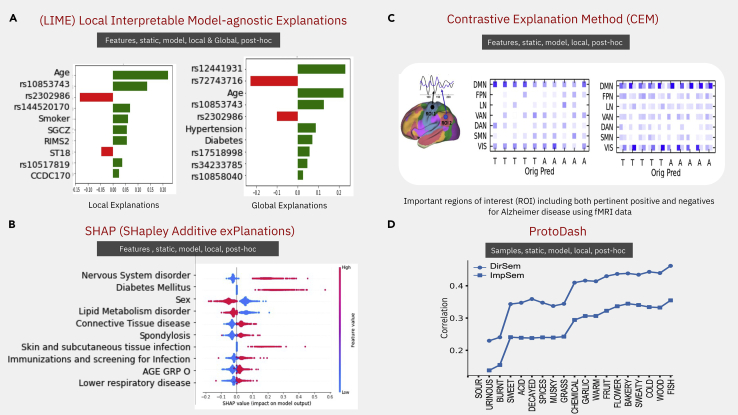
In its objective function, shown in Figure S1A, f is the original complex function that LIME is trying to explain for a given sample, x, and g is the simple explainable model that LIME is trying to learn within that local neighborhood . The main objective function comprises two terms; the first term determines the local faithfulness of the interpretable model within the local neighborhood of x and the local faithfulness of the two functions f and g should be similar, at least within the local neighborhood of x. The second term in the equation controls the complexity of the explanation itself by imposing some regularization on the explanation model. Another point we want to emphasize here is that the input domains of this two functions f and g may be different based on the problem domain. By minimizing the loss function, you learn the new explainable model g, which is much simpler than the original non-explained model f. Another interesting feature of LIME is that it can also generate global summary explanations from the local explanation that it has already generated from the given samples. Sub-modular pick-LIME (SP-LIME) chooses those samples and features that can cover most of the cases in a non-redundant fashion, following a heuristic to generate the global summary explanations from all the local explanations. As an example, we applied SP-LIME to extract insights from an algorithm that predicted the severity of coronavirus disease (COVID-19) based on both clinical and genomic data of patients in a dataset extracted from the UK Biobank (see Figure 3A).65
Figure 3.
Applications of four popular XAI methods
(A) LIME optimizes the local faithfulness and complexity of explanation. It has two versions to find both local and global explanations, which we applied on a COVID-19 longitudinal dataset to represent the clinico-genomic factors associated with COVID-19 severity.65 Local/global importance of single nucleotide polymorphisms, indicated by their chromosomic location and clinical variables relative to the patient outcome, are shown in green or red for positive or negative association, respectively.
(B) SHAP is a more generalized version of six linear-based explainable models using Shapely regression values. The Shapely regression values are applied on a type 2 diabetes longitudinal dataset consisting of electronic health records (EHRs); red dots represent variables negatively influencing and blue dots positively influencing the outcome as shown by the value of the SHAP value.
(C) Contrastive explanation method (CEM) finds the pertinent positive and negative samples that are minimally and sufficiently present and absent for that class, respectively. (Bottom) Shows the pertinent positives and negative regions of interest (ROIs) of the brain related to an fMRI imaging dataset used to differentiate between autistic (A) and neurotypical subjects (T), represented in the columns of the matrices. The raw imaging features were summarized into seven brain regions, represented by the rows in the matrices. Blue hue represents importance of the regions when using the LRB algorithm (left matrix) or CEM (right matrix); see Dhurandhar et al.36
(D) ProtoDash tries to find prototypes samples by summarizing its underlying distribution, which was applied to order the importance of 19 olfactory descriptors used to predict the odor of pure molecules as described by 131 descriptors. Note the descriptors order does not change when using only word embeddings for prediction (ImpSem) or psychophysical olfactory measurements (DirSem).66 For the equation describing each of these methods, see Figure S1B.
Another widely used XAI method is called SHAP (Shapley Additive Explanations).21 Being a post hoc static method that uses a generic version of many different local explainable models to rank feature importance based on their explainability, it is taxonomically similar to LIME (see Figure S1B). SHAP provides the Shapley regression values to rank the features relative to the tasks being learned and provides different techniques to compute very efficiently these values, especially in the presence of multi-collinearity among the features. Also, SHAP provides a theoretical framework to prove, under certain assumptions, the unique existence of such local models. Again, similar to LIME, x and represent two input dimensions; one is for learning the original predictive model and the other one is for the explainable model. Given the original function f and a local sample , the goal of any local method is to learn a new function , where is a local neighborhood of , so that the two functions f and g are similar in this local neighborhood. SHAP generalizes an additive feature attribution method, a linear winner weighted summation of all the feature components φ, where M is the dimension of this feature domain z. This generic framework can easily be cascaded using examples that include LIME,64 DeepLIFT,67 layer-wise relevance propagation,68 and the classic Shapely value estimation.69
Three properties need to be true in order to generate a theoretical solution, using game theory to guarantee that a unique local linear expansion model is always available. The first property, related to the Shapley regression value φ, can be interpreted as a surrogate for feature importance and is trying to determine the difference between the function scores, when any subset of the given features is included. Hence, given a local sample x, the model f first behaves similarly for the simplified input , at least within the local neighborhood of the region. The second property is missingness, such that with the transformed simplified feature space, if is zero, then the shapely regression value φ should be also zero. Finally, the third property is consistency; this property considers that if the contribution of a particular feature is constant regardless of other inputs, when you change the model parameters, then the input attribution φ should not decrease. If these three properties are present, a unique local linear expansion model can be obtained. The shapely regression value is computationally expensive, and approximate algorithms are provided to find these shared values efficiently, namely four different algorithms, the model-agnostic Shapely sampling values, KernelSHAP, MaxSHAP, and DeepSHAP. Intuitively, the SHAP value increases in the predictive performance when including a particular feature into the model framework. Indeed, if the expected predictive performance increases, or follows a similar dimension, then the SHAP values are higher compared with when values are not good and some multi-collinearity is present. Hence, the SHAP values represent feature importance of the task being learned within a local neighborhood.
We applied SHAP to understand time to event predictions of a method predicting, in a cohort extracted from a private claims dataset (Marketscan),70 complications of type 2 diabetes. In this cohort, we identified the first event of type 2 diabetes (T2D) diagnosis and predicted using deep learning models the onset of neuropathy complication, a typical complication associated with this disease. Applying SHAP on the DeepSurv model (Figure 3B) shows that being male or having other nervous system disorders increases the risk of neuropathy whereas patients with disorders of lipid metabolism have lower risk of developing such neuropathy.
The contrastive explanation method (CEM)71,72 is a more recent method that has been successfully applied to many different domains because it provides natural explanations not only in terms of the positive features but also the negative ones (see Figure 3C). It is a local, static, post hoc model, with similar position as SHAP and LIME in the taxonomy (see Figure 1). Two terms are critical to understand this method, pertinent positives and pertinent negatives. The former represents the features that should be minimally and sufficiently present for a classifier to predict from the same class, and the latter defines the features that should be minimally and necessarily absent for the classifier to not predict the opposite class.
To determine pertinent negatives, the loss function of CEM is composed of three parts (Figure S1C). The first is used to perturb a sample by δ to make it belong to any other class but the given class. The confidence parameter k provides an additional separation between the two predicted classes of and . The next term in the loss function is the regularization term, which is an elastic net regularization using and norms, particularly useful for large amounts of data. The last part of the loss function is the auto-encoder reconstruction loss, ensuring that the original given sample and the partner sample are similar, as assessed by the norm reconstruction error of the auto-encoder. A similar loss function can be defined for finding pertinent positives as well, the difference resting in the first part, such that the perturbation is defined as being the union of the same classes of samples, and the perturbed sample will have the same class as the original sample .
Figure 3C shows the application of the CEM algorithm to a brain fMRI imaging dataset used to differentiate between autistic and neurotypical subjects.36 The raw imaging features were summarized into seven brain regions, which are shown in Figure 3C (left), and each row in the middle matrices represents a region of interest, while the columns represent a subject which has to be classified as either autistic or neurotypical. The two heatmaps show the results of CEM algorithm and the right shows the LRB algorithm. As we can see here, the CEM algorithm highlights two different coefficients, one for pertinent positive and another for pertinent negatives, because LRP only highlights feature importance by a single entry in the heatmap. This shows the effectiveness of the CEM algorithm, which can identify both pertinent positives and also pertinent negatives for each of these subjects. Finally, the results are consistent between these two algorithms because two regions of brain, namely DMN (default mode network) and VIS (visual cortex), are mostly related to autism. However, the CEM method provides more detail about how these two regions are related to autism in terms of whether they are pertinent positives or pertinent negatives.
The last method here presented is called ProtoDash,33 a static, local, post hoc model that relies on the data to generate explanations and hence can take any complex model (see Figure 3D). However, it is significantly different from the previous methods, given that instead of generating features, it produces as explanation representative samples from the dataset that summarize its underlying complex distribution. ProtoDash generates these representative samples by assigning non-negative weights of importance, and it not only can find prototypes for a given dataset but also outliers.
The approach can be defined as finding a subset S of a collection V of items; these can be data points and features that maximize a scoring function (see Figure S1D). The whole framework is built on the important property called sub-modularity of the scoring function , such that, given two sets S and V, we want to find a data sample I that does not belong to T and holds the following property: the functional score when sample I is included in a subset increases the score more than when we add that sample I to the superset T. If this condition is maintained, a theoretical property guarantees that you can find sample prototypes very efficiently. However, it is computationally expensive to guarantee the sub-modular property of an algorithm, so a class of approximate sub-modular functions need to be defined to implement an efficient algorithm to search for this kind of prototypes. The scoring function, which holds the sub-modular property, works for any symmetric positive definite matrix. Existing state-of-the-art methods require further conditions imposed on the kernel matrix. In contrast, ProtoDash can generalize those kernels by using only symmetric positive definite kernels, with the expense of forgoing the sub-modularity.
Figure 3D shows the application of ProtoDash to understand the performance of a model that uses semantic embeddings to predict the rating values of 131 olfactory descriptors (violet, pineapple, sour, etc.) for 128 pure molecules from imputed (for 70 molecules) and measured values (for 58 molecules) of 19 olfactory descriptors.66 ProtoDash prioritizes the 19 perceptual descriptors used by the model and shows that as the number of descriptors increases, prediction performance generally increases, also showing that indeed the descriptors were chosen correctly and represent the most informative samples of the dataset
Knowledge transfer for explainability
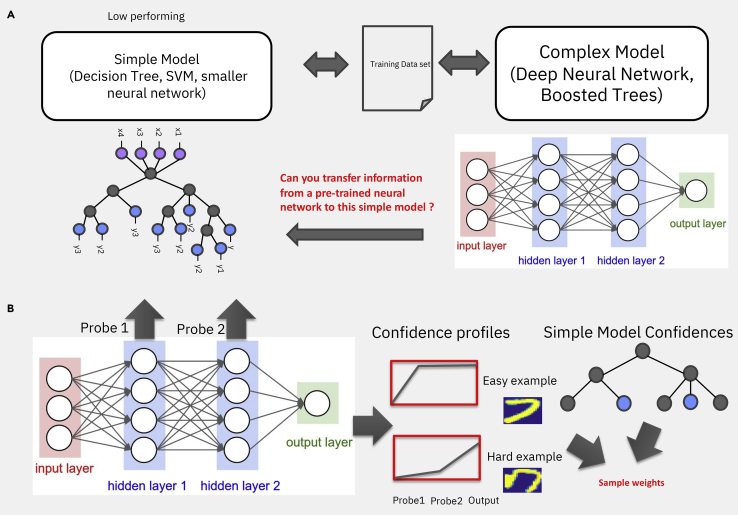
Knowledge transfer methods73, 74, 75 can be used, in the context of explainability, to extract information from a high-performing black-box model to boost the performance of a low-performing interpretable simple model, such as a decision tree or a support vector machine (see Figure 4A). Three situations typically demand this, the first one being when a subject matter expert (SME) wants to use a trusted model they understand and in this case the transfer methods are used to maximize the performance of the SME preferred model. The second case entails a situation with a small amount of data, where a complex black-box model has been trained on a large public or private dataset and has to be applied to a smaller amount of data, over which it is but reasonable to only train a simple model because complicated models could overfit the dataset. The motivation for such approaches derives from the large body of work on transfer learning and more recently on foundational models.76 The third situation arises when computational resources are constrained, such as the model being deployed on a cell phone or an unmanned aerial vehicle (UAV), which elicits strict memory and computational constrains, only allowing a simple model to be deployed.
Figure 4.
Knowledge transfer for explainability
(A) Scheme of transfer learning from a complex model to the right to a simpler one to the left, where a globally explainable model was constructed from local explanations.
(B) (Left) Example of a neural network with two hidden layers and the associated probes, i.e., linear classifiers. To the right are shown (top) an easy example of a number 7 from the MNIST (Modified National Institute of Standards and Technology) dataset to be classified and a hard example on the bottom, with their associated area under the curve (AUC), approximated by taking averages of the classification performance of the two probes. The probe output is an indication of how easy or hard that example is to classify. The easy example obtains good classification from the first layer, but the hard example, as 7, is not well written; it is very hard for the neural network to classify it correctly. Only when reaching higher-level probes, essentially a deep neural network, the performance is high. (Right) The AUC is then used to weight the loss function of a simple model and retrain it. If the simple models do not use weighted loss functions for training, the training set can be re-sampled according to their difficulty following re-weighted ratios.
One of the most popular techniques that transfers from a complex black-box model to a simple model is known as knowledge distillation.73 This method alters the target fitted by the simple model. It does so by fitting soft predictions, i.e., the predicted probability of a data point belonging to one of the classes of the complex model and not the class itself, where the softness can be changed by tuning a temperature parameter. Model compression77 is a specific case where hard labels of the complex model can be fitted to, i.e., the predicted class for a sample point, leading in some cases to improved simple models.
The second class is sample re-weight-based methods, such as ProfWeight78 (Figure 4B), where, instead of changing the target, you re-weight the samples, hence re-weighting the loss function of the simple model. Because generally neural networks are not good estimators of densities, the confidence of the accuracy of their predictions is not reliable. The solution for this entails attaching linear classifiers, which are termed probes, to the intermediate layers of this neural network and calculating for every training sample the probability of classifying it into the correct class. The gradation across probes of the difficulty to classify samples is obtained by averaging across samples and it is used to weight the loss function of a simple model in order to retrain it. If the simple models do not use weighted loss functions for training, the training set can be re-sampled according to their difficulty following re-weighted ratios. SRatio is a more recent example of a re-weighting method and consists of using as a weight the probes average of the performance across samples, divided by primary confidence estimate for the input of the simple models, rather than just the average.75 This approach can be used not only for neural networks but for any type of model.
The third class of knowledge transfer to simple models consists of constructing a globally explainable model from local explanations (Figure 4A, left). SHAP21 and LIME64 can be used to obtain local explanations and then integrated into global explanations (see TreeExplainer79 and model agnostic multilevel explanation [MAME]80). A third method, called the global Boolean formula learning (GBFL) method,81 can take local contrastive or counter-factual explanations and create globally transparent rules, which can then be used to build a classifier. The common point of these three approaches is that they transfer information from the complex black-box model through local explanations, to create a global understanding.
Visualization as an explanation
We have discussed the potential solution of implementing transparent models to meet the concerns that researchers or clinicians may have using black-box models, such as the impression that they learn unreliable signals, amplify existing biases, or simply do not bring real knowledge. However, transparent models need to be understood and often the user does not have the required expertise in data science. Interactive visualizations can be a solution, generating trust and understanding by facilitating the inspection and interpretation of models while easily conducting their analysis.82 We will now present the four different types of visualization that exist and their motivation (Figure 5), while also discussing the use of these types of interactive visualizations for three kinds of personas: researchers/data scientists, clinical researchers, and clinicians (Figure 6).
Figure 5.
Example of what-if analysis tools
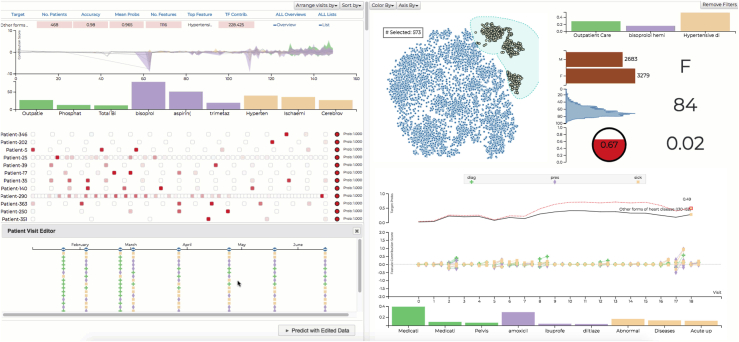
(Left) RETAINVis83 RNN ”RETAIN” model, showing the contribution to the overall outcome of patient visits through feature contribution score, representing drugs (violet), diagnosis (yellow), or physiological markers (green) for each visit. (Bottom) Patient list shows individual patients in a row of rectangles. In the patient list, users can select a patient of interest to view details, shown below, and edit patients to conduct a what-if analysis. (Right top) Dimensionality reduction techniques like t-SNE (t-distributed stochastic neighbor embedding) result in the blue scatterplot to gain an overview and then build patient cohorts using the lasso selection tools and take a look at the distribution for demographic information like biological sex, age, and risk prediction scores (red circle). (Right bottom) Contribution scores for each visit and patient details are shown after the updated results of the what-if analysis. In the middle, an area chart shows aggregated contribution scores of nine medical codes over time. It shows mean and standard deviation as an area. Users can also see the medical codes and their mean contribution scores in bar chart.
Figure 6.
Visualization and explanations for three types of users
(A) ProtoDash and CEM Explorer84 allow data scientists to inspect the trained model using the contrastive explanation method. The examples shown are related to an RNN model predicting in patient re-admission risk based on previous emergency room (ER) visits and variables extracted from insurance claims, such as hospital-acquired condition, vascular-catheter-associated infection, length of stay, number of diagnosis on claims, and number of prior ER visits. Each representative patient (yellow line and yellow dot) is extracted using ProtoDash, and then CEM is used to obtain explanations, as represented by the different colors: red box, inpatient; yellow box, outpatient; and green box, skilled nursing facility (SNF).
(B) DPVis85 helps clinical researchers to understand the disease progression patterns by interacting with multiple, coordinated visualizations. (Top) Represents a diagram of a hidden Markov model (HMM) and the used/unused variables to find disease progression states using different visits over time. The HMM extracts the most probable sequence of states for a specific patient. (Bottom) The waterfall view shows the state progression patterns and time/age for each patient (represented by a line) over time as well as the age distribution at diagnosis for all the cohort (red) and the selected cohort (yellow). Overlap is shown in orange.
(C) RetainVis83 can help clinicians test how (top) an RNN-based model performs on a set of patients by conducting various what-if analyses. (Middle) Single-patient view of the feature contribution score, representing drugs (violet), diagnosis (yellow), or physiological markers (green) for each visit in the treatment pathway. (Bottom) Questions can be answered by editing patient visits, because medical records and update timestamps can be modified for each visit obtaining new predictions and contributions over patients visits by re-running the model. Contribution scores show how much each medical code and visit affects the prediction score at the end. Top contribution scores can be also generated per patient and for multiple patients by aggregating the scores.
The first type of visualization tools is explainers; they provide web-based tools in which users can learn how a model is trained from a dataset and how it makes inferences on new data points. They provide self-paced tutorials with scrollable walkthroughs and more interactive playgrounds. Examples are R2D3 for decision tree models,86 GAN live87 for generative adversarial networks, and CNN Explorer for convolutional neural networks.88 Explainers are gaining traction as more academic and non-academic venues, such as Distill.pub or VISxAI, appear to present such tools.
The second category is visualization for model debugging and inspection, developed to understand machine learning models, inspect any issues, and gain insights on how to improve the model. These visualizations provide interactive visuals tailored for such tasks and show activation of neurons, aggregates of them in different layers, or visualization of attention scores. Users can gain an overview and also query details on demand. Examples of such types of visualization techniques include LSTMVis,89 CNNVis,90 Seq2seqVis,91 and SANVis.92 The third type of visualization tools is designed to support users’ data analysis tasks in their domain study (Figure 5). The views provide information that are useful to answer questions that users have. In particular, users can conduct what-if scenarios by perturbing input data values and checking how decisions are impacted by changes.93 They allow users to compare multiple model instances by showing differences in model outputs from same data input instances. Prominent examples of such what-if models are Clustervision,94 RetainVis,83 FairVis,95 etc. The fourth category consists of visualization methods developed specifically to make more useful explainability methods. SHAP is a great example, because it provides simple toolkits to visualize the results of SHAP learning explainability in an interactive computing environment like Jupyter Notebook (Figure 3B).
Three use cases for three different personas
We now will present applications of visualizations for three different kinds of researchers (Figure 6). For researchers/data scientists, we integrated a visualization tool to the previously described model-agnostic methods of ProtoDash and CEM, so that users can generate and interpret explanations for representative samples from a population dataset (Figure 6A). An example of this tool entails training a model to predict unplanned patient re-admission risks from clinical claims datasets that include millions of patients with more than 300 different features. A recurrent neural network was used to learn sequential and temporal patterns as ProtoDash generates representative examples that best summarize the complex data distribution of the population dataset. For every model decision, CEM also generates pertinent negatives and pertinent positives. In future work, we plan to implement the technique as a full-fledged interactive visualization system and also make CEM and ProtoDAsh more interactive.
For the second user type and through a collaboration with several clinical researchers, we explored what patient information needed to be collected from patient records or observational studies, to study their disease progression trajectories85 (Figure 6B). Overall, clinicians wanted to learn how patients develop diseases over a certain observed period of time and the association between these and the patient characteristics. Because discovering and summarizing trajectories is challenging, especially when they involve evolution of multi-dimensional variables across time, clinical researchers need to participate in the investigation and extraction of disease progression trajectories. We used a hidden Markov model (HMM) to summarize the states of patient’s disease status as a function of their age and other variables, to facilitate the understanding of their progression using these state sequence patterns. In the diagram in Figure 6B, a patient has four different visits where multiple laboratory test results were collected. We selected a set of variables to train the model with a parameter defining the number of states the model learns in an unsupervised manner. HMMs find some most probable state based on the time or age with the values of the selected variables that the model can assign the most probable state sequence by applying the Viterbi algorithm96 on posterior distributions over states. The model can summarize a patient’s disease development using the sequence of states, and the state can be manifested by distribution or by use over the selected variables. However, because it is not easy to interpret HMMs, clinical researchers need visual aids to drive the analysis because they want to interpret the state’s distribution across variables, gain a summary of state transition patterns or subjects, and find the associations between measures and trajectories. DPVis is designed to fulfill the needs of visual support and allows users to visually explore disease progression pathways while understanding state characteristics, build cohorts, and visually compare them. Finally clinicians also want to compare trajectories, genetic profiles, and various characteristics of patients between subgroups. Using a dataset of type 1 diabetes, we explored observational data of 559 patients who were ultimately diagnosed with this disease and observed until ages up to 20 years old. We modeled the HMM using three islet auto-antibodies (IAs) and 11-state models. The goal was to explore the heterogeneous pathways before a type 1 diabetes diagnosis with the evolution of IAs for patients. DPVis has two main views, clinical data matrix and pathway waterfall (see Figure 6B), respectively explaining the states discovered by HMMs by showing distribution of data attribute values and showing the progression patterns for all subjects in cohort. It is also possible to compare two different states based on overall distribution of data values across multiple core features. Figure 6B shows the benefit of visualization in this scenario, where clinical researchers need explanation, exploration, and interaction in order to use HMM for their clinical and analytic goals.
Finally, for clinicians, the third user type, we trained models to predict the health outcomes, such as admission, onset, or death, based on electronic health records, such as diagnosis, medication, and procedures, over time. Electronic health records are sequences over time of medical records, so it makes sense to use recurrent neural networks (RNNs) in order to take into account sequential patterns for computing outcome scores (Figure 6C). However, clinicians want to know why and how the model reached a conclusion, i.e., “what-if” questions. For instance, they ask questions, such as, What was the most influential visit? Which diagnosis was the most influential? Will the outcome be different if the patient had received testing earlier? and How can this be done for multiple patients? So, clinicians want to understand diagnostic risks predicted by the model and want to perform this on multiple patients with various conditions. For this example, we used International Classification of Diseases, Ninth Revision (ICD-9) codes from 5,962 patients’ data with heart failure pulled from health records. We used a modified attention-based RNN model, called reverse time attention model (RETAIN)97 and RETAINVis,83 to support similarity analysis (Figure 6C). “What-if” questions can be answered by editing patient visits, because medical records and update timestamps can be modified for each visit obtaining new predictions and contributions over patients’ visits. When multiple medical codes are collected, feedback can be provided so that the model can actually increase the attention scores for the selected medical codes and see how they affect the decisions for selected patients. Data values for a particular patient can be edited to conduct a what-if analysis. Visual analytic methods like RetainVis best improve explainability of RNNs through visualizations and interactivity for experiments. It is also possible to do a mix initiative of model exploration and improvement methods using humans and AI through visualizations; future work is needed to learn how to minimize false predictions and explain model failure with examples. This work needs to include clinicians for continuous model improvement while taking into account human biases and to learn how to communicate uncertainties and biases of the trained model. We also expect that tools as such, presented here, will help develop subgroup-based explainability in contrast to global/local interpretability, where the explanation is needed for a particular subset of populations (e.g., for a particular ethnic group or gender or age group). So, the explanations will be applicable for a smaller group of individuals instead of global model explanations or a single individual-based interpretation.98
Discussion
In this review, we presented popular approaches to extract explainable knowledge from the kind of black-box machine learning models that are becoming prevalent with the advent of deep learning in both life science research and more clinical-oriented environments. To help navigate the different options in XAI, we propose not only a taxonomy of available model types but also, to help fill the explainability gap, we underline the importance of having a user-centric approach to select the adequate XAI model as well as the format/visualization in which its results will be presented. The purpose was not to perform an exhaustive description of explainability99 but to describe practical uses with examples in life sciences and a focus on user-centered XAI in clinical use cases related to healthcare around the concept of persona.
Although such methods can lead to an increased trust in using AI applications, recent literature also provides critiques of certain class of XAI methods41 as well as how their improper usage can lead to unwarranted trust in underlying AI systems where such methods may not be applicable.54,55 Indeed, although the state of the art changes rapidly and new neural network architectures are been developed to be directly interpretable by design,100 most XAI methods are fundamentally post hoc in nature and perform a global explanation through the aggregation of local ones. Hence, XAI methods cannot compete with directly interpretable methods, where the dependency between the input variables and output predictions is global and transparent. Similarly, visualization in itself cannot be the only method to gather explanations; we advocate that this in conjunction with other dimensions/tools for explainability do indeed form a useful system for the end user to iteratively improve their understanding. Finally, we also think that a back-and-forth argument relative to whether an explanation is sufficient or satisfactory, i.e., interpretable, should be framed in the context of the development of quantitative metrics of XAI to allow direct comparison of different explanations. This has already been done in the context of model selection101,102 and model comparison.103
Conclusions
In life sciences, the open discussion on the explainability and interpretation of innovative black-box solutions has progressed with the popularity of these approaches but in part also through the development of crowdsourcing platforms and competitions.104 For the former, it has happened through the “wisdom of the crowd” solutions that ensure not only the most robust and often the best prediction but also unearth the most frequently used features for prediction present in the datasets, thus generating confidence and global explainability of the prediction results.1 Both approaches help fill the gap between model-directed explainability and user-dependent interpretability because they also focus on communities of interest that have specific domain knowledge. In the healthcare domain, explainability can help generate trust in AI services used, complemented by the implementation of general safety and reliability engineering methodologies, with identification of new AI specific issues and challenges and transparent reporting mechanisms on how services operate and perform. This has to be coupled with stringent tests regarding whether the XAI is robust to variations of inputs or with additional data and for some higher-risk applications with well-designed clinical trials. For now, in research, the same type of procedures also needs to be implemented, i.e., using a variety and diversity of datasets, with the sole goal of generating trust in the knowledge generated by the XAI. The ultimate goal of adopting AI in medical practice and patient care goes beyond explainability and will need the development of extra layers of security and confidence, in particular regarding AI trustworthiness, as XAI transparent systems become prone to attacks that may reveal confidential information, and AI fairness, as systems developed and tested, in particular socio-economic/racial environments, need to be expanded to real-world situations.105,106
Acknowledgments
We thank the editor and reviewers for their useful comments.
Declaration of interests
The authors declare no competing interests.
Biography
About the author
Pablo Meyer is manager of the Biomedical Analytics and Modeling group at IBM research and director of DREAM Challenges, and, as such, has been working on developing models and applying AI to biological/biomedical problems, Systems Biology, Olfaction, and disease. He joined IBM research in 2010; he received his undergraduate degree in Physics from the Universidad Nacional Autonoma de Mexico (UNAM) (2000), master’s degree from the University of Paris VII/XI, and Ph.D. in genetics from Rockefeller University (2005). As director of DREAM Challenges, he looks for including algorithmic and artificial intelligence solutions in Systems Biology/biomedical problems via crowd-sourced competitions.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2022.100493.
Supplemental information
References
- 1.Meyer P., Saez-Rodriguez J. Advances in systems biology modeling: 10 years of crowdsourcing dream challenges. Cell Syst. 2021;12:636–653. doi: 10.1016/j.cels.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with alphafold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.M., Zietz M., Hoffman M.M., et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15:20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson N., Shane E., Chase J., Rowland J., Ries D., Justice N., Zhang J., Chan L., Cao R. Survey of machine learning techniques in drug discovery. Curr. Drug Metab. 2019;20:185–193. doi: 10.2174/1389200219666180820112457. [DOI] [PubMed] [Google Scholar]
- 5.Erickson B.J., Korfiatis P., Akkus Z., Kline T.L. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisaso K.R., Anguzu G.T., Karungi S.A., Kiragga A., Castelnuovo B. A survey of machine learning applications in HIV clinical research and care. Comput. Biol. Med. 2017;91:366–371. doi: 10.1016/j.compbiomed.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium The encode (encyclopedia of dna elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 8.Tomczak K., Czerwińska P., Wiznerowicz M. The cancer genome atlas (tcga): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson A.E., Pollard T.J., Shen L., Lehman L.W., Feng M., Ghassemi M., Moody B., Szolovits P., Celi L.A., Mark R.G. MIMIC-III, a freely accessible critical care database. Sci. Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 12.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly C.J., Karthikesalingam A., Suleyman M., Corrado G., King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. doi: 10.1186/s12916-019-1426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zech J.R., Badgeley M.A., Liu M., Costa A.B., Titano J.J., Oermann E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordling L. A fairer way forward for AI in health care. Nature. 2019;573:S103–S105. doi: 10.1038/d41586-019-02872-2. [DOI] [PubMed] [Google Scholar]
- 16.Oh S.S., Galanter J., Thakur N., Pino-Yanes M., Barcelo N.E., White M.J., de Bruin D.M., Greenblatt R.M., Bibbins-Domingo K., Wu A.H., et al. Diversity in clinical and biomedical research: a promise yet to Be fulfilled. PLoS Med. 2015;12:e1001918. doi: 10.1371/journal.pmed.1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller T. Explanation in artificial intelligence: insights from the social sciences. Artif. Intell. 2019;267:1–38. [Google Scholar]
- 18.Arya V., Bellamy R.K.E., Chen P.Y., Dhurandhar A., Hind M., Hoffman S.C., Houde S., Vera Liao Q., Luss R., Mojsilović A. One explanation does not fit all: a toolkit and taxonomy of ai explainability techniques. Preprint at arXiv. 2019 1909.03012. [Google Scholar]
- 19.Jain S., Wallace B.C. Vol. 1. Long and Short Papers; 2019. Attention is not explanation; pp. 3543–3556. (Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies). [Google Scholar]
- 20.Wiegreffe S., Pinter Y. EMNLP-IJCNLP; 2019. Attention is not not explanation; pp. 11–20. (Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing). [Google Scholar]
- 21.Lundberg S.M., Lee S.I. In: Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R., editors. Vol. 30. 2017. A unified approach to interpreting model predictions; pp. 4765–4774. (Advances in Neural Information Processing Systems). [Google Scholar]
- 22.Kumar I.E., Suresh V., Scheidegger C., Friedler S. International Conference on Machine Learning. PMLR; 2020. Problems with shapley-value-based explanations as feature importance measures; pp. 5491–5500. [Google Scholar]
- 23.Nest C. Google’s ai guru wants computers to think more like brains. 2018. https://www.wired.com/story/googles-ai-guru-computers-think-more-like-brains
- 24.Lakkaraju H., Bastani O. Proceedings of the AAAI/ACM Conference on AI. Ethics, and Society; 2020. “How do i fool you?” manipulating user trust via misleading black box explanations; pp. 79–85. [Google Scholar]
- 25.Hind M. Explaining explainable ai. XRDS: Crossroads, ACM Mag. Students. 2019;25:16–19. [Google Scholar]
- 26.Lipton Z.C. The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery. Queue. 2018;16:31–57. [Google Scholar]
- 27.Liao Q.V., Gruen D., Miller S. Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems. 2020. Questioning the ai: informing design practices for explainable ai user experiences; pp. 1–15. [Google Scholar]
- 28.Chari S., Seneviratne O., Gruen D.M., Foreman M.A., Das A.K., McGuinness D.L. International Semantic Web Conference. Springer; 2020. Explanation ontology: a model of explanations for user-centered ai; pp. 228–243. [Google Scholar]
- 29.Chari S., Seneviratne O., Gruen D.M., Foreman M.A., Das A.K., McGuinness D.L. Explanation ontology in action: a clinical use-case. Preprint at arXiv. 2020 2010.01478. [Google Scholar]
- 30.Arrieta A.B., Díaz-Rodríguez N., Del Ser J., Bennetot A., Tabik S., Barbado A., García S., Gil-López S., Molina D., Benjamins R., et al. Explainable artificial intelligence (xai): concepts, taxonomies, opportunities and challenges toward responsible ai. Inf. Fusion. 2020;58:82–115. [Google Scholar]
- 31.Linardatos P., Papastefanopoulos V., Kotsiantis S. Explainable ai: a review of machine learning interpretability methods. Entropy. 2021;23:18. doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A., Sattigeri P., Balakrishnan A. Variational inference of disentangled latent concepts from unlabeled observations. Preprint at arXiv. 2017 1711.00848. [Google Scholar]
- 33.Gurumoorthy K.S., Dhurandhar A., Cecchi G., Aggarwal C. 2019 IEEE International Conference on Data Mining (ICDM) IEEE; 2019. Efficient data representation by selecting prototypes with importance weights; pp. 260–269. [Google Scholar]
- 34.Hind M., Wei D., Campbell M., Codella N.C.F., Dhurandhar A., Mojsilović A., Ramamurthy K.N., Varshney K.R. Proceedings of the 2019 AAAI/ACM Conference on AI. Ethics, and Society; 2019. Ted: Teaching ai to explain its decisions; pp. 123–129. [Google Scholar]
- 35.Dash S., Günlük O., Wei D. Boolean decision rules via column generation. Preprint at arXiv. 2018 1805.09901. [Google Scholar]
- 36.Dhurandhar A., Chen P.Y., Luss R., Tu C.-C., Ting P., Shanmugam K., Das P. Explanations based on the missing: towards contrastive explanations with pertinent negatives. Preprint at arXiv. 2018 1802.07623. [Google Scholar]
- 37.Ribeiro M.T., Singh S., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016. Why should i trust you?” explaining the predictions of any classifier; pp. 1135–1144. [Google Scholar]
- 38.Wei D., Dash S., Gao T., Gunluk O. International Conference on Machine Learning. PMLR; 2019. Generalized linear rule models; pp. 6687–6696. [Google Scholar]
- 39.Wang T., Rudin C., Doshi-Velez F., Liu Y., Klampfl E., Perry M.N. A bayesian framework for learning rule sets for interpretable classification. J. Machine Learn. Res. 2017;18:2357–2393. [Google Scholar]
- 40.Razavian N., Blecker S., Schmidt A.M., Smith-McLallen A., Nigam S., Sontag D. Population-level prediction of type 2 diabetes from claims data and analysis of risk factors. Big Data. 2015;3:277–287. doi: 10.1089/big.2015.0020. [DOI] [PubMed] [Google Scholar]
- 41.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Machine Intell. 2019;1:206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ustun B., Rudin C. Supersparse linear integer models for optimized medical scoring systems. Machine Learn. 2016;102:349–391. [Google Scholar]
- 43.Ustun B., Rudin C. Learning optimized risk scores. J. Mach. Learn. Res. 2019;20:150–151. [Google Scholar]
- 44.Xie F., Chakraborty B., Ong M.E.H., Goldstein B.A., Liu N. Autoscore: a machine learning–based automatic clinical score generator and its application to mortality prediction using electronic health records. JMIR Med. Inform. 2020;8:e21798. doi: 10.2196/21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kodialam R., Boiarsky R., Lim J., Sai A., Dixit N., Sontag D. Vol. 35. 2021. Deep contextual clinical prediction with reverse distillation; pp. 249–258. (Proceedings of the AAAI Conference on Artificial Intelligence). [Google Scholar]
- 46.Liu N., Hu Q., Xu H., Xu X., Chen M. Med-bert: a pre-training framework for medical records named entity recognition. IEEE Trans. Ind. Inform. 2021 [Google Scholar]
- 47.Zeiler M.D., Fergus R. In: Computer Vision – ECCV 2014. Fleet D., Pajdla T., Schiele B., Tuytelaars T., editors. Springer International Publishing; 2014. Visualizing and understanding convolutional networks; pp. 818–833. [Google Scholar]
- 48.Krause J., Adam P., Ng K. Proceedings of the 2016 CHI conference on human factors in computing systems. 2016. Interacting with predictions: visual inspection of black-box machine learning models; pp. 5686–5697. [Google Scholar]
- 49.Hohman F., Head A., Caruana R., DeLine R., Drucker S.M. Proceedings of the 2019 CHI conference on human factors in computing systems. 2019. Gamut: a design probe to understand how data scientists understand machine learning models; pp. 1–13. [Google Scholar]
- 50.Molnar C. Lulu. com; 2020. Interpretable Machine Learning. [Google Scholar]
- 51.Cutillo C.M., Sharma K.R., Foschini L., Kundu S., Mackintosh M., Mandl K.D. Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. npj Digital Med. 2020;3:47. doi: 10.1038/s41746-020-0254-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunning D., Stefik M., Choi J., Miller T., Stumpf S., Yang G.Z. XAI-Explainable artificial intelligence. Sci. Robot. 2019;4:eaay7120. doi: 10.1126/scirobotics.aay7120. [DOI] [PubMed] [Google Scholar]
- 53.Tonekaboni S., Joshi S., McCradden M.D., Goldenberg A. What clinicians want: contextualizing explainable machine learning for clinical end Use. Preprint at arXiv. 2019 1905.05134:21. [Google Scholar]
- 54.Ghassemi M., Oakden-Rayner L., Beam A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digital Health. 2021;3:e745–e750. doi: 10.1016/S2589-7500(21)00208-9. [DOI] [PubMed] [Google Scholar]
- 55.Babic B., Gerke S., Evgeniou T., Cohen I.G. Beware explanations from ai in health care. Science. 2021;373:284–286. doi: 10.1126/science.abg1834. [DOI] [PubMed] [Google Scholar]
- 56.factsheet. http://aifs360.mybluemix.net/
- 57.Doshi-Velez, Kim B. Towards a rigorous science of interpretable machine learning. Preprint at arXiv. 2017 1702.08608. [Google Scholar]
- 58.Chakraborty P., Kwon B.C., Dey S., Dhurandhar A., Gruen D., Ng K., Sow D., Varshney K.R. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020. Tutorial on human-centered explainability for healthcare; pp. 3547–3548. [Google Scholar]
- 59.Patel V.L., Arocha J.F., Zhang J. Vol. 14. Cambridge University Press; 2005. Thinking and reasoning in medicine; pp. 727–750. (The Cambridge Handbook of Thinking and Reasoning). [Google Scholar]
- 60.Smucny J., Davidson I., Carter C.S. Comparing machine and deep learning-based algorithms for prediction of clinical improvement in psychosis with functional magnetic resonance imaging. Hum. Brain Mapp. 2021;42:1197–1205. doi: 10.1002/hbm.25286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chauhan D., Anyanwu E., Goes J., Besser S.A., Anand S., Madduri R., Getty N., Kelle S., Kawaji K., Mor-Avi V., et al. Comparison of machine learning and deep learning for view identification from cardiac magnetic resonance images. Clin. Imag. 2022;82:121–126. doi: 10.1016/j.clinimag.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park Y.R., Kim Y.J., Ju W., Nam K., Kim S., Kim K.G. Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-95748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Severson K.A., Chahine L.M., Smolensky L., Ng K., Hu J., Ghosh S. Machine Learning for Healthcare Conference. PMLR; 2020. Personalized input-output hidden Markov models for disease progression modeling; pp. 309–330. [Google Scholar]
- 64.Ribeiro M.T., Singh S., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, August 13-17, 2016. 2016. why should I trust you?”: explaining the predictions of any classifier; pp. 1135–1144. [Google Scholar]
- 65.Dey S., Bose A., Chakraborty P., Ghalwash M., Saenz A.G., Ultro F., NG K., Hu J., Parida L., Sow D. Impact of clinical and genomic factors on sars-cov2 disease severity. Preprint at medRxiv. 2021 [PMC free article] [PubMed] [Google Scholar]
- 66.Gutiérrez E.D., Dhurandhar A., Keller A., Meyer P., Cecchi G.A. Predicting natural language descriptions of mono-molecular odorants. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-07439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shrikumar A., Greenside P., Kundaje A. International Conference on Machine Learning. PMLR; 2017. Learning important features through propagating activation differences; pp. 3145–3153. [Google Scholar]
- 68.Bach S., Binder A., Montavon G., Klauschen F., Müller K.R., Samek W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PloS One. 2015;10:e0130140. doi: 10.1371/journal.pone.0130140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipovetsky S., Conklin M. Analysis of regression in game theory approach. Appl. Stoch Model Bus. Ind. 2001;17:319–330. [Google Scholar]
- 70.Marketscan Dataset Truven. https://www.ibm.com/products/marketscan-research-databases
- 71.Dhurandhar A., Chen P.Y., Luss R., Tu C.C., Ting P., Shanmugam K., Das P. Explanations based on the missing: towards contrastive explanations with pertinent negatives. NIPS'18: Proceedings of the 32nd International Conference on Neural Information Processing Systems December 2018. 2018:590–601. [Google Scholar]
- 72.Dhurandhar A., Pedapati T., Balakrishnan A., Chen P.Y., Shanmugam K., Puri R. Model agnostic contrastive explanations for structured data. Preprint at arXiv. 2019 1906.00117. [Google Scholar]
- 73.Hinton G., Vinyals O., Dean J. Distilling the knowledge in a neural network. Preprint at arXiv. 2015 1503.02531. [Google Scholar]
- 74.Bastani O., Kim C., Bastani H. Interpreting blackbox models via model extraction. Preprint at arXiv. 2017 1705.08504. [Google Scholar]
- 75.Dhurandhar A., Shanmugam K., Luss R. International Conference on Machine Learning. PMLR; 2020. Enhancing simple models by exploiting what they already know; pp. 2525–2534. [Google Scholar]
- 76.Bommasani R., Hudson D.A., Adeli E., Altman R., Arora S., von Arx S., Bernstein M.S., Bohg J., Bosselut A., Brunskill E., et al. On the opportunities and risks of foundation models. Preprint at arXiv. 2021 2108.07258. [Google Scholar]
- 77.Buciluǎ C., Caruana R., Niculescu-Mizil A. Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining. 2006. Model compression; pp. 535–541. [Google Scholar]
- 78.Dhurandhar A., Shanmugam K., Luss R., Olsen P. Improving simple models with confidence profiles. Adv. Neural Inf. Process. Syst. 2018 [Google Scholar]
- 79.Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.I. From local explanations to global understanding with explainable ai for trees. Nat. Machine Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karthikeyan Natesan Ramamurthy. Vinzamuri B., Zhang Y., Dhurandhar A. Model agnostic multilevel explanations. Preprint at arXiv. 2020 2003.06005. [Google Scholar]
- 81.Pedapati T., Balakrishnan A., Shanmugam K., Dhurandhar A. Learning global transparent models consistent with local contrastive explanations. Preprint at arXiv. 2020 2002.08247. [Google Scholar]
- 82.Daniel S.W., Bansal G. The challenge of crafting intelligible intelligence. Commun. ACM. 2019;62:70–79. [Google Scholar]
- 83.Kwon B.C., Choi M.J., Kim J.T., Choi E., Kim Y.B., Kwon S., Sun J., Choo J. RetainVis: visual analytics with interpretable and interactive recurrent neural networks on electronic medical records. IEEE Trans. Visual. Comput. Graph. 2019;25:299–309. doi: 10.1109/TVCG.2018.2865027. [DOI] [PubMed] [Google Scholar]
- 84.Kwon B.C., Chakraborty P., CodellaJ., DhurandharA., Sow D., and Ng K. Visually exploring contrastive explanation for diagnostic risk prediction on electronic health records. ICML 2020 Workshop on Human Interpretability in Machine Learning.
- 85.Kwon B.C., Anand V., Severson K.A., Ghosh S., Sun Z., Frohnert B.I., Lundgren M., Ng K. Visual analytics with hidden Markov models for disease progression pathways. IEEE Trans. Visual. Comput. Graph. 2021;27:3685–3700. doi: 10.1109/TVCG.2020.2985689. [DOI] [PubMed] [Google Scholar]
- 86.A Visual Introduction to Machine Learning. http://www.r2d3.us/visual-intro-to-machine-learning-part-1/
- 87.Generative adversarial networks (gans) in your browser! https://poloclub.github.io/ganlab/
- 88.Learn Convolutional Neural Network (Cnn) in Your Browser! https://poloclub.github.io/cnn-explainer/
- 89.Strobelt H., Gehrmann S., Pfister H., Rush A.M. A tool for visual analysis of hidden state dynamics in recurrent neural networks. IEEE Trans. Visual. Comput. Graph. 2017;24:667–676. doi: 10.1109/TVCG.2017.2744158. [DOI] [PubMed] [Google Scholar]
- 90.Liu M., Shi J., Li Z., Li C., Zhu J., Liu S. Towards better analysis of deep convolutional neural networks. IEEE Trans. Visual. Comput. Graph. 2016;23:91–100. doi: 10.1109/TVCG.2016.2598831. [DOI] [PubMed] [Google Scholar]
- 91.Strobelt H., Gehrmann S., Behrisch M., Perer A., Pfister H., Rush A.M. S eq 2s eq-v is: a visual debugging tool for sequence-to-sequence models. IEEE Trans. Visual. Comput. Graph. 2018;25:353–363. doi: 10.1109/TVCG.2018.2865044. [DOI] [PubMed] [Google Scholar]
- 92.Park C., Inyoup N., Jo Y., Shin S., Yoo J., Kwon B.C., Zhao J., Noh H., Lee Y., Choo J. 2019 IEEE Visualization Conference (VIS) 2019. Sanvis: visual analytics for understanding self-attention networks; pp. 146–150. [Google Scholar]
- 93.Wexler J., Pushkarna M., Bolukbasi T., Wattenberg M., Viegas F., Wilson J. The what-if tool: interactive probing of machine learning models. IEEE Trans. Visual. Comput. Graph. 2019;26:56–65. doi: 10.1109/TVCG.2019.2934619. [DOI] [PubMed] [Google Scholar]
- 94.Kwon B.C., Eysenbach B., Verma J., Ng K., De Filippi C., Stewart W.F., Perer A. Clustervision: visual supervision of unsupervised clustering. IEEE Trans. Visual. Comput. Graph. 2018;24:142–151. doi: 10.1109/TVCG.2017.2745085. [DOI] [PubMed] [Google Scholar]
- 95.Alexander Cabrera Á., Epperson W., Hohman F., Kahng M., Morgenstern J., Chau D.H. 2019 IEEE Conference on Visual Analytics Science and Technology (VAST) 2019. Fairvis: visual analytics for discovering intersectional bias in machine learning; pp. 46–56. [Google Scholar]
- 96.Forney G.D. The viterbi algorithm. Proc. IEEE. 1973;61:268–278. [Google Scholar]
- 97.Choi E., Bahadori M.T., Kulas J.A., Schuetz A., Stewart W.F., Sun J. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. Preprint at arXiv. 2016 1608.05745. [Google Scholar]
- 98.Dey S., Cooner J., Delaney C.W., Fakhoury J., Kumar V., Simon G., Steinbach M., Weed J., Westra B.L. Mining patterns associated with mobility outcomes in home healthcare. Nurs. Res. 2015;64:235–245. doi: 10.1097/NNR.0000000000000106. [DOI] [PubMed] [Google Scholar]
- 99.Chari S., Gruen D., Seneviratne O.W., McGuinness D.L. Vol. 47. IOS Press; 2020. Directions for explainable knowledge-enabled systems; p. 245. (Knowledge Graphs for eXplainable Artificial Intelligence). [Google Scholar]
- 100.Puri I., Dhurandhar A., Pedapati T., Shanmugam K., Wei D., Varshney K.R. Advances in Neural Information Processing Systems. 2021. CoFrNets: interpretable neural architecture inspired by continued fractions. [Google Scholar]
- 101.Akaike H. A new look at the statistical model identification. IEEE Trans. Automatic Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 102.Schwarz G. The annals of statistics; 1978. Estimating the Dimension of a Model; pp. 461–464. [Google Scholar]
- 103.Vittadello S.T., Stumpf M.P.H. Model comparison via simplicial complexes and persistent homology. Preprint at arXiv. 2020 doi: 10.1098/rsos.211361. 2012.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saez-Rodriguez J., Costello J.C., Friend S.H., Kellen M.R., Mangravite L., Meyer P., Norman T., Stolovitzky G. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat. Rev. Genet. 2016;17:470–486. doi: 10.1038/nrg.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang F., Preininger A. Ai in health: state of the art, challenges, and future directions. Yearb. Med. Inform. 2019;28:016–026. doi: 10.1055/s-0039-1677908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eshete B. Making machine learning trustworthy. Science. 2021;373:743–744. doi: 10.1126/science.abi5052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.