Abstract
The frequency of isolation of three nonhomologous chlorobenzoate catabolic genotypes (clc, cba, and fcb) was determined for 464 isolates from freshwater sediments and groundwater in the vicinity of the Hyde Park industrial landfill site in the Niagara watershed. Samples were collected from both contaminated and noncontaminated sites during spring, summer, and fall and enriched at 4, 22, or 32°C with micromolar to millimolar concentrations of chlorobenzoates and 3-chlorobiphenyl (M. C. Peel and R. C. Wyndham, Microb. Ecol: 33:59–68, 1997). Hybridization at moderate stringency to restriction-digested genomic DNA with DNA probes revealed the chlorocatechol 1,2-dioxygenase operon (clcABD), the 3-chlorobenzoate 3,4-(4,5)-dioxygenase operon (cbaABC), and the 4-chlorobenzoate dehalogenase (fcbB) gene in isolates enriched from all contaminated sites in the vicinity of the industrial landfill. Nevertheless, the known genes were found in less than 10% of the isolates from the contaminated sites, indicating a high level of genetic diversity in the microbial community. The known genotypes were not enriched from the noncontaminated control sites nearby. The clc, cba, and fcb isolates were distributed across five phenotypically distinct groups based on Biolog carbon source utilization, with the breadth of the host range decreasing in the order clc > cba > fcb. Restriction fragment length polymorphism (RFLP) patterns showed that the cba genes were conserved in all isolates whereas the clc and fcb genes exhibited variation in RFLP patterns. These observations are consistent with the recent spread of the cba genes by horizontal transfer as part of transposon Tn5271 in response to contaminant exposure at Hyde Park. Consistent with this hypothesis, IS1071, the flanking element in Tn5271, was found in all isolates that carried the cba genes. Interestingly, IS1071 was also found in a high proportion of isolates from Hyde Park carrying the clc and fcb genes, as well as in type strains carrying the clcABD operon and the biphenyl (bph) catabolic genes.
Metabolic redundancy in bacteria is a common feature of microbial ecosystems. For example, multiple biochemical pathways encoded by genetically divergent operons may function in the degradation of single, relatively simple organic compounds. Among the aerobic members of the class Proteobacteria alone, toluene degradation is initiated by a range of dioxygenase (tod) and monooxygenase (xyl, tmo, tbm, tbu, and tou) catabolic operons (2, 5, 15, 33, 45). Two or more of these operons may be expressed in a single strain (16). Similarly, two nonhomologous phenol degradation operons occur in aerobic members of the Proteobacteria: the multicomponent phenol hydroxylases represented by the dmp operon (30, 31) and the single-component phenol hydroxylases represented by the pheA or tbuD genes (20, 22). For chlorobenzoic acid (CBA) degradation, there are at least three distinct biochemical pathways found in isolates collected from around the world (Fig. 1) (9, 10, 27, 29, 37, 42). The clc, cba, and fcb operons are nonhomologous and are found at a variety of different loci including chromosomes, plasmids, and transposons in different bacteria (8, 25, 32, 41, 46). A fourth pathway via gentisate has recently been characterized at the biochemical level in Alcaligenes sp. strain L6 (21).
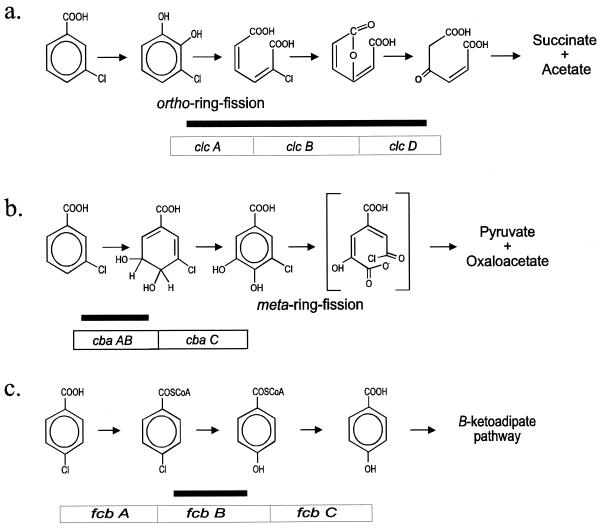
FIG. 1.
Representative metabolic pathways encoded by the nonhomologous CBA-degradative operons clcABD, cbaABC, and fcbBAC. The genes that make up these operons are indicated in open boxes below the metabolic steps they encode. Schematic representations of the locations of the three DNA probes used in this study, with respect to the operons, are shown as black bars. (a) Chlorocatechol ortho-ring-fission pathway. Following initial attack by a nonspecific dioxygenase such as the benzoate (benABC) or toluate (xylXYZ) dioxygenase and dihydrodiol dehydrogenase to form chlorocatechols, the clcABD gene products catalyze ortho-ring fission to open the aromatic ring and continue metabolism. (b) CBA 3,4-(4,5)-dioxygenase pathway. The cbaABC genes encode dioxygenase, reductase, and dehydrogenase enzymes that initiate 3-CBA and 3,4-DCBA (not shown) degradation. Protocatechuic acid is also formed in this pathway. The host’s protocatechuate 4,5-dioxygenase (meta-ring-fission) enzymes then complete the pathway. (c) 4-CBA dehalogenase pathway. The fcbA gene encodes a 4-CBA-CoA ligase; fcbB encodes the dehalogenase; fcbC encodes a 4-hydroxybenzoate-CoA thioesterase. Note that the gene order in the control strain Pseudomonas sp. strain CBS-3 is fcbBAC.
While redundancy of metabolic pathways in microbial ecosystems is recognized, few studies have been designed to show whether pollutants in natural environments select a single, dominant genotype or multiple, redundant genotypes. In addition, we know little about how genotypes found at polluted sites differ from those that occur at pristine sites. These were questions we addressed in this study of the selection of clc, cba, and fcb genotypes at the Hyde Park chemical landfill site. In recent studies of the genotypes responsible for 2,4-dichlorophenoxyacetic acid (2,4-D) degradation in 2,4-D-amended soils, it was found that selection of divergent families of 2,4-D-α-ketoglutarate dioxygenase (tfdA) genes occurred (12, 17, 24). tfdA gene families I and III, representative of the Sphingomonas and broad-host-range pJP4-like operons, respectively, were most commonly selected following long-term 2,4-D exposure. In other work, the distribution of the nonhomologous phenol hydroxylase genes was studied in isolates from freshwater and marine samples (31, 36). Genes homologous to the multicomponent phenol hydroxylase dmp operon were most often detected in isolates from both of these environments. The single-component phenol hydroxylase operon pheBA, which is part of a composite transposon structure, occurred rarely. This property allowed Peters et al. (36) to infer that pheBA had transferred horizontally between bacteria in river water following the introduction of this genotype for bioremediation of phenol contaminants.
In recent studies of enrichments from pristine soils, it was found that the ability to degrade 3-chlorobenzoate (3-CBA) was widespread and that the predominant pathway was via chlorocatechol intermediates (12, 13, 23). Nevertheless, genomic DNA of isolates from these pristine soils failed to hybridize with DNA probes derived from two of the known CBA catabolic operons (clc and cba). Studies using gene probes representative of the range of known CBA metabolic pathways have not been done on samples from environments chronically contaminated by CBA. This is surprising given the central role of CBA degradation in the aerobic metabolism of polychlorinated biphenyls. In a previous study, we determined the 3-CBA, 4-CBA, and 3,4-dichlorobenzoate (3,4-DCBA) degradation potentials of contaminated and noncontaminated freshwater sediment samples taken from three streams at the Hyde Park, Niagara Falls, chemical landfill site (34, 35). The previous study showed that contaminants leaching from the site, which include CBA congeners, chlorobiphenyls (CBPs), chlorinated hydrocarbons, and phenols, have significantly increased the CBA degradation potential relative to that of a noncontaminated control stream nearby. The previous study also revealed seasonal and year-to-year variation in degradation potentials. In the study reported here, we used the same sites to determine the incidence of the clcABD, cbaABC, and fcbB catabolic genotypes among isolates enriched from samples of both contaminated and noncontaminated sources at Hyde Park. Genetic variation at the clc, cba, and fcb loci; the incidence of IS1071, which flanks the cba operon; and phenotypic relationships between the hosts of these genes were determined.
MATERIALS AND METHODS
Isolation of chlorobenzoate-degrading bacteria.
Enrichment cultures were previously described (35). Briefly, samples were taken from the sediment-water interface of several creeks and groundwater seeps in the Niagara watershed, including Bloody Run Creek and Devil’s Hole Creek (both contaminated by Hyde Park leachate) and Fish Creek (a noncontaminated control). Samples from groundwater wells within the perimeter trench of the Hyde Park landfill site and from the sequencing batch reactors used for aerobic bioremediation of the groundwater were provided by the owners (Occidental Chemical Co., Niagara Falls, N.Y.). The Hyde Park site and the content of chlorinated aromatics including CBPs and chlorobenzoates in the groundwater leachate have been described previously (34, 35, 49). Subsamples of 950 μl of suspended sediment were enriched in 1-ml batch cultures initially containing 10 μM substrate, followed by sequential transfers to 100 μM, 600 μM, 1.2 mM, and 1.5 mM 3-CBA, 4-CBA, 3,4-DCBA, or 3-CBP in minimal medium A (35). Each enrichment was shaken for 14 days at 4°C, 10 days at 22°C, or 8 days at 32°C, followed by transfer of 500 μl to the next highest substrate concentration. The 1.5 mM enrichments were serially diluted; spread onto minimal medium A plates containing 1.5 mM appropriate CBA congener (3-CBA for the 3-CBP enrichments) plus 1.6% agar (Difco Bacto Agar); and incubated for 8, 10, and 14 days at 32, 22, or 4°C, respectively. All enrichment culture dilutions were also plated onto medium A without a carbon source to distinguish false-positive growth on agar impurities from growth on CBA. Isolated colonies were transferred on selection plates to ensure purity and then were confirmed to degrade the CBA congeners by high-pressure liquid chromatography as previously described (35). A 20% glycerol stock suspension of each purified enrichment culture was stored at −70°C and used as the source of inoculum in all subsequent experiments.
Genomic DNA and plasmid isolation.
Table 1 lists the bacterial strains and plasmids used in this study. The genotype screening design was based on previous knowledge of the range of CBA congeners degraded by the control strains and their homologues (Table 1) (9, 10, 37, 42). Isolates from all CBA enrichments were screened for the cbaABC and clcABD genotypes, but only the 4-CBA isolates were screened for the fcbB genotype. The 464 CBA-degrading isolates in the collection were pooled in groups of 10 for genotype screening. A single colony from a selection plate of each isolate was resuspended in 5 ml of tryptone-yeast extract broth (1% tryptone, 1% yeast extract, 0.5% NaCl) along with colonies of nine other isolates. The cell suspensions were grown overnight on a shaker-incubator at the original isolation temperature. Genomic DNA was extracted from the pooled cells by using a 10-times-scaled-up version for DNA isolation as described previously (3). Positive hybridization to the pooled genomic DNAs (see below) was followed by isolation of total genomic DNA from each isolate by the same protocol. The methods of extracting pooled isolate genomic DNAs followed by probe hybridization were validated with the control Comamonas testosteroni BR60 and by randomly screening for false negatives among the isolates.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Plasmid and/or phenotypesa | Genotypeb | Reference(s) |
|---|---|---|---|
| E. coli | |||
| DH1 | pDC100 Apr | clcABD | 11 |
| JM109 | pBRH4 Apr | IS1071 | 25, 27–29 |
| JM109 | pBRH2 Apr | cbaABC | 25, 27–29 |
| JM105 | pHUG01 Apr Smr Sur | fcbBAC | 4 |
| Pseudomonas sp. | |||
| CBS3 | 4-CBA+ | fcbBAC | 9 |
| AC866 | pAC27 3-CBA+ 4-CBA+ | clcABD | 11 |
| B356 | BPH+ CBP+ | bph | 14 |
| Burkholderia sp. strain LB400 | BPH+ CBP+ | bph clc | 14 |
| Alcaligenes sp. strain H850 | BPH+ CBP+ | bph | 14 |
| Comamonas sp. strain BR60c | pBRC60 3-CBA+ 3,4-DCBA+ | cbaABC | 25, 27–29 |
Resistance phenotypes: Apr, ampicillin resistance; Smr, streptomycin resistance; Sur, sulfathiazole resistance; growth phenotypes: 3-CBA+, 3-CBA; 4-CBA+, 4-CBA; 3,4-DCBA+, 3,4-DCBA; BPH+, biphenyl; CBP+, polychlorinated biphenyls.
Genotypes: clc, chlorocatechol ortho-ring-fission pathway genes; IS1071, insertion sequence flanking Tn5271; cba, chlorobenzoate 3,4-(4,5)-dioxygenase pathway; fcb, 4-CBA dehalogenase pathway; bph, biphenyl dioxygenase pathway.
The identification of Alcaligenes sp. strain BR60 has been revised to C. testosteroni BR60 based on 100% sequence identity of the 16S ribosomal DNA gene to that of the type strain.
Two plasmid isolation methods were used to screen for high-molecular-weight plasmids in all isolates carrying the known genes, a standard alkaline lysis method (6) and a modified alkaline lysis method for high-molecular-weight plasmids (47). Plasmid DNA from isolates and control strains was detected by electrophoresis in 0.65% agarose at 80 V.
Detection of sibling species with ERIC PCR.
Genomic DNA was obtained from all isolates that hybridized with the clc, cba, or fcb probes (see below) as described above. PCR was used to amplify genomic DNA fragments from 50 ng of purified genomic DNA for each isolate by using the enterobacterial repetitive intergenic consensus (ERIC) sequence primers and following the ERIC PCR conditions specified previously (44). Vent polymerase and 10× Vent buffer (New England Biolabs Canada, Mississauga, Ontario, Canada), deoxynucleoside triphosphates (Boehringer Mannheim Canada, Montreal, Canada), and primers (University of Ottawa, Biotechnology Research Institute, Ottawa, Canada) were used with a Perkin-Elmer Cetus 480 Thermocycler (Foster City, Calif.). After PCR amplification, the products (10 μl) were separated on 1.5% agarose gels with Tris-acetate-EDTA buffer at 105 V for 8 h, stained in ethidium bromide, and photographed under UV illumination. Genomic DNAs from all clc, cba, and fcb isolates were subjected to ERIC PCR amplification two or three times to ensure a reproducible pattern of fragment sizes. Isolates with similar ERIC PCR fragment sizes were considered to be sibling species. All siblings isolated from different sites and at different times were included in the isolate collection as independent isolates. One sibling of each pair was selected at random for inclusion in the phenotype analysis (see below).
Probe preparation.
Plasmid DNA for probe preparation was isolated by alkaline lysis (6). The three probes were prepared from clcABD genes carried on a 4.2-kb EcoRI fragment of pDC100 (corresponding to the original BglII fragment E of pAC27) (11), cbaABC genes carried on the 1.1- plus 1.4-kb EcoRI fragments (pooled) of pBRH2 (27–29), and the 4-CBA coenzyme A (CoA) dehalogenase (fcbB) gene carried on a 1.6-kb SacI-to-SalI fragment of pHUG01 (kindly provided by M. Sylvestre [4]). The fcbB gene alone, rather than the operon, was used as a probe because of the broad substrate specificity of the ligase (fcbA) and thioesterase (fcbC) functions (4, 9).
The probe DNA fragments were separated by electrophoresis on 1.0% low-melting-temperature agarose gels (FMC Bioproducts, Rockland, Maine) submerged in Tris-acetate-EDTA buffer and purified with glass milk according to the protocol supplied by the manufacturer (Gene Clean; Bio 101, Inc., Mississauga, Ontario, Canada). All probes were labeled with digoxigenin-11-dUTP, for 16 to 20 h under reaction conditions recommended by the manufacturer (Boehringer Mannheim Canada).
Genomic DNA restriction fragment length polymorphism (RFLP) analysis and hybridization.
Restriction endonucleases (New England Biolabs) were selected for treatment of genomic DNAs based on the known sequences (4, 11, 28): NdeI for clcABD hybridizations and EcoRI for all cbaABC and fcbB hybridizations. Endonuclease reactions were carried out in 20-μl volumes with 1 μg of DNA according to the manufacturer’s protocol (New England Biolabs Canada). Following digestion, DNA was separated on a 0.7% agarose gel by electrophoresis at 30 V overnight. The DNA in the gels was denatured, neutralized, and transferred to nylon membranes by capillary action with materials and protocols provided by Boehringer Mannheim Canada. These blots were fixed under UV light for 5 min prior to hybridization with the digoxigenin-labeled probes. The blots were prehybridized at 65°C for 2 h and hybridized at 65°C overnight in 5× sodium chloride-sodium citrate–0.1% (wt/vol) N-laurylsarcosine–0.02% (wt/vol) sodium dodecyl sulfate–1% (wt/vol) skim milk powder (Carnation). The membranes were washed and developed by the chemiluminescence protocol described by the manufacturer (Boehringer Mannheim Canada) followed by detection with Kodak XAR-5 film.
All isolates and type strains that hybridized with either the clc, cba, or fcb probes and type strains were screened for the presence of IS1071, the insertion sequence known to flank the cba operon in C. testosteroni BR60 (8, 25). Total genomic DNAs were digested with NheI (having two recognition sites within the 110-bp inverted repeats of IS1071 at positions 43 and 3158), yielding a 3.1-kb fragment that was resolved by gel electrophoresis and hybridized with the HindIII-H4 probe from IS1071 (25) as described above. Further characterization of selected IS1071-containing genomic DNAs was done by digestion with additional restriction enzymes (BamHI, HindIII, BglII, EcoRI, SacI, SalI, and NarI [New England Biolabs Canada]) followed by hybridization with IS1071 probes.
Phenotype analyses of isolates.
The carbon substrate utilization range of each isolate carrying the clc, cba, or fcb genes and the control strains, which were all gram negative, was determined with 96-well Biolog GN (gram negative) plates (Biolog, Inc., Hayward, Calif.) according to the suggestions by Kidd-Haack et al. (18) for the analysis of environmental isolates. The cluster analysis program of SSPC for MS Windows (version 6.0) was used to show phenotypic similarities among the Hyde Park isolates, the archetypal strains, and the type cultures C. testosteroni, Serratia ficaria, Burkholderia pickettii, and Pseudomonas tolaasii, based on carbon substrate utilization range.
RESULTS
Chlorobenzoate-catabolic isolates.
A collection of 464 independent isolates was made following CBA congener enrichment and selective plating from the sampling locations at Hyde Park (35). Each isolate was tested for CBA congener degradation by high-pressure liquid chromatography. The different CBA congeners were generally metabolized to undetectable concentrations (<10 μM) within 14 days at 4°C, 10 days at 22°C, or 8 days at 32°C (data not shown). The isolation frequency of CBA-degrading bacteria from the control sampling sites on unpolluted Fish Creek was low in comparison to that for the contaminated sites (33 of 464 isolates). This was a consequence of the low CBA biodegradation potentials found previously for Fish Creek (35) and of the occurrence of false positives, unable to degrade CBA congeners in liquid culture. Direct 3-CBA enrichment yielded no isolates from samples of Fish Creek; however, prior enrichment on 3-CBP followed by plating on 3-CBA yielded 11 isolates. Another 21 isolates from Fish Creek were enriched on 4-CBA, and 1 isolate was enriched on 3,4-DCBA. The remaining 431 isolates were obtained from the contaminated sites, Bloody Run Creek (206 isolates), Devil’s Hole Creek (177 isolates), and the groundwater and sequencing batch reactor samples from the Hyde Park facility itself (48 isolates). All carbon sources and all temperatures of enrichment yielded isolates from the contaminated sites, with no clear trends in these selection variables between the contaminated sites.
The greatest number of CBA-degrading isolates was obtained by using 4-CBA for enrichment (214 of 464). More isolates from all CBA enrichments were obtained from the 32 and 22°C enrichments (205 of 464 and 167 of 464, respectively) than from the 4°C enrichments (92 of 464). These proportions reflected the yield of fast-growing isolates that retained the CBA-degradative phenotype after subculturing. They do not necessarily reflect the proportions of different populations active in situ.
Genotype screening of isolates.
The clcABD, cbaABC, and fcbB genes were detected among the 464 isolates in the collection (Tables 2 and 3). All isolates carrying these genes were enriched from the contaminated sites adjacent to Hyde Park. The known CBA-degradative operons or genes were not detected among the 33 isolates collected from the control site, Fish Creek. The genomic DNA of the vast majority of isolates (436 of 464) failed to hybridize to the known CBA-degradative probes. No single CBA-degradative genotype was numerically dominant in the isolate collection from the Hyde Park sites. The cbaABC and clcABD genotypes were both found in the 3-CBA and 3-CBA (3-CBP) enrichments. The cbaABC genotype was also represented in 2 of 31 isolates from the 3,4-DCBA enrichments. The clcABD and fcbB genotypes were represented in isolates from 4-CBA enrichments in almost equal proportions. None of the 4-CBA enrichment cultures harbored the cbaABC genotype.
TABLE 2.
Frequencies of isolation of CBA-degrading isolates and the genotype distribution by carbon source
| Carbon sourcea | No. of isolates at enrichment temp (°C):
|
No. of isolates carrying the three genotypesb (%)
|
|||||
|---|---|---|---|---|---|---|---|
| 32 | 22 | 4 | Total | cbaABC | clcABD | fcbB | |
| 3-CBA | 54 | 51 | 22 | 127 | 3 (2.4) | 9 (7.1) | |
| 4-CBA | 91 | 75 | 48 | 214 | 0 (0) | 3 (1.4) | 4 (1.9) |
| 3,4-DCBA | 15 | 10 | 6 | 31 | 2 (6.5) | 0 (0) | |
| 3-CBA (3-CBP) | 45 | 31 | 16 | 92 | 4 (4.3) | 3 (3.3) | |
| Total | 205 | 167 | 92 | 464 | 9 (1.9) | 15 (3.2) | 4 (0.9) |
Abbreviations are as noted for Table 1, with the addition of 3-CBA (3-CBP), isolates from 3-CBA plates of samples previously enriched on 3-CBP.
Genotypes are as described for Table 1. The numbers in parentheses are the percentages of isolates of each genotype relative to the total number of isolates for that carbon source, as listed in column 4. The fcbB genotype was only screened for among the 4-CBA isolates. ERIC PCR revealed four sets of two sibling species among the 15 clcABD isolates (see Materials and Methods). Each sibling was isolated independently from different sites and different sampling times; therefore, they are included in the total for that genotype. No sibling species were found among the cbaABC and fcbB isolates.
TABLE 3.
Numbers of clc, cba, and fcb isolates; their substrate ranges; and genetic properties
| Genotypea | No. of isolates distributed by sampling siteb
|
Substrate rangec
|
RFLP pattern(s)d | HMW plasmide | IS1071f | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | BRC | DHC | Well | GW | SBR | 3-CBA | 4-CBA | 3,4-DCBA | ||||
| clcABD | 15 | 6 | 0 | 3 | 4 | 2 | 12 | 6 | 0 | ii–vi | 8 | 6 |
| cbaABC | 9 | 6 | 2 | 1 | 0 | 0 | 8 | 4 | 8 | i | 4 | 9 |
| fcbB | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | ii–iv | 2 | 3 |
Genotypes are as described for Table 1.
Abbreviations for isolate sources: BRC, isolates from samples of the sediment-water interface of Bloody Run Creek (this site was previously shown to be contaminated by Hyde Park leachate [35]); DHC, isolates from the Devil’s Hole Creek groundwater seep contaminated by Hyde Park leachate (35); Well, isolates from sampling wells drilled into the unconsolidated wastes at Hyde Park; GW, isolates from contaminated groundwater collected from the perimeter trench around the Hyde Park site; SBR, isolates from the sequencing batch reactors used for treating contaminated groundwater at Hyde Park (49).
Numbers of isolates able to degrade selected CBA congeners. Substrate abbreviations are as described in the footnotes to Tables 1 and 2. Some isolates degraded more than one CBA congener.
RFLP patterns grouped according to the fragment sizes following restriction enzyme digestion as follows. For clcABD, NdeI restriction fragments that hybridized with the clcABD probe were as follows: i, 19.5, 9.0, and 1.6 kb (detected in the control strain AC866 only); ii, 7.4 kb; iii, 9.5 and 5.7 kb; iv, 13.3, 10.3, and 5.7 kb; v, 10.3 and 5.7 kb; vi, 7.1 kb. For cbaABC, EcoRI restriction fragments that hybridized with the cbaABC probe represented one pattern only: i, 1.4- and 1.1-kb fragments (as for the control strain BR60). For fcbB, EcoRI restriction fragments that hybridized with the fcbB probe were as follows: i, 2.0 and 1.6 kb (detected in the control strain CBS3 only); ii, 4.9 kb; iii, 11.2 kb; and iv, 3.4 kb.
Number of isolates containing plasmids greater than 30 kb in size. HMW, high molecular weight.
Number of isolates containing the IS1071 3.1-kb NheI restriction fragment detected by hybridization with the HindIII fragment H4 probe.
Sibling pairs were found only among the 15 clcABD isolates by ERIC PCR (data not shown). Each of the four sibling pairs was found in different samples from the same stream or groundwater source. They were independently enriched from different sites along the stream bed, different groundwater wells, or different sequencing batch reactors. They were therefore considered to be independent isolates. Interestingly, in the case of siblings BRC3-2-3A and BRC4-4-3, different enrichment regimens were followed. The former was enriched on 3-CBA directly, while the latter was enriched initially on 3-CBP followed by isolation on 3-CBA agar. All cbaABC and fcbB isolates were nonclonal. No single isolate or sibling pair showed hybridization to more than one of the CBA-degradative gene probes used in this study.
A large proportion of the clcABD isolates (6 of 15 [40%]) was obtained from the 4°C enrichment cultures (data not shown). By contrast, none of the cbaABC isolates was collected from enrichment cultures incubated at 4°C. Isolates containing different CBA-degradative genes were found in the same sample in some cases. For example, isolates WellD1-3A, WellD1-3B, and WellD1-3C, enriched from one sample of Hyde Park groundwater on 3-CBA, at 4, 22, and 32°C, respectively, contained the clcABD genes in the first two isolates but the cbaABC genes in the last (Fig. 2). Another example of the coexistence of different genotypes in the same sample is found with isolates BRC3-2-3A (clcABD) and BRC3-2-3B (fcbB) from a single sample of Bloody Run Creek (Fig. 2).
FIG. 2.
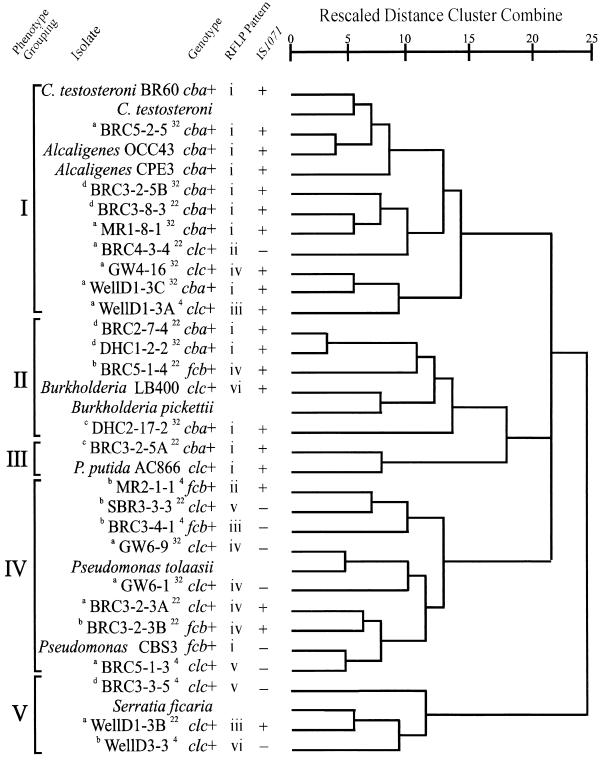
Phenotype distance cluster dendrogram of the clcABD, cbaABC, and fcbB isolates from the Hyde Park landfill based on carbon source utilization (Biolog GN). Control strains (clcABD, cbaABC, and fcbBAC) and representative β- and γ-Proteobacteria members from the Biolog GN database are included for reference. Phenotype grouping, an arbitrary division of isolates into five groups at a rescaled distance of 15. Isolate, the numbers following the source designation (Table 3) in each isolate name give the sampling site, the isolate number, and the sampling trip number (35), respectively. The superscript preceding the isolate name refers to the enrichment carbon source: a, 3-CBA; b, 4-CBA; c, 3,4-DCBA; d, 3-CBA (3-CBP). The superscript after the isolate name gives the enrichment temperature (degrees Celsius). Genotype, as described in the legend to Fig. 1. RFLP pattern, as described in footnote d to Table 3. IS1071, detection of the insertion sequence in the genome by hybridization to NheI digests. Rescaled distance cluster combine, dendrogram created from average linkage (between groups) data (SPSS).
RFLP patterns of target genotypes.
The RFLP patterns of the clcABD-harboring isolates following NdeI digestion and hybridization with the clcABD probe were diverse, and all were different from the RFLP pattern for the control Pseudomonas putida AC866(pAC27). The RFLP types were classified into six groups (Table 3 and Fig. 2). The isolates in RFLP groups iii, iv, and v contained a 5.7-kb fragment in common but showed variation in other fragments. One isolate, BRC4-3-4, had a unique 7.4-kb fragment (group ii). One isolate, WellD3-3, had a single 7.1-kb fragment as found for the control Burkholderia sp. strain LB400 (group vi).
All cbaABC-containing isolates had the same RFLP pattern as C. testosteroni BR60 following EcoRI digestion and hybridization with the cbaABC probe (Table 3 and Fig. 2). The RFLP patterns of the fcbB isolates were diverse, and all were different from the pattern for Pseudomonas sp. strain CBS3 (Table 3 and Fig. 2). Two fcb isolates, BRC5-1-4 and BRC3-2-3B, shared the same RFLP pattern (group iv).
Chlorobenzoate congener degradation and phenotype cluster analysis.
The clcABD isolates degraded 3- and/or 4-CBAs as predicted from previous studies of the specificity of the modified catechol ortho-ring-fission pathway (9, 10) (Table 3). Siblings exhibited the same patterns of CBA substrate utilization (data not shown). No isolates in this group metabolized 3,4-DCBA.
Almost all cbaABC isolates degraded 3-CBA and 3,4-DCBA (Table 3), with the exception of isolate DHC2-17-2, which degraded 4-CBA and 3,4-DCBA, and isolate BRC2-7-4, which degraded only 3-CBA (data not shown). Three isolates degraded all three isomers. These results are also consistent with the biochemical characterization of the pathway (27, 29). Interestingly, while 4-CBA enrichment did not yield isolates carrying the cbaABC genes (Table 2), three of the nine isolates with these genes degraded the 4-CBA substrate (Table 3).
The fcbB collection of four isolates degraded only 4-CBA as predicted from the specificity of the dehalogenase pathway in other isolates (4, 9, 38, 50).
Cluster analysis of the isolates carrying the known genes, based on the ability to utilize 95 carbon sources in Biolog GN plates, is presented in Fig. 2. Several strains from the Biolog database representative of the β- and γ-Proteobacteria were included in this dendrogram as reference strains. The clcABD siblings are represented by only one strain of each pair. Five major phenotypic groups were identified, arbitrarily separated at a rescaled distance of 15. The cbaABC isolates clustered in groups I and II, which included the β-Proteobacteria genera Alcaligenes, Comamonas, and Burkholderia, with a single cbaABC-containing isolate in the fluorescent Pseudomonas (γ-Proteobacteria) group III along with the control P. putida AC866. This isolate was confirmed to be a fluorescent pseudomonad by detection of fluorescence on King’s B agar (19).
The clcABD isolates were broadly distributed in the β-Proteobacteria groups I and II, as well as groups IV and V. Group IV includes P. tolaasii and the fcb control Pseudomonas sp. strain CBS3, while group V contains S. ficaria, all of which belong to the γ-Proteobacteria. Interestingly, the only group not represented in the clcABD isolate collection, group III, contained the clcABD control strain P. putida AC866. Isolates that were grouped by clcABD operon RFLP pattern are widely separated by phenotype. For example, three of four nonsibling isolates in the clcABD RFLP group iv are found in phenotype group IV (Pseudomonas), but the other member of this RFLP group is found in phenotype group I along with Alcaligenes and Comamonas control strains. clcABD RFLP groups v and vi show similar broad distributions in the phenotype cluster analysis. Hosts of the fcbB genotype were primarily found in group IV, with a single isolate from group II. Of additional interest in the group of WellD1-3A, -B, and -C isolates from the same Hyde Park groundwater sample discussed above is the observation that two of these isolates, WellD1-3A and WellD1-3C, are phenotypically very similar (group I [Fig. 2]) but carry different chlorobenzoate-degradative operons (clcABD and cbaABC, respectively). The third isolate in this group, WellD1-3B, is phenotypically very different (group V [Fig. 2]) and yet carries a clcABD operon with the same RFLP pattern as the group I isolate WellD1-3A. Another example of the presence of different CBA-degradative genotypes in similar hosts is the occurrence of the clcABD genes in BRC3-2-3A and of the fcbB gene in BRC3-2-3B, very similar hosts (although not siblings) within phenotype group IV (Fig. 2). In keeping with previous studies of the genus Burkholderia that have emphasized its metabolic versatility, we have found that phenotype group II, which includes this genus, contains isolates with all three CBA-degradative operons (Fig. 2).
Plasmid content and the incidence of IS1071.
High-molecular-weight plasmids (>30 kb) were detected from isolates representative of all three genotypes (Table 3). There was no strong correlation of plasmid content with the genotype, the RFLP group, the temperature of isolation, or the CBA substrate utilization profile. We note that the three clcABD isolates enriched on 3-CBP and transferred to 3-CBA (Table 2) contained high-molecular-weight plasmids that may be involved in either 3-CBP or 3-CBA degradation, or both.
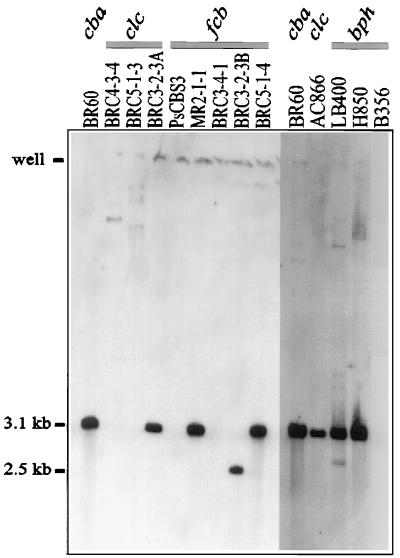
The results of genomic DNA digestion with NheI and hybridization to the insertion sequence IS1071 showed that this element was not limited to the cbaABC collection. All nine cbaABC isolates contained IS1071, and 4 of 11 (nonsibling) isolates carrying the clcABD genes and 3 of 4 isolates carrying the fcbB gene also hybridized to the H4 probe for this element (Table 3 and Fig. 3). All isolates yielded identical 3.1-kb fragment sizes expected for IS1071 with the single exception of one of the fcbB isolates, BRC3-2-3B, which yielded a 2.5-kb fragment (Fig. 3). Additional restriction digestions of genomic DNAs of selected IS1071-containing isolates showed no variation in the expected fragment sizes other than that noted above for isolate BRC3-2-3B (data not shown). Several of the control strains including the clcABD control P. putida AC866 and the biphenyl-degrading controls Burkholderia sp. strain LB400 (clcABD) and Alcaligenes sp. strain H850 contained the IS1071 element. There was no correlation between high-molecular-weight plasmid content and the incidence of IS1071 in any of the isolates or controls, consistent with previous observations of the plasmid and chromosomal location of this element (8, 25).
FIG. 3.
Hybridization of the IS1071 internal probe H4 to genomic DNA treated with NheI for selected isolates that carried clcABD and fcbB genes for chlorobenzoate degradation. Control genomic DNA digests for C. testosteroni BR60 (cbaABC); P. putida AC866 (clcABD); and the biphenyl-CBP-degrading strains Burkholderia sp. strain LB400 (bph clc), Alcaligenes sp. strain H850 (bph), and Pseudomonas sp. strain B-356 (bph) are also shown. The size of hybridizing DNA fragments was determined relative to the mobility of HindIII-digested λ DNA. Note that the copy number of IS1071 is not apparent in these digests because the NheI recognition sites are located in the inverted repeats of the element. For example, the genomic DNA of strain BR60 contains seven copies of IS1071.
DISCUSSION
All three of the well-characterized clc, cba, and fcb genotypes for CBA degradation can be found in roughly equivalent but low proportions among the isolates from the contaminated water sources at Hyde Park. None of these genotypes were enriched from the noncontaminated sites along Fish Creek. This observation in combination with the study of pristine soils by Fulthorpe et al. (12, 13) may indicate that prolonged selection by chloroaromatic pollutants in the environment is required to increase the incidence of these genotypes to detectable levels. Additional evidence for an influence of prior CBA exposure on community composition comes from the 3-CBA selection studies. Almost 30% of isolates from the contaminated sites were obtained by direct enrichment on 3-CBA, whereas no isolates were obtained in this way from Fish Creek. Preenrichment steps on 3-CBP were required in order to isolate 3-CBA degraders from this noncontaminated site, and the known genes were not detected among these isolates.
Evidence presented here shows that multiple genotypes for CBA degradation may occur simultaneously in the bacterial community degrading these pollutants. This conclusion is supported by our observation of multiple genotypes isolated from the same samples, for example, isolates WellD1-3A and -B (clcABD) with WellD1-3C (cbaABC) and also BRC3-2-3A (clcABD) with BRC3-2-3B (fcbB) (Fig. 2). Without additional biochemical and genetic studies on each isolate to show the involvement of each operon in CBA degradation, we cannot be certain of the link between genotype and phenotype. Nevertheless, in all cases the utilization by each isolate of the different CBA congeners matched the expected substrate range for the genotype. Also, for isolates carrying the cbaABC operon the evidence linking the presence of the genes to the degradative phenotype is strong. All of these cbaABC-containing isolates were unstable, showing spontaneous deletion at high frequencies of the operon and loss of the CBA-degradative phenotype (data not shown). We have shown previously that this is due to recombination between the IS1071 copies flanking the cbaABC genes (8, 25).
This study has not addressed particular advantages of one genotype over another at this site, nor the frequency of these genotypes in situ, which may differ substantially from the frequencies reported in Tables 2 and 3. Nevertheless, in combination with data showing enhanced CBA biodegradation potentials as a result of chloroaromatic pollutant exposure (35), the complete study demonstrates that there exist ecological niches for each of the well-characterized CBA degradation operons at the sediment-water interface of the contaminated streams and that they are potentially involved in contaminant removal at these sites. The findings point to the necessity for comprehensive screening of microbial communities involved in contaminant removal in situ in order to assess the overall potential for bioremediation and for bioaugmentation. Screening for single genotypes will in most cases be insufficient. Furthermore, this study has shown that the largest part (>90%) of the isolate collection failed to hybridize with the known clc, cba, and fcb probes, even under relatively relaxed stringency. Clearly, the genetic diversity for CBA degradation is much greater than previously suspected. The results for the 3-CBA (3-CBP) enrichments in which cba and clc genes were found in a total of 8% of the isolates (Table 2) indicate that communities of aerobic bacteria involved in polychlorinated biphenyl degradation also show genetic diversity for CBA degradation, with much of that diversity yet uncharacterized. These findings emphasize the need to continue to characterize new pollutant biodegradation pathways and their genetic determinants.
Some part of the isolate collection that failed to hybridize to the known CBA catabolic genes may carry genes for other CBA degradation pathways. Krooneman et al. have shown that Alcaligenes sp. strain L6 uses a novel 3-CBA degradation pathway via gentisate to grow at low oxygen tensions and dilution rates (21). These environmental conditions may well occur at the sediment-water interface of the sites we sampled; however, genetic probes for this pathway are not yet available. The high proportion of nonhybridizing isolates within the collection may also carry homologues of the clcABD, cbaABC, or fcbBAC operons that have diverged in sequence sufficiently to escape detection by hybridization. Fulthorpe et al. obtained chlorocatechol 1,2-dioxygenase (clcA-like) gene sequences by PCR with redundant primers with 3-CBA-degrading bacteria from pristine soils (12, 13, 23). They showed that the PCR-amplified sequences were less than 60% similar to the clcA gene of P. putida AC866(pAC27) and were more closely related to the tfdC gene from 2,4-D-degrading isolates. As reported by Schlomann (37), this level of sequence divergence suggests that evolution of chlorocatechol dioxygenases has been ongoing for the past 70 to 90 million years.
The fcbBAC operon also shows evidence of extensive sequence divergence. For example, the fcbB (dehalogenase) and fcbA (CoA ligase) genes in the gram-positive isolate Arthrobacter sp. strain SU are 81 and 75% identical, respectively, to the genes in Pseudomonas sp. strain CBS3, while the putative fcbC (thioesterase) genes are nonhomologous (38). The gene orders also differ in these isolates (fcbABC in strain SU).
There is no evidence that similar variation exists in the cbaABC operon. In all isolates examined to date from the Hyde Park site (references 26 and 47 and this study), as well as an isolate from polychlorinated biphenyl-contaminated soils from Italy (8), the cbaABC operon was completely conserved. In the latter study comparing isolates from different continents, the sequence similarity in the cbaA gene was 99.3% and the operons were carried on similar, although not identical, composite transposons. The RFLP data described in Table 3 and additional data for hybridization with other Tn5271-derived probes (not shown) indicate that the nine cbaABC isolates found in this study carry composite transposons similar or identical to Tn5271 (34). The evidence therefore suggests that horizontal transfer has been more important for the recent dissemination of the cbaABC genotype than for the other genotypes we examined. There is now substantial evidence for the involvement of gene mobilization and horizontal transfer in the biodegradation of pollutants in situ, for instance, in the natural attenuation of chlorobenzene contamination of an aquifer at Kelly Air Force Base in Texas (43) and the degradation of coal-tar-derived naphthalene in groundwater seeps (39). The observed noncongruency of the phylogenetic trees of the tfdA and 16S ribosomal DNA genes of 2,4-D-degrading bacteria shows that interspecies gene transfer has been an important factor in their evolution (24).
The clcABD operon was found to be broadly distributed in all five phenotypic groups of CBA-degrading bacteria (Fig. 2). This may reflect the broad distribution of relaxed-specificity benzoate and toluate dioxygenases needed to initiate this pathway. It may also reflect the involvement of this operon in the degradation of other chlorinated aromatic compounds that are converted to chlorocatechols. For instance, the study of chlorobenzene-degrading isolates from a contaminated aquifer at Kelly Air Force Base noted above has shown that genes almost identical to clcABD have combined in situ with chlorobenzene dioxygenase genes in Ralstonia species to naturally attenuate the pollutants at the site (43). Chlorobenzenes and chlorotoluenes are among the contaminants at Hyde Park (49), so that the observed selection of clc genes in a variety of different hosts may have been in response to other chlorinated aromatic pollutants in addition to CBA and CBPs. We have not tested our clc isolate collection for degradation of other chlorinated aromatic compounds.
In contrast to the clc genes, the cba genes appear to be involved only in CBA and CBP degradation (27, 29, 47). The cbaABC host range, which is primarily in the β-Proteobacteria, has previously been correlated with the distribution of the protocatechuate meta-ring-fission pathway (26). This is a consequence of the preferential metabolism of the 3-CBA metabolites protocatechuate and 5-chloroprotocatechuate through the meta-ring-fission pathway in cbaABC hosts (26, 29). There are very few reports of 3,4-DCBA mineralization by pure cultures (8, 29); nevertheless, Acinetobacter sp. strain 4-CB1 carrying an fcbBAC-like dehalogenase operon is able to cometabolize 3,4-DCBA in the presence of 4-CBA (1). The cbaABC operon was the only one associated with 3,4-DCBA-degrading isolates in our collection. Metabolism of this congener through the cbaABC pathway also requires the (5-chloro)-protocatechuate meta-ring-fission pathway (29).
The uniform association of the cbaABC genes with IS1071 in all isolates that carry this operon (Table 3) (8, 25, 34, 46, 47) suggests that horizontal transfer of the composite transposon Tn5271 is the primary mode of dissemination of these genes. An unexpected finding was the occurrence of IS1071 in the genomes of isolates that carried nonhomologous operons (Table 3 and Fig. 2 and 3). Here we show that this element occurs in 6 of 15 (40%) of the clcABD isolates and 3 of 4 (75%) of the fcbB isolates (Table 3) and that it occurs in all five phenotypic groups defined in Fig. 2. In addition, IS1071 was detected in control strains AC866 (clcABD), LB400 (bph clcABD), and H850 (bph) (Fig. 3). A previous review of the distribution of IS1071 linked this element with a diverse collection of genes for the degradation of aliphatic and aromatic contaminants (8). Many of these contain composite transposon structures flanked by direct repeats of IS1071. Recently, an example of this kind of transposon coding for the degradation of 2,4-D has been described (48). In the latter case, the IS1071 elements are both interrupted by a class I insertion sequence, IS1471, inserted at identical positions in the tnpA genes of the flanking elements. A nested insertion of another transposon within IS1071 may explain the 2.5-kb hybridizing fragment that we observed for isolate BRC3-2-3B (Fig. 3) for which the restriction pattern differs from those of all other isolates. Despite the natural instability of IS1071 composite transposons, this element is likely a major contributor to biodegradative gene rearrangements and mobilization in bacteria in contaminated environments.
ACKNOWLEDGMENTS
This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and from the NSERC-Environment Canada, Great Lakes University Research Fund. M.C.P. was the recipient of an NSERC Postgraduate Scholarship.
REFERENCES
- 1.Adriaens P, Focht D D. Cometabolism of 3,4-dichlorobenzoate by Acinetobacter sp. strain 4-CB1. Appl Environ Microbiol. 1991;57:173–179. doi: 10.1128/aem.57.1.173-179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 4.Babbitt, P. C., G. L. Kenyon, B. M. Martin, H. Charest, M. Sylvestre, J. D. Scholten, K.-H. Chang, P.-H. Liang, and D. Dunaway-Mariano. 1993. Genpept accession number 419530 (PIR: locus B42560).
- 5.Bertoni G, Martino M, Galli E, Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–3632. doi: 10.1128/aem.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copley S D, Crooks G P. Enzymatic dehalogenation of 4-chlorobenzoyl coenzyme A in Acinetobacter sp. strain 4-CB1. Appl Environ Microbiol. 1992;58:1385–1387. doi: 10.1128/aem.58.4.1385-1387.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Gioia D, Peel M, Fava F, Wyndham R C. Structures of homologous composite transposons carrying cbaABC genes from Europe and North America. Appl Environ Microbiol. 1998;64:1940–1946. doi: 10.1128/aem.64.5.1940-1946.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fetzner S, Lingens F. Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol Rev. 1994;58:641–685. doi: 10.1128/mr.58.4.641-685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Focht D D. Biodegradation of chlorobenzoates. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 71–80. [Google Scholar]
- 11.Frantz, B., and A. M. Chakrabarty. 1996. Genbank accession no. M16964.
- 12.Fulthorpe R R, Rhodes A N, Tiedje J M. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl Environ Microbiol. 1996;62:1159–1166. doi: 10.1128/aem.62.4.1159-1166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulthorpe R R, Rhodes A N, Tiedje J M. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl Environ Microbiol. 1998;64:1620–1627. doi: 10.1128/aem.64.5.1620-1627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa K. Molecular genetics and evolutionary relationships of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 15.Gibson D T, Hensley M, Yoshioka H, Mabry T J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970;9:1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 16.Johnson G R, Olsen R H. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl Environ Microbiol. 1997;63:4047–4052. doi: 10.1128/aem.63.10.4047-4052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ka J O, Burauel P, Bronson J A, Holben W E, Tiedje J M. DNA probe analysis of the microbial community selected in the field by long-term 2,4-D application. Soil Sci Soc Am J. 1995;59:1581–1587. [Google Scholar]
- 18.Kidd-Haack S, Garchow H, Klug M J, Forney L J. Analysis of factors affecting the accuracy, reproducibility, and interpretation of microbial community carbon source utilization patterns. Appl Environ Microbiol. 1995;61:1458–1468. doi: 10.1128/aem.61.4.1458-1468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocin and fluorescin. J Lab Clin Med. 1954;44:301. [PubMed] [Google Scholar]
- 20.Kivisaar M, Hõrak R, Kasak L, Heinaru A, Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- 21.Krooneman J, Moore E R B, van Velzen J C L, Prins R A, Forney L J, Gottschal J C. Competition for oxygen and 3-chlorobenzoate between two aerobic bacteria using different degradation pathways. FEMS Microbiol Ecol. 1998;26:171–179. [Google Scholar]
- 22.Kukor J J, Olsen R H. Complete nucleotide sequence of tbuD, the gene encoding phenol/cresol hydroxylase from Pseudomonas pickettii PKO1, and functional analysis of the encoded enzyme. J Bacteriol. 1992;174:6518–6526. doi: 10.1128/jb.174.20.6518-6526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leander M, Vallaeys T, Fulthorpe R R. Amplification of putative chlorocatechol dioxygenase gene fragments from α- and β-proteobacteria. Can J Microbiol. 1998;44:482–486. doi: 10.1139/cjm-44-5-482. [DOI] [PubMed] [Google Scholar]
- 24.McGowan C, Fulthorpe R, Wright A, Tiedje J M. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol. 1998;64:4089–4092. doi: 10.1128/aem.64.10.4089-4092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatsu C H, Fulthorpe R R, Holland B A, Peel M C, Wyndham R C. The phylogenetic distribution of a transposable dioxygenase from the Niagara river watershed. Mol Ecol. 1995;4:593–603. doi: 10.1111/j.1365-294x.1995.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 28.Nakatsu, C. H., N. A. Straus, and R. C. Wyndham. 1997. Genbank accession no. U18133.
- 29.Nakatsu C H, Providenti M, Wyndham R C. The cis-diol dehydrogenase cbaC gene of Tn5271 is required for growth on 3-chlorobenzoate but not 3,4-dichlorobenzoate. Gene. 1997;196:209–218. doi: 10.1016/s0378-1119(97)00229-1. [DOI] [PubMed] [Google Scholar]
- 30.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordlund I, Powlowski J J, Hagström A, Shingler V. Conservation of regulatory and structural genes for a multicomponent phenol hydroxylase within phenol-catabolizing bacteria that utilize a meta-cleavage pathway. J Gen Microbiol. 1993;139:2695–2703. doi: 10.1099/00221287-139-11-2695. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa N, Miyashita K. A transposon-like structure carrying genes for the catabolism of 3-chlorobenzoate on a plasmid from Alcaligenes eutrophus strain NH9. Appl Environ Microbiol. 1995;61:3788–3795. doi: 10.1128/aem.61.11.3788-3795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peel M C. Catabolic genotype distributions in the Niagara watershed. Ph.D. thesis. Ottawa, Ontario, Canada: Institute of Biology, Carleton University; 1996. [Google Scholar]
- 35.Peel M C, Wyndham R C. The impact of industrial contamination on microbial chlorobenzoate degradation in the Niagrara watershed. Microb Ecol. 1997;33:59–68. doi: 10.1007/s002489900008. [DOI] [PubMed] [Google Scholar]
- 36.Peters M, Heinaru E, Talpsep E, Wand H, Stottmeister U, Heinaru A, Nurk A. Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Appl Environ Microbiol. 1997;63:4899–4906. doi: 10.1128/aem.63.12.4899-4906.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlomann M. Evolution of chlorocatechol catabolic pathways. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz A, Gartemann K, Fiedler J, Grund E, Eichenlaub R. Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU. Appl Environ Microbiol. 1992;58:4068–4071. doi: 10.1128/aem.58.12.4068-4071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart-Keil K G, Hohnstock A M, Drees K P, Herrick J B, Madsen E L. Plasmids responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB 9816-4. Appl Environ Microbiol. 1998;64:3633–3640. doi: 10.1128/aem.64.10.3633-3640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsoi T V, Zaitsev G M, Plotnikova E G, Kosheleva I A, Boronin A M. Cloning and expression of the Arthrobacter globiformis KZT1 fcbA gene encoding dehalogenase (4-chlorobenzoate-4-bydroxylase) in Escherichia coli. FEMS Microbiol Lett. 1991;81:165–170. doi: 10.1016/0378-1097(91)90298-o. [DOI] [PubMed] [Google Scholar]
- 41.Tsuda M. Catabolic transposons in pseudomonads. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 219–228. [Google Scholar]
- 42.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meer J R, Werlen C, Nishino S, Spain J C. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl Environ Microbiol. 1998;64:4185–4193. doi: 10.1128/aem.64.11.4185-4193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whited G M, Gibson D T. Toluene-4-monoxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyndham R C, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 47.Wyndham R C, Nakatsu C, Peel M, Cashore A, Ng J, Szilagyi F. Distribution of the catabolic transposon Tn5271 in a groundwater bioremediation system. Appl Environ Microbiol. 1994;60:86–93. doi: 10.1128/aem.60.1.86-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia X-S, Aathithan S, Oswiecimska K, Smith A R W, Bruce I J. A novel plasmid pIJB1 possessing a putative 2,4-dichlorophenoxyacetate degradative transposon Tn5530 in Burkholderia cepacia strain 2a. Plasmid. 1998;39:154–159. doi: 10.1006/plas.1997.1332. [DOI] [PubMed] [Google Scholar]
- 49.Ying Y-C, Wnukowski J, Wilde D, McLeod D. Proceedings of the 47th Industrial Waste Conference, 11 May 1992. Ann Arbor, Mich: Lewis Publishers; 1992. Successful leachate treatment in SBR-adsorption system; pp. 501–518. [Google Scholar]
- 50.Zaitsev G M, Tsoi T V, Grisenkov V G, Plotnikova E G, Boronin A M. Genetic control in Arthrobacter globiformis, Corynebacterium sepedonicum and Pseudomonas cepacia strains. FEMS Microbiol Lett. 1991;81:171–176. doi: 10.1016/0378-1097(91)90299-p. [DOI] [PubMed] [Google Scholar]