Abstract
Egg-laying defective nine 1 (EGLN1) functions as an oxygen sensor to catalyze prolyl hydroxylation of the transcription factor hypoxia-inducible factor-1 α under normoxia conditions, leading to its proteasomal degradation. Thus, EGLN1 plays a central role in the hypoxia-inducible factor–mediated hypoxia signaling pathway; however, the posttranslational modifications that control EGLN1 function remain largely unknown. Here, we identified that a lysine monomethylase, SET7, catalyzes EGLN1 methylation on lysine 297, resulting in the repression of EGLN1 activity in catalyzing prolyl hydroxylation of hypoxia-inducible factor-1 α. Notably, we demonstrate that the methylation mimic mutant of EGLN1 loses the capability to suppress the hypoxia signaling pathway, leading to the enhancement of cell proliferation and the oxygen consumption rate. Collectively, our data identify a novel modification of EGLN1 that is critical for inhibiting its enzymatic activity and which may benefit cellular adaptation to conditions of hypoxia.
Keywords: SET7, EGLN1, hypoxia signaling, methylation, gene expression
Abbreviations: EGLN, egg-laying defective nine; FKBP, FK506-binding protein; HA, hemagglutinin; HIF, hypoxia-inducible factor; MS, mass spectrometry; PHD, prolyl hydroxylase domain; PTM, posttranslational modification; sgRNA, single guide RNA; SIAH2, seven in absentia homolog 2
Egg-laying defective nine (EGLN) 1, EGLN2, and EGLN3 (also known as prolyl hydroxylase domain [PHD] enzyme, PHD2, PHD1, and PHD3) are evolutionarily conserved oxygen sensors, which target hypoxia-inducible factor (HIF)-α subunits for hydroxylation and subsequent proteasomal degradation under normoxia, thus playing a central role in hypoxia signaling pathway (1, 2, 3, 4, 5). HIF-mediated hypoxia signal pathway facilitates cell survival and adaptation in response to varying environmental oxygen levels, functioning critically in multiple biological processes, including angiogenesis, erythropoiesis, and tumorigenesis (6, 7, 8, 9, 10, 11, 12).
EGLNs contain a conserved carboxy-terminal catalytic domain and a more variable amino-terminal domain (1). While molecular oxygen, the substrate 2-oxoglutarate (also known as α-ketoglutarate), and the cofactors ferrous iron (Fe2+) and ascorbic acid (vitamin C) are required for PHD-catalyzed hydroxylation reactions, other factors also modulate PHD activity (1). FK506-binding protein (FKBP) 38 has been shown to negatively regulate PHD2 (EGLN1) activity (13), whereas seven in absentia homolog 2 (SIAH2) has been identified to degrade PHD3 (EGLN3) (14). Moreover, mTOR (the mechanistic target of rapamycin downstream kinase P70S6K) mediates PHD2 (EGLN1) phosphorylation on serine 125 (S125), leading to the enhancement of PHD2 protein stability, but PP2A directly dephosphorylates PHD2 on S125, leading to the fine-tuning of HIF1α levels (15). Recently, cystathionine β-synthase was found to persulfidate PHD2, resulting in the augment of PHD2 prolyl hydroxylase activity (16). In response to oxidative stress, PHD2 is dimerized and its activity is inhibited, resulting in the stabilization of HIF1α (17). Given the importance of posttranslational modifications (PTMs) in the regulation of protein functions, other unidentified PTMs of PHDs should affect PHD activity and subsequent hypoxia signaling pathway.
SET7 (also known as SETD7, SET9, and SET7/9) was originally identified as a monomethylase of histone H3 lysine 4, involved in gene activation (18, 19). Lately, SET7 was revealed to monomethylate various nonhistone proteins, either negatively or positively modulating their functions (20, 21, 22, 23, 24, 25, 26). SET7 was also found to regulate HIFα activity, suggesting SET7-mediated lysine monomethylation has impacts on hypoxia signaling (27, 28)
In this study, we found that that EGLN1 contains a SET7 targeting motif (RS/TK). Further assays show that SET7 mediated monomethylation of EGLN1 on lysine 297, leading to the relief of its suppressive role on HIF1α activity and promoting cellular hypoxia adaptation.
Results
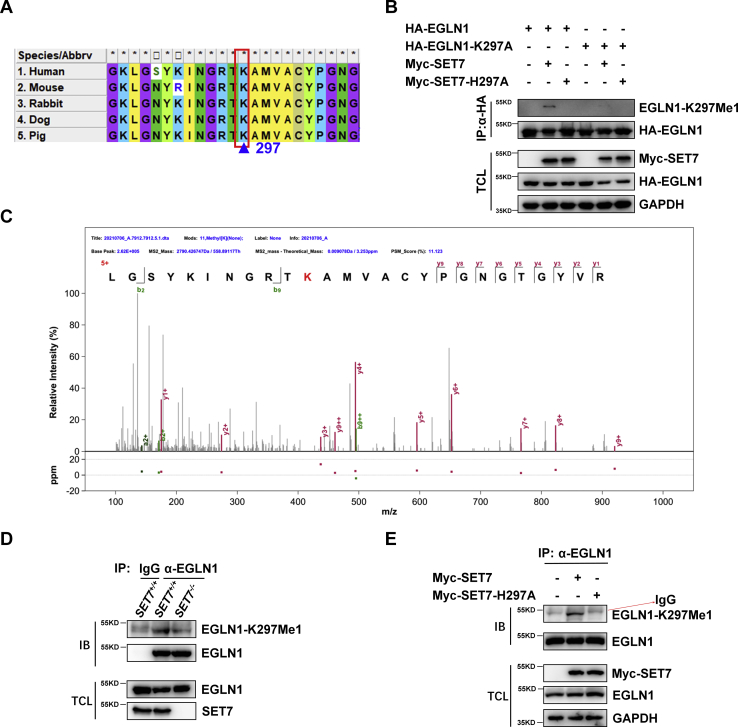
SET7 methylates EGLN1 on lysine 297
In addition to histone H3, SET7 was also found to monomethylate nonhistone proteins by recognizing a conserved core domain ([K/R] [S/T] K) (24, 27, 29, 30, 31) in these proteins. After searching the amino acid sequences of EGLN1 from different organisms, the three “RTK (Argnine-Threonine-Lysine)” residues were identified in the catalytic domain (PH domain) of EGLN1 (Fig. 1A), in which lysine 297 might be methylated by SET7. To confirm this hypothesis, we developed an antibody (anti-EGLN1-K297me1) to specifically recognize lysine 297 of human EGLN1. When WT hemagglutinin (HA)-tagged EGLN1 (HA-EGLN1) was cotransfected with SET7, the methylated EGLN1 was readily detected by anti-EGLN1-K297me1 antibody after the coimmunoprecipitation with anti-HA–conjugated agarose beads (Fig. 1B). However, the methylated EGLN1 was not shown up when the mutant (H297A) was cotransfected (Fig. 1B). To further validate this result, we performed mass spectrometry assay. As shown in Figure 1C, the monomethylated K297 was identified in EGLN1 when SET7 was coexpressed (Fig. 1C). To determine whether endogenous EGLN1 was methylated by SET7, we examined methylation of EGLN1 in SET7+/+ and SET7−/− HEK293T cells after coimmunoprecipitation with anti-EGLN1 antibody. Monomethylation of EGLN1 in SET7+/+ HEK293T cells was higher than that in SET7−/− HEK293T cells (Fig. 1D). Furthermore, we transfected WT SET7 and its enzymatically deficient mutant (H297A) into SET7−/− HEK293T cells and examined methylation of endogenous EGLN1 after coimmunoprecipitation with anti-EGLN1 antibody. Monomethylation of EGLN1 in the cells transfected with WT SET7 was higher in the cells transfected with SET7-H297A (Fig. 1E).
Figure 1.
SET7 methylates EGLN1 on lysine 297.A, sequence alignment of partial EGLN1 proteins (285–307 amino acids) of human, mouse, rabbit, dog, and pig. The red box indicates a conserved consensus motif (R/K-S/T-K) methylated by SET7; the blue arrow indicates the conserved lysine (K297) methylated by SET7. The amino acid is numbered based on human EGLN1 amino acid sequence. B, Western blot analysis of EGLN1 or its mutant (K297A) methylated by WT SET7 or the enzymatic-deficiency mutant of SET7 (H297A) in HEK293T cells transfected with indicated plasmids. Anti-HA antibody–conjugated agarose beads were used for immunoprecipitation and anti-EGLN1-K297Me1 antibody was used for detecting monomethyl EGLN1. C, the methylated residue in EGLN1 identified by mass spectrometry analysis. HEK293T cells were cotransfected with HA-EGLN1 and Myc-SET7 plasmids. Cell lysate was immunoprecipitated with anti-HA antibody–conjugated agarose beads overnight. Immunoprecipitated EGLN1 proteins were subjected to 8% SDS-PAGE gel, and EGLN1 bands were excised from the gel and analyzed by mass spectrometry. D, Western blot analysis of endogenous methylated EGLN1 in SET7-deficient or WT HEK293T cells (SET7−/− or SET7+/+). Anti-EGLN1 antibody was used for immunoprecipitation, and normal rabbit IgG was used as a control. Anti-EGLN1-K297Me1 antibody was used to detect monomethyl EGLN1. E, Western blot analysis of endogenous methylated EGLN1 in SET7-deficient HEK293T cells (SET7−/−) transfected with indicated plasmids. Anti-EGLN1 antibody was used for immunoprecipitation, and anti-EGLN1-K297Me1 antibody was used to detect monomethyl EGLN1. EGLN, egg-laying defective nine; HA, hemagglutinin; IgG, immunoglobulin G.
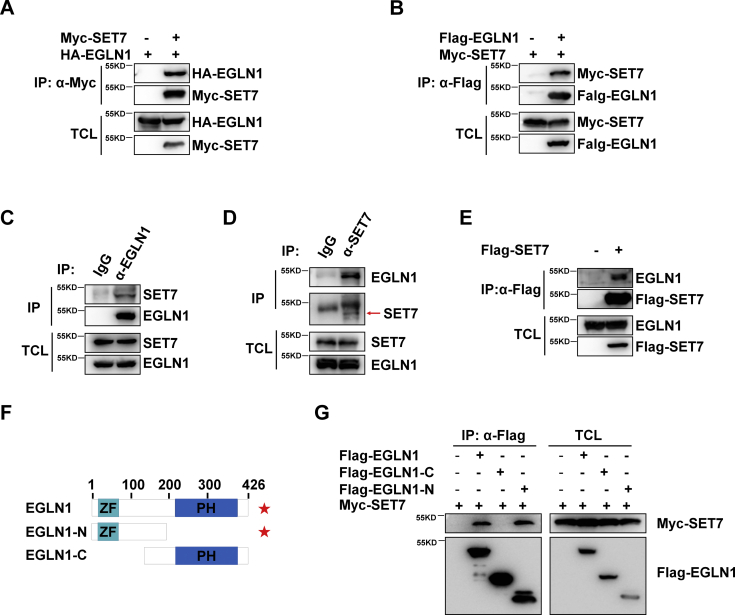
Subsequently, we examined whether EGLN1 interacts with SET7 by coimmunoprecipitation assay. Ectopically expressed EGLN1 interacted with ectopically expressed SET7 (Fig. 2, A and B). Moreover, endogenous SET7 interacted with endogenous EGLN1 in HEK293T cells (Fig. 2, C and D). Ectopically expressed SET7 also interacted with endogenous EGLN1 (Fig. 2E). Domain mapping indicated that SET7 interacted with the N terminus of EGLN1 (Fig. 2, F and G).
Figure 2.
SET7 interacts with EGLN1.A, coimmunoprecipitation of Myc-SET7 with HA-EGLN1. HEK293T cells were cotransfected with indicated plasmids for 24 h. Anti-Myc antibody–conjugated agarose beads were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. B, coimmunoprecipitation of FLAG-EGLN1 with Myc-SET7. HEK293T cells were cotransfected with indicated plasmids for 24 h. Anti-FLAG antibody–conjugated agarose beads were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. C, endogenous interaction between SET7 and EGLN1. Anti-EGLN1 antibody was used for immunoprecipitation, and normal rabbit IgG was used as a control. D, endogenous interaction between EGLN1 and SET7. Anti-SET7 antibody was used for immunoprecipitation, and normal rabbit IgG was used as a control. E, coimmunoprecipitation of FLAG-SET7 with endogenous EGLN1. HEK293T cells were transfected with indicated plasmids for 24 h. Anti-FLAG antibody–conjugated agarose beads were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. F, schematic of EGLN1 domains interacted with SET7. The interaction is indicated by (★) sign. G, coimmunoprecipitation analysis of Myc-SET7 with FLAG-EGLN1–truncated mutants. HEK293T cells were cotransfected with the indicated plasmids. Anti-FLAG antibody–conjugated agarose beads were used for immunoprecipitation, and the interaction was analyzed by immunoblotting with the indicated antibodies. FLAG-EGLN1 fragments (EGLN1-C, 130–426 aa; EGLN1-N, 1–196 aa). EGLN, egg-laying defective nine.
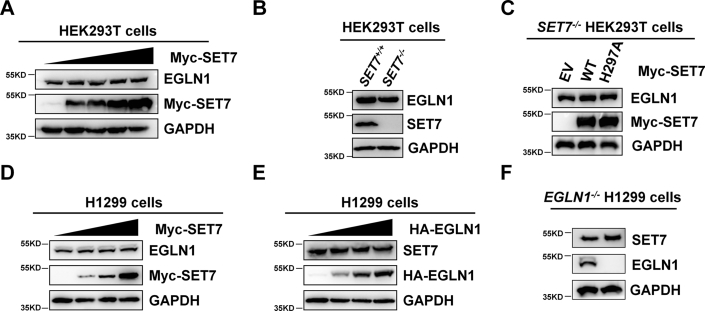
To determine whether SET7 has effects on EGLN1 protein level, we transfected Myc-SET7 into HEK293T cells with an increasing amount and found that endogenous EGLN1 protein level was not changed at all (Fig. 3A). EGLN1 was the same between SET7+/+ and SET7−/− HEK293T cells (Fig. 3B). Similar EGLN1 protein level was also detected in SET7−/− HEK293T cells transfected with empty vector control, WT SET7, and the enzymatically deficient SET7 (H297A) (Fig. 3C). In H1299 cells, overexpression of SET7 also had no effect on EGLN1 protein level (Fig. 3D). Of note, EGLN1 had no effect on SET7 protein level (Fig. 3, D and E). The data indicate that EGLN1 and SET7 do not affect each other at protein level even though they are associated in cells.
Figure 3.
SET7 has no obvious effect on EGLN1 protein level and vice versa.A, Western blot analysis of endogenous EGLN1 expression in HEK293T cells transfected with an increasing amount of Myc-SET7 expression plasmid. B, Western blot analysis of endogenous EGLN1 expression in SET7-deficient or WT HEK293T cells (SET7−/− or SET7+/+). C, Western blot analysis of endogenous EGLN1 expression in SET7-deficient HEK293T cells (SET7−/−) transfected with indicated plasmids. D, Western blot analysis of endogenous EGLN1 expression in H1299 cells transfected with an increasing amount of Myc-SET7 expression plasmid. E, Western blot analysis of endogenous SET7 expression in H1299 cells transfected with an increasing amount of HA-EGLN1 expression plasmid. F, Western blot analysis of endogenous SET7 expression in EGLN1-deficient or WT H1299 cells (EGLN1−/− or EGLN1+/+). EGLN, egg-laying defective nine; HA, hemagglutinin.
Taken together, these findings suggest that SET7 interacts with EGLN1 to catalyze monomethylation of EGLN1 on K297.
The prolyl hydroxylase activity of EGLN1 is attenuated by SET7-mediated methylation on K297
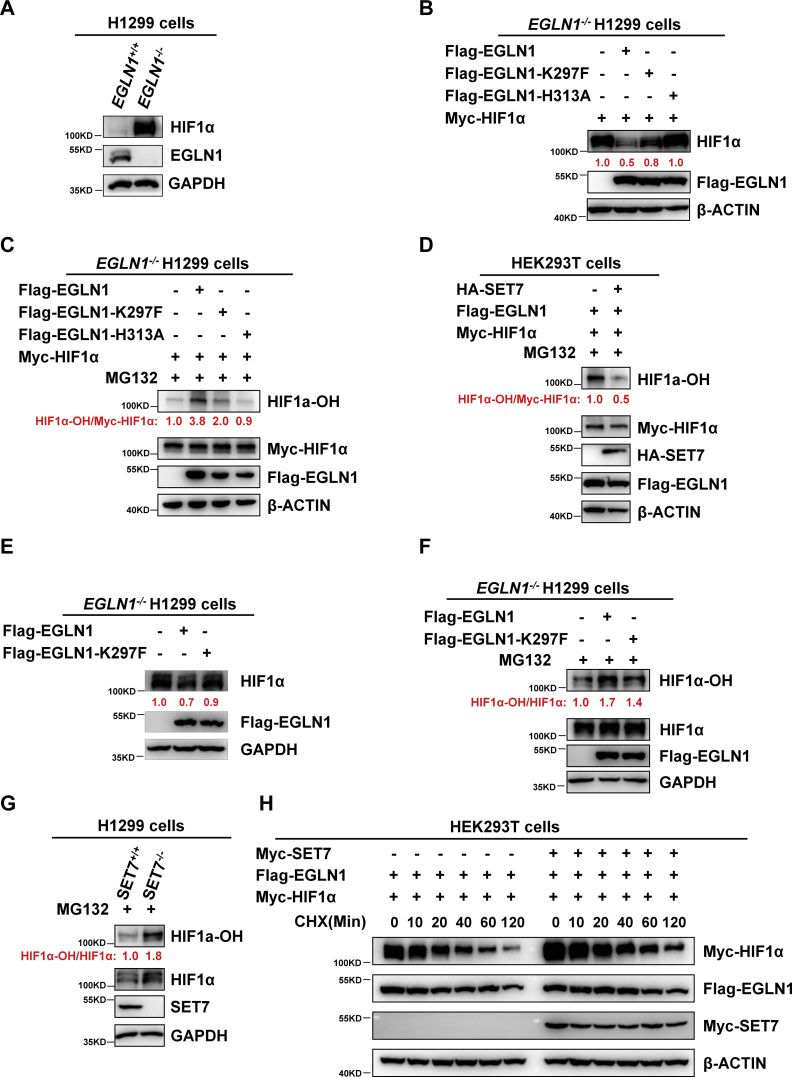
Given the well-defined function of EGLN1 in catalyzing prolyl hydroxylation of HIF1α, we sought to determine whether methylation of EGLN1 by SET7 can affect the prolyl hydroxylase activity of EGLN1 on HIF1α. In EGLN1−/− H1299 cells, as expected, overexpression of WT EGLN1 caused a reduction of cotransfected Myc-HIF1α. However, overexpression of the methylation-mimic mutant of EGLN1 (K297F) recovered the protein level of cotransfected Myc-HIF1α even though it was not as dramatic as overexpression of the enzymatically deficient mutant of EGLN1 (H313A), suggesting that methylation of EGLN1 on K297 caused a reduction of enzymatic activity of EGLN1 (Fig. 4, A and B).
Figure 4.
The prolyl hydroxylase activity of EGLN1 is attenuated by SET7-mediated methylation on K297.A, Western blot analysis of endogenous EGLN1 and HIF1α expression in EGLN1-deficient or WT H1299 cells (EGLN1−/− or EGLN1+/+). B, Western blot analysis of exogenous Myc-HIF1α expression in EGLN1-deficient H1299 cells (EGLN1−/−) transfected with indicated plasmid. The cells were cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the enzymatically deficient mutant (H313A), the methylation-mimic mutant (K297F). FLAG empty was used as a control. The relative intensities of Myc-HIF1α were determined by normalizing the intensities of Myc-HIF1α to the intensities of β-ACTIN. C, Western blot analysis of HIF1α hydroxylation with anti-hydroxy-HIF1α (Pro564) antibody in EGLN1-deficient H1299 cells (EGLN1−/−) transfected with indicated plasmid. The cells were cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the enzymatically deficient mutant (H313A), the methylation-mimic mutant (K297F), followed by MG-132 (20 μM) treatment for 8 h. FLAG empty was used as a control. The relative intensities of HIF1α-OH/Myc-HIF1α were determined by normalizing the intensities of HIF1α-OH to the intensities of Myc-HIF1α. D, Western blot analysis of HIF1α hydroxylation with anti-Hydroxy-HIF1α (Pro564) antibody in HEK293T transfected with indicated plasmid. The cells were cotransfected with Myc-HIF1α, FLAG-EGLN1, and HA-SET7 (HA empty was used as a control), followed by MG-132 (20 μM) treatment for 8 h. The relative intensities of HIF1α-OH/Myc-HIF1α were determined by normalizing the intensities of HIF1α-OH to the intensities of Myc-HIF1α. E, Western blot analysis of endogenous HIF1α expression in EGLN1-deficient H1299 cells (EGLN1−/−) transfected with indicated plasmid. The cells were transfected with WT FLAG-EGLN1 or the methylation-mimic mutant (K297F). FLAG empty was used as a control. The relative intensities of HIF1α were determined by normalizing the intensities of HIF1α to the intensities of GAPDH. F, Western blot analysis of HIF1α hydroxylation with anti-hydroxy-HIF1α (Pro564) antibody in EGLN1-deficient H1299 cells (EGLN1−/−) transfected with indicated plasmid. The cells were transfected with WT FLAG-EGLN1 or the methylation-mimic mutant (K297F) (HA empty was used as a control), followed by MG-132 (20 μM) treatment for 8 h. The relative intensities of HIF1α-OH/HIF1α were determined by normalizing the intensities of HIF1α-OH to the intensities of HIF1α. G, Western blot analysis of HIF1α hydroxylation with anti-hydroxy-HIF1α (Pro564) antibody in SET7-deficient or WT H1299 cells (SET7−/− or SET7+/+) treated with MG-132 (20 μM) for 8 h. The relative intensities of HIF1α-OH/HIF1α were determined by normalizing the intensities of HIF1α-OH to the intensities of HIF1α. H, Western blot analysis of exogenous Myc-HIF1α expression in HEK293T transfected with indicated plasmid. The cells were cotransfected with Myc-HIF1α, FLAG-EGLN1, and HA-SET7 (HA empty was used as a control), followed by CHX (50 μg/ml) treatment for indicated times. Myc empty was used as a control. The relative intensities of Myc-HIF1α were determined by normalizing the intensities of Myc-HIF1α to the intensities of β-actin. CHX, cycloheximide; EGLN, egg-laying defective nine; HA, hemagglutinin; HIF, hypoxia-inducible factor.
Subsequently, we confirmed that compared to EGLN1−/− H1299 cells with overexpression of WT EGLN1, HIF1α hydroxylation was reduced in EGLN1−/− H1299 cells with overexpression of the methylation-mimic mutant of EGLN1 (K297F) (Fig. 4C). Furthermore, we verified that overexpression of SET7 attenuated HIF1α hydroxylation when EGLN1 was coexpressed (Fig. 4D).
Notably, in EGLN1−/− H1299 cells, overexpression of WT EGLN1 caused a more dramatic reduction of endogenous HIF1α than overexpression of the methylation-mimic mutant of EGLN1 (K297F) (Fig. 4E). In agreement, overexpression of WT EGLN1 caused a more dramatic enhancement of HIF1α hydroxylation than overexpression of the EGLN1-K297F mutant (Fig. 4F). In addition, the level of HIF1α hydroxylation was much higher in SET7-null H1299 cells (SET7−/−) than that in SET7-intact H1299 cells (SET7+/+) (Fig. 4G). Consistently, in cycloheximide pulse chase assay, overexpression of SET7 slowed down degradation of HIF1α when EGLN1 was coexpressed (Fig. 4H).
These data suggest that the prolyl hydroxylase activity of EGLN1 is attenuated by SET7-mediated methylation.
Methylation of EGLN1 promotes cellular hypoxia adaptation by attenuating EGLN1 activity on HIF1α
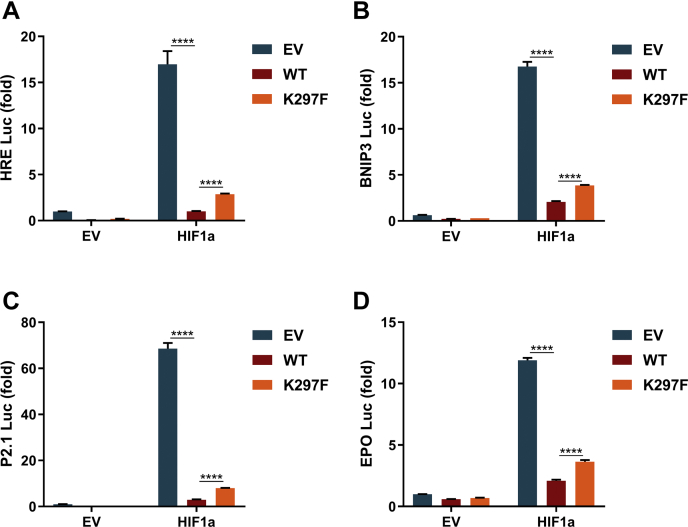
To determine whether methylation of EGLN1 on K297 by SET7 has impacts on HIF1α-mediated hypoxia signaling, we examined the effect of methylation-mimic mutant of EGLN1 (K297F) on HIF1α-induced luciferase reporter activity of typical promoters used for monitoring hypoxia response (27). Overexpression of WT EGLN1 caused a dramatic reduction of all reporter activity, including hypoxia response element reporter, BNIP3 promoter reporter, p2.1 promoter reporter, and EPO promoter reporter, but overexpression of EGLN1-K297F released the suppressive effect of WT EGLN1 on HIF1α significantly (Fig. 5, A–D).
Figure 5.
Methylation of EGLN1 on K297 attenuates its effect on HIF1α-mediated hypoxia signaling.A, hypoxia response element (HRE) reporter activity in HEK293T cells cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the methylation-mimic mutant (K297F). Flag empty (EV) was used as a control. B, BNIP3 promoter activity in HEK293T cells cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the methylation-mimic mutant (K297F). Flag empty (EV) was used as a control. C, P2.1 reporter activity in HEK293T cells cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the methylation-mimic mutant (K297F). Flag empty (EV) was used as a control. D, EPO promoter activity in HEK293T cells cotransfected with Myc-HIF1α and WT FLAG-EGLN1 or the methylation-mimic mutant (K297F). FLAG empty (EV) was used as a control. Data show mean ± SEM; Student’s two tailed t test. EGLN, egg-laying defective nine.
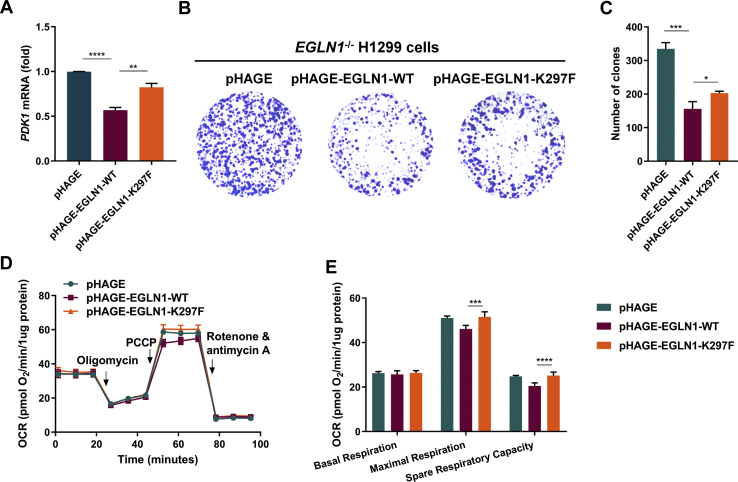
HIF1α is a master regulator for metabolic adaptation under hypoxia (32, 33, 34). Subsequently, we determined the effect of methylation-mimic mutant of EGLN1 (K297F) on cellular hypoxia adaptation. As shown in Figure 6A, overexpression of WT EGLN1 resulted in an overt reduced expression of PDK1, a gatekeeper of glycolysis (35) (Fig. 6A). However, overexpression of EGLN1-K297F recovered this effect significantly (Fig. 6A). In addition, the proliferation rate of EGLN1−/− H1299 cells reconstituted with EGLN1-K297F was higher than EGLN1−/− H1299 cells reconstituted with WT EGLN1 as revealed by colony formation assay (Fig. 6, B and C). Furthermore, the maximal respiration and spare respiratory capacity of EGLN1−/− H1299 cells reconstituted with EGLN1-K297F was higher than EGLN1−/− H1299 cells reconstituted with WT EGLN1 (Fig. 6, D and E).
Figure 6.
Methylation of EGLN1 on K297 promotes cellular hypoxia adaptation.A, quantitative real-time PCR (qPCR) analysis of PDK1 mRNA in EGLN1-deficient H1299 cells (EGLN1−/−) reconstituted with EGLN1 or its mutant by lentivirus. Data show mean ± SEM; Student’s two tailed t test. B and C, colony formation of EGLN1-deficient H1299 cells (EGLN1−/−) reconstituted with EGLN1 or its methylation-mimic mutant (EGLN1-K297F) by lentivirus (n = 3) cultured for 11 days by colony-formation assay. D and E, oxygen consumption rate (OCR) changes were measured by Seahorse XFe24 Extracellular Flux Analyzer in EGLN1-deficient H1299 cells (EGLN1−/−) reconstituted with EGLN1 or its methylation-mimic mutant (EGLN1-K297F) by lentivirus (D). Statistics of basal respiration, maximal respiration, and spare respiratory capacity were presented in (E). EGLN, egg-laying defective nine.
Collectively, these data suggest that methylation of EGLN1 facilitates hypoxia adaptation, which might benefit cell proliferation.
Discussion
Since EGLN1 was first identified as a prolyl hydroxylase of HIF1α, its function and molecular mechanism on hypoxia signaling has been well defined (4, 36, 37). Because of its activity depending on oxygen, it is recognized as an oxygen sensor involved in multiple biological process by targeting different molecules (38, 39, 40). However, whether the activity of EGLN1 is regulated by other factors, particularly by PTMs, is relatively not well understood (15, 16). In this study, we identified that SET7 methylates EGLN1 on K297 to inhibit its enzymatic activity on HIF1α. This result indicates that some PTMs also can affect EGLN1’s activity. To further figure out the effects of PTMs or associated factors of EGLNs on their activity for regulating HIFα may not only enrich our knowledge about the modulation of hypoxia signaling but also help us to understand the fine-tuning mechanisms of cellular hypoxia adaptation.
Notably, even though we provided evidence to show that the methylation of EGLN1 catalyzed by SET7 suppresses the activity of EGLN1 on HIF1α, the methylation-mimic mutant of EGLN1 (in which lysine 297 is mutated into phenylalanine) does not completely lose its enzymatic activity on hydroxylating HIF1α, different from the enzymatically inactive mutant of EGLN1 (EGLN1-H313A). Therefore, the change of lysine to phenylalanine on K297 of EGLN1 might not fully mimic methylation status of EGLN1 catalyzed by SET7 in vivo. Alternatively, other lysine residues might be also methylated by SET7, which are not identified in this study.
For the regulation of hypoxia signaling, in addition to EGLN1 identified here, SET7 is also found to directly methylate HIFα (27, 28). So, SET7 seems to affect hypoxia signaling through multiple mechanisms. Given that the importance of hypoxia signaling in cellular metabolism (2, 3), SET7 appears to play an important role in cellular hypoxia adaptation through influencing the key factors of hypoxia signaling. To further reveal, the effects of SET7 in glycolysis may benefit for understanding the pathological mechanisms of some metabolic disorders.
Experimental procedures
Cell line and culture conditions
HEK293T and H1299 cells originally obtained from American Type Culture Collection were cultured in Dulbecco’s modified Eagle medium (DMEM) (HyClone) with 10% fetal bovine serum. The cells were grown at 37 °C in a humidified incubator containing 5% CO2.
Antibodies and chemical reagents
Antibodies including anti-EGLN1 (#4835), anti-HIF1α (#36169), anti-hydroxy-HIF1α (Pro564) (#3434), anti-SET7 (#2825), and normal rabbit immunoglobulin G (#2729) were purchased from Cell Signaling Technology. Anti-β-actin (#AC026) was purchased from ABclonal. Anti-Flag (#F1804) was purchased from Sigma. Anti-HA (#901515) was purchased from Covance. Anti-Myc (#SC-40) and Anti-GAPDH (#SC-47724) antibody was purchased from Santa Cruz Biotechnology. Anti-SET7 (#ab124708) was purchased from Abcam. MG-132 (#474790) was purchased from Sigma. Cycloheximide (#HY-12320) was purchased from MCE.
Generation of anti-EGLN1-K297Me1 antibody
EGLN1-K297 site-specific methylation antibody (anti-EGLN1-K297Me1 antibody) was generated by using a human EGLN1-methylated peptide (VKERSK(me)AMVACYPGNGC) as an antigen (ABclonal). After purifying the antibodies with excess unmodified peptide (VKERSKAMVACYPGNGC), the specificity of anti-EGLN1-K297Me1 antibody was verified by Western blot analysis.
Immunoprecipitation and Western blot
Coimmunoprecipitation and Western blot analysis were performed as described previously (27). Anti-FLAG antibody–conjugated agarose beads (#A2220), anti-HA antibody–conjugated agarose beads (#A2095), and anti-Myc antibody–conjugated agarose beads (#A7470) were purchased from Sigma. Protein G Sepharose (#17-0618-01) was purchased from GE HealthCare. The Fuji Film LAS4000 mini-luminescent image analyzer was used to photograph the blots.
Identification of EGLN1 methylation site(s) by mass spectrometry
HEK293T cells were cotransfected with HA-EGLN1 and Myc-SET7 plasmids. Cell lysate was immunoprecipitated with anti-HA antibody–conjugated agarose beads overnight. Immunoprecipitated EGLN1 proteins were subjected to 8% SDS-PAGE gel, and EGLN1 bands were excised from the gel and analyzed by mass spectrometry in Protein Gene Biotech. The bands were digested as previously described (41).
The digested peptides were dissolved in 0.1% formic acid, separated on an online nano-flow EASY-nLC 1200 system with a 75 μm × 15 cm analytical column (C18, 3 μm, Thermo Fisher Scientific), and then analyzed on a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Peptides were eluted with the gradient of solvent B (0.1% Formic acid in 80% acetonitrile) increased from 4% to 6% over 1 min, 6% to 10% in 2 min, 10% to 18% in 40 min, 18% to 33% in 10 min, climbed to 90% in 0.5 min, and then held at 90% for 6.5 min, all at a constant flow rate of 300 nl/min. The mass spectrometer was operated in data-dependent acquisition mode with full scans (m/z range of 350–1800) at 60,000 mass resolution using an automatic gain control target value of 3e6. The top 20 most intense precursor ions were selected for following MS/MS fragmentation by higher-energy collision dissociation with normalized collision energy of 28% and analyzed with 15,000 resolution in the Orbitrap. The dynamic exclusion was set to 25 s and the isolation width of precursor ion was set to 1.6 m/z. The maximum injection times were 20 ms and 50 ms for both MS and MS/MS, respectively. The intensity threshold was set to 5000.
The pFind software (version 3.1) (http://pfind.org/software/pFind/index.html) was employed for all MS/MS spectra analysis against the human protein database (https://www.ncbi.nlm.nih.gov/protein) combined with the reverse decoy database and common contaminants (42). Two missed cleavages were allowed for trypsin and open-search algorithm in pFind was used. Methylation (K) was also set as variable modifications. The precursor and fragment ion mass tolerances were 20 ppm and 20 ppm, respectively. Minimum peptide length was set at 6 while the estimated false discovery rate threshold for peptide and protein were specified at maximum 1%.
CRISPR-Cas9 KO cell lines
To generate H1299 or HEK293T knocked-out cell lines of indicated genes, single guide RNA (sgRNA) sequence were ligated into Lenti-CRISPRv2 plasmid and then cotransfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. Six hours after transfection, medium was changed, and viral supernatant was collected and filtered through 0.45-μm strainer. Targeted cells were infected by viral supernatant and selected by 1 μg/ml puromycin for 2 weeks. The sgRNA sequence targeting SET7 is TAGCGACGACGAGATGGTGG. The sgRNA sequence targeting EGLN1 is CCCGCCGCTGTCATTGGCCA.
Luciferase reporter assays
Cells were grown in 24-well plates and transfected with various amounts of plasmids by VigoFect (Vigorous Biotech), as well as with pCMV-Renilla used as an internal control. After the cells were transfected for 18 to 24 h, the luciferase activity was determined by the dual-luciferase reporter assay system (Promega). Data were normalized to Renilla luciferase. Data are reported as mean ± SEM, which are representative of three independent experiments, each performed in triplicate.
Quantitative real-time PCR assay
Total RNAs were extracted using RNAiso Plus (TaKaRa Bio) following the protocol provided by the manufacturer. Complementary DNAs were synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). MonAmp SYBR Green qPCR Mix (high Rox) (Monad Bio) was used for quantitative real-time PCR assays (quantitative PCR). The primers for quantitative real-time PCR assays are listed in Table S1.
Lentivirus-mediated gene transfer
HEK293T cells were transfected with pHAGE empty, pHAGE-EGLN1, or pHAGE-EGLN1-K297F, together with the packaging vectors psPAX2 and pMD2G. Eight hours later, the medium was changed with fresh medium containing 10% fetal bovine serum, 1% streptomycin–penicillin, and 10 μM β-mercaptoethanol. Forty hours later, supernatants were harvested and filtered through 0.45-μm strainer and then used to infect EGLN1-deficient H1299 cells (EGLN1−/−).
Colony formation assay
EGLN1-deficient H1299 cells (EGLN1−/−) reconstituted with EGLN1 or its mutant were seeded in 10 cm cell culture dishes at 3000 cells per dish. After 11 days, the colonies were fixed by methanol, stained with crystal violet (0.5% in methanol) and washed with PBS, and then photographed. Colonies of a suitable size were counted based on the images in each well.
Mitochondrial stress test assay
The oxygen consumption rate under mitochondrial stress test assay was performed using the Seahorse XFe24 Extracellular Flux Analyzer (Agilent Technologies). Mitochondrial stress assay was performed using XF Cell Mito Stress Test kit (Agilent Technologies; #103015-100). The assays were performed according to the manufacturer’s instructions. The H1299 cells (4 × 104 cells/well) were cultured in XF24 cell culture microplate (Agilent Technologies; #102340-100). For mitochondrial stress test assay, oligomycin (1.5 μM), carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (2 μM), and antimycin A and rotenone mixture (0.5 μM) were added to cell culture plate for determining mitochondrial respiration including basal respiration, maximal respiration, and spare respiratory capacity.
Statistical analysis
GraphPad Prism software (7.0) (GraphPad) was used for all statistical analysis. Results with error bars express mean ± SEM. Statistical analysis was performed by using Student’s two-tailed t test. A p value less than 0.05 was considered significant. Statistical significance is represented as follows: ∗p< 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Data availability
Raw mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via iProX partner repository (43) with the dataset identifier PXD032126. The location is http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD032092. All peptide sequences assigned are listed in Table S1.
Supporting information
This article contains supporting information (41, 42).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Drs Peter J. Ratcliffe, William Kaelin, Amato Giaccia, Eric Huang, Navdeep Chandel, Bo Zhong, and Lingqiang Zhang for the generous gifts of reagents.
Author contributions
J. T., X. L., and W. X. conceptualization; J. T. and X. L. methodology; J. T., H. D., Z. W., and X. L. investigation; X. L. data curation; X. L. writing–original draft; X. L. writing–review & editing; J. T., H. D., H. Z., Z. W., Q. L., C. Z., X. C., X. S., S. J., and G. O. visualization; W. X. supervision.
Funding and additional information
This work was supported by NSFC, China [31830101 and 31721005 to W. X.]; the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA24010308 to W. X.); the National Key Research and Development Program of China, China (2018YFD0900602, to W. X.).
Edited by Brian Strahl
Contributor Information
Xing Liu, Email: liuxing@ihb.ac.cn.
Wuhan Xiao, Email: w-xiao@ihb.ac.cn.
Supporting information
References
- 1.Fong G.H., Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- 2.Wong B.W., Kuchnio A., Bruning U., Carmeliet P. Emerging novel functions of the oxygen-sensing prolyl hydroxylase domain enzymes. Trends Biochem. Sci. 2013;38:3–11. doi: 10.1016/j.tibs.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Aragones J., Fraisl P., Baes M., Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell Metab. 2009;9:11–22. doi: 10.1016/j.cmet.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin W.G., Jr., Ratcliffe P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Yang M., Su H., Soga T., Kranc K.R., Pollard P.J. Prolyl hydroxylase domain enzymes: Important regulators of cancer metabolism. Hypoxia (Auckl.) 2014;2:127–142. doi: 10.2147/HP.S47968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 7.Kaelin W.G., Jr. The VHL tumor suppressor gene: Insights into oxygen sensing and cancer. Trans. Am. Clin. Climatol. Assoc. 2017;128:298–307. [PMC free article] [PubMed] [Google Scholar]
- 8.Wheaton W.W., Chandel N.S. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am. J. Physiol. Cell Physiol. 2011;300:C385–C393. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris A.L. Hypoxia--a key regulatory factor in tumour growth. Nat. Rev. Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 10.Semenza G.L. Hypoxia-inducible factor 1 and cardiovascular disease. Annu. Rev. Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corrado C., Fontana S. Hypoxia and HIF signaling: One axis with divergent effects. Int. J. Mol. Sci. 2020;21:5611. doi: 10.3390/ijms21165611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth S., Edlich F., Berchner-Pfannschmidt U., Gneuss S., Jahreis G., Hasgall P.A., Fandrey J., Wenger R.H., Camenisch G. Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 2009;284:23046–23058. doi: 10.1074/jbc.M109.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama K., Gazdoiu S., Abraham R., Pan Z.Q., Ronai Z. Hypoxia-induced assembly of prolyl hydroxylase PHD3 into complexes: Implications for its activity and susceptibility for degradation by the E3 ligase Siah2. Biochem. J. 2007;401:217–226. doi: 10.1042/BJ20061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Conza G., Trusso Cafarello S., Loroch S., Mennerich D., Deschoemaeker S., Di Matteo M., Ehling M., Gevaert K., Prenen H., Zahedi R.P., Sickmann A., Kietzmann T., Moretti F., Mazzone M. The mTOR and PP2A pathways regulate PHD2 phosphorylation to fine-tune HIF1alpha levels and colorectal cancer cell survival under hypoxia. Cell Rep. 2017;18:1699–1712. doi: 10.1016/j.celrep.2017.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey A., Prabhudesai S., Zhang Y., Rao G., Thirugnanam K., Hossen M.N., Dwivedi S.K.D., Ramchandran R., Mukherjee P., Bhattacharya R. Cystathione beta-synthase regulates HIF-1alpha stability through persulfidation of PHD2. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee G., Won H.S., Lee Y.M., Choi J.W., Oh T.I., Jang J.H., Choi D.K., Lim B.O., Kim Y.J., Park J.W., Puigserver P., Lim J.H. Oxidative dimerization of PHD2 is responsible for its inactivation and contributes to metabolic reprogramming via HIF-1alpha activation. Sci. Rep. 2016;6:18928. doi: 10.1038/srep18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C.D., Tempst P., Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Cao R., Xia L., Erdjument-Bromage H., Borchers C., Tempst P., Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 20.Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J., Barlev N.A., Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 21.Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 22.Kontaki H., Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Kurash J.K., Lei H., Shen Q., Marston W.L., Granda B.W., Fan H., Wall D., Li E., Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol. Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian K., Jia D., Kapoor-Vazirani P., Powell D.R., Collins R.E., Sharma D., Peng J., Cheng X., Vertino P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ea C.K., Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X.D., Huang B., Li M., Lamb A., Kelleher N.L., Chen L.F. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., Chen Z., Xu C., Leng X., Cao H., Ouyang G., Xiao W. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res. 2015;43:5081–5098. doi: 10.1093/nar/gkv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y., Nam H.J., Lee J., Park D.Y., Kim C., Yu Y.S., Kim D., Park S.W., Bhin J., Hwang D., Lee H., Koh G.Y., Baek S.H. Methylation-dependent regulation of HIF-1alpha stability restricts retinal and tumour angiogenesis. Nat. Commun. 2016;7:10347. doi: 10.1038/ncomms10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couture J.F., Collazo E., Hauk G., Trievel R.C. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 30.Pradhan S., Chin H.G., Esteve P.O., Jacobsen S.E. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteve P.O., Chin H.G., Benner J., Feehery G.R., Samaranayake M., Horwitz G.A., Jacobsen S.E., Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denko N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 33.Semenza G.L. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb. Symp. Quant. Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 34.Abu-Remaileh M., Aqeilan R.I. Tumor suppressor WWOX regulates glucose metabolism via HIF1 alpha modulation. Cell Death Differ. 2014;21:1805–1814. doi: 10.1038/cdd.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenza G.L. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 36.Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., von Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J., Maxwell P.H., Pugh C.W., Ratcliffe P.J. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 37.Ivan M., Kondo K., Yang H.F., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., Kaelin W.G. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O-2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Wu T., Simon J., Takada M., Saito R., Fan C., Liu X.D., Jonasch E., Xie L., Chen X., Yao X.S., Teh B.T., Tan P., Zheng X.N., Li M.J., et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science. 2018;361:290–295. doi: 10.1126/science.aap8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J.P., Chakraborty A.A., Liu P.D., Gan W.J., Zheng X.N., Inuzuka H., Wang B., Zhang J.F., Zhang L.L., Yuan M., Novak J., Cheng J.Q., Toker A., Signoretti S., Zhang Q., et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353:929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X.N., Zhai B., Koivunen P., Shin S.J., Lu G., Liu J.Y., Geisen C., Chakraborty A.A., Moslehi J.J., Smalley D.M., Wei X., Chen X., Chen Z.M., Beres J.M., Zhang J., et al. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev. 2014;28:1429–1444. doi: 10.1101/gad.242131.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X., Yang M., Liu X., Cheng Z., Ge F. Characterization of lysine monomethylome and methyltransferase in model cyanobacterium Synechocystis sp. PCC 6803. Genomics Proteomics Bioinformatics. 2020;18:289–304. doi: 10.1016/j.gpb.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi H., Liu C., Yang H., Zeng W.F., Wu L., Zhou W.J., Wang R.M., Niu X.N., Ding Y.H., Zhang Y., Wang Z.W., Chen Z.L., Sun R.X., Liu T., Tan G.M., et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- 43.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., Hermjakob H., Zhu Y. iProX: An integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via iProX partner repository (43) with the dataset identifier PXD032126. The location is http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD032092. All peptide sequences assigned are listed in Table S1.