Abstract
Nonalcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases worldwide. However, the molecular mechanisms that promote dysregulation of hepatic triglyceride metabolism and lead to NAFLD are poorly understood, and effective treatments are limited. Leukemia inhibitory factor (LIF) is a member of the interleukin-6 cytokine family and has been shown to regulate a variety of physiological processes, although its role in hepatic triglyceride metabolism remains unknown. In the present study, we measured circulating LIF levels by ELISA in 214 patients with biopsy-diagnosed NAFLD as well as 314 normal control patients. We further investigated the potential role and mechanism of LIF on hepatic lipid metabolism in obese mice. We found that circulating LIF levels correlated with the severity of liver steatosis. Patients with ballooning, fibrosis, lobular inflammation, and abnormally elevated liver injury markers alanine transaminase and aspartate aminotransferase also had higher levels of serum LIF than control patients. Furthermore, animal studies showed that white adipose tissue–derived LIF could ameliorate liver steatosis through activation of hepatic LIF receptor signaling pathways. Together, our results suggested that targeting LIF-LIF receptor signaling might be a promising strategy for treating NAFLD.
Keywords: LIF, LIFR, lipogenesis, nonalcoholic fatty liver disease, obesity
Abbreviations: AMPK, AMP-activated protein kinase; DNL, de novo lipogenesis; ELISA, enzyme-linked immunosorbent assay; eWAT, epididymal white adipose tissue; FA, fatty acid; FFA, free fatty acid; GFP, green fluorescent protein; GTT, glucose tolerance test; HCD, high-carbohydrate diet; HFD, high-fat diet; HG, high-glucose; IL, interleukin; ITT, insulin tolerance test; iWAT, inguinal white adipose tissue; JNK, c-Jun N-terminal kinase; LG, low-glucose; LIF, leukemia inhibitory factor; LIFR, LIF receptor; MAPK, mitogen-activated protein kinase; MCD, methionine-/choline-deficient; MPH, mouse primary hepatocyte; mTOR, mammalian target for rapamycin; NAFL, nonalcoholic fatty liver; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; NASH, nonalcoholic steatohepatitis; NCD, normal chow diet; SREBP-1c, sterol regulatory element–binding protein 1c; STAT, signal transducer and activator of transcription; SVF, stromal vascular fraction; TGs, triglycerides
Nonalcoholic fatty liver disease (NAFLD) has become one of the most common chronic liver diseases worldwide (1), causing a heavy burden to society. It is characterized by accumulation of excessive triglycerides (TGs) in the hepatocytes and closely related to insulin resistance and cardiometabolic risk factors (i.e., abdominal obesity, dyslipidemia, and hyperglycemia) (2, 3, 4, 5). NAFLD can further develop into nonalcoholic steatohepatitis (NASH) characterized by persistent liver injury and inflammation, which can progress to hepatic fibrosis, liver cirrhosis, and hepatocellular carcinoma (1, 6, 7). However, the molecular mechanisms of NAFLD remain to be further determined. Furthermore, it is important to seek new potential therapeutic targets.
Leukemia inhibitory factor (LIF) is a member of the interleukin (IL)-6 cytokine family, which plays important roles in the regulation of many different physiological and pathological processes. It induces activation of signal transducer and activator of transcription (STAT) and mitogen-activated protein kinase (MAPK) pathways by binding to its receptor—LIF receptor (LIFR) (8). Notably, previous studies have shown that LIF can regulate the self-renewal of embryonic stem cells and the tumorigenesis (9, 10). For instance, LIF secreted from pancreatic stellate cells acts on cancer cells to regulate cancer cell differentiation, and changes in the circulating LIF levels are significantly correlated with tumor response to therapy (11). In addition, recent studies suggest that LIF might be involved in metabolic regulation (12, 13, 14, 15). It has been shown that central LIF gene therapy suppresses food intake and decreases body weight and serum leptin in animal models (13). Florholmen et al. reported that LIF stimulates glucose transport in isolated cardiomyocytes (16). Besides, Edwin et al. found that LIF could induce human β-cell differentiation in response to increased metabolic demands (17). However, its role in the regulation of hepatic TG homeostasis has not been explored. In the present study, through human studies and animal experiments, we found that serum LIF levels were significantly associated with severity of NAFLD and LIF treatment could alleviate fatty liver through binding to LIFR and activating the STAT3 signaling pathway. Therefore, our results may provide a potential therapeutic target for the treatment of fatty liver and related metabolic disorders.
Results
Circulating LIF concentration is associated with NAFLD/NASH
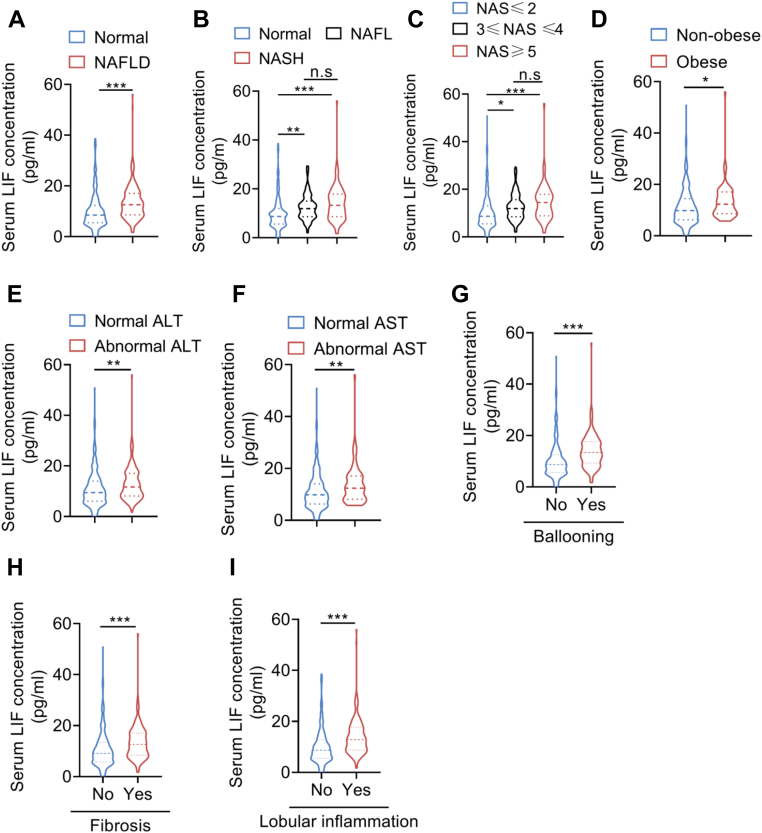
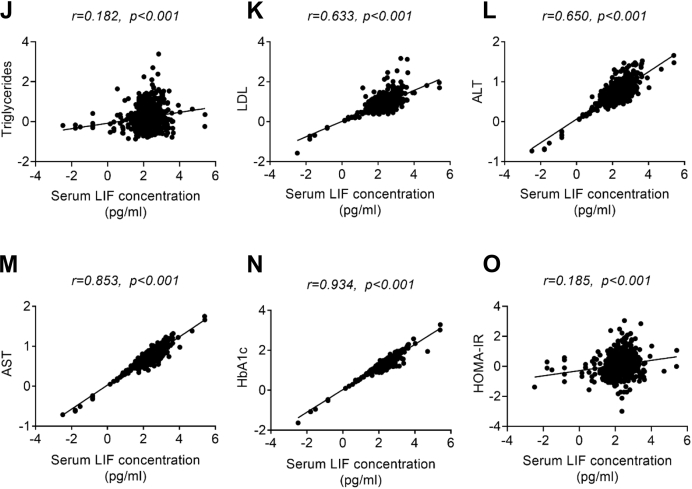
Although the potential functions of LIF on energy metabolism has been recently studied (14, 18), its role in hepatic lipid metabolism is still unknown. Therefore, we first explored the association between serum LIF levels and NAFLD. A total of 528 human subjects, including 314 normal subjects and 214 biopsy-proven NAFLD patients, were recruited in the current study. The clinical and biochemical variables of subjects are summarized in Table 1. Interestingly, patients with NAFLD had higher serum LIF levels than the non-NAFLD subjects (Table 1 and Fig. 1A). Furthermore, patients with simple steatosis and NASH had higher levels of serum LIF than their healthy controls. There was no significant difference between patients with simple steatosis and those with NASH (p > 0.05) (Table 2 and Fig. 1B). Further analysis showed that patients with NAFLD activity score (NAS) NAS≥3 had higher levels of serum LIF than those with NAS≤2 (Fig. 1C). It is worth noting that serum LIF concentration was higher in obese patients than those in nonobese patients (Fig. 1D). Compared with patients with normal serum alanine transaminase (ALT) and aspartate aminotransferase (AST), patients with elevated ALT and AST levels had higher levels of serum LIF (Fig. 1, E and F). Likewise, patients with ballooning, fibrosis, or lobular inflammation had elevated serum LIF levels compared with their controls (Fig. 1, G–I). Furthermore, Spearman correlation analysis showed that serum LIF levels were positively associated with serum TG, low-density lipoprotein (LDL), ALT, AST, glycated hemoglobin, and homeostasis model assessment of insulin resistance (HOMA-IR) (Fig. 1, J–O). These results indicated that LIF might be involved in the development of NAFLD.
Table 1.
Clinical characteristics of patients with NAFLD and healthy subjects
| Variables | Control | NAFLD | p-value | p-valuea |
|---|---|---|---|---|
| Sample size | 314 | 214 | − | |
| Age (years) | 43.0±11.9 | 42.4±11.1 | 0.577 | − |
| Gender (female n, %) | 201 (64.0) | 42 (19.6) | <0.001 | − |
| BMI (kg/m2)b | 21.9 (20.3–24.0) | 26.5 (24.6–28.7) | <0.001 | − |
| Systolic BP (mmHg) | 112.8±11.6 | 128.9±14.6 | <0.001 | <0.001 |
| Diastolic BP (mmHg) | 74.5±10.9 | 84.1±10.2 | <0.001 | <0.001 |
| Triglycerides (mmol/L)b | 0.90 (0.72–1.19) | 1.91 (1.36–2.93) | <0.001 | <0.001 |
| Total cholesterol (mmol/L) | 4.48±0.62 | 5.25±1.21 | <0.001 | <0.001 |
| LDL-c (mmol/L) | 2.50±0.53 | 3.03±0.93 | <0.001 | <0.001 |
| HDL-c (mmol/L) | 1.55±0.38 | 0.99±0.21 | <0.001 | <0.001 |
| FPG (mmol/L)b | 4.7 (4.4–5.0) | 5.3 (4.9–6.1) | <0.001 | <0.001 |
| HbA1c (%)b | 5.3 (5.1–5.4) | 5.8 (5.4–6.6) | <0.001 | <0.001 |
| HOMA-IRb | 0.79 (0.60–1.04) | 1.84 (1.19–2.83) | <0.001 | <0.001 |
| ALT (U/L)b | 15 (12–20) | 48 (28–79) | <0.001 | <0.001 |
| AST (U/L)b | 19 (17–23) | 32 (24–49) | <0.001 | <0.001 |
| ALP (U/L) | 60 (49–72) | 79 (67–93) | <0.001 | <0.001 |
| GGT (U/L)b | 15 (10–21) | 53 (33–85) | <0.001 | <0.001 |
| LIF (pg/ml)b | 8.54 (5.53–12.38) | 12.64 (8.62–17.06) | <0.001 | <0.001 |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; BMI, body mass index; BP, blood pressure; FGP, fasting plasma glucose; GGT, γ-glutamyl transpeptidase; HbA1c, glycated hemoglobin; HDL-c, high-density lipoprotein cholesterol; HGB, hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; LDL-c, low-density lipoprotein cholesterol; LIF, leukemia inhibitory factor; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Data are presented as the mean ± SD or median (interquartile range).
adjusted for age gender and BMI.
Analysis performed on log-transformed data.
Figure 1.
Serum LIF levels are elevated in patients with NAFLD/NASH.A, serum LIF concentration in normal subjects (n = 314) and NAFLD subjects (n = 214). B, serum LIF concentration in normal subjects (n = 314), NAFL subjects (n = 107), and NASH subjects (n = 107). C, comparison of serum LIF levels in patients with NAFLD with different grades (NAS≤2 [n = 338], 3≤NAS≤4 [n = 122], NAS≥5 [n = 68]). D, serum LIF concentration in nonobese subjects (n = 457) and obese subjects (n = 71). E, serum LIF levels in subjects with (n = 401) and without (n = 127) normal ALT. F, serum LIF levels in subjects with (n = 459) and without (n = 69) normal AST. G, concentration of serum LIF in subjects with the presence (n = 361) or absence (n = 167) of ballooning. H, concentration of serum LIF in subjects with the presence (n = 387) or absence (n = 141) of fibrosis. I, concentration of serum LIF in subjects with the presence (n = 345) or absence (n = 183) of lobular inflammation. J–O, association between serum LIF concentration and triglycerides, LDL, ALT, AST, HbA1c, and HOMA-IR (n = 528). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗ p < 0.001. For I–N, spearman correlation analysis was used. ALT, alanine transaminase; AST, aspartate aminotransferase; HbA1c, glycated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; LDL, low-density lipoprotein; LIF, leukemia inhibitory factor; NAFL, nonalcoholic fatty liver; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; NASH, nonalcoholic steatohepatitis.
Table 2.
Clinical characteristics of patients with NAFLD and healthy subjects
| Variables | Control | Non-NASH | NASH |
|---|---|---|---|
| Sample size | 314 | 107 | 107 |
| Age (years) | 43.0±11.9 | 42.3±11.4 | 42.5±11.0 |
| Gender (female n, %) | 201 (64.0) | 21 (19.6)a | 21 (19.6)a |
| BMI (kg/m2)b | 21.9 (20.3–24.0) | 26.0 (24.5–28.1)a | 27.1 (24.6–29.4)a |
| Systolic BP (mmHg) | 112.8±11.6 | 128.1±15.2a | 129.8±14.0a |
| Diastolic BP (mmHg) | 74.5±10.9 | 83.4±10.0a | 84.8±10.4a |
| Triglycerides (mmol/L)b | 0.90 (0.72–1.19) | 1.79 (1.18–2.50)a | 2.18 (1.63–3.10)a |
| Total cholesterol (mmol/L) | 4.48±0.62 | 4.89±1.04a | 5.61±1.26a |
| LDL-c (mmol/L) | 2.50±0.53 | 2.83±0.79a | 3.24±1.02a |
| HDL-c (mmol/L) | 1.55±0.38 | 1.00±0.21a | 0.99±0.20a |
| FPG (mmol/L)b | 4.7 (4.4–5.0) | 5.2 (4.9–6.0)a | 5.4 (4.9–6.2)a |
| HbA1c (%)b | 5.3 (5.1–5.4) | 5.8 (5.4–6.6)a | 5.9 (5.4–6.6)a |
| HOMA-IRb | 0.79 (0.60–1.04) | 1.52 (1.01–2.31)a | 2.20 (1.38–2.06)a |
| ALT (U/L)b | 15 (12–20) | 39 (25–62)a | 61 (38–90)a |
| AST (U/L)b | 19 (17–23) | 29 (23–36)a | 38 (27–56)a |
| ALP (U/L) | 60 (49–72) | 74 (63–92)a | 85 (70–96)a |
| GGT (U/L)b | 15 (10–21) | 44 (27–78)a | 58 (41–109)a |
| LIF (pg/ml)b | 8.54 (5.53–12.38) | 11.98 (8.62–15.02)a | 13.28 (8.62–17.92)a |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; BMI, body mass index; BP, blood pressure; FGP, fasting plasma glucose; GGT, γ-glutamyl transpeptidase; HbA1c, glycated hemoglobin; HDL-c, high-density lipoprotein cholesterol; HGB, hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; LDL-c, low-density lipoprotein cholesterol; LIF, leukemia inhibitory factor; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Data are presented as the mean ± SD or median (interquartile range).
p< 0.01 compared with control.
Analysis performed on log-transformed data.
Identification of LIF as a white fat–enriched protein
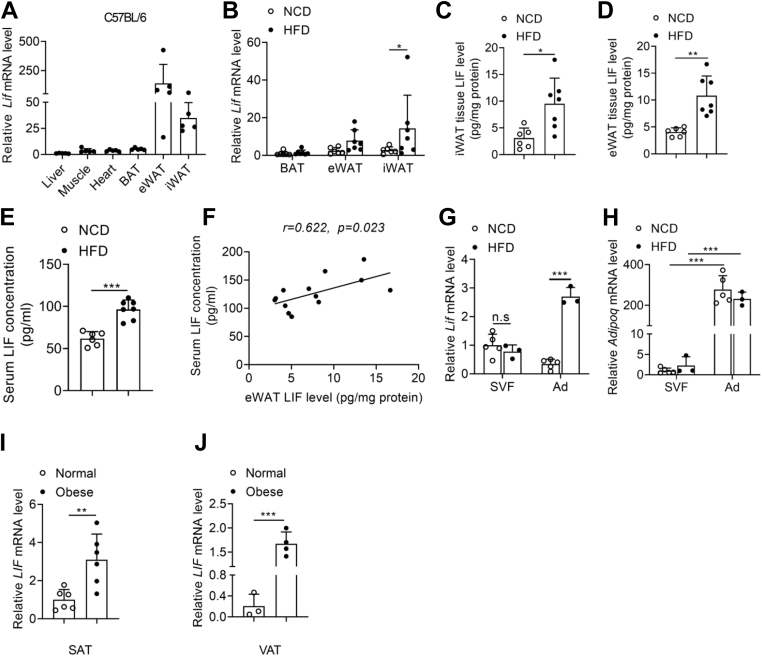
To further investigate the role of LIF in lipid metabolism and fatty liver, the tissue expression of LIF in C57BL/6 mice was detected by quantitative PCR (qPCR) analysis. qPCR analysis showed that LIF mRNA expression was enriched in epididymal white adipose tissue (eWAT) and presented at a marginally high level in inguinal white adipose tissue (iWAT) (Fig. 2A). In contrast, its expression in other tissues, including the liver, brown adipose tissue, skeletal muscle, and heart, was much lower than in WAT (Fig. 2A). To determine LIF expression in obesity-associated NAFLD, the mRNA and protein levels of LIF in the adipose tissues of mice fed a high-fat diet (HFD) for 16 weeks were evaluated. Compared with mice fed a normal chow diet (NCD), the mRNA (Fig. 2B) and protein (Fig. 2, C and D) levels of LIF were significantly increased in the iWAT and eWAT of HFD mice. Similarly, LIF expression in adipose tissue was markedly increased in ob/ob (leptin-deficient) mice compared with lean mice (Fig. S1, A−C). Furthermore, serum LIF concentrations were significantly elevated in HFD and ob/ob mice (Figs. 2E and S1D). Pearson correlation analysis showed that protein levels of LIF in WAT were positively correlated with circulating LIF concentrations (Figs. 2F and S1E). In isolated adipocytes and stromal vascular fractions (SVFs) from eWAT, similar pattern of differential expression of LIF was identified. In isolated mature adipocytes, the expression of LIF was dramatically higher in HFD mice than in NCD mice (Fig. 2, G and H). In agreement, the LIF mRNA levels were significantly elevated in subcutaneous adipose tissue and visceral adipose tissue of obese individuals (Fig. 2, I and J). Collectively, elevated serum LIF levels may be largely attributed to its increased expression in WAT in obesity.
Figure 2.
Serum and adipose tissue LIF levels are increased in obese mice and human.A, relative mRNA levels of Lif in tissues of C57BL/6J mice (n = 5). (B–F) C57BL/6J mice were fed normal chow diet (NCD, n = 6) or high-fat diet (HFD, n = 7) for 16 weeks. B, relative mRNA levels of Lif in brown adipose tissue (BAT), inguinal white adipose tissue (iWAT), and epididymal white adipose tissue (eWAT) in NCD and HFD mice. C, protein levels of LIF in iWAT determined by ELISA in NCD and HFD mice. D, protein levels of LIF in eWAT determined by ELISA in NCD and HFD mice. E, serum LIF concentration in NCD and HFD mice. F, association between serum LIF concentration and eWAT LIF levels in mice (n = 13, including 6 NCD mice and seven HFD mice). G–H, relative mRNA levels of Lif in stromal vascular fraction (SVF) and adipocyte fraction (Ad) isolated from eWAT from C57BL/6J mice were fed NCD (n = 5) or HFD for 12 weeks (n = 3). I, relative mRNA levels of LIF in subcutaneous adipose tissue (SAT) from normal subjects and obese patients (n = 6). J, relative mRNA levels of LIF in visceral adipose tissue (VAT) from normal subjects (n = 3) and obese patients (n = 4). Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. For F, spearman correlation analysis was used. LIF, leukemia inhibitory factor.
Overexpression of LIF in adipose tissue protects mice from NAFLD
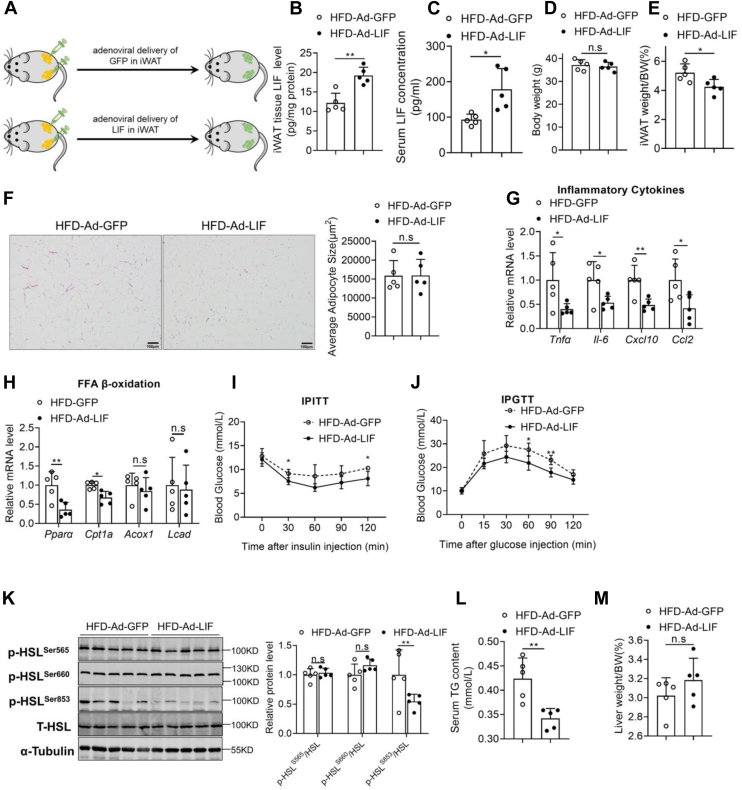
To further address whether increased LIF level in WAT could affect the development of obesity-associated fatty liver, adenovirus particles carrying LIF gene were multipoint-injected into iWAT on both sides of HFD-induced obese mice (Fig. 3A) to mimic the fold change of circulating LIF levels of obese mice. As shown in the Figure 3B, forced overexpression of LIF was observed in iWAT of HFD mice, but not in other metabolic organs (Fig. S2A). HFD-Ad-LIF mice exhibited a significant increase in circulating LIF concentration (Fig. 3C). There was no significant difference in body weight between the two groups, but the mass of iWAT significantly decreased in HFD-Ad-LIF mice (Fig. 3, D and E). Furthermore, histological examination revealed that the inflammation in iWAT was significantly improved in the Ad-LIF mice compared to the Ad-GFP mice (Fig. 3F). Additionally, metabolic status and inflammatory responses in the adipose tissues were assessed. Consistent with the hematoxylin and eosin (H&E) results, several proinflammatory cytokines such as Tnfα, Il-6, Cxcl10, and Ccl2 were significantly decreased in the iWAT of Ad-LIF mice (Fig. 3G). The fatty acid β oxidation genes, including Pparα and Cpt1a, were also dramatically decreased in Ad-LIF mice compared to those in the control condition, while the expressions of Acox1 and Lcad genes were not significantly changed (Fig. 3H). These results suggest that LIF may reduce WAT mass and relieve inflammation and lipotoxic damage (19). Chronic inflammation in adipose tissue has been well established to promote insulin resistance in obesity (20). Glucose tolerance test (GTT) and insulin tolerance test (ITT) revealed that overexpression of LIF ameliorated glucose tolerance and insulin sensitivity (Fig. 3, I and J). Lipolysis is defined as the sequential hydrolysis of triacylglycerol stored in cell lipid droplets to supply fatty acids (FAs) and glycerol as energy substrates for other organs and associated with hyperlipidemia, insulin resistance, and obesity (21, 22). The phosphorylation of hormone-sensitive lipase, a key enzyme for lipolysis, at Ser853 in iWAT was significantly decreased in Ad-LIF mice, while the Ser565 and Ser660 phosphorylation were unaffected (Fig. 3K). Furthermore, the serum free fatty acid (FFA) concentration was unchanged (Fig. S2C). These results suggest that lipolysis might not be the main factor that mediates the actions of LIF. In addition, serum TG and total cholesterol levels were also significantly reduced in Ad-LIF mice (Figs. 3L and S2D).
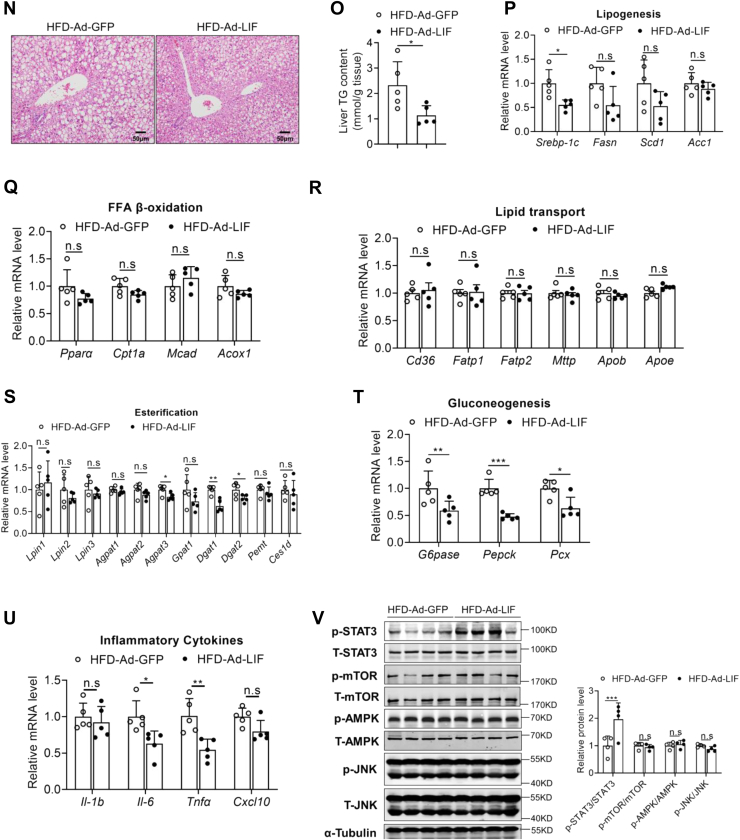
Figure 3.
Overexpression of LIF in adipose tissue protects mice from NAFLD. C57BL/6J mice fed a NCD or HFD for 12 weeks were administrated with adenovirus containing GFP or LIF through multipoint-injected into iWAT and sacrificed in a fed state at day 14 post injection (n = 5). A, illustration of the strategy used to achieve modulation of LIF levels in iWAT via adenoviral delivery. B, protein levels of LIF determined by ELISA in iWAT of HFD mice administrated with Ad-GFP or Ad-LIF. C, serum LIF levels in two groups of HFD mice. D–E, body weight (D) and iWAT weight (E) in two groups of HFD mice. F, representative H&E staining of iWAT sections of HFD mice (100 × magnification). G–H, relative mRNA levels of genes involved in inflammation (G) and FFA β-oxidation (H) in iWAT. I, intraperitoneal insulin tolerance test (IPITT). Mice were fasted for 6 h before the experiment at day 5 post injection, and all mice were treated with intraperitoneal injection of 1 U/kg insulin. J, intraperitoneal glucose tolerance test (IPGTT). Mice were fasted for 16 h before the experiment at day 10 post injection, and all mice were treated with intraperitoneal injection of 1 g/kg glucose. K, protein levels of key lipolytic proteins in iWAT from Ad-GFP and Ad-LIF mice. L, serum TG levels in two groups of HFD mice. M, liver weight in two groups of HFD mice. N, representative H&E staining of liver sections of HFD mice (200 × magnification). O, liver TG contents in two groups of HFD mice. P–U, expression of genes involved in de novo synthesis (P), β-oxidation (Q), lipid transport (R), esterification process (S), gluconeogenesis (T), and inflammation (U) in the livers of two groups. V, Western blot analysis of expression of STAT3, mTOR, AMPK, and JNK pathways in the livers of two groups. Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. AMPK, AMP-activated protein kinase; FFA, free fatty acid; GFP, green fluorescent protein; iWAT, inguinal white adipose tissue; H&E, hematoxylin and eosin; HFD, high-fat diet; JNK, c-Jun N-terminal kinase; LIF, leukemia inhibitory factor; mTOR, mammalian target for rapamycin; NAFLD, nonalcoholic fatty liver disease; NCD, normal chow diet; STAT3, signal transducer and activator of transcription 3; TG, triglyceride.
To further explore the reason behind the improved metabolism after LIF overexpression, we examined the metabolic changes in the liver. Although there was no significant difference in liver weight between the two groups (Fig. 3M), Ad-LIF-injected HFD mice exhibited marked improvement on hepatosteatosis as shown by H&E staining (Fig. 3N) and liver TG contents (Fig. 3O). Hepatic TG storage was regulated by the balance of de novo lipogenesis (DNL), FFA β-oxidation, and lipid uptake and release (23). The expression levels of genes for DNL including Srebp-1c, Fasn, Scd1, and Acc1 were decreased in the Ad-LIF-injected HFD (Fig. 3P); the expression levels of genes for FFA oxidation including Pparα, Cpt1a, Mcad, and Acox1 and lipid transport genes including Cd36, Fatp1, Fatp2, Mttp, Apob, and Apoe showed no significant differences (Fig. 3, Q–R). Besides, expression levels of genes involved in esterification process, including Lpin1, Lpin2, Lpin3, Agpat1, Agpat2, Agpat3, Dgat1, Dgat2, Gpat1, Pemt, and Ces1d, remained unaltered or minorly changed (Fig. 3S). Consistently, overexpression of LIF significantly suppressed gluconeogenic enzymes like G6pase, Pepck, and Pcx and inflammatory genes expression in the liver (Fig. 3, T–U).
High-carbohydrate diets (HCDs) provide a large number of substrates for the de novo fat synthesis (24). To further explore the mechanism of LIF in the regulation of DNL, adenovirus particles carrying LIF gene were multipoint-injected into iWAT on both sides in HCD-induced mice. Forced overexpression of LIF was observed in the iWAT of HCD mice (Fig. S3A). The circulating LIF concentration was significantly increased in the HCD-Ad-LIF mice (Fig. S3B). There was no significant difference in body weight between the two groups (Fig. S3C). The LIF mRNA expression was not upregulated in Ad-LIF mice compared with the controls (Fig. S3D). Although there was no significant difference in liver weight between the two groups (Fig. S3E), HCD-Ad-LIF mice showed improved hepatic steatosis in H&E staining (Fig. S3F) and reduced liver TG contents (Fig. S3G). Consistently, hepatic Srebp-1c expression was significantly reduced in the HCD-Ad-LIF mice (Fig. S3H).
LIF activates the STAT3 signaling pathway
To further explore the potential mechanism of LIF, several signaling pathways were tested by Western blotting. LIF treatment resulted in marked activation of STAT3 signaling pathway in the livers of mice with iWAT-overexpression LIF, as shown by enhanced phosphorylated STAT3; however, the AMP-activated protein kinase (AMPK), mammalian target for rapamycin (mTOR), and c-Jun N-terminal kinase (JNK) signaling pathways were unaffected (Fig. 3V). Likewise, overexpression of LIF activated the STAT3 signaling pathway in the HCD-feeding mice (Fig. S3I). Thus, these results indicated that forced expression of LIF in iWAT could ameliorate liver steatosis by activating the STAT3 signaling pathway.
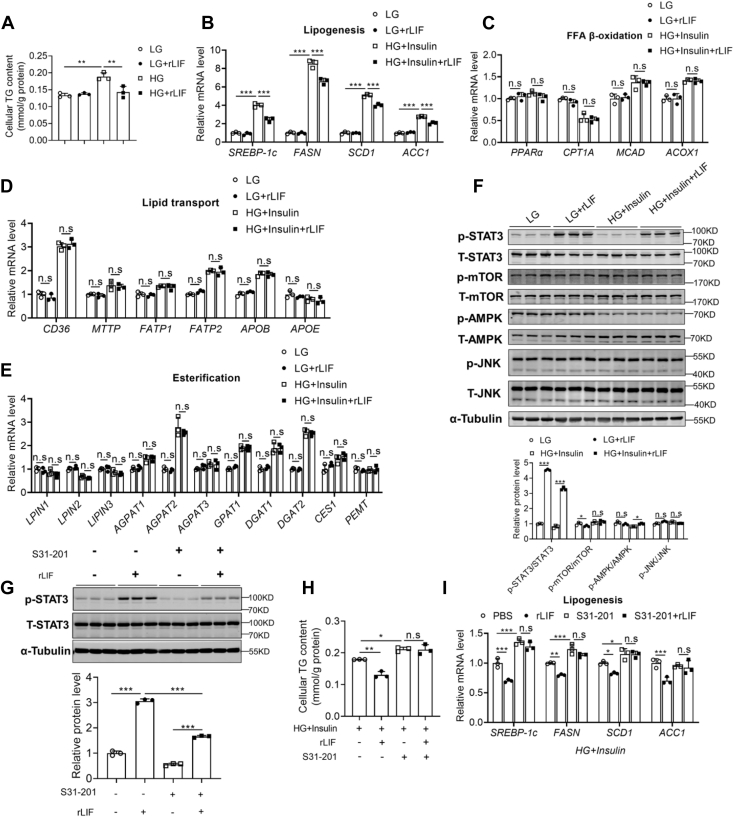
To confirm the in vivo findings, the effects of recombinant LIF protein on cellular TG accumulation in HepG2 cells and mouse primary hepatocytes (MPHs) were examined. The effect of LIF on the SREBP-1c mRNA was determined in HepG2 cells under low-glucose (LG) or high-glucose (HG) plus insulin condition which promotes de novo synthesis. In agreement, recombinant LIF protein significantly reduced cellular TG contents induced by HG plus insulin in HepG2 and MPHs (Figs. 4A and S4A). The expression levels of SREBP-1c gene were reduced by LIF treatment (Figs. 4B and S4B). Meanwhile, the related genes involved in β-oxidation, lipid transporters, and esterification process were not affected (Fig. 4, C–E). Consistently, LIF treatment resulted in marked activation of STAT3 signaling pathway, as shown by enhanced phosphorylated STAT3, while the AMPK, mTOR, and JNK signaling pathways were unaffected (Figs. 4F and S4C).
Figure 4.
LIF activates the STAT3 signaling pathway in vitro. A, cellular TG contents. HepG2 cells were exposed to high glucose (HG, 30 mM) plus insulin (100 nM) or low glucose (LG, 5 mM) as a control for 48 h and then were treated with recombinant human LIF protein (10 ng/ml) or phosphate-buffered saline (PBS) as a vehicle control for 1.5 h. B–E, expression of genes involved in de novo synthesis (B), β-oxidation (C), lipid transport (D), and esterification process (E) in HepG2 cells. HepG2 cells were treated with recombinant LIF protein (10 ng/ml) or PBS for 1.5 h, with preincubation with HG (30 mM) plus insulin (100 nM) or LG (5 mM) for 48 h. F, Western blot analysis of expression of STAT3, mTOR, AMPK, and JNK pathways. HepG2 cells were treated with recombinant LIF protein (10 ng/ml) or PBS for 0.5 h, with preincubation with HG (30 mM) plus insulin (100 nM) or LG (5 mM) for 48 h. G, Western blot analysis of expression of STAT3. HepG2 cells were treated with recombinant LIF protein (10 ng/ml) or PBS for 0.5 h, with or without preincubation with S31 to 201 (100 μM), a STAT3 inhibitor, for 24 h. H and I, cellular TG contents (H) and expression of genes involved in de novo synthesis (I). HepG2 cells were treated with recombinant LIF protein (10 ng/ml) or PBS for 1.5 h, with preincubation with HG (30 mM) plus insulin (100 nM) for 24 h, with or without preincubation with S31 to 201 (100 μM) for 24 h. n = 3 per each group. Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. AMPK, AMP-activated protein kinase; JNK, c-Jun N-terminal kinase; LIF, leukemia inhibitory factor; mTOR, mammalian target for rapamycin; STAT3, signal transducer and activator of transcription 3.
Previous studies have demonstrated that the antisteatotic effect of STAT3 is mediated, at least in part, through inhibition of sterol regulatory SREBP-1c, a master transcription factor in the control of FA and TG synthesis (25). The mRNA levels of SREBP-1c were also inhibited in HepG2 cells treated with LIF and in the livers of LIF-overexpressing mice (Figs. 3P and 4B). To determine whether STAT3 activation was indispensable for the antisteatotic effects of LIF, S31-201, a STAT3 inhibitor, was used to block STAT3 function. S31-201 significantly inhibited LIF-induced activation of STAT3 in vitro (Fig. 4G). These data showed that S31-201 abrogated the effects of LIF on cellular TG contents and lipogenic gene expression (Fig. 4, H and I). Furthermore, either S31-201 or LIF showed little effects on β-oxidation, lipid transporters, and esterification process (Fig. S5, A–C).
LIF alleviates hepatic lipid accumulation via LIFR
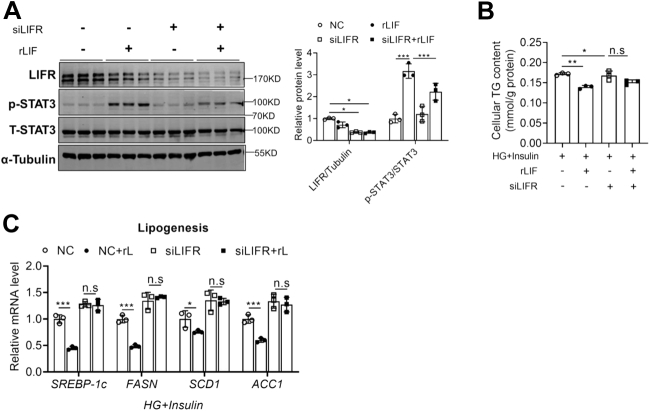
To explore whether LIF regulates lipogenesis through LIFR, which is the receptor for LIF, an siRNA against LIFR were transfected in HepG2 cells. Knockdown of LIFR significantly inhibited the phosphorylation of STAT3 (Fig. 5A) and eliminated the decline of cellular TG levels caused by recombinant LIF protein (Fig. 5B). Consistently, lipogenic genes were downregulated after LIF treatment and significantly upregulated after siLIFR intervention (Fig. 5C). In addition, there were no obvious effects on β-oxidation genes, lipid transport genes, and esterification process (Fig. S6, A–C). These findings suggest that LIF could activate the STAT3 pathway by binding to LIFR, thereby inhibiting the de novo FA synthesis pathway.
Figure 5.
LIF alleviates hepatic lipid accumulation via LIFR.A, Western blot analysis expression of STAT3 pathway in HepG2 cells transfected with LIFR siRNA or NC siRNA for 48 h and treated with recombinant LIF protein (10 ng/ml) or PBS for 0.5 h. B and C, cellular TG contents (B) and expression of genes involved in de novo synthesis (C). HepG2 cells were transfected with LIFR siRNA or NC siRNA for 48 h, treated with HG (30 mM) plus insulin (100 nM) for 24 h, with recombinant LIF protein (10 ng/ml) or PBS for 1.5 h. n = 3 per each group. Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. HG, high-glucose; LIF, leukemia inhibitory factor; LIFR, LIF receptor; NC, nitrocellulose; STAT3, signal transducer and activator of transcription 3; TG, triglyceride.
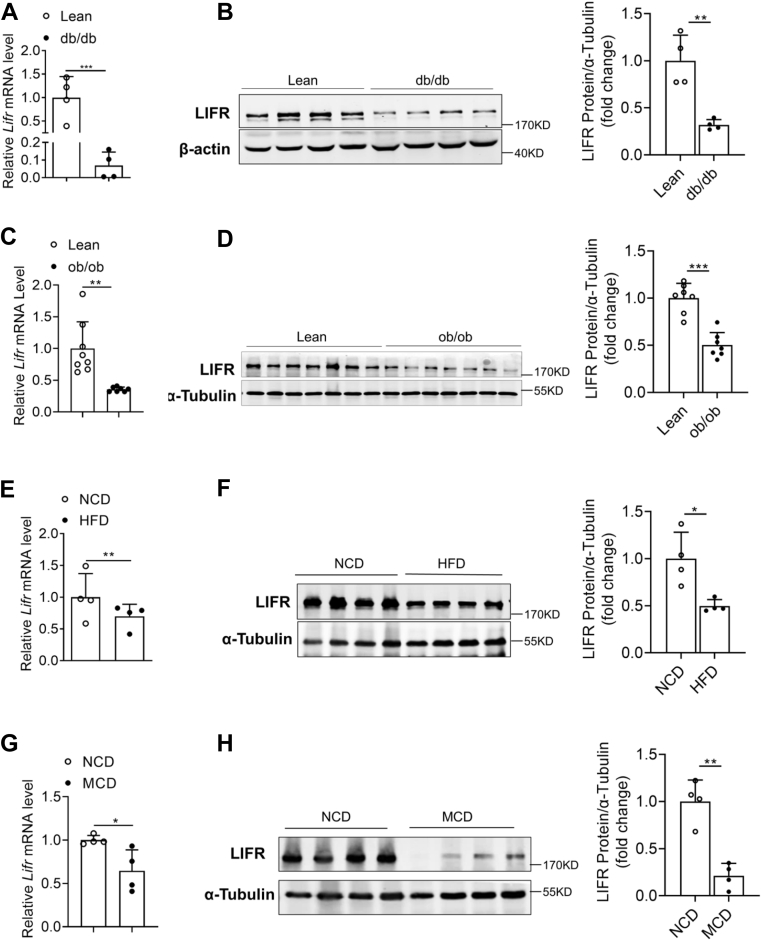
Hepatic LIFR expression is reduced in obese mice
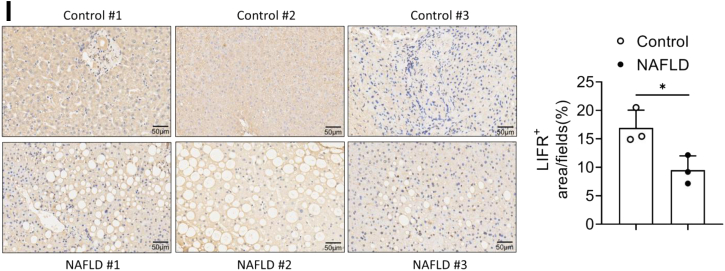
The critical role of hepatic LIFR in lipid metabolism was further investigated. The mRNA and protein levels of LIFR were significantly downregulated in the livers of db/db mice, compared with that in the lean mice (Fig. 6, A and B). To explore whether the downregulation of hepatic LIFR represents a more general feature of obesity-related hepatosteatosis, its expressions in ob/ob mice and HFD-induced obese mice were also examined. In agreement, the mRNA and protein levels of LIFR were markedly decreased in the livers of ob/ob and HFD mice compared with their corresponding controls (Fig. 6, C–F). Furthermore, LIFR expression was also downregulated both at mRNA and protein levels in the liver of methionine/choline-deficient diet mice (Fig. 6, G and H). The hepatic expression of LIFR in human samples was analyzed using immunohistochemistry. Compared with normal controls, the significantly lower LIFR expression was observed in the liver sections of NAFLD patients (Fig. 6I). Collectively, these data suggest that the downregulated expression of hepatic LIFR is involved in the development of NAFLD.
Figure 6.
Hepatic LIFR expression is reduced in obese mice.A and B, relative mRNA levels (A) and protein levels (B) of LIFR in the livers of lean and db/db mice aged 12 weeks (n = 4). C and D, relative mRNA levels (C) and protein levels (D) of LIFR in the livers of lean and ob/ob mice aged 12 weeks (n = 7). E and F, relative mRNA levels (E) and protein levels (F) of LIFR in the livers of C57BL/6 mice fed an NCD or HFD for 16 weeks (n = 4). G and H, relative mRNA levels (G) and protein levels (H) of LIFR in the livers of C57BL/6 mice fed an NCD or methionine-/choline-deficient diet (MCD) for 4 weeks (n = 4). I, immunohistochemistry staining of LIFR in human normal and NAFLD liver tissues (200 × magnification). Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. HFD, high-fat diet; LIFR, leukemia inhibitory factor receptor; NCD, normal chow diet.
Hepatic LIFR protects obese mice against hepatic steatosis
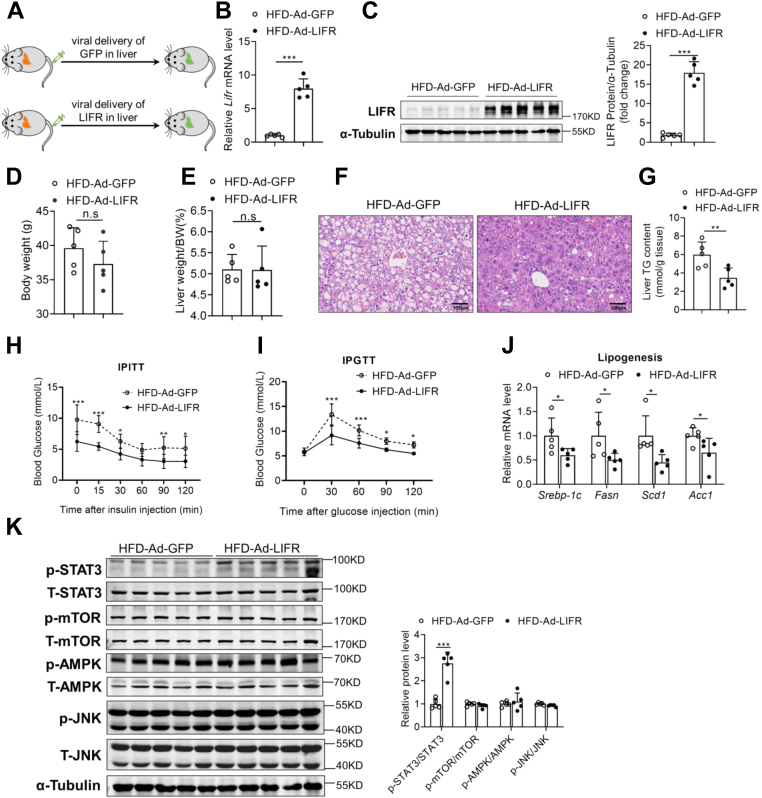
Since overexpression of endogenous LIF alleviated hepatic lipid accumulation in obese mice, whether hepatic LIFR overexpression had similar effects was assessed. To test this, HFD mice were injected with LIFR (HFD-Ad-LIFR) or control (HFD-Ad-GFP) adenovirus via tail vein injection (Fig. 7A). As shown in Figure 7, B and C, forced overexpression of LIFR was observed in livers of HFD mice, but not in other metabolic organs (Fig. S7A). Body weight and liver weight were not altered between the two groups (Fig. 7, D and E). Compared to Ad-GFP controls, Ad-LIFR-injected HFD mice exhibited marked improvement on hepatosteatosis as shown by H&E staining (Fig. 7F). Furthermore, LIFR overexpression dramatically decreased hepatic TG accumulation (Fig. 7G). Serum total cholesterol levels were also significantly reduced in Ad-LIFR-injected HFD mice (Fig. S7B). To explore systemic insulin sensitivity, the ITT and GTT were performed and revealed a significant improvement in insulin resistance and glucose intolerance in HFD-Ad-LIFR mice (Fig. 7, H and I). Consistently, overexpression of LIFR in the livers of HFD mice significantly suppressed the lipogenic genes (Fig. 7J), as well as inflammatory genes (Fig. S5B), without obvious effects on β-oxidation genes and lipid transport genes (Fig. S7, C–E). In line with the improvement of blood glucose, expression levels of gluconeogenic genes were significantly downregulated in Ad-LIFR-injected HFD mice (Fig. S7F).
Figure 7.
LIFR overexpression improves lipid metabolism and hepatic steatosis. C57BL/6J mice were fed an NCD or HFD for 12 weeks. Mice were administrated with adenovirus containing GFP or LIFR through tail vein injection and sacrificed in a fed state at day 14 post injection (n = 5). A, illustration of the strategy used to achieve modulation of LIFR levels in liver. B and C, relative mRNA levels (B) and protein levels of LIFR (C) in the livers of Ad-GFP and Ad-LIFR mice. D and E, body weight (D) and liver weight (E) in two groups of HFD mice. F, representative H&E staining of liver sections (200 × magnification). G, liver TG contents in two groups of HFD mice. H, IPITT. Mice were fasted for 6 h before the experiment at day 5 post injection, and all mice were treated with intraperitoneal injection of 1 U/kg insulin. I, IPGTT. Mice were fasted for 16 h before the experiment at day 10 post injection, and all mice were treated with intraperitoneal injection of 1 g/kg glucose. J, relative mRNA levels of genes involved in de novo synthesis in two groups of mice. K, Western blot analysis of expression of STAT3, mTOR, AMPK, and JNK pathway in the livers of Ad-GFP and Ad-LIFR mice. Data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗ p < 0.001. AMPK, AMP-activated protein kinase; GFP, green fluorescent protein; H&E, hematoxylin and eosin; HFD, high-fat diet; IPGTT, intraperitoneal glucose tolerance test; IPITT, intraperitoneal insulin tolerance test; JNK, c-Jun N-terminal kinase; LIFR, leukemia inhibitory factor receptor; mTOR, mammalian target for rapamycin; NCD, normal chow diet; STAT3, signal transducer and activator of transcription 3.
Consistent with the results observed in the HFD mice, ob/ob mice administrated with Ad-LIFR also displayed significantly reduced liver and serum TG contents (Fig. S8, A–H). Similarly, overexpression of LIFR significantly improved insulin sensitivity and reduced the expression of genes involved in lipogenesis, gluconeogenesis, and inflammation (Fig. S8, I–O). Consistently, the STAT3 pathway was significantly activated; meanwhile, Srebp-1c expression was inhibited after overexpression of LIFR (Fig. S8, K and P). Together, these results demonstrated that hepatic overexpression of LIFR could improve obesity-associated liver steatosis.
Discussion
The essence of NAFLD is the imbalance between the utilization of FAs and the secretion of very-low-density lipoprotein, that is, the progression of DNL is enhanced to form excessive TGs in the liver, FA oxidation is impaired, or very-low-density lipoprotein is abnormally secreted (26, 27). SREBP-1c and carbohydrate response element–binding protein are the key transcription factors in the DNL process, and SREBP-1c is the main regulatory factor which upregulates genes involved in TG synthesis (3). Besides, the expression of SREBP-1c can be mediated by several signaling pathways, including STAT3 signaling (25, 28). Importantly, in obese subjects, the STAT3 phosphorylation is suppressed and SREBP-1c expression is upregulated in the liver (25). In the present study, we identified LIF as a white fat–enriched secreted protein that attenuated hepatic steatosis through activation of hepatic STAT3 signaling. Firstly, the levels of LIF in WAT and serum were increased in obese mice models and patients with NAFLD. Furthermore, circulating LIF levels were significantly and positively correlated with metabolic risk factors. Secondly, overexpression of LIF in WAT ameliorated hepatosteatosis and insulin resistance in obese mice. Thirdly, LIF inhibited SREBP-1c expression through activating the STAT3 signaling pathway, which in turn attenuated TG accumulation in the liver. Therefore, our results indicated that LIF might protect against obesity-associated NAFLD.
Previous studies have shown that LIF regulates many biological processes (9, 10, 17, 29). It has been documented that LIF was highly expressed in tumor tissues and circulating LIF levels were significantly increased in patients with pancreatic cancer and nasopharyngeal carcinoma (11, 30). Additionally, LIF was also identified as a tumor-secreted molecule that promotes adipose tissue and body weight loss in cachectic settings (15). Interestingly, our studies showed that increased WAT expression and circulating LIF levels were associated with the obese state. We found that the expression of LIF in WAT were significantly upregulated at both mRNA and protein levels, and serum LIF level was also significantly increased in ob/ob mice and HFD mice. In humans, we provide novel clinical evidence that circulating LIF concentrations were significantly increased in subjects with NAFLD and were significantly associated with severity of NAFLD and metabolic risk factors. Moreover, our data demonstrated that there was wide variation of circulating LIF levels in mice and humans because of the differences in different species. Besides, we performed additional analysis showing that LIF levels were correlated with the NAS. These results suggest that changes in LIF levels may occur in the early stage of NAFLD. Further studies are needed to explore the effect of LIF in the NASH model. In addition, we found that the expression of LIF in human WAT was upregulated. Notably, despite high endogenous levels in obese mice, elevated circulating LIF concentrations improved fatty liver and insulin resistance in vitro and in vivo. Given that LIF is enriched in WAT, we injected adenoviruses expressing LIF directly into iWAT of HFD mice. To mimic the fold change of circulating LIF levels in the disease state, we chose a relatively low dose of injected adenovirus. As a result, LIF overexpression in iWAT ameliorated HFD-induced adipose inflammation, inhibited FA oxidation in iWAT, reduced insulin resistance, elevated serum LIF, and alleviated hepatic steatosis. Although hepatic LIF mRNA expression was not upregulated in Ad-LIF mice, the results suggested that the improvements of liver metabolic homeostasis would be mainly due to the elevated serum LIF levels after overexpression of LIF in iWAT. Furthermore, recombinant LIF protein ameliorated cellular TG accumulation and lipogenic gene expression induced by HG plus insulin. Thus, these results suggested that LIF might affect metabolic processes via autocrine, paracrine, or endocrine signaling.
LIF signals to target cells through LIFR, which is composed of a signal peptide followed by three main domains. LIFR binds with several IL-6 cytokine family members, including LIF, oncostatin-M, cardiotrophin-1, ciliary neurotrophic factor, neuropoietin, and cardiotrophin-like cytokine factor 1 (31). Currently, LIFR has received extensive attention mainly in the field of tumors, and it plays a role in inhibiting the proliferation and metastasis of cancer (31). LIFR signal is an important link between IL-6 family cytokines and systemic metabolic regulation. Our data demonstrated that LIFR was significantly downregulated in the livers of obese mice and patients with NAFLD. Overexpressing LIFR in obese mice activated the STAT3 signaling pathway, improved fatty liver and insulin resistance, and downregulated the expression of hepatic inflammatory genes. Moreover, knockdown of LIFR in HepG2 cells weakened the beneficial effects on cellular lipid accumulation of exogenous LIF. Therefore, our data indicated that LIF-LIFR signal may play a role in the obesity-associated NAFLD.
In addition, our data showed that levels of LIF in WAT and serum were elevated in obese ob/ob mice, HFD mice, and obese human. In contrast, hepatic expression of LIFR was reduced in NAFLD models. Therefore, our data suggested that reduced expression of LIFR in the liver more likely contributed to elevated circulating LIF levels in vivo. Elevated serum LIF levels in obesity may be attributed to a compensatory response to reduced LIFR expression in the liver, which is similar to FGF21 as previously reported (32, 33, 34). Previous studies showed that serum FGF21 levels were elevated in patients with NAFLD and obese individuals because of the reduced expression of FGF receptors in liver and WAT (32, 33, 34); meanwhile, exogenous FGF21 administered at pharmacologic doses improves metabolic parameters and induces weight loss in obese mice (32). Our findings suggested that obesity-related NAFLD may be a state of “LIF resistance" in that hepatic LIFR expression was decreased. Furthermore, our results showed that LIF mRNA expression was enriched in WAT and protein levels of LIF were positively correlated with circulating LIF concentration in an animal model. These results indicated that WAT might be an important source of LIF. However, we cannot exclude the possibility that LIF derived from other tissues might also be involved in the regulation of hepatic lipid homeostasis.
It has been reported that hypothalamic anti-LIF antibody treatment impaired glucose tolerance (18), while recombinant LIF protein treatment increased muscle glucose uptake (14). In consistency, our data suggested that LIF-LIFR enhanced glucose tolerance and insulin sensitivity in HFD and ob/ob mice, at least in part, via suppression of gluconeogenesis. Further research needs to determine the role and mechanism of LIF-LIFR in the process of glucose metabolism. At the mechanistic level, it is known that LIF-LIFR can activate many signaling pathways, including STAT3, MAPK, AKT, ERK1/2, and mTOR pathways (35, 36). Our study showed that LIF-LIFR activated the STAT3 signaling pathway to suppress SREBP-1c and other downstream target genes related to TG synthesis, contributing to reduce TG contents in the liver of obese mice. However, our data indicated that LIF treatment had no effect on the other signaling pathways in hepatocytes. Thus, the biological functions of LIF and signaling pathways activated by LIF might be variable in different tissues and disease states. Although our study showed that LIF could improve the metabolic phenotype, further studies are warranted to explore the long-term beneficial and potential adverse effects of LIF in NAFLD due to its carcinogenic effects (11, 15, 30). On the other hand, activation of LIFR might be an alternative therapeutic target for the treatment of NAFLD.
Our research has several limitations. First, in this study, we injected adenovirus expressing LIF into iWAT of HFD mice to promote the secretion of LIF instead of using adipose tissue–specific transgenic mice. However, we successfully achieved LIF overexpression in iWAT which resulted in increased circulating LIF levels. Second, although LIF only activated the STAT3 signaling pathway in hepatocytes, we could not exclude the possibility that LIF induces the activation of other pathways. Label-free phosphor-proteomic analysis might help to explore novel signaling in response to LIF treatment. Third, the population study was based on cross-sectional data with a relatively limited sample size; further prospective cohort studies with larger sample size need to determine the causal association between circulating LIF and NAFLD and metabolic disorders.
Conclusions
The present study demonstrated that LIF was an important regulator of liver TG homeostasis. Downregulation of hepatic LIFR expression may contribute to the increased circulating LIF levels in obesity. Exogenous LIF attenuated hepatic steatosis and insulin resistance in a diet-induced mouse model of NAFLD. Therefore, our findings may provide a new therapeutic target for NAFLD. In view of the carcinogenic effects of LIF, it may be impractical to directly use LIF as a therapeutic target. However, it may be applied to target LIFR into the treatment of NAFLD if new compounds that targets LIFR can be developed to avoid carcinogenic effects of LIF.
Experimental procedures
Animal experiments
Male C57BL/6J mice aged 8 to 10 weeks, leptin-deficient ob/ob and leptin receptor–deficient db/db mice were obtained from Nanjing Biomedical Research Institute of Nanjing University (Nanjing). For HFD feeding, C57BL/6J mice were fed a diet containing 60% fat, 20% carbohydrate, and 20% protein (D12492, Research Diets). For HCD feeding, C57BL/6J mice were fed a diet containing 10% fat, 70% carbohydrate, and 20% protein (M21040805, MolDiets) for 8 weeks. For NASH diet feeding, C57BL/6J mice were fed a methionine- and choline-deficient diet (A02082002B, Research Diets) for 4 weeks. The mice were sacrificed in a fed state. The animal protocol was reviewed and approved by the Animal Care Committees of Southern Medical University.
Human samples
A total of 214 patients with biopsy-proven NAFLD and 314 healthy controls were included in our study. The NAFLD group consisted of 107 patients with nonalcoholic fatty liver (NAFL) and 107 patients with NASH. The diagnosis of NAFLD was based on the following four histopathological features, including hepatic macrovesicular steatosis, hepatocellular ballooning, lobular inflammation, and stage of fibrosis. The NAS was calculated according to the Kleiner scoring system. Serum samples were obtained from healthy controls and patients with NAFLD. The HOMA-IR measures were conducted in Nanfang Hospital of Southern Medical University. Serum insulin concentrations were measured using electrochemiluminescence immunoassay (Roche Elecsys Insulin Test, Roche Diagnostics). The HOMA-IR value was calculated as the level of fasting glucose (measured in millimoles per liter) × the level of fasting insulin (measured in microunits per milliliter) divided by 22.5. For analysis of hepatic LIFR expression in humans, liver tissues were collected from the normal cohort and NAFLD. The subcutaneous adipose tissue and visceral adipose tissue tissues were recruited from additional cohort at Nanfang Hospital of Southern Medical University. The clinical characteristics of participants are presented in Table 1. All subjects provided written informed consent for this study. The human study was approved by institutional review boards of the First Affiliated Hospital of Wenzhou Medical University and Nanfang Hospital of Southern Medical University, in line with the ethical guidelines of the 1975 Declaration of Helsinki. The methods were carried out in accordance with the approved guidelines.
Immunohistochemical assessment
Human liver biopsies (0.5–1 cm3) were fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin. The sections were deparaffinized and rehydrated, and endogenous peroxidase activity and background binding were inhibited using 3% hydrogen peroxide in methanol and 1% bovine serum, respectively. The slides were immunoprobed with LIFR antibodies (ab202847, Abcam) in a dilution of 1:300. Images were taken using an inverted microscope under × 200 magnification (each group contained three samples).
Measurement of serum and tissue LIF
Human serum LIF levels were measured using a commercially available enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer’s protocol (DLF00B, R&D). The sensitivity of the assay is 5.36 pg/ml. Mouse serum LIF levels were measured using a commercially available ELISA kit according to the manufacturer’s protocol (ml002284, ML Bio). The minimum detectable dose of mouse LIF is typically less than 1.0 pg/ml. For ELISA measurements of tissue LIF, eWAT and iWAT were rinsed with ice-cold phosphate-buffered saline (PBS; 0.01 M, pH=7.4) to remove excess blood thoroughly. Fresh-frozen samples were homogenized in PBS buffer containing antiprotease cocktail (Beyotime). The volume depends on the weight of the tissue. Ten microliters of PBS would be appropriate to 1 mg of tissue pieces. Homogenates were snap frozen, thawed on ice, and centrifuged for 5 min at 5000g to get the supernatant at 4 °C.
Mature adipocytes and SVF separation
eWAT was dissected from lean and HFD-fed obese C57BL/6J mice. The tissues were minced into ∼2- to 3-mm pieces and digested in PBS supplemented with 10 mM CaCl2, 2.4 U/ml Dispase Ⅱ, and 2 mg/ml type II collagenase for 30 min at 37 °C with gentle agitation. Digested tissues were filtered through 100-μm nylon filters into 50-ml conical tubes followed by centrifugation at 500g for 5 min. The precipitation part containing the SVF was transferred into another tube using a needle syringe. The combined precipitation was centrifuged at 4000 rpm for 5 min to obtain cell pellets containing the SVF for RNAs. The top layer containing mature adipocytes was washed again with Dulbecco's modified Eagle's medium (DMEM; Gibco) buffer and used for total RNA isolation. After the final removal of infranatant, the adipocyte fraction was lysed for RNAs.
Primary hepatocyte isolation
MPHs were isolated from 6- to 8-week-old male C57BL/6J mice by collagenase perfusion. Freshly prepared hepatocytes were suspended in DMEM supplemented with 10% fetal bovine serum (FBS, Excell) and 1% penicillin–streptomycin and then seeded at a final density of 0.5 × 106 cells/well in 6-well plates. After 6 h, cells were washed with PBS, and the medium was replaced with fresh 10% FBS HG DMEM complete medium.
Cell culture
HepG2 cells and MPHs were cultured in DMEM supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin and incubated at 37 °C in a 5% CO2 atmosphere. Cells were evenly seeded onto 6-well plates and incubated for 24 h. For the in vitro model of cellular steatosis, HepG2 cells and MPHs were incubated in glucose-free DMEM supplemented with 0.25% bovine serum albumin overnight and exposed to high glucose (30 mM) plus insulin (100 nM) to induce cellular TG accumulation or low glucose (5 mM) as a control for 24 to 48 h and then treated with 10 ng/ml recombinant human LIF protein (Sino Biological, 14,890-H08H) or recombinant mouse LIF protein (10 ng/ml, Sino Biological, 50,755-M08H). To further investigate the role of STAT3 activation, HepG2 cells were treated with S31 to 201 (100 μM) (S1155, Selleck), a STAT3 inhibitor, to block STAT3 function and then exposed to recombinant LIF protein or vehicle control. To silence LIFR expression in HepG2 cells, a negative control siRNA was purchased from RIBOBIO (siN0000001, Guangzhou) and siRNA targeting LIFRs (siLIFRs) were synthesized by RIBOBIO (Guangzhou) with the following sequences: siLIFR 5′-CGATCACAATCAACAA TTT-3’. siRNA was transferred into cells using advanced transfection reagent (AD600025, ZETA LIFE) according to the manufacturer’s instructions.
Adenovirus
Adenoviruses using CMV promoter expressing murine LIFR gene or green fluorescent protein (GFP) (Ad-LIFR and Ad-GFP) were constructed by Genechem Company. Overexpression of LIFR or GFP in the liver of HFD mice were achieved by means of tail vein injection of Ad-LIFR or Ad-GFP (1 × 109 plaque-forming units [pfu] for each mouse). Adenovirus particles carrying LIF or GFP (Ad-LIF and Ad-GFP) were multipoint-injected into iWAT on both sides of HFD-induced obese mice (3 × 107 pfu on each side, Genechem Company). Mice were sacrificed at day 14 post injection in the fed state.
Glucose and insulin tolerance tests
C57BL/6J mice were fed HFD for 12 weeks and then given the adenovirus constructs. The ITTs were performed on the fifth and the GTTs were performed on the 10th day after the adenovirus injection. For ITT, mice were injected intraperitoneally with insulin after a 6-h fast. For GTT, mice were injected intraperitoneally with glucose after a 16-h fast. Blood glucose levels were determined by a glucose meter (ACCU-CHEK, Roche) by tail venipuncture immediately before (0 min) and after (30, 60, 90, and 120 min) insulin or glucose injection.
Quantitative real-time PCR
Total RNA was isolated from cells or tissue using TRIzol (Thermo Fisher Scientific). The reverse transcription products of total RNA from each sample were prepared using the RT reagent kit (Takara) according to the manufacturer’s protocol. The SYBR Real-time PCR Master Mix kit (Yeason) was used for mRNA amplification, and the amplification reactions were run on a LightCycler 480 Instrument (Roche). Data calculated by quantitative real-time PCR were analyzed by the 2-ΔΔCt method. Primers used for the analyses are shown in Table S1.
Western blot
Tissues or cells were homogenized with RIPA buffer (Beyotime) on ice, and supernatants were harvested. Cell lysate protein was quantified by the BCA Protein Assay Kit (Pierce) and separated on 10% polyacrylamide gels, which were then transferred to nitrocellulose membranes (Millipore, Billerica, MA). After blocking with 5% skim milk, the membranes were incubated with the indicated primary antibodies overnight at 4 °C and then with secondary antibodies for 1 h at room temperature (RT) in dark. Finally, the protein expression signals were visualized by Odyssey imaging systems (LI-COR). Antibodies used are shown in Table S2.
Serum lipid and liver TG assays
Blood was collected from the mice via the orbital venous plexus. Serum TG (Applygen Technologies Inc), total cholesterol (Nanjing Jiancheng Bio), and serum FFA (ML Bio) levels were determined according to the manufacturer’s instructions. The intracellular and liver lipids were extracted using a triglyceride assay kit (Applygen Technologies Inc). Briefly, approximately 20 mg of liver tissue was homogenized and then the homogenate was centrifuged at 3000 rpm for 5 min. The subsequent step followed the manufacturer’s instructions.
Liver slice collection and histopathological analysis
Apportion of freshly extracted liver tissue was cut into ∼3-mm slices and washed in PBS at RT, fixed in 4% w/v paraformaldehyde for 10 min at RT, and then incubated for 6 h at 4 °C. After a PBS rinse, the samples were dehydrated in 80%, 90%, 95%, and 100% ethanol solutions for 1 h each at 4 °C and finally in dimethyl benzene at RT. Paraffin immersion was at 60 °C for 2 h and then embedding at 4 °C. A rotary microtome was used for cutting 5-μm sections at 4 °C. The sections were then deparaffinized and prepared for H&E staining (Servicebio). Images were acquired under a Leica microscope (Leica DMi8-M).
Statistical analysis
All data were analyzed using SPSS 22.0 (IBM Corp). Data from mice are presented as mean ± standard deviation (SD); for data on human subjects, data are presented as means ± SD or median (interquartile range) for continuous variables or number and percentage for categorical variables. Data that were not normally distributed were logarithmically transformed before analysis. When comparing two groups, unpaired two-tailed Student’s t test was used to determine statistical significance. When more than two groups were compared, two-way analysis of variance (ANOVA) followed by multiple comparisons was applied. Group differences were evaluated using the general linear model for continuous variables and the chi-squared test for categorical variables. Multivariable logistic regression models were used to examine the association of circulating LIF levels with risks of NAFLD. Correlation analysis was performed using the Pearson correlation. Two-sided values of p <0.05 were considered statistically significant. ∗, ∗∗, and ∗∗∗ represent p < 0.05, p < 0.01, and p < 0.001, respectively.
Data availability
This study includes no data deposited in external repositories.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Y. L. and H. Z. conceptualization; Y. Y., K. L., F. T., W. W., B. Z., X. Z., J. L., X. Y., Y. D., W. L., S. L., P. Z., and D. L. investigation; Y. Y., K. L., and F. T. formal analysis; M. Z., J. L., Y. L., and H. Z. methodology; M. Z. and J. L. data curation; Y. Y., K. L., Y. L., and H. Z. writing-original draft; K. L. writing-review and editing; H. Z. and Y. L. supervision.
Funding and additional information
This study was supported by the National Key Research and Development Project (No.2018YFA0800404), Natural Science Foundation and Key-Area Research and Development Program of Guangdong Province (No.2018B030311031), National Natural Science Foundation of China (Nos.81970736, 81974119 and 82070588), and the Program of Shanghai Academic Research Leader by Shanghai Municipal Science and Technology Committee (No. 21XD1423400). Dr Huijie Zhang was partially supported by Distinguished Young Scholars Training Program of Nanfang Hospital.
Edited by Qi-Qun Tang
Contributor Information
Yan Lu, Email: lu.yan2@zs-hospital.sh.cn.
Huijie Zhang, Email: huijiezhang2005@126.com.
Supporting information
References
- 1.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M., George J., Bugianesi E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Khan R.S., Bril F., Cusi K., Newsome P.N. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatol. (Baltimore, Md.) 2019;70:711–724. doi: 10.1002/hep.30429. [DOI] [PubMed] [Google Scholar]
- 4.Cai J., Zhang X.J., Ji Y.X., Zhang P., She Z.G., Li H. Nonalcoholic fatty liver disease pandemic fuels the upsurge in cardiovascular diseases. Circ. Res. 2020;126:679–704. doi: 10.1161/CIRCRESAHA.119.316337. [DOI] [PubMed] [Google Scholar]
- 5.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., Wai-Sun Wong V., Yilmaz Y., George J., Fan J., Vos M.B. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatol. (Baltimore, Md.) 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 6.Francque S., Szabo G., Abdelmalek M.F., Byrne C.D., Cusi K., Dufour J.F., Roden M., Sacks F., Tacke F. Nonalcoholic steatohepatitis: The role of peroxisome proliferator-activated receptors. Nat. Rev. Gastroenterol.Hepatol. 2020;18:24–39. doi: 10.1038/s41575-020-00366-5. [DOI] [PubMed] [Google Scholar]
- 7.Rotman Y., Sanyal A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66:180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., Liu J., Wang J., Hu W., Feng Z. The Emerging Role of Leukemia Inhibitory Factor in Cancer and Therapy. Pharmacol. Ther. 2020;221:107754. doi: 10.1016/j.pharmthera.2020.107754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicola N.A., Babon J.J. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahéron L., Opitz S.L., Zaehres H., Lensch M.W., Andrews P.W., Itskovitz-Eldor J., Daley G.Q. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells (Dayton, Ohio) 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., Gao W., Lytle N.K., Huang P., Yuan X., Dann A.M., Ridinger-Saison M., DelGiorno K.E., Antal C.E., Liang G., Atkins A.R., Erikson G., Sun H., Meisenhelder J., Terenziani E., et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569:131–135. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson J.O., Movérare-Skrtic S., Berndtsson A., Wernstedt I., Carlsten H., Ohlsson C. Leukemia inhibitory factor reduces body fat mass in ovariectomized mice. Eur. J. Endocrinol. 2006;154:349–354. doi: 10.1530/eje.1.02082. [DOI] [PubMed] [Google Scholar]
- 13.Beretta E., Dhillon H., Kalra P.S., Kalra S.P. Central LIF gene therapy suppresses food intake, body weight, serum leptin and insulin for extended periods. Peptides. 2002;23:975–984. doi: 10.1016/s0196-9781(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 14.Brandt N., O'Neill H.M., Kleinert M., Schjerling P., Vernet E., Steinberg G.R., Richter E.A., Jørgensen S.B. Leukemia inhibitory factor increases glucose uptake in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2015;309:E142–E153. doi: 10.1152/ajpendo.00313.2014. [DOI] [PubMed] [Google Scholar]
- 15.Arora G.K., Gupta A., Narayanan S., Guo T., Iyengar P., Infante R.E. Cachexia-associated adipose loss induced by tumor-secreted leukemia inhibitory factor is counterbalanced by decreased leptin. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florholmen G., Thoresen G.H., Rustan A.C., Jensen J., Christensen G., Aas V. Leukaemia inhibitory factor stimulates glucose transport in isolated cardiomyocytes and induces insulin resistance after chronic exposure. Diabetologia. 2006;49:724–731. doi: 10.1007/s00125-006-0150-6. [DOI] [PubMed] [Google Scholar]
- 17.Rosado-Olivieri E.A., Aigha, Kenty J.H., Melton D.A. Identification of a LIF-responsive, replication-competent subpopulation of human β cells. Cell Metab. 2020;31:327–338.e6. doi: 10.1016/j.cmet.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Fioravante M., Bombassaro B., Ramalho A.F., Dragano N.R., Morari J., Solon C., Tobar N., Ramos C.D., Velloso L.A. Inhibition of hypothalamic leukemia inhibitory factor exacerbates diet-induced obesity phenotype. J. Neuroinflam. 2017;14:178. doi: 10.1186/s12974-017-0956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carobbio S., Hagen R.M., Lelliott C.J., Slawik M., Medina-Gomez G., Tan C.Y., Sicard A., Atherton H.J., Barbarroja N., Bjursell M., Bohlooly Y.M., Virtue S., Tuthill A., Lefai E., Laville M., et al. Adaptive changes of the Insig1/SREBP1/SCD1 set point help adipose tissue to cope with increased storage demands of obesity. Diabetes. 2013;62:3697–3708. doi: 10.2337/db12-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane H., Lynch L. Innate immune control of adipose tissue homeostasis. Trends Immunol. 2019;40:857–872. doi: 10.1016/j.it.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Wu T., Liu Q., Li Y., Li H., Chen L., Yang X., Tang Q., Pu S., Kuang J., Li R., Huang Y., Zhang J., Zhang Z., Zhou J., Huang C., et al. Feeding-induced hepatokine, Manf, ameliorates diet-induced obesity by promoting adipose browning via p38 MAPK pathway. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmadian M., Wang Y., Sul H.S. Lipolysis in adipocytes. Int. J. Biochem. Cell Biol. 2010;42:555–559. doi: 10.1016/j.biocel.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne C.D., Targher G. NAFLD: A multisystem disease. J. Hepatol. 2015;62:S47–64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Yki-Järvinen H., Luukkonen P.K., Hodson L., Moore J.B. Dietary carbohydrates and fats in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2021;18:770–786. doi: 10.1038/s41575-021-00472-y. [DOI] [PubMed] [Google Scholar]
- 25.Ueki K., Kondo T., Tseng Y.H., Kahn C.R. Central role of suppressors of cytokine signaling proteins in hepatic steatosis, insulin resistance, and the metabolic syndrome in the mouse. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10422–10427. doi: 10.1073/pnas.0402511101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarei M., Pizarro-Delgado J., Barroso E., Palomer X., Vázquez-Carrera M. Targeting FGF21 for the treatment of nonalcoholic steatohepatitis. Trends Pharmacol. Sci. 2020;41:199–208. doi: 10.1016/j.tips.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z., Yu Y., Cai J., Li H. Emerging molecular targets for treatment of nonalcoholic fatty liver disease. Trends Endocrinol. Metab. 2019;30:903–914. doi: 10.1016/j.tem.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Inoue H., Ogawa W., Ozaki M., Haga S., Matsumoto M., Furukawa K., Hashimoto N., Kido Y., Mori T., Sakaue H., Teshigawara K., Jin S., Iguchi H., Hiramatsu R., LeRoith D., et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat. Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 29.Graf U., Casanova E., Cinelli P.J.G. The role of the leukemia inhibitory factor (LIF) - pathway in derivation and maintenance of murine pluripotent stem cells. Genes (Basel) 2011;2:280–297. doi: 10.3390/genes2010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S.C., Tsang N.M., Chiang W.C., Chang K.P., Hsueh C., Liang Y., Juang J.L., Chow K.P., Chang Y.S. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J. Clin. Invest. 2013;123:5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auernhammer C.J., Melmed S. Leukemia-inhibitory factor-neuroimmune modulator of endocrine function. Endocr. Rev. 2000;21:313–345. doi: 10.1210/edrv.21.3.0400. [DOI] [PubMed] [Google Scholar]
- 32.Fisher F.M., Chui P.C., Antonellis P.J., Bina H.A., Kharitonenkov A., Flier J.S., Maratos-Flier E. Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes. 2010;59:2781–2789. doi: 10.2337/db10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Fang Q., Gao F., Fan J., Zhou J., Wang X., Zhang H., Pan X., Bao Y., Xiang K., Xu A., Jia W. Fibroblast growth factor 21 levels are increased in nonalcoholic fatty liver disease patients and are correlated with hepatic triglyceride. J. Hepatol. 2010;53:934–940. doi: 10.1016/j.jhep.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Gallego-Escuredo J.M., Gómez-Ambrosi J., Catalan V., Domingo P., Giralt M., Frühbeck G., Villarroya F. (2015) Opposite alterations in FGF21 and FGF19 levels and disturbed expression of the receptor machinery for endocrine FGFs in obese patients. Int. J. Obes. 2005;39:121–129. doi: 10.1038/ijo.2014.76. [DOI] [PubMed] [Google Scholar]
- 35.Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Yang Q., Yu H., Wu L., Zhao Y., Zhang C., Yue X., Liu Z., Wu H., Haffty B.G., Feng Z., Hu W. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget. 2014;5:788–801. doi: 10.18632/oncotarget.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study includes no data deposited in external repositories.