Abstract
Using consensus regions in gene sequences encoding the two forms of nitrite reductase (Nir), a key enzyme in the denitrification pathway, we designed two sets of PCR primers to amplify cd1- and Cu-nir. The primers were evaluated by screening defined denitrifying strains, denitrifying isolates from wastewater treatment plants, and extracts from activated sludge. Sequence relationships of nir genes were also established. The cd1 primers were designed to amplify a 778 to 799-bp region of cd1-nir in the six published sequences. Likewise, the Cu primers amplified a 473-bp region in seven of the eight published Cu-nir sequences. Together, the two sets of PCR primers amplified nir genes in nine species within four genera, as well as in four of the seven sludge isolates. The primers did not amplify genes of nondenitrifying strains. The Cu primers amplified the expected fragment in all 13 sludge samples, but cd1-nir fragments were only obtained in five samples. PCR products of the expected sizes were verified as nir genes after hybridization to DNA probes, except in one case. The sequenced nir fragments were related to other nir sequences, demonstrating that the primers amplified the correct gene. The selected primer sites for Cu-nir were conserved, while broad-range primers targeting conserved regions of cd1-nir seem to be difficult to find. We also report on the existence of Cu-nir in Paracoccus denitrificans Pd1222.
Biological denitrification is a respiratory process defined as the enzymatic, stepwise reduction of nitrogen oxides associated with electron transport phosphorylation and evolution of the gases nitric oxide (NO), nitrous oxide (N2O), and molecular itrogen (N2). The ability to denitrify is a facultative trait spread among a wide variety of physiological and taxonomic groups. Nearly 130 denitrifying bacterial species are found within more than 50 genera (32). The denitrification pathway gives them a competitive advantage in low-oxygen environments. However, some also denitrify aerobically. Denitrifying bacteria prosper in practically all habitats, and the reductive process is of global concern. Denitrification causes nitrogen loss in agricultural soils, and emitted N2O destroys the ozone layer and contributes to global warming. Still, denitrification fills an important function in waste treatment by removing excess nitrogen in local environments and by anaerobically degrading organic pollutants.
The key step in the denitrification pathway is the reduction of nitrite by nitrite reductase (Nir). This reaction distinguishes denitrifiers from nitrate respirers. Denitrification includes two known and distinct types of Nir enzymes: one with heme c and heme d1 (cd1-Nir) and the other containing copper (Cu-Nir) (2, 4, 12). The two Nir types are functionally and physiologically equivalent, which is indicated by the fact that the Cu-nir gene from Pseudomonas aureofaciens can be expressed in a P. stutzeri mutant lacking the gene encoding cd1-Nir (10). Whereas cd1-Nir is the predominant reductase in denitrifying bacteria, copper reductases show greater variation in molecular weight and immunological reactions and are present in more taxonomically unrelated strains (5). It has also been demonstrated that the two reductases are mutually exclusive in any given strain, although the Nir type may differ within the same genera and even within the same species (5). This is particularly true for Alcaligenes faecalis isolates.
The classic approach, using 16S rRNA gene sequences to detect and analyze bacterial communities in environmental samples without isolation and cultivation, is not possible when studying denitrifying bacteria, except at the species or genus level. Instead, the phylogenetic diversity of denitrifying bacteria suggests the use of functional probes and PCR primers based on structural nir genes to detect denitrifying bacteria in general. This implies sufficient genetic homology of the structural gene. DNA probes have been more or less successfully used to detect cd1- and Cu-nir genes in culturable denitrifying bacteria, enrichment cultures, and a variety of environmental samples (9, 15, 26, 29, 31). One attempt has been made to PCR amplify nir fragments from denitrifying bacteria (29). That study was, however, limited to experiments with culturable strains, and the primers used were based on three sequences of the cd1-Nir-encoding gene derived from two species (13, 25, 26).
Since then, the number of available nir sequences has increased, facilitating the construction of more reliable and general primers for the study of denitrifying populations. We designed two sets of PCR primers to amplify fragments of the gene coding for the two Nir enzymes using consensus regions in published sequences for the structural nir gene. Our primers were evaluated by screening DNAs from denitrifying strains from culture collections and unidentified denitrifying isolates from different wastewater treatment plants, as well as DNA extracts from activated sludge. Sequence relationships of nir genes were established by Southern blotting and sequencing.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Designations and sources of all of the defined bacterial strains used in this study are listed in Table 1. Isolates of denitrifying bacteria from different activated-sludge plants in Stockholm, Sweden, were kindly provided by Gunnel Dalhammar, Royal Institute of Technology, Stockholm, Sweden (Table 2). Strains and isolates were cultivated aerobically in nutrient broth (Oxoid) at 30°C and stored in 15% glycerol at −70°C.
TABLE 1.
Bacterial strains used in this study and results of PCR amplification of cd1- and Cu-nir genes and Southern blot hybridization of PCR products
| Bacterial strain | Source or referencea | Nir typeb (reference) | Result of:
|
|||
|---|---|---|---|---|---|---|
| PCRc with nir primers
|
Hybridizationd with nir probes
|
|||||
| cd1 | Cu | cd1 | Cu | |||
| Denitrifying strains | ||||||
| Pseudomonas stutzeri ATCC 14405 | ATCC | cd (13) | + | 0 | + | − |
| P. stutzeri CCUG 29240 | CCUG | ? | + | − | + | − |
| P. fluorescens ATCC 33512 | ATCC | cd (5) | + | − | − | − |
| P. fluorescens Mi32 | SLU | ? | + | − | + | − |
| P. aeruginosa CCUG 241 | CCUG | ? | + | 0 | + | − |
| P. aeruginosa Mi11 | SLU | ? | + | − | + | − |
| P. putida CCUG 2479 | CCUG | ? | 0 | + | − | − |
| P. aureofaciens ATCC 13985 | ATCC | Cu (5) | − | − | − | − |
| P. denitrificans ATCC 13867 | CCUG | Cu (5) | 0 | + | − | + |
| P. denitrificans CCUG 2519 | CCUG | ? | 0 | − | − | − |
| Paracoccus denitrificans ATCC 19367 | CCUG | cd (5) | 0 | − | − | − |
| P. denitrificans Pd1222 | A. H. Stouthamer | cd (6) | 0 | + | − | + |
| Ralstonia eutrophae CCUG 13724 | CCUG | ? | + | − | + | − |
| Alcaligenes eutrophus ATCC 17699 | ATCC | cd (24) or Cu (31) | 0 | 0 | − | − |
| A. faecalis ATCC 8750 | ATCC | ? | 0 | + | − | + |
| A. faecalis ATCC 19018 | CCUG | ? | − | 0 | − | + |
| A. denitrificans ATCC 15173 | CCUG | ? | − | − | − | − |
| Achromobacter cycloclastes ATCC 21921 | ATCC | Cu (5) | − | + | − | + |
| Nondenitrifying strains | ||||||
| Escherichia coli TG1 | 22 | ? | − | − | − | − |
| Staphylococcus aureus 8325-4 | 18 | ? | − | − | − | − |
ATCC, American Type Culture Collection; CCUG, Culture Collection of the University of Gothenburg; SLU, Swedish University of Agricultural Sciences (U. Granhall); A. H. Stouthamer, Vrije Universiteit, Amsterdam, Amsterdam, The Netherlands.
?, no data found in the literature.
+, visible band of the expected size; 0, band of any other size; −, no visible band.
A positive hybridization signal (+) was defined as a visible band of the expected size (45°C).
Formerly known as A. eutrophus.
TABLE 2.
Results of PCR amplification of cd1- and Cu-nir genes in bacterial isolates from activated sludge
| Bacterial isolatea | Groupb | Nitrite reduc-tionc | PCRd result obtained with nir primer:
|
|
|---|---|---|---|---|
| cd1 | Cu | |||
| 80g | α-Purple, rhodobacter group | + | + | − |
| k7g | γ-Purple, pseudomonas under-group | + | + | − |
| 89 | NDe | − | + | − |
| 2:99g | β-Purple, rubrivivax group | + | 0 | − |
| R51g | β-Purple, rubrivivax group | + | − | + |
| 110 | β-Purple, rubrivivax group | + | − | − |
| 123 | β-Purple, rubrivivax group | + | − | − |
Isolates were provided by G. Dalhammar, Royal Institute of Technology, Stockholm, Sweden.
Determined by Magnusson et al. (16).
Determined by measurements of nitrite reduction rates (G. Dahlhammar).
+, visible band of the expected size; 0, band of any other size; −, no visible band (this study).
ND, not determined.
DNA extraction.
Bacteria were grown aerobically on an orbital shaker (100 rpm). Cells were harvested after overnight growth from 6 ml of a bacterial suspension by centrifugation for 10 min at 19,000 × g. The pellets were suspended in 2 ml of 0.15 M NaCl–0.10 M EDTA–0.01 M Tris-HCl (pH 7.9) with lysozyme and RNase A added to final concentrations of 1 mg ml−1 and 50 μg ml−1, respectively. After the cells were incubated for 1 h at 37°C, 0.1 ml of a saturated solution of sodium dodecyl sulfate (SDS) in 45% ethanol was added. After 5 min at 37°C, 0.65 ml of 5 M NaClO4 was added. DNA was recovered in the water phase after extraction with 3 ml of chloroform and 0.15 ml of isoamyl alcohol. The DNA was precipitated by adding 2 volumes of ice-cold 95% ethanol. The precipitate was washed once in 70% ice-cold ethanol, air dried, and dissolved in 200 μl of water.
Genomic DNA was extracted from activated-sludge samples collected from different experimental lines at the Kungsängen municipal wastewater treatment plant in Uppsala. The lines were operated as single-sludge systems with predenitrification. Sludge samples (1 ml) were centrifuged for 10 min at 19,000 × g, and the pellets were resuspended in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). After washing the pellets twice with TE buffer and suspending them in 100 μl of TE buffer with 1% Tween 80, we lysed the cells by boiling the samples for 1 min and then freezing them at −70°C for 1 min. The thawing-freezing procedure was repeated three times. The supernatants were collected after centrifugation (19,000 × g, 10 min), and DNA originating from 3 ml of activated sludge was purified and concentrated by using an Elutip-d minicolumn (Schleicher & Schuell) attached to a 0.45-μm-pore-size cellulose acetate prefilter. DNA was concentrated by ethanol precipitation, dried, and suspended in 100 μl of water.
PCR amplification of nir genes.
Primers for PCR were based on cd1- and Cu-nir genes from denitrifying bacteria and amplified nir fragments of approximately 800 and 473 bp, respectively (Fig. 1). The oligonucleotides were purchased from Pharmacia Biotech. PCR amplification of nir fragments was carried out with 50-μl reaction mixtures in 0.5-ml Eppendorf tubes containing 10 to 50 ng of template DNA, 1.25 U of Taq polymerase (Pharmacia Biotech) with the manufacturer’s reaction buffer at 1.5 mM MgCl2, 10 nmol of each deoxynucleotide triphosphate, and 50 pmol of each primer. The reaction mixtures were covered with mineral oil and placed in a thermocycler (Perkin Elmer). The PCR was run with initial denaturation of the DNAs at 94°C for 3 min followed by 35 cycles of 30 s at 94°C, 1 min at 57°C, and 1 min at 73°C. The reaction was completed after 10 min at 75°C. We determined an optimal annealing temperature in the interval between 50 and 65°C.
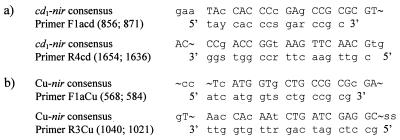
FIG. 1.
Consensus regions and PCR primers for the gene encoding Nir. Uppercase letters represent total homology, lowercase letters point to one deviating nucleotide, and hooks (˜) indicate no consensus (R, A and G; S, C and G; Y, C and T). (a) cd1 primers and alignments of cd1-nir genes. The positions of the first and last nucleotides in each primer corresponding to the gene sequence of P. stutzeri ATCC 14405 are in parentheses. (b) Cu primers and alignments of Cu-nir genes. The positions of the first and last nucleotides in each primer corresponding to the gene sequence of A. faecalis S-6 are in parentheses.
Nucleotide sequencing and computer analysis.
PCR products recovered from an agarose gel and purified with GenElute Minus EtBr Spin Columns (Supelco) were sequenced on both strands by using the Thermo Sequenase dye terminator cycle sequencing premix kit (Amersham Life Science) with 50 to 100 ng of template DNA. The original PCR primers (Fig. 1) and two other oligonucleotides were used as sequencing primers (Table 3). An ABI PRISM 377 (Perkin-Elmer) automated DNA sequencer was used for sequencing. DNA sequences were analyzed with the PCGene program package (IntelliGenetics). Pairwise similarity was calculated with the NALIGN module by the method of Myers and Miller (17), while multiple-sequence alignments were determined with the Clustal V software (11). Phylogenetic trees were constructed from the alignments by using the neighbor-joining method of Saitou and Nei (22). Tree topology was evaluated by bootstrap analysis using 10,000 replicates.
TABLE 3.
Oligonucleotides used for DNA probe construction and sequencing
| Primer | Use | Sequencea (5′ to 3′) |
|---|---|---|
| FPs | cd1 probe (P. stutzeri) | CAT CGT CGC TTC GCA CAT |
| RPs | cd1 probe (P. stutzeri) | GGT GAT GAC GCG CTT GAG |
| FAf | Cu probe (A. faecalis) | GGC AAC TCG CTG CAT TAC GAC C |
| RAf | Cu probe (A. faecalis) | CCA GGC TGC TTG AAG GTG TAG AGC |
| IcdR | Internal cd1-nir sequence | GGT CTT GAT RAA CAG CGW GCC |
| IIcdF | Internal cd1-nir sequence | GGC WCG CTG TTY ATC AAG ACC |
R, A and G; W, A and T; Y, C and T.
DNA probes for detection of nir genes.
DNA probes for detection of nir genes were created by amplifying nir fragments from P. stutzeri (ATCC 14405) and A. faecalis (ATCC 8750) with internal primers (Table 3). The cd1 probe was a 736-bp fragment starting at 882 bp from the 5′ end within the amplified 799-bp region of the nir gene of P. stutzeri (ATCC 14405). The Cu probe was a 398-bp fragment within the 473-bp region of the nir gene of A. faecalis (ATCC 8750). After PCR amplification, the fragments were separated by agarose gel electrophoresis, purified with GenElute Minus EtBr Spin Columns (Supelco), and then labeled with [α-32P]CTP by random priming using the Multiprime DNA labeling system kit (Amersham). The labeled probes were separated from unincorporated nucleotides on a Sepharose CL-6B (Pharmacia Biotech) spin column and used immediately.
Southern blotting and hybridization conditions.
The PCR-amplified fragments were transferred to Hybond-N+ nylon membranes (Amersham) with 20× SSC (3 mol of NaCl liter−1 and 0.3 mol of Na citrate liter−1) by vacuum blotting (VacuGene XL; Pharmacia) and cross-linked by UV light in a UV oven (Hoefer Pharmacia Biotech). The blots were prehybridized at 45°C for 2 h in tubes containing 50 ml of a hybridization solution containing 6× SSC, 0.5% SDS, and 3× Denhardt’s reagent (0.6 g each of Ficoll, polyvinylpyrrolidone, and bovine serum albumin liter−1). The blots were hybridized for 17 h at 45°C in 15 ml of hybridization solution with the respective, previously heat-denatured probe (4 × 105 cpm ml−1). After hybridization, the filters were washed twice for 15 min each time in 2× SSC–0.1% SDS at room temperature and then twice for 15 min each time in 0.1× SSC–0.1% SDS at 45, 50, 55, 60, and 65°C, respectively, in consecutive steps. The hybridization signals were visualized on a PhosphorImager (Molecular Dynamics) after each washing step.
Nucleotide sequence accession numbers.
The partial nir gene nucleotide sequences of A. faecalis ATCC 8750, Achromobacter cycloclastes ATCC 21921, Paracoccus denitrificans Pd1222, Ralstonia eutropha CCUG 13724, P. aeruginosa CCUG 241, P. aeruginosa Mi11, and P. fluorescens Mi32 have been submitted to GenBank under accession no. AF114786 to AF114792.
RESULTS
Selection of PCR primers and optimization of annealing temperature.
We created PCR primers to amplify nir fragments coding for cd1- and Cu-Nir (Fig. 1). To do this, we used consensus regions in sequences for the structural genes encoding Nir. These sequences were retrieved from the GenBank, EMBL, and DDBJ databases. The primers were evaluated by searching the databases with the NCBI sequence similarity search tool BLAST. Single primers had similarity to other sequences, but for the pairwise primers, BLAST predicted only genes encoding Nir. The cd1 primers were designed to amplify a 778 to 799-bp region of the nir gene in the six published sequences, i.e., P. stutzeri ATCC 14405 (13; X56813), P. stutzeri JM300 (26; M80653), Alcaligenes eutrophus H16 (21; X91394), P. aeruginosa NCTC 6750 (25; X16452), P. denitrificans Pd1222 (6; U05002), and P. denitrificans LMD 92.63 (U75413). Likewise, the Cu primers amplified a 473-bp region in seven Cu-nir sequences, Achromobacter cycloclastes (Z48635), A. faecalis S-6 (18; D13155), Pseudomonas sp. strain G-179 (31; M97294), Rhodobacter sphaeroides ATCC 17025 (28; U62291), Alcaligenes xylosoxidans NCIB 11015 (AF051831), Bradyrhizobium japonicum USDA 110 (AJ002516), and Rhizobium “hedysari” HCNT1 (27; U65658). The nucleotide sequence of the gene encoding Cu-Nir from P. aureofaciens ATCC 13985 (10; Z21945) did not share consensus regions of suitable sizes with the other seven Cu-nir sequences and was excluded when we designed the Cu primers. The Cu primers have 100% homology with the Cu-nir sequence of R. “hedysari” HCNT1, but nitrite reduction is not coupled to energy conservation in this strain (27). All four PCR primers include wobbling at one to three positions to cover some of the variation found in the published sequences.
Annealing temperatures in the interval between 50 and 65°C were evaluated, and the optimum was determined to be 57°C. At temperatures below 57°C, nonspecific binding of the primers resulted in a noticeable number of amplification products from all strains. Although fewer bands were observed at higher temperatures, some of the fragments of the expected size were lost. We decided to allow some nonspecific binding in order to obtain as many different fragments of the expected size as possible. Nevertheless, it was not possible to amplify the cd1-nir fragment in A. eutrophus ATCC 17699 at an annealing temperature of 57°C. The 800-bp fragment was only visible when the annealing temperature was 50°C.
Detection of cd1- and Cu-nir genes in different strains and sewage sludge.
The two sets of PCR primers for cd1-nir and Cu-nir genes amplified nir genes in several of the defined strains (Table 1; Fig. 2), as well as in bacterial isolates from activated sludge (Table 2). The primers did not amplify genes of nondenitrifying strains, although cd1-nir from isolate 89, which cannot reduce nitrite, was amplified. The cd1 primers were not able to amplify 800-bp nir fragments from the two strains of P. denitrificans, even though the gene from P. denitrificans Pd1222 has been sequenced and includes the primer sites (6). However, the Cu-nir primers amplified a 473-bp fragment from P. denitrificans Pd1222.
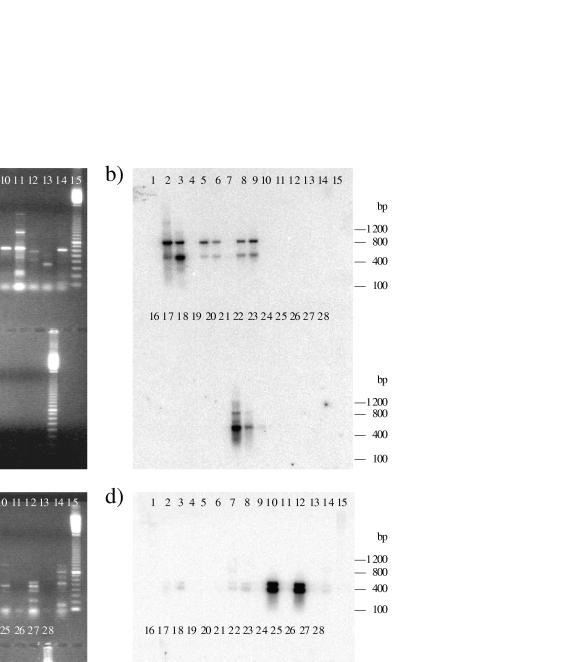
FIG. 2.
Agarose gels of PCR fragments and Southern blot hybridizations. Arrows indicate fragments of the expected sizes. Panels: a, cd1-nir primers; b, cd1 probe; c, Cu-nir primers; d, Cu probe. The upper row has molecular size markers in lane 1, P. stutzeri ATCC 14405 in lane 2, P. stutzeri CCUG 29240 in lane 3, P. fluorescens ATCC 33512 in lane 4, P. fluorescens Mi32 in lane 5, R. eutropha CCUG 13724 in lane 6, A. eutrophus ATCC 17699 in lane 7, P. aeruginosa CCUG 241 in lane 8, P. aeruginosa Mi11 in lane 9, P. denitrificans Pd1222 in lane 10, P. denitrificans ATCC 19367 in lane 11, P. denitrificans ATCC 13867 in lane 12, P. denitrificans CCUG 2519 in lane 13, P. putida CCUG 2479 in lane 14, and molecular size markers in lane 15. The lower row has molecular size markers in lane 16, A. faecalis ATCC 8750 in lane 17, A. faecalis ATCC 19018 in lane 18, P. aureofaciens ATCC 13985 in lane 19, A. denitrificans ATCC 15173 in lane 20, A. cycloclastes ATCC 21921 in lane 21, sludge L1 in lane 22, sludge L2 in lane 23, sludge P1 in lane 24, E. coli TG1 in lane 25, S. aureus 8325-4 in lane 26, a negative control in lane 27, and molecular size markers in lane 28. The molecular size markers consisted of a 100-bp ladder with the 800-bp fragment twice as intense as the other bands (Pharmacia).
Fragments of the expected sizes were obtained for both the cd1- and Cu primers by using chromosomal DNA isolated from activated sludge as the template (Fig. 2). DNA extracted from 13 different sludge samples was tested, and in five of these samples, 800-bp nir fragments were amplified with the cd1 primers. The Cu primers amplified a 470-bp fragment in all sludge samples. After staining with ethidium bromide, PCR fragments amplified with Cu primers tended to be visually more intense than PCR fragments amplified with cd1 primers.
Similarity of amplified nir fragments.
The DNA probes hybridized to almost every PCR product of the expected size, including the fragments amplified from the sludge samples (Fig. 2). The cd1-nir probe derived from P. stutzeri ATCC 14405 hybridized to all 800-bp fragments but one from P. fluorescens ATCC 33512. A second band of approximately 450 bp also hybridized to the probe. The hybridization signal from the smaller fragment was even stronger in the sludge samples. The Cu-nir probe derived from A. faecalis ATCC 8750 hybridized to all of the amplified 473-bp fragments. The probe hybridized to a 473-bp fragment in A. faecalis ATCC 19018 that was not detectable on an ethidium bromide-stained agarose gel. This probe also detected a second, smaller fragment of 400 bp in all of the strains that displayed the 473-bp fragment. This fragment was clearly visible in the control, A. faecalis ATCC 8750, with a short exposure time. The strong signals visualized after washing at 45°C were still detectable after washing at 65°C, but they were not as distinct as the hybridization signals seen in the controls.
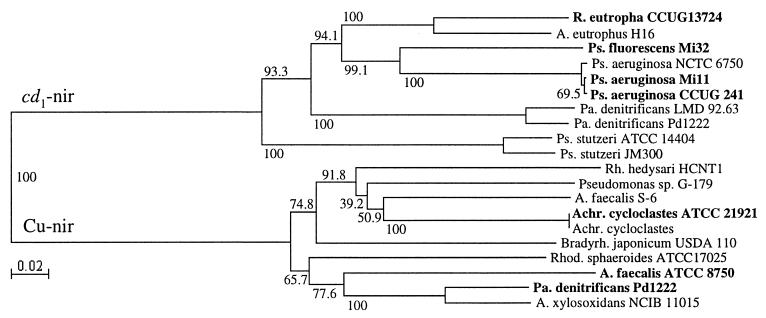
A subset of the PCR fragments of the expected sizes (473 and 800 bp) was sequenced. The sequences were similar to those of other cd1- and Cu-nir genes, demonstrating that the primers amplified the correct genes (Fig. 3). The degree of pairwise similarity was the same within the cd1- and Cu-nir gene fragments, respectively (data not shown). The cd1-nir fragments divided into two major clusters (Fig. 3). One cluster contained only the two published sequences from P. stutzeri, and the other cluster included all of the other sequences. The three nir fragments from P. aeruginosa strains were closely related, and the new sequence from R. eutropha CCUG 13724, formerly known as A. eutrophus, was most similar to the A. eutrophus H16 sequence. The Cu-nir fragments were distributed in two heterogeneous clusters. The two strains of A. cycloclastes were identical, with 100% sequence similarity.
FIG. 3.
Neighbor-joining analysis of partial nir gene sequences. Bootstrap values (percentages) are presented at the nodes. New sequences from this study are in boldface.
DISCUSSION
Our strategy was to design PCR primers general enough to amplify denitrifying bacteria regardless of their genus or species affiliation. General PCR primers permit the study of denitrifying bacteria without prior cultivation. Denitrifying bacteria have at least Nir in common, and we therefore based the primers on the structural nir genes. Because there are two structurally different reductases, no conserved regions for both genes were found. However, the use of two sets of PCR primers to distinguish between cd1 heme-type Nir and copper-dependent Nir will enhance resolution when analyzing denitrifying populations in environmental samples.
Both sets of primers were able to amplify the correct fragment of the nir gene in different strains and in a total DNA extract from sewage sludge as well. In some cases, such as the two P. denitrificans strains and A. eutrophus, the primers did not amplify the cd1-nir gene under the conditions used in this study, although cd1-Nir exists in these strains. This might be explained by a tendency for the target DNA to form secondary structures, which makes it difficult for the primers to anneal. Amplification of a Cu-nir fragment in P. denitrificans Pd1222 surprised us. To our knowledge, the existence of Cu-nir in P. denitrificans Pd1222 has not been reported before. This strain has been shown to carry a nir gene of the cd1 type (6). A Cu-nir fragment of the expected size in this strain hybridized with a strong signal to the DNA probe derived from A. faecalis ATCC 8750, and the sequence of that fragment was similar to the other sequences of copper-containing nir genes. The second band visualized in all Southern blot hybridizations could be a mutated copy of the cd1- or Cu-nir gene, but there is no published evidence that several copies exist.
Researchers have used nir genes to construct DNA probes that identify denitrifying bacteria in general. The detection of cd1-nir genes has been largely based on sequences from different P. stutzeri strains. One DNA probe, constructed from P. stutzeri JM300 for the detection of heme-type denitrifiers, identified a great number of defined denitrifying strains and found nir genes in bioreactors, aquifer microcosms, and toluene-degrading enrichment cultures (9, 26). Nevertheless, this particular probe did not hybridize to DNA extracted from soil or wetland sediments. The distribution of denitrifying bacteria in soil was investigated by Linne von Berg and Bothe (15). The cd1-nir probe used in that study was fairly large and contained additional sequences not encoding the structural gene, which caused nonspecific hybridization. Marine denitrifying isolates were detected with cd1-nir probes derived from P. stutzeri ATCC 14405 (29). Ye et al. (31) constructed a Cu-nir probe from Pseudomonas sp. which was then used to detect Cu-nir in denitrifying strains and enrichment cultures (9). DNA probes based on genes encoding cd1-Nir in P. aeruginosa and Cu-Nir in A. xylosoxidans were used to screen Hyphomicrobium isolates to separate denitrifiers from nondenitrifiers (7, 14).
Ward (29) suggested that PCR amplification is a more specific test than DNA hybridization, since PCR primers are dependent on a high degree of similarity in two separate 15 to 25-bp regions. The regions we selected for use as primer sites for the Cu-nir genes appear well conserved. The amplified fragment is exactly 473 bp in the available sequences and the sequenced fragments presented in this report, which further indicates that these regions are conserved in the Cu-nir genes. Ward (29) concluded that the functional gene encoding cd1 heme-type Nir was variable enough to support species-specific identification with DNA probes or primers and that a general probe for denitrifiers requires a different approach. Her conclusions were based on information from three sequences in two different species. The primer sites used by Ward et al. (30) are less conserved than the regions suggested in this report, although we agree that true broad-range primers for amplification of cd1-nir are more difficult to construct than Cu-nir primers.
By using an annealing temperature of 57°C, we allowed some nonspecific binding of the primers. This was done to obtain as many different nir fragments of the expected sizes as possible. If the next step is to separate and analyze the PCR products, it is important not to lose potential nir genes in crude DNA extracts. Although the primers amplified the correct gene segment in many different strains, the primers were constructed on the basis of nir sequences in culturable denitrifying bacteria. Since less than 1% of all bacteria are considered culturable (1), the amplified cd1- and Cu-nir fragments may not reflect the dominant denitrifiers present in the environment. Even though current research results are still fragmentary, evidence from rRNA gene sequencing of clone libraries generated from DNA extracted from soil indicates the existence of great unexplored bacterial diversity (3).
It is uncertain whether nir gene diversity is related to the phylogenetic diversity of denitrifying bacteria. Ohkubo et al. (20) showed that the grouping in a phylogenetic tree based on 5S rRNA sequences was almost compatible with the type of nitrite reductase in denitrifying bacteria. Otherwise, the relationship among denitrifying isolates derived from hybridization to a DNA probe for a nir gene was not correlated with restriction fragment length polymorphism patterns of ribosomal genes (29). Even if the diversity of the structural gene is not correlated with phylogenetic relationships, the nir gene still reveals the diversity of the gene encoding the key enzyme in denitrifying bacteria.
Primers capable of amplifying genes from all denitrifiers may prove to be impossible to design. Perhaps several pairs of primers will better detect more denitrifiers, in particular, those carrying cd1-Nir. New molecular approaches, such as denaturing gradient gel electrophoresis of PCR-amplified DNA fragments, can be used to examine the ecological importance of denitrifiers, as well as detect and identify new denitrifying bacteria. It might also be possible to quantify denitrifying bacteria with a competitive PCR (8). The use of the primers reported here will further our understanding of denitrifier complexity and diversity in the environment.
ADDENDUM
After submission of the manuscript, a similar report by Braker et al. (3a) was published. We acknowledge their work.
ACKNOWLEDGMENTS
Financial support for this work was partially provided by the National Board for Industrial and Technical Development in Sweden, the Swedish Environmental Protection Agency, and the Swedish Council for Forestry and Agricultural Research.
We thank Lars Frykberg for constructive discussions. Thanks are also extended to Martin Nilsson for assistance with vacuum blotting, as well as to Marianne Boysen, Nora Ausmees, and Karin Jacobsson for help with the sequencing.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 3.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brittain T, Blackmore R, Greenwood C, Thomson A J. Bacterial nitrite-reducing enzymes. Eur J Biochem. 1992;209:793–802. doi: 10.1111/j.1432-1033.1992.tb17350.x. [DOI] [PubMed] [Google Scholar]
- 5.Coyne M S, Arunakumari A, Averill B A, Tiedje J M. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl Environ Microbiol. 1989;55:2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer A P N, Reijnders W N M, Kuenen J G, Stouthamer A H, van Spanning R J M. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction in Paracoccus denitrificans. Antonie van Leeuwenhoek. 1994;66:111–127. doi: 10.1007/BF00871635. [DOI] [PubMed] [Google Scholar]
- 7.Fesefeldt A, Kloos K, Bothe H, Lemmer H, Gliesche C G. Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can J Microbiol. 1998;44:181–186. [Google Scholar]
- 8.Förster E. Rapid generation of internal primers for competitive PCR by low stringency primer annealing. BioTechniques. 1994;16:1006–1008. [PubMed] [Google Scholar]
- 9.Fries M R, Zhou J, Chee-Sandford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glockner A B, Jüngst A, Zumft W G. Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome cd1-free background (NirS−) of Pseudomonas stutzeri. Arch Microbiol. 1993;160:18–26. doi: 10.1007/BF00258141. [DOI] [PubMed] [Google Scholar]
- 11.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comp Appl Biosci. 1991;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 12.Hochstein L I, Tomlinson G A. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- 13.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 14.Kloos K, Fesefeldt A, Gliesche C G, Bothe H. DNA-probing indicates the occurrence of denitrification and nitrogen fixation genes in Hypohomicrobium. Distribution of denitrifying and nitrogen fixing isolates of Hypohomicrobium in a sewage treatment plant. FEMS Microbiol Ecol. 1995;18:205–213. [Google Scholar]
- 15.Linne von Berg K-H, Bothe H. The distribution of denitrifying bacteria in soils monitored by DNA-probing. FEMS Microbiol Ecol. 1992;86:331–340. [Google Scholar]
- 16.Magnusson G, Edin H, Dahlhammar G. Characterisation of efficient denitrifying bacterial strains isolated from activated sludge by 16S-rDNA analysis. Water Sci Technol. 1998;38:63–68. [Google Scholar]
- 17.Myers E W, Miller W. Optimal alignments in linear space. Comp Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Nishiyama M, Suzuki J, Kukimoto M, Ohnuki T, Horinouchi S, Beppu T. Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J Gen Microbiol. 1993;139:725–733. doi: 10.1099/00221287-139-4-725. [DOI] [PubMed] [Google Scholar]
- 19.Novick R P. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–156. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 20.Ohkubo S, Iwasaki H, Hori H, Osawa S. Evolutionary relationship of denitrifying bacteria as deduced from 5S rRNA sequences. J Biochem. 1986;100:1261–1267. doi: 10.1093/oxfordjournals.jbchem.a121832. [DOI] [PubMed] [Google Scholar]
- 21.Rees E, Siddiqui R A, Köster F, Schneider B, Friedrich B. Structural gene (nirS) for the cytochrome cd1 nitrite reductase of Alcaligenes eutrophus H16. Appl Environ Microbiol. 1997;63:800–802. doi: 10.1128/aem.63.2.800-802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sann R, Kostka S, Friedrich B. A cytochrome cd1-type nitrite reductase mediates the first step of denitrification in Alcaligenes eutrophus H16. Arch Microbiol. 1994;161:453–459. doi: 10.1007/BF00307765. [DOI] [PubMed] [Google Scholar]
- 25.Silvestrini M C, Galeotti C L, Gervais M, Schininà E, Barra D, Bossa F, Brunori M. Nitrite reductase from Pseudomonas aeruginosa: sequence of the gene and the protein. FEBS Lett. 1989;254:33–38. doi: 10.1016/0014-5793(89)81004-x. [DOI] [PubMed] [Google Scholar]
- 26.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toffanin A, Wu Q, Maskus M, Caselia S, Abruna H D, Shapleigh J P. Characterization of the gene encoding nitrite reductase and the physiological consequences of its expression in the nondenitrifying Rhizobium “hedysari” strain HCNT1. Appl Environ Microbiol. 1996;62:4019–4025. doi: 10.1128/aem.62.11.4019-4025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tosques I E, Kwiatkowski A V, Shi J, Shapleigh J P. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:1090–1095. doi: 10.1128/jb.179.4.1090-1095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward B B. Diversity of culturable denitrifying bacteria. Arch Microbiol. 1995;163:167–175. [Google Scholar]
- 30.Ward B B, Cockcroft A R, Kilpatrick K A. Antibody and DNA probes for detection of nitrite reductase in seawater. J Gen Microbiol. 1993;139:2285–2293. doi: 10.1099/00221287-139-9-2285. [DOI] [PubMed] [Google Scholar]
- 31.Ye R W, Fries M R, Bezborodnikov S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zumft W G. The denitrifying procaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. New York, N.Y: Springer Verlag; 1992. pp. 554–582. [Google Scholar]