Abstract
Techniques based on amplification of 16S rRNA genes for comparing bacterial communities are now widely used in microbial ecology, but calibration of these techniques with traditional tools, such as cultivation, has been conspicuously absent. In this study, we compared levels of bacterial community diversity in two pinyon rhizosphere soil samples and two between-tree (interspace) soil samples by analyzing 179 cultivated bacterial isolates and 801 16S rRNA genes amplified from extracted soil DNA. Phylotypes were defined by performing a restriction fragment length polymorphism analysis of 16S rRNA gene sequences with the enzymes RsaI and BstUI. The average level of 16S rRNA gene sequence similarity of members of a phylotype was 86.6% based on an analysis of partial sequences. A total of 498 phylotypes were identified among the 16S ribosomal DNA (rDNA) clones, while 34 phylotypes occurred among the cultivated isolates. Analysis of sequences from a subset of the phylotypes showed that at least seven bacterial divisions were represented in the clone libraries, whereas the isolates represented only three. The phylotype richness, frequency distribution (evenness), and composition of the four culture collections and the four clone libraries were investigated by using a variety of diversity indices. Although cultivation and 16S rRNA cloning analyses gave contradictory descriptions of the relative phylotype richness for one of the four environments, the two methods identified qualitatively consistent relationships when levels of evenness were compared. The levels of phylotype similarity between communities were uniformly low (15 to 31%). Both methods consistently indicated that one environment was distinct from the other three. Our data illustrate that while 16S rDNA cloning and cultivation generally describe similar relationships between soil microbial communities, significant discrepancies can occur.
Bacterial communities have traditionally been compared by analyzing isolates cultivated on plates. Clustering isolates into operational taxonomic units based on phenotypic or genotypic characteristics allows comparisons of the following three elements of diversity in a sample: the types of bacteria present (composition), the number of types (richness), and the frequency distribution or relative abundance of types (structure). Evaluating these elements for collections of cultivated isolates provides relative measures of community diversity but not accurate descriptions of community diversity in situ. For example, the potential for cultivable organisms to enter the viable-but-noncultivable state (32) can dramatically distort the relative abundance of an organism observed in a culture collection. Nonetheless, for samples that are treated in a uniform manner, measures of community composition, richness, and structure derived from a sample of cultivated isolates can identify meaningful differences between bacterial communities.
Methods that rely on direct amplification and analysis of 16S rRNA gene (rDNA) sequences are rapidly replacing cultivation as a way to compare the composition, richness, and structure of microbial communities. Such methods include denaturing gradient gel electrophoresis (13, 19, 23, 37, 49), terminal restriction fragment analysis (4, 6, 28), and 16S rDNA cloning (2, 8, 15, 38). Like cultivation, amplification of rDNA can distort the apparent structure of a community as a result of biases in cellular rDNA copy number (11), DNA extraction (33, 48), and PCR amplification (40, 46) but may still provide meaningful comparisons of bacterial communities. Methods in which direct amplification and analysis of rDNA are used allow more comprehensive sampling of microbial communities than cultivation. However, thus far such methods have not been compared in parallel with cultivation methods for assessing similarities between communities.
Numerous studies have investigated the phylogenetic overlap between organisms obtained by cultivation and organisms identified by direct amplification and cloning of 16S rDNA (2, 5, 38, 45, 47, 50). These studies have consistently demonstrated that the two methods generally sample different fractions of bacterial communities. In some cases, no overlap was observed between culture collections and 16S rDNA clone libraries (2, 47). In other cases, as much as 41% of the phylotypes identified in a culture collection were also identified in 16S rDNA clone libraries (5, 50). The amount of overlap between culture collections and clone libraries appears to depend on several factors, such as the complexity of the environment being examined, the discrepancy between plate counts and direct counts, and, importantly, the sample size of 16S rDNA clones. Of greater interest than the overlap between culture and 16S rDNA clone libraries is the extent to which these two methods describe similar relationships between different microbial communities despite the biases inherent in each method.
We used cultivation and 16S rDNA cloning to study the diversity of pinyon pine rhizosphere communities and interspace (between-tree) communities in two arid southwestern United States soils. One soil is in the hot, extremely dry, 900-year-old cinder field of an extinct volcano, while the other is a sandy loam soil 19 km away that is typical of the region. This is the first report describing the use of both cultivation and 16S rDNA clone libraries to compare microbial communities in different samples. We used restriction fragment length polymorphism (RFLP) analysis of 16S rDNA sequences of cultivated isolates and 16S rDNA clones to define phylotypes and then compared the phylotype richness, distribution, and composition of four culture collections and four clone libraries. Additionally, partial or full-length sequences were obtained from representatives of a subset of the RFLP patterns for phylogenetic analysis. Based on comparisons of a variety of diversity indices, cultivation and 16S rDNA cloning generally provided similar assessments of the relationships among the four soil environments, although some notable exceptions occurred.
MATERIALS AND METHODS
Field sites.
Soil samples were collected from two field sites 19 km apart in northern Arizona that have similar plant communities (pinyon pine-juniper woodlands), elevations, and general weather patterns but differ dramatically in soil type (7, 24). One site is located in the Coconino National Forest near the town of Cosnino. The other site is located 19 km due north on the eastern edge of Sunset Crater National Monument. At Cosnino the soil is a light sandy loam (18), and the areas between widely spaced trees (interspaces) are sparsely covered with grass and forb species. In contrast, the Sunset Crater soil consists of black, coarse-textured, well-drained cinders, and the interspaces are largely barren. Although the soil pH values (pH 7.13 to 7.57) and nitrate and ammonium nitrogen concentrations are similar at the two sites, the available organic matter, phosphorus, potassium, calcium, and magnesium concentrations are significantly lower in the Sunset Crater cinders than in the Cosnino soil (25).
Soil samples.
Soil samples were collected in April 1994 as previously described (24). Rhizosphere samples were collected from one mature tree at each site; the trees at the two sites were matched for age (160 years). For each tree, 10 50-cm3 subsamples were collected at evenly spaced locations around the drip line at a depth of 10 to 15 cm and then pooled to create a single composite sample. Composite interspace samples were similarly created by combining subsamples taken at a depth of 10 to 15 cm across an approximately 1,000-ft2 area surrounding each tree. The composite samples were mixed well, immediately placed on ice for transport, and frozen at −70°C after 24 h.
DNA extraction from soil and cinders.
Nucleic acids were extracted from two 10-g aliquots of each of the four soil samples collected in April 1994 as previously described (24). Briefly, the extraction procedure consisted of incubation at 70°C in TENS buffer (50 mM Tris [pH 8.0], 20 mM disodium EDTA, 100 mM NaCl, 1% [wt/vol] sodium dodecyl sulfate), bead mill homogenization (Biospec Products), three cycles of freezing and thawing, and nucleic acid precipitation with ethanol. The extracted DNA was purified by phenol-chloroform-isoamyl alcohol extraction (41) and passage through Sephadex G-200 spin columns (48), reprecipitated with ethanol, and dissolved in TE buffer (41).
Small-subunit rDNA libraries.
Clone libraries were created from 16S rDNA sequences amplified from extracted DNA (24). PCR amplicons approximately 1.5 kb long were ligated into the pGEM-T plasmid vector (Promega, Madison, Wis.) and transformed into Escherichia coli DH10B Electromax cells (Gibco, BRL, Gaithersburg, Md.). Two hundred clones containing inserts of the correct size were stored in 15% glycerol at −70°C for each of the four soil samples. Each clone was designated C0 (Cosnino sandy loam interspace), C1 (Cosnino sandy loam rhizosphere), S0 (Sunset Crater cinder interspace), or S1 (Sunset Crater cinder rhizosphere), followed by the clone number (1 to 200).
Bacterial culture collections.
Culture collections were established from bacterial isolates cultivated from the composite soil samples used for DNA extraction. For each soil sample, 10 g of soil was vortexed with 50 ml of sterile, distilled water for 5 min, rocked horizontally for 5 min, and then serially diluted. Appropriate dilutions were spread onto 0.1× Trypticase soy agar (Difco Laboratories, Inc., Detroit, Mich.). After incubation at 26°C for 3 days, plates containing between 30 and 300 colonies were examined. For each sample, 50 randomly selected colonies were purified by streaking them onto fresh medium. Purified isolates were stored in 15% glycerol at −70°C. Each isolate was designated iC0, iC1, iS0, or iS1, followed by the isolate number (1 to 50).
RFLP analysis of 16S rDNA.
16S rDNA sequences of cultured isolates and 16S rDNA clones were amplified either directly from glycerol stock preparations by using primers pA (5′-AGAGTTTGATCCTGGCTCAG; E. coli bases 8 to 27) (10) and PC5B (5′-TACCTTGTTACGACTT; E. coli bases 1507 to 1492) (51) or from cells subjected to three cycles of freezing and thawing. Each 50-μl reaction mixture contained 30 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2 (39), each deoxynucleoside triphosphate at a concentration of 50 μM, 50 pmol of each primer, and 1.9 U of Taq polymerase (AmpliTaq; Perkin-Elmer, Foster City, Calif.). Extracted DNA (1 ng) was used as the template in PCR for isolates which were not amenable to direct 16S rDNA amplification from glycerol stock preparations or from cells subjected to three freeze-thaw cycles.
Following PCR amplification, 8 or 10 μl of 16S rDNA from each of the clones and isolates was digested separately with 2.5 U of RsaI and 2.5 U of BstUI (New England Biolabs, Beverly, Mass.) in 12-μl reaction mixtures as recommended by the manufacturer. RsaI and BstUI digests were electrophoresed in 3 and 4% Metaphor (FMC, Rockland, Maine) gels, respectively, with TBE buffer (41). The gels were stained with ethidium bromide and then photographed under UV light.
DNA fragment sizes were determined by using Kodak Digital Image analysis software (Kodak, Rochester, N.Y.). The quality of the numerical data was checked by comparing the sum of fragment sizes in each restriction pattern with the original product size (approximately 1,499 bp). Gel images of patterns with size discrepancies exceeding the acceptable margin of error (1,499 ± 100 bp) were carefully reanalyzed and corrected. Digitized restriction patterns were sorted with Microsoft Excel, version 5.0 (Microsoft Corp.), in order to group similar patterns. Following each sorting, patterns were compared, and groups of indistinguishable patterns were recorded. Each phylotype was defined as a group of sequences that had indistinguishable RsaI and BstUI restriction patterns.
DNA sequencing.
16S rDNA templates for DNA sequencing reactions were amplified directly from glycerol stock preparations of environmental isolates or cloned 16S rDNA by using primers pA and PC5B for environmental isolates and primers M13-20 (5′-GTAAAACGACGGCCAGT) and M13-24 (5′-AACAGCTATGACCATG) for 16S rDNA clones. The PCR conditions were the same as those described above. Amplified DNA was purified by using a QIAQUICK PCR cleanup kit (Qiagen, Inc., Chatsworth, Calif.), and DNA concentrations were estimated by gel electrophoresis and ethidium bromide staining. Approximately 100 ng of 16S rDNA was used as the template in dye terminator cycle sequencing reactions (ABI PRISM dye terminator cycle sequencing kit; Perkin-Elmer). Primer p3MODrc (5′-GGACTACHAGGGTATCTAAT; E. coli positions 806 to 787) was used in sequencing reactions to obtain partial DNA sequences. Nearly full-length sequences were obtained for a subset of partially sequenced clones by using primers M13-20, M13-24, P3MOD (5′-ATTAGATACCCTDGTAGTCC; E. coli positions 787 to 806) (51), P3MODrc, and 533 forward (5′-CCAGCSGCCGCGGTAA; E. coli positions 519 to 533) (26) in sequencing reactions. Reaction mixtures were electrophoresed through 4.0% polyacrylamide gels by using a model 373A Stretch DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.).
DNA distance analysis.
To determine the average level of 16S rDNA sequence similarity of organisms that produced the same RFLP pattern, partial 16S rDNA sequences of two to four representatives of each of 12 RFLP groups were compared. The sequences were aligned on the basis of primary- and secondary-structure considerations by using GDE sequence editing software (http://rdp.life.uiuc.edu) (30). Similarity values (corrected evolutionary distances) were calculated by using the DNADIST program in PHYLIP (version 3.5; distributed by J. Felsenstein, University of Washington, Seattle) and the Kimura two-parameter model of sequence evolution.
Phylogenetic analysis.
16S rDNA sequences were compared with sequences obtained from the Ribosomal Database Project (RDP), version 7.0 (30), by using the SIMILARITY_RANK program to obtain Sab values with database sequences. Database sequences with less than 307 nucleotides for comparison were excluded from the analysis. Sequences were assigned to recognized bacterial divisions (or were given “uncertain” status for Sab values less than 0.50) based on the affiliation of the nearest-neighbor sequences from the RDP.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the NCBI database under accession no. AF128631 to AF128768.
RESULTS
RFLP phylotypes.
Each phylotype from the clone and culture libraries was defined by RFLP patterns obtained from RsaI- and BstUI-digested 16S rDNA. A comparison of the RsaI-BstUI RFLP patterns of 801 clones and 179 isolates resulted in the identification of 526 patterns (Table 1). A total of 498 patterns were identified among the clones, while 34 patterns occurred among the isolates. Phylotypes defined by RFLPs obtained with two tetrameric enzymes have been reported previously to represent organisms with a median genetic distance of 95.6% (34). We determined the phylogenetic discrimination of the RsaI-BstUI phylotypes in our collections by comparing partial 16S rDNA sequences of two to four representatives from each of 12 randomly selected RFLP patterns. The 12 RFLP patterns represented organisms belonging to four bacterial divisions, namely, the Acidobacterium division, the proteobacteria, the gram-positive bacteria, and the Bacteroides-Cytophaga-Flexibacter division (as determined by analysis of partial or full-length sequences [see below]). The average level of similarity of 16S rDNA sequences having identical RFLP patterns was 86.88% (median, 89.89%; range, 52.16 to 99.85%) based on an analysis of 427 nucleotides.
TABLE 1.
Numbers of RsaI-BstUI RFLP phylotypes in 16S rDNA clone libraries and culture collections from four soil environments
| Parameter | Clone libraries
|
Culture collections
|
Clones | Isolates | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C0 | C1 | S0 | S1 | iC0 | iC1 | iS0 | iS1 | ||||
| Total no. of patterns | 154 (196)a | 161 (212) | 134 (203) | 150 (190) | 8 (46) | 14 (37) | 8 (49) | 18 (47) | 498 (801) | 34 (179) | 526 (980) |
| No. of unique patternsb | 107 (110) | 106 (118) | 98 (114) | 114 (130) | 2 (6) | 7 (7) | 5 (7) | 9 (10) | 425 (472) | 23 (30) | 448 (502) |
| % of unique patterns | 69 (56) | 66 (56) | 73 (56) | 76 (68) | 25 (13) | 50 (19) | 63 (14) | 50 (21) | 85 (59) | 68 (17) | 85 (51) |
| No. of rare phylotypesc | 132 | 136 | 106 | 130 | 3 | 8 | 3 | 13 | |||
The values in parentheses are the numbers of clones or isolates.
Unique patterns were found in only one clone library or culture collection.
Rare phylotypes were phylotypes that were represented by only a single clone in a clone library or a single isolate in a culture collection.
Phylogenetic diversity among RFLP phylotypes.
To evaluate the phylogenetic diversity represented by the 526 RFLP patterns identified for the 16S rDNA clone libraries and the culture collections, 203 partial (typically about 600-bp) or full-length 16S rDNA sequences were obtained and used for phylogenetic analysis; sequences of 56 clones analyzed previously (24) were also included. The 203 sequences obtained in this study represented 154 of the 498 RFLP patterns identified for the clone libraries and 33 of the 34 RFLP patterns identified for the culture collections. Of the 168 sequences of cloned 16S rDNA, only 20 (12%) had Sab values greater than 0.85 with sequences obtained from the RDP (median Sab value, 0.68; Sab value range, 0.19 to 0.97). Since the RDP database currently contains the 56 C0, C1, S0, and S1 sequences described previously (24), many of the Sab values greater than 0.50 obtained with our set of 168 sequences resulted from matches with the previously described set of 56 sequences, which increased the median Sab value. Seventeen (49%) of the 35 partial sequences obtained from cultivated isolates had Sab values greater than 0.85 with RDP sequences (median Sab value, 0.84; Sab value range, 0.32 to 0.98). Only 33% of the 16S rDNA clone library sequences with Sab values greater than 0.50 had nearest neighbors that were named, cultivated organisms. In contrast, 89% of the sequences from the culture collections had nearest neighbors in the RDP database that were named, cultivated organisms. These data demonstrated that the organisms represented by the cultivated isolates were more similar to previously identified bacteria than the organisms represented by the cloned 16S rDNA were.
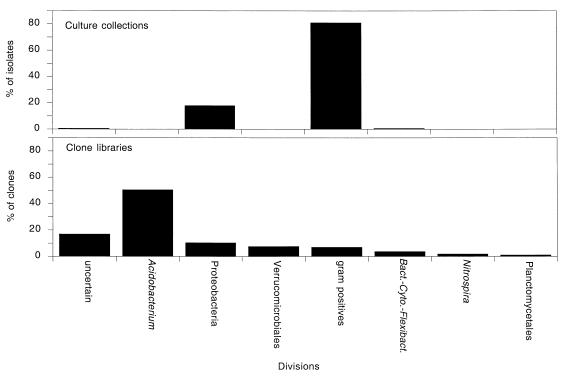
Seven bacterial divisions were identified from the sequences representing RFLP patterns from the clone libraries (Fig. 1). The distribution of clones among different bacterial divisions was uneven. As shown in Fig. 1, four divisions—the Acidobacterium division, the proteobacteria, the Verrucomicrobiales, and the gram-positive bacteria—accounted for 76% of the clones examined. The Acidobacterium division was the most abundant phylogenetic group both in terms of the variety of RFLP patterns and in terms of the number of clones. Members of this division accounted for 27% of the 154 RFLP patterns (51% of the 356 clones represented by the 154 patterns). The proteobacteria and the Verrucomicrobiales were the second and third most abundant phylogenetic groups found in the clone libraries, accounting for 16 and 7% of the 154 patterns (10 and 8% of the clones), respectively. The gram-positive bacteria comprised 12% of the patterns but only 7% of the clones. Thirty-six sequences (representing 17% of the clones) were phylogenetically ambiguous (Sab values, <0.50; median Sab value, 0.40) and represented either chimeras, deeply branching members of previously described bacterial divisions, or members of new, undescribed divisions. The combined data demonstrated that the clone libraries represented a phylogenetically broad spectrum of organisms.
FIG. 1.
Division level phylogenetic diversity identified in 16S rDNA clone libraries and culture collections. Division level affiliations were determined by phylogenetic analysis of partial or nearly full-length 16S rDNA sequences from 168 cloned 16S rDNA obtained primarily from the S0 and C0 clone libraries and from 35 bacterial isolates from the four culture collections. Bact.-Cyto.-Flexibact., Bacteroides-Cytophaga-Flexibacter.
In contrast to the clone libraries, only three divisions—the gram-positive bacteria, the proteobacteria, and the Bacteroides-Cytophaga-Flexibacter group—were identified among 178 isolates in the culture collections. Based on an analysis of partial 16S rDNA sequences obtained from isolates representing 33 of the 34 RFLP patterns identified among the four culture collections, 81% of the isolates were gram-positive organisms, primarily Bacillus species. Proteobacterial species accounted for 18% of the isolates (12 of the 33 RFLP patterns). One isolate belonged to the Cytophaga group, and one isolate was phylogenetically undefined. Thus, the phylogenetic breadth of the cultivated isolates was restricted compared to the diversity observed among 16S rDNA clones.
Phylotype richness and distribution.
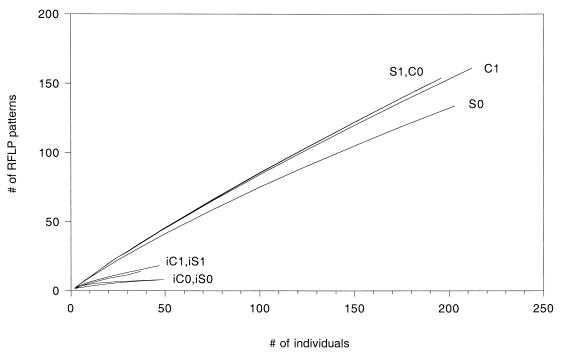
The number of phylotypes (richness) and the frequency distribution of the phylotypes (evenness) in each of the clone libraries and culture collections were evaluated by using a variety of standard diversity indices. In most cases a phylotype probably does not represent a single species. However, phylotypes are nonetheless discrete units of biological information that should be suitable for traditional analyses of information complexity regardless of the taxonomic level of discrimination. Since the libraries differed in size, estimated phylotype richness [E(S)] was calculated by rarefaction (21, 43) for smaller sample sizes to allow standardized comparisons. As shown in Fig. 2, the 16S rDNA cloning and plate culture techniques provided different assessments of diversity in the Cosnino interspace compared to the other environments. According to values obtained with the 16S rDNA clone libraries, the Sunset Crater interspace had a relatively low level of phylotype richness, whereas the Cosnino interspace, Cosnino rhizosphere, and Sunset Crater rhizosphere environments had similarly high richness values. For a sample size of 190 clones, the S1, C0, and C1 libraries had estimated diversities of 150, 150, and 147 phylotypes, respectively, while only 127 phylotypes were estimated to occur in the S0 library (Table 2). In contrast, the culture collection values suggested that the interspace environments at Sunset Crater and at Cosnino were substantially less diverse than the rhizosphere environments (Fig. 2). Approximately one-half as many phylotypes were detected in the interspace culture collections (iS0 and iC0) as in the rhizosphere collections (iS1 and iC1). For a sample size of 37 isolates per collection, 14 patterns occurred in the iC1 collection and 15 patterns were estimated to occur in the iS1 collection, while only 7 were estimated to occur in the iC0 and iS0 (interspace) collections (Table 2). Interestingly, a much larger number of 16S rDNA clones had to be analyzed in order to detect a significant difference between the Sunset Crater interspace and the other environments. No significant differences were apparent among the clone libraries when phylotype richness was estimated for a sample size of 37 clones [E(S) ranged from 32 to 35 patterns for the clone libraries, and the average varianceE(S) was 2.6].
FIG. 2.
Phylotype richness curves for clone and culture libraries. Sampling curves were calculated by rarefaction (21, 43).
TABLE 2.
Diversity indices based on RsaI-BstUI RFLP phylotypes in 16S rDNA clone libraries and culture collections from four soil environments
| Index | Clone libraries
|
Culture collections
|
||||||
|---|---|---|---|---|---|---|---|---|
| C0 | C1 | S0 | S1 | iC0 | iC1 | iS0 | iS1 | |
| E(S)a | 150 (1.3) | 147 (4.3) | 127 (2.3) | 150 (NA)f | 7 (0.5) | 14 (NA) | 7 (0.6) | 15 (1.6) |
| Hb | 7.067 | 7.092 | 6.612 | 7.018 | 2.405 | 3.254 | 1.541 | 3.337 |
| Evennessc | 0.972 | 0.967 | 0.936 | 0.971 | 0.802 | 0.855 | 0.514 | 0.800 |
| Dd | 0.991 | 0.990 | 0.981 | 0.990 | 0.757 | 0.859 | 0.448 | 0.843 |
| D/Dmaxe | 0.997 | 0.997 | 0.989 | 0.997 | 0.865 | 0.925 | 0.512 | 0.893 |
E(S) was calculated by rarefaction (21, 43) for a standardized sample size of 190 clones for each clone library and 37 isolates for each culture collection. The values in parentheses are the variances arising from inherent statistical uncertainty during the estimation of phylotype richness for sample sizes smaller than the actual sample size.
H was calculated as follows (31): H = − Σ (pi)(log2pi) where pi is the proportion of the ith phylotype.
Evenness (E) (31) was calculated from H as follows: E = H/Hmax, where Hmax = log2(S) and S is the total number of phylotypes.
D was calculated as follows (44): D = 1 − Σ (pi)2. The scale for D ranges from 1 to Dmax, where Dmax = 1 − 1/(S).
Since the scale for D varies between communities according to S, we standardized the measure by dividing by Dmax. This provided an evenness index conceptually similar to the index derived from the Shannon-Weiner function.
NA, not applicable.
As shown in Table 2, other diversity measures that emphasize phylotype richness revealed relationships between clone libraries and between culture collections similar to those illustrated in Fig. 2. When either the Shannon-Weiner index (H) or Simpson’s index (D) was used, the C0 clone library appeared to be as diverse as the C1 and S1 libraries, whereas the iC0 culture collection appeared to be more similar to the iS0 collection since it had noticeably lower diversity values compared to the iC1 and iS1 collections. These data demonstrated that both 16S rDNA cloning and cultivation can detect differences between environments. However, the two methods may in some cases provide contradictory views of species richness in communities.
Although 16S rDNA cloning and cultivation provided slightly inconsistent descriptions of relative phylotype richness, these two methods described roughly similar relationships among the four communities when the equability (or evenness) of phylotype distribution in each library was measured (Table 2). Evenness values were calculated by using derivations of H and D (Table 2). By either measure, evenness in three of the environments was relatively high, while the distribution in the Sunset Crater interspace environment appeared to be more skewed.
Community composition.
Only 15% of the phylotypes (78 of 526 patterns) identified in the 16S rDNA clone libraries and culture collections combined were found in more than one library or collection. As a result, the similarity values obtained from pairwise comparisons of the compositions of clone libraries or culture collections were very low (Table 3). The most dissimilar 16S rDNA clone libraries were the S1 and S0 libraries (11% similarity), while the C1 and C0 clone libraries were the most similar (22%). Comparisons of Sunset Crater clone libraries with Cosnino libraries yielded intermediate similarity values, which ranged from 13 to 16%. The lowest similarity values arose from comparisons of the S0 clone library with other libraries, suggesting that the Sunset Crater interspace environment was the most divergent. According to the values in Table 3, the S1 and S0 libraries each exhibited greater similarity to a 16S rDNA clone library produced from soil obtained 19 km away (the C1 library in particular) than to one another despite having been produced from soil samples collected no more than 33 m apart.
TABLE 3.
Similarity matrix for phylotype composition for clone libraries and culture collections
| Clone library or culture collection | % Similaritya
|
||||||
|---|---|---|---|---|---|---|---|
| iS0 | iS1 | iC0 | iC1 | S0 | S1 | C0 | |
| iS1 | 23 | ||||||
| iC0 | 25 | 31 | |||||
| iC1 | 15 | 31 | 31 | ||||
| S0 | 1 | 4 | 4 | 4 | |||
| S1 | 0 | 1 | 0 | 1 | 11 | ||
| C0 | 0 | 0 | 0 | 0 | 13 | 15 | |
| C1 | 1 | 3 | 1 | 2 | 16 | 16 | 22 |
Pairwise similarity values were calculated as follows: [(2 × common types)/(total types)] × 100. Boldface percentages are values referred to in the text.
Like the comparison of clone libraries, comparison of the phylotype compositions of the culture collections indicated that the S0 environment was the most divergent. The iC0, iC1, and iS1 collections were equally similar (31%). Pairwise comparisons of these collections with the iS0 collection yielded lower values, the lowest of which was obtained from the comparison of the iC1 and iS0 collections (15%). While the clone libraries and culture collections both indicated that the level of similarity between the C0 and C1 environments was relatively high, this was the only consistent pairwise comparison. In the absence of replication, the significance of the similarities and discrepancies between specific pairwise comparisons could not be reliably interpreted.
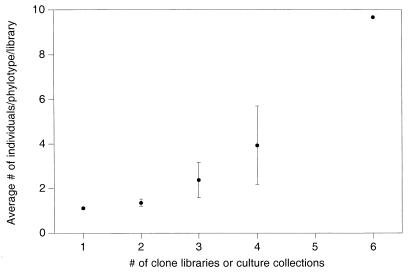
In an attempt to ascertain whether the low similarity values were a true reflection of community similarities or merely the result of an overabundance of rare types with low detection probabilities, we constructed a frequency table to examine the probability of a phylotype occurring in more than one library (data not shown). For example, for the 16S rDNA clone libraries, phylotypes represented by only one individual in a library had a 22% probability of appearing in a second library. In contrast, phylotypes represented by at least five individuals in one library had a 92% chance of appearing in a second library. As shown in Fig. 3, the number of libraries (including culture collections in this case) in which a phylotype appeared was correlated with the average abundance of the phylotype. This relationship was evident not only when clone libraries and culture collections were analyzed together but also when the two data sets were analyzed separately (data not shown). Such a relationship would be expected for replicate samples or for very similar communities but not for communities that are substantially different. Thus, the four environments may be more similar than the pairwise similarity values (Table 3) suggest.
FIG. 3.
Relationship between phylotype abundance and detection frequency for 16S rDNA clone libraries and culture collections. The average abundance of each phylotype was calculated across all eight libraries. Phylotypes were then grouped according to the number of libraries in which each phylotype appeared. The mean phylotype abundance for each group was calculated by using the average abundance values previously calculated for individual phylotypes. Only one phylotype appeared in six libraries. The error bars indicate 95% confidence intervals.
DISCUSSION
We investigated community diversity in four soil environments by determining phylotype richness, distribution, and similarity for approximately 50 isolates and nearly 200 16S rDNA clones from each environment. Both plating and 16S rDNA cloning suffer from biases that can distort community composition, richness, and structure. We assumed that the biases operated uniformly for our four environmental samples and that therefore the soil samples could be compared. Although our data were not replicated, they provided a starting point for assessing the merits of 16S rDNA cloning compared to cultivation. The data demonstrated that the 16S rDNA clone libraries provided more comprehensive sampling of the phylogenetic diversity in the four soil environments than the culture collections provided. Use of the two methods to assess community diversity demonstrated that the methods provided contradictory measures of relative phylotype richness in the Cosnino interspace environment, but the methods identified roughly similar relationships among the four communities when phylotype distributions were compared (that is, evenness was high in three of the environments but was comparatively lower in the Sunset Crater interspace environment). Both methods also identified the Cosnino interspace and rhizosphere environments as the environments that were most similar in terms of phylotype composition and the Sunset Crater interspace as the most dissimilar environment.
The ability of 16S rDNA cloning to sample the phylogenetic diversity in natural communities more comprehensively than cultivation is characteristic of the method. Our results are entirely consistent with this characteristic. The phylogenetic breadth of any one of our clone libraries is comparable to the phylogenetic breadth observed in previous studies in which 16S rDNA cloning was used to characterize soil microbial communities (3, 12, 27, 29, 45). An extensive phylogenetic analysis of 56 clones from our libraries and a comparison of the data with data obtained in other studies of cloned 16S rDNA sequences from soil have been described previously (24). In addition to multiple representatives of well-known bacterial divisions, members of a new division (now called the Acidobacterium division) were described. The Acidobacterium division was the most abundant and diverse group in our 16S rDNA clone libraries. This division was also the most abundant division in a clone library established from Wisconsin soil (3) despite the use of different PCR primers (primer 530 forward instead of primer 8-27 forward) and DNA extraction procedures (see reference 24 for placement of the Wisconsin sequences in the Acidobacterium division). Phylogenetic information for an additional 112 clones obtained in this study has not substantially altered the division level diversity noted previously (24). Sequences affiliated with the genus Nitrospira have been identified, thus expanding the number of divisions detected in the four soil samples from six to seven. Additional divisions may be represented by the 36 sequences (placed in uncertain category [Fig. 1]) that had extremely low Sab values with RDP sequences. For example, an RDP sequence representing candidate division OP9 (20) had an Sab value of 0.474 with one of the S0 16S rDNA clones. We are currently collecting full-length sequences from some of the 36 rDNA clones in order to accurately define their phylogenetic affiliations. The phylogenetic data obtained in the current study augment the impressive diversity described previously (24). When compared with data in other reports, our data demonstrate results typical of 16S rDNA cloning.
The phylogenetic diversity observed among our four culture collections is also typical of the cultivation method. Gram-positive bacteria and proteobacteria are the predominant bacteria cultivated from soil on general-purpose, aerobic-growth media (1). The skewed abundance of gram-positive isolates may have been due in part to the fact that isolates were selected after plates were incubated for only 3 days. In a study of two soils in Japan, 71% of the isolates that formed visible colonies within 18 h of inoculation were members of gram-positive species (22). As the length of the incubation period increased, the relative abundance of gram-positive organisms in the total collection of isolates decreased. With two soils from New Mexico the proportion of cultivated gram-positive organisms decreased from 96 to 67% in a collection obtained after 24 h of incubation and then augmented after 3 weeks of incubation (9). Gram-positive organisms have also been found to predominate in the outer layer of soil aggregates (17) and are preferentially recovered during extraction procedures that do not adequately disrupt aggregates (through sonication, for example). Nonetheless, since the culture collections used in the current study were established in the same way from the four communities, they should allow comparisons between communities despite their narrow phylogenetic scope.
Measures of diversity among the clone libraries and culture collections revealed similar relationships among three of the four environmental samples. The results obtained with both methods used indicated that the levels of diversity in the Sunset Crater and Cosnino rhizosphere environments were comparable, while phylotype richness was lower and more skewed in the Sunset Crater interspace. Relatively similar values for phylotype richness (S, D, and H) and evenness (H/Hmax and D/Dmax) were obtained for the rhizosphere environments compared to the Sunset Crater interspace (Table 1). Culture collections from samples obtained in September 1994 exhibited the same pattern of diversity as the culture collections obtained in April 1994 (data not shown), suggesting that the diversity in the Sunset Crater interspace was in fact less than the diversity in the rhizosphere environments. This observation is consistent with the idea that pinyon pine roots (matched for age) provide a homogeneous environment for microbial growth and community development and modulate the effects of different soil types. The lower diversity observed in the Sunset Crater interspace was likewise consistent with our knowledge of the physical characteristics of this environment. The Sunset Crater interspaces are devoid of macroscopic plant life. The black cinder gravel that constitutes the soil at Sunset Crater creates a hot, exceptionally arid environment due to low water retention. The apparently stressful conditions in the interspace are expected to reduce microbial community diversity, and the low moisture content and rapid water drainage in the cinder gravel should limit horizontal migration of bacteria from the rhizosphere environment to the interspace.
The 16S rDNA cloning and plate cultivation methods provided inconsistent assessments of relative phylotype richness in the fourth environment, the Cosnino interspace. Unlike the barren Sunset Crater interspaces, the interspaces at Cosnino are covered by grass and forb species, and the sandy loam soil at Cosnino retains more water than the Sunset Crater cinder gravel. Grass roots are abundant in the Cosnino interspace soil. Phylotype richness appeared to be as high in this environment as it was in the pinyon pine rhizospheres when it was measured by 16S rDNA cloning. However, based on plate cultivation, the Cosnino interspace appeared to have approximately one-half as many phylotypes (like the Sunset Crater interspace) as the rhizosphere environments (Fig. 2). This result was surprising in light of the physical characteristics of the Cosnino interspace. Interestingly, 16S rDNA cloning and plate cultivation both demonstrated that the frequency distribution of phylotypes (evenness) in the Cosnino interspace was relatively similar to that observed in the pinyon pine rhizosphere environments. In the absence of replicated data and larger sample sizes, it is difficult to accurately interpret the discrepancy between the 16S rDNA cloning and plate cultivation data. The most probable explanation, however, is that the discrepancy arose from sampling different fractions of the microbial community. Whereas the C0 clone library contained a wide variety of eubacteria, the iC0 culture collection contained primarily fast-growing, cultivable, aerobic heterotrophs belonging to the gram-positive and proteobacteria divisions. Thus, it seems reasonable to assume that one subcomponent of the interspace community may have lower species richness but that the total community diversity in the Cosnino interspace is probably similar to the community diversity in the rhizosphere environment.
Interpretation of phylotype composition similarity between communities was more problematic than analysis of phylotype richness or distribution. The similarity values were uniformly low (11 to 31%). However, the low values obtained from standard similarity comparisons may have been largely the result of each library containing a large proportion of rare phylotypes that had a low probability of being detected. Indeed, the average similarity of the culture collections (which contained smaller proportions of rare phylotypes) was higher than the average similarity of the clone libraries. The detection frequency of a phylotype in two or more libraries was positively correlated with phylotype abundance. We observed this relationship among the clone libraries as well as among the culture collections, although in the latter case the small sample size (34 phylotypes) provided a marginal number of data points. This relationship was expected for replicate samples but not for samples which had substantially different compositions. For replicate or very similar samples, abundant phylotypes should have greater probabilities of being resampled than rare phylotypes have. If community similarity values were calculated by using only the subset of phylotypes that appeared in at least two libraries, the average levels of similarity of the clone libraries and culture collections rose from 15 to 53% and from 26 to 52%, respectively. These data suggest that the phylotype compositions of the four environments may be substantially similar but that 16S rDNA clone libraries and culture collections document the similarities inadequately due to the large abundance of rare phylotypes typical for such collections.
In summary, 16S rDNA cloning appears to be as valid as plate cultivation for investigating diversity in environments despite the numerous biases that can occur in the 16S rDNA cloning method. Although the two approaches in some cases provide different assessments of relative community diversity, the discrepancies probably arise from sampling different segments of microbial communities. Identifying consistent relationships between environments based on comparisons of culture collections and culture-independent techniques may be highly dependent on the habitats sampled due to the limited ability of a single cultivation method to survey the bacterial domain and the influence of bacterial physiology in situ on the success of cultivation in the laboratory. Previous studies comparing the compositions of microbial communities by analysis of 16S rDNA clone libraries have been confined almost exclusively to comparisons of marine environments (2, 8, 14, 15, 36, 38, 42). Overall community similarities have not been described. However, identical or nearly identical (<1% mismatch) 16S rDNA sequences have been retrieved from marine samples obtained thousands of miles apart (8, 14, 16, 36, 42). Thus far, such sequences have not been identified in clone libraries derived from different soil samples. Analyses of community diversity by using clone libraries have not been replicated to date. Key questions about sampling probabilities and the range of composition similarity need to be addressed by using replicate samples in order to facilitate the interpretation of clone diversity in different environments. Other techniques based on direct 16S rDNA amplification that are more amenable to replication should provide powerful tools for rapidly investigating diversity in communities despite the inability of such techniques to accurately assess community structure.
REFERENCES
- 1.Atlas R M. Diversity of microbial communities. Vol. 7. New York, N.Y: Plenum Press; 1984. [Google Scholar]
- 2.Benlloch S, Rodriguez-Valera F, Martinez-Murcia A J. Bacterial diversity in two coastal lagoons deduced from 16S rDNA PCR amplification and partial sequencing. FEMS Microbiol Ecol. 1995;18:267–280. [Google Scholar]
- 3.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E R. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler D P, Li S, Spadoni C M, Drake G R, Balkwill D L, Fredrickson J K, Brockman F J. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol Ecol. 1997;23:131–144. [Google Scholar]
- 6.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 7.Cobb N S, Mopper S, Gehring C A, Caouette M, Christensen K M, Whitham T G. Increased moth herbivory associated with environmental stress of pinyon pine at local and regional levels. Oecologia (Berlin) 1997;109:389–397. doi: 10.1007/s004420050098. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E F, Franks E G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 9.Dunbar, J., and C. R. Kuske. 1998. Unpublished data.
- 10.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete determination of entire genes. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felske A, Wolterink A, Lis R V, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field K G, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannoni S J, Vritschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 17.Hattori T. Microbial life in the soil: an introduction. New York, N.Y: Marcel Dekker; 1973. [Google Scholar]
- 18.Hendricks D M. Arizona soils. Tucson: University of Arizona Press; 1985. [Google Scholar]
- 19.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara Y, Hattori T. Analysis of bacterial populations in a grassland soil according to rates of development on solid media. FEMS Microbiol Ecol. 1991;86:95–102. [Google Scholar]
- 23.Kowalchuk G A, Stephen J R, Boer W D, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuske, C. R., J. D. Busch, D. L. Adorada, K. Banton, and L. O. Ticknor. Unpublished data.
- 26.Lane D. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 27.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margalef R. Information theory in ecology. Gen Syst. 1958;3:36–71. [Google Scholar]
- 32.McDougald D, Rice S A, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation? FEMS Microbiol Ecol. 1998;25:1–9. [Google Scholar]
- 33.More M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller-Dombois D, Ellenberg J. Aims and methods of vegetation ecology. New York, N.Y: Wiley; 1974. [Google Scholar]
- 36.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 37.Ovreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen K, Arlinger J, Ekendahl S, Hallbeck L. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Aspo hard rock laboratory, Sweden. FEMS Microbiol Ecol. 1996;19:249–262. [Google Scholar]
- 39.Ponce M R, Michol J L. PCR amplification of long DNA fragments. Nucleic Acids Res. 1992;20:623. doi: 10.1093/nar/20.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reysenbach A, Giver L H, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simberloff D. Use of rarefaction and related methods in ecology. In: Dickson K L, Cairns J J, Livingston R J, editors. Biological data in water pollution assessment: quantitative and statistical analyses. West Conshohocken, Pa: American Society for Testing and Materials; 1978. pp. 150–165. [Google Scholar]
- 44.Simpson E H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 45.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki M T, Rappe M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai Y L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 50.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson K H, Blitchington R B, Green R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]