Abstract
The glyceraldehyde-3-phosphate dehydrogenase (gpd) promoter was used to drive expression of lip2, the gene encoding lignin peroxidase (LiP) isozyme H8, in primary metabolic cultures of Phanerochaete chrysosporium. The expression vector, pUGL, also contained the Schizophyllum commune ura1 gene as a selectable marker. pUGL was used to transform a P. chrysosporium Ura11 auxotroph to prototrophy. Ura+ transformants were screened for peroxidase activity in liquid cultures containing high-carbon and high-nitrogen medium. Recombinant LiP (rLiP) was secreted in active form by the transformants after 4 days of growth, whereas endogenous lip genes were not expressed under these conditions. Approximately 2 mg of homogeneous rLiP/liter was obtained after purification. The molecular mass, pI, and optical absorption spectrum of rLiPH8 were essentially identical to those of the wild-type LiPh8 (wt LiPH8), indicating that heme insertion, folding, and secretion functioned normally in the transformant. Steady-state and transient-state kinetic properties for the oxidation of veratryl alcohol between wtLiPH8 and rLiPH8 were also identical.
The white rot basidiomycete Phanerochaete chrysosporium has been the focus of numerous studies on the degradation of lignin (6, 15, 22) and aromatic pollutants (5, 17). Two peroxidases, manganese peroxidase (MnP) and lignin peroxidase (LiP), along with an extracellular H2O2-generating system, are thought to be the major extracellular components of the lignin-degrading system (14, 18, 22) of this organism. Both MnP and LiP occur as a series of isozymes encoded by a family of genes which are expressed under secondary metabolic growth conditions (9, 12, 14). The major isozymes, MnP1 (H3) and LiPH8, have been characterized in detail (14), and the X-ray structures of MnP1 (38) and LiPH8 (30, 31) have been reported. In addition, a homologous expression system (28) and several heterologous expression systems for MnP have been established (37, 41), allowing structure-function studies of mutant MnPs (24, 25, 42).
In contrast, the efficient expression of active recombinant LiPH8 (rLiPH8) has not been achieved. The use of Escherichia coli as a LiP expression host has resulted in expression; however, refolding of denatured LiP from E. coli inclusion bodies resulted in the isolation of active rLiPH8 (10) and rLiPH2 (29) in relatively low yield. In addition, neither isozyme was glycosylated and, in one case, the recombinant protein contained seven extra N-terminal amino acids (10).
In this paper, we report the first successful homologous expression of rLiPH8 in P. chrysosporium and the characterization of the recombinant enzyme.
MATERIALS AND METHODS
Organisms.
P. chrysosporium wild-type strain OGC-101 (3), auxotrophic strain OGC316-7 (Ura11) (1), and prototrophic transformants were maintained as described previously (2). E. coli DH5α/F′ was used for subcloning of plasmids.
Construction of the Ura transformation plasmid.
A 1.5-kb blunt-ended BspMI-EcoRI fragment of pEF1 (1, 11), containing the Schizophyllum commune ura1 gene, was ligated into the blunt-ended EcoO109 site of pUC18 (GibcoBRL) to obtain pUB. This P. chrysosporium transformation plasmid contains the complete pUC18 plasmid and the full S. commune ura1 coding region, including 200 bp of the promoter region.
Construction of pUGL.
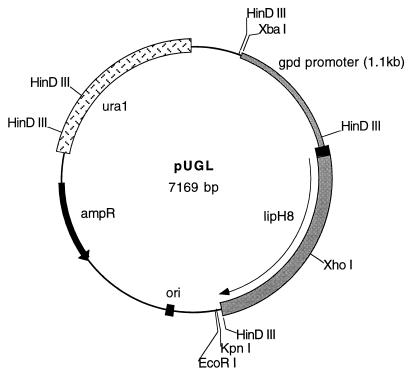
The promoter from the P. chrysosporium gpd gene (28) and the P. chrysosporium lip2 gene (32) were fused at their TATA box sites and subcloned into the multiple cloning site of pUB. The 1.1-kb gpd promoter fragment was prepared by PCR using Vent DNA Polymerase (Biolabs, Inc.), pAGM1 (28) as the template, a forward primer (5′-AATTAACCCTCACTAAAGGG) 1.15 kb upstream of the gpd translation start site, and a reverse primer (5′-AAGGTTTTCGTCATCGATTGG) starting immediately 5′ of the gpd TATA box. The lip2 fragment was prepared by PCR, using a forward primer (5′-TATAAAAGGGACGATGCG) from and including the lip2 TATA box and a reverse primer (5′-TCACGCAGAAAGCATCC) within the coding region of lip2 with pLH8 (32) as the template. The gpd promoter PCR fragment was cut with XbaI at a site 55 bp from the 5′ end. The lip2 coding region PCR fragment was cut with XhoI 25 bp upstream of the 3′ end. Both fragments were phosphorylated and then subcloned into Bluescript II SK-XbaI–HindIII, together with a 1-kb XhoI-HindIII coding region fragment from pLH8 (32) in a four-fragment ligation, to create a blunt junction between the gpd promoter fragment and the lip PCR fragment, to obtain pGL1. Subsequently, a 3.0-kb XbaI-KpnI fragment from pGL1 encompassing the gpd promoter and the lip2 gene was subcloned into pUB to obtain the pUGL expression vector (Fig. 1). The ligation sites and newly synthesized coding sequence were verified by sequencing.
FIG. 1.
Restriction map of the LiPH8 expression vector pUGL, containing the ura1 gene and the gpd promoter fused to the lip gene at their TATA box sites.
Transformation of the P. chrysosporium uracil auxotroph Ura11.
Protoplasts of P. chrysosporium Ura11 were transformed with EcoRI-linearized pUGL or pUB as described previously (1, 4). Prototrophic transformants were transferred to minimal medium, screened, and purified by isolating single basidiospores (3, 4).
Screening for expression of recombinant LiP isozyme H8 (rLiPH8).
Conidia from slants of pUGL prototrophic transformants were used to inoculate 25 ml of high-carbon high-nitrogen (HCHN) medium (23) containing 2% glucose, 20 mM sodium 2,2-dimethyl succinate, 24 mM ammonium tartrate, and 3 mM veratryl alcohol (VA) at pH 4.5 in stationary flasks. After 3 days at 28°C, the extracellular medium was assayed periodically for LiP activity with the diammonium 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonate) (ABTS) assay (13) using 0.1 mM H2O2 and 0.5 mM VA.
Production of rLiPH8.
Selected pUGL transformants were grown for 2 days at 37°C from conidial inocula as stationary cultures in 1-liter flasks containing 80 ml of HCHN medium with 0.2% tryptone. The mycelial mats were homogenized and used as inocula for 1-liter HCHN cultures, containing 3 mM VA and 0.1% Tween 80. The 1-liter cultures were incubated at 28°C for 4 days at 150 rpm on a rotary shaker.
Purification of rLiPH8.
The filtrate obtained from seven 1-liter cultures was concentrated to ∼400 ml at 4°C by using a hollow-fiber filter system (10-kDa molecular mass cutoff; Amicon). (NH4)2SO4 was added to a final concentration of 1.5 M, the mixture was subjected to centrifugation for 1 h at 18,000 × g, and the pellet was discarded. All subsequent steps were performed at 4°C.
Phenyl Sepharose chromatography.
The concentrated culture filtrate was applied to a Phenyl Sepharose CL-4B (Pharmacia) column (100 ml) equilibrated with 20 mM sodium acetate (pH 4.5) containing 1.5 M (NH4)2SO4. The column was washed with 200 ml of 20 mM sodium acetate (pH 4.5) containing 0.8 M (NH4)2SO4, and the protein was eluted with a gradient of 0.8 to 0.2 M (NH4)2SO4 in 20 mM sodium acetate (pH 4.5). Fractions with LiP activity were pooled and concentrated to ∼2 ml by membrane ultrafiltration.
Size exclusion chromatography.
The Phenyl Sepharose fraction was applied to a 100-ml Sephadex G-100 column equilibrated with 20 mM sodium succinate buffer (pH 4.5), and protein was eluted with the same buffer. Fractions with LiP activity were desalted and concentrated.
Anion-exchange FPLC.
The pooled, concentrated Sephadex G-100 protein fraction was applied to a Mono Q HR 5/5 column (Pharmacia) equilibrated with 10 mM sodium acetate (pH 6.0) in a fast protein liquid chromatography (FPLC) system. The protein was eluted with a nonlinear gradient (0.01 to 0.5 M sodium acetate; pH 6.0). Fractions containing LiP activity were desalted and concentrated.
SDS-PAGE and IEF.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a 12% Tris-glycine gel system (27) and a Miniprotean II apparatus (Bio-Rad). The gels were stained with Coomassie blue (16). The Bio-Rad SDS-PAGE Low Range standard mix was used for comparison. Isoelectric focusing (IEF) electrophoresis was performed using the Pharmacia Phastsystem with IEF Phastgels (pH 3 to 9). The gels were stained with Coomassie blue. The Sigma IEF MIX 3.6-6.6 marker kit was used as a standard.
Spectroscopic and kinetic procedures.
Enzyme absorption spectra and assays were determined with a Shimadzu UV-260 spectrophotometer at room temperature, using a 1-cm light path cuvette. LiP oxidation of VA to veratraldehyde was performed as described previously (15, 22) and followed at 310 nm. LiP oxidation of ABTS was carried out in sodium succinate (pH 3.0) in the presence of 0.1 mM H2O2 and 0.5 mM VA and followed at 415 nm as described previously (13). For steady-state kinetic measurements, VA oxidation was determined in the presence of various H2O2 concentrations at a constant VA concentration (0.5 mM) or with various VA concentrations at a constant H2O2 concentration (0.1 mM) in 20 mM sodium-succinate (pH 3.0) with 1 μg of enzyme/ml.
Transient-state kinetics.
Kinetic measurements were conducted at 25°C using an Applied Photophysics stopped-flow reaction analyzer (SX.18MV) with sequential mixing. LiP compound I formation was measured at 397 nm. Native LiP (2 μM) was mixed with a 10- to 50-fold excess of H2O2 in 20 mM sodium succinate (pH 3.0). LiP compound I reduction was measured by first mixing 4 μM enzyme and 1 equiv of H2O2 in H2O. Then, VA in 40 mM sodium succinate (pH 3.0) was added, and compound I reduction was measured at 416 nm. LiP compound II reduction was measured at 397 nm by sequential mixing of 4 μM enzyme, 1 eq of ferrocyanide, and 1 eq of H2O2 in H2O and then by adding VA in 40 mM sodium succinate (pH 3.0).
RESULTS
Expression of rLiP.
Transformation of the Ura11 auxotroph with 2 μg of the linearized LiPH8 expression construct pUGL (Fig. 1) resulted in the isolation of 35 Ura+ transformants. Twelve transformants were grown in HCHN stationary cultures and screened for extracellular LiP activity using the ABTS assay in the presence of VA. Total RNA was extracted from 4-day-old cells, and Northern blotting was carried out on the same sample of transformants. Hybridization with a lip H8 cDNA probe revealed that LiP mRNA was present in all transformants, although in various amounts (data not shown). Two of the pUGL transformants with the highest rLiP activity in this initial screening were purified by isolating single basidiospores (3) and were analyzed further in large liquid shake cultures. A Southern blot of DNA from one of the transformants probed with pUC18 showed that the transforming DNA was chromosomally integrated (data not shown).
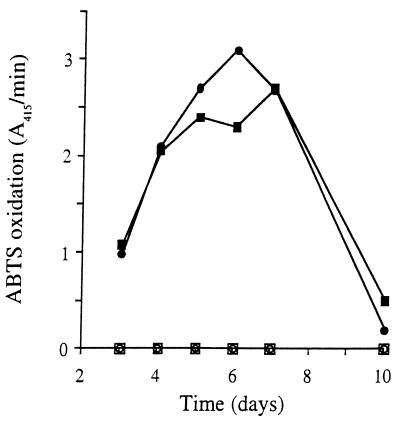
The time courses for the appearance of LiP activity in shaken HCHN cultures of basidiospore purified transformants and controls are shown in Fig. 2. Extracellular LiP activity was detected only in the cultures of pUGL-transformed strains, and maximal activity was reached after 6 days. The wild-type strain OGC-101 and the Ura11 auxotroph transformed with the transformation vector, pUB, exhibited no LiP activity even after 10 days of incubation. This is the period during which endogenous LiP is expressed in low-nitrogen cultures (9, 15, 22). These observations suggest that the activity observed in the transformants was rLiP.
FIG. 2.
LiP activity in the extracellular medium of primary metabolic cultures of P. chrysosporium. UGL10 (●) and UGL11 (■) were pUGL transformants, UB1.7 (○) was a pUB transformant, and OGC-101 (□) was the wild-type strain. Agitated cultures were grown under high-carbon, high-nitrogen conditions, and ABTS oxidation rates were measured as described in the text.
The effect of several culture parameters on the expression of rLiP was examined. The addition of 3 mM VA to the cultures is essential for efficient rLiP expression. In contrast to the results obtained with rMnP expression (28), where an initial pH of 6.5 yielded optimal expression, pH 4.5 was found to be optimal for rLiP expression.
The rLiPH8 was purified from large shake cultures of the transformant UGL10. Successive steps of concentration and hydrophobic-interaction, size exclusion, and anion-exchange chromatography were performed. Mono Q anion-exchange chromatography demonstrated that rLiP activity eluted as a single peak corresponding to that of wild-type LiPH8 (wtLiPH8) (data not shown).
The Rz value (A407/A280) of the purified rLiPH8 was ∼4.2. The specific activity of rLiPH8 for VA was 25.4 U/mg, which is similar to the wild-type wt LiPH8 specific activity of 27.6 U/mg. Approximately 2 mg of purified rLiP was obtained from 1 liter of extracellular culture fluid, which corresponds to a yield of ∼60%. An SDS-polyacrylamide gel (Fig. 3A) of rLiPH8 showed a single band with a molecular mass of 42 kDa, which was identical to that of the wtLiPH8. An IEF gel (Fig. 3B) also showed a single band of rLiPH8 protein with a pI of 3.3, which was identical to that of wtLiPH8.
FIG. 3.
(A) SDS-PAGE of rLiPH8 and wtLiPH8. A total of 2 μg of each protein was loaded onto a 12% Tris-glycine-polyacrylamide gel. The gel was stained with Coomassie brilliant blue. The positions of the BRL low-molecular-mass markers are indicated. (B) IEF of rLiPH8 and wtLiPH8. A total of 2 μg of each protein was loaded onto a Pharmacia Phastgel with a pH gradient of 3 to 9. The positions of the Sigma IEF MIX 3.6-6.6 markers are indicated.
Spectral and kinetic properties.
The absorption spectrum of rLiPH8 (Fig. 4) exhibited a Soret maximum at 407 nm and visible bands at 500 and 635 nm. The shapes and intensities of the absorption bands of rLiPH8 were very similar to those of wtLiP, suggesting that the heme environments of wtLiP and rLiP are very similar.
FIG. 4.
Comparison of the absorption spectra of rLiP (–––) and wtLiP (——). The spectra were determined in the presence of 20 mM sodium succinate (pH 3.0) as described in the text.
Under steady-state conditions, linear Lineweaver-Burke plots were obtained for 1/v versus 1/H2O2 and 1/v versus 1/VA over a range of H2O2 and VA concentrations. The calculated Km, kcat, and kcat/Km for rLiP and wtLiP were similar (Table 1).
TABLE 1.
Steady-state kinetic parameters for rLiPH8 and wtLiPH8a
| Enzyme | Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| rLiP | H2O2 | 53.1 | 21.2 | 4.0 × 105 |
| wtLiP | H2O2 | 47.7 | 23.0 | 4.8 × 105 |
| rLiP | VA | 89.4 | 23.2 | 2.6 × 105 |
| wtLiP | VA | 92.8 | 22.9 | 2.5 × 105 |
Reactions were carried out in 20 mM sodium succinate (pH 3.0). Apparent Km and kcat values for VA were determined by using 0.1 mM H2O2. Apparent Km and kcat values for H2O2 were determined by using 0.5 mM VA.
Transient-state kinetic measurements were determined for each step in the catalytic cycle. The calculated second-order rate constants (k1 app) for compound I formation under transient-state conditions are shown in Table 2. The reduction of compound I for both the wild-type and recombinant enzymes followed similar second-order kinetics. The calculated second-order rate constants (k2 app) are also listed in Table 2. For compound II reduction by VA, the plots of the observed rate constants kobs versus [VA] were hyperbolic, indicating saturation kinetics. The first-order rate constant k3 app and the apparent dissociation constant K3 were calculated from the least-squares fit of kobs versus [VA] and are shown in Table 2. The initial slope of the hyperbolic curve could be used to estimate a second-order rate constant k3 app for compound II reduction.
TABLE 2.
Transient-state kinetic parameters for rLiPH8 and wtLiPH8a
| Enzyme | Compound I formation
|
Compound I reduction by VA
|
Compound II reduction by VA
|
||
|---|---|---|---|---|---|
| k1 app (M−1s−1) | k2 app (M−1s−1) | k3 app (s−1) | K3 (μM) | k3 app (M−1s−1) | |
| rLiP | 4.7 × 105 | 1.03 × 105 | 31 | 242 | 1.3 × 105 |
| wtLiP | 4.5 × 105 | 0.97 × 105 | 40 | 224 | 1.8 × 105 |
Reactions were carried out in 20 mM sodium succinate (pH 3.0) with 1 μM enzyme as described in the text. Compound I formation was monitored at 397 nm. Compound I reduction and compound II reduction in the presence of VA were monitored at 416 and 397 nm, respectively.
DISCUSSION
LiP is a major extracellular component of the lignin-degrading system of P. chrysosporium as well as a variety of other white rot fungi (9, 14, 19, 22). The enzyme is capable of oxidizing lignin and nonphenolic aromatics with redox potentials beyond the reach of other peroxidases (17, 20, 22, 34). Extensive spectroscopic and kinetic studies have been carried out on LiP (14, 21, 22, 26, 35, 40). However, a variety of questions concerning the mechanism of this enzyme are still under discussion. For example, the amino acids involved in the substrate binding site have not been determined (31, 36). Secondly, the role of VA in the catalytic cycle and its presumed role as a mediator in LiP reactions (8, 20, 21, 26, 35) warrant further research. Studies on site-directed mutant forms of LiP will shed light on these questions.
Previously, we developed a homologous expression system for MnP in P. chrysosporium (28). Here, a similar strategy using a Ura− strain (1) and the Ura biosynthetic gene (11) as a selectable marker has enabled the expression of rLiP under primary metabolic conditions during which endogenous lip genes are not expressed (14).
Ectopic integration of the transformation construct required screening of Ura+ transformants for optimal lip expression. Differences in mRNA levels and enzyme activity were used as criteria to select a transformant which expressed LiP efficiently. Neither strain OGC-101 (wild-type) nor the transformant UB1.7 (control plasmid) expressed detectable LiP under primary metabolic conditions, strongly suggesting that the expressed protein was rLiP.
The addition of 3 mM VA to the growth medium was required for the recovery of rLiP. Since lip expression is under the control of the gpd promoter in these experiments, it is unlikely that VA is involved in regulation of lip expression, supporting previous conclusions (7). It is more likely that VA is involved in the stabilization or protection of the enzyme from H2O2 inactivation (39, 40).
Our current yield of ∼3 mg of rLiPH8/liter in the crude extracellular medium was sufficient for structural and kinetic studies. While we obtain ∼10 mg of LiP activity/liter in wild-type cultures, this activity represents all of the isozymes of LiP; therefore, the LiPH8 yields in the two systems are comparable.
Purification of rLiP was achieved by sequential hydrophobic interaction, gel filtration, and anion-exchange chromatographies. SDS-PAGE analysis indicates that rLiP and wtLiP are nearly identical in molecular mass at about 42 kDa. IEF shows that the rLiPH8 is a homogeneous isolate and that it has the same isoelectric point as wtLiPH8 (Fig. 3A and B). This suggests that the LiP protein expressed is encoded by the introduced gene rather than by an endogenous gene. This also suggests that both proteins undergo very similar posttranslational processing, including cleavage of signal and propeptide sequences (33), folding, and glycosylation. The successful homologous expression of both MnP (28) and LiP suggests further that factors that positively regulate the expression of these proteins during secondary metabolism act at the transcriptional level and are mediated by the promoter regions of these genes, since all other factors such as translation, processing, and secretion appear to function during primary metabolic growth.
The wild-type and recombinant enzymes also have identical UV-visible spectral features (Fig. 4), indicating that the insertion, environment, and orientation of the heme are similar. Homologously expressed rLiP exhibits Km, kcat, and kcat/Km values for VA and H2O2 that are very similar to those of the wild-type enzyme (Table 2), suggesting that the substrate binding and catalytic efficiency of rLiP and wtLiP are similar. Furthermore, transient-state kinetic analysis indicates that the rates of the three steps in the LiP catalytic cycle for the two proteins are very similar (Table 1 and 2).
Taken together, these results indicate that the rLiP and wtLiP are very similar, and they suggest that this expression system will enable the generation of site-directed mutant proteins which will be folded and processed in a manner identical to that of wtLiP but will be altered only at the designated site. Structure-function studies on such LiPH8 mutant proteins using this expression system are in progress.
ACKNOWLEDGMENTS
This work was supported by grants to M.H.G. from the Division of Energy Biosciences of the U.S. Department of Energy (DE-FG03-96ER20235) and the National Science Foundation (MCB-9506338).
REFERENCES
- 1.Akileswaran L, Alic M, Clark E K, Hornick J L, Gold M H. Isolation and transformation of uracil auxotrophs of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Curr Genet. 1993;23:351–356. doi: 10.1007/BF00310898. [DOI] [PubMed] [Google Scholar]
- 2.Alic M, Gold M H. Genetic recombination in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1985;50:27–30. doi: 10.1128/aem.50.1.27-30.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alic M, Letzring C, Gold M H. Mating system and basidiospore formation in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1987;53:1464–1469. doi: 10.1128/aem.53.7.1464-1469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alic M, Mayfield M B, Akileswaran L, Gold M H. Homologous transformation of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Curr Genet. 1991;19:491–494. doi: 10.1007/BF00310898. [DOI] [PubMed] [Google Scholar]
- 5.Bumpus J A, Aust S D. Biodegradation of environmental pollutants by the white rot fungus Phanerochaete chrysosporium: involvement of the lignin-degrading system. Bioessays. 1987;6:166–170. [Google Scholar]
- 6.Buswell J A, Odier E. Lignin biodegradation. Crit Rev Biotechnol. 1987;6:1–60. [Google Scholar]
- 7.Cancel A M, Orth A B, Tien M. Lignin and veratryl alcohol are not inducers of the ligninolytic system of Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:2909–2913. doi: 10.1128/aem.59.9.2909-2913.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candeias L P, Harvey P J. Lifetime and reactivity of the veratryl alcohol radical cation. Implications for lignin peroxidase catalysis. J Biol Chem. 1995;270:16745–16748. doi: 10.1074/jbc.270.28.16745. [DOI] [PubMed] [Google Scholar]
- 9.Cullen D. Recent advances on the molecular genetics of lignolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 10.Doyle W A, Smith A T. Expression of lignin peroxidase H8 in Escherichia coli: folding and activation of the recombinant enzyme with Ca2+ and haem. Biochem J. 1996;315:15–19. doi: 10.1042/bj3150015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froeliger E H, Ullrich R C, Novotny C P. Sequence analysis of the URA1 gene encoding orotidine-5′-monophosphate decarboxylase of Schizophyllum commune. Gene. 1989;83:387–393. doi: 10.1016/0378-1119(89)90127-3. [DOI] [PubMed] [Google Scholar]
- 12.Gettemy J M, Ma B, Alic R, Gold M H. Reverse transcription PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol. 1998;64:569–574. doi: 10.1128/aem.64.2.569-574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenn J K, Gold M H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 14.Gold M H, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold M H, Wariishi H, Valli K. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. ACS (Am Chem Soc) Symp Ser. 1989;389:127–140. [Google Scholar]
- 16.Hames B D, Rickwood D. Gel electrophoresis of proteins: a practical approach. Oxford, England: IRL Press; 1981. [Google Scholar]
- 17.Hammel K E. Oxidation of aromatic pollutants by lignin-degrading fungi and their extracellular peroxidases. In: Sigel H, Sigel A, editors. Metal ions in biological systems. Vol. 28. New York, N.Y: Marcel Dekker; 1992. pp. 41–60. [Google Scholar]
- 18.Hammel K E, Jensen K A, Jr, Mozuch M D, Landucci L L, Tien M, Pease E A. Ligninolysis by a purified lignin peroxidase. J Biol Chem. 1993;268:12274–12281. [PubMed] [Google Scholar]
- 19.Hatakka A. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1994;13:125–135. [Google Scholar]
- 20.Joshi D K, Gold M H. Oxidation of dimethoxylated aromatic compounds by lignin peroxidase from Phanerochaete chrysosporium. Eur J Biochem. 1996;237:45–57. doi: 10.1111/j.1432-1033.1996.0045n.x. [DOI] [PubMed] [Google Scholar]
- 21.Khindaria A, Yamazaki I, Aust S D. Stabilization of the veratryl alcohol cation radical by lignin peroxidase. Biochemistry. 1996;35:6418–6424. doi: 10.1021/bi9601666. [DOI] [PubMed] [Google Scholar]
- 22.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 23.Kirk T K, Schultz E, Connors W J, Lorenz L F, Zeikus J G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol. 1978;177:277–285. [Google Scholar]
- 24.Kishi K, Hildebrand D P, Kusters-van Someren M, Gettemy J, Mauk A G, Gold M H. Site-directed mutations at phenylalanine-190 of manganese peroxidase: effects on stability, function, and coordination. Biochemistry. 1997;36:4268–4277. doi: 10.1021/bi962627t. [DOI] [PubMed] [Google Scholar]
- 25.Kishi K, Kusters-van Someren M, Mayfield M B, Sun J, Loehr T M, Gold M H. Characterization of manganese(II) binding site mutants of manganese peroxidase. Biochemistry. 1996;35:8986–8994. doi: 10.1021/bi960679c. [DOI] [PubMed] [Google Scholar]
- 26.Koduri R S, Tien M. Oxidation of guaiacol by lignin peroxidase. Role of veratryl alcohol. J Biol Chem. 1995;270:22254–22258. doi: 10.1074/jbc.270.38.22254. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Mayfield M B, Kishi K, Alic M, Gold M H. Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:4303–4309. doi: 10.1128/aem.60.12.4303-4309.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie G, Reading N S, Aust S D. Expression of the lignin peroxidase H2 gene from Phanerochaete chrysosporium in Escherichia coli. Biochem Biophys Res Commun. 1998;249:146–150. doi: 10.1006/bbrc.1998.9106. [DOI] [PubMed] [Google Scholar]
- 30.Piontek K, Glumoff T, Winterhalter K. Low pH crystal structure of glycosylated lignin peroxidase from Phanerochaete chrysosporium at 2.5 A resolution. FEBS Lett. 1993;315:119–124. doi: 10.1016/0014-5793(93)81146-q. [DOI] [PubMed] [Google Scholar]
- 31.Poulos T L, Edwards S L, Wariishi H, Gold M H. Crystallographic refinement of lignin peroxidase at 2 Å. J Biol Chem. 1993;268:4429–4440. doi: 10.2210/pdb1lga/pdb. [DOI] [PubMed] [Google Scholar]
- 32.Ritch T G, Jr, Gold M H. Characterization of a highly expressed lignin peroxidase-encoding gene from the basidiomycete Phanerochaete chrysosporium. Gene. 1992;118:73–80. doi: 10.1016/0378-1119(92)90250-s. [DOI] [PubMed] [Google Scholar]
- 33.Ritch T G, Jr, Nipper V J, Akileswaran L, Smith A J, Pribnow D G, Gold M H. Lignin peroxidase from the basidiomycete Phanerochaete chrysosporium is synthesized as a preproenzyme. Gene. 1991;107:119–126. doi: 10.1016/0378-1119(91)90304-t. [DOI] [PubMed] [Google Scholar]
- 34.Schoemaker H E. On the chemistry of lignin biodegradation. Recl Trav Chim Pays-Bas. 1990;109:255–272. [Google Scholar]
- 35.Sheng D, Gold M H. Irreversible oxidation of ferricytochrome c by lignin peroxidase. Biochemistry. 1998;37:2029–2036. doi: 10.1021/bi972198e. [DOI] [PubMed] [Google Scholar]
- 36.Smith A T, Veitch N C. Substrate binding and catalysis in heme peroxidases. Curr Opin Chem Biol. 1998;2:269–278. doi: 10.1016/s1367-5931(98)80069-0. [DOI] [PubMed] [Google Scholar]
- 37.Stewart P, Whitwam R E, Kersten P J, Cullen D, Tien M. Efficient expression of a Phanerochaete chrysosporium manganese peroxidase gene in Aspergillus oryzae. Appl Environ Microbiol. 1996;62:860–864. doi: 10.1128/aem.62.3.860-864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaramoorthy M, Kishi K, Gold M H, Poulos T L. The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-A resolution. J Biol Chem. 1994;269:32759–32767. [PubMed] [Google Scholar]
- 39.Tonon F, Odier E. Influence of veratryl alcohol and hydrogen peroxide on ligninase activity and ligninase production by Phanerochaete chrysosporium. Appl Environ Microbiol. 1988;54:466–472. doi: 10.1128/aem.54.2.466-472.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wariishi H, Gold M H. Lignin peroxidase compound III. Mechanism of formation and decomposition. J Biol Chem. 1990;265:2070–2077. [PubMed] [Google Scholar]
- 41.Whitwam R, Tien M. Heterologous expression and reconstitution of fungal Mn peroxidase. Arch Biochem Biophys. 1996;333:439–446. doi: 10.1006/abbi.1996.0413. [DOI] [PubMed] [Google Scholar]
- 42.Whitwam R E, Brown K R, Musick M, Natan M J, Tien M. Mutagenesis of the Mn2+-binding site of manganese peroxidase affects oxidation of Mn2+ by both compound I and compound II. Biochemistry. 1997;36:9766–9773. doi: 10.1021/bi9708794. [DOI] [PubMed] [Google Scholar]