Abstract
Diversity of the nitrous oxide reductase (nosZ) gene was examined in sediments obtained from the Atlantic Ocean and Pacific Ocean continental shelves. Approximately 1,100 bp of the nosZ gene were amplified via PCR, using nosZ gene-specific primers. Thirty-seven unique copies of the nosZ gene from these marine environments were characterized, increasing the nosZ sequence database fourfold. The average DNA similarity for comparisons between all 49 variants of the nosZ gene was 64% ± 10%. Alignment of the derived amino acid sequences confirmed the conservation of important structural motifs. A highly conserved region is proposed as the copper binding, catalytic site (CuZ) of the mature protein. Phylogenetic analysis demonstrated three major clusters of nosZ genes, with little overlap between environmental and culture-based groups. Finally, the two non-culture-based gene clusters generally corresponded to sampling location, implying that denitrifier communities may be restricted geographically.
Denitrification is the reductive respiration of nitrate or nitrite to N2 or N2O and generally occurs under anaerobic conditions. In the marine environment, denitrification is important because it may remove over 50% of the nitrogen inputs to the oceans as a whole (5, 15). Nitrous oxide reduction is the final step in the denitrification pathway and is catalyzed by the enzyme nitrous oxide reductase. The gene encoding nitrous oxide reductase (nosZ) is largely unique to denitrifying bacteria and has recently been used for detection of denitrifier-specific DNA in environmental samples (13). The staff at our laboratory is interested in the ecology of denitrifying bacteria in marine and coastal systems and assessing whether denitrifier diversity plays a role in determining in situ denitrification rates. Given that most bacteria are as yet unculturable (2, 17) and the paucity of nosZ sequence data, it is difficult at present to ascertain whether different geographic areas contain unique suites of denitrifiers or if certain species of denitrifiers are ubiquitous. Furthermore, it is unclear whether the denitrifying strains that have been well studied in culture are also the denitrifying bacteria that predominate in the environment. At the onset of this research 12 different nosZ genes, 9 of them from denitrifiers in culture, had been reported to GenBank. This paper presents a comparison of 37 nosZ genes from sediments collected at two sites, in the Atlantic and Pacific Oceans, with those previously reported sequences.
MATERIALS AND METHODS
Environmental sites.
Atlantic sediment samples were collected in June 1996 from a long-term ecosystem observatory site (LEO-15) that has been described in detail elsewhere (19). Sediment cores at 15-m depth were retrieved and stored frozen at −20°C until analysis. Pacific Ocean sediments were obtained in July 1996 from a site 30 m offshore of San Clemente Island, Calif. (32°58′ N, 118°32′ W). The samples were collected in a sea grass bed at 3-m depth and also stored frozen.
Extraction and amplification of DNA.
Total genomic DNA was extracted from approximately 100 mg (wet weight) of sediment, as described previously (13). The CAMP-Nos661F and CAMP-Nos1773R (Table 1) primers were used to amplify ∼1,100 bp of the nosZ gene. PCR conditions were one cycle at 94°C for 5 min followed by 35 cycles of 95°C for 0.5 min, 56°C for 1.5 min, and 72°C for 2 min and a final extension step at 72°C for 10 min (13).
TABLE 1.
Oligonucleotides used for cloning and sequencing of nosZ gene fragments
| Oligonucleotidea | Sequenceb |
|---|---|
| Cloning primers | |
| CAMP-Nos661F | 5′CUACUACUACUACGGCUGGGGGCUGACCAA |
| CAMP-Nos1773R | 5′CAUCAUCAUCAUAURUCGAUCARCUGBUCGUU |
| Sequencing primers | |
| NosGrpA-For | 5′CGAYGCCGACTATCAGGG |
| NosGrpA-Rev | 5′CCCTGATAGTCGGCRTCG |
| NosGrpB-For | 5′GCCTGGCARGTSATVGTBGACGG |
| NosGrpB-Rev | 5′CCGTCVACBATSACYTGCCAGGC |
| NosGrpC-For | 5′CAYGAAGCRCCRYTGGTCAA |
| NosGrpC-Rev | 5′TTGACCARYGGYGCTTCRTG |
| Nos1319R | 5′GTBGGBGAVARCTTGCC |
All numbers are relative to the nosZ sequence of P. stutzeri Zobell.
B = C, G or T; R = A or G; S = C or G; V = A, C or G; Y = C or T.
Cloning of PCR products.
PCR products were cloned by using the CLONEAMP pAMP1 System (Gibco BRL, Life Technologies, Gaithersburg, Md.) with slight modifications (the volume of PCR product was increased to 3 μl and that of vector DNA was decreased to 1 μl). Unique clones were identified by HaeIII digestion of complete recombinant plasmids. HaeIII is a restriction endonuclease with a 4-bp recognition sequence and would cut each recombinant molecule approximately 20 times (i.e., 4,118 bp [vector] + 1,134 bp [insert]/256). Plasmids demonstrating unique HaeIII banding patterns were identified by using 2% Metaphor (FMC BioProducts, Rockland, Maine) agarose gels. For sequence analysis, plasmids were repurified by using the FlexiPrep Kit (Pharmacia, Piscataway, N.J.) and sequenced on an ABI 373 automated sequencer (Perkin-Elmer/ABI, Foster City, Calif.).
Sequence analysis.
Complete sequences were analyzed by using the Auto Assembler (Perkin-Elmer/ABI), SeqNavigator (Perkin-Elmer/ABI), and BLASTN (1) and FASTA (11) programs. Phylogenetic tree reconstruction used the Genetic Data Environment (16) with the neighbor-joining distance (Jukes-Cantor correction [12]) and maximum-likelihood (6) methods.
Nucleotide sequence accession numbers.
Nucleotide sequences generated in this study were filed in GenBank under the accession nos. AF119918 through AF119954. Accession numbers for previously published nosZ gene sequences used in this study are as follows: Achromobacter cycloclastes, AF047429; Bradyrhizobium japonicum, AJ002531; Paracoccus denitrificans, X74792; Pseudomonas aeruginosa, X65277; Pseudomonas stutzeri Zobell, M22628; Ralstonia eutropha, X65278; Sinorhizobium meliloti, U47133; Thiosphaera pantotropha, AF016058; S321195A, AF016055; S321195B, AF016056; and S321195C, AF016057.
RESULTS
Cloning and sequencing of PCR products.
Twenty novel nosZ genes from the LEO-15 sample were identified and were given designations beginning with 696. The San Clemente Island sample yielded 17 additional nosZ genes that were given designations starting with Pro. Although plasmids yielding similar HaeIII banding patterns were found to contain identical nosZ genes, this screening method may underestimate the actual diversity present in the clonal library.
Percent similarities for all nosZ genes at the DNA level ranged from a low of 33% (similarity between R. eutropha and clone 696E) to a high of 99% (similarity between clones ProS and ProJ) (data not shown). The average DNA percent similarity for comparisons between all 49 variants of the nosZ gene was 64% ± 10%. The marine sediment nosZ clones were no more than 69% similar to any of the nosZ genes of cultured denitrifiers, with the exception of ProL (which had 99% similarity to T. pantotropha).
Percent similarities at the derived amino acid level were slightly higher than those at the DNA level and averaged 66% ± 17%. In most instances, percent similarities increased at the amino acid level, as would be expected to result from the translation of ambiguous codons into amino acid residues. One notable exception to this trend was R. eutropha, which showed 46% average similarity to all other nosZ sequences at the DNA level but had derived amino acid sequence similarity of only 25%.
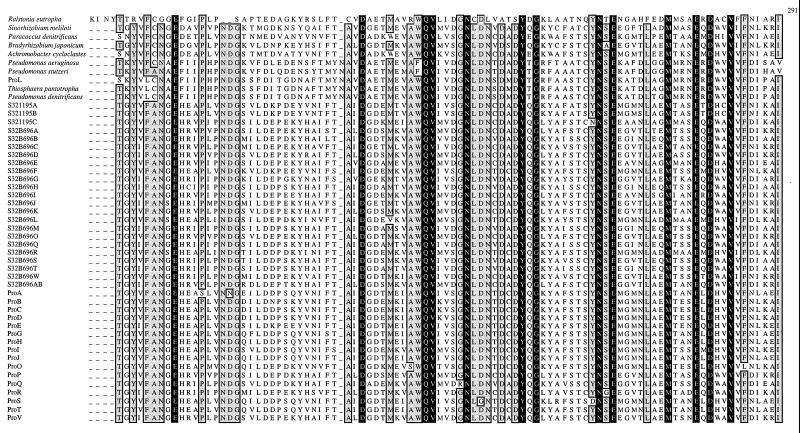
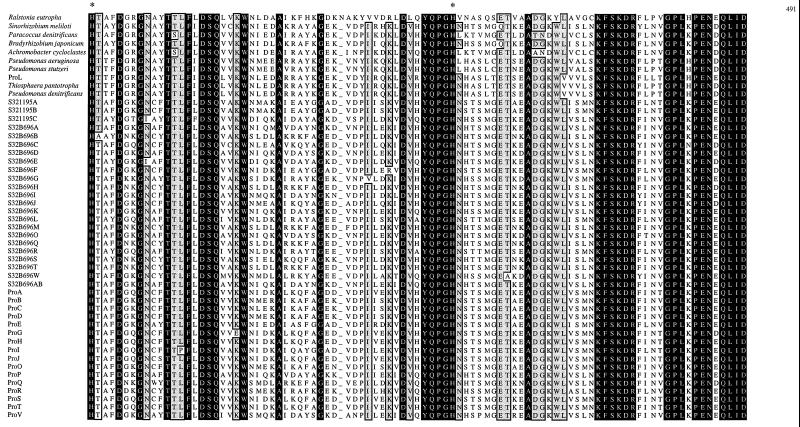
The derived amino acid sequences of all 49 nosZ gene sequences were aligned (Fig. 1). Approximately 22% of the 395 amino acid residues were conserved across all nosZ genes. This region includes 7 of the 11 conserved histidine residues reported previously (13). Despite large differences at both the DNA and amino acid sequence levels, all seven of these histidine residues remained conserved.
FIG. 1.
Alignment of the derived amino acid sequences from 49 nosZ genes. White letters on a black field indicate amino acids conserved across all 49 protein sequences. Conserved histidine residues are highlighted by an asterisk. Sites with at least 94% identical residues (46 or more of the 49 residues) are indicated by boxing and light shading. The proposed CuZ site is indicated by a line beginning at residue 132 (numbering is per the sequence from P. stutzeri). Amino acid sequences were aligned with CLUSTAL (8) and a Microsoft Excel coloring Macro (7).
Phylogenetic analyses.
A preliminary phylogenetic analysis based on ∼420 bp was used to identify high-level groups of nosZ genes for the design of internal sequencing primers (Table 2). These internal primers were used to obtain the full sequences of the 1.1-kb nosZ fragments for further phylogenetic analysis.
TABLE 2.
Groups of nosZ gene fragments used to determine sequencing primers
| Sequencing group | nosZ clones |
|---|---|
| Group A | 696C, 696J, 696G, ProR |
| Group B | |
| Subgroup 1 | 696B, 696H, 696M, 696Q, 696S, 696T, 696W, ProQ |
| Subgroup 2 | S321195C, 696A, 696D, 696E, 696I, 696K, 696O, 696AB, ProP, ProV |
| Group C | |
| Subgroup 1 | ProO, ProB, ProC, ProD, ProI, 696F |
| Subgroup 2 | ProA, ProE, ProG, ProH, ProS, ProT |
| Subgroup 3 | 696L, 696R |
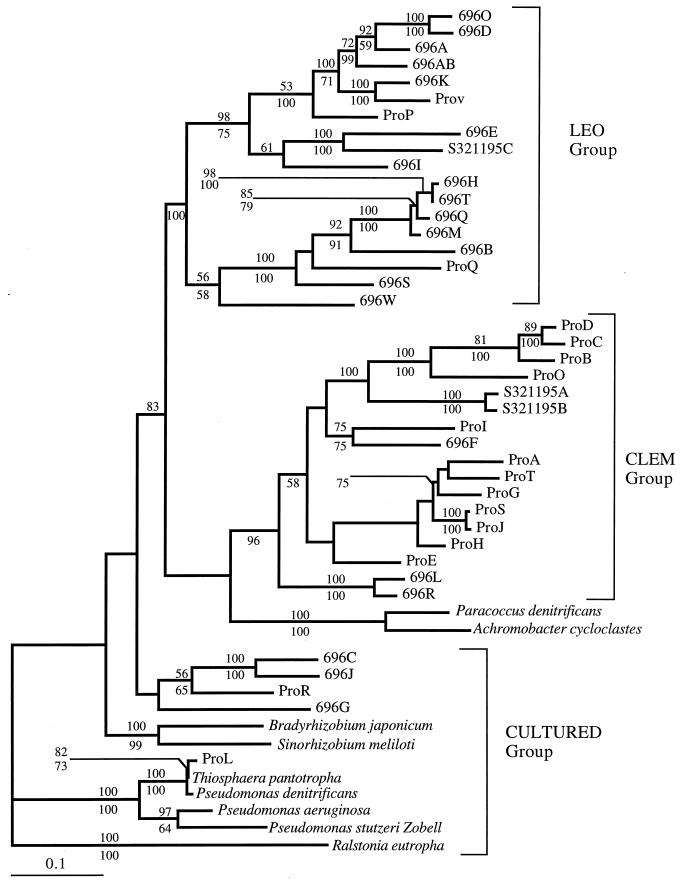
Phylogenetic tree reconstruction of the 1,100-bp nosZ genes indicates there are three broad groups of nosZ genes in the areas examined (Fig. 2). The CULTURED group contains all but two of the denitrifiers in culture collections. The two environmental clusters correlate generally with geography. The LEO group contains 18 genes, 16 of which are from the Atlantic Ocean sample. The CLEM group consists of 17 genes, 12 of which are from the Pacific Ocean sample.
FIG. 2.
Phylogenetic tree of 49 nosZ genes from different species of denitrifiers. The tree was generated by using the maximum likelihood method, based on the alignment of 1,053 bp of the nosZ gene fragments. The best topology was chosen by using the maximum likelihood “loop” algorithm. Bootstrap values (percentages of 100 bootstraps) for maximum likelihood are indicated above the line, and those for neighbor joining analyses are indicated below the line. Only values greater than 50% are shown. The bar labeled 0.1 represents 10% sequence divergence.
Based on bootstrap values, both the maximum-likelihood and neighbor-joining phylogenies support the idea that the LEO group is separate from both the CLEM group and the CULTURED group. These phylogenies did not strongly support the concept that the CULTURED group is a distinct cluster (<50% bootstrap values); however, these genes are clearly divergent from those obtained from the environment.
DISCUSSION
Sequencing of the 37 environmental copies of the nosZ gene demonstrates the inherent diversity of this gene within coastal systems. The levels of similarity between different pairs of nosZ genes ranged from 33 to 99%, and the question arises as to what level of sequence difference results from actual diversity in the sample and what level is a PCR artifact. If we assume a high nucleotide misincorporation rate for Taq polymerase, i.e., one error per 104 bases (3), then for a given round of the PCR an ∼1,100-bp fragment would incorporate ∼0.1 erroneous nucleotides. This would result in a similarity between the correct template and the “corrupted” copy of 99.99%. Assuming that this error rate is perpetuated over each of 35 PCR cycles, the final percent similarity between the correct product and the corrupt PCR product would be 99.7% (i.e., 0.999935). Therefore, any two nosZ amplicons having 99.7% or greater similarity must be considered identical (i.e., with sequence difference not above the noise level of the polymerase). In this study, genes with similarity approaching 99% at the DNA level were included separately when it was clear that these differences were affecting codon usage. For instance, ProS and ProJ were considered different nosZ genes even though their DNA percent similarity was 99.3%, because the similarity at the amino acid level decreased to 97.8%.
In addition to the diversity, our data indicate that lateral transfer of certain nosZ genes via plasmid exchange may not be widespread in the continental shelf environment. Recent studies have examined the genetic organization of denitrification enzymes (10, 21) in a variety of isolates. The nosZ gene has been found to be borne on large plasmids in Rhodobacter sphaeroides (115 kb), R. eutropha (450 kb), and S. meliloti (1.4 Mb) (4, 14, 21).
In this study, a low degree of homology between these various plasmid-borne nosZ genes and those found in the marine environment was observed; i.e., very few of the environmental nosZ clones cluster with either the R. eutropha or S. meliloti sequences. In addition, the R. eutropha nosZ sequence was found to be remarkably unlike all nosZ genes from denitrifying isolates and from the environmental clones; the average amino acid percent similarity was only ∼25% (Fig. 1). The average percent similarity between the S. meliloti gene and the other nosZ variants was ∼60%, which is similar to the overall average percent similarity of 64%. Given that the nosZ genes of R. eutropha and S. meliloti are both plasmid borne, this may indicate that different sources for the two genes exist, that there are different selection pressures, or that perhaps a great deal of time has passed, allowing for divergence of the two sequences. Our data support the conclusion of Vollack et al. (18) that denitrification is not likely to be an accessory function arising from gene transfer events, with the exception of T. pantotropha as described previously (13). Of course, there is a possibility that some of the cloned nosZ genes arise from plasmids different than those of R. eutropha and S. meliloti. However, given the diversity of sequences found within each of the study sites, large-scale homogenization of nosZ genes arising from transfer events is not observed.
Although there is great diversity observed in environmental nosZ genes, alignment of the 49 derived amino acid sequences (Fig. 1) highlights conservation of specific residues and regions that may serve to identify important structural motifs. For instance, it has been reported that the protein encoded by nosZ contains two binuclear copper centers and that certain conserved histidine residues are involved in copper binding of the mature protein (9; for a review, see reference 21). The two binuclear centers are termed CuA, the entry site for electrons, and CuZ, the substrate binding center. The CuA center is well described and corresponds to the location of the Nos2230R primer (13). However, there is little structural information regarding the CuZ, or catalytic, center. Zumft et al. (20) regard eight conserved histidine residues as likely candidates for involvement in the coordination of copper in the protein encoded by nosZ. Our data support this contention, since the seven of these eight histidine residues which fall within the range of the Nos661F/1773R priming set are shown to be conserved in the products of all 49 nosZ genes in this study (Fig. 1). The conservation of these histidine residues suggests that the basic structural motifs necessary for proper functioning of the enzyme are maintained despite wide variability of both the amino acid sequences and the DNA sequences.
Furthermore, if we use the model of the CuA site presented by Zumft (21) as a starting point in searching for the CuZ site, there is only one region in the amino acid alignment which demonstrates the proper spacing and sulfur-containing amino acid side chains to coordinate the additional binuclear copper center. In the CuA site, histidines at positions 583 and 626 coordinate with two cysteines (at positions 618 and 622) and a methionine (at position 629) to bind the copper atoms (numbers are relative to the sequence of P. stutzeri). We propose that the region between amino acids 132 and 178 is the catalytic, CuZ, site (Fig. 1). This site contains four of the seven conserved histidines observed in all 49 nosZ genes. Additionally, two histidine residues are 46 amino acids apart. This spacing is nearly identical to the spacing of the histidine residues in the CuA site. The requisite sulfur atoms could be provided by two nearly conserved methionine residues at positions 133 and 163 and/or by the highly conserved cysteine residues at positions 160 and 165. (Note that for the nosZ genes of Pseudomonas-like strains the conserved methionine is shifted to position 135.)
The conservation of structural motifs suggests that the cloned genes are most likely functional nosZ genes and do not arise from duplications or related genes. Pseudogenes arising from duplications, or other nonfunctioning genes, are under no selective pressure and would not be expected to maintain the structural elements needed for a functional protein. However, techniques such as Northern analysis or reverse transcription-PCR are needed to conclusively determine whether a specific sequence is functionally active (i.e., transcribed).
This study supports our previous finding (13) that there is substantial diversity among the nosZ genes that is not represented in culture collections. Our phylogenetic analyses also indicate that there are a minimum of three broad groups of nosZ sequences, i.e., the CULTURED group, the LEO group, and the CLEM group (Fig. 2). Essentially no overlap between the nosZ sequences in the CULTURED group and the environmental sequences was observed, indicating that the denitrifiers used as model organisms in the laboratory are poorly represented in the continental shelf environment. This may be due to biases in the culturing techniques used or to a general lack of attempts to culture denitrifiers from the shelf environment. What is clear, however, is that results obtained from such culture-based studies may not have a direct relevance to denitrification in the continental shelf environment. It was also observed that each site is characterized by its own unique set of nosZ genes. The LEO group is predominantly Atlantic Ocean shelf sequences, while the CLEM group consists largely of Pacific Ocean sequences. In addition, these analyses indicate a splitting of the LEO and CLEM cluster into the sequence subgroups listed in Table 2. If this general trend holds for additional shelf sites, it would suggest evolution of site-specific denitrifier assemblages. However, it may be that temperature or temporal effects are equally important in controlling denitrifier diversity. For instance, three nosZ sequences obtained from LEO-15 in November 1995, when analyzed by using an earlier PCR primer set (13), were split between the CLEM and LEO groups. Further examination over both spatial and temporal scales will be necessary to decide this issue.
Finally, the correlation between the sequencing groups and phylogenetic clusters indicates that these internal sequencing primers may also serve as group-specific probes. Although extensive testing of this is required, preliminary results are encouraging. For example, clone ProB was successfully sequenced by using the appropriate group C sequencing primer, but no sequencing product resulted from use of either group A or group B primers (data not shown). Furthermore, the division of the sequencing groups into phylogenetically coherent subgroups may indicate that probes with higher resolution than those presently used are possible.
ACKNOWLEDGMENTS
The work was supported in part from funds supplied by the Institute of Marine and Coastal Sciences, New York Sea Grant/Hudson River National Estuarine Research Reserve Fellowship to D.J.S. and NOAA grant NA46RU0149 to L.J.K. and Sybil P. Seitzinger.
We thank Robert Sherrell for the San Clemente Island sediment samples and Rose Petrecca and the IMCS divers for the LEO-15 samples.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha R S, Thilly W G. Specificity, efficiency, and fidelity of PCR. Genome Res. 1993;3:S18–S29. doi: 10.1101/gr.3.3.s18. [DOI] [PubMed] [Google Scholar]
- 4.Chan Y-K, Wheatcroft R. Detection of a nitrous oxide reductase structural gene in Rhizobium meliloti strains and its location on the nod megaplasmid of JJ1c10 and SU47. J Bacteriol. 1993;175:19–26. doi: 10.1128/jb.175.1.19-26.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen J P, Murray J W, Devol A H, Codispoti L A. Denitrification in continental shelf sediments has major impact on the oceanic nitrogen budget. Global Biogeochem Cycles. 1987;1:97–116. [Google Scholar]
- 6.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 7.Haygood M. Spreadsheet Macros for coloring sequence alignments. Biotechniques. 1993;15:1084–1089. [PubMed] [Google Scholar]
- 8.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoeren F U, Berks B C, Ferguson S J, McCarthy J E G. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans: new and conserved structural and regulatory motifs. Eur J Biochem. 1993;218:49–57. doi: 10.1111/j.1432-1033.1993.tb18350.x. [DOI] [PubMed] [Google Scholar]
- 10.Holloway P, McCormick W, Watson R J, Chan Y-K. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J Bacteriol. 1996;178:1505–1514. doi: 10.1128/jb.178.6.1505-1514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Scala D J, Kerkhof L J. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett. 1998;162:61–68. doi: 10.1111/j.1574-6968.1998.tb12979.x. [DOI] [PubMed] [Google Scholar]
- 14.Schwintner C, Sabaty M, Berna B, Cahors S, Richaud P. Plasmid content and localization of the genes encoding the denitrification enzymes in two strains of Rhodobacter sphaeroides. FEMS Microbiol Lett. 1998;165:313–321. doi: 10.1111/j.1574-6968.1998.tb13163.x. [DOI] [PubMed] [Google Scholar]
- 15.Seitzinger S P, Giblin A E. Estimating denitrification in North Atlantic continental shelf sediments. Biogeochemistry. 1996;35:235–260. [Google Scholar]
- 16.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vollack K-U, Xie J, Hartig E, Romling U, Zumft W G. Localization of denitrification genes on the chromosomal map of Pseudomonas aeruginosa. Microbiology. 1998;144:441–448. doi: 10.1099/00221287-144-2-441. [DOI] [PubMed] [Google Scholar]
- 19.von Alt C J, Grassle J F. LEO-15 an unmanned long term environmental observatory. Proc Oceans. 1992;2:849–854. [Google Scholar]
- 20.Zumft W G, Dreusch A, Lochelt S, Cuypers H. Derived amino acid sequences of the nosZ-gene (respiratory N2O reductase) from Alcaligenes eutrophus, Pseudomonas aeruginosa, and Pseudomonas stutzeri reveal potential copper binding residues—implications for the CuA site of N2O reductase and cytochrome-c oxidase. Eur J Biochem. 1992;208:31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]
- 21.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]