Abstract
Systemic JIA (SJIA) is distinguished from other forms of JIA by the prevalence of the severe, life-threatening complications macrophage activation syndrome (SJIA-MAS) and lung disease (SJIA-LD). Alternative therapeutics are urgently needed, as disease pathogenesis diverges from what is observed in SJIA, and currently available biologics are insufficient. SJIA-MAS, defined by a cytokine storm and dysregulated proliferation of T-lymphocytes, and SJIA-LD which presents with lymphocytic interstitial inflammation and pulmonary alveolar proteinosis, are both thought to be driven by IFNs, in particular the type II IFN-γ. Involvement of IFNs and a possible crosstalk of type I IFNs with existing biologics indicate a distinct role for the JAK-STAT signalling pathway in the pathogenesis of SJIA-MAS and SJIA-LD. Here, we review this role of JAK-STATs and IFNs in SJIA complications and discuss how new insights of ongoing research are shaping future therapeutic advances in the form of JAK inhibitors and antibodies targeting IFNs.
Keywords: systemic JIA, macrophage activation syndrome, lung disease, interferon, JAK-STAT
Rheumatology key messages.
Available biologics are insufficient to treat the severe complications SJIA-MAS and SJIA-LD.
Interferons, signaling via the JAK-STAT pathway, play a major role in SJIA-MAS and SJIA-LD.
Targeting interferons and the JAK-STAT pathway may shape the next generation of therapeutics for SJIA-MAS/LD.
SJIA and current therapies
JIA describes a group of heterogeneous childhood-onset diseases with unknown aetiology. The different JIA disease subtypes have diverging pathophysiologic origins and mechanisms, ranging from various degrees of adaptive and innate immune dysfunction, production of autoantibodies, and dysregulation of immune cell populations such as T cells and monocytes. However, all JIA subtypes are unified by the presence of chronic childhood arthritis [1, 2]. In systemic JIA (SJIA), arthritis can sometimes play a minor role at disease onset and instead systemic inflammation is predominant, with symptoms including fever, rash, lymphadenopathy, hepatosplenomegaly and serositis [3]. Systemic JIA is further set apart from other forms of JIA by its prevalence for the serious complications macrophage activation syndrome (SJIA-MAS) and lung disease (SJIA-LD) [4, 5].
Treatments for pediatric rheumatologic diseases such as SJIA have significantly evolved in the past two decades. Synthetic glucocorticoids that suppress inflammation were developed >60 years ago, and have since been widely used in various chronic diseases, including SJIA [6]. DMARDs such as methotrexate were the first non-steroidal medications shown to significantly improve arthritis in JIA, and together with NSAIDs and glucocorticoids formed the mainstay of treatment in the pre-biologic era [7, 8]. However, methotrexate proved largely ineffective for the systemic features of SJIA, and adverse events, including steroid side effects, had significant impact on pediatric patients [6, 9].
With better understanding of specific inflammatory cytokines and proteins in SJIA disease pathology, including IL-1, IL-6, TNFα, IL-18 and S100 proteins [10], biologic agents in the form of recombinant monoclonal antibodies or recombinant proteins, directed at single cytokines, were developed over the past two decades. The first biologic approved for clinical use in JIA was etanercept in 1999, which binds to circulating TNFα and prevents its interaction with cell surface receptors [11]. Other biologics have since been developed and used and/or approved specifically for SJIA, including anakinra (a recombinant IL-1 receptor antagonist, approved by the European Medicines Agency (EMA) in 2018) and canakinumab (a recombinant monoclonal IL-1β antibody, approved by the EMA and US Food and Drug Administration (FDA) in 2013), and tocilizumab (a recombinant antibody against IL-6, approved by the EMA and FDA in 2011) [7, 12]. Since biologic drugs have been introduced in the clinical practice and management of JIA, the prognosis for pediatric patients has dramatically improved [13–15].
Despite this success, between 20% and 40% of SJIA patients fail to respond to anti-cytokine biologics or develop adverse events during treatment [16, 17]. Some treatments can also lose efficacy over time, resulting in the need for novel therapeutic strategies. For example, in the randomized controlled trial of canakinumab, 38% of SJIA patients do not achieve an adapted JIA ACR criteria (JIA-ACR) score of at least 50 [18]. Additionally, the potentially life-threatening complications SJIA-MAS and SJIA-LD are not prevented by currently available therapies, as they instead seem to be driven by IFNs and IL-18 (discussed below) [4, 5, 19]. While individual pro-inflammatory cytokines present an obvious target in many rheumatologic diseases, evidence has accumulated for the significance of the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signalling pathway in disease pathogenesis. In particular, the key role of this pathway in IFN signalling highlights the importance of JAK-STAT signalling as a potential therapeutical target for SJIA and its complications.
Interferons and JAK-STAT signalling
The JAKs compose a family of four intracellular tyrosine kinases JAK1, JAK2, JAK3 and tyrosine kinase (TYK) 2. When cytokines bind to membrane receptors, JAKs are activated, recruited and in turn phosphorylate the receptors. This allows the selective binding and phosphorylation of members of the STAT family to induce downstream genes transcription [6]. Seven STATs have been identified in mammals: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6 [20].
Overall, >50 cytokines signal via the JAK-STAT pathway, including IL-2, IL-6 and IL-10 family members, to regulate cell homeostasis, proliferation and differentiation, as well as control the immune system and inflammatory response. However, the JAK-STAT pathway also plays a central role in mediating the cellular response to the IFN family cytokines.
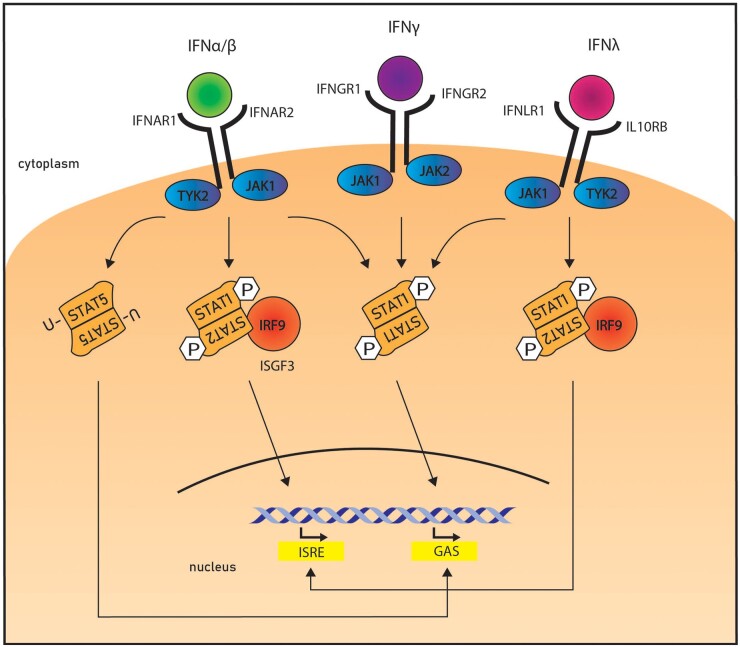
IFN type I, consisting of 13 subtypes of IFNα and one IFNβ, binds to the IFN alpha receptor (IFNAR) 1 and IFNAR2 heterodimer, signalling downstream via JAK1 and TYK2. In response, STAT1 and STAT2 are phosphorylated, heterodimerize, and are transported into the nucleus, where they then associate with IFN regulatory factor (IRF) 9 to form a transcriptionally active complex termed the IFN stimulatory gene factor (ISGF) 3 (Fig. 1). ISGF3 then recognizes specific sequences (IFN-stimulated response elements, ISRE) in IFN-stimulated genes (ISGs), where it binds and facilates their transcription [21].
Fig. 1.
Interferon-induced JAK-STAT signalling pathways
IFN-α/β, IFN-γ and IFNλ signal via distinct receptors, but can share downstream JAK-STAT pathways and signalling molecules. Canonically, IFN-α/β and IFNλ induce assembly of the interferon-stimulated gene factor 3 (ISGF3) which promotes transcription by recognizing and binding to interferon-stimulated response elements (ISREs). IFN-γ canonically leads to the assembly of STAT1 homodimers, which migrate to the nucleus and bind to interferon gamma activation sites (GASs), inducing transcription of ISGs. Unphosphorylated STATs (U-STAT) are also able to facilate transcription via GAS.
Type II IFN, IFN-γ, binds to the IFN gamma receptor (IFNGR) heterodimer IFNGR1/IFNGR2 and utilizes JAK1 and JAK2 to induce phosphorylation of STAT1. STAT1 forms homodimers, which in the nucleus bind to Gamma-IFN sites (GAS) to activate transcription of ISGs (Fig. 1). Finally, type III IFNs, composed of IFNλ1-3, use the same signalling pathway as type II IFN with the exception of binding to a heterodimeric receptor of IL-28Rα and IL10Rβ [21].
While the general mode of IFN-mediated JAK-STAT signal transduction is known, various forms of non-canonical signalling have been discovered, along with different combinations of STAT homo- and heterodimers to convey potentially very different transcription signals. For example, type I IFNs are also able to induce homodimers of STAT1 or STAT3 instead of utilizing the ISGF3 for transcriptional signalling. These IFN type I induced STAT homodimers bind to GAS to induce gene expression [22]. Curiously, while type I and type III IFN both utilize the ISGF3, their ISGF3-induced gene expression profile is not identical [23]. This indicates that mere activation of the same transcriptionally active complexes does not convey which genes are expressed, suggesting that either IFN dosage or presence of other factors can divert ISGF3 to other loci [24]. Type I IFNs also induce responses by different subtypes alone, and patterns of STAT activation can vary depending on cell types [25]. To complicate the matter, unphosphorylated STATs can play a role in nuclear gene expression. Unphosphorylated STAT1 and 3 (U-STAT1, U-STAT-3) (Fig. 1), for example, can enter the nucleus and modulate gene transcription by binding to DNA or to transcription factors such as NFκB [26].
Assessment of the IFN response in disease
To date, it remains difficult to distinguish the specific roles of type I and type II IFN. Direct serum IFN measurements using ELISA have proven to be challenging due to the presence of heterophilic serum proteins that nonspecifically bind to the capture and detection antibodies in the assay, as well as extremely low biological levels of type I IFN in serum [27, 28]. Only recently have new assays been developed that can detect attomolar, physiological levels of IFNα [28]. Therefore, the measuring of IFN-induced chemokines CXCL9, CXCL10 and CXCL11 is widely accepted. CXCL9 is induced by IFN-γ but not IFN-α or -β [29], CXCL10 is induced by IFN-γ and to a lesser degree by IFN-α/β [30] and CXCL11 is similarly induced by IFN-γ and IFN-β, and weakly by IFN-α [31]. Notably, only CXCL9 is completely dependent on IFN-γ and can thus be used as secondary measurement for presence of type II IFN [32]. CXCL10, CXCL11 and neopterin, however, are not selectively induced by IFN-γ despite often being reported as such [33, 34]. Together, this may result in a certain bias towards the role of IFN-γ, while IFN-α and IFN-β responses are not properly assessed. In addition, IFN gene score signatures are used to evaluate the role of IFNs in diseases by assessing the expression of ISGs compared with healthy individuals, and have particularly been utilized in interferonopathies or IFN driven diseases such as systemic lupus erythematosus (SLE). Assessing the IFN gene score by RNA level is a more sensitive method to pick up IFN activity as compared with protein levels, particularly when compared with direct measurement of IFNs. Distinct IFN gene signatures have been developed for type I and type II IFNs, though overlapping transcriptional activation as well as a dynamic IFN signatures that evolve over time continue to make interpretation challenging [35–38].
Role of IFNs and JAK-STAT in SJIA
Polyarticular and oligoarticular JIA have been associated with high plasma levels of IFN-γ, as well as elevated CXCL9/10 in both plasma and synovial fluid [39].
However, SJIA (when not complicated by MAS or LD) is not generally associated with a robust type I or type II IFN signature. Independent microarray gene expression studies using PBMCs did not detect any IFN-induced signature in SJIA [40–42], with the exception of one study on a Japanese SJIA patient cohort [43]. Recent bulk RNA-Seq analysis of SJIA patient monocytes also failed to reveal evidence of an IFN-induced signature [44].
No recent studies have reported detectable type I IFN levels in serum of SJIA patients, owing to the lack of specific, highly sensitive methods to detect low IFN-α/β in serum [27, 28, 45].
In contrast, reports on IFN-γ and CXCL9/10 in SJIA have been contradictory. Gattorno et al. reported that CXCL10 and IFN-γ were modestly elevated compared with controls but did not display the control levels. De Jager et al. also showed that while IFN-γ was not elevated in SJIA patients with longstanding disease, CXCL9 and CXCL10 were, albeit at significantly lower levels than those seen in oligoarticular or polyarticular JIA [39, 46]. Similarly, a recent study reported relatively low but increased CXCL9 levels in SJIA patients that respond to canakinumab vs non-responders [47]. On the other hand, more recent studies have not observed significantly elevated IFN-γ in SJIA patients’ serum [39, 45], or increased IFN-γ production following SJIA PBMC stimulation compared with healthy controls [48]. Similarly, Bracaglia et al. did not observe significantly elevated levels of IFN-γ or IFN-γ inducible chemokines CXCL9, CXCL10 or CXCL11 in active SJIA patients without MAS [49]. SJIA patients’ natural killer (NK) cells in fact have been shown to have a specific defect in IL18-induced IFN-γ production, at least in part caused by a defective phosphorylation of the IL 18 receptor beta [50, 51].
Intriguingly, however, a subset of SJIA patients without MAS treated with anti-IL-1 inhibitors were shown to upregulate a type I IFN gene signature [37, 52]. Monocytes from SJIA patients naïve to biological therapy are hyporesponsive to IFN-γ and lack an IFN-induced gene expression signature, but anti-IL1b treatment can increase their basal IFN signal and also increase monocyte responsiveness to IFN-γ, potentially facilitated by increased levels of IFNGR expression [44, 53, 54].
Role of IFNs and JAK-STAT in the complications SJIA-MAS and SJIA-LD
In contrast to SJIA without MAS, substantial evidence supports a key role for type II IFNs in SJIA-MAS. MAS is a severe and potentially fatal hyperinflammatory complication arising in up to 30% of SJIA patients [5, 55]. Key features of MAS are overactivation of T lymphocytes causing highly elevated production of IFN-γ, which drives activation of hemophagocytic macrophages. What rheumatologists call MAS is also called secondary hemophagocytic lymphohistiocytosis (HLH) when resulting after infection or other triggers [56]. Primary HLH on the other hand is an autosomal recessive disorder caused by deficiencies in genes involved in the cytolytic activity, such as perforin, resulting in impaired NK cell and cytotoxic T-cell function and inability to limit proliferation and expansion of T cells and macrophages [55, 57]. While the pathophysiology is not completely understood, it is thought that risk for MAS is at least partially driven by a genetic component, and indeed several studies have found SJIA-MAS patients carrying heterozygous variants in causative genes for primary HLH [58–60].
Because IFN-γ production plays a key role in MAS, high levels of IFN-γ and IFN-γ inducible chemokines CXCL9 and CXCL10 in serum are characteristic for MAS patients [45, 49, 61]. Bracaglia et al. found that IFN-γ and CXCL9, CXCL10 and CXCL11 are highly elevated in SJIA-MAS patients [49], and another study showed MAS is characterized by IFN-γ production from CD8+ lymphocytes [61]. Murine MAS models have further increased evidence for the pivotal role of IFN-γ in MAS [62–65]. Recently, single-cell RNA-sequencing of bone marrow macrophages in early SJIA-MAS revealed activated subpopulations with altered transcriptomes including upregulated IFN-γ response pathways [44].
IL-18, known for its strong IFN-γ inducing capacities, is also highly increased in SJIA-MAS [36, 47, 49]. Hyperproduction of IL-18 drives the overactivation of Th1 lymphocytes and macrophages. Massive hypersecretion of IFN-γ from these cells is in fact likely a result of high IL-18 levels setting the milieu for MAS [10, 36, 45]. In a murine MAS model, mice lacking the natural inhibitor of IL-18, IL-18 binding protein, had a more severe clinical manifestation of MAS compared with wildtype mice, highlighting the critical role for this protein in driving disease pathogenesis [62, 66].
Another SJIA-associated complication with links to IL-18 and IFN activation is interstitial lung disease (SJIA-LD) [4, 19]. SJIA-LD is characterized by lymphocytic interstitial inflammation as well as accumulation of lipoproteinaceous material and lipid-laden macrophages in the lungs. These are features shared with pulmonary alveolar proteinosis (PAP), where dysfunction of alveolar macrophages by lack of GM-CSF signalling leads to accumulation of pulmonary surfactant in alveolar spaces [4, 67]. In SJIA-LD, GM-CSF signalling remains intact, but substantial Th1-driven lung inflammation is present, along with highly increased IL-18 serum levels, IL-18 and IFN-γ-induced chemokines in the lungs, and pulmonary gene expression reflecting IFN-driven activation [4].
The majority of patients with SJIA-LD were diagnosed after 2000, have refractory systemic disease with MAS, and are being treated with various biological therapies. Saper and colleagues have hypothesized that the timeline of increase in SJIA-LD aligning with the rise of cytokine-directed therapies may indicate a connection of both [19]. The underlying pathogenesis of SJIA-LD remains unknown, and considerations include persistently active SJIA-MAS perhaps modified by biologic therapy, a delayed hypersensitivity reaction to anti-IL1 or anti-IL-6 agents, or an autoinflammatory reaction towards cryptic antigens exposed by persistent inflammation during SJIA-MAS [68, 69].
Therapeutic approaches targeting IFNs in SJIA, SJIA-MAS and SJIA-LD
While IFN-γ appears activated in polyarticular and oligoarticular JIA, it is likely not the main driver of disease. However, in SJIA-MAS, SJIA-LD and HLH, therapy directed at IFN-γ appears much more promising. As noted above, MAS mouse models using repeated CpG DNA induced TLR9 stimulation have shown that direct IFN-γ blockade or JAK/STAT inhibition alleviates inflammation [62, 64,65, 70]. Another MAS mouse model using animals transgenic for human IL-6 challenged by LPS provided evidence that direct IFN-γ inhibition with monoclonal antibodies may be effective in SJIA-MAS [65].
Emapalumab is a monoclonal antibody against human IFN-γ. A recent open-label phase 2–3 study evaluating the efficacy and safety of emapalumab in children with HLH found the treatment efficacious without any organ toxicity, although infections, including severe infections, occurred frequently [32]. Thus far, emapalumab treatment for SJIA-MAS is based on anecdotal clinical experience; an open-label clinical trial is ongoing (Clinicaltrials.gov: NCT03311854), though the preliminary data is promising [71]. There are case reports of emapalumab in secondary HLH/MAS, including a young adult patient with adult-onset Still’s disease and MAS successfully treated with emapalumab following seven infusions within 2 months [72]. Emapalumab was also effective in treating a patient with refractory Epstein–Barr (EBV)-associated secondary HLH despite severe pre-existing comorbidities [73]. However, it remains to be seen if neutralization of IFN-γ alone by emapalumab is an effective therapy outside of HLH, where it was mainly utilized as a bridge to bone marrow transplant [32, 74].
Inhibition of type I IFN has not been investigated in clinical trials for JIA or SJIA patients, as there is no direct evidence for IFN type I contribution to the pathophysiology at this time. Therapy targeting type I IFN, however, has been investigated in other rheumatic diseases. Studies of sifalimumab, an anti-IFN alpha monoclonal antibody, were discontinued despite promising phase 2 b results in systemic lupus erythematosus (SLE), an autoinflammatory disease marked by high elevation of type I IFN gene signatures [75]. Instead, anifrolumab, a monoclonal antibody directed against type I IFN receptor subunit 1 that has recently been found effective in a phase 3 trial in SLE is now FDA approved [76]. Should future studies be able to overcome the difficulties of disentangling type I and II IFN effects and provide evidence of IFN type I driven immunity in JIA or particularly in SJIA-MAS and SJIA-LD, alternative therapies such as anifrolumab should be considered.
Indirect inhibition of IFNs: JAK inhibitors in SJIA, SJIA-MAS and SJIA-LD
In contrast to recombinant antibodies directly targeting IFNs or their receptors, much more is known about the effectiveness of JAK inhibitors (jakinibs), a novel group of oral small molecule inhibitors.
Jakinibs target both type I and type II IFN (and type III, though not further discussed here) pathways, as well as other cytokines, by interfering with the JAK-STAT pathway. JAKs provide high biological plausibility as efficacious targets in diseases where either IFNs, other cytokines signalling via the JAK-STAT pathway, or both are driving the disease. These small molecule inhibitors use a novel mechanism of action by affecting intracellular signalling pathways instead of targeting a specific cytokine or its receptor. The competitive interaction of the Jakinib with the JAK region constituting the ATP binding site significantly interferes with JAK and STAT phosphorylation required for downstream signalling [77]. Various jakinibs with different selectivity towards JAK1, JAK2, JAK3 and TYK2 have been developed, though the homology within the JAK family structures makes it challenging to target just one specific JAK selectively [77]. Ruxolitinib and baricitinib are more selective towards JAK1 and JAK2 over the other JAKs. Tofacitinib, on the other hand is most effective at blocking JAK3 [77, 78]. A recent study found that tofacitinib, baricitinib, and two other mainly JAK3 inhibitors named upadacitinib and filgotinib exhibit similar cytokine receptor inhibition profiles in vitro despite different IC50 values for each JAK [79]. Fenwick et al. tested two different JAK inhibitor compounds and their effectiveness at inhibiting CXCL9, CXCL10 and CXCL11 secretion from chronically obstructive pulmonary disease (COPD) patients’ airway epithelial cells. They found both compounds suppressed the chemokines and STAT1 phosphorylation, but one compound (PF1367550) was more effective than the other [80]. Thus, whether subtle differences in structure and selectivity could translate to clinical differences between these JAK inhibitors remains to be seen.
While the Jakinib tofacitinib has been recently FDA approved for treatment of polyarticular course JIA, and there is an ongoing clinical trial for SJIA with systemic features (Clinicaltrials.gov: NCT03000439), currently only case reports support these medications for SJIA or its complications, MAS and SJIA-LD. In fact, use of jakinibs in SJIA-MAS and SJIA-LD is extrapolated from murine data and from clinical trials with similar diseases that are driven by IFN.
Jakinibs have been assessed in monogenic interferonopathies such as SAVI [stimulator of IFN genes-associated (STING-associated) vasculopathy with onset in infancy] and CANDLE (chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperatures), with results showing clinical and laboratory improvement of disease [81, 82]. As noted above, murine models have also shown beneficial effects of JAK/STAT inhibitors in HLH and MAS. Ruxolitinib significantly reduced the clinical and laboratory manifestations of both primary HLH (PRF-/- mice infected with LCMV) and MAS/secondary HLH (mice repeatedly stimulated with CpG DNA) [64, 70, 83].
One report describes using ruxolitinib in an EBV-induced secondary HLH patient, resulting in decreased disease markers, including ferritin. However, with worsening clinical status, treatment was ceased after only 7 days and the patient expired [84]. On the other hand, a pilot study of a 28-day treatment course of oral ruxolitinib in 12 children with secondary HLH showed encouraging results, with two-thirds of patients achieving complete response and maintaining this status for >6 months [85].
A case report in SJIA and another study reporting a case of SJIA-MAS and SJIA-LD have described significant clinical improvement or even complete remission within 3 months of tofacitinib treatment [86, 87]. In another recent case report, a 4-year-old SJIA-LD patient was successfully treated with ruxolitinib, achieving significant clinical improvement that also allowed steroid tapering [88]. Two other juvenile patients with refractory dermatomyositis and associated LD responded remarkably to tofacitinib [89].
Further case reports of adult patients with dermatomyositis-associated LD describe improvement with tofacitinib treatment [90], and in line with this a single-center, open-label clinical study (Chinese Clinical Trial Registry number, ChiCTR-1800016629) evaluating the efficacy of tofacitinib in patients with dermatomyositis-LD showed improved survival and respiratory symptoms [91].
IFN crosstalk can affect SJIA therapeutics
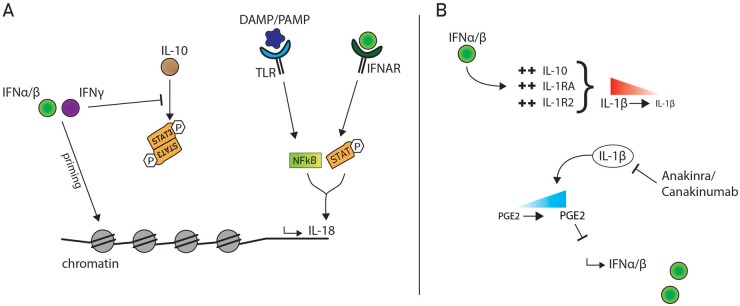
A key consideration when using agents such as jakinibs is the significant crosstalk between Toll-like receptors (TLRs), type I and II IFNs, NFκB and STAT signalling. Type I and type II IFNs are able to prime the chromatin structure to enable robust transcriptional responses to TLR signalling, inducing sustained transcription factor binding to TNFα, IL-6 and IL12 gene loci [92–94]. Furthermore, IFN-γ can augment TLR responses (such as production of TNFα and IL-6) by IFN-γ-induced suppression of the anti-inflammatory cytokine IL-10 and STAT3 [95] (Fig. 2).
Fig. 2.
IFN crosstalk and signalling pathway modulation
(A) Type I and II IFNs modulate the immune response by epigenomic changes. IFN-γ inhibits the anti-inflammatory response of IL-10 by interfering with STAT3 signalling. IFNα/β signalling via IFNAR and DAMPs/PAMPs signalling via TLRs can work together to induce transcriptional regulation of IL-18. (B) IFNα/β downregulates IL-1α/β by upregulation of IL-10, IL1RA and the decoy IL-1R2 receptor. IL-1 suppresses IFNα/β production via Prostaglandin E2 (PGE2) upregulation. Anakinra/Canakinumab, which blocks IL-1 signalling, can thus promote increased IFNα/β levels.
Another study showed type I IFNs and NFκB have overlapping antiviral functions and NFκB is able to mediate effective ISG induction independent of type I IFNs [96]. Two previous studies have also suggested that unconventional transcription initiation complex assembly by both STAT and NFκB regulate nitric oxide synthase expression [97] and IL-18 gene expression [86] (Fig. 2). The latter study suggests that co-induction of IL-18 by IFN-α/β, and not IFN-γ-mediated STAT signalling and DAMP or PAMP mediated TLR-NFκB signalling could have significant implications for the disease pathogenesis in SJIA-MAS and SJIA-LD.
Of interest is the cross-regulation of IL-1β and type I IFNs, particularly as IL1-inhibition has now become the mainstay of treatment in SJIA patients. Type I IFNs attenuate IL-1α/β signalling through induction of anti-inflammatory IL-10, IL1RA (the natural IL1 receptor antagonist) and by regulating the decoy receptor IL-1R2. On the other side, IL-1α and IL-1β limit type I IFN production through direct transcriptional downregulation and Prostaglandin E2 production [98, 99]. Inhibition of IL-1 by canakinumab or the recombinant IL-1 receptor antagonist anakinra could thus lead to increased levels of type I IFN signalling (as previously observed in subsets of SJIA patients [37, 52]) which could in turn augment IL-18 production, setting the stage for massive IFN-γ driven hyperinflammation and MAS and LD [86] (Fig. 2). In fact, it is conceivable that in this subset of patients, dysregulation of signal transduction crosstalk contributes to the pathogenesis of hyperinflammation.
Conclusion
With jakinibs and monoclonal therapies targeting IFNs, promising therapies for SJIA and its complications SJIA-MAS and SJIA-LD may be on the horizon. Current ongoing trials in JIA and SJIA will provide more data about their efficacy and safety. The potential implications for the role of biological therapies in contributing to the development of SJIA-LD have the power to dramatically reshape the treatment landscape for SJIA and its complications. The subset of patients affected will have to be closely investigated to understand what sets them apart, and further study is required to evaluate whether they have an increased risk of developing SJIA complications. More so, there is urgent need for unravelling the origin of pathogenesis and particularly the distinct roles of type I and II IFNs in SJIA-MAS and LD to guide future decisions for novel alternative treatments.
Acknowledgements
E.L.V. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Project number 448863690. G.S.S. is supported by NIAMS/NIH K08-AR072075.
Funding: No specific funding was received from any bodies in the public, commercial or not-for-profit sectors to carry out the work described in this article.
Disclosure statement: G.S.S. has received consulting fees from Novartis and SOBI. E.L.V. has declared no conflicts of interest.
Data availability statement
No new data were generated in support of this paper.
References
- 1. Nieuwenhove E, Van Lagou V, Eyck L, Van Dooley J. et al. Machine learning identifies an immunological pattern associated with multiple juvenile idiopathic arthritis subtypes. Ann Rheum Dis 2019;78:617–28. [DOI] [PubMed] [Google Scholar]
- 2. Prakken B, Albani S, Martini A.. Juvenile idiopathic arthritis. Lancet 2011;377:2138–49. [DOI] [PubMed] [Google Scholar]
- 3. Petty RE, Southwood TR, Manners P. et al. International League of Associations for Rheumatology Classification of Juvenile Idiopathic Arthritis : second Revision, Edmonton, 2001. J Rheumatol 2004;31:390–2. [PubMed] [Google Scholar]
- 4. Schulert GS, Yasin S, Carey B. et al. Systemic juvenile idiopathic arthritis–associated lung disease: characterization and risk factors. Arthritis Rheumatol 2019;71:1943–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ravelli A, Grom AA, Behrens EM, Cron RQ.. Macrophage activation syndrome as part of systemic juvenile idiopathic arthritis: diagnosis, genetics, pathophysiology and treatment. Genes Immun 2012;13:289–98. [DOI] [PubMed] [Google Scholar]
- 6. Hardy RS, Raza K, Cooper MS.. Therapeutic glucocorticoids: mechanisms of actions in rheumatic diseases. Nat Rev Rheumatol 2020;16:133–44. [DOI] [PubMed] [Google Scholar]
- 7. Cimaz R, Maioli G, Calabrese G.. Current and emerging biologics for the treatment of juvenile idiopathic arthritis. Expert Opin Biol Ther 2020;20:725–40. [DOI] [PubMed] [Google Scholar]
- 8. Giannini EH, Brewer EJ, Kuzmina N. et al. Methotrexate in resistant juvenile rheumatoid arthritis- results of the U.S.A.-U.S.S.R. double-blind, placebo-controlled trial . N Engl J Med 1992;326:1043–9. [DOI] [PubMed] [Google Scholar]
- 9. Woo P, Southwood TR, Prieur AM. et al. Randomized, placebo-controlled, crossover trial of low-dose oral methotrexate in children with extended oligoarticular or systemic arthritis. Arthritis Rheum 2000;43:1849–57. [DOI] [PubMed] [Google Scholar]
- 10. Yasin S, Schulert GS.. Systemic juvenile idiopathic arthritis and macrophage activation syndrome. Curr Opin Rheumatol 2018;30:514–20. [DOI] [PubMed] [Google Scholar]
- 11. Lovell DJ, Giannini EH, Reiff A. et al. Etanercept in children with polyarticular juvenile rheumatoid arthritis. N Engl J Med 2000;342:763–9. [DOI] [PubMed] [Google Scholar]
- 12. Hausmann JS. Targeting cytokines to treat autoinflammatory diseases. Clin Immunol 2019;206:23–32. [DOI] [PubMed] [Google Scholar]
- 13. Lee JJY, Schneider R.. Systemic juvenile idiopathic arthritis. Pediatr Clin North Am 2018;65:691–709. [DOI] [PubMed] [Google Scholar]
- 14. Haar NM, ter Dijkhuizen EHP, van Swart JF. et al. Treatment to target using recombinant interleukin-1 receptor antagonist as first-line monotherapy in new-onset systemic juvenile idiopathic arthritis: results from a five-year follow-up study. Arthritis Rheumatol 2019;71:1163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malattia C, Ruperto N, Pederzoli S. et al. Tocilizumab may slow radiographic progression in patients with systemic or polyarticular-course juvenile idiopathic arthritis: post hoc radiographic analysis from two randomized controlled trials. Arthritis Res Ther 2020;22:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glerup M, Rypdal V, Arnstad ED. et al. Long-term outcomes in juvenile idiopathic arthritis: eighteen years of follow-up in the population-based Nordic Juvenile Idiopathic Arthritis Cohort. Arthritis Care Res 2020;72:507–16. [DOI] [PubMed] [Google Scholar]
- 17. Chhabra A, Robinson C, Houghton K. et al. Long-term outcomes and disease course of children with juvenile idiopathic arthritis in the ReACCh-Out cohort: a two-centre experience. Revmatol 2020;59:3727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruperto N, Brunner HI, Quartier P. et al. Canakinumab in patients with systemic juvenile idiopathic arthritis and active systemic features: results from the 5-year long-term extension of the phase III pivotal trials. Ann Rheum Dis 2018;77:1710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saper VE, Chen G, Deutsch GH. et al. Emergent high fatality lung disease in systemic juvenile arthritis. Ann Rheum Dis 2019;78:1722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Shea JJ, Holland SM, Staudt LM.. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med 2013;368:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Majoros A, Platanitis E, Kernbauer-Hölzl E. et al. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol 2017;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005;5:375–86. [DOI] [PubMed] [Google Scholar]
- 23. Zhou Z, Hamming OJ, Ank N. et al. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol 2007;81:7749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Au-Yeung N, Mandhana R, Horvath CM.. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2013;2:e23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Boxel-Dezaire AHH, Rani MRS, Stark GR.. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 2006;25:361–72. [DOI] [PubMed] [Google Scholar]
- 26. Yang J, Stark GR.. Roles of unphosphorylated STATs in signaling. Cell Res 2008;18:443–51. [DOI] [PubMed] [Google Scholar]
- 27. Redondo MJ, Gottlieb PA, Motheral T. et al. Heterophile anti-mouse immunoglobulin antibodies may interfere cytokine measurements in patients with HLA alleles protective for type 1a diabetes. Diabetes 1999;48:2166–70. [DOI] [PubMed] [Google Scholar]
- 28. Lamot L, Niemietz I, Brown KL.. Methods for type I interferon detection and their relevance for clinical utility and improved understanding of rheumatic diseases. Clin Exp Rheumatol 2019;37:1077–83. [PubMed] [Google Scholar]
- 29. Farber JM. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol 1997;61:246–57. [PubMed] [Google Scholar]
- 30. Qian C, An H, Yu Y, Liu S, Cao X.. TLR agonists induce regulatory dendritic cells to recruit Th1 cells via preferential IP-10 secretion and inhibit Th1 proliferation. Blood 2007;109:3308–15. [DOI] [PubMed] [Google Scholar]
- 31. Rani MR, Foster GR, Leung S. et al. Characterization of β-R1, a gene that is selectively induced by interferon β (IFN-β) compared with IFN-α. J Biol Chem 1996;271:22878–84. [DOI] [PubMed] [Google Scholar]
- 32. Locatelli F, Jordan MB, Allen C. et al. Emapalumab in children with primary hemophagocytic lymphohistiocytosis. N Engl J Med 2020;382:1811–22. [DOI] [PubMed] [Google Scholar]
- 33. Groom JR, Luster AD.. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 2011;89:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sghiri R, Feinberg J, Thabet F. et al. Gamma interferon is dispensable for neopterin production in vivo. Clin Diagn Lab Immunol 2005;12:1437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiche L, Jourde-Chiche N, Whalen E. et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type i and type ii interferon signatures. Arthritis Rheumatol 2014;66:1583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. AA de J, Hou Y, Brooks S. et al. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J Clin Invest 2020;130:1669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rice GI, Melki I, Frémond M-L. et al. Assessment of type I interferon signaling in pediatric inflammatory disease. J Clin Immunol 2017;37:123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barrat FJ, Crow MK, Ivashkiv LB.. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol 2019;20:1574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Jager W, Hoppenreijs EPAH, Wulffraat NM. et al. Blood and synovial fluid cytokine signatures in patients with juvenile idiopathic arthritis: a cross-sectional study. Ann Rheum Dis 2007;66:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fall N, Barnes M, Thornton S. et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum 2007;56:3793–804. [DOI] [PubMed] [Google Scholar]
- 41. Ogilvie EM, Khan A, Hubank M, Kellam P, Woo P.. Specific gene expression profiles in systemic juvenile idiopathic arthritis. Arthritis Rheum 2007;56:1954–65. [DOI] [PubMed] [Google Scholar]
- 42. Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J.. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med 2005;201:1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishikawa S, Mima T, Aoki C. et al. Abnormal expression of the genes involved in cytokine networks and mitochondrial function in systemic juvenile idiopathic arthritis identified by DNA microarray analysis. Ann Rheum Dis 2009;68:264–72. [DOI] [PubMed] [Google Scholar]
- 44. Schulert GS, Pickering AV, Do T. et al. Monocyte and bone marrow macrophage transcriptional phenotypes in systemic juvenile idiopathic arthritis reveal TRIM8 as a mediator of IFN-γhyper-responsiveness and risk for macrophage activation syndrome. Ann Rheum Dis 2021;80:617–9. [DOI] [PubMed] [Google Scholar]
- 45. Put K, Avau A, Brisse E. et al. Cytokines in systemic juvenile idiopathic arthritis and haemophagocytic lymphohistiocytosis: tipping the balance between interleukin-18 and interferon-γ. Rheumatology 2015;54:1507–17. [DOI] [PubMed] [Google Scholar]
- 46. Gattorno M, Piccini A, Lasigliè D. et al. The pattern of response to anti-interleukin-1 treatment distinguishes two subsets of patients with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum 2008;58:1505–15. [DOI] [PubMed] [Google Scholar]
- 47. Hinze T, Kessel C, Hinze CH, Seibert J, Gram H, Foell D.. A dysregulated interleukin-18/interferon-γ/CXCL9 axis impacts treatment response to canakinumab in systemic juvenile idiopathic arthritis. Rheumatology 2021;60:5165--74. [DOI] [PubMed] [Google Scholar]
- 48. Lasigliè D, Traggiai E, Federici S. et al. Role of IL-1 beta in the development of human TH17 cells: lesson from NLPR3 mutated patients. PLoS One 2011;6:e20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bracaglia C, Graaf K, De Marafon DP, Guilhot F. et al. Elevated circulating levels of interferon-γ and interferon-γ-induced chemokines characterize patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann Rheum Dis 2017;76:166–72. [DOI] [PubMed] [Google Scholar]
- 50. W De J, Vastert SJ, Beekman JM. et al. Defective phosphorylation of interleukin-18 receptor ß causes impaired natural killer cell function in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum 2009;60:2782–93. [DOI] [PubMed] [Google Scholar]
- 51. Put K, Vandenhaute J, Avau A. et al. Inflammatory gene expression profile and defective interferon-γ and Granzyme K in natural killer cells from systemic juvenile idiopathic arthritis patients. Arthritis Rheumatol 2017;69:213–24. [DOI] [PubMed] [Google Scholar]
- 52. Quartier P, Allantaz F, Cimaz R. et al. A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial). Ann Rheum Dis 2011;70:747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sikora KA, Fall N, Thornton S, Grom AA.. The limited role of interferon-γ in systemic juvenile idiopathic arthritis cannot be explained by cellular hyporesponsiveness. Arthritis Rheum 2012;64:3799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Macaubas C, Wong E, Zhang Y. et al. Altered signaling in systemic juvenile idiopathic arthritis monocytes. Clin Immunol 2016;163:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grom AA, Mellins ED.. Macrophage activation syndrome: advances towards understanding pathogenesis. Curr Opin Rheumatol 2010;22:561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carcillo JA, Simon DW, Podd BS.. How we manage hyperferritinemic sepsis related MODS/macrophage activation syndrome/secondary HLH. 2015;16:598–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grom AA, Horne A, De Benedetti F.. Macrophage activation syndrome in the era of biologic therapy. Nat Rev Rheumatol 2016;12:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vastert SJ, Wijk R, van D’Urbano LE. et al. Mutations in the perforin gene can be linked to macrophage activation syndrome in patients with systemic onset juvenile idiopathic arthritis. Rheumatology 2010;49:441–9. [DOI] [PubMed] [Google Scholar]
- 59. Kaufman KM, Linghu B, Szustakowski JD. et al. Whole exome sequencing reveals overlap between macrophage activation syndrome in systemic juvenile idiopathic arthritis and familial hemophagocytic lymphohistiocytosis. Arthritis Rheumatol 2014;66:3486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang M, Behrens EM, Atkinson TP. et al. Genetic defects in cytolysis in macrophage activation syndrome. Curr Rheumatol Rep 2014;16:439. [DOI] [PubMed] [Google Scholar]
- 61. Billiau AD, Roskams T, Van D-LR, Matthys P, Wouters C.. Macrophage activation syndrome: Characteristic findings on liver biopsy illustrating the key role of activated, IFN-γ-producing lymphocytes and IL-6- and TNF-α-producing macrophages. Blood 2005;105:1648–51. [DOI] [PubMed] [Google Scholar]
- 62. Behrens EM, Canna SW, Slade K. et al. Repeated TLR9 stimulation results in macrophage activation syndrome–like disease in mice. J Clin Invest 2011;121:2264–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weiss ES, Girard-Guyonvarc’h C, Holzinger D. et al. Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood 2018;131:1442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Das R, Guan P, Sprague L. et al. Janus kinase inhibition lessens inflammation and ameliorates disease in murine models of hemophagocytic lymphohistiocytosis. Blood 2016;127:1666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prencipe G, Caiello I, Pascarella A. et al. Neutralization of IFN-γ reverts clinical and laboratory features in a mouse model of macrophage activation syndrome. J Allergy Clin Immunol 2018;141:1439–49. [DOI] [PubMed] [Google Scholar]
- 66. Girard-Guyonvarc'h C, Palomo J, Martin P. et al. Unopposed IL-18 signaling leads to severe TLR9-induced macrophage activation syndrome in mice. Blood 2018;131:1430–41. [DOI] [PubMed] [Google Scholar]
- 67. Kimura Y, Weiss JE, Haroldson KL. et al. Pulmonary hypertension and other potentially fatal pulmonary complications in systemic juvenile idiopathic arthritis. Arthritis Care Res 2013;65:745–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hollenbach J, Ombrello M, Mellins ED.. Hypersensitivity reactions to IL-1 and IL-6 inhibitors are linked to common HLA-DRB115 alleles. 2020;6736:1–24. medRxiv:2020.08.10.20172338, preprint: not peer reviewed. [Google Scholar]
- 69. Fabro AT, Silva PHRQ, da Zocolaro WS. et al. The Th17 pathway in the peripheral lung microenvironment interacts with expression of collagen V in the late state of experimental pulmonary fibrosis. Immunobiology 2015;220:124–35. [DOI] [PubMed] [Google Scholar]
- 70. Albeituni S, Verbist KC, Tedrick PE. et al. Mechanisms of action of ruxolitinib in murine models of hemophagocytic lymphohistiocytosis. Blood 2019;134:147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Benedetti F, Brogan P, Bracaglia C. et al. Emapalumab (anti-interferon-gamma monoclonal antibody) in patients with macrophage activation syndrome (MAS) complicating systemic juvenile idiopathic arthritis (sJIA) [abstract 009]. Ann Rheum Dis 2020;79:180. [Google Scholar]
- 72. Gabr J, Ben Liu E, Mian S. et al. Successful treatment of secondary macrophage activation syndrome with emapalumab in a patient with newly diagnosed adult-onset Still’s disease: case report and review of the literature. Ann Transl Med 2020;8:887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lounder DT, Bin Q, De Min C, Jordan MB.. Treatment of refractory hemophagocytic lymphohistiocytosis with emapalumab despite severe concurrent infections. Blood Adv 2019;3:47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ehl S, Greenwood T von B, Bergsten E. et al. Is neutralization of IFN-γ sufficient to control inflammation in HLH? Pediatr Blood Cancer 2021;68:e28886. [DOI] [PubMed] [Google Scholar]
- 75. Khamashta M, Merrill JT, Werth VP. et al. Sifalimumab, an anti-interferon-α monoclonal antibody, in moderate to severe systemic lupus erythematosus: a randomised, double-blind, placebo-controlled study. Ann Rheum Dis 2016;75:1909–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Morand EF, Furie R, Tanaka Y. et al. Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med 2020;382:211–221. [DOI] [PubMed] [Google Scholar]
- 77. Clark JD, Flanagan ME, Telliez JB.. Discovery and development of Janus kinase (JAK) inhibitors for inflammatory diseases. J Med Chem 2014;57:5023–5038. [DOI] [PubMed] [Google Scholar]
- 78. Kerrigan SA, McInnes IB.. JAK inhibitors in Rheumatology: implications for paediatric syndromes? Curr Rheumatol Rep 2018;20:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dowty ME, Lin TH, Jesson MI. et al. Janus kinase inhibitors for the treatment of rheumatoid arthritis demonstrate similar profiles of in vitro cytokine receptor inhibition. Pharmacol Res Perspect 2019;7:e00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fenwick PS, Macedo P, Kilty IC, Barnes PJ, Donnelly LE.. Effect of JAK inhibitors on release of CXCL9, CXCL10 and CXCL11 from human airway epithelial cells. PLoS One 2015;10:e0128757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Montealegre Sanchez GA, Reinhardt A, Ramsey S. et al. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J Clin Invest 2018;128:3041–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhao Z, Ye C, Dong L.. The off-label uses profile of tofacitinib in systemic rheumatic diseases. Int Immunopharmacol 2020;83:106480. [DOI] [PubMed] [Google Scholar]
- 83. Maschalidi S, Sepulveda FE, Garrigue A, Fischer A, de Saint Basile G.. Therapeutic effect of JAK1/2 blockade on the manifestations of hemophagocytic lymphohistiocytosis in mice. 2016;128:60–72. [DOI] [PubMed] [Google Scholar]
- 84. Sin JH, Zangardi ML.. Ruxolitinib for secondary hemophagocytic lymphohistiocytosis: first case report. Hematol Oncol Stem Cell Ther 2019;12:166–170. [DOI] [PubMed] [Google Scholar]
- 85. Zhang Q, Wei A, Ma H-H. et al. A pilot study of ruxolitinib as a front-line therapy for 12 children with secondary hemophagocytic lymphohistiocytosis. Haematologica 2021;106:1892–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Verweyen E, Holzinger D, Weinhage T. et al. Synergistic TLR/IFNa/b-signaling facilitates escape of IL-18 expression from endotoxin tolerance. Am J Respir Crit Care Med 2020;201:526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang Z, Lee PY, Yao X, Zheng S, Li T.. Tofacitinib treatment of refractory systemic juvenile idiopathic arthritis. Pediatrics 2019;143;e20182845. [DOI] [PubMed] [Google Scholar]
- 88. Bader-Meunier B, Hadchouel A, Berteloot L. et al. Effectiveness and safety of ruxolitinib for the treatment of refractory systemic idiopathic juvenile arthritis like associated with interstitial lung disease: a case report. Ann Rheum Dis 2020;doi: 10.1136/annrheumdis-2020-216983. [DOI] [PubMed] [Google Scholar]
- 89. Sabbagh S, De Jesus AA, Hwang S. et al. Treatment of anti-MDA5 autoantibody-positive juvenile dermatomyositis using tofacitinib. Brain 2019;142:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wendel S, Venhoff N, Frye BC. et al. Successful treatment of extensive calcifications and acute pulmonary involvement in dermatomyositis with the Janus-Kinase inhibitor tofacitinib – A report of two cases. J Autoimmun 2019;100:131–136. [DOI] [PubMed] [Google Scholar]
- 91. Chen Z, Wang X, Ye S. Tofacitinib in amyopathic dermatomyositis – associated interstitial lung disease. N Engl J Med 2019;381:291–293. [DOI] [PubMed] [Google Scholar]
- 92. Park SH, Kang K, Giannopoulou E. et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol 2017;18:1104–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qiao Y, Giannopoulou GE, Chan CH. et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 2013;39:454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM. et al. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol 2009;183:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hu X, Chen J, Wang L, Ivashkiv LB.. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol 2007;82:237–43. [DOI] [PubMed] [Google Scholar]
- 96. Rubio D, Xu R-H, Remakus S. et al. Crosstalk between the type 1 interferon and nuclear factor kappa B pathways confers resistance to a lethal virus infection. Cell Host Microbe 2013;13:701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Farlik M, Reutterer B, Schindler C. et al. Nonconventional initiation complex assembly by STAT and NF-κB transcription factors regulates nitric oxide synthase expression. Immunity 2010;33:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mayer-Barber KD, Yan B.. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol 2017;14:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mayer-Barber KD, Andrade BB, Oland SD. et al. Host-directed therapy of tuberculosis based on interleukin-1 and type i interferon crosstalk. Nature 2014;511:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated in support of this paper.