Abstract
We isolated Saccharomyces cerevisiae yeast strains that are able to carry out the second fermentation of sparkling wine from spontaneously fermenting musts in El Penedès (Spain) by specifically designed selection protocols. All of them (26 strains) showed one of two very similar mitochondrial DNA (mtDNA) restriction patterns, whereas their karyotypes differed. These strains showed high rates of karyotype instability, which were dependent on both the medium and the strain, during vegetative growth. In all cases, the mtDNA restriction pattern was conserved in strains kept under the same conditions. Analysis of different repetitive sequences in their genomes suggested that ribosomal DNA repeats play an important role in the changes in size observed in chromosome XII, whereas SUC genes or Ty elements did not show amplification or transposition processes that could be related to rearrangements of the chromosomes showing these sequences. Karyotype changes also occurred in monosporidic diploid derivatives. We propose that these changes originated mainly from ectopic recombination between repeated sequences interspersed in the genome. None of the rearranged karyotypes provided a selective advantage strong enough to allow the strains to displace the parental strains. The nature and frequency of these changes suggest that they may play an important role in the establishment and maintenance of the genetic diversity observed in S. cerevisiae wild populations.
El Penedès is the major sparkling-wine-producing region of Spain. The traditional method of sparkling-wine elaboration was first developed in La Champagne (France) in the 18th century. It requires addition of sucrose and preconditioned yeast cells (the so-called pied de cup) to young wine for a second fermentation, which takes place in the characteristic sparkling-wine bottles for several months. In a previous work, we presented the analysis and characterization of the mycoflora associated with the three traditional grape varieties from El Penedès (16). This analysis helped us to isolate naturally occurring Saccharomyces cerevisiae yeast strains capable of carrying out the different processes in the sequence leading from must to sparkling wine under the conditions demanded by the wine industry. We refer to these strains herein as “sparkling-wine yeasts.”
Karyotype profiles are relatively consistent within a single yeast species. They serve as systematic criteria to distinguish between related yeast species from the genus Saccharomyces (7, 10). However, different strains of S. cerevisiae show a considerable variation of their karyotypes (2, 30). In addition, dramatic changes in karyotype occurring during vegetative growth have been reported for wild strains (1, 13, 14). One of the most intriguing findings from our previous work (16) was the observation of strains with the same mitochondrial DNA (mtDNA) patterns and similar phenotypic characteristics but different karyotypes. We interpreted these strains as originating from a preexistent population of different, though related, yeast clones (16).
We present here a further characterization of the sparkling-wine yeast strains isolated by the selection scheme described in reference 16. When we analyzed the stability of the karyotypes of these strains during vegetative growth, we observed a considerable karyotype instability. The frequency and nature of the karyotype changes depended on the genetic composition of the strain as well as on the medium in which it grew. We did not find any strong indication for a selective advantage of the rearranged karyotypes relative to those of the parental strains, such as the displacement of the parental strain by any of its rearranged derivatives. Our data suggest that karyotype rearrangements that occur during vegetative growth may play an important role in the establishment and maintenance of the genetic variability observed in wild yeast populations from different wine-producing regions.
MATERIALS AND METHODS
Plasmids and strains.
The S. cerevisiae strain W303a was obtained from the Yeast Stock Center, Berkeley, Calif. Plasmids pRB117, containing the SUC2 sequences, and p29, containing Ty1 sequences (3, 18), were a generous gift from T. Benítez, Departamento de Genética, Universidad de Sevilla, Seville, Spain. Plasmid pTK701, containing Ty2 sequences, was a gift from E. Martin-Rendon (University of Oxford, Oxford, United Kingdom).
Culture medium and conditions.
All strains were propagated in yeast peptone dextrose (YPD) (5 g of yeast extract/liter, 10 g of peptone/liter, 20 g of glucose/liter) at 30°C with continuous shaking. YS is similar to YPD but contains sucrose (20 g/liter) instead of glucose. Synthetic medium with ethanol (SE) contained 6.7 g of yeast nitrogen base without amino acids (Difco) per liter, 20 g of sucrose/liter, and 5 ml of ethanol/liter. WS medium consisted of regular wine from the firm Nadal (containing 105 ml of ethanol/liter) plus 16 g of sucrose/liter. Serial cultures were grown for different periods (depending on the medium) either at 30°C in a roller (YS) or at 17°C without shaking (SE and WS). When cultures reached saturation, new flasks were inoculated with the previous culture to an optical density at 600 nm of 0.025. Sporulation was performed in plates with 1% potassium acetate–0.1% yeast extract–0.05% glucose–2% agar (28) at 22°C for several weeks. Spores were isolated in a Tetrad Dissection Microscope (Micro Video Instruments, Inc., Avon, Mass.) after digestion of the ascus wall with Zymoliase 20T.
Sampling of yeast strains.
The general procedure for yeast strain sampling has already been published (16). Strains used in the present work were isolated from musts from grapes of the three traditional varieties: Macabeu, Xarel.lo, and Parellada (harvests from the years 1993 to 1996). The grapes came from the vineyards of the firm Nadal, located in El Pla del Penedès, 50 km southwest of Barcelona (Spain). They were separately pressed, clarified, and allowed to ferment in 20,000-liter tanks. Samples from the surface, the center, and the bottom of each of the three tanks were taken at different stages of fermentation, as monitored by the change in density of the fermenting must. Yeast cells present in the samples were spun down, resuspended in YPD, and frozen at −80°C after the addition of glycerol to 50%. Starting samples were streaked on YPD plates, and several isolated colonies from each plate were picked, grown in YPD, and frozen as described above.
Isolation of sparkling-wine yeast strains.
The yeast strain isolation method is described in reference 16. Combinations of the frozen yeast stocks were used to inoculate 50-ml flasks containing mixture A (740 ml of wine–65 g of sucrose per liter) at 17°C with gentle shaking. The ethanol concentration of mixture A was 80 ml/liter at inoculation and 120 ml/liter when all sugar had been consumed. These cultures were used to inoculate a second set of flasks with mixture B (830 ml of wine–49 g of sucrose per liter), which was designed to have a starting ethanol concentration of 90 ml/liter and an ending concentration of 120 ml/liter. These flasks were again incubated at 17°C until the consumption of all sugar available. After the last round of selection strains were tested for their fermenting capacity at 17°C by using inverted Durham tubes and checking for the appearance of gas bubbles in mixture A. Strains showing strong fermenting activity were stored at −80°C as indicated.
mtDNA analysis.
Total DNA extraction and restriction pattern analysis of mtDNA were performed as described previously (25). Yeast DNA was digested with HinfI or RsaI and analyzed in TBE (100 mM Tris–hydroxymethylaminomethane borate–5 mM EDTA [pH 8.4])–1% agarose gels.
Karyotype analysis.
Yeast cells from late exponential phase cultures were embedded in low-melting-point agarose and digested first with Zymoliase 20T (Seikagaku, Kyogo, Japan) and then with proteinase K (Sigma) as described previously (9). Yeast chromosomes were separated by pulsed-field gel electrophoresis (PFGE) in a Hula-Gel (Hoeffer) at 200 V, using a pulse ramp ranging from 60 to 150 s, for a total of 50 h, in 0.5× TBE buffer at 12°C.
Southern blots.
Chromosomes separated by PFGE were depurinized by soaking the gels in 50 mM HCl for 15 min and were then denatured with 1 M NaOH–1.5 M NaCl for 30 min. DNA was blotted onto nylon filters (Hybond-N, Amersham) by capillarity in 20× SSPE (1× SSPE is 180 mM NaCl, 10 mM sodium phosphate, and 1 mM EDTA [pH 7.7]). Filters were afterwards baked for 2 h at 80°C. Prehybridization was performed in 5× SSPE plus 5× Denhardt’s solution (2% Ficoll, 2% polyvinylpyrrolidone, and 2% bovine serum albumin) and 20 μg of single-stranded salmon sperm DNA/ml at 65°C for more than 2 h. DNA probes were labeled with 32P by the random primer (Ready-to-Go; Pharmacia) protocol. Hybridization was carried out at 65°C overnight in the prehybridization solution plus the labeled DNA probe. Filters were then washed three times with 2× SSPE–0.1% sodium dodecyl sulfate at 65°C for 30 min each time and then once with 0.2× SSPE–0.1% sodium dodecyl sulfate at 65°C for 10 min. Filters were exposed with Kodak X-OMAT AR films with intensifying screens at −80°C. The following probes were used: for SUC, a 0.9-kb fragment from pRB117 (18); for rDNA, a 1-kb EcoRI-HindIII genomic fragment encoding part of the 18S rRNA gene (24) (a gift from A. Rodriguez-Campos); for Ty1, a 1.3-kb EcoRI-SalI fragment from plasmid p29 (18); and for Ty2, a 1.7-kb fragment from plasmid pTK701 (18).
DNA content measurements.
Relative DNA contents were measured by flow cytometry. Cells from 1 ml of late exponential phase cultures were washed with distilled water and fixed in 70% ethanol for 30 min at −20°C. About 20 × 106 fixed cells were spun down and resuspended in 0.5 ml of sterile 50 mM sodium citrate. RNA was removed by addition of 5 μl of RNase A (Sigma) (concentration, 10 mg/ml) and incubated for 2 h at 37°C. Cells were stained by addition of one volume of a solution containing 50 mM sodium citrate plus 10 μg of propidium iodide (Sigma) per ml and incubated at room temperature for 30 min. Stained samples were kept at 4°C in the dark. Samples were analyzed by a Coulter Epics Elite flow cytometer (Serveis Tècnics, Universitat de Barcelona, Barcelona, Spain) with a blue argon laser (at 488 nm and 15 mW). Fluorescence was detected at 665 to 685 nm.
RESULTS
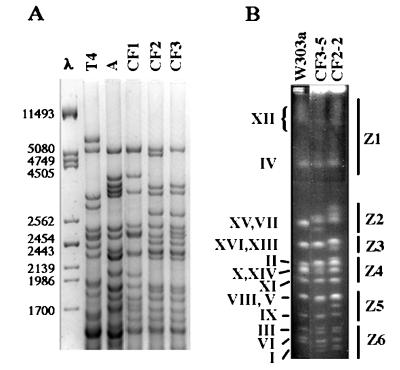
Analysis of a natural population of sparkling-wine yeasts in fermenting musts from El Penedès. Analysis of mtDNA patterns of the 29 yeast strains obtained by our selection scheme revealed five different mtDNA patterns, three of them found only once (Table 1). Among all strains we have isolated either directly from the musts or by our selection scheme, only strains showing one of the two mtDNA patterns CF2 and CF3 were indeed able to carry out the second fermentation of sparkling wine when tested in the cellar. Analysis of mtDNA patterns from 277 strains isolated from musts revealed that only three strains had a CF2 or CF3 mtDNA pattern (Table 1), suggesting that sparkling-wine yeast strains constituted a small subset (about 1%) within the natural yeast mycoflora. As shown in Fig. 1, CF2 and CF3 restriction patterns were very similar, suggesting that the strains that carry them are probably closely related. Considering their mtDNA restriction patterns (Fig. 1A) as well as their metabolic behavior (data not shown), we concluded that both CF2- and CF3-carrying strains belonged to S. cerevisiae (see below and reference 10). We reached the same conclusion after we compared their chromosomal profiles with that of the haploid S. cerevisiae laboratory strain W303a (Fig. 1B).
TABLE 1.
Distribution of mtDNA patterns in yeast strains from fermenting musts and in sparkling-wine yeast strains
| mtDNA patterna | No. (%) of clones isolated from:
|
|
|---|---|---|
| Must | Sparkling wine | |
| A | 50 (18.1) | 1 (3.4) |
| T4 | 1 (0.4) | 1 (3.4) |
| CF1 | 0 (0.0) | 1 (3.4) |
| CF2 | 2 (0.7) | 6 (20.7) |
| CF3 | 1 (0.4) | 20 (69.0) |
| Other | 223 (80.5) | 0 (0.0) |
| Total | 277 (100.1) | 29 (99.9) |
mtDNA patterns are designated as described in reference 16.
FIG. 1.
(A) mtDNA patterns from sparkling-wine yeasts. The column labeled λ shows phage 1 DNA cut with PstI, which was used as a size marker (sizes are shown on the left, expressed in base pairs). Note the strong similarity between patterns CF2 and CF3. (B) Karyotype profiles of strains CF2-2 (a CF2 strain) and CF3-5 (a CF3 strain) and the laboratory strain W303a. Roman numerals shown on the left indicate the ascription of each band to the different yeast chromosomes (as described in reference 19). Z1 through Z6 shown on the right indicate the different chromosome regions into which the karyotypes of the sparkling-wine yeasts were subdivided as described previously (16).
DNA contents of different sparkling-wine yeast strains were measured by flow cytometry. By taking as a standard the DNA content of strain W303a, sparkling-wine yeast strains were found to contain an amount of DNA equivalent to 1.6 to 2.3 haploid genomes (Table 2). Taking into account the standard deviations of the measured values, we concluded that sparkling-wine yeast strains had DNA content very close to 2C (1C being the DNA content of the haploid genome). From the karyotype profiles, it was relatively easy to ascribe the different bands to 16 pairs of homologous chromosomes, taking into account the relative intensities of the different bands and by comparison to the profile of W303a (Fig. 1B). However, although our strains behave essentially as diploids, we found indications of a low degree of aneuploidy in some cases (see below).
TABLE 2.
DNA contents of sparkling-wine yeasts and of their monosporidic derivatives
| Strain or derivative | DNA contenta |
|---|---|
| Strain | |
| CF2-2 | 2.28 ± 0.21 |
| CF2-18 | 2.09 ± 0.35 |
| CF2-19 | 2.05 ± 0.11 |
| CF3-5 | 1.92 ± 0.17 |
| CF3-6 | 1.98 ± 0.05 |
| CF3-22 | 1.59 ± 0.02 |
| Derivative | |
| CF3-5.1D | 1.86 ± 0.30 |
| CF3-5.5A | 1.85 ± 0.03 |
DNA content obtained by flow cytometry is expressed as the ratio relative to the DNA content of the laboratory strain W303a. Values are expressed as the means ± standard deviations from at least two independent determinations.
Karyotypic analysis of sparkling-wine yeasts.
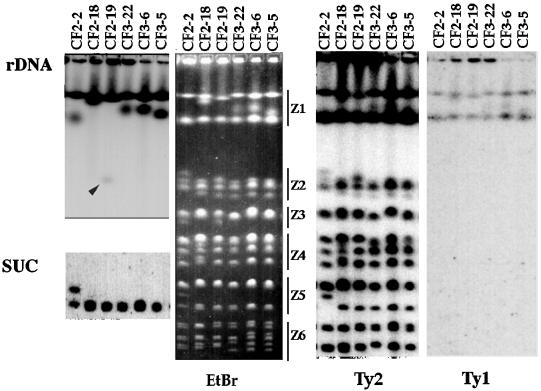
We have previously observed strains isolated from musts showing identical mtDNA patterns but different karyotype profiles (16). Figure 2 shows that this was also the case for sparkling-wine yeast strains with CF2 and CF3 mtDNA patterns. As described previously (16), the highest variability among strains showing the same mtDNA pattern corresponded to a low-mobility band in region Z1 (defined as described in reference 16), presumably chromosome XII (see below). However, other chromosome regions also showed differences (Fig. 2).
FIG. 2.
Karyotypes of several sparkling-wine yeast strains. The figure shows three CF2 strains (CF2-2, CF2-18, and CF2-19) and three CF3 strains (CF3-22, CF3-6, and CF3-5). The panel labeled EtBr shows a PFGE gel stained with ethidium bromide. The other panels show Southern blots obtained with different probes, rDNA, SUC, Ty1, and Ty2, as indicated for each panel. The two lefthand panels are only fragments of the total pictures, aligned on the corresponding positions. The arrowhead in the panel labeled rDNA points to a band, with relatively high mobility, which contained rDNA sequences.
High variability of the size of chromosome XII appears to be a common feature of yeast strains (4, 20), and it probably originates from the presence of several (up to 200) repeats of the rRNA-encoding genes in this chromosome. Other DNA repetitive sequences may also have an important role in variations in chromosome size. We have explored the presence of some of the repetitive DNA sequences known to be present in the yeast genome in sparkling-wine yeasts, to check whether at least part of the observed differences in karyotype between related strains could be related to changes on these sequences. We tested genes for rRNA, the SUC loci, and the transposon-like elements Ty1 and Ty2.
Genes coding for the different rRNAs (hereinafter referred as rDNA) are organized in pairs of 20 to 200 copies in chromosome XII (8, 23, 24, 27). Natural yeast strains are known to have hypervariable chromosomal bands coinciding in size with the expected molecular mass for chromosome XII (4, 9, 20). The blot in Fig. 2 shows that rDNA was indeed located both in this low-mobility band and in a faster, hypervariable band. Although it is possible that this band could correspond to size variants of chromosome XII, we have data suggesting that this band has an abnormal mobility and probably a peculiar structure. In some cases we have observed very small versions of this band (for example, see the high-mobility band in the rDNA blot in Fig. 2). In any case, the band that we propose contained the bona fide chromosome XII (the uppermost band that hybridized to the rDNA probe shown in Fig. 2) did show some variations in length (see Fig. 4), very possibly related to increases and decreases in the total number of rDNA repeats (4, 20).
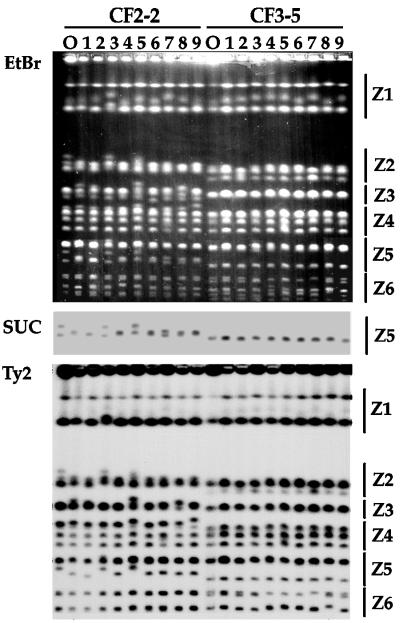
FIG. 4.
Changes in the karyotype profiles of CF2-2 and CF3-5 strains during vegetative growth. The columns labeled O show the karyotype of the original clone. The other tracks show karyotypes of different clones obtained after 100 doublings in SE medium. On the right are indicated the chromosome groups or zones we refer to throughout the text. The upper panel shows the ethidium bromide (EtBr)-stained gel, and the middle and the lower panels present the corresponding Southern blots hybridized with the SUC probe (only the region Z5 is shown) and Ty2, respectively.
Yeast strains contain variable numbers of the SUC gene, which encodes for invertase (19), an enzyme essential for sucrose utilization. Although must does not contain substantial amounts of it, the ability to ferment sucrose vigorously is a key feature for sparkling-wine yeast strains: sucrose is the only sugar available for the sparkling-wine second fermentation. For this reason, we added the ability to ferment sucrose vigorously in wine-plus-sucrose mixtures to the selection criteria for isolating sparkling-wine yeast strains. Therefore, we were interested in checking the number and distribution of SUC genes in sparkling-wine yeasts.
Figure 2 shows that all strains but CF2-2 showed a single band hybridizing with the SUC probe, which corresponded in size to chromosome IX. The simplest explanation is that the sparkling-wine yeasts had only the so-called SUC2 locus as a source of invertase—the usual localization for SUC genes in laboratory strains (19). Strain CF2-2 showed two bands hybridizing with the SUC probe; we interpreted these bands as two homologous chromosomes IX with different sizes.
The yeast retrotransposons Ty1 and Ty2, as well as their long terminal repeats (called δ elements), are usually present in many copies dispersed throughout the yeast genome (24). As shown in Fig. 2, sparkling-wine yeasts contained very few copies of Ty1 elements, and a single copy was probably present in each of the two low-mobility chromosomes. In contrast, Ty2 elements were much more abundant and distributed throughout most chromosomes (Fig. 2). Yeasts isolated directly from the musts, without selection, showed the more usual prevalence of Ty1 sequences over Ty2 sequences (data not shown). The distribution of Ty2 elements among the chromosomal bands of the sparkling-wine yeasts was not even. For example, the second-fastest chromosome band (which would correspond to chromosome VI in laboratory strains) did not contain Ty2 sequences in any of the sparkling-wine yeast strains checked so far (Fig. 2, and see Fig. 4). Our Ty probes were designed not to hybridize to δ elements, which are assumed to occur in more than 100 copies interspersed in the yeast genome. Most likely, δ sequences would be found in all yeast chromosomes in our strains.
From the bands shown in Fig. 1B and 2, it was evident that a considerable degree of polymorphism between homologous chromosomes was present in our sparkling-wine yeast strains. This was particularly evident in the four highest-mobility bands, presumably corresponding to chromosomes IX, III, VI, and I, from top to bottom (Fig. 2, and see 4). Although the sparkling-wine yeasts proved to sporulate with an extremely low efficiency (unpublished observation), we obtained several monosporidic derivatives from three strains, i.e., CF3-5, CF3-6, and CF2-18 (Fig. 3). The analysis of these different monosporidic derivatives provides information on the degree of aneuploidy of our strains. We have not been able to obtain a single complete tetrad from any of our strains; however, it was possible to follow the segregation of the different chromosomal size variants. For example, the three bands observed in region Z2 segregate as predicted for two pairs of homologous chromosomes, if the intermediate, double band containing one chromosome of each pair is considered (Fig. 3). Analogously, the six bands of region 6 in strain CF3-5 show the segregation pattern predicted for three pairs of homologous chromosomes of different sizes each. From these and similar considerations for other chromosomal bands, we concluded that most, if not all, of these bands showed a completely regular segregation, indicating that the degree of aneuploidy of our strains was low.
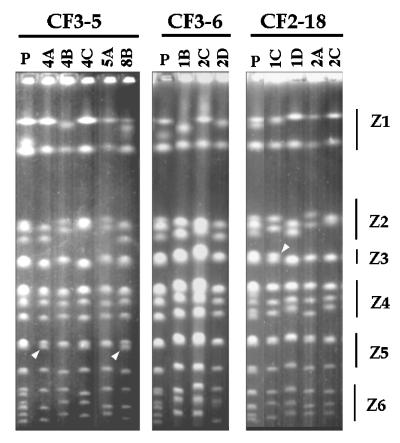
FIG. 3.
Karyotype profiles from monosporidic derivatives from strains CF3-5, CF3-6, and CF2-18. Lanes labeled P contain the parental strain. None of the dissected tetrads gave four viable spores. Numbers on the top refer to independent tetrads, and letters refer to different spores from a given tetrad. The different chromosome zones are indicated on the right. Arrowheads indicate bands, possible products of meiotic recombination, that were not in the parental strain.
The only size variants that did not show a normal segregation were the bands we showed to contain rDNA sequences and that presumably corresponded to chromosome XII. This suggests meiotic rearrangements of the rDNA repeats, as previously reported (8, 22, 29) (region Z1, Fig. 3). Besides the abnormal segregation observed in region Z1, we observed some chromosomal bands in the monosporidic derivatives that differed from the corresponding bands in the parental strains (Fig. 3). We interpreted them as indicative of meiotic chromosome rearrangements. Therefore, some of the karyotypic variability observed among strains with identical mtDNA patterns may have arisen from chromosomal rearrangements during meiosis, as previously reported for baker’s yeast strains (6).
Karyotype instability of sparkling-wine yeast strains.
Our starting hypothesis was that strains with identical mtDNA patterns differing in their karyotypic profiles were probably genetically related. To explore how close this relationship could be, we checked the karyotypic variability of different strains during vegetative growth. Figure 4 shows the karyotypic profiles of nine independent clones picked from a culture of the CF2-2 (left) and CF3-5 (right) strains after 100 doublings in the SE media. Chromosomal profiles of the original isolates are also shown. Chromosomal rearrangements were apparent in many clones for both strains. They occurred in essentially all chromosome zones, although they were most evident in the zones Z1 and Z2, in the upper part of the gel, as well as in Z5 and Z6, which correspond to the high-mobility chromosomal bands. Although the distribution of changes among the different zones varied somewhat from one experiment to another, we have always observed changes both in the upper part and in the lower part of the gel, indicating that chromosome rearrangements were restricted neither to the small chromosomes nor to the highly repetitive chromosome XII. Typically, all rearranged clones were different, that is, the only repeated chromosomal pattern observed after 100 generations was the original one. We interpreted these data as indicating that none of the rearranged clones was able to displace the original clone from the culture.
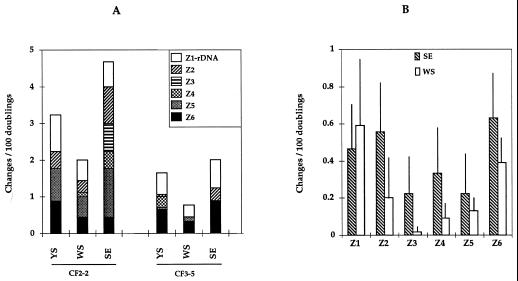
Figure 5A shows a quantitation of the number of observed chromosomal rearrangements observed after 100 doublings of strains CF2-2 and CF3-5 in three media: SE, WS, and YS. Strain CF2-2 seemed somewhat more variable than strain CF3-5 in all three media. In both cases, WS medium was the one giving the least variability, whereas SE medium was the one giving the most. Similar results have been obtained with all CF2 and CF3 strains tested so far. Figure 5B shows the combined data for the six sparkling-wine yeast strains analyzed in WS and SE media. Although the standard deviations of the results are high, probably due to the genetic heterogeneity among these six strains, it is clear that WS media gave significantly fewer chromosomal changes than the SE media.
FIG. 5.
(A) Quantitative analysis of the distribution of chromosome rearrangements in strains CF2-2 and CF3-5 during vegetative growth in YS, WS, and SE media. Figures indicate numbers of changes on chromosomal bands per 100 doublings observed in PFGE gels. Nine clones were analyzed in each experiment; duplicated experiments gave comparable results. The different boxes on each histogram indicate the values for the different chromosome regions or zones, as defined in Fig. 1. (B) Zone distribution of chromosomal changes after 100 doublings in SE or WS media, calculated as described above. Data are the mean values for the six strains for which karyotypes are shown in Fig. 2. Lines indicate standard deviations. The data are derived from the analysis of more than 100 individual clones.
Figure 4 shows a blot of the gel hybridized with the SUC probe (center) and the Ty2 probe (bottom). These blots gave some insights about the nature of the observed chromosome rearrangements. For example, the two SUC-containing bands of CF2-2 change with very high frequency, especially the band with lower mobility (Fig. 4). In some cases, these two bands merge at a mobility similar to that of the putative chromosome IX of the other strains. We consider that these two bands of CF2-2 correspond to two mobility variants of chromosome IX. By comparing the intensities of labeling of the two bands corresponding to chromosome IX, we concluded that they contain the same number of copies of SUC2, presumably a single copy. The changes in mobility of these bands did not appear to result from amplifications or deletions of the SUC genes (Fig. 4). In addition, we found that only the low-mobility band for chromosome IX contained Ty2 sequences in the strain CF2-2 (Fig. 2 and 4). Figure 4 shows that for the strains in which the two bands corresponding to chromosome IX merge, the Ty2 hybridizing sequences were present in the resulting band, even when its size coincided with the lower original band, which did not contain Ty2 sequences (Fig. 4, lower panel, third track from the left). We interpreted these data as suggesting that merging of the two bands was not a consequence of the substitution of one of the homologous chromosomes for the other. We have reached the same conclusion from the analysis of similar cases in other chromosomes in different strains (data not shown).
We did not find any obvious relationship between the presence or the amount of Ty sequences and the variability of a given chromosomal band. For example, the second-fastest-migrating pair of chromosomal bands, possibly corresponding to chromosome VI, contained no Ty1 or Ty2 sequences, but it showed approximately the same rate of changes as the bands immediately above and below it (these bands might correspond to chromosomes III and I), which contained Ty2 (Fig. 4 and 6). On the other hand, when only one of a pair of homologous chromosomes contained Ty2, we observed in some cases that this band changed with higher frequency than its counterpart which did not contain Ty2. This is the case, for example, for the upper band corresponding to chromosome IX of CF2-2 (Fig. 4). In any case, we have not observed any evidence of either Ty amplification or Ty transposition.
FIG. 6.
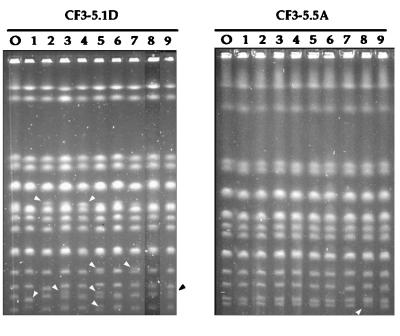
Changes in the karyotype profiles of two monosporidic derivatives from the CF3-5 strain, CF3-5.1D (left) and CF3-5.5A (right), during vegetative growth. Columns labeled O show the karyotypes of the original clones. The other tracks show those of different clones obtained after 100 doublings in YS.
In contrast to the frequent variations we observed in the karyotypes of the strains we examined, the mtDNA patterns remained very stable during vegetative growth. From more than 150 clones picked after 100 doublings in different media, we detected no changes in the mtDNA restriction pattern (data not shown).
Karyotype changes in monosporidic derivatives.
In all CF2 and CF3 strains we tested so far, karyotype instability was accompanied by a somewhat high level of chromosomal polymorphism (Fig. 2 and 3). A conceivable source of chromosomal size changes might be recombination between homologous chromosomes of different sizes, giving products of sizes different from those of the two parental bands. A direct way to check this hypothesis is to analyze the behavior of completely homozygous strains, where such chromosomal exchanges should produce no changes in chromosome sizes. Under our electrophoretic conditions, the chromosomal patterns of these derivatives contain 13 or 14 chromosomal bands, and 4 or 3 of them (respectively) are apparently doublets (Fig. 3 and 6). This is remarkably similar to the pattern of the haploid strain W303a, which contains 13 chromosomal bands, 4 of them corresponding to the doublets formed by chromosomes XV plus VII, XVI plus XIII, X plus XIV, and V plus VIII (Fig. 1B and reference 19). Double bands in Z3, Z4, and Z5 from the monosporidic derivatives showed a mobility similar to that of the corresponding doublets in W303a (Fig. 1B and data not shown). We concluded that monosporidic derivatives with a 2C DNA content contained exactly 16 pairs of homologous chromosomes.
We analyzed different clones obtained after 100 doublings in YS of two monosporidic derivatives from CF3-5, CF3-5.1D (Fig. 6, left panel) and CF3-5.5A (Fig. 6, right panel). These two derivatives proved to have a 2C DNA content (Table 2) and were homozygous for all observed chromosomal bands, with no bands that could be attributed to missegregation or meiotic rearrangements (Fig. 6). This notwithstanding, they did also show a detectable level of chromosomal instability upon vegetative growth (Fig. 6). The observed changes may implicate either one or both of the members of a given chromosome pair; in the first case, the rearranged karyotype showed an increased number of total chromosome bands relative to the parental one (see tracks 3, 5, 7, and 9 on the left gel of Fig. 6). From the relative intensities of the new chromosomal bands, we concluded that the rearranged clones are still diploids. Figure 6 also shows that the frequencies of chromosomal changes differed substantially among monosporidic derivatives: CF3-5.5A showed a frequency of changes about 10 times lower than that of CF3-5.1D. These observations, together with similar observations obtained from up to 20 derivatives from three different sparkling-wine yeast strains, suggest to us that the karyotype instability may be linked to a relatively small number of genes (3a).
DISCUSSION
Sparkling-wine yeast strains belong to the S. cerevisiae species, although they show some specific phenotypic traits (17). In our search for yeasts with these characteristics in the natural yeast population of El Penedès, we included resistance to ethanol and capacity for vigorous fermentation under conditions of high ethanol and low oxygen content as selective criteria (16). We have isolated 29 independent clones, 26 of them showing two related mtDNA patterns, CF2 and CF3. These patterns were found in a minor proportion of the total yeast population of clones directly isolated from fermenting musts (3 of 277 clones [16 and unpublished results]). These data reinforce our hypothesis that mtDNA patterns are indicative of the presence of distinct subpopulations of the natural yeast mycoflora, perhaps as a result of adaptations to specific microenvironments (16). In this context, sparkling-wine yeasts represented a very minor fraction of the natural mycoflora.
Analysis of several CF2 and CF3 strains indicated that they had a 2C DNA content. Their karyotype patterns were very similar to that of a S. cerevisiae laboratory strain, although there was a considerable degree of polymorphism for homologous chromosomes. The analysis of the segregation of these polymorphic chromosomes suggested that CF2 and CF3 strains have a rather low degree of aneuploidy, in contrast to the massive aneuploidy observed for several yeast strains found in wine (2). A distinctive characteristic was a large prevalence of Ty2 sequences over Ty1 sequences, a feature these strains share with flor yeast strains from sherry wines (11). Other strains isolated from the same musts from which the CF2 and CF3 strains were isolated showed the usual prevalence of Ty1 sequences (data not shown). It is also remarkable that, although CF2 and CF3 strains were selected by their vigorous fermentation of sucrose, they apparently attained this capability without amplification of the SUC genes.
A striking feature of the sparkling-wine yeasts is the natural variability of their karyotypes. Our data suggest that at least part of this variability could result from chromosomal rearrangements during vegetative growth. Such changes have been observed in several wine yeast populations (1, 13). Our data showed that the rate of chromosomal changes may be influenced by the medium in which the strains grow, although we did not observe any obvious cause-effect relationship. Ethanol, or its first metabolite, acetaldehyde, may cause lesions to both mtDNA (5, 12) and chromosomal DNA (26). We do not consider it probable that this was the case in our experiments, as it was precisely the medium with the highest ethanol concentration (wine plus sucrose; WS medium) that produced the fewest changes in chromosome size. This is specific for sparkling-wine yeasts, because a similar experiment using must strains showed the highest proportion of chromosomal rearrangements in the WS medium (unpublished observations). This might be related to the adaptation of CF2 and CF3 strains to growth in wine-plus-sugar mixtures.
Although the chromosomal changes might conceivably have an adaptative meaning (1), we do not have any clear indication for such an adaptation. For example, all karyotype patterns that were different from the parental ones were observed only once in all experiments. Should a given chromosomal rearrangement provide a selective advantage, the affected strain would have displaced both the parental one and the other karyotype variants from the yeast population. Following this reasoning, we cannot rule out some kind of selective advantage for specific chromosomal changes, the proportion of which changed considerably from one medium to the other. For example, this may be the case for the homozygous upper band of region Z2, which was found in six of the nine clones from the CF2-2 strain grown in SE medium (Fig. 3) but was much rarer in clones from the same strain grown in other media. Nevertheless, we consider our data to suggest that most chromosomal rearrangements in wild populations are selectively neutral. However, in the natural populations they may well provide a significant source of genetic variability that could be important for the adaptation of a given clone to changing environmental conditions.
Our data provided some hints about the mechanisms that may be implicated in the observed karyotype variability. A possible mechanism may be recombination, either reciprocal or nonreciprocal, between homologous chromosomes of different sizes, giving products that might migrate at different positions relative to the parental bands. Although this mechanism is possible, given the high degree of polymorphism between homologous chromosomes in our strains, it cannot be the only one, for we have observed chromosomal rearrangements in homozygous derivatives, in some cases leading to the appearance of new chromosomal bands in heterozygosity.
Repetitive sequences interspersed in the yeast genome are possible sources for genome instability. In this regard, we have a strong indication that amplifications, deletions, and rearrangements of the rDNA repeats may be the most important source of the variation in size of the putative chromosome XII we observed in almost all clones we have examined. On the contrary, changes on size of putative chromosome IX were not due to amplification of the locus SUC2.
Ty1 and Ty2 are assumed to be by far the most frequent transposon-like elements in S. cerevisiae. From the analysis of their distribution in rearranged clones, we concluded that these chromosomal rearrangements were not related to the mobilization of Ty elements. In addition, we did not observe any obvious relationship between their distribution and the rate of changes in different chromosomes. This is in striking contrast to the published results for meiotic rearrangements in baker’s yeasts, where mobilization and amplification of Ty elements seems to play an important role (6). Interestingly, we have found a relatively low frequency of meiotic rearrangements. We take these data to suggest that meiotic and mitotic chromosomal rearrangements are independent phenomena and that their relative contributions to the observed variability of the wild yeast genomes vary widely among the different yeast populations.
Taking into account the data set forth above, we consider it likely that a main source of mitotic chromosomal instability in our strains might be recombination between nonallelic loci (ectopic recombination) (21). This phenomenon could be triggered by the presence of repeated sequences interspersed in the genome, most likely Ty and δ elements, but also perhaps Y′ subtelomeric sequences, as reported for meiotic reorganization of baker’s yeast chromosomes (6). Our preliminary results indicate that chromosome rearrangements occurred very rarely, if ever, in haploid monosporidic derivatives of our strains (unpublished observations); therefore, we propose that most of these recombination events should occur between homologous chromosomes.
We have observed no changes in the mtDNA pattern during vegetative growth, even in strains with very high rates of chromosomal rearrangements. These data accord absolutely with our previous results acquired from analysis of yeast strains from El Penedès (16), where karyotypic analysis revealed a rate of variability much higher than that of the mtDNA restriction pattern. This is probably also the case in baker’s yeast strains (5). Natural yeast populations from grape musts show a considerable degree of heterozygosity (reference 15 and our unpublished results). The data shown here suggest that at least part of this heterozygosity may result from chromosomal rearrangement during vegetative growth and that this can be an important source of genetic variability in the natural yeast populations.
ACKNOWLEDGMENTS
We thank Enric Bartra (INCAVI, Vilafranca del Penedès, Spain), Rafael Oliva (University of Barcelona), Amparo Querol, Daniel Ramón (IATA-CSIC, Valencia, Spain), and Gemma Marfany and Montserrat Aguadé (Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Barcelona, Spain) for their advice and useful comments. We also thank Tahía Benítez (Departamento de Genética, Universidad de Sevilla, Seville, Spain), Antonio Rodriguez-Campos (Centre d’Investigació i Desenvolupament, Consejo Superior de Investigaciones Científicas, Barcelona, Spain), and Encarna Martin-Rendon (University of Oxford, Oxford, United Kingdom) for their gifts of different plasmids.
This work has been supported by grants from the Spanish Ministry of Education and Science (PB92-0051, PB95-0433, and 95-0012-OP), by additional support from the Generalitat de Catalunya (GRQ93-8024), and by the Alexander von Humboldt Stiftung (Germany) to B.P. D.N. has been partially supported by fellowships (RE93-05 and RI94-20) from the Generalitat de Catalunya. The firm Ramón Nadal Giró also acknowledges a grant from the Generalitat de Catalunya (IT94/214).
REFERENCES
- 1.Adams J, Puskas R S, Simlar J, Wilke C M. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr Genet. 1992;22:13–19. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- 2.Bakalinsky A T, Snow R. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast. 1990;6:367–382. doi: 10.1002/yea.320060503. [DOI] [PubMed] [Google Scholar]
- 3.Carlson M, Celenza J L, Eng F J. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985;5:2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Carro, D., D. Nadal, and B. Piña. Unpublished data.
- 4.Chindamporn A, Iwaguchi S, Nakagawa Y, Homma M, Tanaka K. Clonal size-variation of rDNA cluster region on chromosome XII of Saccharomyces cerevisiae. J Gen Microbiol. 1993;139:1409–1415. doi: 10.1099/00221287-139-7-1409. [DOI] [PubMed] [Google Scholar]
- 5.Codon A C, Benitez T. Variability of the physiological features and of the nuclear and mitochondrial genomes of bakers’ yeasts. Syst Appl Microbiol. 1995;18:343–352. [Google Scholar]
- 6.Codon A C, Benitez T, Korhola M. Chromosomal reorganization during meiosis of Saccharomyces cerevisiae baker’s yeasts. Curr Genet. 1997;32:247–259. doi: 10.1007/s002940050274. [DOI] [PubMed] [Google Scholar]
- 7.De Jonge P, De Jongh F, Meijers R, Steensma H, Scheffers W. Orthogonal-field-alternation gel electrophoresis banding patterns of DNA from yeasts. Yeast. 1986;2:193–204. doi: 10.1002/yea.320020307. [DOI] [PubMed] [Google Scholar]
- 8.Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 9.Gerring S, Connelly C, Hieter P. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 1991;194:55–57. doi: 10.1016/0076-6879(91)94007-y. [DOI] [PubMed] [Google Scholar]
- 10.Guillamón J M, Barrio E, Huerta T, Querol A. Rapid characterization of four species of the Saccharomyces sensu stricto complex according to mitochondrial DNA pattern. Int J Syst Bacteriol. 1994;44:708–714. doi: 10.1099/00207713-44-4-708. [DOI] [PubMed] [Google Scholar]
- 11.Ibeas J I, Jimenez J. Genomic complexity and chromosomal rearrangements in wine-laboratory yeast hybrids. Curr Genet. 1996;30:410–416. doi: 10.1007/s002940050150. [DOI] [PubMed] [Google Scholar]
- 12.Ibeas J I, Jimenez J. Mitochondrial DNA loss caused by ethanol in Saccharomyces flor yeasts. Appl Environ Microbiol. 1997;63:7–12. doi: 10.1128/aem.63.1.7-12.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longo E, Vezinhet F. Chromosomal rearrangements during vegetative growth of a wild strain of Saccharomyces cerevisiae. Appl Environ Microbiol. 1993;59:322–326. doi: 10.1128/aem.59.1.322-326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miklos I, Varga T, Nagy A, Sipiczki M. Genome instability and chromosomal rearrangements in a heterothallic wine yeast. J Basic Microbiol. 1997;37:345–354. doi: 10.1002/jobm.3620370507. [DOI] [PubMed] [Google Scholar]
- 15.Mortimer R K, Romano P, Suzzi G, Polsinelli M. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10:1543–1552. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- 16.Nadal D, Colomer B, Piña B. Molecular polymorphism distribution in phenotypically distinct populations of wine yeast strains. Appl Environ Microbiol. 1996;62:1944–1950. doi: 10.1128/aem.62.6.1944-1950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naumov G. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol. 1996;17:295–302. [Google Scholar]
- 18.Naumov G, Naumova E, Lantto R, Louis E, Korhola M. Genetic homology between Saccharomyces cerevisiae and its sibling species S. paradoxus and S. bayanus: electrophoretic karyotypes. Yeast. 1992;8:599–612. doi: 10.1002/yea.320080804. [DOI] [PubMed] [Google Scholar]
- 19.Olson M. Genome structure and organization in Saccharomyces cerevisiae. In: Broach J, Pringle J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 1–39. [Google Scholar]
- 20.Pasero P, Marilley M. Size variation of rDNA clusters in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol Gen Genet. 1993;236:448–452. doi: 10.1007/BF00277147. [DOI] [PubMed] [Google Scholar]
- 21.Petes T, Malone R, Symington L. Recombination in yeast. In: Broach J, Pringle J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 22.Petes T D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980;19:765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- 23.Petes T D. Yeast ribosomal DNA genes are located on chromosomes XII. Proc Natl Acad Sci USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippsen P, Stotz A, Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 25.Querol A, Barrio E, Ramón D. A comparative study of different methods of yeast strain characterization. Syst Appl Microbiol. 1992;15:439–446. [Google Scholar]
- 26.Ristow H, Seyfarth A, Lochmann E-R. Chromosomal damages by ethanol and acetaldehyde in Saccharomyces cerevisiae as studied by pulsed field gel electrophoresis. Mutat Res. 1995;326:165–170. doi: 10.1016/0027-5107(94)00165-2. [DOI] [PubMed] [Google Scholar]
- 27.Rustchenko-Bulgac E P, Sherman F. Physical constitution of ribosomal genes in common strains of Saccharomyces cerevisiae. Yeast. 1994;10:1157–1171. doi: 10.1002/yea.320100904. [DOI] [PubMed] [Google Scholar]
- 28.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 29.Szostak J W, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 30.Vezinhet F, Hallet J-N, Valade M, Poulard A. Ecological survey of wine yeast strains by molecular methods of identification. Am J Enol Vitic. 1992;43:83–86. [Google Scholar]