Abstract
Introduction
Recent studies have revealed that several deubiquitinating enzymes (DUBs) play important roles in hepatocellular carcinoma (HCC) progression, but the roles of Otubain 2 (OTUB2) in HCC remain obscure.
Methods
In this study, we investigated the expression of OTUB2 in HCC based on clinical samples and a public online database (ENCORI), and its roles and working mechanisms were further explored by in vitro experiments.
Results
It was found that the expression of OTUB2 was significantly up-regulated in HCC tissues, and correlated with poor prognosis of HCC patients. Functionally, the overexpression of OTUB2 could promote malignant proliferation and metastasis of HCC cells, while knockdown of OTUB2 exerted the opposite results. Using two bioinformatics tools, PJA1 was identified as a potential gene regulated by OTUB2. Mechanistically, it was found that OTUB2 promoted the stabilization of PJA1 by deubiquitylation, based on immunoprecipitation (IP) and cycloheximide (CHX) assays. Moreover, the suppressive effects of OTUB2 depletion on the malignant phenotypes of HCC cells could be reversed by overexpressing PJA1.
Conclusion
In conclusion, our study indicated that OTUB2 could promote the malignant proliferation and migration of HCC cells by increasing the stability of PJA1 via deubiquitylation.
Keywords: Otubain 2, Hepatocellular carcinoma, Deubiquitylation, PJA1
Introduction
According to the recent report, liver cancer remains one of the most common malignancies, which brings a global health burden with a poor prognosis. Hepatocellular carcinoma (HCC) accounts for approximately 90% of primary liver cancer, which is usually at an advanced stage upon diagnosis.32 Due to the late diagnosis of HCC, it is difficult to treat HCC and the poor prognosis of those patients is usually poor.36 Even though the pathological mechanism of HCC has been investigated in the last decades, the overall survival and prognosis of HCC patients are still not favorable. Therefore, it is necessary to further clarify the exact mechanism of HCC onset and progression and develop novel targeted drugs for the treatment of HCC patients.
The roles of post-translational modifications (PTMs) in the pathogenesis of various diseases has become a hot topic globally. PTMs of proteins participate in many essential biological processes by targeting the proteins involved in complex assembly, transport, and degradation. Ubiquitylation and deubiquitylation are two dynamic modalities of PTMs in cells.5 Ubiquitylation mainly regulates protein degradation, which plays an important role in various cellular regulation events.12,29 Many studies have revealed that abnormal ubiquitylation of pathways is implicated in the onset and progression of multiple cancers,6,11 including HCC.2 Given that deubiquitinating enzymes (DUBs) can reverse the ubiquitylation of target proteins and promote the stability of downstream target proteins, DUBs have emerged as promising targets in cancer therapy.1
According to sequence and structural similarity, DUBs could be classified into six subfamilies.15 As the second-largest member of DUBs, the ovarian tumor (OTU) subfamily is identified to have 16 members, which is further divided into four categories, including OTUBs, OTUDs, A20s, and OTULINs.33 Otubain 2 (OTUB2) is a DUB belonging to OTUBs, the functions and underlying mechanisms of which are largely unknown.24 In recent years, several studies have revealed that OTUB2 may play a role in tumorigenesis. Li et al.20 found that OTUB2 may activate the AKT/mTOR pathway and Warburg effect to drive the tumorigenesis of lung cancer by reversing the ubiquitylation of U2AF2. OTUB2 has also been proven to promote breast cancer metastasis by deubiquitinating YAP and TAZ in vivo.38 The depletion of OTUB2 has been shown to effectively repress the proliferation of HCC cells in a recent study.13 OTUB2 expression was positively correlated with the phosphorylation levels of NF-κB p65, and OTUB2 silencing suppressed the activation of NF-κB p65, thereby exerting antitumor effects on HCC cells. However, the deubiquitylation role and mechanisms of OTUB2 in HCC progression have not yet been reported.
In this study, the roles and molecular mechanism of OTUB2 in HCC were investigated, the expression and functions of OTUB2 in tumorigenesis were analyzed, and the interaction between OTUB2 and PJA1 was further investigated. It was found that OTUB2 played a crucial role in proliferation and migration of HCC cells. Moreover, it was revealed that the OTUB2 promoted proliferation and migration of HCC cells as a specific DUB for PJA1. Collectively, OTUB2 was a potential molecular target for the treatment of HCC via enhancing PJA1 stability.
Materials and Methods
Clinical Sample Collection
Clinical HCC and adjacent noncancerous tissue samples were collected from the resected tissues of HCC patients. The samples were all harvested after informed consent by the patients, and stored at − 80 °C before use. The procedure of this research was reviewed and approved by Strategic Support Force Characteristic Medical Center.
Cell Culture and Cell Transfection
The human HCC cell line MHCC-97h was obtained from the Liver Cancer Institute of Fudan University (Shanghai, China), and Huh7 was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI 1640-medium containing 10% FBS (Gibco, NY, USA) at 37 °C in a 5% CO2 incubator.
PJA1 and OTUB2 were cloned into pcDNA3.1 to construct PJA1 and OTUB2 overexpression vectors, respectively. The expression of OTUB2 and PAJ1 in cells was knocked down by using the short hairpin RNAs (sh-RNAs) against OTUB2 (sh-OTUB2-1: 5′-TCCGTTTACCTGCTCTATAAA-3′; sh-OTUB2-2: 5′-CCTATGTGTCACTGGATTATT-3′) and sh-PJA1 (sh-PJA1-1: 5′-GCCTCACAGTCGTCTTCTGAA-3′; sh-PJA1-2: 5'-GAGGGATACCGCCAATGACAA-3'). Indicated plasmids were transfected into cells by using Lipofectamine® 2000 (Invitrogen, CA, USA) according to the manufacturer instructions. 48h later, the cells were collected for subsequent experiments.
Cell Counting Kit-8 (CCK-8) Assay
Cells were seeded into 96-well plates at 5 × 103 per well, and incubated for 24, 48, and 72 h, respectively. Medium in each well was replaced with fresh medium containing CCK-8 reagent (10%, v/v) after incubation. The cells were further incubated for 1.5h, and their absorbance at 450mm was recorded.
Clone Formation Assay
Five hundred cells were plated into each well of 6-well plates. After 14 days of cultivation, cells were fixed with 4% paraformaldehyde, followed by staining with 0.1% crystal violet. Finally, stained colonies were photographed and counted under a microscope.
Transwell Assay
5 × 104 cells suspended in 200 μL serum-free medium were added to the top chamber with a filter membrane of 8 mm pores. Meanwhile, 700 μL medium containing 10% FBS was added to the bottom chamber. After 24 h incubation, cells passing through the membrane were fixed and stained with 0.1% crystal violet for 10 min. Finally, the migrated cells on the underside of the membrane were photographed (× 100 magnification) and counted in six random fields under a microscope.
Wound Healing Assay
Briefly, cells were seeded into 6-well plates and grew until reaching approximately 90% confluence. Then, a 10 μL pipette tip was used to scratch monolayer cells. The wound was rinsed with PBS, incubated with medium containing 1% FBS, and photographed under a microscope (× 100 magnification) at 0 h (used as a baseline) and 24 h. The width of the wound was measured, and the cell migration rate was measured as follows: cell migration rate = (1 − wound width (24 h)/wound width (0 h)) × 100%. The duration of the microscopic procedures was maintained identical to minimize the environmental condition-related bias in wound healing responses.
Western Blot
Collected tissues were lysed in RIPA buffer (Beyotime, Shanghai, China), and the total protein was harvested. Proteins were quantified by BCA Protein Assay Kit (Beyotime, Shanghai, China), subsequently separated by SDS-PAGE and transferred onto PVDF membranes. Then, the membrane was blocked in 5% skimmed milk at room temperature for 1 h, and incubated with primary antibodies (OTUB2, cat.no ab74371, Abcam; PJA1, cat.no 17687-1-AP, Proteintech) overnight. On the next day, the membrane was washed three times with PBST, and then incubated with the secondary antibody (Anti-Rabbit IgG H&L (HRP), cat.no ab6721, Abcam) for 1 h. BeyoECL Plus (Beyotime, Shanghai, China) was used to visualize the protein bands. GADPH was used as an internal reference for normalization of the expressions of OTUB2 and PJA1, and the protein band intensity was analyzed using Image J 6.0 software.
Co-immunoprecipitation (Co-IP)
Co-IP analysis was performed as reported previously.10 Briefly, total protein was collected and quantified before incubated with magnetic bead-conjugated specific antibody for 2 h with gentle rotation at 4 °C. Then, cell lysis buffer was used to wash the beads four times. Bound proteins were subsequently eluted and analyzed by Western blot.
Ubiquitylation and Protein Half-Life Assay
In vitro ubiquitylation was performed as Linares et al.22 described. Flag-OTUB2, HA-PJA1, and His-ubiquitin (His-ub) were transfected into 293T cells. Before collecting the transfected cells, MG132 (cat.no C2211, Sigma-Aldrich, MO, USA), a proteasome inhibitor, was added to inhibit proteasome-mediated protein degradation. 48 h later, the cells were harvested for ubiquitylation determination based on Western blot analysis.
For protein half-life assay, transfected cells were treated with 15 mM cycloheximide (CHX, Sigma Aldrich). Cell lysate was prepared at 0, 2, 4, and 6 h after treatment, respectively, and analyzed by Western blot.
Bioinformatics Analysis
The expression of OTUB2 in HCC tissues and normal tissues was assessed based on ENCORI (http://starbase.sysu.edu.cn/)21 online database.
The prognostic roles of OTUB2 in different subgroups of patients with HCC were evaluated using an online interactive web tool, Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html).35 Patients with HCC were divided into high and low expression groups based on the median values of OTUB2 expression.
OTUB2 was input into STRING (https://string-db.org/)34 to identify its potential interacted proteins. In the meantime, GENEMANIA (http://genemania.org/)37 was used to explore internal correlations of OTUB2 with other genes. Afterwards, the genes identified by two tools were intersected to obtain the potential targets of OTUB2.
Statistical Analysis
Each experiment was performed for three times. All data were expressed as mean ± SD. Students t-test was used for statistical analysis using GraphPad Prism software version 8.0. p < 0.05 was considered statistically significant.
Results
OTUB2 is Up-regulated and Associated with Poor Prognosis of HCC Patients
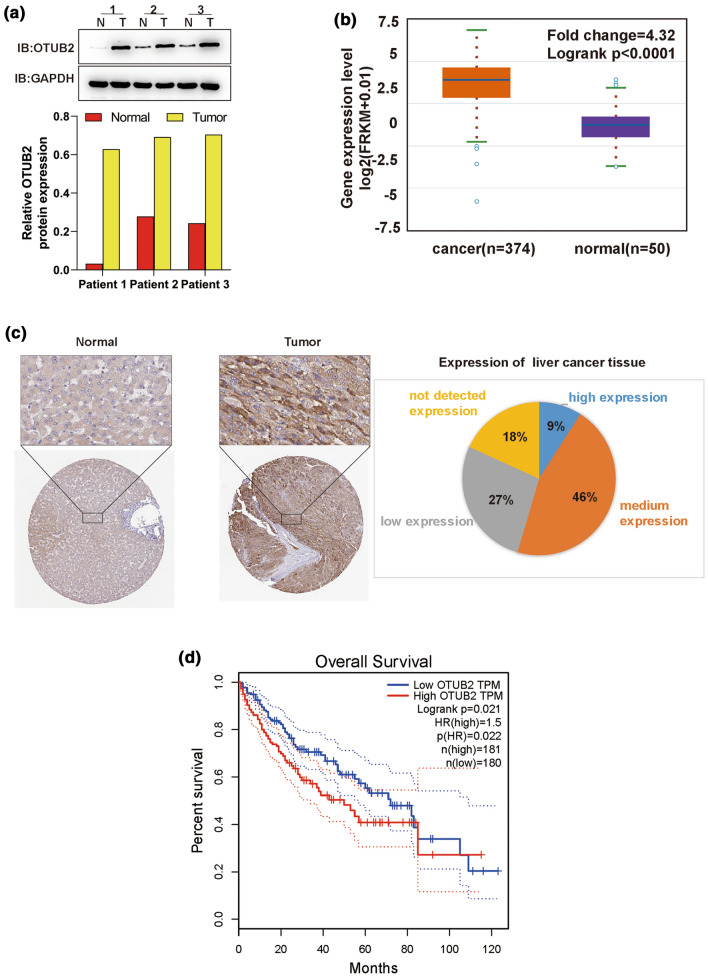
Based on determination of OTUB2 protein expression in three pairs of clinical HCC and adjacent noncancerous tissue samples, and the ENCORI online database, it was found that OTUB2 was obviously up-regulated in HCC tissues (Figs. 1a and 1b), consistent with the data from Human Protein Atlas (HPA, https://www.proteinatlas.org) (Fig. 1c). Afterwards, the correlation between OTUB2 expression and overall survival of HCC patients was analyzed based on another public online database (GEPIA). HCC patients with high expression of OTUB2 displayed a significantly lower 5-year survival rate (Fig. 1d). All results suggested that OTUB2 is highly expressed in HCC, and may act as an oncogene to regulate HCC progression.
Figure 1.
OTUB2 is highly expressed in HCC and indicates poor prognosis of HCC patients. (a) The protein expression of OTUB2 in three paired HCC and the adjacent non-cancerous tissues detected by Western blot. (b) The expression of OTUB2 in HCCOTUB2 in HCC tumor tissues (n = 374) and normal controls (n = 50) based on ENCORI (http://starbase.sysu.edu.cn/). (c) Representative images of immunohistochemistry for OTUB2 expression in HCC downloaded from HPA (https://www.proteinatlas.org) database. (d) The over survival analysis for OTUB2 expression in HCC based on GEPIA (http://gepia.cancer-pku.cn/index.html); patients with HCC were divided into high and low expression groups based on the median values of OTUB2 expression, and the dotted lines indicated 95% Confidence Interval.
OTUB2 Promotes HCC Cell Proliferation
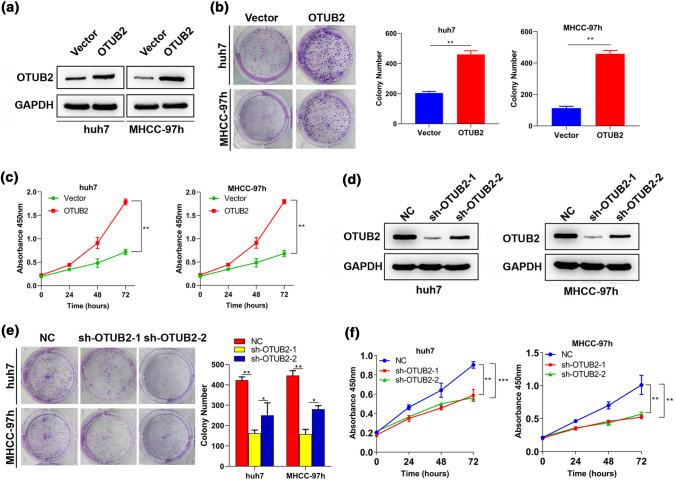
In order to assess whether OTUB2 regulates the proliferation of HCC cells, OTUB2 was overexpressed or knocked down in Huh 7 and MHCC-97h cells in colony formation and CCK-8 assays. The results of Western blot revealed that OTUB2 was successfully overexpressed or knocked down in Huh 7 and MHCC-97h cells (Figs. 2a and 2d). Compared with cells transfected with empty vectors, significantly increased cell colonies were observed in OTUB2-overexpressed cells (Fig. 2b). CCK-8 assay indicated that OTUB2 overexpression significantly promoted proliferation of Huh 7 and MHCC-97h cells compared with empty vectors (Fig. 2c). These data hinted that OTUB2 may be involved in HCC cell proliferation, as was subsequently corroborated by the same functional analysis on OTU2-silenced Huh 7 and MHCC-97h cells (Figs. 2e and 2f).
Figure 2.
Effects of OTUB2 on proliferation of HCC cells. Huh7 and MHCC-97h cells were transfected with OTUB2 overexpression vectors (OTUB2) or empty vectors (Vector) for 48 h before analyzed. (a) The transfection efficiency of OTUB2 in Huh7 and MHCC-97h cells was examined by Western blot. The proliferation of OTUB2 overexpressing cells was detected by (b) colony formation (n = 3) and (c) CCK-8 (n = 3) assays. Huh7 and MHCC-97h cells were transfected with short hairpin RNAs against OTUB2 (sh-OTUB2-1 or sh-OTUB2-2) or the corresponding negative control RNA (NC) for 48 h before analyzed. (d) The expression of OTUB2 protein in sh-OTUB2-1 or sh-OTUB2-2 expressing cells was examined by Western blot to determine transfection efficiency. The proliferation of sh-OTUB2-1/2 expressing cells was detected by (e) colony formation (n = 3) and (f) CCK-8 (n = 3) assays. Quantitative data are presented as mean ± SD of three replicates. *p < 0.05 and **p < 0.01, compared with the NC or vector group.
OTUB2 Enhances Migration of HCC Cells
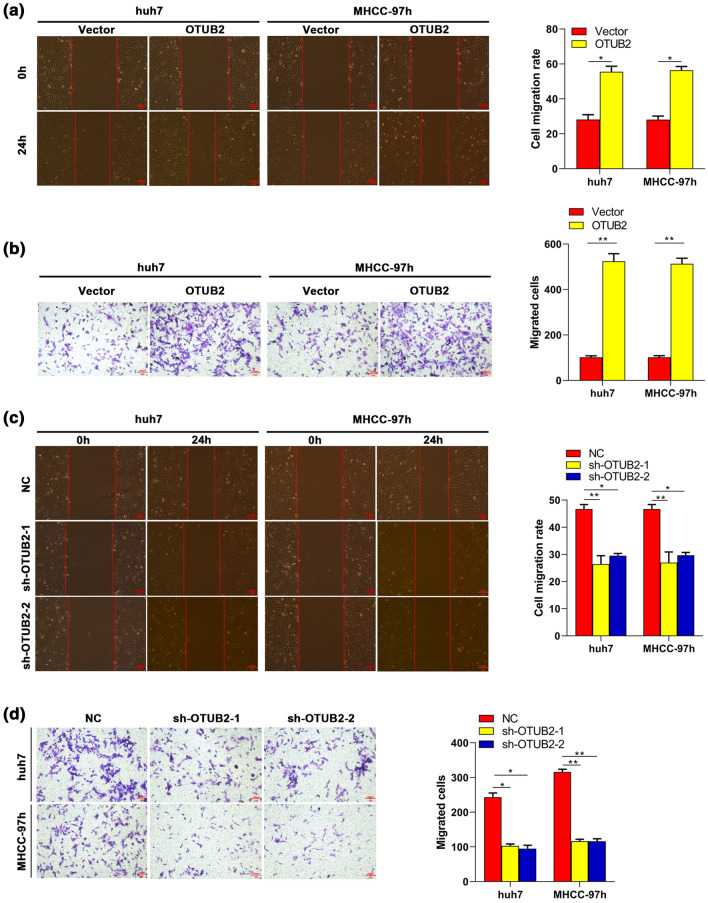
As is known, metastasis indicates HCC progression and poor prognosis of HCC patients, while cell migration is a key step of metastasis. Therefore, whether OTUB2 regulated migration of HCC cells was also addressed. Wound healing assay showed that migration of Huh 7 and MHCC-97h cells was significantly enhanced by OTUB2 overexpression, while impaired by OTUB2 silencing (Figs. 3a and 3c). As expected, transwell assay showed a similar trend to the result of wound healing (Figs. 3b and 3d). These results collectively suggested that OTUB2 was involved in migration of HCC cells.
Figure 3.
Effects of OTUB2 on migration of HCC cells. After transfected OTUB2 or vectors into Huh7 and MHCC-97h cells for 48 h, the cell migration was assessed by (a) wound healing (n = 3) and (b) transwell (n = 3) assays. After transfected sh-OTUB2-1/2 or the NC into Huh7 and MHCC-97h cells for 48 h, the cell migration was detected by (c) wound healing (n = 3) and (d) transwell (n = 3) assays. Scale bar, 100 μm. Quantitative data are presented as mean ± SD of three replicates. *p < 0.05 and **p < 0.01, compared with the NC or vector group.
OTUB2 Plays a Deubiquitylation Role in PJA1 and Promotes Its Stability
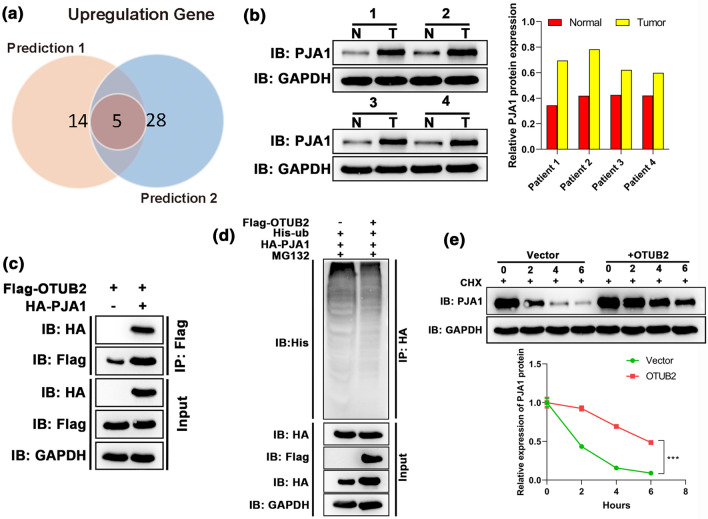
Given that OTUB2 is a DUB, it was speculated that OTUB2 reverses the ubiquitylation of a potential oncogene and regulates HCC progression. Thus, we predicted the potential targets of OTUB2 with the help of a bioinformatics tool to further explore the mechanism behind the roles of OTUB2 in HCC. After intersecting the prediction of two databases, five potential targets were obtained, which included ATXN3, RFFL, PJA1, TRIM55, and TRIM54 (Figs. 4a and 4b). Based on a previous finding that PJA1 is involved in metastasis of HCC cells and is closely related to the prognosis of HCC patients,4 PJA1 was selected as the downstream gene of OTUB2 for further analysis. The expression profile of PJA1 in clinical samples demonstrated that PJA1 was highly expressed in HCC, which is consistent with the previous study.4 Co-immunoprecipitation (Co-IP) assay was performed on 293T cells to confirm the interaction between OTUB2 with PJA1. As expected, the result of Co-IP revealed OTUB2 efficiently interacted with PJA1 (Fig. 4c). Moreover, ubiquitylation assay showed that exogenous OTUB2 overexpression could decrease ubiquitylation of PJA1 (Fig. 4d), and the cycloheximide (CHX) also demonstrated that OTUB2 overexpression could effectively promote the protein stability of PJA1 (Fig. 4e). Hence, PJA1 is a downstream effector gene for OTUB2, and its stability could be regulated by OTUB2 deubiquitylation.
Figure 4.
OTUB2 interacted with PJA1, and regulated its stability via deubiquitylation. (a) 19 and 33 genes which potentially interacted with OTUB2 were predicted based on STRING and GeneMANIA databases, respectively, and five potential targets were obtained after the intersection. (b) The protein expression of PJA1 in four paired HCC and the adjacent non-cancerous tissues was detected by Western blot. (c) The interaction between OTUB2 and PJA1 was determined by Co-IP assay (n = 3); cells were transfected with Flag-OTUB2 alone or together with HA-PJA1 expression plasmids, and the interaction between OTUB2 and PJA1 was determined by immunoprecipitation with α-Flag beads followed by immunoblotting with α-HA or α-Flag antibody. (d) Ubiquitylation assay confirmed the deubiquitylation effect of OTUB2 on PJA1 expression (n = 3); Flag-OTUB2, HA-USP38, and His-Ub expression plasmids were co-transfected into 293T cells for 48 h. The cells were treated with MG132 for 6 h before the extraction of total protein, PJA1 was immunoprecipitated with anti-HA agarose beads, and the ubiquitylation of PJA1 was examined by WB using anti-His antibody. (e) The effect of OTBU2 on PJA1 stability was determined by protein half-life assay (n = 3); the PJA1 expression levels in OTBU2 overexpressing cells were detected after treated with 15 mM CHX for 0, 2, 4 and 6 h, respectively. Quantitative data are presented as mean ± SD of three replicates. *p<0.05, compared with the vector group.
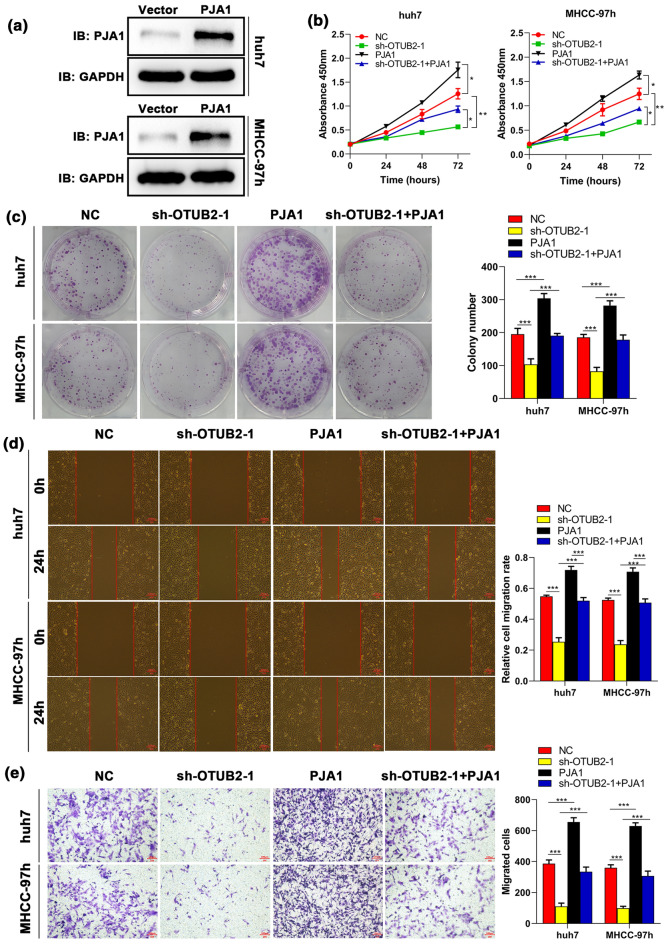
Suppressive Effects Induced by OTUB2 Silencing on Proliferation and Migration of HCC Cells can be Reversed by PJA1
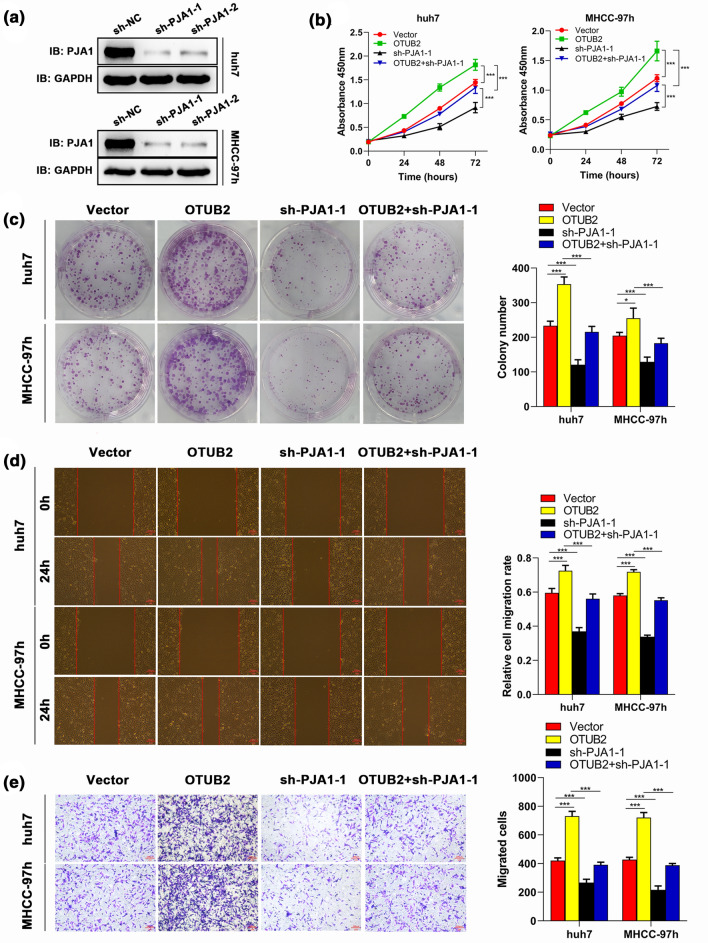
To delineate the role of PJA1 in OTUB2 mediated-HCC progression, we transfected sh-OTUB2 and PJA1 overexpression vectors alone or both into Huh7 and MHCC-97h cell lines, respectively. Western blot analysis demonstrated PJA1 overexpression vectors could effectively up-regulate PJA1 in both Huh7 and MHCC-97h cell lines (Fig. 5a). The results of proliferation analysis showed that compared with the control group, OTUB2 silencing could obviously inhibit cell growth, while this inhibitory effect could be partly neutralized by PJA1 overexpression (Figs. 5b and 5c). Meanwhile, wound healing indicated that suppressed migration of HCC cells induced by sh-OTUB2 was significantly countered by co-transfecting PJA1 overexpression vectors (Fig. 5d). This finding was confirmed by transwell assay (Fig. 5e). Furthermore, it was found that PJA1 knockdown could reverse the promoting role in proliferation and migration of HCC cells mediated by OTUB2 overexpression (Figs. 6a–6e). All the results suggested that PJA1 participates in HCC cell proliferation and migration regulated by OTUB2.
Figure 5.
PJA1 reverses the effect of OTUB2 knockdown on the proliferation and migration of HCC cells. Huh7 and MHCC-97h cells were transfected with sh-OTUB2-1, PJA1 overexpression vectors (PJA1), or both. (a) The transfection efficiency of PJA1 was detected by Western blot (n = 3). After 48 h transfection, cell proliferation was examined by (b) CCK-8 (n = 3) and (c) colony formation (n = 3) assays; cell migration was evaluated by (d) wound healing (n = 3) and (e) transwell (n = 3) assays. Scale bar, 100 μm. Quantitative data are presented as mean ± SD of three replicates. *p < 0.05 and **p < 0.01, compared with the NC group; #p < 0.05, compared with the sh-OTUB2-1 group
Figure 6.
PJA1 silencing blocks the effect of OTUB2 overexpression on proliferation and migration of HCC cells. Huh7 and MHCC-97h cells were transfected with OTUB2 OE, short hairpin RNAs against PJA1 (sh-PJA1), or both. (a) The transfection efficiency of sh-PJA1-1 and sh-PJA1-2 was detected by Western blot (n = 3), and sh-PJA1-1 was selected for the subsequent experiment. After 48 h transfection, cell proliferation was examined by (b) colony formation (n = 3) and (c) CCK-8 (n = 3) assays; cell migration was evaluated by (d) wound healing (n = 3) and (e) transwell (n = 3) assays. Scale bar, 100 μm. Quantitative data are presented as mean ± SD of three replicates. *p < 0.05 and **p < 0.01, compared with the vector group; #p < 0.05, compared with the OTUB2 OE group.
Discussion
Ubiquitylation and deubiquitylation are the most effective post-translational modifications of protein, as they could control the fate of proteins and thus regulate a variety of cellular processes. During the disease development, DUBs could separate ubiquitin from proteins that are highly related to pathologies to reduce the degradation of these proteins, thereby driving the development of diseases. Accumulating studies have revealed that DUBs are involved in tumorigenesis in several ways. Some DUBs, including BAP1, CYLD, and USP8, have been reported as oncogenes or tumor suppressors with intrinsic activities.8,16 Besides, multiple DUBs, such as USP28 and USP9, regulate the protein levels of various oncogenes or tumor suppressors via deubiquitylation.14,19 Therefore, DUBs are emerging as novel targets for the treatment of specific cancers. Kuo et al.18 demonstrated that PR-619, a pan-DUB inhibitor, effectively enhanced the antitumor effect of cisplatin and inhibited tumor growth of cisplatin-resistant bladder urothelial carcinoma. Even though we have known more about the roles of DUBs in cancers, their regulatory mechanisms remain to be further investigated in clinical research.
OTUB2 has recently been shown to participate in the development of many diseases including HCC.13 In our study, it was confirmed that OTUB2 was up-regulated in HCC tissues and indicated a poor prognosis of the patients. A recent study demonstrated that OTUB2 knockdown significantly decreased HCC cell viability.13 Similarly, our functional experiment showed that OTUB2 was involved in HCC cell proliferation. Moreover, our data also revealed that OTUB2 was positively correlated with HCC cell migration in vitro. These data hinted that OTUB2 plays an important role in the malignant behaviors of HCC cells. To explore the downstream regulatory mechanism of OTUB2 in HCC, proteins that potentially interacted with OTUB2 were predicted by the bioinformatics tools. Given that OTUB2 is a DUB that may target protein to stabilize its expression by deubiquitylation, we screened out five target genes (ATXN3, RFFL, PJA1, TRIM55, and TRIM54) that might be up-regulated by OTUB2 by using STRING and GeneMANIA. Previous studies documented that both the mRNA and protein expressions of PJA1 were significantly increased in HCC compared to normal liver.4,30 It has recently been reported that PJA1 can promote cell proliferation via TGF-β/SMAD3/SPTBN1 signaling in HCC.3,25 High expression levels of TGF-β are closely related to tumor invasiveness in advanced-stage or metastatic tumors.26 Consistent with the literature, our study showed that the protein expression of PJA1 in HCC tissues was higher than that in adjacent noncancerous tissues, suggesting the oncogenic role of PJA1 in HCC. It has been reported that OTUB2 acts as a pivotal driver in non-small cell lung cancer tumorigenesis by stabilizing U2AF2 via deubiquitylation.20 Therefore, we proposed that OTUB2 may up-regulate the protein expression of PJA1 to enhance proliferation and migration of HCC cells, thereby promoting HCC progression. The Co-IP assay of PJA1 and OTUB2 in HCC cells confirmed a direct interaction between PJA1 and OTUB2. Several studies demonstrated that DUBs regulated diverse types of cancer via deubiquitylation.9,28 YAP expression has been proven to be associated with cancer metastasis, poor prognosis, chemo-resistance, and relapse.17 A recent study demonstrated that elevated YAP expression in colorectal cancer is attributed to deubiquitylation of USP47 in promoting the protein stability of YAP,27 highlighting DUBs as a therapeutic target for cancer management. In our study, ubiquitylation and protein half-life assay revealed that OTUB2 could reduce the ubiquitylation of PJA1 and prolong the PJA1 half-life, suggesting that OTUB2 promotes the stability of PJA1 via deubiquitylation of PJA1, thereby increasing its expression levels in HCC.
To verify that OTUB2 promotes HCC cell proliferation and migration partly due to up-regulation of PJA1 protein expression, a rescue experiment was further performed. As expected, our study showed that the inhibitory effect induced by OTUB2 silencing on HCC cell proliferation and migration could be rescued by increasing the expression of PJA1. It has been found that PJA1 is an E3 ubiquitin ligase, which exerts the opposite molecular function of DUBs.7,31 It has been reported that PJA1 serves as a tumor suppressor in lung adenocarcinoma by promoting ubiquitin-mediated degradation of FOXR2.23 Work from Chen's lab recently revealed that PJA1 promoted SMAD3 degradation via by ubiquitylation, thereby suppressing TGF‐β signaling in HCC.4 OTUB2 has been reported to participate in HCC progression via suppressing NF-κB pathway.13 TGF-β and NF-κB signal pathway cross-talk has been widely reported in various studies on cancer development. Therefore, the role of the OTUB2/PJA1 axis in HCC might be attributed to TGF‐β/NF-κB signaling pathways. Furthermore, whether PJA1 is responsible for OTUB2 ubiquitylation and whether they form a feedback loop in HCC remain to be further explored in future studies.
Taken together, our study implied that OTUB2 contributed to the progression of HCC by promoting HCC cell proliferation and metastasis via PJA1 deubiquitylation. Our finding hence provides a clue of OTUB2 as a novel target for the treatment of HCC. Nevertheless, there are also certain shortcomings in this study. First, our findings would be more representative if more types of HCC cell lines were used. Besides, in vivo experiments are required to validate the present findings. Further, the present study is preliminary in nature, and in-depth understanding of the molecular mechanisms behind OTUB2/PJA1 axis regulating HCC is still required in future.
Collectively, our study revealed that OTUB2 expression was up-regulated in HCC tissues and closely related to patient prognosis. The OTUB2 promoted HCC cell proliferation and migration by deubiquitylation of PJA1. Our findings suggested that OTUB2 may a potential target for the treatment of HCC.
Acknowledgments
None.
Funding
No funding was received.
Conflict of interest
Gang Hu, Jianwu Yang, Hongwen Zhang, Zhen Huang and Heming Yang declare that they have no conflicts of interest.
Ethical Approval
The procedure of this research was reviewed and approved by Strategic Support Force Characteristic Medical Center.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gang Hu, Email: doctorhuhu@126.com.
Jianwu Yang, Email: yangjianwu306@sohu.com.
Hongwen Zhang, Email: 875217989@qq.com.
Zhen Huang, Email: hz8669@sohu.com.
Heming Yang, Email: yangheming051@163.com.
References
- 1.Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers (Basel) 2020;12:1579. doi: 10.3390/cancers12061579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan DW, Chan CY, Yam JW, Ching YP, Ng IO. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology. 2006;131:1218–1227. doi: 10.1053/j.gastro.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Gingold JA. Dysregulated PJA1-TGF-β signaling in cancer stem cell-associated liver cancers. Oncoscience. 2020;7:88–95. doi: 10.18632/oncoscience.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Mitra A, Li S, Song S, Nguyen BN, Chen JS, Shin JH, Gough NR, Lin P, Obias V, He AR, Yao Z, Malta TM, Noushmehr H, Latham PS, Su X, Rashid A, Mishra B, Wu RC, Mishra L. Targeting the E3 ubiquitin ligase PJA1 enhances tumor-suppressing TGFβ signaling. Cancer Res. 2020;80:1819–1832. doi: 10.1158/0008-5472.CAN-19-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28:591–605. doi: 10.1038/s41418-020-00708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consalvi S, Brancaccio A, Dall'Agnese A, Puri PL, Palacios D. Praja1 E3 ubiquitin ligase promotes skeletal myogenesis through degradation of EZH2 upon p38α activation. Nat. Commun. 2017;8:13956. doi: 10.1038/ncomms13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, Wu J, Phu L, Katavolos P, LaFave LM, Abdel-Wahab O, Modrusan Z, Seshagiri S, Dong K, Lin Z, Balazs M, Suriben R, Newton K, Hymowitz S, Garcia-Manero G, Martin F, Levine RL, Dixit VM. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 10.Franke FC, Müller J, Abal M, Medina ED, Nitsche U, Weidmann H, Chardonnet S, Ninio E, Janssen KP. The tumor suppressor SASH1 interacts with the signal adaptor CRKL to inhibit epithelial–mesenchymal transition and metastasis in colorectal cancer. Cell Mol. Gastroenterol. Hepatol. 2019;7:33–53. doi: 10.1016/j.jcmgh.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 13.Gu ZL, Huang J, Zhen LL. Knockdown of otubain 2 inhibits liver cancer cell growth by suppressing NF-κB signaling. Kaohsiung J. Med. Sci. 2020;36:399–404. doi: 10.1002/kjm2.12187. [DOI] [PubMed] [Google Scholar]
- 14.Haq S, Das S, Kim DH, Chandrasekaran AP, Hong SH, Kim KS, Ramakrishna S. The stability and oncogenic function of LIN28A are regulated by USP28. Biochim Biophys. Acta Mol. Basis Dis. 2019;1865:599–610. doi: 10.1016/j.bbadis.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo KL, Liu SH, Lin WC, Chow PM, Chang YW, Yang SP, Shi CS, Hsu CH, Liao SM, Chang HC, Huang KH. The deubiquitinating enzyme inhibitor PR-619 enhances the cytotoxicity of cisplatin via the suppression of anti-apoptotic Bcl-2 protein: in vitro and in vivo study. Cells. 2019;8:1268. doi: 10.3390/cells8101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Cheng Z, Raghothama C, Cui Z, Liu K, Li X, Jiang C, Jiang W, Tan M, Ni X, Pandey A, Liu JO, Dang Y. USP9X controls translation efficiency via deubiquitination of eukaryotic translation initiation factor 4A1. Nucleic Acids Res. 2018;46:823–839. doi: 10.1093/nar/gkx1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, Geng Q, Pan H, Yan M, Yao M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–195. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z, Ye X, Cheng Y, Li F, Shou F, Wang G. E3 ubiquitin ligase PJA1 regulates lung adenocarcinoma apoptosis and invasion through promoting FOXR2 degradation. Biochem. Biophys. Res. Commun. 2021;556:106–113. doi: 10.1016/j.bbrc.2021.03.137. [DOI] [PubMed] [Google Scholar]
- 24.Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–788. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohshiro K, Chen J, Srivastav J, Mishra L, Mishra B. Alterations in TGF-β signaling leads to high HMGA2 levels potentially through modulation of PJA1/SMAD3 in HCC cells. Genes Cancer. 2020;11:43–52. doi: 10.18632/genesandcancer.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padua D, Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 27.Pan B, Yang Y, Li J, Wang Y, Fang C, Yu FX, Xu Y. USP47-mediated deubiquitination and stabilization of YAP contributes to the progression of colorectal cancer. Protein Cell. 2020;11:138–143. doi: 10.1007/s13238-019-00674-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto-Fernandez A, Kessler BM. DUBbing cancer: deubiquitylating enzymes involved in epigenetics, DNA damage and the cell cycle as therapeutic targets. Front. Genet. 2016;7:133. doi: 10.3389/fgene.2016.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J. Biol. Chem. 2002;277:22541–22546. doi: 10.1074/jbc.M109728200. [DOI] [PubMed] [Google Scholar]
- 32.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 33.Sivakumar D, Kumar V, Naumann M, Stein M. Activation and selectivity of OTUB-1 and OTUB-2 deubiquitinylases. J. Biol. Chem. 2020;295:6972–6982. doi: 10.1074/jbc.RA120.013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 37.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Du J, Wang S, Shao L, Jin K, Li F, Wei B, Ding W, Fu P, van Dam H, Wang A, Jin J, Ding C, Yang B, Zheng M, Feng XH, Guan KL, Zhang L. OTUB2 promotes cancer metastasis via hippo-independent activation of YAP and TAZ. Mol. Cell. 2019;73:7–21.e27. doi: 10.1016/j.molcel.2018.10.030. [DOI] [PubMed] [Google Scholar]