Abstract
Introduction
This study aimed to develop and validate an interpretable machine-learning model based on clinical features for early predicting in-hospital mortality in critically ill patients with sepsis.
Methods
We enrolled all patients with sepsis in the Medical Information Mart for Intensive Care IV (MIMIC-IV, v.1.0) database from 2008 to 2019. Lasso regression was used for feature selection. Seven machine-learning methods were applied to develop the models. The best model was selected based on its accuracy and area under curve (AUC) in the validation cohort. Furthermore, we employed the SHapley Additive exPlanations (SHAP) method to illustrate the effects of the features attributed to the model, and to analyze how the individual features affect the output of the model, and to visualize the Shapley value for a single individual.
Results
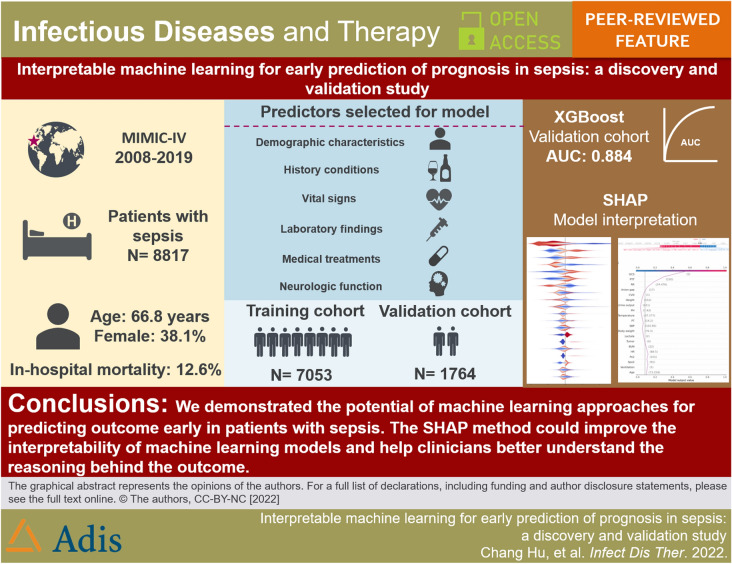
In total, 8,817 patients with sepsis were eligible for participation, the median age was 66.8 years (IQR, 55.9–77.1 years), and 3361 of 8817 participants (38.1%) were women. After selection, 25 of a total 57 clinical parameters collected on day 1 after ICU admission remained associated with prognosis and were used for developing the machine-learning models. Among seven constructed models, the eXtreme Gradient Boosting (XGBoost) model achieved the best performance with an AUC of 0.884 and an accuracy of 89.5% in the validation cohort. Feature importance analysis showed that Glasgow Coma Scale (GCS) score, blood urea nitrogen, respiratory rate, urine output, and age were the top 5 features of the XGBoost model with the greatest impact. Furthermore, SHAP force analysis illustrated how the constructed model visualized the individualized prediction of death.
Conclusions
We have demonstrated the potential of machine-learning approaches for predicting outcome early in patients with sepsis. The SHAP method could improve the interpretability of machine-learning models and help clinicians better understand the reasoning behind the outcome.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-022-00628-6.
Keywords: Machine learning, Algorithm, Sepsis, Critically ill, Mortality
Key Summary Points
| Why carry out this study? |
| Sepsis is a common cause for hospitalization associated with a high mortality rate and morbidity. |
| Early detection of septic patients with potential for acute deterioration has been proven effective in improving clinical outcomes. However, relatively few constructed models have been applied for practical use due to the black box in machine learning. |
| This study aimed to develop and validate an interpretable machine-learning model based on clinical features for early predicting in-hospital mortality in critically ill patients with sepsis by using the SHapley Additive exPlanations (SHAP) method. |
| What was learned from the study? |
| We demonstrated the potential of machine-learning approaches for early predicting prognosis in patients with sepsis. |
| The SHAP method could improve the interpretability of machine-learning models and help clinicians better understand the reasoning behind the outcome. |
Introduction
Sepsis is a common dysregulated systemic response to infection and usually results in significant morbidity and mortality [1, 2]. A cross-section survey study from China showed that sepsis accounted for 20.6% of all cases admitted to intensive care unit (ICU), and around 35.5% of whom were dead at 90 days after ICU admission in 2015 [3]. Despite advances in management and therapy, sepsis is still recognized as a global health priority [4].
Recent evidence suggested that early detection of critically ill patients with potential for acute deterioration has been proven effective at improving clinical outcomes [5]. It is generally accepted that machine-learning (ML) approaches can help identify septic patients at high risk for in-hospital mortality [6–9]. Although numerous ML methods have demonstrated a remarkable performance in medicine, surprisingly, few constructed models have been used for clinical practice [10]. A major barrier to this progress is the black box in ML [11, 12]. Therefore, an understanding of why and how decisions are made by the algorithms are crucial to enable the algorithm to be used in clinical practice. However, many of the current establishment of prognosis models pay particular attention to accuracy, but lack transparency for sepsis [6, 13]. These will limit the model’s application in clinical use. Fortunately, a new interpretation model in ML, called the SHapley Additive exPlanations (SHAP), was first articulated by Lundberg and Lee to explain individual predictions for both kernel-based approaches and tree-based models [14]. Additionally, this explainable ML has been successfully applied to a variety of medical fields, such as predictions for the prevention of hypoxemia during surgery [15] and precision medicine in acute myeloid leukemia [16]. However, this advanced method for model interpretation has not yet been used in sepsis mortality prediction.
Hence, the objective of this study was to develop and validate seven ML models based on clinical features for early prediction of in-hospital mortality in critically ill patients with sepsis. The best performing model was selected for further mortality prediction and interpretability by using the SHAP method.
Methods
Study Design
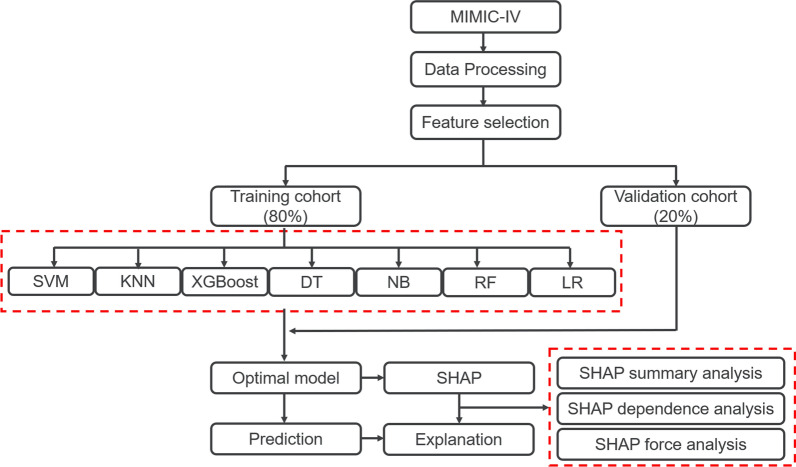
In this retrospective modeling study, the development, validation and interpretation of the models proceeded in three steps. First, we developed the models using seven ML methods. Next, we evaluated and compared the model performance of the ML methods in a validation cohort. Finally, we interpreted why and how decisions are made in an optimal algorithm by using the SHAP method.
Data
Data for this study were collected using the Medical Information Mart for Intensive Care IV (MIMIC-IV, v.1.0) database. MIMIC-IV is a large, single-center, freely accessible database with clinical data from patients admitted to an ICU at a large tertiary care hospital in the US between 2008 and 2019, and contains over 70,000 ICU admissions for adult patients (aged 18 years old or older) [17]. One member in our team has finished the Collaborative Institutional Training Initiative (CITI) examination (Record ID: 47460147 for CH) and has been approved to access the MIMIC-IV database. This project was approved by the Institutional Review Boards of Beth Israel Deaconess Medical Center (Boston, MA, USA) and the Massachusetts Institute of Technology (Cambridge, MA, USA). Requirements for individual patient consent and an ethical approval statement were waived because the project did not impact clinical care and all patients in database were deidentified [18]. Additionally, our study complies with the Declaration of Helsinki and was performed according to Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis guidelines [19].
Participants
We enrolled all adult patients (aged 18 years old and older) with sepsis in the MIMIC-IV database. Patients were diagnosed with sepsis using the Sepsis-3 criteria [2] in the first 24 h of ICU admission: (1) with confirmed or suspected infection, and (2) with 2 or more Sequential Organ Failure Assessment (SOFA) [20] points (Supplementary Figure S1). Exclusion criteria for participation in the study were: (1) patients had multiple ICU admissions (only the first admission was included for analysis), and (2) patients with ICU length of stay less than 24 h. The primary clinical outcome was in-hospital mortality.
Variable Selection
Following the Deshmukh et al. [21] procedure for variable selection, we selected 57 candidate variables based on their association with the outcome. These included demographic variables, comorbidities, vital signs, laboratory findings, medical treatments, urine output, and the Glasgow Coma Scale (GCS) score. Demographic variables were included for age, gender, body weight, and height. Comorbidities were included for hypertension, diabetes, congestive heart failure, cerebrovascular disease, chronic pulmonary disease, liver disease, renal disease, tumor, and acquired immune deficiency syndrome. For vital signs, we selected the mean values in the ICU for the following variables: heart rate, systolic blood pressure, diastolic blood pressure, mean arterial pressure, respiratory rate, and body temperature. For laboratory findings, we selected the maximum value for the following variables: blood glucose, lactate, pH, PCO2, base excess, white blood cell, anion gap, bicarbonate, blood urea nitrogen, serum calcium, serum chloride, serum creatinine, serum sodium, serum potassium, serum fibrinogen, international normalized ratio, prothrombin time, partial thromboplastin time, alanine aminotransferase, alkaline phosphatase, aspartate aminotransferase, total bilirubin, amylase, creative phosphokinase, creatine kinase MB, and lactate dehydrogenase. The minimum value was selected for PaO2, SPO2, PaO2/FiO2 ratio, hematocrit, hemoglobin, platelets, and albumin. Medical treatments were included for use of ventilation, antibiotic, and vasopressor. Finally, the cumulative urine output and the minimum value of GCS score were also included (Supplementary Table S1). For each variable above, we extracted the value recorded within the first 24 h of ICU admission. Features with > 30% missing were dropped (Supplementary Figure S3), and multiple imputation by chained equations (MICE) was used for missing value imputation in the remaining feature space. Considering the large number of features still present in the cohort, least absolute shrinkage and selection operator (LASSO) regression, which could effectively prevent overfitting, was utilized in variable selection. The list of input variables used in the model development are presented in Supplementary Table S2.
Statistical Analysis
Categorical data are presented as counts and percentages. Between-group differences are compared by the χ2 test or the Fisher test. Continuous data are presented as means with standard deviations (SDs) or medians with interquartile ranges (IQRs) depending on the distribution of the data. We used analysis of variance or the Mann–Whitney U test for continuous data to compare the differences between groups.
Missing data with < 30% in each feature were handled by multiple imputation with ‘mice’ package in R. LASSO regression was used for feature selection. After that, the population was divided randomly into two parts, with 80% as the derivation data and 20% as the validation data. We employed a median absolute deviation based robust similarity metric to eliminate the outliers. Additionally, the continuous variables were placed on a z scale with a mean of 0 and SD of 1 to estimate the model parameters. Seven common ML methods [Support Vector Machine (radial bias function) (SVM), k-Nearest Neighbors (KNN), eXtreme Gradient Boosting (XGBoost), Decision Tree (DT), Naive Bayes (NB), Random Forest (RF) and logistic regression (LR)] were applied to develop the models in training cohort. The quantitative performance of models was compared with area under curve (AUC) and accuracy in validation cohort. The optimized model with the best performance for mortality prediction in validation cohort defined as the final model. The illness severity scoring systems including SOFA, SAPS-II, and qSOFA were assessed to predict the risk of death and were compared with our final model in validation cohort.
Furthermore, we employed SHAP method to improve the interpretability for the final model. Firstly, the SHAP summary plot was used to illustrate the effects of the features attributed to the model. Secondly, the SHAP dependence plot was utilized to analyze the importance of individual features affects the output of the model. Finally, the SHAP force plot was employed to visualize the impact of key features on the final model in individual patients.
All analyses were carried out using Python (v.3.6.6) and R (v. 3.6.1, R Foundation for Statistical Computing). Two-tailed P values < 0.05 were labeled as significant.
Results
Participants
Of 76,540 critically ill patients assessed from the MIMIC-IV database, 12,292 with sepsis were screened. Of these, 2657 were excluded because of multiple ICU admission (only the first ICU admission was included for analysis), and 818 were excluded due to their length of ICU stay being less than 24 h. Finally, a total of 8817 patients were eligible for participation (Supplementary Figure S2).
Baseline characteristics of the eligible participants are shown in Table 1. The median age was 66.8 years (IQR, 55.9–77.1 years), and 3361 of 8817 participants (38.1%) were women. Hypertension was the most common (4256 of 8817, 48.3%) comorbidities, followed by diabetes (2528 of 8817, 28.7%) and congestive heart failure (2365 of 8817, 26.8%). Those non-survivors were more likely to be older [70.2 (58.5–79.9) vs. 66.4 (55.5–76.6), P < 0.001], have a higher proportion of medical treatments, such as ventilation use (68.5% vs. 58.2%, P < 0.001), vasopressor use (51.3% vs. 38.2%, P < 0.001), and have higher severity of illness scores (P < 0.001), compared with survivors.
Table 1.
Demographic and clinical characteristics at baseline
| Characteristics | Survivors (n = 7710) | Non-survivors (n = 1107) | P value |
|---|---|---|---|
| Demographic | |||
| Age, year | 66.4 (55.5–76.6) | 70.2 (58.5–79.9) | < 0.001 |
| Gender | 0.001 | ||
| Male, n (%) | 4819 (62.5) | 637 (57.5) | |
| Female, n (%) | 2891 (37.5) | 470 (42.5) | |
| Weight, kg | 81 (69–95) | 80 (67–95) | 0.003 |
| Height, cm | 170 (163–178) | 168 (160–178) | < 0.001 |
| Comorbidities | |||
| Hypertension, n (%) | 3815 (49.5) | 441 (39.8) | < 0.001 |
| Diabetes, n (%) | 2208 (28.6) | 320 (28.9) | 0.853 |
| Congestive heart failure, n (%) | 1968 (25.5) | 397 (35.9) | < 0.001 |
| Cerebrovascular disease, n (%) | 996 (12.9) | 238 (21.5) | < 0.001 |
| Chronic pulmonary disease, n (%) | 1875 (24.3) | 325 (29.4) | < 0.001 |
| Liver disease, n (%) | 794 (10.3) | 292 (26.4) | < 0.001 |
| Renal disease, n (%) | 1278 (16.6) | 276 (24.9) | < 0.001 |
| Tumor, n (%) | 883 (11.5) | 215 (19.6) | < 0.001 |
| Acquired immune deficiency syndrome, n (%) | 21 (0.3) | 1 (0.1) | 0.512 |
| Vital signs on day 1 | |||
| Heart rate, bpm | 83 (75–94) | 89 (76–102) | < 0.001 |
| Systolic blood pressure, mmHg | 115 (107–124) | 111 (102–124) | < 0.001 |
| Diastolic blood pressure, mmHg | 60 (55–66) | 60 (53–67) | 0.259 |
| Mean arterial pressure, mmHg | 76 (71–82) | 75 (69–83) | < 0.001 |
| Respiratory rate | 18 (16–21) | 21 (18–24) | < 0.001 |
| Body temperature, °C | 36.9 (36.6–37.2) | 36.8 (36.4–37.2) | < 0.001 |
| SPO2, % | 93 (91–95) | 92 (88–95) | < 0.001 |
| Laboratory findings on day 1 | |||
| Blood glucose, mg/dL | 170 (140–206) | 188 (146–257) | < 0.001 |
| Lactate, mmol/L | 2.2 (1.5–3.2) | 2.9 (1.7–5.7) | < 0.001 |
| pH | 7.43 (7.39–7.47) | 7.41 (7.36–7.46) | < 0.001 |
| PO2, mmHg | 88 (63–122) | 63 (42–91) | < 0.001 |
| PCO2, mmHg | 46 (41–52) | 47 (39–56) | 0.293 |
| PaO2/FiO2 ratio | 192 (118–276) | 129 (77–223) | < 0.001 |
| Base excess | 1 (0–3) | 0 (− 3 to 2) | < 0.001 |
| Hematocrit, % | 29 (25–34) | 30 (25–34) | 0.030 |
| Hemoglobin, g/dL | 9.8 (8.4–11.3) | 9.6 (8.2–11.4) | 0.123 |
| Platelets, × 103/µL | 157 (113–216) | 154 (88–227) | 0.001 |
| White blood cell, × 103/µL | 13.8 (10.4–18.2) | 16.0 (11.3–21.5) | < 0.001 |
| Anion gap | 14 (12–17) | 18 (15–22) | < 0.001 |
| Bicarbonate, mmol/L | 25 (23–27) | 23 (20–26) | < 0.001 |
| Blood urea nitrogen, mg/dL | 19 (14–28) | 32 (20–50) | < 0.001 |
| Serum calcium, mmol/L | 8.4 (8.0–8.9) | 8.5 (8.0–9.0) | < 0.001 |
| Serum chloride, mmol/L | 108 (104–111) | 106 (101–111) | < 0.001 |
| Serum creatinine, mg/dL | 1.0 (0.8–1.4) | 1.6 (1.0–2.6) | < 0.001 |
| Serum sodium, mmol/L | 140 (138–142) | 141 (137–144) | 0.001 |
| Serum potassium, mmol/L | 4.4 (4.1–4.8) | 4.6 (4.1–5.2) | < 0.001 |
| International normalized ratio | 1.3 (1.2–1.5) | 1.5 (1.2–2.1) | < 0.001 |
| Prothrombin time, s | 14.6 (12.9–16.6) | 16.2 (13.5–22.8) | < 0.001 |
| Partial thromboplastin time, s | 32.4 (28.3–40.8) | 38.9 (30.3–63.0) | < 0.001 |
| Medical treatments | |||
| Antibiotic, n (%) | 5543 (71.9) | 764 (69.0) | 0.047 |
| Ventilation, n (%) | 4484 (58.2) | 758 (68.5) | < 0.001 |
| Vasopressor, n (%) | 2942 (38.2) | 568 (51.3) | < 0.001 |
| Urine output on day 1, mL | 1763 (1195–2526) | 1035 (464–1797) | < 0.001 |
| Severity of illness scores | |||
| GCS | 14 (10–15) | 9 (3–14) | < 0.001 |
| SOFA | 5 (3–8) | 10 (6–13) | < 0.001 |
| OASIS | 34 (28–40) | 44 (37–50) | < 0.001 |
| SAPS-II | 35 (28–44) | 50 (39–61) | < 0.001 |
Data are reported as no. (%) or median (IQR)
GCS Glasgow Coma Scale, SOFA Sequential Organ Failure Assessment, OASIS Oxford Acute Severity of Illness Score, SAPS-II Simplified Acute Physiology Score II
Model Development and Validation
A total of 57 clinical variables collected during the first 24 h after ICU admission, 10 of which with > 30% missing were dropped (Supplementary Figure S3). MICE was used to impute missing data in the remaining feature space. LASSO regression was employed to identify signature variables for hospital mortality in patients with sepsis. Ultimately, 25 out of 47 clinical parameters remained associated with prognosis, and these results are presented in Supplementary Figure S4 and Table S2. We have constructed seven ML binary classifiers in predicting the risk of death for sepsis: eXtreme Gradient Boosting (XGBoost), Random Forest (RF), Naive Bayes (NB), Logistic Regression (LR), Support Vector Machine (radial bias function) (SVM), k-Nearest Neighbors (KNN), and Decision Tree (DT) (Fig. 1). Figure 2A describes the performance of these predictive models and shows that the XGBoost model could provide a relatively better model fit performance with an area under the curve (AUC) of 0.884 and an accuracy of 0.895 in the validation cohort, compared with the other ML models (AUC: RF, 0.882; NB, 0.856; LR, 0.845; SVM, 0.763; KNN, 0.651; DT, 0.655; Accuracy: RF, 0.891; NB, 0.862; LR, 0.890; SVM, 0.872; KNN, 0.868; DT, 0.842) (Table 2). Additionally, Fig. 2B shows that the AUCs of the analyzed scores and the comparison of the curves between each other. The XGBoost model was superior to other severity of illness scores (AUC: XGBoost, 0.884; SOFA, 0.770; SAPS-II, 0.766; qSOFA, 0.647). The calibration curve for the XGBoost model showed that the predicted risk is in good agreement with the actual risk. The predicted value of the model is close to the actual probability of the outcome (Supplementary Figure S5). Therefore, XGBoost was selected for further prediction in this study.
Fig. 1.
Flowchart of this study. MIMIC-IV Medical Information Mart for Intensive Care IV
Fig. 2.
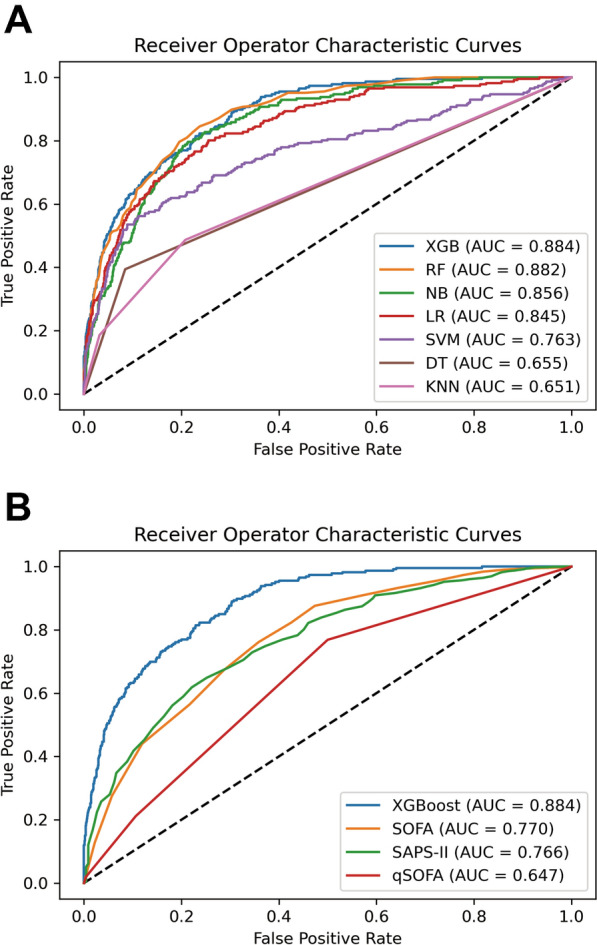
Receiver operator characteristic (ROC) curves for the ML models and the traditional severity of illness scores to predict in-hospital mortality (validation cohort). A ROC curves for the seven ML models to predict in-hospital mortality; B ROC curves for the traditional severity of illness scores to predict in-hospital mortality. XGB eXtreme Gradient Boosting, RF Random Forest, SVM Support Vector Machine (radial bias function), LR Logistic Regression, NB Naive Bayes, KNN k-Nearest Neighbors, DT Decision Tree, OASIS Oxford Acute Severity of Illness Score, SAPS-II Simplified Acute Physiology Score II, AUC area under the curve
Table 2.
Performances of the seven machine learning models for predicting in-hospital mortality
| ML | Accuracy | AUC |
|---|---|---|
| XGBoost | ||
| Training set | 1.000 | 1.000 |
| Validation set | 0.895 | 0.884 |
| RF | ||
| Training set | 1.000 | 1.000 |
| Validation set | 0.891 | 0.882 |
| SVM | ||
| Training set | 0.875 | 0.747 |
| Validation set | 0.872 | 0.763 |
| LR | ||
| Training set | 0.882 | 0.833 |
| Validation set | 0.890 | 0.845 |
| NB | ||
| Training set | 0.856 | 0.836 |
| Validation set | 0.862 | 0.856 |
| KNN | ||
| Training set | 0.904 | 0.940 |
| Validation set | 0.868 | 0.651 |
| DT | ||
| Training set | 1.000 | 1.000 |
| Validation set | 0.842 | 0.655 |
ML machine learning, XGBoost eXtreme Gradient Boosting, RF Random Forest, SVM Support Vector Machine (radial bias function), LR Logistic Regression, NB Naive Bayes, KNN k-Nearest Neighbors, DT Decision Tree, AUC the area under curve
Model Explanation
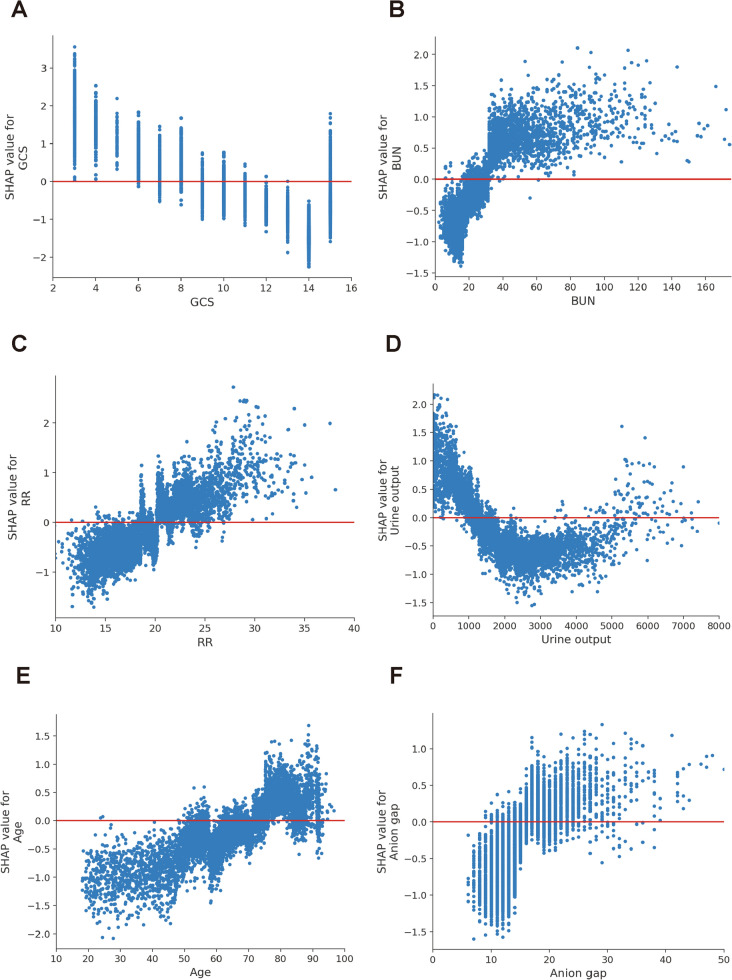
We calculated the feature importance using the SHAP value for XGBoost, which had the greatest discriminatory ability in the validation cohort. Figure 3A presents the top 20 clinical features according to the average absolute SHAP value. Figure 3B provides an overview of the (positive or negative) impact of factors on the XGBoost model. The features specific to death included a lower GCS score, urine output, PO2, and systolic blood pressure, and each of these had a negative SHAP value and therefore drove the prediction toward mortality. Additionally, elevated blood urea nitrogen, respiratory rate, age, and anion gap also supported a prediction of mortality. The impact of top 6 factors on the XGBoost model’s mortality risk prediction were further explored using the the SHAP dependency plot. Figure 4 shows that a higher level of blood urea nitrogen, higher respiratory rate, and older and higher anion gap were associated with a significant increase in mortality risk. Conversely, as the GCS score increased, the risk of mortality decreased. Interestingly, we found that there was a U-shaped curve association between urine output and mortality (Fig. 4D).
Fig. 3.
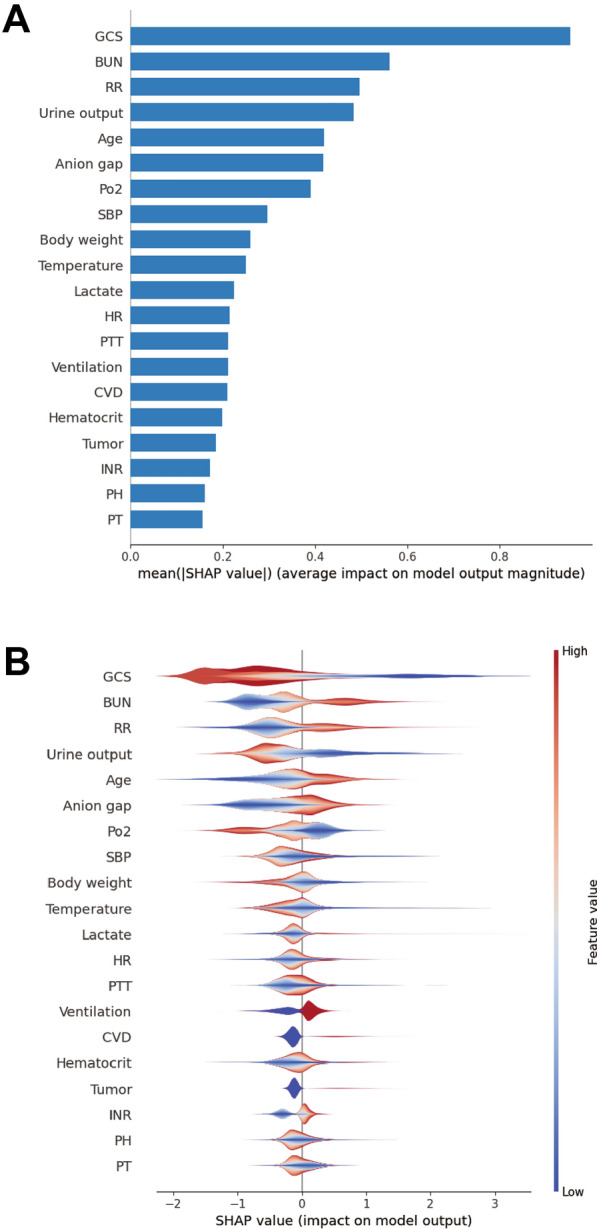
SHAP summary plot for the top 20 clinical features contributing to the XGBoost model. A SHAP feature importance measured as the mean absolute Shapley values. This matrix plot depicts the importance of each covariate in the development of the final predictive model. B The attributes of the features in the model. The position on the y-axis is determined by the feature and on the x-axis by the Shapley value. The color represents the value of the feature from low to high. SHAP SHapley Additive explanation, XGBoost eXtreme Gradient Boosting. GCS Glasgow Coma Scale, BUN blood urea nitrogen, RR respiratory rate, HR heart rate, PTT partial thromboplastin time, CVD cerebrovascular disease, MAP mean arterial pressure, WBC white blood cell, PT prothrombin time
Fig. 4.
SHAP dependency plot for the top 6 clinical features contributing to XGBoost model. A GCS, B BUN, C RR, D urine output, E age, F anion gap. SHAP values for specific features exceed zero, representing an increased risk of death. GCS Glasgow Coma Scale, BUN blood urea nitrogen, RR respiratory rate, SHAP SHapley Additive explanation, XGBoost eXtreme Gradient Boosting
Model Application
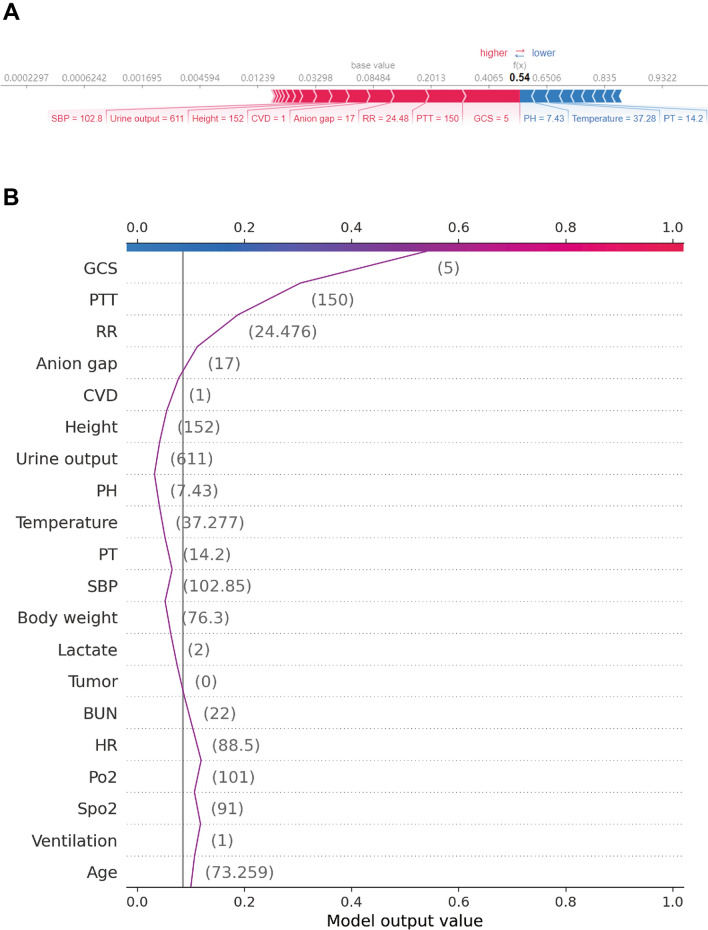
To further explore the contribution of the features on a certain individual patient and clinical application for the XGBoost model, we randomly selected one patient from the validation cohort. Figure 5 provides a visual interpretation of the XGBoost model. The constructed model predicted the probability of death in this patient to be 54% (Fig. 5A). The result suggested that the GCS score of 5, partial thromboplastin time value of 150, and respiratory rate of 25/min were the top 3 contributors to this prediction. Figure 5B presents the specific impact of how these factors contributed to the prediction.
Fig. 5.
SHAP force plot for explaining of individual’s prediction results in the validation cohort. Screenshot of the death prediction in patients with sepsis. A, B Model predictions by randomly drawing a single sample from the validation cohort. Redder sample points indicate that the value of the feature is larger, and bluer sample points indicate that the value of the feature is smaller. GCS Glasgow Coma Scale, BUN blood urea nitrogen, RR respiratory rate, CVD cerebrovascular disease
Discussion
Seven ML models have been developed and validated, based on 25 clinical variables collected in the first 24 h after ICU admission, to predict the prognosis for patients with sepsis admitted to ICU. The XGBoost model demonstrated the best performance. Feature importance analysis showed that the GCS score, blood urea nitrogen, respiratory rate, urine output, age, and anion gap were the top 6 features of the XGBoost model, with the greatest impact in predicting mortality. Furthermore, we have described and captured how these features affected the XGBoost model. Finally, we used a SHAP force analysis to illustrate how the XGBoost model visualized the individualized prediction of death for patients with sepsis. These findings should make an important contribution to a deeper understanding of ML in patients with sepsis.
Recently, ML approaches have become a powerful tool to distinguish and predict prognoses in patients with sepsis. Several previous studies have constructed ML models with good performance to predict mortality in sepsis populations admitted to the ICU, compared with traditional regression model [6, 9, 13, 22]. For instance, Yao et al. [6] revealed that the XGBoost model displayed a better performance in discrimination and calibration than that of the conventional logistic regression model and SOFA score to early recognize septic patients who are at high risk of death (AUC: 0.835 vs. 0.737 and 0.621, respectively). Hou et al. [13] concluded that ML-based XGBoost possessed a significant predictive value to estimate the risk of 30-day mortality in sepsis patients, compared with a traditional logistic regression model (AUC were 0.857 and 0.819; respectively). Kong et al. [22] developed four ML methods to predict in-hospital mortality of sepsis patients, and found that a gradient boosting machine showed the best performance in discrimination (AUC: 0.845). In accordance with the previous results, our study demonstrated that ML methods were more accurate in predicting death than logistic regression among patients with sepsis. Meanwhile, we compared the optimized model with other existing severity of illness scores commonly applied to predict in-hospital mortality. These results demonstrated that the XGBoost model could effectively improve the prediction of in-hospital mortality in patients with sepsis.
However, even the success of algorithms in this field, one of the current challenges is opening the black box of ML [12]. In other words, accuracy is important but not sufficient to engender trust. Before an algorithm can be used in clinical practice, care providers need to understand why and how decisions are made by the algorithm. SHAP is a game-theoretic technique developed by Lundberg and Lee [15] to successfully overcome the black box nature of ML by providing consistent interpretability. Nevertheless, none of the previous models have utilized SHAP to interpret the results of ML algorithms in predicting the death of patients with sepsis. We first used SHAP in the XGBoost model to achieve the best predictive effect and interpretability. Additionally, individual explanations constructed by SHAP force analysis can help doctors understand why the model makes specific recommendations for high-risk decisions. All of these have contributed to a better understanding of the decision-making process of the predictive model for users.
To further confirm how input factors contribute to the model, we calculated SHAP feature importance and feature effects. The importance of variables showed that the GCS score, blood urea nitrogen, respiratory rate, urine output, and age were the most important input parameters that contribute to the predicted risk of death in patients with sepsis. The GCS score, as a level of consciousness scale ranging from 3 to 15, could obviously indicate neurological deterioration (with lower scores indicating worse condition). Previously constructed models had confirmed that the GCS score was associated with the risk of mortality in the sepsis group [9, 23]. Consistently, in the current study, the GCS score accounted for the biggest weight in the XGBoost model, and indicated that it was the most important predictor to predict the mortality of patients with sepsis. Our study also found that the blood urea nitrogen was closely related to mortality in sepsis. Blood urea nitrogen is excreted mainly by the kidney. The elevated blood urea nitrogen level indicated renal lesions, while the latter has been determined to cause adverse outcomes [24]. The respiratory rate was another important effector in predicting outcomes for sepsis. To date, several studies have identified a link between the respiratory rate and worse outcomes [25]. Additionally, we found that urine output was associated with better outcomes, which was consistent with those of previous studies [26]. In the present study, age was shown to be a predictor of mortality in critically ill patients with sepsis. An observational study in Australia and New Zealand also showed that the mortality of sepsis was less than 5% in the absence of comorbidities and older age [27]. Finally, we explored the association between the features’ dynamic changes and outcomes in patients with sepsis. Interestingly, we found that there was a non-linear, U-shaped association between urine output and mortality in patients with sepsis. In brief, the values of urine output between approximately 2000 and 4000 ml/L on day 1 after ICU admission were associated with the lowest mortality in patients with sepsis. This finding was consistent with the observation from a previous study that there may exist an optimal range of urine output on day 1 after ICU admission for critically ill patients with sepsis [28].
However, there were also some limitations in this study. Firstly, the current research was a single-center retrospective modeling study using the MIMIC-IV database, which restricted us from identifying the causal relationship between features and outcomes. Thus, further prospective randomized clinical trials are required to validate the effectiveness for our model. Secondly, many unmeasured confounders may contribute to the impact on the prognosis of death for septic patients, such as racial and treatment strategies. We may therefore have neglected some important features used for the XGBoost model due to the limitation of MIMIC-IV. Thirdly, the imputation and feature selection were performed before the split of the training and validation sets, which may have influenced the final models. Fourthly, our study lacked external validation by an independent cohort from other regions or other countries, thus further research is needed to verify our results. Finally, our modeling study only recruited adults, and the predictive effectiveness of the XGBoost model on the prognosis for children with sepsis remains unclear. This conclusion should be interpreted with caution and needs more evidence to confirm it in the near future.
Conclusions
We have demonstrated the potential of ML approaches for predicting outcomes early in patients with sepsis. The SHAP method could improve the interpretability of ML models and help clinicians better understand the reasoning behind the outcomes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
This work and the journal’s Rapid Service Fee were funded by the Chinese Medical Information and Big Data Association (Bo Hu, No. Z-2019–1-003), and by Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (Bo Hu, No. ZNJC202011).
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
Conceptualization: Chang Hu, Lu Li, Weipeng Huang; Methodology: Chang Hu, Tong Wu; Formal analysis and investigation: Qiancheng Xu; Writing: Chang Hu, Juan Liu, Bo Hu. Funding acquisition: Bo Hu; Supervision: Juan Liu, Bo Hu. All authors read and approved the final manuscript.
Disclosures
Chang Hu, Lu Li, Weipeng Huang, Tong Wu, Qiancheng Xu, Juan Liu and Bo Hu have nothing to disclose.
Compliance with Ethics Guidelines
The establishment of this database was approved by the Massachusetts Institute of Technology (Cambridge, MA) and Beth Israel Deaconess Medical Center (Boston, MA), and consent was obtained for the original data collection. Therefore, the ethical approval statement and the need for informed consent were waived for this manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Digital Features
This article is published with digital features, including [list digital features available e.g. a video abstract and slide deck], to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.19375553.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chang Hu and Lu Li contributed equally and share the first authorship.
Contributor Information
Chang Hu, Email: huchang@whu.edu.cn.
Lu Li, Email: lulu189vip@163.com.
Weipeng Huang, Email: huang7@whu.edu.cn.
Tong Wu, Email: annie_wutong@foxmail.com.
Qiancheng Xu, Email: qianchengxu@whu.edu.cn.
Juan Liu, Email: liujuan@whu.edu.cn.
Bo Hu, Email: hobbier1979@163.com.
References
- 1.Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet. 2018;392(10141):75–87. doi: 10.1016/S0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J, Wang H, Kang Y, et al. The epidemiology of sepsis in Chinese ICUs: a national cross-sectional survey. Crit Care Med. 2020;48(3):e209–e218. doi: 10.1097/CCM.0000000000004155. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, Finfer S. Recognizing sepsis as a global health priority—a WHO resolution. N Engl J Med. 2017;377(5):414–417. doi: 10.1056/NEJMp1707170. [DOI] [PubMed] [Google Scholar]
- 5.Alam N, Hobbelink EL, van Tienhoven AJ, van de Ven PM, Jansma EP, Nanayakkara PW. The impact of the use of the Early Warning Score (EWS) on patient outcomes: a systematic review. Resuscitation. 2014;85(5):587–594. doi: 10.1016/j.resuscitation.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Yao RQ, Jin X, Wang GW, et al. A machine learning-based prediction of hospital mortality in patients with postoperative sepsis. Front Med (Lausanne) 2020;7:445. doi: 10.3389/fmed.2020.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doorn W, Stassen PM, Borggreve HF, et al. A comparison of machine learning models versus clinical evaluation for mortality prediction in patients with sepsis. PLoS ONE. 2021;16(1):e0245157. doi: 10.1371/journal.pone.0245157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46(4):547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Z, Bo L, Xu Z, et al. An explainable machine learning algorithm for risk factor analysis of in-hospital mortality in sepsis survivors with ICU readmission. Comput Methods Progr Biomed. 2021;204:106040. doi: 10.1016/j.cmpb.2021.106040. [DOI] [PubMed] [Google Scholar]
- 10.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 11.Watson DS, Krutzinna J, Bruce IN, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ. 2019;364:l886. doi: 10.1136/bmj.l886. [DOI] [PubMed] [Google Scholar]
- 12.The Lancet Respiratory M Opening the black box of machine learning. Lancet Respir Med. 2018;6(11):801. doi: 10.1016/S2213-2600(18)30425-9. [DOI] [PubMed] [Google Scholar]
- 13.Hou N, Li M, He L, et al. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J Transl Med. 2020;18(1):462. doi: 10.1186/s12967-020-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg SM, Nair B, Vavilala MS, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2(10):749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SI, Celik S, Logsdon BA, et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun. 2018;9(1):42. doi: 10.1038/s41467-017-02465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S, Zeng Z, Wei H, Sha T, An S. Early combination of albumin with crystalloids administration might be beneficial for the survival of septic patients: a retrospective analysis from MIMIC-IV database. Ann Intensive Care. 2021;11(1):42. doi: 10.1186/s13613-021-00830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 20.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22(7):707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 21.Deshmukh F, Merchant SS. Explainable machine learning model for predicting GI bleed mortality in the intensive care unit. Am J Gastroenterol. 2020;115(10):1657–1668. doi: 10.14309/ajg.0000000000000632. [DOI] [PubMed] [Google Scholar]
- 22.Kong G, Lin K, Hu Y. Using machine learning methods to predict in-hospital mortality of sepsis patients in the ICU. BMC Med Inform Decis Mak. 2020;20(1):251. doi: 10.1186/s12911-020-01271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane DJ, Wunsch H, Saskin R, et al. Epidemiology and patient predictors of infection and sepsis in the prehospital setting. Intensive Care Med. 2020;46(7):1394–1403. doi: 10.1007/s00134-020-06093-4. [DOI] [PubMed] [Google Scholar]
- 24.Hoste EA, Bagshaw SM, Bellomo R, et al. Epidemiology of acute kidney injury in critically ill patients: the multinational AKI-EPI study. Intensive Care Med. 2015;41(8):1411–1423. doi: 10.1007/s00134-015-3934-7. [DOI] [PubMed] [Google Scholar]
- 25.Barthel P, Wensel R, Bauer A, et al. Respiratory rate predicts outcome after acute myocardial infarction: a prospective cohort study. Eur Heart J. 2013;34(22):1644–1650. doi: 10.1093/eurheartj/ehs420. [DOI] [PubMed] [Google Scholar]
- 26.Ren Y, Zhang L, Xu F, et al. Risk factor analysis and nomogram for predicting in-hospital mortality in ICU patients with sepsis and lung infection. BMC Pulm Med. 2022;22(1):17. doi: 10.1186/s12890-021-01809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaukonen KM, Bailey M, Suzuki S, Pilcher D, Bellomo R. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000–2012. JAMA. 2014;311(13):1308–1316. doi: 10.1001/jama.2014.2637. [DOI] [PubMed] [Google Scholar]
- 28.Heffernan AJ, Judge S, Petrie SM, et al. Association between urine output and mortality in critically ill patients: a machine learning approach. Crit Care Med. 2022;50(3):e263–e271. doi: 10.1097/CCM.0000000000005310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.