Abstract
Bacterial community structure and diversity in rhizospheres in two types of grassland, distinguished by both plant species and fertilization regimen, were assessed by performing a 16S ribosomal DNA (rDNA) sequence analysis of DNAs extracted from triplicate soil plots. PCR products were cloned, and 45 to 48 clones from each of the six libraries were partially sequenced. Phylogenetic analysis of the resultant 275 clone sequences indicated that there was considerable variation in abundance in replicate unfertilized, unimproved soil samples and fertilized, improved soil samples but that there were no significant differences in the abundance of any phylogenetic group. Several clone sequences were identical in the 16S rDNA region analyzed, and the clones comprised eight pairs of duplicate clones and two sets of triplicate clones. Many clones were found to be most closely related to environmental clones obtained in other studies, although three clones were found to be identical to culturable species in databases. The clones were clustered into operational taxonomic units at a level of sequence similarity of >97% in order to quantify diversity. In all, 34 clusters containing two or more sequences were identified, and the largest group contained nine clones. A number of diversity, dominance, and evenness indices were calculated, and they all indicated that diversity was high, reflecting the low coverage of rDNA libraries achieved. Differences in diversity between sample types were not observed. Collector’s curves, however, indicated that there were differences in the underlying community structures; in particular, there was reduced diversity of organisms of the α subdivision of the class Proteobacteria (α-proteobacteria) in improved soils.
Land use in the United Kingdom has undergone considerable change over the last decade due to both economic and political pressure and increasing public concern regarding the quality of the environment. This has led to more extensive grazing regimens in the uplands of Britain and reductions in fertilizer applications. While considerable information concerning the effect of this extensification on the vascular plant community is available, the effect on soil bacteria is not understood. Plant and bacterial activities are closely linked through microbial utilization of root exudates, dead cells, and litter, and soil bacterial diversity may therefore be influenced by plant diversity and community structure.
The traditional approach to analysis of bacterial diversity involves identification of pure cultures isolated on laboratory media. There is strong evidence that this approach detects only a small proportion (estimated to be less than 1% [38]) of the bacteria present due to the selectivity of growth media and conditions. Analysis of respiration in individual cells extracted from soil, however, has indicated that virtually all bacteria in soil are metabolically active (38). Analysis of DNA extracted from environmental samples has allowed workers to investigate bacterial communities without cell extraction and laboratory cultivation. Broad-scale techniques, such as DNA reassociation and reannealing, provide a measure of total microbial diversity and have revealed the influence of environmental parameters, such as pollution (1) and agricultural exploitation, on microbial diversity (38).
More detailed analyses can be performed by using 16S ribosomal DNA (rDNA)-based techniques, and a range of methods targeting both rRNA and rDNA are now used routinely in microbial ecology (reviewed in references 8, 14, and 40). These methods include detection by PCR, in situ hybridization, sequence analysis, and denaturing gradient gel electrophoresis. Primers and probes having different specificities, ranging from universal to species specific, can be used with combinations of these techniques to provide a comprehensive understanding of bacterial community structure. 16S rDNA-based analyses of terrestrial samples from a range of geographical locations (3, 4, 10, 16, 17, 27, 29, 45) have demonstrated that there is considerable bacterial diversity in natural environments. Sequences cloned from environmental samples are rarely identical to sequences of cultured bacteria represented in gene databases, and all investigations have recovered clones belonging to a new bacterial group, which is represented by the cultured species Acidobacterium capsulatum and Holophaga foetida (16, 22). Several other clusters of sequences have been identified, and these sequences appear to be widespread in soils with very diverse physicochemical properties (3, 4, 17, 29).
The aim of this project was to apply quantitative measures of diversity to 16S rDNA data and to use the information obtained to determine the effects of grass species and other influences associated with improvement (namely, fertilization and grazing) on the diversity and community structure of rhizobacteria by using samples from the same geographical location.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Rhizosphere samples from two characteristic vegetation types, designated unimproved and improved (U4a and MG6 of the National Vegetation Classification, respectively) (31), were collected from Sourhope Research Station (map reference NT 850 205) in the Borders Region of Scotland. The unimproved plots (brown forest soil; pH 5.5 to 6.9; 0.33 to 0.37 μg of C g [dry weight]−1) represented natural grassland dominated by the grass species Agrostis capillaris and Festuca ovina. They had never received fertilizer treatment but were grazed by sheep during the summer months. The improved plots (brown forest soil; pH 6.1 to 6.8; 0.21 to 0.26 μg of C g [dry weight]−1) were reseeded with Lolium perenne (perennial ryegrass) and Trifolium repens (clover) approximately 25 years ago. These plots received dressings consisting of 50 kg of N ha−1 in March and August and fertilizer containing N, P, and K (40:20:20) in May. They were also grazed by sheep from spring to mid-November (weather permitting) at a sward height of 4 cm. At each sampling site, three 5- by 5-m quadrats were randomly located, and 50 cores (diameter, 3.5 cm; depth, 5 cm) were collected, combined, and sieved (mesh size, <2 mm) to remove plant material. Due to the density of the grass root systems, all soil was assumed to be in contact with plant roots and was considered rhizosphere. Subsamples of soil used for molecular analyses were stored at −70°C.
DNA was extracted by C. D. Clegg (Scottish Crop Research Institute, Invergowrie, United Kingdom), who used the protocol of Clegg et al. (6). This method involved incubation of 1-g samples of soil in the presence of polyvinylpolypyrrolidone and lysozyme, followed by three rounds of freezing and thawing to facilitate cell disruption. Crude DNA was purified by two rounds of cesium chloride centrifugation, which produced DNA of high purity (6).
PCR amplification, cloning, and sequencing.
PCR amplification of bacterial rDNA was carried out by using the primers Bf (20) and 1390r (7). Amplification was performed in a 50-μl (total volume) reaction mixture containing ∼10 ng of soil DNA, 1 U of Taq DNA polymerase (Promega UK Ltd., Southampton, United Kingdom), an appropriate dilution of the manufacturer’s buffer, each deoxynucleoside triphosphate at a concentration of 250 μM, each primer at a concentration of 0.4 μM, and 1 μl of a bovine serum albumin solution (20 mg ml−1; Boehringer Mannheim Diagnostics and Biochemicals Ltd., Lewes, United Kingdom). Thirty cycles of amplification were carried out with a model Omn-E thermal cycler (Hybaid Ltd., Teddington, United Kingdom) as follows: one cycle consisting of 95°C for 10 min, 50°C for 1 min, and 72°C for 2 min; nine cycles consisting of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min; and 20 cycles consisting of 92°C for 30 s, 50°C for 30 s, and 72°C for 2.5 min. This was followed by a final step consisting of incubation at 72°C for 30 min. Products were visualized on an agarose gel stained with ethidium bromide (1%, wt/vol), bands were excised, and DNA was purified from gel slices by using a QIAEX II gel extraction kit (Qiagen Ltd., Crawley, United Kingdom).
Purified amplification products were cloned into the vector pCR 2.1 from an Original TA Cloning kit (Invitrogen BV, Groningen, The Netherlands) by using a vector/insert ratio of 1:1. Ligations were transformed into supercompetent Escherichia coli XL1-Blue MRF′ Kan (Stratagene Ltd., Cambridge, United Kingdom). White colonies were screened directly for inserts by performing colony PCR with T7 and M13 reverse primers. The amplification conditions were the same as those described above except that bovine serum albumin was not included in the mixture and the final step consisted of incubation for 10 min at 72°C. The PCR products were visualized by agarose gel electrophoresis, and the products obtained from randomly selected positive clones were purified by adding an equal volume of chloroform-isoamyl alcohol (24:1), followed by vortexing and centrifugation for 5 min at 10,000 × g. Radiolabeled primer cycle sequencing was carried out by using a Thermo Sequenase cycle sequencing kit (Amersham International plc., Slough, United Kingdom) and primer 537r (7) according to the manufacturer’s instructions. Reaction products were analyzed by standard polyacrylamide gel electrophoresis and autoradiography.
Sequence analysis.
A total of 281 partial clone sequences were compared with sequences in the Ribosomal Database Project (RDP) database (18) by using the SEQUENCE_SIMILARITY function and with sequences in the GenBank database by performing FastA searches with Genetics Computer Group software (12) installed in the Seqnet node of the BBSRC Daresbury Laboratory (Warrington, United Kingdom). Sequences were also checked for chimeric properties by using CHIMERA_CHECK of the RDP. On the basis of the results of database searches, sequences were aligned with representative bacterial sequences from the RDP and GenBank databases by using the Genetic Data Environment running in ARB (37). Phylogenetic trees were constructed by using the Jukes-Cantor model (15) and neighbor joining (32) with PHYLIP, version 3.5 (9). Data sets were bootstrapped by using SEQBOOT (PHYLIP, version 3.5). The levels of similarity of clones were assessed by using the GAP algorithm of the Genetics Computer Group, the clones were clustered into operational taxonomic units (OTUs) at a level of sequence similarity of >97%, and a number of diversity indices were calculated. These indices included (i) library coverage, the portion of a clone library of infinite size that is sampled (13, 25); (ii) species richness, the total number of OTUs (5); (iii) the Shannon diversity index, a general diversity index which considers both species richness and evenness (28); (iv) evenness, which describes the distribution of abundance of clone types (28); and (v) dominance, which describes the extent of dominance by individual OTUs (28). For comparisons of library coverage, the Shannon diversity index, dominance, and evenness, libraries with sample sizes of more than 45 were artificially reduced to a sample size of 45 by randomly removing clone sequences. For species richness, normalization of library size to 45 was achieved by rarefaction (5). Diversity indices for unimproved and improved sites were also calculated by using the pooled data from triplicate libraries whose combined data set sizes were reduced to 135 as described above. The maximum and minimum values for each index were also determined. Similarity coefficients, which reflected the proportions of shared OTUs, were calculated by performing pairwise comparisons of libraries (28). Finally, collector’s curves or species abundance curves (28) (the number of species detected plotted versus the number of clones analyzed) were constructed to compare the diversities of major phylogenetic groups. Pooled data for unimproved and improved grasslands used as the data sets obtained from individual libraries were too small to allow meaningful comparisons.
Nucleotide sequence accession numbers.
The partial clone sequences determined in this study have been deposited in the GenBank database under accession no. AF078179 to AF078453.
RESULTS
Construction of rDNA libraries.
Bacterial diversity in triplicate soil samples from rhizospheres from unimproved and improved grassland pastures was analyzed by PCR amplification of total DNA extracts with bacterial primers (6). Products from three separate amplifications were pooled prior to cloning in order to minimize PCR drift (39). For each of the six clone libraries obtained, 45 to 48 clones were selected randomly for sequencing of 321 to 466 bases, including variable regions V2 and V3 at the 5′ end of the rRNA gene (26). In all, 281 partial sequences were obtained; 6 of these were considered putative chimeras on the basis of the results of a CHIMERA_CHECK analysis and were omitted from further analyses.
Identification and distribution of clones.
Phylogenetic analysis of the 275 clones studied and their closest relatives in the RDP and GenBank databases and FastA and SEQUENCE_SIMILARITY searches were used to assign each environmental clone to a major bacterial group, when possible. Two individual clones and six clone clusters comprising two to seven sequences were not related to cultured or uncultured representatives of the sequence databases, formed distinct phylogenetic branches, and were considered novel groups. Several clone sequences were identical in the region analyzed. These sequences included eight pairs of duplicate clones and two sets of triplicate clones. While seven of these sets, including one set of triplicate clones, contained clones from a single unimproved soil library, the remaining three groups comprised clones from both unimproved and improved soil libraries. In addition, we identified three clones whose sequences were identical, as determined by FastA searches, in the region sequenced to the sequences of culturable species in the databases (namely, Agrobacterium tumefaciens, Rhizobium loti, and Staphylococcus succinus).
Table 1 lists the broad phylogenetic distribution of clones within each library and within all of the clones analyzed, along with average values for each grassland type. Many clones belonged to major previously characterized groups, including the α subdivision of the class Proteobacteria (α-proteobacteria), β-proteobacteria, γ-proteobacteria, flavobacteria, and actinomycetes, and many belonged to relatively recently recognized groups, such as the Acidobacterium, Holophaga, and Verrucomicrobium groups. Clones related to Deinococcus radiodurans and the green sulfur bacteria were also obtained. Eight clusters of sequences, accounting for 9.3% of all of the clones, were not related to culturable organisms in the databases (level of sequence similarity, <81%). These clusters were designated unclassified Sourhope groups 1 to 8. A novel group which clustered with the β- and γ-proteobacteria was also observed, and as the phylogeny of this group is uncertain, it is referred to below as the β-/γ-proteobacteria.
TABLE 1.
Relative abundance of clones from triplicate unimproved and improved grassland soil samples and belonging to a number of bacterial phylogenetic groups
| Phylogenetic groupa | Relative clone abundance (%)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Library from unimproved grassland
|
Library from improved grassland
|
Grassland mean value
|
Pooled data set (improved and unimproved grasslands) | ||||||
| SAF1 | SAF2 | SAF3 | SL1 | SL2 | SL3 | Unimproved | SL improved | ||
| Proteobacteria | |||||||||
| α-Proteobacteria | 52.1 | 24.4 | 53.3 | 22.2 | 43.5 | 37.0 | 43.3 (±18.5)b | 34.2 (±12.3) | 39.6 |
| β-Proteobacteria | 8.3 | 11.1 | 2.2 | 2.2 | 2.2 | 6.5 | 7.2 (±5.1) | 3.6 (±2.8) | 5.6 |
| γ-Proteobacteria | 2.1 | 0 | 4.4 | 6.7 | 2.2 | 6.5 | 2.2 (±2.5) | 5.1 (±2.9) | 3.7 |
| β-/γ-Proteobacteria | 4.2 | 11.1 | 4.4 | 0.0 | 2.2 | 0 | 6.6 (±4.4) | 0.7 (±1.4) | 3.7 |
| Flavobacteria | 0 | 2.2 | 0 | 17.8 | 0 | 4.3 | 0.7 (±1.5) | 7.4 (±10.5) | 4.1 |
| Green sulfur bacteria | 0 | 0 | 2.2 | 0 | 0 | 0 | 0.7 (±1.5) | 0 (ND)c | 0.4 |
| Actinomycetes | 2.1 | 17.8 | 4.4 | 13.3 | 19.6 | 22.2 | 8.1 (±9.6) | 18.2 (±4.9) | 13.3 |
| Low-G+C-content bacteria | 0 | 0 | 2.2 | 0 | 0 | 2.2 | 0.7 (±1.5) | 0.7 (±1.4) | 0.7 |
| Deinococcus-Thermus group | 0 | 0 | 2.2 | 0 | 0 | 0 | 0.7 (±1.5) | 0 (ND) | 0.4 |
| Acidimicrobium-Microthrix group | 2.1 | 15.6 | 6.7 | 6.7 | 8.7 | 0 | 8.1 (±7.8) | 5.1 (±5.1) | 6.7 |
| Rubrobacter | 6.3 | 4.4 | 0 | 11.1 | 4.3 | 2.2 | 3.6 (±3.6) | 5.9 (±5.3) | 4.8 |
| Holophaga | 4.2 | 8.9 | 0 | 4.4 | 4.3 | 0 | 4.4 (±5.0) | 2.9 (±2.9) | 3.7 |
| Acidobacterium | 4.2 | 0 | 0 | 6.7 | 4.3 | 2.2 | 1.4 (±2.7) | 4.4 (±2.5) | 3.0 |
| Verrucomicrobium | 0 | 2.2 | 6.7 | 4.4 | 0.0 | 4.3 | 3.0 (±3.8) | 2.9 (±2.9) | 3.0 |
| Unclassified groupsd | |||||||||
| Sourhope group 1 | 4.2 | 2.2 | 2.2 | 0 | 0 | 2.2 | 2.9 (±1.3) | 0.7 (±1.4) | 1.9 |
| Sourhope group 2 | 2.1 | 0 | 0 | 0 | 2.2 | 0 | 0.7 (±1.4) | 0.7 (±1.4) | 0.7 |
| Sourhope group 3 | 4.2 | 0 | 0 | 0 | 0 | 2.2 | 1.4 (±2.7) | 0.7 (±1.4) | 1.1 |
| Sourhope group 4 | 0 | 0 | 0 | 0 | 0 | 2.2 | 0 (ND) | 0.7 (±1.4) | 0.4 |
| Sourhope group 5 | 4.2 | 0 | 0 | 2.2 | 2.2 | 0 | 1.4 (±2.7) | 1.5 (±1.4) | 1.5 |
| Sourhope group 6 | 0 | 0 | 4.4 | 0 | 0 | 0 | 1.5 (±2.9) | 0 (ND) | 0.7 |
| Sourhope group 7 | 0 | 0 | 4.4 | 2.2 | 4.3 | 4.3 | 1.5 (±2.9) | 3.6 (±1.4) | 2.6 |
| Sourhope group 8 | 0 | 0 | 0 | 0 | 0 | 2.2 | 0 (ND) | 0.7 (±1.4) | 0.4 |
The numbers of clones in the libraries and data set were as follows: SAF1, 48; SAF2, 45; SAF3, 45; SL1, 45; SL2, 46; SL3, 46; and pooled data set, 275.
The values in parentheses are 95% confidence limits.
ND, not determined.
Unclassified Sourhope groups 1 through 8 are defined in the phylogenetic tree in Fig. 4.
Sequences belonging to members of the α-proteobacteria were most abundant, were recovered in all libraries, and comprised 43.5 and 34.2% (grassland means) of the unimproved and improved soil clone sequences, respectively. Actinomycete sequences were the next most abundant clone sequences recovered for both the unimproved and improved grassland types (mean values, 8.1 and 18.2%, respectively) and for all of the improved soil replicate samples (13.3 to 22.2%). Considerable variability in the triplicate libraries is evident in Table 1. For example, while actinomycete sequences were the second most abundant sequence type in library SAF2 (accounting for 17.8% of the sequences), these sequences accounted for only 2.1 and 4.4% of the sequences in the SAF1 and SAF3 libraries, respectively. Similarly, sequences of flavobacteria accounted for 0, 4.4, and 17.8% of the sequences in the replicate libraries from the improved grassland soils. Analysis of the relative abundance of each group in unimproved and improved soil replicates by using the Student t test did not reveal any statistically significant differences between the means for the grasslands for any phylogenetic group (P ≥ 0.05). The greatest apparent differences between the grassland types were the differences for Sourhope group 1 and the β-/γ-proteobacteria, which were more prevalent in the unimproved soils (P = 0.091 and P = 0.112, respectively). In addition, there was some suggestion that there were slight excesses of actinomycete sequences (P = 0.163) and sequences of members of the Acidobacterium group (P = 0.189) in improved soils.
Phylogenetic analysis.
A phylogenetic analysis was carried out with sequence data for four groups (Fig. 1 through 4). The first three groups, which formed distinct lineages on the bacterial 16S rRNA tree with strong bootstrap support, were the α-proteobacteria (100% support), the β- and γ-proteobacteria (100%), and the actinomycetes (97%). The fourth group contained all of the other Sourhope clones and included clones belonging to the flavobacteria, to the novel lineage containing A. capsulatum and H. foetida, and to the eight unclassified groups. Alignments consisting of 327, 373, 379, and 276 bases were analyzed for the α-proteobacteria, the β- and γ-proteobacteria, the actinomycetes, and all other clones, respectively. Bootstrap values were obtained for the branch topologies with >50% support in the bootstrap analysis of 100 replicates. Clones that exhibited >97% sequence similarity were clustered on the trees (Fig. 1 through 4, solid boxes), unless high levels of similarity to other sequences made this impossible.
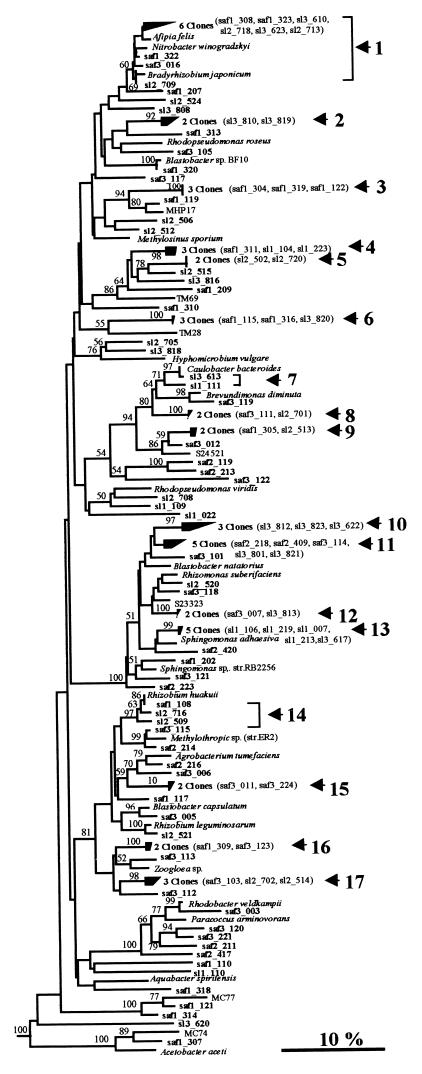
FIG. 1.
Neighbor-joining tree showing the relationship of grassland rhizosphere clones to reference members of the α-proteobacteria based on analysis of 327 bases of aligned 16S rDNA sequences. Clones exhibiting >97% sequence similarity are included in numbered clusters. The horizontal scale represents the extent of variation within each cluster, and brackets indicate clones belonging to the same group. Bootstrap values are shown for nodes that had >50% support in a bootstrap analysis of 100 replicates. Sequences obtained from unimproved and improved grassland soils are designated by the prefixes SAF and SL, respectively, followed by replicate numbers (SAF1, SAF2, etc.). The clones obtained during other direct analyses of environmental samples are MHP17 (21), MC74 (36), MC77 (36), TM69 (30), and TM28 (30). Sequences whose designations begin with the prefix S represent bacterial isolates (23). For convenience, the tree was pruned from a larger tree containing additional sequences from reference bacteria. The scale bar indicates an estimated change of 10%.
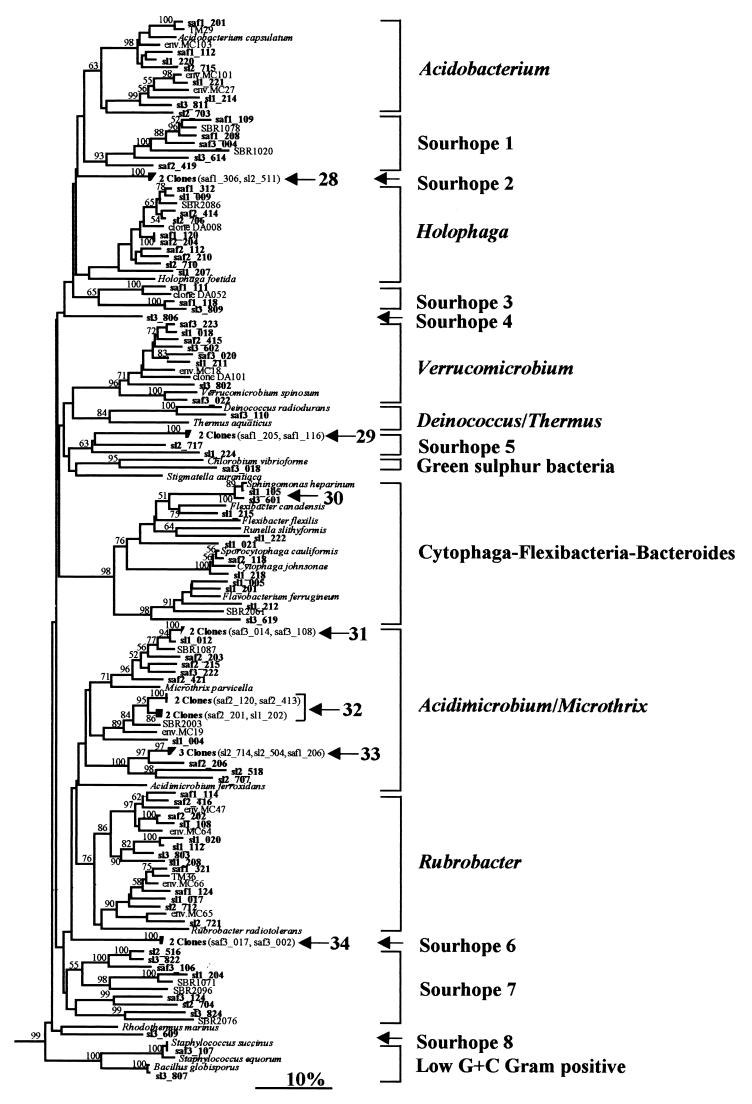
FIG. 4.
Neighbor-joining tree showing the relationship of grassland rhizosphere clones to reference bacteria based on analysis of 276 bases of aligned 16S rDNA sequences. The positions of all other clones, which are associated with the α-, β-, and γ-proteobacteria and the actinomycetes, are shown in Fig. 1 to 3. The clones obtained during other direct analyses of environmental samples are MC18 (17), all other sequences whose designations begin with the prefix MC (36), and sequences whose designations begin with the prefixes TM (30) DA (10), and SBR (2). Sequences whose designations begin with the prefix S represent bacterial isolates (23). For other sequence nomenclature and branch labeling, see the legend to Fig. 1.
In all, 34 clusters containing two or more sequences with >97% sequence homology were observed (Fig. 1 through 4, arrows), and these clusters comprised 94 of the 275 clone sequences examined. The clusters generally comprised only two or three sequences, although the largest group (cluster 1), which contained nine clones (Fig. 1), and three clusters containing five clones (clusters 11, 13, and 19) (Fig. 1 and 2) were also observed in the proteobacterial region. Approximately one-half of the clusters (19 of the 34 clusters) comprised sequences from either unimproved or improved soil libraries, although no cluster contained representatives of all three replicate samples. In addition, 13 of these clusters consisted of sequences from single libraries. The 15 remaining clusters contained sequences from both improved and unimproved grassland libraries.
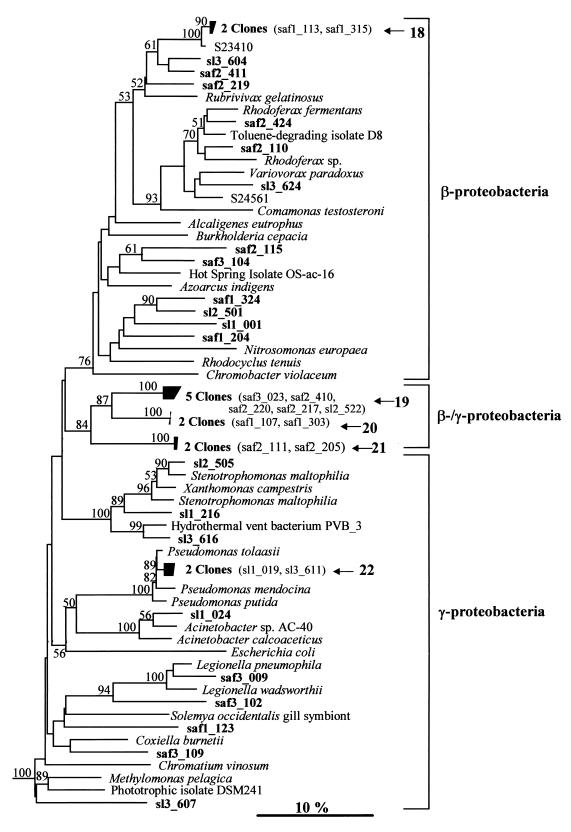
FIG. 2.
Neighbor-joining tree showing the relationship of grassland rhizosphere clones to reference members of the β- and γ-proteobacteria based on analysis of 373 bases of aligned 16S rDNA sequences. The sequence designated PVB_3 is a clone obtained during another direct analysis of an environmental sample (24). Sequences whose designations begin with the prefix S represent bacterial isolates (23). For other sequence nomenclature and branch labeling, see the legend to Fig. 1.
(i) α-Proteobacterial clones.
The α-proteobacterial clones (Fig. 1) were the most abundant clones obtained, accounting for 109 (40%) of the 275 clones with 100% support in the bootstrap analysis and representing 17 of the 34 clusters. Three of the four largest clusters, which contained five and nine sequences, fell in this group. In addition, 5 of the 10 groups of identical sequences, including both sets of triplicate clones, were found in the α-proteobacteria. Many sequences were closely associated with culturable organisms and clustered, with strong support in the bootstrap analysis, with a range of common rhizosphere and soil bacteria, including Sphingomonas spp., Rhizobium spp., and A. tumefaciens. In addition, the group of clones containing saf3_012 and cluster 9 clones and clones which occurred close to Rhizomonas suberafaciens on Fig. 1, including cluster 12 clones, were related to cultures obtained from paddy field soils (23). Cluster 1 contained a range of physiologically distinct, cultured members of the α-proteobacteria, including Afipia felis, Nitrobacter winogradskyi, and Bradyrhizobium japonicum, which highlighted the difficulties in inferring the physiology of these and most other clones obtained during this study.
Within the α-proteobacteria, several clones clustered with clones obtained in other studies. For example, saf1_121, saf1_314, and saf1_307 clustered with environmental clone sequences MC77 and MC74, which originally were isolated from an Australian acid soil (36). In addition, a group of four clones which shared a lineage on this tree with the methane oxidizer Methylosinus sporium was closely related to environmental clone MHP17, exhibiting 94% bootstrap support. This peat clone is believed to be a member of a novel, acidophilic group of methane oxidizers (21). Other Sourhope clones which fell in the α-proteobacteria were related to clones TM69 and TM28 recovered from peat (30).
(ii) β- and γ-proteobacteria.
Clones falling in the β-proteobacteria formed a cluster with 76% bootstrap support (Fig. 2). All of the other clones on this tree belonged either to the γ-proteobacterial group or to clusters 19 to 21, which formed a deeply branching group (84% bootstrap support) designated the β-/γ-proteobacteria (Table 1). Although the presence of these sequences in the β- or γ-proteobacteria was strongly supported (100% of bootstrap trees), the phylogeny of this group is unclear. An analysis of the signature nucleotides of β- and γ-proteobacterial species (43, 44) was performed by using partial sequence data when possible. Cluster 21 sequences had a signature nucleotide thought to be unique to the majority of γ-proteobacterial sequences (44). No other β- or γ-proteobacterial signature nucleotides were detected in any of these clones, but many of the signature nucleotides were specific to the 3′ region of the rRNA gene, which was not analyzed in this study.
Most of the clones on the β- and γ-proteobacterial tree were not closely related to culturable organisms; the exceptions were two clones which fell in the fluorescent Pseudomonas group of the γ-proteobacteria (cluster 22) and a clone closely related to Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia). A group of deeply branching clones shared a lineage on this tree with the ammonia oxidizer Nitrosomonas europaea, although there was not strong support for this relationship.
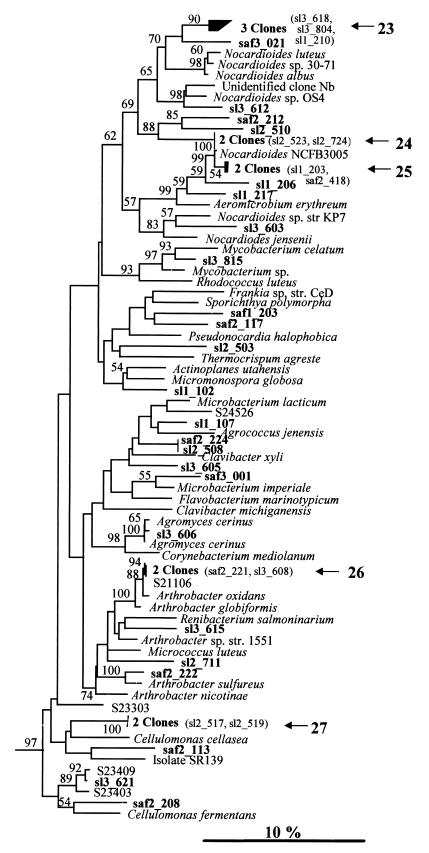
(iii) Actinomycete clones.
The actinomycete clone types recovered were diverse, although five clusters contained two or three clones (clusters 23 to 27) (Fig. 3). Most clones were not closely related to cultured representatives of the actinomycetes; the exceptions were five clones related to Arthrobacter oxidans (cluster 26), Nocardioides sp. strain NCFB3005 (cluster 25), and Agromyces cerinus (sl3_606). In addition, clone sl3_621 and cluster 26 were very closely related (>99% related) to bacterial cultures obtained from paddy field soil (23).
FIG. 3.
Neighbor-joining tree showing the relationship of grassland rhizosphere clones to reference actinomycetes based on analysis of 379 bases of aligned 16S rDNA sequences. Sequences whose designations begin with the prefix S represent bacterial isolates (23). For other sequence nomenclature and branch labeling see the legend to Fig. 1.
(iv) Other clones.
The majority of the clones which did not cluster with the α-, β-, or γ-proteobacteria or with the actinomycetes were most closely related to environmental clones of uncultivated bacteria obtained in other studies (2, 10, 17, 30, 36) and belonged to groups containing only one or a few cultured representatives (Fig. 4). Many clones fell in the Acidobacterium and Holophaga groups, which are believed to belong to a novel lineage on the bacterial tree (16, 22). Other clone groups (e.g., Sourhope groups 1 and 2) appeared to belong to this lineage, although the relationships between the clusters were unstable and the bootstrap values were low. Similarly, many clones were recovered in other recently described groups, such as the Verrucomicrobium group, which may share ancestry with members of the genus Chlamydia and the family Planctomycetaceae (17, 41), and the Acidimicrobium and Rubrobacter groups, which have been described as deeply branching actinomycetes (36). Sourhope group 7 comprises four strongly supported lineages (≥98% support in bootstrap analysis), although the clustering of these sequences as a single group was not strongly supported by the bootstrap analysis. Sourhope groups 4 through 6 and 8 contained only clones isolated in this study, and the phylogenetic position of these groups is unclear. The positions of clones which are closely related to culturable isolates belonging to the low-G+C-content gram-positive bacterial group and the Cytophaga-Flavobacterium group are also shown in Fig. 4. In particular, clones saf3_107 and sl3_807 exhibited >99% sequence similarity to the cultured species Staphylococcus equorum and Bacillus globisporus (both members of the low-G+C-content gram-positive group), respectively, and clones sl1_105 and saf2_118 exhibited >99% sequence similarity to the cultured species Sphingobacterium heparinum and Sporocytophaga cauliformis (both members of the Cytophaga-Flexibacterium-Bacteroides group), respectively.
Diversity indices.
Diversity indices were calculated by using sequence data obtained from each library and from each grassland type. Clones which showed >97% sequence similarity, as indicated in Fig. 1 to 4, were clustered into OTUs after normalization of sample sizes in order to directly compare individual libraries and pooled data for each grassland type. Table 2 shows the diversity indices obtained, along with the minimum and maximum values possible when the number of clones was 45 (n = 135 for pooled data) (i.e., when all sequences were identical and different, respectively). The coverage within each library (13, 25) was low, ranging from 6.7 to 15.6%, and the Shannon diversity index and species richness values were high for all libraries. The evenness and dominance values approximated the maximum possible values as most sequence types were recovered only once and the majority of sequence clusters comprised only two clones. Differences between the diversities of the unimproved and improved soil libraries were not evident, although the coverage, Shannon diversity index, and evenness values were more varied for the unimproved soil samples than for the improved soil samples. In addition, although the values for individual libraries were similar, when the pooled data for each grassland type were considered, all indices except the dominance index indicated that the diversity in unimproved soil samples was slightly greater than the diversity in improved soil samples.
TABLE 2.
Diversity indices obtained for rDNA libraries from triplicate unimproved and improved grassland soil samples
| Diversity indexa | Library from unimproved grassland
|
Library from improved grassland
|
Index range (n = 45)b
|
Pooled data
|
Index range (n = 135)b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAF1 | SAF2 | SAF3 | SL1 | SL2 | SL3 | Minimum | Maximum | Unimproved | Improved | Minimum | Maximum | |
| % Coverage | 15.56 | 13.33 | 6.67 | 8.89 | 15.56 | 11.1 | 100 | 0 | 15.56 | 17.78 | 100 | 0 |
| Species abundance | 37.8 | 39.0 | 42.0 | 41.0 | 37.3 | 40.2 | 1 | 45 | 113.7 | 112.5 | 1 | 135 |
| Shannon | 1.549 | 1.563 | 1.613 | 1.550 | 1.560 | 1.586 | 0 | 1.653 | 2.024 | 2.007 | 0 | 2.130 |
| Evenness | 0.981 | 0.976 | 0.994 | 0.961 | 0.987 | 0.984 | 0.028 | 1.000 | 0.984 | 0.981 | 0.009 | 1.000 |
| Dominance | 0.031 | 0.028 | 0.025 | 0.029 | 0.029 | 0.030 | 1 | 0.022 | 0.011 | 0.011 | 1 | 0.007 |
For all indices except species abundance, numbers of clones in individual libraries and pooled data sets were reduced to 45 and 135, respectively, by randomly removing sequence data from larger libraries. Species abundance was calculated by rarefaction, where the expected number of species in a library containing 45 clones or a data set containing 135 clones was determined by using complete data sets.
Values are the minimum and maximum values which could be obtained for libraries containing n sequences at minimum diversity and maximum diversity.
The similarity coefficients, calculated by performing pairwise comparisons of libraries (Table 3), were generally low, ranging from 0 to 0.132. Interestingly, the libraries which shared the greatest number of OTUs were obtained from different grassland types (e.g., the similarity coefficient for the SAF1 and SL2 libraries was 0.132, and the similarity coefficient for the SAF3 and SL2 libraries was 0.125), although two improved libraries, SL1 and SL2, also had a relatively high similarity coefficient (0.123). Several libraries, including replicate libraries from the same grassland (e.g., SAF1 and SAF2, as well as SL1 and SL2), contained no common species. A comparison of pooled data from each grassland also resulted in a low similarity coefficient, 0.132.
TABLE 3.
Pairwise comparisons of species compositions of rDNA libraries from triplicate unimproved and improved grassland soil samples
| Soil samples | Library | Similarity coefficienta
|
||||
|---|---|---|---|---|---|---|
| Libraries from unimproved grassland
|
Libraries from improved grassland
|
|||||
| SAF1 | SAF2 | SAF3 | SL1 | SL2 | ||
| Unimproved | SAF1 | |||||
| SAF2 | 0 | |||||
| SAF3 | 0.075 | 0.049 | ||||
| Improved | SL1 | 0.025 | 0.025 | 0 | ||
| SL2 | 0.132 | 0.026 | 0.125 | 0 | ||
| SL3 | 0.051 | 0.051 | 0.073 | 0.123 | 0.026 | |
The similarity coefficient (S) was determined as follows: S = 2 C/(A +B), where A and B are the numbers of OTUs in libraries A and B, respectively, and C is the number of shared OTUs.
Collector’s curves.
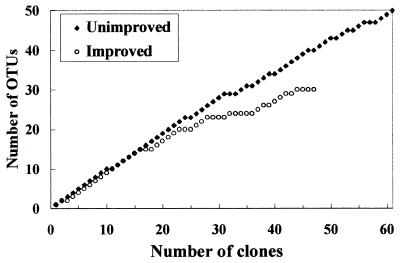
Collector’s curves were constructed for clones belonging to the most abundant phylogenetic groups obtained during this study by plotting the number of OTUs (level of sequence similarity, >97%) as a function of the number of sequences sampled. Full coverage of a library would be expected to give a plateau-shaped curve. The collector’s curves derived from pooled data for each grassland type for clones belonging to the α-proteobacteria (Fig. 5) diverged, indicating that the recovery of diversity was greater in the unimproved soil libraries than in the improved soil libraries. No single library was found to be responsible for this result, and similar curves were obtained for individual libraries or pairs of replicate libraries (data not shown). In addition, although considerably more α-proteobacterial sequences were obtained from unimproved soil libraries (60 clones) than from improved soil libraries (30 clones), the library coverage was considerably lower in the unimproved libraries. Collector’s curves were also constructed for actinomycete and Acidimicrobium-Microthrix clones, which were the next most abundant groups in the total sequence data set, but no differences were observed. No other phylogenetic groups were present in sufficient amounts in unimproved and improved soil samples to allow informative comparisons to be made.
FIG. 5.
Collector’s curves for α-proteobacterial clones from triplicate unimproved and improved soil libraries. Clones were grouped into OTUs at a level of sequence similarity of >97%.
DISCUSSION
The aim of this study was to use 16S rDNA sequence analysis to characterize the diversity and community structure of rhizobacteria associated with upland grass pastures, which provide the mainstay for grazing in the United Kingdom, and to assess differences between unimproved and improved pastures. These pastures differed in grass species diversity and composition and in land management regimen, factors which may influence the genetic diversity of microbial communities. While several other rDNA or rRNA approaches are available which may provide more rapid analysis (e.g., denaturing gradient gel electrophoresis-temperature gradient gel electrophoresis and amplified ribosomal restriction analysis), the great strength of sequence analysis is the generation of additive and retrievable data which can be used to generate phylogenetic probes and primers for use in further studies.
The sequences amplified from extracted DNA represented a wide range of bacterial phylogenetic groups, and sequences related to mitochondrial or chloroplast rDNA were not detected. Phylogenetic analysis of the sequences provided no evidence that there were differences in diversity between unimproved and improved soils or differences in the relative incidence of particular phylogenetic groups. In all of the libraries, the sequences of clones belonging to the α-proteobacteria were the most abundant sequences (22 to 53%), and these sequences occurred at levels similar to those found in an acid forest soil (50%) (17), a peat sample (42%) (30), and soil from arctic tundra (21%) (45). In contrast, in the studies of Borneman and Triplett (3) and Kuske et al. (16), α-proteobacterial clones accounted for only 4 and 3%, respectively, of the clones in libraries obtained from Amazonian soils and pinyon-juniper woodland soils. Actinomycete clones were abundant, particularly in the improved soil libraries. Borneman and Triplett (3) also found more high-G+C-content gram-positive sequences in soil from an active pasture than in mature forest soil. However, a comparison of the results of different studies was hindered by variations both in the sample type (e.g., soil pH, location, land use, and climate) and in the method used (e.g., DNA extraction technique and PCR primers). Nevertheless, 16S rDNA analyses clearly showed that terrestrial systems and other natural environments have extremely diverse bacterial populations. Considerable variation was also observed in replicate samples from both the unimproved plots and the improved plots, both in the relative distributions of different groups and in diversity measurements, as might be expected from the low coverage of individual libraries.
Large proportions of sequences in environmental samples are only distantly related to database sequences of cultured organisms, and many of the major clusters of sequences obtained in this study have only deeply branching cultured representatives. These clusters include clones associated with the recently described prosthecate organism Verrucomicrobium spinosum, Acidimicrobium ferroxidans, Rubrobacter radiotolerans, and the acidophile A. capsulatum (2, 10, 17, 30, 36). The depth of branching in these clusters prevents inference by physiological characteristics of the organisms from which the clones were derived. Although representatives of these groups were first cloned from acid Australian soils several years ago (17, 36), closely related pure-culture representatives have not been obtained yet, and their significance in natural environments remains unclear. Analysis of ribosomes extracted from peat, however, has indicated that bacteria related to clone DA079, which is affiliated with the Acidimicrobium line of descent, are probably physiologically active in the environment (11).
Duplicate sequences in soil libraries have rarely been recovered in other rDNA-based studies (3, 27, 45), but we obtained several groups of identical sequences. Felske et al. (10) proposed that the presence of duplicate sequences in libraries derived from acid, peaty soil (10) and peat bog samples (29) implied that the diversity was reduced due to selection of particular microorganisms by low pH, which is a feature of our soils, while Marilley et al. (19) observed reduced 16S rDNA diversity in rhizosphere compared to bulk soil. Identical 16S rDNA sequences from cultured organisms and environmental clones are also rarely reported but three partial sequences identical to sequences of cultured species were recovered from Sourhope libraries. This allowed us to infer physiological properties of the bacteria from which the clones were derived, although closely related organisms often have very different physiologies (33, 35).
This is the first study to apply diversity indices to 16S rDNA sequence abundance data obtained from environmental samples. Marilley et al. (19) analyzed soil microbial diversity by using restriction patterns of 25 16S rDNA clones and clustering of clones into OTUs at a level of sequence similarity of >96%. A value of >97% was used in our study as this value generally discriminates between bacterial species previously defined on the basis of DNA-DNA reassociation values (34). Great diversity was detected in improved and unimproved rhizospheres, and the Shannon indices approximated the maximum possible values. In contrast, Marilley et al. (19) found that diversity was considerably reduced in the rhizospheres of monocultures of L. perenne and T. repens compared to the diversity in bulk soil. The greater diversity of plant species may be responsible in part for the greater bacterial diversity in the Sourhope plots.
Low library coverage may have been responsible for the great diversity detected in the six rDNA libraries, and in order to obtain collector’s curves which reached a plateau, considerably more sequence data would be required, a costly and time-consuming prospect. Despite this, the underlying differences in evenness and dominance, as determined by clustering OTUs at >95 and 90%, suggest that the improved soil samples may have been slightly less diverse than the unimproved soil samples (data not shown). Similarly, indices calculated by using pooled data indicate that the diversity was slightly greater in the unimproved soil libraries than in the improved soil libraries, which may have reflected greater plant diversity at unimproved sites or selection of bacterial communities by specific plants, fertilizer addition, and grazing in the improved soil samples.
Biases associated with the use of molecular techniques may under- or overestimate diversity (42). For example, inclusion of chimeric or heteroduplex sequences and divergent sequences from single bacteria which possess multiple rRNA copy numbers could result in overestimation of diversity. In all, 6 of 281 sequences were discarded as chimeras in this study. Conversely, underestimation of diversity may have occurred through lysis bias, unequal binding of PCR primers to different bacterial groups, or PCR selection. It must be noted, however, that all samples were treated identically to ensure, as far as possible, that any biases occurred to the same degree throughout the analysis. In addition, steps were taken to reduce bias; these steps included addition of low template DNA concentrations to PCR amplification mixtures to minimize PCR selection and pooling of multiple PCR products prior to cloning to minimize the effect of PCR drift (39).
Although differences in total bacterial diversity between unimproved and improved soil libraries were not observed, variation in the community structure of some bacterial groups was apparent. In particular, α-proteobacterial clones were more diverse in the unimproved soil samples than in the improved soil samples. Although there were probably multiple factors resulting in this difference, it is interesting that many α-proteobacterial clones isolated in this study are closely related to nitrogen-fixing bacteria, particularly Bradyrhizobium and Rhizobium spp. The improved Sourhope plots are dominated by L. perenne and the leguminous plant species T. repens, while the unimproved plots are dominated by nonleguminous species, such as A. capillaris, suggesting that selection for nitrogen-fixing bacteria may have occurred in the former plots.
In summary, our 16S rDNA sequence analysis did not detect significant differences in bacterial diversity in agriculturally important grassland soils which differed in constituent grass species, grazing intensity, and fertilizer application but did indicate that there were underlying differences in specific components of the populations which may have been related to differences in community function.
ACKNOWLEDGMENTS
We thank Christopher Clegg (Scottish Crop Research Institute, Invergowrie, United Kingdom) for providing DNA samples.
This work was carried out as part of the MICRONET project funded by the Scottish Office Agriculture, Environment and Fisheries Department (SOAEFD).
REFERENCES
- 1.Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microb Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 2.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer A, Williamson M. A new relationship for rarefaction. Biodiv Conserv. 1994;3:373–379. [Google Scholar]
- 6.Clegg C D, Ritz K, Griffiths B S. Direct extraction of microbial community DNA from humified upland soils. Lett Appl Microbiol. 1997;25:30–33. doi: 10.1046/j.1472-765x.1997.00166.x. [DOI] [PubMed] [Google Scholar]
- 7.Embley T M. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol. 1991;13:171–174. doi: 10.1111/j.1472-765x.1991.tb00600.x. [DOI] [PubMed] [Google Scholar]
- 8.Embley T M, Stackebrandt E. The use of 16S rRNA sequences in microbial ecology. In: Pickup W, Saunders J R, editors. Molecular approaches in environmental microbiology. London, United Kingdom: Ellis-Horwood; 1996. pp. 39–62. [Google Scholar]
- 9.Felsenstein J. PHYLIP: phylogeny inference package. Seattle: University of Washington; 1993. [Google Scholar]
- 10.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 12.Genetics Computer Group. Program manual for the Wisconsin Package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 13.Good I J. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–262. [Google Scholar]
- 14.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal rRNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 15.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 16.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak B L, Larsen N, McCaughey J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marilley L, Vogt G, Blanc M, Aragno M. Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil. 1998;198:219–224. [Google Scholar]
- 20.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 21.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 22.McVeigh H P, Munro J, Embley T M. Molecular evidence for the presence of novel actinomycete lineages in a temperate forest soil. J Ind Microbiol. 1996;17:197–204. [Google Scholar]
- 23.Mitsui H, Gorlach K, Lee H, Hattori R, Hattori T. Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J Microbiol Methods. 1997;30:103–110. [Google Scholar]
- 24.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 26.Neefs J-M, de Peer Y V, Rijik P D, Chapelle S, Wachter R D. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odum E P. Fundamentals of ecology. Philadelphia, Pa: Saunders College Publishing; 1971. Principles and concepts pertaining to organization at the community level; pp. 140–161. [Google Scholar]
- 29.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 30.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 31.Rodwell J S. British plant communities. Vol. 3. Cambridge, United Kingdom: Cambridge University Press; 1992. [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Seewaldt E, Schleifer K H, Bock E, Stackebrandt E. The close phylogenetic relationship of Nitrobacter and Rhodopseudomonas palustris. Arch Microbiol. 1982;131:287–290. [Google Scholar]
- 34.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 35.Stackebrandt E, Wehmeyer U, Schink B. The phylogenetic status of Pelobacter acidigallici, Pelobacter venetianus and Pelobacter carbinolicus. Syst Appl Microbiol. 1989;11:257–260. [Google Scholar]
- 36.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 37.Strunk O, Ludwig W. ARB: a software environment for sequence data, 2.1.1 ed. Munich, Germany: Department of Microbiology, Technical University of Munich; 1996. [Google Scholar]
- 38.Torsvik V, Sorheim R, Goksoyr J. Total bacterial diversity in soil and sediment communities: a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 39.Wagner A, Blackstone N, Cartwright P, Dick M, Misof B, Snow P, Wagner G P, Bartels J, Murtha M, Pendleton J. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst Biol. 1994;43:250–261. [Google Scholar]
- 40.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:220–286. [Google Scholar]
- 41.Ward-Rainey N, Rainey F A, Schlesner H, Stackebrandt E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology. 1995;141:3247–3250. [Google Scholar]
- 42.Wintzingerode F V, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 43.Woese C R, Weisburg W G, Paster B J, Hahn C M, Tanner R S, Krieg N R, Koops H-P, Harms H, Stackebrandt E. The phylogeny of the purple bacteria: the beta-subdivision. Syst Appl Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- 44.Woese C R, Weisburg W G, Hahn C M, Paster B J, Zablen L B, Lewis B J, Macke T J, Ludwig W, Stackebrandt E. The phylogeny of the purple bacteria: the gamma-subdivision. Syst Appl Microbiol. 1985;6:25–33. [Google Scholar]
- 45.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]