Abstract
To assess the distribution and diversity of members of the recently identified bacterial kingdom Acidobacterium, members of this kingdom present in 43 environmental samples were surveyed by PCR amplification. A primer designed to amplify rRNA gene sequences (ribosomal DNAs [rDNAs]) from most known members of the kingdom was used to interrogate bulk DNA extracted from the samples. Positive PCR results were obtained with all temperate soil and sediment samples tested, as well as some hot spring samples, indicating that members of this kingdom are very widespread in terrestrial environments. PCR primers specific for four phylogenetic subgroups within the kingdom were used in similar surveys. All four subgroups were detected in most neutral soils and some sediments, while only two of the groups were seen in most low-pH environments. The combined use of these primers allowed identification of a novel lineage within the kingdom in a hot spring environment. Phylogenetic analysis of rDNA sequences from our survey and the literature outlines at least six major subgroups within the kingdom. Taken together, these data suggest that members of the Acidobacterium kingdom are as genetically and metabolically diverse, environmentally widespread and perhaps as ecologically important as the well-known Proteobacteria and gram-positive bacterial kingdoms.
The use of rRNA sequence-based analysis of microbial populations has allowed study of complex communities in the environment without the requirement for laboratory cultivation of organisms. This approach has revealed astonishing diversity in many environments (3, 7, 12, 13, 17, 21, 23, 24, 30, 39). Several studies have analyzed over 100 sequences from a single environmental sample, and yet, remarkably, very little repetition has been seen among sequences, and virtually all previous studies have revealed novel groups with few or no known cultivated members. Most of these studies have identified new genera and even kingdoms (also termed “divisions” or “phyla” by other authors; “kingdoms” is here used in accordance with Woese [42]) of microorganisms, strongly suggesting that we as yet understand little about the total diversity of microbes in the environment (30).
One approach to studying complex ecosystems is to study subsets of the taxa present in hopes of better understanding the roles of individual groups within the larger community. During a recent survey of soil bacteria associated with pinyon-juniper woodlands in the southwestern United States, we recovered numerous 16S ribosomal DNA (rDNA) sequences that clustered into a diverse, novel group having only a single known cultivated member, Acidobacterium capsulatum (21). Analysis of “novel” sequences reported in studies of other soil samples (4, 23, 35, 37) revealed that many of them also clustered within this large group, suggesting that it was likely to be widespread in the environment. In addition, three extensive rDNA sequence analyses have indicated that the group is probably as phylogenetically diverse and distinct as previously recognized bacterial kingdoms and constitutes a previously unknown, major bacterial lineage (17, 21, 24). Such phylogenetically coherent groups are amenable to study by rDNA sequence-based techniques, because their relationships facilitate the design of group- or species-specific PCR primers and oligonucleotide probes for determination of their diversity, distribution, abundance, and other properties.
We have undertaken a PCR-based survey of a wide variety of samples representing terrestrial, marine, aerosol, subsurface, hot spring, and animal environments in order to understand more about the distribution and diversity of members of the kingdom Acidobacterium. Based on available 16S rDNA sequences, a PCR primer was designed and tested that specifically amplifies rDNAs from this group when used with a conserved reverse primer. Four additional primers, targeting four phylogenetic subgroups identified in our original study (21), also were designed and evaluated. These five primers were then used to interrogate bulk community DNA extracted from diverse environments. Concurrently, an analysis of sequences reported in the literature was performed in order to map the known phylogenetic diversity of the group, facilitate primer evaluation, and further investigate the environmental distribution of members of the four subgroups targeted in the PCR survey.
MATERIALS AND METHODS
Environmental sample collection and extraction of nucleic acids.
Marine snow and marine picoplankton (collected and purified by Ed DeLong, Monterey Bay Aquarium Research Institute, Monterey, Calif.) (34) and animal fecal DNAs (horse, pig and ostrich feces; extracted by Connie Gebhart, University of Minnesota) were provided as purified nucleic acids. Hot spring and temperate sediment samples were collected with sterile spatulas and corers into 50-ml polypropylene tubes, frozen immediately on dry ice, and stored at −70°C. Soil samples (approximately 1 kg) were collected into clean plastic bags for shipment to the laboratory. Upon receipt, soil samples were stored under refrigeration or frozen on dry ice until transferred to a freezer (−70°C) for long-term storage. Sample sources and descriptions are listed in Table 1. Chemical analysis of soil samples was performed by the Soil, Water and Air Testing Laboratory at New Mexico State University.
TABLE 1.
Presence of members of subgroups in environmental samples
| Source locationa | Source type | Sample typeb | pH | Reaction with primerc:
|
|||
|---|---|---|---|---|---|---|---|
| Y | O | G | A | ||||
| Soils | |||||||
| Sunset Crater, Ariz. | Volcanic | Cinders | 7.1 | + | + | + | + |
| Sunset Crater, Ariz. | Pinyon rhizosphere | Cinders | 7.1 | + | + | + | + |
| Cosnino, Ariz. | Pinyon woodland | Sandy loam | 7.6 | + | + | + | + |
| Cosnino, Ariz. | Pinyon rhizosphere | Sandy loam | 7.3 | + | + | + | + |
| Las Cruces, N. Mex. | Chile field | Sandy loam | 7.4 | + | + | + | + |
| Raleigh, N.C. | Meadow | Clay loam | 4.5 | + | + | + | + |
| Elk River, Minn. | Garden “black soil” | Organic | 6.5 | + | + | + | + |
| Paineville, Ohio | Agricultural | Sandy loam | 6.8 | + | + | + | + |
| Elk River, Minn. | Aged cow manure | Organic | 6.1 | + | + | + | + |
| Oslo, Norway | Boreal forest | Organic | NDd | + | +/− | + | + |
| Antelope Valley, Calif. | Citrus grove | Sandy loam | 6.1 | + | − | + | + |
| Dugway, Utah | Semiarid desert | Sandy clay loam | 7.7 | + | − | + | + |
| Elk River, Minn. | Gravel pit “white peat” | Silt organic | 7.2 | + | − | + | +/− |
| Niceville, Fla. | Pine forest | Sandy | 3.1 | − | − | + | + |
| Poncha Pass, Colo. | Roadside fill | Sandy | 2.5 | − | − | + | + |
| Berkeley Heights, N.J. | Hardwood forest | Organic | 4.0 | − | − | + | + |
| Ohio | Cranberry bog | Muck; rifle peat | 5.4 | − | − | +/− | + |
| Wilmington, N.C. | Coastal pine forest | Sandy loam | 3.2 | − | − | − | + |
| Sediments and mats | |||||||
| Los Alamos, N. Mex. | Marsh (anaerobic) | Organic | 6.5 | + | + | + | + |
| Abiquiu, N. Mex. | River (anaerobic) | Organic | ND | + | +/− | + | +/− |
| Los Alamos, N. Mex. | Shallow lake | Organic | 5.8 | + | − | + | + |
| Santa Rosa I., Fla. | Shallow marine | Sandy | 7.4 | − | − | + | +/− |
| Pine Barrens, N.J. | Cedar swamp | Organic | Acidic | − | − | − | + |
| Pine Barrens, N.J. | Hardwood forest | Streambed | Acidic | − | − | − | + |
| YNP | 71°C hot spring | Sediment | 7 | − | + | − | + |
| YNP | 67°C hot spring | Mat | 7 | − | + | − | − |
| YNP | 64°C hot spring | Mat | 7 | − | + | − | − |
| YNP | 67°C hot spring | Mat | 9 | − | − | − | − |
| YNP | 77°C hot spring | Sediment | 8 | − | − | − | − |
| YNP | 75°C hot spring | Sediment | 7 | − | − | − | − |
| YNP | 72°C hot spring | Sediment | 1 | − | − | − | − |
| YNP | 70°C hot spring | Mat | 4 | − | − | − | − |
| YNP | 18°C spring | Mat | 1.5 | − | − | − | − |
| Other samples | |||||||
| Minneapolis, Minn. | Horse, pig, ostrich | Feces | ND | − | − | − | − |
| North Central Pacific Ocean | Water | Marine snow | ND | − | − | − | − |
| North Central Pacific Ocean | Water | Picoplankton | ND | − | − | − | − |
| Dugway, Utah | Aerosol | Filtrate | ND | − | − | − | − |
| Los Alamos, N. Mex. | Aerosol | Filtrate | ND | − | − | − | − |
| Albuquerque, N. Mex. | Aerosol | Filtrate | ND | − | − | − | − |
| Artesia, N. Mex. | Aerosol | Filtrate | ND | − | − | − | − |
| White Sands, N. Mex. | Aerosol | Filtrate | ND | − | − | − | − |
| Lechugilla Cave, N. Mex. | Cave wall rock | Corrosion residue | ND | − | − | − | − |
YNP, Yellowstone National Park, Wyo. Additional source information available from the authors upon request.
Mat, photosynthetic microbial mat. Marine snow and picoplankton samples are described in reference 34; the cave corrosion residue is described in reference 6.
+, visible PCR product band present on ethidium bromide-stained agarose gel; +/−, faint band present, or visible band present only in reamplification reactions; −, no product band of correct size visible in any reaction.
ND, not determined.
Nucleic acids were extracted and purified from approximately 0.5-g (or 0.5-ml) aliquots of each sample by using procedures described by Kuske et al. (20). Each sample was placed in a 2-ml screw-cap tube, and 0.3 g of each of three sizes of glass beads (710 to 1,180, 425 to 600, or 106 μm in diameter; Sigma, St. Louis, Mo.) was added to each sample. TENS buffer (50 mM Tris [pH 8.0], 20 mM disodium EDTA, 100 mM NaCl, 1% [wt/vol] sodium dodecyl sulfate) was added to fill each tube, vortexed to mix thoroughly, and incubated in a 70°C water bath for 40 min. Samples were mixed well at 10-min intervals during the incubation. After incubation, samples were homogenized at 5,000 rpm for 3 min in a mini bead beater cell disrupter (type BX-4; Bio-Spec Products, Bartlesville, Okla.) and then centrifuged at 12,000 × g for 10 min to pellet the bead mix. The supernatant was transferred, and the bead pellet was washed once with 1 ml of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) and centrifuged again. The wash supernatant was pooled with the original supernatant. Nucleic acids were precipitated from the solution by using 0.1 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol, incubation on ice, and centrifugation for 30 min at 12,000 × g. Precipitated nucleic acids were suspended in TE. DNA concentration was estimated in ethidium bromide-stained 3% SeaKem agarose gels (FMC Bioproducts, Rockland, Maine) by using lambda DNA as a calibration standard or by solution quantitation with PicoGreen dye (Molecular Probes, Eugene, Oreg.) (20). DNA was purified away from contaminants by using Sephadex G-200 spin columns (in 96-well coarse polypropylene filter plates; Advanced Genetic Technology Corp., Gaithersburg, Md.) equilibrated in TE, as described previously (20). The clear column eluate containing DNA was precipitated and suspended in TE buffer, and the DNA concentration was determined as described above. Negative control samples were prepared with TENS buffer alone, with no sample addition, and proceeding with all extraction and purification steps described above.
PCR amplification of small subunit rRNA genes from plasmids and environmental DNAs.
Preparation of plasmid DNAs from rDNA clones from two soils of the arid Southwest United States is described by Kuske et al. (21). The forward primer sequences used in amplification reactions are listed in Table 2, together with optimum annealing temperatures used in thermal cycling reactions. Annealing temperatures were optimized by amplification of 56 Acidobacterium kingdom and other bacterial kingdom rDNA clones characterized in our previous study (21). The optimum temperature was determined to be the lowest temperature at which amplification of target group clones, and no amplification of nontarget clones, was observed. For all reactions, the reverse primer used was the “universal” 1492R primer (22). Amplification reaction mixtures contained 30 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 5 μg of bovine serum albumin (Boehringer Mannheim), 200 μM (each) deoxynucleoside triphosphates, 25 pmol of each primer, and 1 U of Taq polymerase (AmpliTaq LD; Perkin-Elmer, Foster City, Calif.) in a final reaction volume of 25 μl. For primer testing, 100 pg of plasmid DNA per reaction was used. For amplification of environmental DNAs, approximately 0.1 to 1 ng of bulk DNA (where purified DNA concentrations were sufficient) was used. PCR was conducted with a Perkin-Elmer 9600 thermal cycler as follows: 2 min of denaturation at 94°C, followed by 30 cycles of 30 s at the optimized annealing temperature (Table 2), 60 s at 72°C (extension), and 5 s at 94°C (denaturation), with a final 5-min 72°C extension step after cycling was complete. For reamplification reactions, products from reactions with primer set 31F-1492R were diluted in serial 10-fold dilutions, and 1 μl each of the 101 and 102 dilutions was used to template reamplification reactions with one or more subgroup-specific primers. Five microliters of each reaction mixture was analyzed on 1 or 1.5% SeaKem agarose gels, and DNA was visualized by ethidium bromide staining and UV illumination. All samples were tested at least twice with each primer set to confirm results.
TABLE 2.
Forward rDNA PCR primers used in the survey of environmental DNA samples
| Phylogenetic group | Primer (reference)a | Nucleotide sequence (5′→3′)b | Target regionc | Optimum annealing temp (°C)d |
|---|---|---|---|---|
| All bacteria | pA (10) | AGAGTTTGATCCTGGCTCAG | 8–27 | 42 |
| Kingdom Acidobacterium | 31F | GATCCTGGCTCAGAATC | 15–31 | 42 |
| Subgroupse | ||||
| A | A | GCCTGAGAGGGCRC | 293–306 | 50 |
| G | G | CGCAAGCCTGACGAC | 379–393 | 60 |
| O | O | CGACGGTACCTTGCGT | 480–497 | 57 |
| Y | Y | GGTACYGTTTGTAAGSTC | 484–503 | 57 |
All primers, except pA, were designed in this study.
R, mixture of G and A (1:1); Y, mixture of C and T (1:1); S, mixture of C and G (1:1).
Positions correspond to E. coli nucleotide numbering (14).
Optimum PCR annealing temperature for specificity, when used with the 1492R reverse primer.
Subgroups as indicated in Fig. 1.
rDNA gene clones from a Yellowstone hot spring sample.
A clone library of small subunit rRNA gene copies was generated from a photosynthetic mat sample from the unnamed hot spring designated “GFP.” PCR products (1.5 kb) from 31F-1492R amplification reactions were ligated into pGEM-T plasmid vectors (Promega, Madison, Wis.) using T4 DNA ligase and overnight incubation at 16°C, according to the manufacturer’s protocols. Recombinant plasmids were transformed into Escherichia coli JM109 competent cells (Promega), and colonies containing plasmids with inserts were identified by blue/white color selection on agar plates (32). Individual white colonies were subjected to PCR of insert sequences to select clones with the correct size insert (11). Clones were screened by restriction fragment length polymorphism (RFLP) analysis of PCR products, using the restriction enzymes HhaI and RsaI (28). PCR products from seven clones, representative of each of seven RFLP types identified, were purified with PCR Prep columns (Qiagen, Inc., Chatsworth, Calif.). One microgram of purified DNA was used as a template in cycle sequencing reactions with fluorescent dye-labeled terminators (ABI PRISM dye terminator cycle sequencing kit; Perkin-Elmer). Primers used for sequencing (Table 2) were pA (9), 533FU (22), P3MOD (5′-ATTAGATACCCTDGTAGTCC-3′; E. coli bases 787 to 806) (40), the reverse complement of P3MOD, EC910/931-RC (5′-CTCAAAGGAATTGACGGGGGC-3′; E. coli nucleotides [nt] 931 to 910), and 1492R. Electrophoresis of sequencing reactions was performed by using 4.0% polyacrylamide gels on a 373A Stretch DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.). The nearly full-length sequence of clone GFP1 was analyzed for secondary structure anomalies and submitted to the CHECK_CHIMERA program of the Ribosomal Database Project (RDP) (25) to detect the presence of possible chimeric artifacts. Sequence data for clone GFP1 have been deposited in the NCBI database under accession no. AF130858.
Phylogenetic analysis of sequence data.
Sequences were obtained from GenBank, the RDP, and unpublished sources. In some cases, affiliation with the kingdom Acidobacterium was tested with SIMILARITY_RANK (25) and FASTA (Wisconsin Package version 9.1; Genetics Computer Group [GCG], Madison, Wis.) analyses of the RDP and GenBank databases, respectively. Alignment of sequences was accomplished with Clustal W (36) and manually with the GDE multiple sequence editor (25), by using conserved primary sequence regions and secondary structure folding as guidelines. Sequence regions which could not be aligned with confidence were excluded from analyses.
Maximum likelihood (ML) analyses were performed by using fastDNAml (version 1.1; distributed by the RDP [29]) with empirical base frequencies, and a transition/transversion ratio (T) of 1.5, optimized by comparing likelihoods under T = 1.0 to 2.0. The tree of highest likelihood was found by repeated tree building with random sequence input orders and optimized by global rearrangement of branches. Distance matrix (DM) analysis of sequences was performed with PAUPSearch* (a version of D. Swofford’s PAUP, distributed with the Wisconsin Package version 9.1), by using the hky85 distance correction. Unweighted maximum parsimony (MP) analyses were also performed with PAUPSearch*. DM and MP trees were constructed by using 10 rounds of random sequence addition order and optimized by tree bisection-reconstruction branch swapping. Bootstrap analysis by MP and DM methods was performed with 100 resampled data sets. Insertion of partial sequences into an optimized ML tree of longer sequences was performed 100 times with the “restart” script of fastDNAml (29), with random addition of taxa, without rearrangement of branches in the starting tree. The tree of highest likelihood from this analysis is shown in Fig. 1. The alignment used to generate the tree of Fig. 1 is available online (23a).
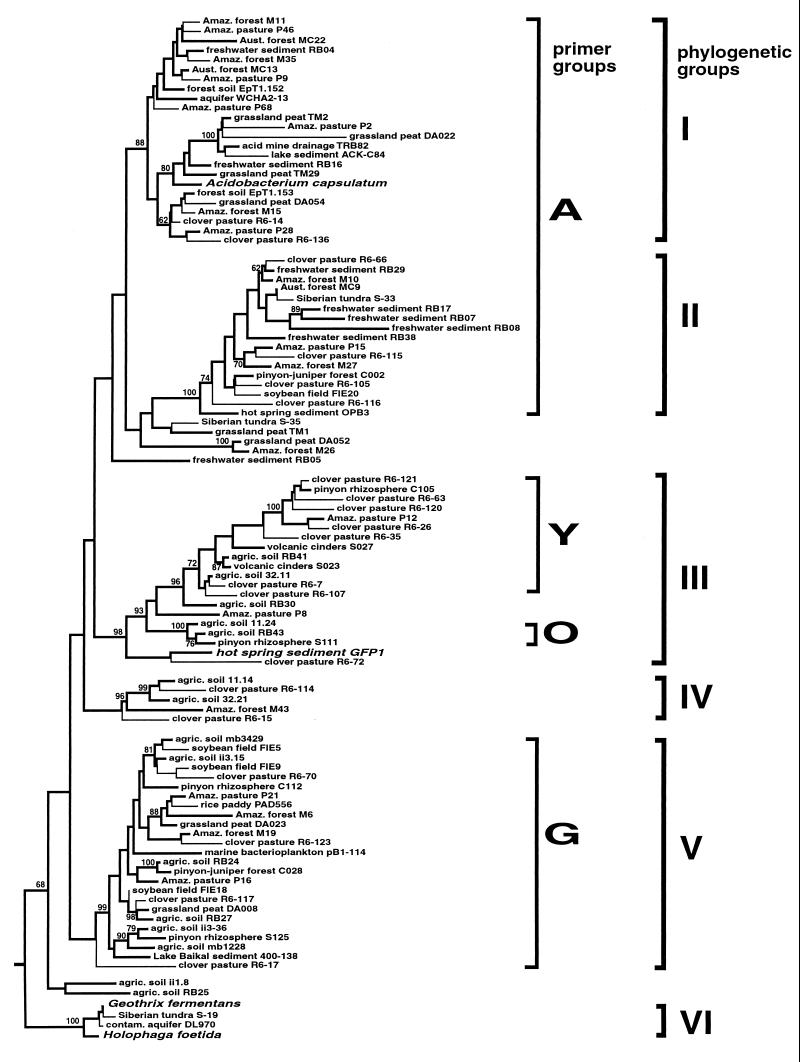
FIG. 1.
Phylogenetic tree representing the known diversity of the kingdom Acidobacterium. Sequences (other than GFP1) were obtained from databases and unpublished sources and analyzed as described in the text. The source references for the sequences shown are as follows: Amaz. (Amazon) forest/Amaz. pasture, 5; Aust. (Australian) forest, 23 and 35; freshwater sediment, 41; forest soil, 26; grassland peat (TM sequences), 31; grassland peat (DA sequences), 10; acid mine drainage, 8; lake sediment, 15; clover pasture, 4; Siberian tundra, 43; pinyon-juniper forest/pinyon rhizosphere/volcanic cinders, 21; hot spring sediment (OP), 17; agricultural soil, 24; rice paddy, 37; and contaminated aquifer, 1. Others are unpublished sequences from GenBank. Bootstrap values for MP analysis are given for branches having >60% support. Phylogenetic groups are designated based on primary branching clades appearing in all (ML, MP, and DM) optimal trees and having >85% bootstrap support, based on long (>900 nt) sequences (heavy lines). Partial sequences (thin lines) represent sequences added to the optimized ML tree without global rearrangement. The tree was rooted with the rRNA sequence of E. coli. The primer groups are those sequences expected to amplify with the primers listed in Tables 1 and 2.
RESULTS AND DISCUSSION
Design and specificity of Acidobacterium kingdom- and subgroup-specific primers.
The initial design of the PCR primers used in this study was guided by available kingdom Acidobacterium rDNA sequences. A signature nucleotide (G) for the Acidobacterium kingdom was identified, corresponding to nt 31 of the E. coli sequence (14). This specificity was confirmed by using the CHECK_PROBE function of the RDP (25). Amplification of archaeal or eukaryotic rDNA sequences by a primer targeting this region is very unlikely, due to numerous mismatches with sequences in these groups. A forward PCR primer (designated 31F) was designed, utilizing position 31 at its 3′ end. Using 1492R as the reverse primer, 31F was tested against 56 rDNA clones representative of several bacterial kingdoms (21). The results (not shown) indicated that the primer is specific and inclusive for clones of the kingdom Acidobacterium, producing a single strong product in positive reactions and no product of any size with clones from outside the kingdom. In a similar fashion, primers for four subgroups of the kingdom identified in our original analysis (designated Y, O, A, and G) (21) were tested against the clone collection and found to be specific (data not shown). Sequences and optimized annealing temperatures for kingdom and subgroup primers are given in Table 2.
Distribution of members of the kingdom Acidobacterium.
Our PCR-based survey of diverse environmental samples demonstrated that members of the kingdom Acidobacterium are widespread in the environment. Environmental DNAs were first tested for their ability to support PCR. All extracted and purified DNAs were found to support PCR in positive control reactions using primers pA and 1492R, with the exception of a red clay which also yielded no visible DNA by agarose gel analysis (data not shown), possibly due to binding of DNA to clay particles during extraction. Negative control extractions (no input sample) gave no product when amplified with any primer set.
Amplification of 43 environmental DNAs with the Acidobacterium kingdom primer set (31F-1492R) yielded either a single visible band of the anticipated size (approximately 1.5 kb) or no visible product when analyzed on agarose gels (not shown). All soils and freshwater and marine sediments tested gave positive results with this primer set. In addition, several hot spring sediments and mats, from springs of neutral to alkaline pH and up to 71°C in temperature, were also positive with the kingdom-specific primer set. These soils and sediments were of diverse lithology, chemistry, temperature, and location, from both pristine and agricultural settings (Table 1). Soils and sediments that were chemically analyzed varied considerably in their texture, pH (Table 1), percent organic matter (0.5 to 84%), and nitrate nitrogen (1.5 to 497 ppm). Concentrations of mono- and divalent cations also varied over a wide range (Mg, 0.2 to 534 meq/liter; Ca, 0.21 to 9,510 meq/liter; Na, 0 to 137 meq/liter). Samples which did not yield products with the kingdom-specific primer (listed in the bottom section of Table 1) include acidic hot spring sediments, hot spring sediments with temperatures over 71°C, aerosol samples from five locations in the Southwest, animal feces (horse, pig, and ostrich), Pacific Ocean marine snow and marine picoplankton (34), and cave “corrosion residues” (6).
Members of this group have also been detected in several studies of soils and sediments by other researchers (4, 10, 15, 23, 24, 28, 35, 37, 41, 43). Additional environments reported in the literature from which Acidobacterium sequences have been obtained include lake snow and activated sludge (24), and benzene-contaminated aquifer material (1). This ubiquity supports the suggestion (24) that members of this kingdom are capable of growth in many different environments. Although it is conceivable that these organisms occupy the same microniche in many different habitats, the broad phylogenetic diversity of rDNA sequences in the Acidobacterium kingdom, comparable to that seen in members of the Proteobacteria and other bacterial kingdoms, indicates that members of this group are likely to be quite diverse physiologically and ecologically. Temperature gradient gel electrophoresis (10) and in situ hybridization with rRNA-targeted probes (24) have indicated that these organisms may be abundant and active in some environments as well.
Distribution of phylogenetic subgroups in environmental samples.
To map the distribution of members of four phylogenetic subgroups identified in our original analysis (21), primers Y, O, G, and A were designed for these groups. All environmental DNAs, regardless of their reaction with the 31F primer, were screened with these primers (Table 1). Those which gave no product with the 31F-1492R primer pair were also negative with the subgroup primers, as anticipated. In order to confirm negative subgroup reactions, reamplification of products from positive 31F-1492R (kingdom-specific) reactions was performed with the subgroup primers. Three soils produced faint products when reamplified with subgroup primers; however, in most cases, reamplification did not change the previous results.
The results given in Table 1 indicate that members of the A subgroup are possibly ubiquitous in soils and marine and freshwater sediments and are also present in some hot spring communities. Members of the G subgroup are also very widespread, but were not found in the hot spring samples analyzed. Fewer samples produced amplification products with the Y and O primers. These results concur with other cultivation and rDNA sequence-based surveys of microbial distribution, which have both found that phylogenetically related groups of organisms vary in their ubiquity (13, 33). As a result, some groups of organisms are widely distributed in the environment, while others are highly restricted to particular habitat types.
For those groups of more limited apparent distribution, as measured by our PCR survey, it may be possible to infer some physiological properties based on the nature of their habitats. For instance, members of the Y and O subgroups are largely absent from acidic soils and sediments (<pH 6); thus, neutrophily may be a common property of these groups. Some members of the O group are evidently thermophilic or thermotolerant, because they are present in hot spring environments up to 71°C. The observed distribution of members of these groups is probably determined to some extent by the phylogenetic breadth of PCR primer sequences as well. Mapping of the target sequence distribution onto the tree of Fig. 1 shows that the A group primer is likely to amplify rDNA sequences from a much more diverse (phylogenetically and probably physiologically) selection of organisms than the individual G, O, and Y primers. This is unlikely to explain all of the results observed, however, because the sequences of the combined Y and O groups appear as diverse as those of the G group, and yet are seen in fewer environments. In general, however, it is likely that the organisms detected by the primers used in this study were too diverse to be restricted to distinct habitat types.
The PCR approach to survey for the presence of specific groups of organisms is subject to the biases which may affect all rDNA-based studies of environmental samples (19, 38). Most relevant to this study is differential lysis of diverse cell types (20, 27), which may contribute to negative results in this survey. However, since members of the kingdom Acidobacterium are probably gram negative in cell type (18) and an aggressive lysis technique was used to obtain DNA (20), it seems likely that negative PCR results with environmental samples truly indicate the absence of members of the target group.
Identification of a novel lineage in a hot spring sediment.
DNA from the 67°C hot spring GFP gave strong positive results in kingdom Acidobacterium primer set (31F-1492R) amplifications, but was negative with all four subgroup primers. To investigate this result further, PCR products from the 31F-1492R reaction were cloned, and seven clones with distinct RFLP patterns were partially sequenced and found to be >99% identical through the approximately 400 nt sequenced. The nearly complete (approximately 1,400 nt) sequence of one clone, GFP1, was generated. Phylogenetic analysis of GFP1 places it as a deep branch within the kingdom Acidobacterium, related to group IV. This subtractive PCR approach has thus enabled targeted identification of new phylogenetic diversity within the kingdom, without prior information on what form that diversity might take.
Phylogenetic analysis of kingdom Acidobacterium rDNA sequences.
An analysis of sequences representative of the currently known phylogenetic and environmental diversity of the kingdom Acidobacterium is presented in Fig. 1. For initial analysis, sequences of greater than 900 nt were selected (represented by thick lines on Fig. 1). These were used to construct and optimize trees by the DM, MP, and ML methods and to perform bootstrap analyses. Groups that were moderately to well supported by bootstrap evaluations (>70% support) were also seen in all optimal trees. The tree of longer sequences was used as a backbone into which shorter sequences (thin lines in Fig. 1) were placed without rearrangement of the starting tree. These shorter sequences were chosen to span the phylogenetic diversity of sequences obtained in other studies. From this analysis, it can be seen that most of the partial sequences cluster within the primary groups outlined in the starting tree, although a few deeply branching sequences (Amazon pasture P68; Siberian tundra S-35; and clover pasture R6-15, R6-17, and R6-72) may constitute distinct lineages.
Our initial study of two arid soils (21) recovered 31 kingdom Acidobacterium rDNA sequences, all of which could be grouped into four phylogenetic subgroups having moderate to good bootstrap support. Most sequences in Fig. 1 still appear to cluster within the four groups originally identified, although the overall topology of the tree has changed somewhat. A minimum of six primary groups (I to VI in Fig. 1) within the kingdom Acidobacterium are now apparent. The branching order of these within the larger kingdom cannot be resolved from this analysis, except for the Holophaga-Geothrix lineage (group VI), which was consistently the deepest group. Several additional deeply branching lineages of variable placement were observed (grassland peat TM1, grassland peat DA052-Amazon forest M26, agriculture soil RB05, and agriculture soil RB25/ii 1.8) as well, and these may represent primary groups in their own right. Doubtless the number and diversity of such groups will continue to grow rapidly in the near future as rDNA sequences from additional environments accumulate. However, there is already evidence that the kingdom Acidobacterium harbors at least as much genetic, and probably metabolic, diversity as any previously identified bacterial kingdom.
Since our initial report (2) and the initiation of the studies described here, many additional sequences from the kingdom Acidobacterium have been obtained from diverse environments. Remarkably, over 250 such sequences are now known. Comparison of the sequence set used in construction of the tree of Fig. 1 with the sequences of the kingdom and subgroup primers indicates that several deeply branching groups would not be detected by these primers. Most significant of these is the Holophaga-Geothrix group. Although phylogenetic analyses have indicated membership of this group within the kingdom Acidobacterium (16, 17, 24), albeit as the deepest branch, these sequences lack the novel residue responsible for the specificity of the 31F primer. Addition of new sequences to the known kingdom diversity has not, however, changed the specificity of the four subgroup primers.
The kingdom Acidobacterium is an example of a vast group of organisms that were virtually unknown prior to rDNA sequence-based surveys. Suddenly, we are aware that they are diverse and widespread and may be very important in many environments. It has become evident that the microbial communities present in soils and sediments are too diverse to be comprehensively described by general 16S rDNA cloning approaches. Yet it is equally evident that such communities may be dominated by organisms that are only distantly related to species which have been studied to any extent in the laboratory. Molecular sequence-based techniques, therefore, will likely play an important role in providing information about diversity and function in these communities.
ACKNOWLEDGMENTS
We thank the following generous people who provided samples from far and wide for the PCR survey: Martha Barns, Steven Koch, Jody Davis, Kaysie Banton, Darryl Ricke, George Schneider, Ed DeLong, Connie Gebhart, Jack Kuske, Ann O’Leary, Patricia Maurice, Paul Jackson, Diana Northup, and Phil Hugenholtz. We also thank John Dunbar and Karen Hill for helpful comments on the manuscript.
This work was supported by a J. Robert Oppenheimer Postdoctoral Fellowship to S.M.B. and the U.S. Department of Energy, Office of Health and Environmental Research Program for Ecosystem Research.
REFERENCES
- 1.Anderson R T, Rooney-Varga J N, Gaw C V, Lovely D R. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum-contaminated aquifers. Environ Sci Technol. 1998;32:1222–1229. [Google Scholar]
- 2.Barns S M, Busch J D, Kuske C R. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Molecular characterization of pinyon pine-associated soil microbial communities at Sunset Crater, AZ, abstr. N-198; p. 413. [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham K I. Organic and inorganic composition of colored corrosion residues: Lechuguilla Cave: preliminary report. NSS News. 1991;49:252–254. [Google Scholar]
- 7.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, K. J., B. M. Goebel, T. M. Rodgers, M. O. Schrenk, T. M. Gihring, M. M. Cardona, B. Hu, M. M. McGuire, R. J. Hamers, N. R. Pace, and J. F. Banfield. Geomicrobiology of pyrite (FeS2) dissolution: a case study at Iron Mountain, California. Geomicrobiol. J., in press.
- 9.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete nucleotide determination of entire genes. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frothingham R, Allen R L, Wilson K H. Rapid 16S ribosomal DNA sequencing from a single colony without DNA extraction or purification. BioTechniques. 1991;11:40–44. [PubMed] [Google Scholar]
- 12.Fuhrman J A, McCallum K, Davis A A. Novel marine archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni S J, Rappe M S, Gordon D, Urbach E, Suzuki M, Field K G. Ribosomal RNA and the evolution of bacterial diversity. In: Roberts D M, Sharp P, Alderson G, Collins M A, editors. Evolution of microbial life. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 63–85. [Google Scholar]
- 14.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiorns W D, Methé B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishimoto N, Kosako Y, Tano T. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol. 1991;22:1–7. doi: 10.1007/BF01576009. [DOI] [PubMed] [Google Scholar]
- 19.Komatsoulis G A, Waterman M S. A new computational method for detection of chimeric 16S rRNA artifacts generated by PCR amplification from mixed bacterial populations. Appl Environ Microbiol. 1997;63:2338–2346. doi: 10.1128/aem.63.6.2338-2346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuske C R, Banton K L, Adorada D L, Stark P C, Hill K K, Jackson P J. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl Environ Microbiol. 1998;64:2463–2472. doi: 10.1128/aem.64.7.2463-2472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 23.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Los Alamos National Laboratory. 19 March 1999, expected posting date. Sequences. [Online.] http://www.ls.lanl.gov/lsdiv/LS7/Acidobacterium/Acidoseqs.html.
- 24.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McVeigh H P, Munro J, Embley T M. Molecular evidence for the presence of novel actinomycete lineages in a temperate forest soil. J Ind Microbiol. 1996;17:197–204. [Google Scholar]
- 27.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 30.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 31.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schelgel H G, Jannasch H W. Prokaryotes and their habitats. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 75–125. [Google Scholar]
- 34.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap weighting and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 38.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 39.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiol Rev. 1990;75:105–116. doi: 10.1111/j.1574-6968.1990.tb04088.x. [DOI] [PubMed] [Google Scholar]
- 40.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Davey M E, Figueras J B, Rivdina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]