Abstract
Background
Omics approaches are widely applied in the field of biology for the discovery of potential CAZymes including whole genome sequencing. The aim of this study was to identify protein encoding genes including CAZymes in order to understand glycans-degrading machinery in the thermophilic Caldicoprobacter algeriensis TH7C1T strain.
Results
Caldicoprobacter algeriensis TH7C1T is a thermophilic anaerobic bacterium belonging to the Firmicutes phylum, which grows between the temperatures of 55 °C and 75 °C. Next generation sequencing using Illumina technology was performed on the C. algeriensis strain resulting in 45 contigs with an average GC content of 44.9% and a total length of 2,535,023 bp. Genome annotation reveals 2425 protein-coding genes with 97 ORFs coding CAZymes. Many glycoside hydrolases, carbohydrate esterases and glycosyltransferases genes were found linked to genes encoding oligosaccharide transporters and transcriptional regulators; suggesting that CAZyme encoding genes are organized in clusters involved in polysaccharides degradation and transport. In depth analysis of CAZomes content in C. algeriensis genome unveiled 33 CAZyme gene clusters uncovering new enzyme combinations targeting specific substrates.
Conclusions
This study is the first targeting CAZymes repertoire of C. algeriensis, it provides insight to the high potential of identified enzymes for plant biomass degradation and their biotechnological applications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-022-01818-0.
Keywords: Caldicoprobacter algeriensis TH7C1T; CAZymes, CGC; Biotechnological applications
Background
The Carbohydrate Active Enzymes (CAZymes) are enzymes involved in the assembly, modification or deconstruction of carbohydrates [1]. Based on amino acid sequence similarities, CAZymes are divided into several classes, including glycosyltransferases (GT) [2, 3], glycoside hydrolases (GH) [4–6], polysaccharide lyases (PL) [7, 8], carbohydrate esterases (CE) [8], and auxiliary activities (AA) [9] that have been stored in the CAZy database. The huge diversity of natural glycans and their complexity has boosted studies uncovering novel CAZymes. Thus, the number of CAZymes families increases exponentially by about four new GH families per year [10]. This broad diversity has allowed their use in plenty of industrial applications as they have been described to offer attractive opportunities in a wide range of biotechnological applications such as animal feed, biocatalysis, agriculture, biorefinery, glycoengineering and biobleaching industries [11–16]. Along with classical methods, various omics approaches are presently applied in the field of biology for the discovery of potential CAZymes. This “omics” technologies include proteomics, transcriptomics, metagenomics, metabolomics and whole genome sequencing [13, 17, 18].
The systematic genome sequencing has largely fueled the discovery of novel plant biomass degrading enzymes [10]. Studies have shown that bacteria and fungi are the main producers of CAZymes in nature. Among them extremophilic microorganisms have received special attention because of their capacity to live in extreme conditions such as high temperature, pressure, alkalinity, acidity, or salinity, thanks to their corresponding extremozymes [19]. Owing to their robustness, extremozymes are capable to function under harsh conditions more effectively than enzymes from other microorganisms [20]. Accordingly, thermophilic enzymes offer great potential for application in biotechnology, opening the possibility of performing biocatalysis at higher temperatures that can be more beneficial in some industrial settings [21, 22]. Thus, the study of thermophilic microorganisms have emerged during recent years including genome profiling and exploration of CAZymes content [23]. It has been demonstrated that carbohydrate acting enzymes works in conjunction with other CAZymes and proteins forming clusters of physically linked genes called polysaccharide utilization loci (PULs) [24–26]. These clusters that occur in bacteria of bacteroidetes phylum have been progressively identified in firmicutes phylum [27].
The thermophilic anaerobic Caldicoprobacter algeriensis TH7C1T strain was isolated from the hydrothermal hot spring of Guelma. It was classified as a novel species in Calidicoprobacter genus [28] and was demonstrated to produce some thermophilic enzymes [29, 30]. However, its exploitation, in particularly discovery of enzymes content such as CAZymes, was hampered by culture limitations, anaerobic and high optimal temperature (65 °C).
In order to understand plant biomass-degrading machinery and to discover new potential interesting CAZymes for biotechnological applications, we report, for the first time, the genome sequence of C. algeriensis TH7C1T. Furthermore, we report the prediction of CAZyme encoding genes as well as the identification of clusters acting on polysaccharides.
Results
Genome sequence and analysis
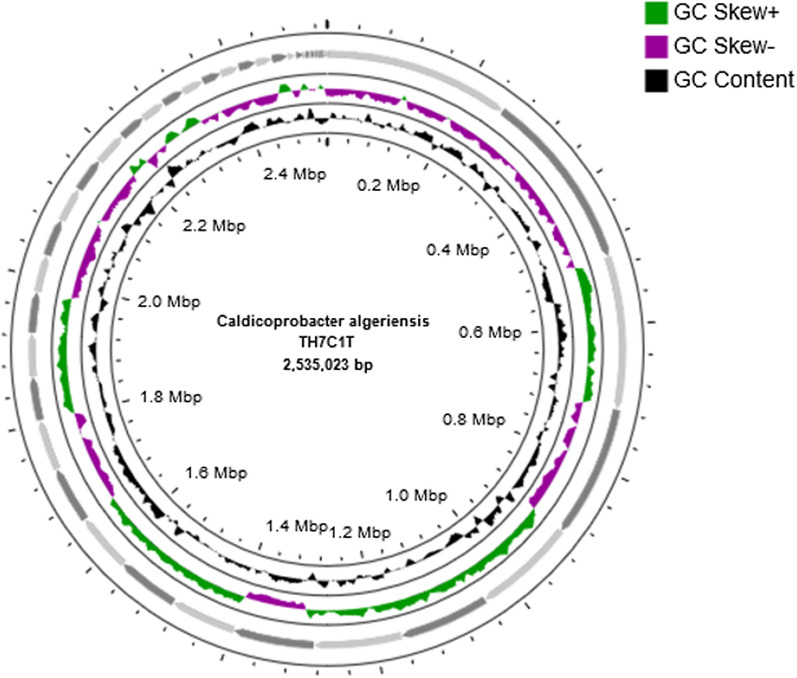
The genome sequencing of C. algeriensis TH7C1T rendered 473,434 Illumina reads with an average coverage of 34.55x. The de novo assembly resulted in 45 contigs and a total length of 2,535,023 bp (Accession number PRJNA743054) with an overall GC content of 44.9%. A circular genome map of C. algeriensis was constructed, showing contigs, GC content, and GC skew (Fig. 1).
Fig. 1.
Graphical Circular genome map of Caldicoprobacter algeriensis TH7C1T generated using CGView. From outside to inside, ring 1 represents the 45 assembled contigs. Ring 2 shows the GC skew with green indicating positive values and purple indicating negative values. The GC content is represented by the inner most ring
The overall genome statistics of C. algeriensis are close to those from Caldicoprobacter faecalis, Caldicoprobacter oshimai and Caldicoprobacter guelmensis (Table 1).
Table 1.
Comparison of genome features between C. algeriensis, C. faecalis, C. oshimai and C. guelmensis
| Caldicoprobacter species | C. algeriensis | C. faecalis | C. oshimai | C. guelmensis |
|---|---|---|---|---|
| Total Sequence Length (bp) | 2,535,023 | 2,579,145 | 2,693,766 | 2,398,524 |
| Number of contigs | 45 | 87 | 427 | 20 |
| Longest Sequences (bp) | 255,969 | 158,877 | – | 505,220 |
| N50 (bp) | 121,172 | 78,525 | 10,681 | 394,366 |
| GC content (%) | 44.9 | 44.9 | 45.4 | 43.7 |
| Number of CDSs | 2425 | 2337 | 2618 | 2150 |
| Average Protein Length | 297.1 | – | – | – |
| Number of RNAs | 53 | 62 | 71 | 60 |
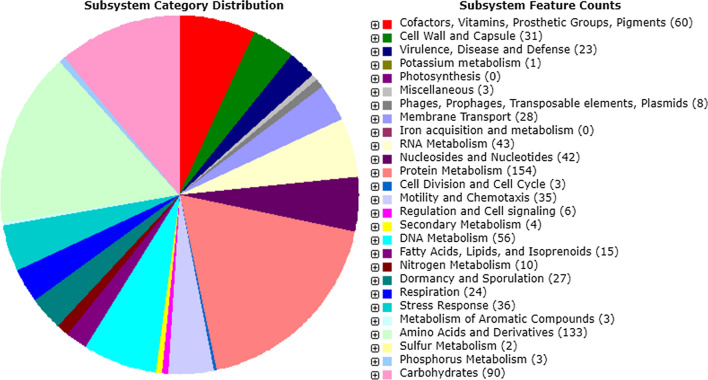
Gene prediction performed with the RAST server resulted in 2720 features including 2666 protein coding sequences (CDSs) classified in 226 SEED subsystems and 53 RNA genes. Figure 2 shows the subsystem category distribution following RAST annotation. The largest part of this subsystem is allocated to the Amino Acids and Derivatives, and Carbohydrate metabolism with 15.83% and 10.71%, respectively. Dfast annotation revealed 2425 protein coding sequences with CDSs and 53 RNA genes covering 85.3% of the genome, the average length of the CDSs is 297 bp.
Fig. 2.
An overview of the RAST annotation and subsystems distribution for the C. algeriensis genome
Analysis of genome stability using RAST and the CRISPRCasFinder server revealed two CRISPR array sequences located in contig 13 (with the evidence level of 4) and contig 2 (1 evidence level). This analysis revealed also three Cas cluster sequences detected in contig13 and related to CAS, CAS-TypeIIID and CAS-TypeIB. Another Cas cluster belonging to CAS-Type IE is located in contig 12.
CAZymes annotation
Sequences submitted to the dbcan server allowed the automated annotation of CAZymes using the HMMER3.0 package and the dbcan CAZyme database (see Additional file 1: Table S1). This analysis resulted in 97 genes associated with glycan assembly and breakdown. The most abundant enzymatic family predicted in this genome was glycoside hydrolases with 57 CAZyme encoding genes divided into 32 different families.
The highest number of glycoside hydrolases found in C. algeriensis was related to GH109 with 9 predicted encoding genes, followed by GH3 with 6 genes and GH2/GH13 with 4 genes. GH109 family, which contains members involved in the deconstruction of galactomannans was widely represented in this genome. Interestingly, CAZymes belonging to this family had not been identified as major catalysts in previous studies highlighting biomass-degrading potential in hot spring. The GH3 family is represented by 6 predicted enzymes for hemicellulose hydrolyzing and debranching activities such as glucosidase, xylosidase and glucanase. Interestingly, GH3 has been reported as the most abundant GH family for oligosaccharides degradation in hot spring ecosystems [31]. The other abundant glycoside hydrolases predicted in this genome were identified to belong to the GH2 and GH13 families catalyzing the degradation of oligosaccharides and starch, respectively.
The second most frequent enzyme family contained in this genome is the glycosyltransferases GT family (20 encoding genes). GTs are known to catalyze the transfer of sugar residues from activated donor molecules to saccharide or non-saccharide acceptor molecules to form glycosidic linkages. The finding corroborates the results of biomass-degrading enzyme potential exploration in hot spring ecosystems previously reported [31] demonstrating that glycoside hydrolases and glycosyltransferases are widespread groups of CAZymes present in thermophilic microbial communities.
The output from dbCAN2 also included multiple hits corresponding to carbohydrate esterases (CEs) represented with 6 predicted genes attributed to CE1, CE4 and CE9 families. CEs are enzymes acting on ester bonds in carbohydrates accelerating the degradation of polysaccharides and facilitating the access of glycoside hydrolases. The most abundant CEs in C. algeriensis genome belong to CE4 family acting on acetylated xylan and chitin. Members of CE1 and CE9 families are involved in xylan and acetylglucosamine hydrolyzing, respectively.
The remaining putative CAZyme detected has been attributed to polysaccharide lyases (PL) represented by only one predicted gene. This genome also encodes 14 carbohydrate-binding modules (CBM). The majority of predicted CBMs belong to CBM4 and CBM50. CBM4 encodes specific modules that recognize xylan, 1,3-glucan, 1,3-1,4-glucan, 1,6-glucan, and amorphous cellulose, while CBM50 proteins are responsible for binding of enzymes having cleavage activity of chitin or peptidoglycan. They were found associated to GH genes or other CBMs. CAZyme genes prediction as well as the protein encoding genes sequences are available in supplementary (Additional files 1: Table S1 and 2).
Fast blast hit of CAZyme encoding genes in the CAZy database was performed by querying the genome against DIAMOND from dbcan meta-server. This analysis showed an identity between 35 and 83% with their nearest neighbors (Table 2).
Table 2.
Comparison of predicted CAZymes of C. algeriensis with those available in CAZy database using DIAMOND tool in dbCAN
| Query ID | Best Blast hit ID | % identical | Length | Query Start | Query End | Subject Start | Subject End | E Value |
|---|---|---|---|---|---|---|---|---|
| NODE_1_98 | AFM44649.1|GH3|3.2.1.37|3.2.1.37 | 69.6 | 782 | 4 | 782 | 9 | 788 | 0.0e+00 |
| NODE_1_142 | QBQ55251.1|CBM50| | 41.6 | 878 | 1 | 867 | 19 | 889 | 4.3e−173 |
| NODE_2_26 | CAN65674.1|GH28| | 51.4 | 552 | 73 | 618 | 111 | 642 | 2.2e−158 |
| NODE_2_32 | AWP04987.1|GT13| | 37.9 | 939 | 10 | 854 | 100 | 1037 | 6.2e−172 |
| NODE_2_34 | AUS96821.1|GH4| | 83.7 | 436 | 3 | 438 | 2 | 437 | 4.1e−225 |
| NODE_2 _43 | QAV32893.1|GH3| | 65.8 | 719 | 6 | 717 | 4 | 722 | 4.0e−278 |
| NODE_2_48 | QTM98178.1|GH35| | 40.1 | 770 | 8 | 770 | 2 | 757 | 1.1e−172 |
| NODE_2_73 | QEX52072.1|GH29| | 70.4 | 422 | 1 | 422 | 3 | 424 | 2.3e−196 |
| NODE_2_91 | QBQ55156.1|CBM50| | 36.6 | 658 | 164 | 820 | 62 | 707 | 2.3e−112 |
| NODE_2_93 | ADH60031.1|GH67| | 76.9 | 684 | 5 | 686 | 10 | 690 | 0.0e+00 |
| NODE_2_112 | ADH61394.1|GH127| | 58.5 | 650 | 5 | 651 | 8 | 648 | 1.7e−235 |
| NODE_2_119 | AZR73767.1|GH1| | 63.7 | 443 | 6 | 447 | 6 | 445 | 2.3e−178 |
| NODE_2_195 | QTH41126.1|GH30_1| | 62.0 | 447 | 1 | 445 | 2 | 447 | 1.0e−175 |
| NODE_2_205 | QGG57161.1|GH2| | 63.2 | 560 | 1 | 560 | 1 | 550 | 4.9e−221 |
| NODE_3_3 | AQQ10327.1|CBM38|GH32| | 47.2 | 557 | 11 | 538 | 201 | 751 | 9.7e−152 |
| NODE_3_5 | BBH23071.1|GH151| | 37.5 | 677 | 4 | 672 | 2 | 670 | 1.1e−147 |
| NODE_3_18 | AEE96970.1|GH109| | 78.1 | 375 | 3 | 377 | 2 | 374 | 1.2e−180 |
| NODE_3_20 | AEE96984.1|GH148|GH30| | 62.0 | 900 | 28 | 922 | 1290 | 2181 | 0.0e+00 |
| NODE_3_28 | QHT63269.1|GH28| | 44.3 | 422 | 19 | 438 | 31 | 450 | 1.1e−103 |
| NODE_3_33 | AEE96972.1|GH106| | 53.6 | 1009 | 1 | 1007 | 1 | 999 | 0.0e+00 |
| NODE_3_34 | ALS29147.1|GH140| | 76.0 | 437 | 3 | 439 | 8 | 442 | 1.3e−213 |
| NODE_3_ 39 | AEM77894.1|GH2| | 60.4 | 835 | 1 | 834 | 1 | 823 | 0.0e+00 |
| NODE_3_88 | AHO16406.1|GH13_11| | 61.0 | 367 | 9 | 371 | 3 | 367 | 7.4e−123 |
| NODE_3_137 | SNX53392.1|GH42| | 72.2 | 684 | 5 | 688 | 3 | 686 | 0.0e+00 |
| NODE_3_151 | AEE96723.1|GH3| | 56.8 | 368 | 119 | 486 | 101 | 468 | 1.1e−120 |
| NODE_4_38 | SMX54901.1|GH38| | 59.6 | 1026 | 1 | 1024 | 1 | 1022 | 0.0e+00 |
| NODE_4_77 |
CCO21038.1|GH11|3.2.1.8| 3.2.1.8 |
52.3 | 622 | 5 | 625 | 24 | 634 | 3.7e−195 |
| NODE_4_111 | QNB44994.1|CBM34|GH13_39| | 60.7 | 646 | 6 | 650 | 5 | 649 | 2.6e−247 |
| NODE_4_112 | QCX33357.1|GT5| | 63.8 | 475 | 2 | 476 | 1 | 472 | 6.2e−182 |
| NODE_4_113 | QCX33360.1|CBM48|GH13_9| | 70.6 | 622 | 8 | 629 | 4 | 620 | 2.1e−278 |
| NODE_5_24 | AXM88146.1|GH65| | 52.0 | 782 | 1 | 777 | 1 | 778 | 1.1e−241 |
| NODE_5_25 | QUH28797.1|GH13_18| | 56.8 | 555 | 18 | 572 | 8 | 553 | 7.1e−185 |
| NODE_5_27 | AUS95782.1|GH51| | 73.7 | 490 | 1 | 490 | 1 | 490 | 4.9e−235 |
| NODE_5_125 | AZN38808.1|PL33_1| | 48.1 | 626 | 1 | 626 | 1 | 623 | 4.6e−185 |
| NODE_6_132 | QAT62855.1|GT8| | 64.6 | 268 | 1 | 268 | 1 | 268 | 2.6e−103 |
| NODE_7_1 | AEF18092.1|GH106| | 65.9 | 1005 | 1 | 1001 | 1 | 1001 | 0.0e+00 |
| NODE_7_17 | AWZ17133.1|GH13_30| | 41.4 | 1205 | 4 | 1159 | 406 | 1579 | 1.2e−247 |
| NODE_8_2 | AJO67863.1|GH3| | 71.2 | 712 | 20 | 731 | 11 | 721 | 0.0e+00 |
| NODE_8_11 | AEE96371.1|GT51| | 43.5 | 839 | 6 | 830 | 13 | 822 | 1.7e−183 |
| NODE_8_27 | ABB14581.1|GT51| | 40.4 | 774 | 38 | 801 | 11 | 757 | 2.1e−153 |
| NODE_8_48 | QCW79542.1|GH101| | 54.6 | 504 | 12 | 515 | 583 | 1068 | 1.4e−155 |
| NODE_9_39 | QSL64261.1|GT58| | 39.1 | 504 | 15 | 480 | 202 | 683 | 4.7e−105 |
| NODE_9_41 | AEE95239.1|GH38| | 61.6 | 921 | 19 | 938 | 42 | 961 | 0.0e+00 |
| NODE_9_43 | AEE97446.1|GH151| | 53.7 | 659 | 4 | 657 | 6 | 662 | 1.5e−215 |
| NODE_9_45 | QGT51146.1|GT28| | 55.1 | 365 | 1 | 364 | 1 | 365 | 3.1e−117 |
| NODE_9_60 | ACX65278.1|GH33| | 56.9 | 334 | 1 | 328 | 1 | 323 | 1.5e−110 |
| NODE_10_1 | ADU28965.1|CBM4|GH16_3| | 43.2 | 844 | 5 | 729 | 500 | 1333 | 8.0e−177 |
| NODE_10_ 30 | QVK18514.1|GH0| | 30.4 | 828 | 50 | 855 | 61 | 856 | 3.9e−110 |
| NODE_10_34 | AXR85444.1|GH9|3.2.1.151|3.2.1.4|3.2.1.6|3.2.1.151|3.2.1.4|3.2.1.6 | 54.9 | 539 | 153 | 685 | 1 | 533 | 4.1e−171 |
| NODE_10_35 | AEE95681.1|GH10| | 80.6 | 340 | 35 | 374 | 39 | 378 | 1.5e−175 |
| NODE_10_41 | QSQ07709.1|GH94|GT84| | 60.2 | 2866 | 1 | 2865 | 1 | 2857 | 0.0e+00 |
| NODE_11_1 | AEE95836.1|GH116| | 73.2 | 880 | 1 | 879 | 1 | 876 | 0.0e+00 |
| NODE_14_60 | AEV67980.1|GT4| | 70.3 | 391 | 2 | 392 | 1 | 391 | 1.4e−163 |
| NODE_17_1 |
QCT01556.1|CBM4|CBM54| GH16_3| |
71.5 | 256 | 199 | 454 | 440 | 695 | 1.1e−111 |
| NODE_17_20 | AXG41146.1|GH6| | 35.3 | 842 | 6 | 828 | 12 | 840 | 5.5e−128 |
| NODE_17_35 |
AFM44649.1|GH3|3.2.1.37 |3.2.1.37 |
68.6 | 784 | 4 | 784 | 9 | 789 | 0.0e+00 |
| NODE_18_30 | AXG41146.1|GH6| | 38.4 | 852 | 6 | 782 | 12 | 848 | 1.3e−128 |
| NODE_18_36 | BBH86227.1|GH2| | 60.4 | 1038 | 10 | 1041 | 11 | 1025 | 0.0e+00 |
| NODE_19_14 | AEE95407.1|GH2| | 73.7 | 582 | 1 | 582 | 1 | 571 | 2.1e−269 |
| NODE_21_5 | SNV81985.1|GT28| | 56.5 | 352 | 2 | 353 | 3 | 353 | 1.3e−117 |
| NODE_22_2 | BAZ22920.1|GT4| | 60.8 | 357 | 1 | 357 | 451 | 807 | 1.3e−131 |
| NODE_23_13 | AUS96695.1|GH31| | 78.6 | 774 | 1 | 774 | 1 | 773 | 0.0e+00 |
| NODE_25_1 | QUI22575.1|CBM50| | 38.5 | 589 | 7 | 593 | 34 | 621 | 9.5e−124 |
| NODE_25_5 | QMV43391.1|GH95| | 54.6 | 799 | 13 | 804 | 8 | 787 | 1.2e−259 |
| NODE_26_23 | AEE96522.1|GH50| | 51.8 | 467 | 1 | 464 | 65 | 528 | 4.2e−143 |
| NODE_30_9 | AEM77732.1|GH3| | 74.1 | 509 | 1 | 508 | 17 | 525 | 3.5e−220 |
| NODE_31_9 | VUW72999.1|AA1| | 51.0 | 451 | 166 | 612 | 182 | 620 | 8.4e−115 |
PUL annotation and CGC prediction
To examine the presence of Gram-positive polysaccharide utilization loci (gpPUL) in the genome of C. algeriensis, we used nucleotide Basic Local Alignment search tool (BLASTX) available in dbCAN-PUL. This tool uses the repository as a database to query sequences against PUL proteins in dbCAN-PUL. This analysis resulted in a huge number of sequence similarities (11,320) (see Additional file 1: Table S2) including 36 CAZymes, 21 transporters (TCs) and 6 signal transduction proteins (STP). The PUL showing the highest number of hits to our query sequences is PUL0390 with a total of 10 hits. This PUL is predicted to be capable of degrading acetylated glucuronoxylan.
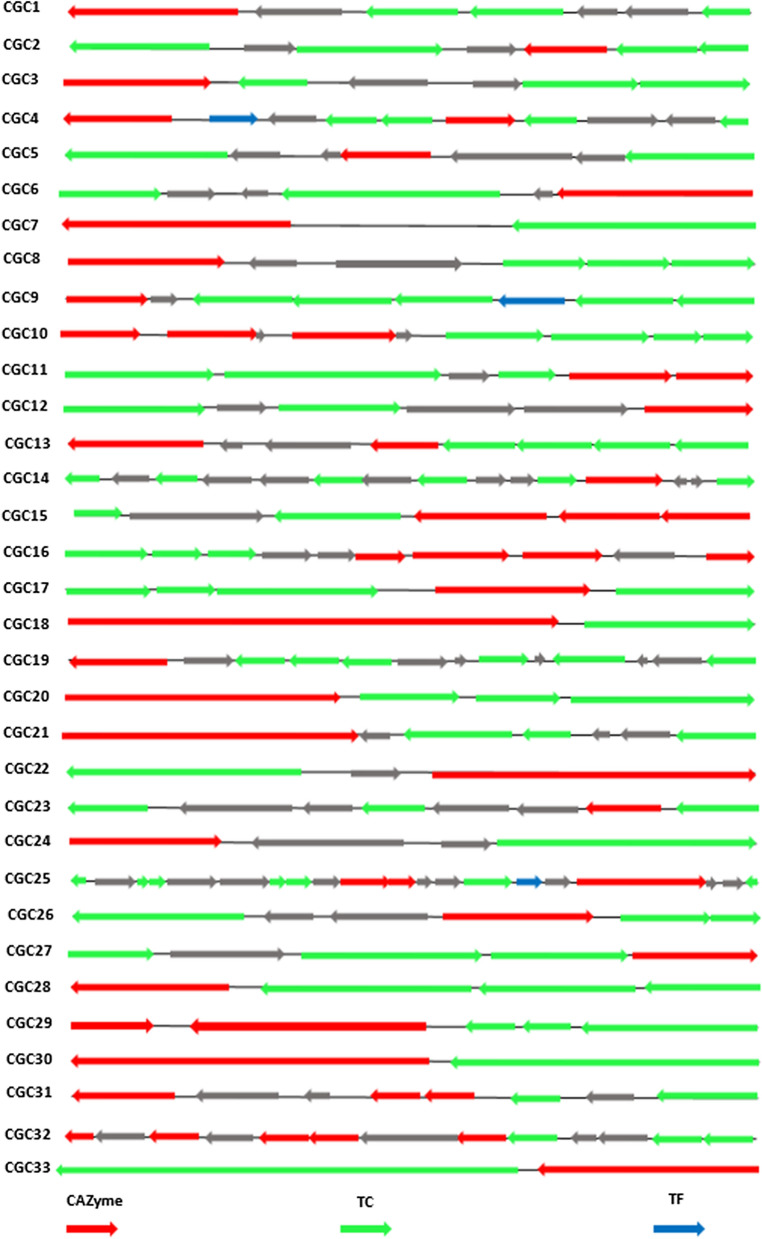
CAZyme gene clusters (CGC) prediction via the dbCAN2 with the CGC-Finder unveiled 33 CGCs defined by the presence of at least one CAZyme, one transporter and one transcription factor encoding genes (Fig. 3 and Additional file 1: Table S3).
Fig. 3.
Schematic representation of the predicted 33 CAZyme Gene Clusters (CGCs) showing organization of genes in each cluster. CAZymes genes are colored red, TC (Transporters Classification) are colored green, TF (Transcription Factor) are colored blue. Non-signature genes, which can be inserted between signature genes, are colored gray
CAZymes gene labels are based on CAZyme domain assignment, TC genes were predicted by searching against the TCDB and TF genes searched against the transcription factor families in Pfam and Superfamily. Genes organization of predicted clusters is shown in Fig. 3.
Results of sequence similarities were used for the determination of carbohydrate utilization ecotypes. Among the predicted CGCs, 20 of them contain CAZymes with no similarity with proteins in the repository. Based on enzymes combination in predicted CGCs and genes similarities with those available in dbCAN-PUL database, we predict a specific polysaccharide for each cluster (Table 3). The determination of carbohydrate utilization ecotypes provides insight to their biotechnological potential.
Table 3.
Targeted substrates predicted for CAZymes genes clusters
| CGC | Predicted CAZyme family | Targeted substrates |
|---|---|---|
| 1 | GH3 | Xylan |
| 2 | CE19 | Pectin |
| 3 | GH4 | Melibiose, raffinose |
| 4 | GH3-GH35 | Xylan, galactan |
| 5 | CE1 | Xylan |
| 6 | GH67 | Arabinoxylan |
| 7 | CBM25 | Starch |
| 8 | GH30-1 | Beta glucan, xylan |
| 9 | GH2 | Hemicellulose |
| 10 | CBM66-GH32-GH151 | Fructans |
| 11 | GH109-GH109 | – |
| 12 | GH3 | Chitin, xylan |
| 13 | GH109-GH109 | – |
| 14 | GH38 | Alpha mannan |
| 15 | GH13_39/CBM34-GT5-GH13_9/CBM48 | Starch, glycogen |
| 16 | GH109-GH65-GH13_18-GH51 | Arabinan |
| 17 | PL33-1 | N-glycan |
| 18 | GH106 | Rhamnose |
| 19 | AA1 | – |
| 20 | GH3 | Xyloglucan |
| 21 | GT51 | – |
| 22 | GT51 | – |
| 23 | GT2 | – |
| 24 | GH109 | – |
| 25 | GH5-25/GH9-GH10-GT84/GH94 | Cellulose, hemicellulose |
| 26 | GH109 | – |
| 27 | CBM50 | – |
| 28 | GH109 | – |
| 29 | GH109-GH2 | Hemicellulose |
| 30 | GH2 | Hemicellulose |
| 31 | GT4 | – |
| 32 | GT2-GT4-GT4-GT2 | Polysaccharides, O-antigen |
| 33 | GH23 | peptidoglycan |
Discussion
Extremophilic microorganisms are of prime interest for biotechnological applications. They possess great potential to degrade plant biomass thanks to their corresponding enzymes [20]. Previous studies have shown that they are efficient producers of CAZymes [32, 33]. In the present work, we gained insight into the profile of genes involved in the carbohydrate metabolism (CAZomes) in the thermophilic and anaerobic Caldicoprobacter algeriensis TH7C1T. This strain classified as novel species in the Caldicoprobacter genera, was isolated from a hot spring. Owing to its harsh culture conditions, we proceeded with the whole genome sequencing to unveil the capability of C. algeriensis strain for polysaccharides utilization using complex machineries including efficient carbohydrate active enzymes. The C. algeriensis TH7C1T genome consists of 2,535,023 bp with 44.9% GC content, which is similar to already sequenced Caldicoprobacter species, namely faecalis, oshimai and guelmensis.
In this study, we report for the first time, CAZymes repertoire of a thermophilic bacteria assigned to the Caldicoprobacter genera. The CAZymes prediction via the dbCAN server using predicted amino acid sequences of C. algeriensis unveiled the presence of 97 CDSs belonging to CAZymes representing 4% of protein coding genes. This percentage is within the range of CAZymes encoding-genes estimated for all microorganisms genomes [1] and genomes of previously reported thermophilic Firmicutes, such as BZ3 isolated from a new thermophilic compost-derived consortium (4%) [34], the thermophilic bacterium Caldanaerobacter sp. strain 1523vc isolated from a hot spring of Uzon Caldera (3,6%) [35]. Among predicted CAZymes, the most abundant class was glycosides hydrolases (GH), about 58% of CAZymes showing the highest percentage of Glycosidases reported in genomes and metagenomes from hot spring ecosystems. C. algeriensis also stands out for being the richest in diversity of GHs families (32) compared to other thermophilic genomes [34, 36, 37]. These GHs include the major families for hemicellulose and cellulose metabolism. Based on this, we speculate that C. algeriensis possess great potential to degrade carbohydrates much more effectively than other strains described previously.
When examining Glycosides hydrolase families by relative abundance, the maximum representation was from the families GH109 and GH3 genes. These two families are responsible for hemicelluloses and oligosaccharides biomass degrading respectively. As reported previously in thermophilic microbial consortia and hot spring samples, the other abundant class of CAZymes was glycosyl transferases (GT), 20% of predicted CAZymes. This large diversity of biomass degrading-related genes encoded by the C. algeriensis genome supports studies showing the importance of Firmicutes phylum in deconstruction of structural plant polysaccharides [27]. It has been demonstrated that this group of bacteria among the 6 predominant phyla in hot spring ecosystems [36, 38]. Given that they are nutritionally pecialized [27], they develop a battery of endo- and exo-acting Carbohydrate Active Enzymes and transporters, responsible for the cleavage of particular carbohydrates. Earlier studies reported that these genes are organized in clusters involved in polysaccharides degradation and transport forming Gram-positive polysaccharide Utilization Loci (gpPUL) [27]. In our study, we report for the first time the existing of CAZymes gene clusters in this group of Caldicoprobacteraceae.
PULs were analyzed based on genes homology with PULs available in dbCAN-PUL database. Results showed 11,320 gene similarities in CAZymes, transporters and signal transduction proteins across all PULs in the dbCAN repository, displaying an identity between 18.7% and 80.7%. To further analyze carbohydrate utilization ability of C. algeriensis, we performed CAZymes gene cluster analysis via the CGC finder in dbCAN2 meta server. We obtained 33 CAZymes gene clusters. Among them, 22 CGCs including 19 GH families, were predicted to be involved in cellulose and hemicellulose hydrolysis (GH3/GH5/GH2/GH10/GH30/GH35/GH38/GH4), glycogen degradation (debranching enzymes), (GH3/GH13_9/GH67/GH94) and starch utilization (GH13_39). The most abundant CAZyme identified in CGCs was related to the GH109 family. Nine genes, which typically encode α-N-acetylgalactosaminidase and β-N-acetylhexosaminidase, were found in seven clusters (CGC11, 13, 16, 24, 26, 28 and 29). GH109 genes were combined to other GH families genes, GH65/13/51 and GH2 in CGC 16 and 29 respectively, supporting that synergistic action of many CAZymes is required for polysaccharides cleavage [39]. Interestingly, analysis of GH109 genes similarities against genes from PULs available in the database, revealed no significant similarity. Thus, we suggest that C. algeriensis encodes new gene clusters not identified previously. Indeed, few studies reporting characterization of GH109 family members were performed [20] and CAZy database lists only 7 GH109 nagalases as characterized. Members of this family are particularly interesting for their ability to convert RBC A-antigens into H-antigens, turning type-A blood into universal donor type-O blood [40, 41].
The C. algerinsis also encodes six CAZymes genes clusters including members of GT families. As reported previously in extremophilic ecosystems, most of GT genes belonged to GT2 and GT4 families [20, 36, 42]. These two families have been reported to perform the synthesis of alpha and beta glycans and glycoconjugates. The GT4 contains a large variety of enzymes that are involved in lipopolysaccharide and antibiotic avilamycin A synthesis [43]. Owing the difficulty on purifying and investigating the biochemical features of these membrane associated enzymes, a few number of GTs has been characterized. Nevertheless, they have been described to offer potential opportunities in biotechnological applications such as biomedicine, cell biology field and pharmaceutical industry. Consequently, an in depth analysis of genes belonging to this family is very important.
Carbohydrates esterases are also identified in two CAZymes genes clusters (CGC2 and CGC5), related to CE1 and CE19 families. CE1 constitute the largest family of esterases including 5062 entries listed in CAZy database. Members of family CE1 were known to target xylan while CE19 family members are involved in pectin degradation. Recently, Carbohydrate esterases have shown great potential in several industrial applications such as food industry, pulp and paper industry, biofuel production, animal feed, medical and pharmacological industry [44, 45].
Genes Similarity analysis has shown 11 other genes, in addition to GH109, with no homologous in PULs database, including genes belonging to GT2, GT4 and CE19 CAZymes families. Thus, the C.algeriensis genome could be a source of novel and original thermophilic enzymes with strong potential for biotechnological applications.
Conclusions
The present work constitutes the first study targeting CAZymes repertoire of bacteria belonging Caldicoprobacteraceae group based on whole genome sequencing. CAZyme encoding genes prediction results highlighted the high potential of C.algeriensis bacteria for the degradation of structural plant polysaccharides. Detailed analysis of predicted genes unveiled complex machineries involved in the metabolization of these major components of the plant cell wall and put the emphasis on newly identified enzymes. The in depth characterization of the specificity of each of these enzymes is the next challenge that will allow the understanding at the molecular level of the involvement of these loci in carbohydrates metabolism and their potential industrial applications.
Methods
Sampling and DNA extraction
Strain C. algeriensis TH7C1T was isolated from the hydrothermal hot spring of Guelma [28]. Genomic DNA was extracted as previously described [46] with some modifications. Briefly, cells harvested in the exponential phase were suspended in TRIS–HCl (pH 8.0), EDTA, NaCl and incubated in the presence of lysozyme at 37 °C. Sodium dodecyl sulfate was added to 1% and the incubation continued until clarification was complete. Chloroform extractions were carried out and followed by ethanol precipitation. The DNA was drawn out of solution by being wound around a glass rod.
Sequencing and functional annotation
The isolated DNA from C. algeriensis TH7C1T was used to generate Illumina shotgun paired-end sequencing libraries, which were sequenced with a MiSeq instrument and the MiSeq reagent kit version 3 (2 × 250 bp paired-end reads), as recommended by the manufacturer (Illumina, San Diego, CA, USA) at IBISBA CSIC-CellFactory_MM platform. Quality filtering using Trimmomatic version 0.36 resulted in 473,434 paired-end reads rendering an approximate genome coverage of 30x. The sequence was assembled using the SPAdes Genome Assembler version 3.15.2. Assembled contigs were submitted to the Rapid Annotation Server (RAST) (http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/RapidAnnotationServer) [47] and the DFAST server (https://dfast.nig.ac.jp/) for protein coding sequences (CDSs) prediction. The Circular Genome Viewer (CGView server) [48] was used to construct a circular graphical map of C. algeriensis TH7C1T. Carbohydrate-active enzyme (CAZyme) searches were performed using HMMER3.0 package (http://hmmer.org/) available from dbCAN (http://csbl.bmb.uga.edu/dbCAN/) [49], this search is run against Pfam Hidden Markov Models (HMMs). DIAMOND available from the dbcan CAZyme database was used for fast blast hits in the CAZy database.
Polysaccharides Utilization Loci (PULs) were analyzed via the dbcanPUL meta server [50]. CAZyme gene cluster (CGC) Finder in the database was used for carbohydrate-active enzyme clusters annotation. CGCs were defined as genomic regions containing at least one CAZyme gene, one transporter (TC) gene, and one transcription factor (TF) gene. Genome sequence has been submitted to the public genomic NCBI database under accession number PRJNA743054.
Prediction of CRISPR-Cas sequence (Clustered Regularly Interspaced Short Palindromic Repeats) in the genome was performed using the CRISPRCasFinder server) [51].
Supplementary Information
Additional file 1: Table S1. Predicted CAZyme genes using the HMMER3.0 package and the dbcan CAZyme database. Table S2. Sequence similarities with PUL proteins in dbCAN-PUL. Table S3. Predicted CAZyme gene clusters (CGC) via the dbCAN2.
Additional file 2. Protein encoding genes sequences.
Acknowledgements
Not applicable.
Abbreviations
- CAZyme
Carbohydrate Active Enzyme
- GT
Glycosyltransferases
- GH
Glycoside hydrolases
- CE
Carbohydrate esterases
- PL
Polysaccharide lyases
- AA
Auxiliary activities
- SPAdes
St. Petersburg genome assembler
- RAST
Rapid Annotations using Subsystems
- DFAST
DDBJ Fast Annotation and Submission Tool
- CRISPR-Cas
Clustered Regularly Interspaced Short Palindromic Repeats
- PUL
Polysaccharide Utilization Locus
- gpPUL
Gram-positive Polysaccharide Utilization Locus
- CGC
CAZyme Gene Cluster
- CBM
Carbohydrate-binding module
- TC
Transporter Classification
- TCDB
Transporter Classification Database
- STP
Signal Transduction Protein
- TF
Transcription Factor
- Pfam
Protein families database
- HMMs
Hidden Markov Models
- CDS
Coding sequence
- dbCAN
DataBase for automated Carbohydrate-active enzyme ANnotation
Author contributions
Conceptualization, SB, AB and JB; methodology, RA, NZJ, SM, NP, MM and JLG; software, RA, JLG and JB; validation, SB, AB and JB; formal analysis, SB; investigation, RA, NZJ, SN and JLG; data curation, RA, AB and JB; writing, RA and JLG; review and editing, SB and JB; visualization, RA and JLG; supervision, SB, AB and JB; project administration, SB and JB; funding acquisition, SB and JB. All authors read and approved the final manuscript.
Funding
This work was supported by the Tunisian Ministry of Higher Education and Scientific Research (contract program LBMIE-CBS, code: LR15CBS06) and the Algerian-Tunisian R&I Cooperation for the Mixed Laboratories of Scientific Excellence 2021–2024 (Hydro-BIOTECH, code LABEX/TN/DZ/21/01). The high throughput sequencing, assembly and annotation work was supported by the IBISBA1.0 H2020 project 730976 (https://www.ibisba.eu/) at its CSIC Cell Factory node.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files. The C.algeriensis Genome has been deposited in the public genomic NCBI database with accession code: PRJNA743054, (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA743054).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amel Bouanane Derenfed, Email: amelbouanane@gmail.com.
Nathalie Pradel, Email: nathalie.pradel@ird.fr.
Jorge Barriuso, Email: jbarriuso@cib.csic.es.
Samir Bejar, Email: samir.bejar@cbs.rnrt.tn, Email: samirbejar@gmail.com.
References
- 1.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochemical Journal. 1997;326:929. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 7.Garron M-L, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- 8.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 9.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:1–14. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garron M-L, Henrissat B. The continuing expansion of CAZymes and their families. Curr Opin Chem Biol. 2019;53:82–87. doi: 10.1016/j.cbpa.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Mhiri S, Bouanane-Darenfed A, Jemli S, Neifar S, Ameri R, Mezghani M, Bouacem K, Jaouadi B, Bejar S. A thermophilic and thermostable xylanase from Caldicoprobacter algeriensis: recombinant expression, characterization and application in paper biobleaching. Int J Biol Macromol. 2020;164:808–817. doi: 10.1016/j.ijbiomac.2020.07.162. [DOI] [PubMed] [Google Scholar]
- 12.Pallister E, Gray CJ, Flitsch SL. Enzyme promiscuity of carbohydrate active enzymes and their applications in biocatalysis. Curr Opin Struct Biol. 2020;65:184–192. doi: 10.1016/j.sbi.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Chettri D, Verma AK, Verma AK. Innovations in CAZyme gene diversity and its modification for biorefinery applications. Biotechnol Rep. 2020;28:e00525. doi: 10.1016/j.btre.2020.e00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandi CK, Agrawal A, Chundawat SP. Carbohydrate-Active enZyme (CAZyme) enabled glycoengineering for a sweeter future. Curr Opin Biotechnol. 2020;66:283–291. doi: 10.1016/j.copbio.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Rajeswari G, Jacob S, Chandel AK, Kumar V. Unlocking the potential of insect and ruminant host symbionts for recycling of lignocellulosic carbon with a biorefinery approach: a review. Microb Cell Fact. 2021;20:1–28. doi: 10.1186/s12934-021-01597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karuppiah V, Zhixiang L, Liu H, Vallikkannu M, Chen J. Co-culture of Vel1-overexpressed Trichoderma asperellum and Bacillus amyloliquefaciens: an eco-friendly strategy to hydrolyze the lignocellulose biomass in soil to enrich the soil fertility, plant growth and disease resistance. Microb Cell Fact. 2021;20:1–14. doi: 10.1186/s12934-021-01540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raupach MJ, Amann R, Wheeler QD, Roos C. The application of “-omics” technologies for the classification and identification of animals. Org Divers Evol. 2016;16:1–12. doi: 10.1007/s13127-015-0234-6. [DOI] [Google Scholar]
- 18.Häkkinen M, Arvas M, Oja M, Aro N, Penttilä M, Saloheimo M, Pakula TM. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb Cell Fact. 2012;11:1–26. doi: 10.1186/1475-2859-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumorné K, Córdova DC, Astorga-Eló M, Renganathan P. Extremozymes: a potential source for industrial applications. J Microbiol Biotechnol. 2017;27(4):649–659. doi: 10.4014/jmb.1611.11006. [DOI] [PubMed] [Google Scholar]
- 20.Strazzulli A, Cobucci-Ponzano B, Iacono R, Giglio R, Maurelli L, Curci N, Schiano-di-Cola C, Santangelo A, Contursi P, Lombard V. Discovery of hyperstable carbohydrate-active enzymes through metagenomics of extreme environments. FEBS J. 2020;287:1116–1137. doi: 10.1111/febs.15080. [DOI] [PubMed] [Google Scholar]
- 21.Barnard D, Casanueva A, Tuffin M, Cowan D. Extremophiles in biofuel synthesis. Environ Technol. 2010;31:871–888. doi: 10.1080/09593331003710236. [DOI] [PubMed] [Google Scholar]
- 22.Irla M, Drejer EB, Brautaset T, Hakvåg S. Establishment of a functional system for recombinant production of secreted proteins at 50 C in the thermophilic Bacillus methanolicus. Microb Cell Fact. 2020;19:1–16. doi: 10.1186/s12934-020-01409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan M, Sathya T. Extremozymes from metagenome: Potential applications in food processing. Crit Rev Food Sci Nutr. 2018;58:2017–2025. doi: 10.1080/10408398.2017.1296408. [DOI] [PubMed] [Google Scholar]
- 24.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HM, Bernalier-Donadille A, Duncan SH, O'Toole PW, Scott KP, Flint HJ. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genomics. 2016;2:e000043. doi: 10.1099/mgen.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouanane-Darenfed A, Fardeau M-L, Grégoire P, Joseph M, Kebbouche-Gana S, Benayad T, Hacene H, Cayol J-L, Ollivier B. Caldicoprobacter algeriensis sp. nov. a new thermophilic anaerobic, xylanolytic bacterium isolated from an Algerian hot spring. Curr Microbiol. 2011;62:826–832. doi: 10.1007/s00284-010-9789-9. [DOI] [PubMed] [Google Scholar]
- 29.Amel B-D, Nawel B, Khelifa B, Mohammed G, Manon J, Salima K-G, Farida N, Hocine H, Bernard O, Jean-Luc C. Characterization of a purified thermostable xylanase from Caldicoprobacter algeriensis sp. nov. strain TH7C1T. Carbohydr Res. 2016;419:60–68. doi: 10.1016/j.carres.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Bouacem K, Bouanane-Darenfed A, Jaouadi NZ, Joseph M, Hacene H, Ollivier B, Fardeau M-L, Bejar S, Jaouadi B. Novel serine keratinase from Caldicoprobacter algeriensis exhibiting outstanding hide dehairing abilities. Int J Biol Macromol. 2016;86:321–328. doi: 10.1016/j.ijbiomac.2016.01.074. [DOI] [PubMed] [Google Scholar]
- 31.Reichart NJ, Bowers RM, Woyke T, Hatzenpichler R. High potential for biomass-degrading enzymes revealed by hot spring metagenomics. Front Microbiol. 2021;12:668238. doi: 10.3389/fmicb.2021.668238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coker JA. Extremophiles and biotechnology: current uses and prospects. F1000Res. 2016;5:F1000. doi: 10.12688/f1000research.7432.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhtar S, Aslam M. Biofuel synthesis by extremophilic microorganisms. In Biofuels production–sustainability and advances in microbial bioresources. Springer; 2020: 115–138.
- 34.Lemos LN, Pereira RV, Quaggio RB, Martins LF, Moura L, da Silva AR, Antunes LP, da Silva AM, Setubal JC. Genome-centric analysis of a thermophilic and cellulolytic bacterial consortium derived from composting. Front Microbiol. 2017;8:644. doi: 10.3389/fmicb.2017.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korzhenkov A, Toshchakov S, Podosokorskaya O, Patrushev M, Kublanov I. Data on draft genome sequence of Caldanaerobacter sp. strain 1523vc, a thermophilic bacterium, isolated from a hot spring of Uzon Caldera, (Kamchatka, Russia) Data Brief. 2020;33:106336. doi: 10.1016/j.dib.2020.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaushal G, Kumar J, Sangwan RS, Singh SP. Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int J Biol Macromol. 2018;119:882–895. doi: 10.1016/j.ijbiomac.2018.07.196. [DOI] [PubMed] [Google Scholar]
- 37.Zayulina KS, Elcheninov AG, Toshchakov SV, Kublanov IV. Complete genome sequence of a hyperthermophilic archaeon, Thermosphaera sp. Strain 3507, isolated from a Chilean Hot Spring. Microbiol Resour Announc. 2020;9:e01262–e11220. doi: 10.1128/MRA.01262-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghelani A, Patel R, Mangrola A, Dudhagara P. Cultivation-independent comprehensive survey of bacterial diversity in Tulsi Shyam Hot Springs, India. Genomics Data. 2015;4:54–56. doi: 10.1016/j.gdata.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park Y-J, Jeong Y-U, Kong W-S. Genome sequencing and carbohydrate-active enzyme (CAZyme) repertoire of the white rot fungus Flammulina elastica. Int J Mol Sci. 2018;19:2379. doi: 10.3390/ijms19082379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu QP, Sulzenbacher G, Yuan H, Bennett EP, Pietz G, Saunders K, Spence J, Nudelman E, Levery SB, White T. Bacterial glycosidases for the production of universal red blood cells. Nat Biotechnol. 2007;25:454–464. doi: 10.1038/nbt1298. [DOI] [PubMed] [Google Scholar]
- 41.Rahfeld P, Sim L, Moon H, Constantinescu I, Morgan-Lang C, Hallam SJ, Kizhakkedathu JN, Withers SG. An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nat Microbiol. 2019;4:1475–1485. doi: 10.1038/s41564-019-0469-7. [DOI] [PubMed] [Google Scholar]
- 42.Amin K, Tranchimand S, Benvegnu T, Abdel-Razzak Z, Chamieh H. Glycoside hydrolases and glycosyltransferases from hyperthermophilic archaea: Insights on their characteristics and applications in biotechnology. Biomolecules. 2021;11:1557. doi: 10.3390/biom11111557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Fleites C, Proctor M, Roberts S, Bolam DN, Gilbert HJ, Davies GJ. Insights into the synthesis of lipopolysaccharide and antibiotics through the structures of two retaining glycosyltransferases from family GT4. Chem Biol. 2006;13:1143–1152. doi: 10.1016/j.chembiol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Kameshwar AKS, Qin W. Structural and functional properties of pectin and lignin–carbohydrate complexes de-esterases: a review. Bioresourc Bioprocess. 2018;5:1–16. doi: 10.1186/s40643-017-0187-z. [DOI] [Google Scholar]
- 45.Li X, Dilokpimol A, Kabel MA, de Vries RP. Fungal xylanolytic enzymes: diversity and applications. Biores Technol. 2022;344:126290. doi: 10.1016/j.biortech.2021.126290. [DOI] [PubMed] [Google Scholar]
- 46.Fouet A, Sonenshein AL. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ausland C, Zheng J, Yi H, Yang B, Li T, Feng X, Zheng B, Yin Y. dbCAN-PUL: a database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res. 2021;49:D523–D528. doi: 10.1093/nar/gkaa742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EP, Vergnaud G, Gautheret D, Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Predicted CAZyme genes using the HMMER3.0 package and the dbcan CAZyme database. Table S2. Sequence similarities with PUL proteins in dbCAN-PUL. Table S3. Predicted CAZyme gene clusters (CGC) via the dbCAN2.
Additional file 2. Protein encoding genes sequences.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files. The C.algeriensis Genome has been deposited in the public genomic NCBI database with accession code: PRJNA743054, (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA743054).