Abstract
A method for studying bacteria that are attached to carcass surfaces would eliminate the need for exogenous sampling and would facilitate understanding the interaction of potential human food-borne pathogens with food animal tissue surfaces. We describe such a method in which we used a bioluminescent reporter strain of Escherichia coli O157:H7 that was constructed by transformation with plasmid pCGLS1, an expression vector that contains a complete bacterial luciferase (lux) operon. Beef carcass surface tissues were inoculated with the bioluminescent strain, and adherent bacteria were visualized in real time by using a sensitive photon-counting camera to obtain in situ images. The reporter strain was found to luminesce from the tissue surfaces whether it was inoculated as a suspension in buffer or as a suspension in a bovine fecal slurry. With this method, areas of tissues inoculated with the reporter strain could be studied without obtaining, excising, homogenizing, and culturing multiple samples from the tissue surface. Use of the complete lux operon as the bioluminescent reporter eliminated the need to add exogenous substrate. This allowed detection and quantitation of bacterial inocula and rapid evaluation of adherence of a potential human pathogen to tissue surfaces. Following simple water rinses of inoculated carcass tissues, the attachment duration varied with different carcass surface types. On average, the percent retention of bioluminescent signal from the reporter strain was higher on lean fascia-covered tissue (54%) than on adipose fascia-covered tissue (18%) following water washing of the tissues. Bioluminescence and culture-derived viable bacterial counts were highly correlated (r2 = 0.98). Real-time assessment of microbial attachment to this complex menstruum should facilitate evaluation of carcass decontamination procedures and mechanistic studies of microbial contamination of beef carcass tissues.
Understanding the mechanisms of microbial attachment and adherence to various surfaces is critical to finding new methods for inactivating or removing attached microorganisms from those surfaces. This problem includes microbial attachment to the surfaces of meat animal and poultry carcasses and food-processing surfaces.
Microbial attachment to or association with animal and poultry carcasses has been studied by using a variety of techniques, most of which involve surface sampling, culturing, and back-extrapolation of the resulting counts to the original surface area. This approach and the data derived from it are predicated on the efficiency of the sampling method used, as well as the recovery of culturable organisms. Both destructive and nondestructive sampling methods are inherently variable in terms of the efficiency of removing organisms from food surfaces (11). Alternatively, direct microscopic observation by light microscopy of sample sections and by electron microscopy of fixed and stained specimens has been used (36). Scanning confocal microscopy of nonfixed samples has revealed the actual spatial arrangement of the natural menstruum and has permitted optical sectioning below the surface (18). All three microscopic approaches allow observation of only a small percentage of the total surface area and rely on some form of sampling.
Ideally, microbial attachment processes are best observed under natural or undisturbed conditions without sampling, fixation, or any treatment detrimental to the biological or environmental integrity of the specimen. Nondestructive, in situ investigations of microbial associations have been performed by using bioluminescent and fluorescent reporter gene systems with a myriad of biological systems (5, 6, 8, 20, 29, 38), as well as food contaminated with microbes (1, 3, 10, 12, 14, 16, 19, 28, 31, 34, 35, 37). The biochemical basis of bioluminescence has been reviewed previously; examples of applications abound in the literature and are cited but not reviewed in this paper (15, 21, 22, 23, 24, 26).
The power of bioluminescent tagging of pathogens was demonstrated by an approach in which the infection process could be monitored in living mammalian hosts (4). Using an intensified charge-coupled device (ICCD) camera, workers visualized light from viable bacteria directly through the viscera, skin, and fur of intact, live animals as the infection spread, and thus this technique provided both spatial and temporal information (5, 6). Antibiotic inhibition of bacterial disease progression in the gastrointestinal tract was demonstrated in situ by the loss of luminescence. This noninvasive, in situ approach for studying microbial processes provides important real-time information that could not otherwise be obtained and has direct applications to the study of food contamination.
A similar approach for assessing the association or attachment of food-borne pathogens, such as Escherichia coli O157:H7, to beef carcass surface tissue in situ would have several advantages. First, assessing the presence of inoculated bacteria on a carcass surface would not rely on any type of sampling that would only partially estimate the microbial population. Second, larger, complete, contiguous areas of surface tissue could be examined simultaneously, thereby eliminating the need for multiple sampling of smaller areas of surface tissue and extrapolation of the results to larger areas. Third, the presence or partial removal of the inoculum, as well as the physical location of the inoculum, could be monitored in real time, eliminating the need for repeated sampling and retrospective culture data. And finally, specific gene expression could be monitored and potentially quantified in situ if the proper promoter lux gene fusions were used (13, 26).
Association of the enterohemorrhagic E. coli serotypes, including the most prevalent serotypes of this class (O157:H7 and O157:nonmotile), with products of cattle origin (17, 25) has led to implementation of zero-tolerance rules for visible fecal matter on beef carcasses and to legislation classifying E. coli O157 as an adulterant of ground beef in the United States. The source of E. coli O157 and other gram-negative enteropathogens is feces. Regardless of whether the feces come directly from an animal’s gastrointestinal tract (e.g., from a ruptured gut, which occurs infrequently) or are part of the hide and hoof soil load, fecal sources have been implicated in postharvest carcass contamination. Decontamination research has focused on antimicrobial treatment of the carcass surface, which either inactivates or removes pathogenic E. coli and other pathogens (36). Such research has traditionally relied on inoculating carcass surface tissues, treating them, sampling them, and then counting the pathogens by a quantitative culture enumeration method requiring from 24 to 48 h. While this approach is valuable for carcass antimicrobial intervention research, it has not been able to address the localization of contamination on the superficial fascia of the beef carcass surface or to determine preferential binding to different tissue surface types on the carcass. Thus, there are still questions concerning possible preferential attachment or association of bacterial pathogens to the carcass surface.
In this study, we used a strain of bioluminescent E. coli O157:H7 to study the association of this organism with beef carcass surface tissue macroscopically by using a very sensitive ICCD camera. Images of bacterial inocula in buffer suspensions, as well as in bovine feces, were obtained for a variety of beef carcass surface tissue types to determine the level of detection by this method. Our data indicate that real-time monitoring of the duration of attachment of a bioluminescent bacterial strain during simulated carcass washing is possible without exogenously sampling the tissue surface and that there are differences in the adherence of this organism to different carcass surface types.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli O157:H7 strain B6-914 (= ATCC 43888) was used in this study. This parent strain was screened to determine its stx1, stx2a, and stx2b gene content by PCR (Life Technologies PCR reagent system) by using a previously described primer set (27) and the method described by Nancy Strockbrine, Centers for Disease Control and Prevention, Atlanta, Ga. (35a); E. coli O157:H7 strain B6-914 was negative for the stx toxin genes as determined by this assay (data not shown).
Plasmid pCGLS1 was a gift from K. H. Nealson (University of Wisconsin—Milwaukee) in host strain E. coli DH5α (13). This plasmid contains a complete lux operon, luxCDABE (bioluminescence genes), from the nematode symbiont bacterium Photorhabdus luminescens (previously known as Xenorhabdus luminescens [39]) in a pUC18 backbone (13). The native P. luminescens promoter is included in this construct and efficiently directs expression of the lux genes in gram-negative organisms (13). Plasmid pCGLS1 was purified from the host strain by using standard procedures and then was used to transform E. coli O157:H7 by the CaCl2 transformation technique (40). Transformants were selected initially on the basis of ampicillin resistance and then on the basis of visible bioluminescence. The bioluminescent signal intensities of transformant clones were initially determined with a cooled charge-coupled device camera (ChemiImager 4000; Alpha Innotech, San Leandro, Calif.). The brightly luminescent strain selected for use in this study was designated E. coli O157:H7 strain L-2.
All of the transformed strains were propagated at 37°C on Luria agar or in Luria broth containing 100 μg of ampicillin per ml. In the case of broth propagation, culture tubes that were one-third filled with medium were rotated with a tube rotator at approximately 100 rpm to obtain stationary-phase cells for inoculation. Transformed strains were tested for any major phenotypic differences from the parent strains with regard to attachment to beef carcass surface tissue, turbidimetric growth rate, and a 95-carbon-source utilization metabolic profile (Biolog, Haywood, Calif.). There were no apparent differences between the tagged and parent strains in these assays (data not shown). The presence of a plasmid of the proper size was confirmed by MluI (Promega, Madison, Wis.) digestion of plasmid DNA obtained from an alkaline lysis miniprep procedure, followed by agarose gel electrophoresis (40). All transformants were maintained as part of the culture collection of the Roman L. Hruska U.S. Meat Animal Research Center (Clay Center, Nebr.) as glycerol suspensions at −20°C.
Imaging.
As described previously (4), photons emitted from inoculated specimens and cultures were collected and imaged with an ICCD camera (model C2400-32; Hamamatsu, Hamamatsu City, Japan) fitted with a 50-mm f-1.2 Nikkor lens (Nikon, Tokyo, Japan). For all experiments, spatial reference images (grayscale) were collected in dim light immediately before photon emission data were collected in total darkness. Bioluminescent signals were represented as pseudocolor images, where color was indicative of light intensity (red represented the most intense signal, and blue represented the least intense signal). The pseudocolor images were overlaid on the reference images to reveal the spatial distribution of the bioluminescent bacteria and the signal intensities on tissue surfaces. Five-minute integration times were used unless otherwise noted. The images were processed with an Argus 20 image processor (Hamamatsu) and were transferred to a Macintosh Power PC (model 8100-100; Apple Computer, Cupertino, Calif.). Images were superimposed by using Adobe Photoshop, version 4.0 (Adobe Systems, Inc., Seattle, Wash.), and annotations were introduced by using Canvas, version 5.0 (Deneba Systems Software, Miami, Fla.). The bit range settings for pseudocolor images varied between 0 to 3 and 2 to 9 for appropriate display.
Beef carcass surface tissues.
Each type of tissue section (lean or adipose) used in these experiments was excised from several different carcasses. The tissues used in each triplicate experiment were from the same tissue section. The tissues collected were in similar states of hydration.
Beef carcass surface tissues were collected primarily from the flank area, but for comparison, samples were also obtained from the rump, flank, brisket, cranial back-neck, caudal back, and foreshank areas. All samples were obtained from postrigor beef carcasses and were frozen for storage and transport. Assignment of the lean and adipose designations was based only on the appearance of tissue sites. Whether they were primarily lean or adipose, all tissues were covered with intact fascia tissue. Additional beef carcass surface tissues were excised from sites near the flank area that had a combination of lean and adipose tissues. These transitional-site tissues encompassed actual areas of tissue where the lean-to-adipose change was readily observable and were, as were the other tissues unless noted, covered with the intact fascia of the bovine carcass. All tissues were thawed overnight and allowed to equilibrate to room temperature before inoculation.
Tissue inoculation and washing.
E. coli O157:H7 strain L-2 was propagated as described above. For larger tissue samples (6 by 6 cm), 5 ml of a diluted culture in Luria broth containing 100 μg of ampicillin per ml was placed in a plastic weigh boat (7 by 7 cm), and the beef carcass surface tissue was placed surface side down in the inoculum. Following incubation at room temperature for 15 min, the tissue section was lifted, allowed to drain and drip directly onto a sterile gauze pad, and then placed face up in a clean dry weigh boat for imaging. For fecal inoculation, a diluted aliquot of a strain L-2 culture was mixed with a 1:10 slurry of bovine feces that had been strained through a glass fiber filter in a filtered stomacher bag (Spiral Biotech, Bethesda, Md.) and was inoculated directly onto the tissue surface. For comparisons of surface tissues obtained from different carcass sites, 100-μl portions of a bacterial suspension were applied to the surfaces of tissue sections (2 by 6 cm) and spread with the round fire-polished end of a solid glass spreading rod over the entire surface. After 15 min of incubation at room temperature, any remaining liquid was allowed to drip or drain off, and the inoculated section was placed in a clean dry weigh boat.
Two washing techniques, an immersion-agitation water wash method and a spray method, were used. The immersion method involved either placing an entire tissue section (2 by 6 cm) into a 50-ml conical centrifuge tube or placing a larger tissue section (6 by 6 cm) into a sterile jar. Distilled water was added (10 ml was added to the tubes, and 100 ml was added to the jars), and the samples were vigorously shaken by hand for 15 s. The tissue sections were removed and allowed to drip or drain prior to imaging. In the spray wash method, a tissue section was held on one end with tissue forceps against the wall of a plastic wash tub, approximately 50 ml of distilled water was sprayed over the surface by using a narrow-tipped squirt bottle held 6 in. from the tissue section for 15 s, and the contaminated runoff was collected. Following this wash step the tissue section was allowed to drip or drain, and the inoculated section was placed in a clean petri dish prior to imaging.
Enumeration of bacteria on tissues.
Tissue sections were placed in a filtered stomacher bag along with 25 ml of buffer consisting of 0.1% (wt/vol) Tween 20 in buffered peptone water (Difco Laboratories, Detroit, Mich.). After 2 min of treatment with a Colworth stomacher at the normal or midrange setting, samples from the filtered side of the bag were diluted and plated by the track dilution method (32) onto Luria agar containing 100 μg of ampicillin per ml. The plates were incubated for 24 h at 37°C before enumerating and calculating the number of CFU per square centimeter of tissue surface.
Statistical analyses.
Bioluminescent signal data was normalized by calculating the signal intensity (I) on a per-pixel-area (A) basis (I/A) by using the quantitation feature of the Argus 20 image processor (Hamamatsu). The fraction of the bioluminescent signal (I/A) or the number of CFU per square centimeter remaining after the wash procedure was determined (means of triplicate determinations were used when replicate data could be obtained) and analyzed by using the InStat, version 3.00, statistical analysis package (GraphPad Software, San Diego, Calif.).
RESULTS
Correlation between bioluminescence and cell numbers.
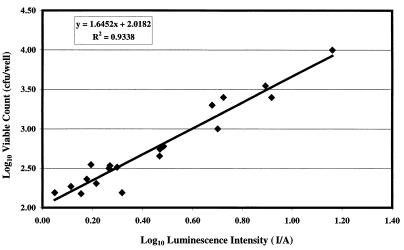
The relationship between the signal intensity (I/A) and viable cell numbers, as determined in a solid-white, flat-bottom microtiter plate by using serial dilutions of cells in phosphate-buffered saline, indicated that there was a high degree of linear correlation in a 30-min exposure experiment conducted at 37°C (r2 = 0.97) (Fig. 1) or a 5-min exposure experiment conducted at room temperature (r2 = 0.93). As determined with microtiter plates, the minimum number of strain L-2 cells with observable luminescence that was at least two times the background luminescence was approximately 50 CFU.
FIG. 1.
Correlation of luminescent signals (log I/A) and viable plate counts of bioluminescent E. coli O157:H7 strain L-2(pCGLS1) in microtiter plate wells. Twofold serial dilutions of strain L-2 were prepared in phosphate-buffered saline in triplicate with 50 μl (final volume) per well. The total luminescence in each well was determined by using the ICCD camera with a 30-min integration time. Samples from each well were plated, and the number of CFU per well was determined. The correlation between viable cell number and light intensity is linear.
Luminescence from E. coli O157:H7 strain L-2 was readily observed when cells suspended in bovine fecal slurries were used. Again, a high degree of correlation between cell number and quantifiable luminescence in fecal slurries was found (r2 = 0.96) based on 5-min integration times at room temperature. The light emission appeared to be less in the fecal suspensions than in the bacterial suspensions in buffer. The calculated regression equation curve for bioluminescence versus viable counts in feces (log CFU/well = 1.44 log I/A + 3.08) had a slope similar to that of the curve generated from buffer suspensions of strain L-2 (log CFU/well = 1.65 log I/A + 2.02). Notably different intercepts for these regression lines would have indicated that the emission patterns on a per-cell basis were similar but that there was some loss of total signal, probably due to absorbance and light scattering by particulate material in the fecal slurry.
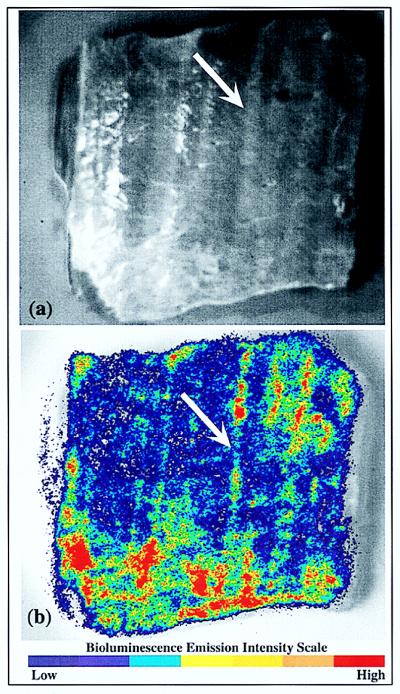
E. coli O157:H7 strain L-2 expressing the lux operon could be readily visualized macroscopically directly on beef carcass surface tissue (Fig. 2). Emitted photons were detected immediately over the entire inoculated surface area of the tissue. After a 5-min integration of the signal, it became apparent that there were different signal intensities in different areas of the tissue surface. The differences appeared to be associated with topographic features (Fig. 2, arrows), which suggested that there was differential adherence of bacterial cells to various surface types.
FIG. 2.
Beef carcass surface tissue inoculated with bioluminescent E. coli O157:H7 strain L-2 by inversion in 5 ml of an ∼107-CFU/ml culture. (a) Grayscale reference image. (b) Bioluminescence. The arrows indicate areas of concentrated residual bioluminescence along tissue striations of the overlying fascia.
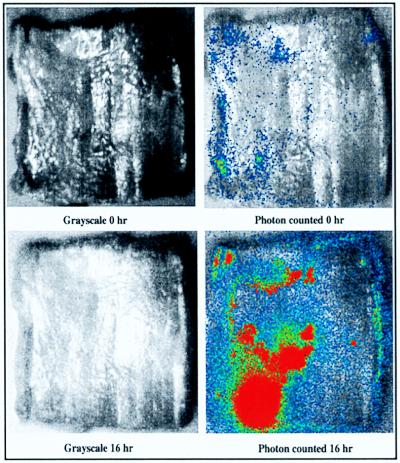
Tissues that were inoculated with lower levels of bacteria (1:1,000 culture dilution) and were incubated aerobically in a weigh boat covered with a noncontacting loose-fitting sheet of Saran Wrap for 16 h at 37°C exhibited increased bioluminescence, which indicated that there were more cells directly on the tissue surface (Fig. 3).
FIG. 3.
Growth of bioluminescent E. coli O157:H7 strain L-2 on beef carcass surface tissue inoculated by inversion in 5 ml of an ∼106-CFU/ml culture (resulting in approximately 103 to 104 CFU/cm2). (Top panels) Grayscale alone (left) and bioluminescence superimposed on grayscale (right), obtained immediately after inoculation. (Bottom panels) Grayscale alone (left) and bioluminescence superimposed on grayscale (right), obtained following 16 h of incubation at 37°C.
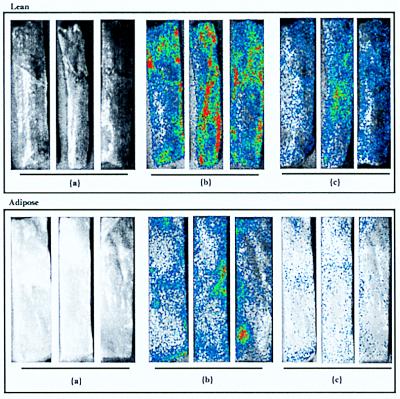
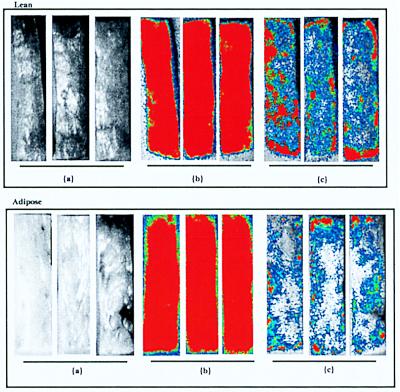
A comparison of the light intensities per unit of area before and after the wash step obtained for lean and adipose tissues showed that E. coli O157:H7 cells preferentially adhered to the surfaces of lean tissues rather than adipose tissues when samples were inoculated by the inversion inoculation method (Fig. 4). The average fraction of the prewash bioluminescence signal (I/A) remaining on the lean tissue after the wash step was 44.9%, compared with only 23.2% for similarly treated adipose tissue sections (three sections of each tissue type were examined). When the cell concentrations of the inocula were 10 times greater, the signal intensities again revealed that there was differential adherence, with 45.9 and 23.5% of the prewash signal remaining for lean and adipose covered tissue sections, respectively (three sections of each tissue type were examined) (data not shown). Inoculation of tissues with bacteria in a fecal slurry and subsequent water washing resulted in similar results, with greater signal intensities on lean tissue surfaces than on adipose tissue surfaces (Fig. 5). Quantitation of signal intensities confirmed this observation (Table 1).
FIG. 4.
Inoculation of beef carcass surface tissue with bioluminescent E. coli O157:H7 strain L-2 suspended in buffered peptone water. Lean (top) and adipose (bottom) tissue sections were inoculated by spreading 100-μl diluted culture aliquots, which resulted in approximately 1.6 × 105 CFU/cm2 (1.9 × 106 total CFU over the entire surface area), and water washed. Washing reduced the signal intensities obtained for both lean and adipose tissues, but the inoculum in buffered peptone water was removed more efficiently from adipose tissue than from lean tissue. (a) Grayscale prewash. (b) Bioluminescence prewash. (c) Bioluminescence postwash.
FIG. 5.
Effects of bovine feces on adherence of E. coli O157:H7 strain L-2 to lean and adipose beef carcass surface tissues. Three lean tissue sections (top) and three adipose tissue sections (bottom) were inoculated with a dilution of E. coli O157:H7 strain L-2 in a bovine feces slurry (approximately 107 total CFU over the entire surface area). (a) Grayscale prewash. (b) Bioluminescence prewash. (c) Bioluminescence postwash.
TABLE 1.
Percentages of the prewash bioluminescence signal and viable counts and after bioluminescent E. coli O157:H7 strain L-2-inoculated beef carcass surface tissues were washed with water
| Tissue type | Inoculation method | Wash method | Inoculum (log10 CFU/cm2) | Inoculation menstruum | % of prewash bioluminescence signal (I/A) remaining | % of prewash viable count recovered |
|---|---|---|---|---|---|---|
| Lean | Inversion | Immersion | 6.23 | Feces | 50 | NDa |
| Lean | Inversion | Immersion | 5.23 | Feces | 51 | ND |
| Lean | Inversion | Immersion | 4.23 | Feces | 70 | ND |
| Adipose | Inversion | Immersion | 6.23 | Feces | 11 | ND |
| Adipose | Inversion | Immersion | 5.23 | Feces | 15 | ND |
| Adipose | Inversion | Immersion | 4.23 | Feces | 23 | ND |
| Lean | Inversion | Immersion | ∼6.0 | Feces | 46 | ND |
| Adipose | Inversion | Immersion | ∼6.0 | Feces | 24 | ND |
| Lean | Inversion | Immersion | ∼5.0 | Feces | 45 | ND |
| Adipose | Inversion | Immersion | ∼5.0 | Feces | 23 | ND |
| Lean | Spread | Flow | 3.90 | Buffer | 61 | 86 |
| Adipose | Spread | Flow | 3.90 | Buffer | 10 | 4 |
ND, not determined.
To further define the differential adherence of bacterial cells to tissues, sections that were predominantly lean or adipose in surface appearance but were covered with an intact fascia were inoculated with bacteria either in fecal suspensions or in buffer alone. Inoculations were performed by spreading bacterial suspensions over the surfaces of the tissue sections with a glass rod. The tissue surfaces were then washed by the spray method. The tendency for lean tissues to retain more bacterial cells after the water rinse and the tendency for adipose tissues to lose a higher percentage of the reporter organism were evident when bioluminescent imaging was used (Fig. 4 and 5). Similar results were obtained when viable counts were determined for these tissues concomitantly (Table 1).
Bacterial bioluminescence on the surface of lean-adipose transitional flank tissue inoculated with sterile bovine fecal suspensions of E. coli O157:H7 strain L-2 also revealed that the inocula adhered preferentially to lean surfaces (Fig. 6). Greater bioluminescence from lean surface tissue than from adipose surface tissue after water washing (spray method) was apparent (Fig. 6). In the case of the tissue section shown in Fig. 6, greater residual bioluminescence was detected along a crevice near the transition between the surface types.
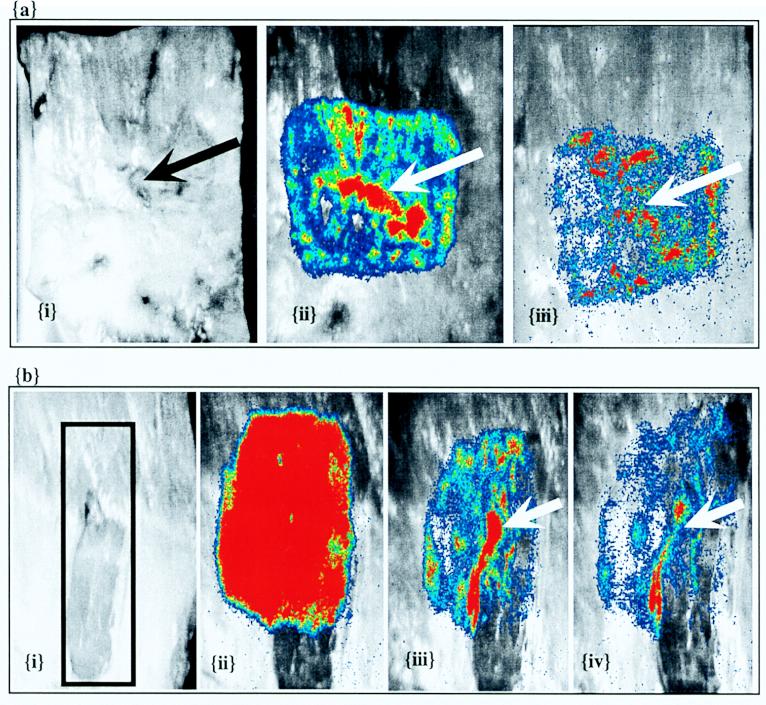
FIG. 6.
Inoculation of mixed surfaces. Bioluminescent E. coli O157:H7 strain L-2 was inoculated (9.2 × 107 total CFU) onto carcass surface tissues that included lean and adipose surfaces across the tissue sections. (Images i) Prewash grayscale. (Images ii) Prewash bioluminescence superimposed on grayscale. (Images iii) Bioluminescence superimposed on grayscale after a single wash. (Image iv) Bioluminescence after a second wash. A transitional tissue section with the overlying fascia tissue intact (a) and a transitional tissue section with the overlying fascia tissue removed by scalpel trimming (b) (indicated by the box in panel b, image i) were inoculated. In panel b inoculation included the section that had intact fascia and the section from which the fascia was removed (indicated by the box in image i). The arrows in panel a indicate the line of tissue topography transition. The arrows in panel b indicate the excision line between the intact fascia and the excised fascia.
In order to study the possible role of the superficial fascia tissue in association or adherence of the reporter bacterial strain to carcass tissue, a section of the fascia (thickness, approximately 1 mm) was removed with a scalpel, which exposed the underlying muscle. The excised area cut across a lean-adipose transition line; this line was subsequently inoculated with a fecal suspension of strain L-2. The inoculated area included lean and adipose surfaces both with and without the overlying fascia. An overall decrease in signal intensity after washing was observed, and greater bioluminescence intensity was observed along the cut line where fascia was removed (Fig. 6b, images iii and iv).
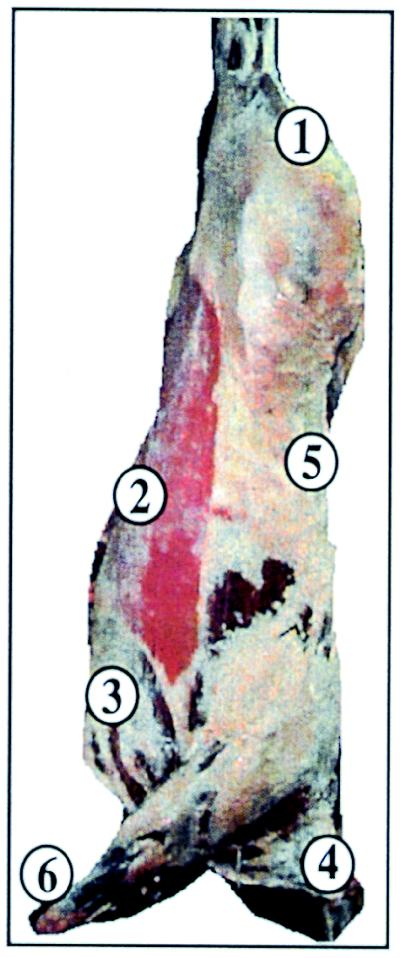
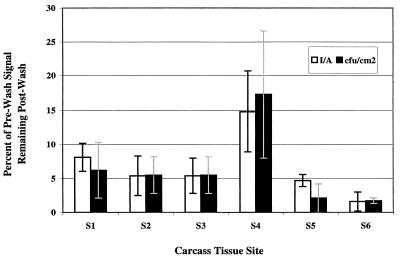
To examine differences in attachment to or association with carcass surface tissue types that were visually different, tissue sections were obtained from six distinct sites on a beef carcass, including sites in the rump area (site 1), the flank area (site 2), the brisket area (site 3), the cranial back-neck area (site 4), the caudal back area (site 5), and the foreshank area (site 6) (Fig. 7). Tissues from each site were inoculated, washed, and imaged. The bacteria were enumerated by the plate counting method after the final imaging step. The percentage of prewash signal detected by both methods indicated that there was a good correlation between photon counting imaging and viable counts (P < 0.05) (Fig. 8). Most notable was the greater retention (or less efficient removal) of the signal from cranial back-neck area tissues (Fig. 7, site 4). This was apparent from an examination of the bioluminescence pseudocolor image (data not shown) and the numerical plate count data (Fig. 8). Although covered with intact fascia, the surface at tissue site 4 (Fig. 7) was characteristically rough (data not shown). The least signal retention, as determined by both indices (bioluminescence and plate counting), was observed with inoculated foreshank tissue (Fig. 7, site 6). The foreshank site was actually stripped of much of the fascia tissue that covered most of the carcass from the rump to the head; instead, homogeneous, tendonous, smooth tissues associated with the lower foreleg and ankles predominated at this site.
FIG. 7.
Sites of sampling for various beef carcass surface tissues. The diagram is representative of a typical grain-fed beef carcass and indicates the locations of tissues used in this experiment. The numbers indicate sampling sites (see text).
FIG. 8.
Effects of washing on the adherence of E. coli O157:H7 strain L-2 to inoculated tissues from different carcass regions. The fractions of signals (bioluminescence or viable count) remaining on various beef carcass surface tissues obtained from six different sites (Fig. 7) following a flow water wash are shown. The data are the means of triplicate values obtained for tissue sections (2 by 6 cm) that were inoculated with an average of 2.1 × 107 CFU of strain L-2.
DISCUSSION
Spatiotemporal analyses of the interaction between E. coli O157:H7 and beef carcass tissue surfaces were conducted by using transformed bacteria which expressed the complete lux operon of P. luminescens (13) as an optical tag. The interaction of the tagged bacterium with tissues was monitored by using bioluminescence as a real-time indicator. The complete lux operon includes genes that encode the biosynthetic enzymes for the substrate, and thus, exogenous substrate addition is not required. Although addition of an exogenous aldehyde, such as decanal, may increase the output of the luxAB genes compared to the output when the substrate is supplied through expression of all of the lux genes (luxCDABE) (2), a system that does not require exogenous substrate addition removes any variability due to substrate availability and streamlines the assay. Moreover, any potential effects of exogenous substrate addition on the physical or temporal location of bacterial cells on the surface tissues are eliminated. The P. luminescens luciferase genes have the advantage of functioning at a higher temperature (up to 45°C) than other luciferases that have been characterized (2), a trait which may be useful in future in vivo applications of the bioluminescent strain of E. coli O157:H7.
The utility and validity of using luminescent bacterial strains to study biological processes have been reviewed previously (33). Data from this study and elsewhere indicate that there is a strong correlation between the viable counts of bioluminescent bacteria and light emission (3).
We observed that as few as 50 cells of E. coli O157:H7 strain L-2 could be detected by bioluminescence and that there was an approximately 10-fold decrease in sensitivity when bacteria were suspended in bovine fecal slurries compared with bacteria suspended buffer or broth. The slopes of the curves in plots of intensity versus cell number indicated that the sensitivity of detection was probably affected more by the optical properties of the fecal slurry than by direct effects of feces components on light output per cell. Bioluminescence from mouse fecal suspensions inoculated with Yersinia enterocolitica containing luxAB (16) and luxCDABE-bearing Salmonella typhimurium (4) and bioluminescence from bovine feces from cows fed luxAB-bearing Y. enterocolitica have been reported previously (16).
Initial inoculation of E. coli O157:H7 strain L-2 onto tissues revealed that bacteria could be visualized directly on a large area of beef carcass surface tissue. The luminescence appeared to be especially concentrated along muscle striations on the surface of the tissue, which differed in appearance, composition, and topography (Fig. 2). In a separate experiment, increases in bioluminescence were observed after incubation on a section of beef tissue (Fig. 3). A similar observation was reported by Chen and Griffiths for luxAB-bearing Salmonella enteritidis inoculated onto chicken meat following incubation (3).
Water washing of beef tissues inoculated with bioluminescent E. coli O157:H7 strain L-2 by the immersion-shaking method or by using a bottle to squirt water onto the inoculated surface revealed that the effects of both decontamination methods could be visualized in real time, whether the surface tissue was lean or adipose. Although these washing procedures were not entirely comparable in terms of pressure and volume to the spray washing procedures used by the meat industry or during pilot-scale research (7), our results suggest that imaging bioluminescent bacterial reporter strains can be used to assess carcass decontamination processes.
Differences in the attachment of food-borne bacterial pathogens to various tissue surfaces are an important issue in food microbiology and food processing. Attachment to tissue-specific molecules or polymers (30) and attachment to or detachment from carcass surface tissues and excised sections of meat are typically determined by sampling and by performing plate count experiments (9). The ability to monitor the process of attachment of pathogens to or association of pathogens with carcass surfaces in real time without exogenous sampling should accelerate such studies and has specific advantages. Unlike using physical biochemical indicators, using luciferase as the reporter ensures that the signal observed is from viable, metabolically active cells which can be detected as they exist in or on a food matrix.
Data presented in this paper indicate that bacterial association with the beef carcass surface may be influenced by the physical topography and structure of the surface. We observed that following a water wash, tissues that were rougher retained more bioluminescence (which correlated with the viable counts) than tissues that were relatively smooth and homogeneous. It has been reported that bacterial removal from or decontamination of beef carcass surface tissue is influenced by the level of lean or adipose tissue in the underlying tissue menstruum (7, 9). In these previous studies, excised samples of treated tissues were more likely to retain bacterial inocula if the surface tissue was lean than if it was adipose. Our bioluminescence data are in agreement with these observations; however, our bioluminescence method generated both spatial and quantitative information in real time. Although bioluminescence monitoring will not entirely replace culture methods in the study of attachment and decontamination, obviating the need for excision sampling, it has distinct advantages as a screening tool for selecting decontamination protocols for validation.
The observation that increased bioluminescence indicative of bacterial growth occurs on tissue surfaces after incubation has broad implications for studying decontamination processes. Since small numbers of tagged organisms (approximately 50 cells) can be detected on tissue surfaces and since luciferase-based photon counting imaging has a broad dynamic range of many logs, low levels of microbial contamination can be monitored over time. Moreover, spatially resolved analyses of bacterial survival and growth patterns after antimicrobial treatments could be rapidly performed by using the molecular biophotonic method described here, which would eliminate the need for exogenous sampling and culturing.
Eliminating back-extrapolation of sample plate count data would greatly improve the validity of carcass surface microbial count data. Biophotonics eliminates the need for the assumption that bacterial loads are homogeneously distributed across the surfaces of carcasses. Extrinsic factors that influence attachment of microorganisms to fascia-covered animal carcasses, such as the degree of hydration and composition, as well as putative intrinsic attachment factors, such as cell surface receptors, host tissue components (e.g., hyaluronan, collagen, and chondroitin sulfate [36]), could be systematically examined in situ over significantly larger areas of carcass tissue surface than the areas previously examined to determine their roles in attachment by using the biophotonic system described in this study.
Cloning promoters of interest upstream of a complete lux operon could provide indicator strains that could report metabolic, regulatory, or gene expression activity under different environmental conditions found in food processing. A similar approach has been described by Dodd et al., who used a spvA-lux fusion in S. enteritidis to monitor levels of the RpoS product in cells in the presence and absence of a competitive microflora (10).
Since bioluminescence has been used as a real-time genetic reporter in live animal models (4–6), the growth and survival of E. coli O157:H7 and other enterohemorrhagic E. coli organisms could be evaluated in animal models involving human infection and carcass processing. Specifically, expression of genotypic traits that influence survival in animals, such as the factors which determine the minimum infectious dose in human hosts, and transfer from the animal or gastrointestinal tract to the carcass during processing could be studied by this biophotonic procedure.
Our findings demonstrate that real-time macroscopic imaging of bacteria on beef carcass tissue surfaces is possible if bacterial strains with a lux gene reporter are used. We present evidence showing that different tissue surfaces, especially lean and adipose surfaces, retain different levels of contaminating bacteria. The magnitude of the differences is consistently less than 1 log10 unit of inoculated population per unit of area. Therefore, any practical implications of this observation are yet to be determined. Previously, however, Thomas and McMeekin (36a) documented the role of the water microlayer on poultry skin, which has a very significant effect on microbial attachment. It is not unlikely that in our experiments adipose tissues retained less water on their surfaces (due to their inherent hydrophobicity) and thus retained fewer bacteria after simulated spray washing. The clear differences between the prewashed and postwashed tissues indicate that a molecular biophotonic approach to studying microbial association with carcass tissue surfaces offers a truly in situ strategy for understanding a problem previously approached only retrospectively (i.e., through sampling, culturing, and subsequent back-calculation of microbial densities on tissue surfaces).
ACKNOWLEDGMENTS
We thank Carole J. Smith (USDA, Agricultural Research Service) for her expert technical assistance and for tissue collection and preparation. We are grateful to Kenneth H. Nealson (Center for Great Lakes Studies, University of Wisconsin—Milwaukee) for the generous gift of pCGLS1 and to R. O. Elder (USDA Agricultural Research Service) for providing stx primer sets.
REFERENCES
- 1.Baker J M, Griffiths M W, Collins-Thompson D L. Bacterial bioluminescence: applications in food microbiology. J Food Prot. 1992;55:62–70. doi: 10.4315/0362-028X-55.1.62. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee J, Meighen E A. Biotechnological applications of bacterial bioluminescence (lux) genes. Photochem Photobiol. 1995;62:641–650. [Google Scholar]
- 3.Chen J, Griffiths M W. Luminescent Salmonella strains as real time reporters of growth and recovery from sublethal injury in food. Int J Food Microbiol. 1996;31:27–43. doi: 10.1016/0168-1605(96)00941-5. [DOI] [PubMed] [Google Scholar]
- 4.Contag C H, Contag P R, Mullins J I, Spilman S D, Stevenson D K, Benaron D A. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 5.Contag C H, Spilman S D, Contag P R, Oshiro M, Eames B, Dennery P, Stevenson D K, Benaron D A. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 6.Contag P R, Olomu I N, Stevenson D K, Contag C H. Bioluminescent indicators in living mammals. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 7.Cutter C N, Siragusa G R. Efficacy of organic acids against Escherichia coli O157:H7 attached to beef carcass tissue using a pilot scale model carcass washer. J Food Prot. 1994;57:97–103. doi: 10.4315/0362-028X-57.2.97. [DOI] [PubMed] [Google Scholar]
- 8.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Dickson J S, Cutter C N, Siragusa G R. Antimicrobial effects of trisodium phosphate against bacteria attached to beef tissue. J Food Prot. 1994;57:952–955. doi: 10.4315/0362-028X-57.11.952. [DOI] [PubMed] [Google Scholar]
- 10.Dodd E R, Sharman R L, Bloomfield S F, Booth I R, Stewart G S A B. Inimical processes: bacterial self-destruction and sub-lethal injury. Trends Food Sci Technol. 1997;8:238–241. [Google Scholar]
- 11.Dorsa W J, Cutter C N, Siragusa G R. Evaluation of six sampling methods for recovery of bacteria from beef carcass surfaces. Lett Appl Microbiol. 1996;22:39–41. doi: 10.1111/j.1472-765x.1996.tb01104.x. [DOI] [PubMed] [Google Scholar]
- 12.Ellison A, Anderson W, Cole M B, Stewart G S A B. Modelling the thermal inactivation of Salmonella typhimurium using bioluminescence data. Int J Food Microbiol. 1994;23:467–477. doi: 10.1016/0168-1605(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 13.Frackman S, Anhalt M, Nealson K H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172:5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill P J, Stewart G S A B. Use of lux genes in applied biochemistry. J Biolumin Chemilumin. 1994;9:211–215. doi: 10.1002/bio.1170090315. [DOI] [PubMed] [Google Scholar]
- 15.Kado C I. Live time quantification of bacterial interactions in various environments. In: Adolph K W, editor. Microbial genome methods. Boca Raton, Fla: CRC Press; 1996. pp. 179–196. [Google Scholar]
- 16.Kaniga K, Sory M P, Delor I, Saegerman C, Limet J N, Cornelis G R. Monitoring of Yersinia enterocolitica in murine and bovine feces on the basis of the chromosomally integrated luxAB marker gene. Appl Environ Microbiol. 1992;58:1024–1026. doi: 10.1128/aem.58.3.1024-1026.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K Y, Frank J F, Craven S E. Three-dimensional visualization of Salmonella attachment to poultry skin using confocal scanning laser microscopy. Lett Appl Microbiol. 1996;22:280–282. doi: 10.1111/j.1472-765x.1996.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 19.Kodikara C P, Crew H H, Stewart G S A B. Near on-line detection of enteric bacteria using lux recombinant bacteriophage. FEMS Microbiol Lett. 1991;83:261–266. doi: 10.1016/0378-1097(91)90486-t. [DOI] [PubMed] [Google Scholar]
- 20.Legocki P R, Legocki M, Baldwin T O, Szalay A A. Bioluminescence in soybean root nodules: demonstration of a general approach to assay gene expression in vivo by using bacterial luciferase. Proc Natl Acad Sci USA. 1986;83:9080–9084. doi: 10.1073/pnas.83.23.9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindow S E. The use of reporter genes in the study of microbial ecology. Mol Ecol. 1995;4:555–566. [Google Scholar]
- 22.Meighen E A. Enzymes and genes from the lux operons of bioluminescent bacteria. Annu Rev Microbiol. 1988;42:151–176. [Google Scholar]
- 23.Meighen E A. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 24.Meighen E A. Genetics of bacterial bioluminescence. Annu Rev Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- 25.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nealson K H, Hastings J W. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newland J W, Neill R J. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J Clin Microbiol. 1988;26:1291–1297. doi: 10.1128/jcm.26.7.1292-1297.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips-Jones M K. Bioluminescence lux expression in the anaerobe Clostridium perfringens. FEMS Microbiol Lett. 1993;106:265–270. doi: 10.1111/j.1574-6968.1993.tb05974.x. [DOI] [PubMed] [Google Scholar]
- 29.Prosser J I, Killham K, Glover L A, Rattray E A S. Luminescence-based systems for detection of bacteria in the environment. Crit Rev Biotechnol. 1996;16:157–183. doi: 10.3109/07388559609147420. [DOI] [PubMed] [Google Scholar]
- 30.Sanderson K, Thomas C J, McMeekin T A. Molecular basis of the adhesion of Salmonella serotypes to chicken muscle fascia. Biofouling. 1991;5:89–101. [Google Scholar]
- 31.Siragusa G R. The effectiveness of carcass decontamination systems for controlling the presence of pathogens on the surfaces of meat animal carcasses. J Food Safety. 1995;15:229–238. [Google Scholar]
- 32.Siragusa, G. R. Statistical validation of the track dilution plating method from ground beef and carcass surface samples. Submitted for publication.
- 33.Stewart G S A B. Challenging food microbiology from a molecular perspective. Microbiology. 1997;143:2099–2108. doi: 10.1099/00221287-143-7-2099. [DOI] [PubMed] [Google Scholar]
- 34.Stewart G S A B, Williams P. Lux genes and the applications of bacterial bioluminescence. J Gen Microbiol. 1992;138:1289–1300. doi: 10.1099/00221287-138-7-1289. [DOI] [PubMed] [Google Scholar]
- 35.Stewart G S A B, Loessner M J, Sherer S. The bacterial lux gene bioluminescent biosensor revisited. ASM News. 1996;62:297–301. [Google Scholar]
- 35a.Strockbrine, N. Personal communication.
- 36.Thomas C J, McMeekin T A. Attachment of Salmonella spp. to chicken muscle surfaces. Appl Environ Microbiol. 1981;42:130–134. doi: 10.1128/aem.42.1.130-134.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Thomas C J, McMeekin T A. Effect of water immersion on the microtopography of the skin of chicken carcasses. J Sci Food Agric. 1982;33:549–554. [Google Scholar]
- 37.Tomicka A, Chen J, Barbut S, Griffiths M W. Survival of bioluminescent Escherichia coli O157:H7 in a model system representing fermented sausage production. J Food Prot. 1997;60:1487–1492. doi: 10.4315/0362-028X-60.12.1487. [DOI] [PubMed] [Google Scholar]
- 38.White D, Leifert C, Ryder M H, Killham K. Lux gene technology—a strategy to optimize biological control of soil-borne diseases. New Phytol. 1996;133:173–181. [Google Scholar]
- 39.Xi L, Cho K W, Tu S C. Cloning and nucleotide sequences of lux genes and characterization of luciferase of Xenorhabdus luminescens from a human wound. J Bacteriol. 1991;173:1399–1405. doi: 10.1128/jb.173.4.1399-1405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zyskind J W, Bernstein S I. Recombinant DNA laboratory manual. San Diego, Calif: Academic Press, Inc.; 1989. [Google Scholar]