Abstract
Invasive fungal infections continue to increase as at-risk populations expand. The high associated morbidity and mortality with fungal diseases mandate the continued investigation of novel antifungal agents and diagnostic strategies that include surrogate biomarkers. Biologic markers of disease are useful prognostic indicators during clinical care, and their use in place of traditional survival end points may allow for more rapid conduct of clinical trials requiring fewer participants, decreased trial expense, and limited need for long-term follow-up. A number of fungal biomarkers have been developed and extensively evaluated in prospective clinical trials and small series. We examine the evidence for these surrogate biomarkers in this review and provide recommendations for clinicians and regulatory authorities.
Keywords: diagnosis, fungal infections, mycology
The morbidity and mortality of invasive fungal diseases has fostered interest in surrogate biomarkers. These are used for diagnosis and may convey prognostic information. We review current evidence for the use of biomarkers for invasive fungal infections.
The use of surrogate end points through the use of indirect markers known as biomarkers has the potential to replace clinical end points in large-scale licensing therapeutic trials and to provide additional prognostic information when used during routine clinical care. Use of biomarkers can avoid the delay of days to weeks for fungal growth in blood or tissue and may obviate the need for invasive diagnostic procedures. To be useful as surrogate end points, however, there must be evidence that these biomarkers correlate with clinically meaningful outcomes. Substantial evidence has been generated pertaining to the use of fungal biomarkers over the last decade. We review the available evidence on the use of fungal biomarkers as surrogate end points in this manuscript.
CRYPTOCOCCOSIS
Surrogate biomarkers for cryptococcosis can be used for diagnosis, prognosis, and treatment decisions. Standard diagnostics are India ink microscopy, cryptococcal antigen (CrAg), and culture [1]. Culture and microscopy techniques date to 1894 [2] and are still used. India ink testing for visualization of the capsule can detect ~103 CFU yeasts/mL in centrifugated cerebrospinal fluid (CSF) and has a sensitivity of ~60%–80% in HIV-infected patients [3] and ~50% in non-HIV cryptococcal meningitis. The change in quantitative CSF microscopy yeast counts correlates poorly with changes in quantitative culture and cannot be used as a surrogate marker [4].
Cryptococcal Antigen
Cryptococcal antigen tests using latex agglutination, enzyme immunoassay (EIA), or lateral flow assays (LFAs), have been integrated into clinical practice and have >95% sensitivity and specificity in CSF, serum, plasma, and whole blood [5, 6]. The CrAg LFA by IMMY (Norman, OK, USA) has the best diagnostic performance of any current point-of-care assay [3], with detection of both C. neoformans and C. gattii species but without the ability to distinguish between species. Internationally, other non–Food and Drug Administration (FDA)–approved CrAg LFAs exist with substantially worse diagnostic performance [7–9]. Serum or plasma CrAg LFAs are >99.9% sensitive for detecting meningitis in HIV-infected persons with meningitis, although if central nervous system symptoms are present despite negative serum testing assessment of the CSF should be pursued [10, 11]. False-negative tests are more common with CrAg latex agglutination and in HIV-negative persons, where ~20% may have a false-negative CrAg in serum with disseminated disease [3, 12]. False-negative CrAg LFAs are possible due to a prozone effect at CrAg titers >1:1 million by serial dilution; however, a new semiquantitative CrAg LFA has been developed that eliminates prozone [11]. The ability to provide a semiquantitative CrAg titer provides prognostic information, as high titers are correlated with high burden of disease and predict poor prognosis [13, 14]. However, serial changes in CSF titers or serum CrAg titers following treatment are not associated with antifungal activity, change in CSF quantitative culture, or survival [15, 16]; thus, longitudinal CrAg titer change is not a useful surrogate biomarker.
Early Fungicidal Activity
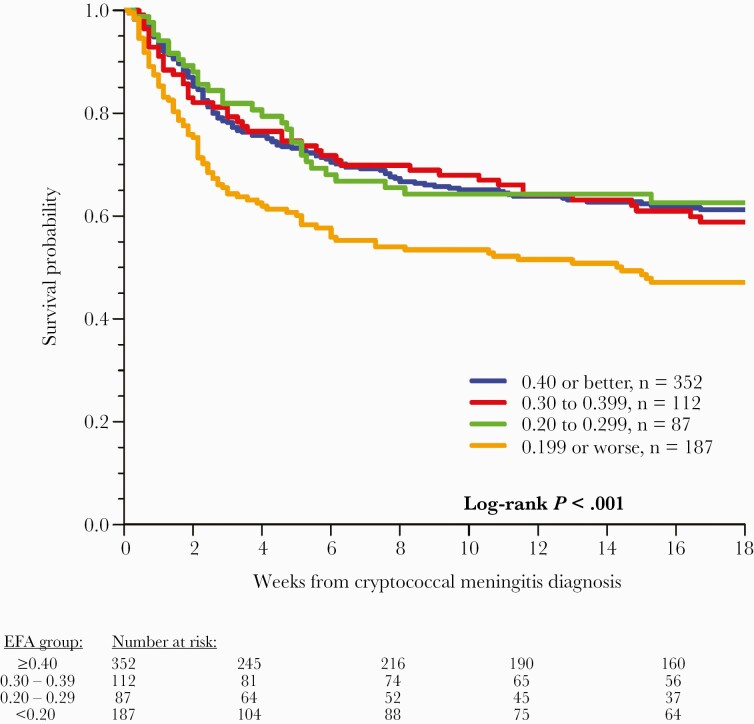
The slope of cryptococcal yeast clearance rate of serial CSF quantitative cultures, termed early fungicidal activity (EFA) [17, 18], is now a frequently used tool in clinical trials as a direct marker of antifungal activity and a surrogate marker for mortality [1, 19, 20]. In a 2020 meta-analysis of 738 cryptococcal meningitis participants from 3 sequential trials who received the same background induction regimen of amphotericin B deoxycholate and fluconazole, those with an EFA <0.20 log10 CFU/mL/d had an increased 18-week mortality compared with those with an EFA >0.20 (Figure 1) [19].
Figure 1.
Association between EFA of CSF Cryptococcus yeast clearance rate and mortality. Increased mortality was observed among those with an EFA <0.20 log10 CFU/mL CSF/d when receiving amphotericin B combination therapy in Uganda. Above this EFA threshold could be a target for phase II trials. Reproduced with permission from Pullen et al. [19]. Abbreviations: CFU, colony-forming units; CSF, cerebrospinal fluid; EFA, early fungicidal activity.
Furthermore, the association of EFA with mortality in HIV-associated cryptococcal meningitis is built on previous separate prospective studies. Bicanic and colleagues used pooled data from 4 trials in persons with HIV-associated cryptococcal meningitis undergoing induction with amphotericin B-based combination therapy or fluconazole monotherapy and found that a slower rate of clearance was associated with 10-week mortality [20]. Those who survived >10 weeks had mean EFAs of 0.41 log10 CFU/mL/d, vs 0.27 log10 CFU/mL/d in those who died. A 2016 systematic review of 27 trials including 2854 participants (EFAs measured with different statistical techniques) also found an association between mean EFA and 10-week survival [21], although this analysis excluded inferior regimens with poor EFAs, such as fluconazole monotherapy. While there are many contributors to cryptococcal meningitis mortality, EFA is a validated surrogate marker for antifungal activity in humans and for all-cause mortality, particularly when the EFA is <0.20 log10 CFU/mL/d. As Cryptococcus quantitative cultures take 3–7 days to grow on standard fungal media [18], EFA is a delayed metric that cannot be used in real time for clinical decision-making. Quantitative CSF culture methodology has been published and cross-validated [18].
Other Potential Biomarkers
Insufficient data exist for other surrogate markers for cryptococcal meningitis. Both CSF (1→3)-β-D-glucan (BDG) and FilmArray polymerase chain reaction (PCR) testing (BioFire, Salt Lake City, UT, USA) are potential diagnostic markers in cryptococcosis, but both are suboptimal compared with CrAg for diagnosis [22–25]. The rapid decay of CSF BDG and the change in PCR cycle threshold (Ct) value for C. neoformans DNA during antifungal therapy make these interesting potential surrogate biomarkers.
Summary and Recommendations for Surrogate Biomarkers in Cryptococcosis
CrAg is an established diagnostic biomarker that is more sensitive than culture. CrAg LFA is most sensitive. CrAg is preferred over BDG or PCR testing for diagnosis, but sequential CSF CrAg titers should not be used for monitoring response to therapy. The CSF clearance rate of Cryptococcus from serial quantitative CSF cultures (EFA) is a direct marker of antifungal activity and a surrogate biomarker of mortality and is now routinely used to evaluate novel antifungal strategies in cryptococcal meningitis clinical trials (Table 1).
Table 1.
Summary of Fungal Biomarkers and Their Use for Diagnosis, Prognosis, or as Surrogate End Points in Clinical Trials
| Fungus | Diagnostic Biomarkers | Prognostic Biomarkers | Surrogate End Points for Treatment Trials |
|---|---|---|---|
| Cryptococcus | CrAg, PCR |
CrAg titer Quantitative CSF culture CSF BDG |
Early fungicidal activity of serial quantitative CSF cultures is an established end point |
| Candida | PCR, T2Candida BDG (nonspecific) |
None validated | Possible: change in BDG, change in PCR Ct value, or time to T2Candida negativity |
| Aspergillus | Blood galactomannan, blood LFA for galactomannan, BDG, PCR |
Quantitative galactomannan value | Possible: change in blood galactomannan |
| Other molds | None | None | None |
| Blastomyces | Serum or urine antigen | Quantitative antigen possible | Possible: change in antigen or CRP |
| Coccidioides | Serology, PCR, antigen |
CF IgG titer | Possible: change in CF IgG titer |
| Histoplasma | Serum or urine antigen | Quantitative antigen | Possible: change in antigen titer |
| Paracoccidioides | IgG detection, antigen |
IgG titer | Possible: change in IgG titer |
| Sporothrix | PCR | None | None |
| Talaromyces | Serum or urine antigen PCR |
Quantitative blood culture | Early fungicidal activity based on serial quantitative blood cultures |
Abbreviations: BDG, 1,3-β-D-glucan; CF, complement fixation; CrAg, cryptococcal antigen; CRP, c-reactive protein; IgG, immunoglobulin G; LFA, lateral flow assay; PCR, polymerase chain reaction.
INVASIVE CANDIDIASIS
Invasive candidiasis includes bloodstream infections by Candida spp. (candidemia), which may be complicated by end-organ infections from hematogenous dissemination, and noncandidemic infections of organs or submucosal tissues. Blood culture analysis is the gold standard and most common approach for identifying candidemia. Noncandidemic invasive candidiasis (deep-seated candidiasis) is typically diagnosed by culture, staining, and/or histopathology of samples acquired by biopsy or aspiration of involved tissue. Newer technologies and commercially available methods have been developed for more rapid diagnosis than the delay observed with cultures.
The average time to detection for culture-based systems ranges from 14 to 38 hours and varies depending on the culture conditions used (most Candida spp. grow more rapidly in aerobic than in anaerobic bottles) [26], the Candida species, and the number of circulating yeast per mL of sample. Moreover, limitations in the sensitivity, reliability, and timeliness of automated blood culture methods exist. The sensitivity of these assays decreases with the inocula (79% sensitivity with 1000 yeast cells/bottle, 73% with 100 yeast cells/bottle, and 70% sensitivity with 10 yeast cells/bottle). The mean detection time for C. albicans is 20–30 hours depending on bottle type and inoculum concentration [27]. Non-albicans species such as C. glabrata may take considerably longer for growth (~80 hours).
Using culture-based diagnostic molecular assays, clinicians are now able to decrease the time to organism identification and improve antimicrobial agent selection and patient outcomes [28]. For example, matrix-assisted laser desorption/ionization mass spectroscopy (MALDI-TOF MS) is based on characteristic mass spectral fingerprints unique to different microorganisms. This technique requires growth of the organism on solid culture media before assessment. Other recently developed rapid diagnostics performed directly on positive blood culture samples include Biofire Direct from Blood Culture, GenMark Yeast ID from Blood Culture, Accelerate Pheno, and PNA Fish [29–31]. The FilmArray blood culture identification panel, a multiplexed assay able to identify 24 different pathogens (BioFire Diagnostics, Salt Lake City, UT, USA), provides results in ~1 hour with reported sensitivity of 96%–100% and specificity of 88%–100% [31, 32].
Limitations of culture techniques have spurred development of culture-independent diagnostic tests for invasive candidiasis, although uncertainty about test performance, costs, and utility remains [33]. Traditional culture-based methods may be less sensitive than newer methodologies and thus result in the erroneous interpretation that newer methodologies are less specific than the traditional “gold standard” cultures despite detection of more true-positive results (Table 2). Data are most robust for 3 culture-independent diagnostic tests: (1→3)-β-D-glucan, Candida polymerase chain reaction (PCR), and T2Candida assays of blood-based samples. Culture-independent diagnostic test–guided patient management strategies have not been validated in clinical trials. We propose that patient populations within the dark lines in Table 2 might be targeted for such future trials to identify whether patients with “false-positive” culture-independent diagnostic tests that are culture negative benefit from antifungal therapy and thus are true positives. Below we discuss how these assays can be rationally incorporated into patient care using current knowledge.
Table 2.
Performance Characteristics of Culture-Independent Diagnostic Tests for Candidemia as Compared With Culture as the Imperfect Reference Standard
| Prevalence | Corresponding Patient Populations | Beta-D-Glucan (60%/80%) |
Beta-D-Glucan (80%/80%) | PCR, T2Candida (70%/90%) |
PCR, T2Candida (90%/90%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| PPV, % | NPV, % | PPV, % | NPV, % | PPV, % | NPV, % | PPV, % | NPV, % | ||
| ~0.4% | Any patient for whom a blood culture is collected | 1 | 99.8 | 1 | 99.9 | 3 | 99.8 |
3 | >99.9 |
| ~1% | Patient in ICU with fever | 3 | 99.5 | 4 | 99.7 | 7 | 99.7 | 8 | 99.9 |
| ~3% | Patients with sepsis, septic shock, in ICU for >3–7 d |
8.5 | 98.5 | 11 | 99.2 | 18 | 99 | 22 | 99.6 |
| ~10% | ICU patient at increased risk for candidemia based on clinical prediction score |
25 | 94.7 | 31 | 97 | 44 | 96 | 50 | 98.8 |
Table 2 adapted from Clancy and Nguyen [34]. BDG sensitivity/specificity for diagnosing candidemia are taken from meta-analyses cited in the text. PCR and T2Candida sensitivity/specificity for diagnosing candidemia are taken from a meta-analysis and DIRECT and DIRECT2 clinical trials [40, 42, 43]. In order to make rational use of culture-independent diagnostic tests, clinicians must be familiar with test performance and prevalence of various types of invasive candidiasis locally.
Culture-independent diagnostic test–guided patient management strategies have not been validated in clinical trials. We propose that patient populations with PPV >15% might be targeted for such trials to better identify the clinical benefit of response to antifungal therapy. These “false positives” among high-risk patient populations may represent deep-seated invasive candidiasis that is negative by blood and other cultures.
Abbreviations: BDG: 1,3-beta-D-glucan; ICU, intensive care unit; NPV, negative predictive value; PCR, Candida polymerase chain reaction; PPV, positive predictive value.
(1→3)-β-D-Glucan (BDG)
BDG is a cell wall constituent of Candida and most pathogenic fungi. There is no conclusive evidence for the superiority of an assay by any particular manufacturer [34]. The majority of the data are for the FDA-approved Fungitell assay (Associates of Cape Cod, Cape Cod, MA, USA). Positive BDG results do not identify Candida species or distinguish between Candida and other fungi. Performance improves if 2 or more sequential samples are positive for BDG (>80 pg/mL) [35]. False positives can occur with intravenous immunoglobulin, albumin, beta-lactam antibiotics, or bacterial infections [36].
In meta-analyses, serum BDG sensitivity and specificity for invasive candidiasis (primarily candidemia) are ~75%–80% and ~60%–80%, respectively [37–40]. Performance varies greatly in individual reports, with sensitivities ranging from 27% to 100% and specificities ranging from 0% to 100% [41]. In studies of deep-seated candidiasis (predominantly intra-abdominal candidiasis), sensitivities and specificities (interquartile range [IQR]) were 65% (60%–77%) and 75% (64%–75%), respectively (Table 3) [42–46]. A recent Cochrane report was unable to perform a meta-analysis or estimate BDG diagnostic accuracy for invasive candidiasis due to heterogeneity in performance and study design [41]. Serial BDG measurements are not generally assessed in invasive candidiasis, although a negative slope (eg, decline) in BDG levels has been correlated with successful treatment outcomes [47]; more data on longitudinal BDG change are needed.
Table 3.
Anticipated Performance of BDG, PCR, and T2Candida in Various Populations at Risk for Intra-abdominal Candidiasis
| BDG (65%/75%) | PCR, T2Candida (45%/95%) | ||||
|---|---|---|---|---|---|
| Prevalence, % | Representative Patient Populations | PPV, % | NPV, % | PPV, % | NPV, % |
| 5 |
- Low- to moderate-risk peritoneal dialysis patient with peritonitis |
12 | 97.6 | 37 | 97 |
| 10 | - Patient with emergent surgery for intra-abdominal infection - Patient with colonic perforation |
22 | 95 | 56 | 94 |
| 20 | - Patient with high-risk severe acute or necrotizing pancreatitis - Patient with small bowel perforation - Patient with emergent surgery for nosocomial intra-abdominal infection |
39 | 90 | 74 | 87 |
| 30 | - Patient who has undergone high-risk GI/hepatobiliary surgery - Patient with a biliary leak - Patient with a gastric/duodenal perforation |
53 | 83 | 83 | 80 |
Table 3 adapted from Clancy and Nguyen [34]. BDG sensitivity/specificity values for diagnosing intra-abdominal candidiasis (the most common type of deep-seated candidiasis) are median values from studies cited in the text. PCR and T2Candida sensitivity and specificity for diagnosis are representative of recently published studies, as cited in the text. In order to make rational use of culture-independent diagnostic tests, clinicians must be familiar with test performance and prevalence of various types of invasive candidiasis locally.
PPVs and NPVs >15–30% and >85%, respectively, signify patients in whom nonculture testing may have the greatest clinical utility, assuming that antifungal treatment is justified at a threshold likelihood of candidemia ≥15%–30%.
Abbreviations: BDG: 1,3-beta-D-glucan; GI, gastrointestinal tract; NPV, negative predictive value; PCR, Candida polymerase chain reaction; PPV, positive predictive value.
PCR
PCR offers potential advantages over BDG by specifically targeting Candida spp. and allowing concurrent identification to the species level. Most PCR assays target C. albicans, C. glabrata, C. parapsilosis complex, C. tropicalis, and C. krusei, which account for >95% of cases of invasive candidiasis at most centers. In a meta-analysis, PCR sensitivity was 95% and specificity was 92% for proven/probable invasive candidiasis (predominantly candidemia) [48]. Sensitivity and specificity for proven/probable/possible invasive candidiasis were 73% and 95% respectively. Higher sensitivity was observed with whole blood (as opposed to serum), and Candida- or fungal-specific rather than broader multiplex assays. There was a trend toward lower specificity among patients colonized by Candida [48]. Another meta-analysis investigated LightCycler SeptiFast (Roche Diagnostics, Mannheim, Germany), a real-time multiplex PCR assay for the 5 most common Candida spp. and 20 other bacterial pathogens, among patients with presumed sepsis [49]. In this study, the sensitivity and specificity for candidemia were 61% and 99%, respectively. In studies of deep-seated candidiasis (predominantly intra-abdominal candidiasis), PCR sensitivity and specificity vary from 25% to 91% and 33% to 97%, respectively [42–45].
T2Candida
T2Candida (T2 Biosystems, Lexington, MA, USA) is an FDA-approved diagnostic for candidemia, which detects the 5 most common Candida species within whole blood by an automated process in which amplified DNA targets are detected by T2 magnetic resonance. In multicenter studies, T2Candida was 89%–91% sensitive and 98% specific for candidemia by targeted species [50, 51]. The mean time to Candida detection and species identification was 4.4 ± 1.0 hours. Data for T2Candida in clinical practice have shown this methodology as 65%–83% sensitive and 93%–96% specific for candidemia [52, 53]. For the diagnosis of deep-seated candidiasis (predominantly intra-abdominal candidiasis), studies have found this system 33%–45% sensitive and 93%–96% specific [53–55].
In small studies of intensive care unit patients at risk for invasive candidiasis, treatment strategies based on serum BDG and T2Candida results were effective in shortening the time to appropriate treatment of invasive candidiasis, safely reducing unnecessary empiric antifungal use, and reducing costs [56–60]. In a multicenter study from Spain of patients treated for candidemia, positive T2Candida (but not BDG) within the first 5 days after a positive Candida blood culture was an independent risk factor for either attributable mortality or development of deep-seated candidiasis [61]. In a companion multicenter study, positive T2Candida after institution of empiric therapy was an independent predictor of death or proven candidiasis, whereas BDG results were not predictive of mortality [62]. Clinical trials of culture-independent diagnostic test–guided therapeutic strategies are needed to validate impact on patient outcomes, antifungal utilization, and health care costs. The results of the CandiSep trial (NCT02734550, discussed below) will hopefully increase our understanding of these tests and their impact on antifungal prescribing.
Culture-independent diagnostic tests have the potential to be useful as inclusion criteria for candidiasis trials. It is apparent that culture alone detects only a subset of clinical disease. Culture positivity should be an important a priori subgroup in clinical trials, but limiting enrollment to culture-positive cases leads to delays in enrollment and artificial populations not reflective of current clinical practice. The CandiSep prospective randomized multicenter trial is evaluating whether the decision to prescribe antifungals guided by (1,3)-β-D-glucan in comparison with standard of care shortens time to antifungal therapy and reduces mortality. The results of this trial should inform inclusion criteria for future candidiasis trials. Most probably, culture positivity/negativity should be a standard a priori subgroup analysis in future clinical trials, not restriction for an inclusion criterion.
There is also a need to validate the clinical role of serial changes of surrogate markers such as BDG or PCR Ct value, or time to T2Candida negativity. With the exception of 1 BDG study, associations are lacking between faster reduction in surrogate markers and clinical survival [47]. Validation of surrogate marker kinetics would allow for smaller phase II trials for new agents or combination therapies. A starting point would be to use existing cohorts with stored specimens to study longitudinal changes in biomarkers.
Summary and Recommendations for Surrogate Biomarkers in Candidiasis
Cultures remain the mainstay of candidiasis diagnostics yet should not be the limiting inclusion criterion for invasive candidiasis trials. Documented clearance of candidemia is a marker of successful antifungal treatment. We recommend using diagnostic molecular assays on positive cultures to accurately and rapidly identify organisms to species level. Culture-independent diagnostic tests may be useful in selected clinical scenarios to initiate or de-escalate antifungal therapy. Positive PCR or T2Candida testing in patients with risk factors for candidiasis warrants enrollment in clinical trials. Multiple possible biomarkers exist that could be used in phase II trials to quantify antifungal activity, but these biomarkers have not been validated.
ASPERGILLOSIS
Aspergillus spp. are ubiquitous in the environment and frequently inhaled into the airways [63]. Diagnosis of invasive aspergillosis requires an appropriate host and compatible clinical syndrome when cultures from the airway return positive to differentiate colonization from infection. Only culture of the organism from a sterile site or demonstration of the organism with hyphal invasion on histopathology is confirmatory of proven disease [64]. The Aspergillus cell wall contains polysaccharides, BDG, chitin, and other components that may be utilized for detection in patient samples. Galactomannan and BDG are the most well studied of these cell wall constituents.
Galactomannan Kinetics
The kinetic profile of serum galactomannan is complex and depends on multiple factors including production and secretion, the fungal burden, the presence and function of neutrophils, the patient’s renal and hepatic function, and exposure to antifungal agents with activity against Aspergillus spp. Galactomannan presence in the circulation correlates with invasive growth of Aspergillus through the pulmonary capillaries, and angio-invasion has been correlated with fungal burden and galactomannan production [65, 66]. Together these findings help explain the variable performance of serum galactomannan in different patient populations (Table 4) [67]. For example, serum galactomannan is detectable in patients with hematological malignancies and allogeneic hematopoietic stem cell transplant recipients who have a higher burden of disease [68]. In contrast, the performance of serum galactomannan for the diagnosis of invasive aspergillosis (IA) in solid organ transplant patients, who typically have a lower burden of disease, is relatively poor [69, 70].
Table 4.
Summary of Galactomannan Performance Characteristics in Serum or Plasma Using a Cutoff ODI of 1.0
| Population | Sensitivity, % | Specificity, % |
|---|---|---|
| Hematologic malignancy | 58 | 95 |
| Hematopoietic stem cell transplantation | 65 | 65 |
| Solid organ transplant | 41 | 85 |
Table modified from Scott et al. [146].
Abbreviation: ODI, optical density index.
There has also been interest in determining if baseline serum galactomannan values correlate with patient outcome and whether the slope of serial galactomannan optical density index readings correlates with survival. Multiple studies have evaluated the role of baseline serum galactomannan as a predictor of treatment response and outcome of aspergillosis. In a recent review of these studies, Mercier et al. found a correlation between serum galactomannan level and survival at 42 and 180 days of treatment among patients with positive serum galactomannan (defined as cutoff >0.5 optical density index), while serum galactomannan ≥2.0 optical density index at baseline was associated with poor outcome [71]. In a recent randomized controlled trial comparing anidulafungin in combination with voriconazole vs voriconazole alone, baseline serum galactomannan was also an independent predictor of 6-week survival [72].
The role of measuring serial galactomannan in patients with IA has been evaluated in several studies [68, 73–76]. Despite the heterogenicity among these studies regarding outcome definition and galactomannan evaluation time points, the data suggest that a declining serum galactomannan value is a predictor of survival, whereas persistently elevated serum galactomannan correlates with death. One study reported that every 0.1-unit increase in galactomannan optical density index from baseline to week 2 corresponded to a 21% higher rate of an unsatisfactory clinical response [77].
Data on the effect of specific antifungal agents on serum galactomannan kinetics are conflicting and remain a subject of investigation. Some studies have shown a distinct kinetic profile depending on the antifungal agent, while others have shown no effect [78–80].
Galactomannan kinetics in bronchoalveolar lavage (BAL) fluid are poorly understood and should be interpreted with caution, as this test is plagued with caveats including sampling error, nonstandardized BAL fluid collection, and site of infection [81].
Based on the available data, current Infectious Diseases Society of America guidelines recommend the use of serial serum galactomannan only in patients with hematologic malignancies and hematopoietic stem cell transplant recipients who have an elevated galactomannan at baseline to monitor disease progression, therapeutic response, and prediction of patient outcomes with IA [82].
The development of lateral flow assays has aided in the diagnosis of aspergillosis, although their role in sequential sampling has not been defined thus far [83–85].
(1→3)-β-D-Glucan (BDG)
The utility of BDG testing in the therapeutic monitoring of patients with IA has been poorly characterized. Pazos et al. first evaluated BDG compared with galactomannan testing in a retrospective cohort of 40 high-risk neutropenic patients (5 proven, 3 probable, and 3 possible) [86]. The sensitivity, specificity, and positive and negative predictive values of galactomannan and BDG testing were almost identical; however, BDG was positive earlier than galactomannan. In the evaluation of BDG testing as a prognostic tool, plasma concentrations showed a consistent increase before clinical and microbiologic evidence of disease but decreased and became negative following a response to antifungal treatment. In contrast, those nonresponsive to antifungal therapy showed a persistence of BDG positivity. Others have similarly observed a decrease in BDG values following a response to therapy [87].
Others have compared BDG and galactomannan directly in the prognosis of IA. In a study of 51 evaluable patients, there was an association between changes from baseline to week 12 when BDG and galactomannan were summed; however, the use of BDG alone was not statistically significant. Others have described similar findings, with most patients having BDG values through 6–12 weeks persistently >80 pg/mL, and with the trajectory not predictive of 6- or 12-week outcomes [88]. BDG kinetics are slow and may require >4–6 weeks after treatment initiation to appreciably change [75]. These limitations, and the intrapatient variability of BDG testing, prompt the authors to not recommend serial BDG [89].
PCR
Availability of Aspergillus PCR testing in the United States remains limited; PCR is performed in reference laboratories only (although Aspergillus PCR testing is commercially available in Europe) [90]. The reported performance characteristics of PCR vary given the different technologies, cycle thresholds for positivity, microbial targets, and study populations that have been assessed [91]. A meta-analysis evaluating 25 studies found a pooled sensitivity and specificity of 84% and 76%, respectively [92]. In patients with 2 positive PCR results, the sensitivity was found to be 64%, although specificity improved to 95% [93].
Few studies have evaluated sequential PCR testing for prognosis; however, quantitative PCR as copies/mL of serum at the time of diagnosis has been found to be predictive of 90-day mortality. The persistence of positivity or the development of a negative test was predictive of 30- or 90-day mortality. Those who became PCR negative between days 14 and 20 were all alive at day 30, while those with PCR persistence exhibited 80% mortality at day 30 [94]. Time to PCR negativity with weekly sampling as an end point may be a future area to investigate.
Aspergillus fumigatus quantitative PCR from BAL has been evaluated for the diagnosis and prognosis of both invasive and noninvasive aspergillosis. In a 4-year retrospective study assessing 613 at-risk patients, the fungal burden (copies/mL) at time of diagnosis was predictive of 90-day mortality with 23% mortality for patients with <500 copies/mL, vs 68% for patients with >500 copies/mL (P < .05) [95].
Summary and Recommendations for Surrogate Biomarkers in Aspergillosis
Galactomannan, BDG, and PCR are standard diagnostic tests used in clinical practice, and some PCR assays have the advantage of also detecting mutations conveying antifungal drug resistance. BDG has the least specificity and is insufficient as a diagnostic biomarker for enrollment of patients into clinical trials. For prognostic markers of response to antifungals, the change in blood galactomannan values over time is a promising serial surrogate marker. PCR negativity over time is also promising for use as a surrogate marker of patient outcomes.
OTHER MOLDS
Non-Aspergillus invasive mold infections caused by Mucorales, Fusarium, Scedosporium spp., and others are increasingly encountered in an expanding population of severely immunosuppressed patients, accounting for 10%–25% of invasive mold infections in autopsy studies [96], and are more common among those receiving antifungal prophylaxis [97]. These molds are characterized by resistance to multiple antifungals, high potential for tissue invasion, and early dissemination with poor outcomes [98]. Although host-related factors (activity of underlying disease, comorbidities) are important for mortality [99], early diagnosis can prevent the evolution of the infection to multifocal or disseminated infection.
As symptoms, signs, and radiologic findings of non-Aspergillus invasive mold infections lack sensitivity and specificity, diagnosis is often delayed, resulting in delayed treatment and poor outcomes. The importance of early effective therapy in these mold infections is poorly understood, with the exception of mucormycosis, for which early therapy with anti-Mucorales agents has been shown to improve survival [100]. Conventional diagnostic confirmation requires invasive procedures such as biopsy or BAL, which are often not feasible in ill patients with severe pancytopenia [101].
The performance of commercially available biomarkers for these pathogens is also problematic. In addition to Aspergillus species, galactomannan is produced by several non-Aspergillus molds [102], particularly other hyalohyphomycetes such as Fusarium spp. [103]. Galactomannan is negative in patients with mucormycosis. Most non-Aspergillus invasive mold infections in high-risk patients are breakthrough infections in those already receiving anti-Aspergillus antifungals [104]. Finally, while BDG cannot identify organisms to the species level, a positive BDG can suggest the presence of molds. More data are needed on longitudinally prognostic data in non-Aspergillus molds [105, 106].
The development of culture-independent biomarkers for the early diagnosis of these important organisms is critically important. The field of PCR-based diagnostics in serum and/or BAL that target Mucorales are the most developed. Several small studies show promising high sensitivity and specificity that could lead to earlier diagnosis, although detailed comparisons to standard microbiologic methods are lacking [107]. In a small study using targeted screening for the detection of circulating Mucorales DNA by real-time quantitative PCR in seriously ill burn patients to identify invasive wound mucormycosis, a mortality benefit was suggested [108]. Other innovative approaches are needed and may involve serologic assessment, detection of volatile organic compounds or other metabolomics, and validation of antigen testing, immunohistochemistry, PCR, and metagenomic sequencing in selected cases of non-Aspergillus invasive mold infections.
The best example of diagnostic and prognostic use to date was in the Exserohilum rostratum meningitis outbreak, where PCR and BDG were utilized for CSF diagnosis and CSF BDG levels could be used to monitor response to antifungal therapy [106, 109]. More data are needed on response to therapy for other molds.
Summary and Recommendations for Surrogate Biomarkers in Non-Aspergillus Molds
Diagnostics for non-Aspergillus molds require further development, and specific biomarkers for these organisms would be helpful in disease management. Insufficient data exist on the utility of using any surrogate end points in clinical trials.
BLASTOMYCOSIS
End points for successful outcomes of treatment in blastomycosis classically have been clinical improvement together with resolution of skin lesions or radiologic findings. A standard assay for the diagnosis of probable blastomycosis is a Blastomyces antigen, which measures the cell wall polysaccharide (galactomannan) of Blastomyces dermatitidis (Mira Vista Laboratories, Indianapolis, IN, USA) in urine, serum, or body fluids (Table 5) [64, 110–112]. However, longitudinal antigen measurement to follow the course of infection has been infrequently reported [113–115]. Three children with blastomycosis had serial urine antigen tests performed during treatment [113]. Fall in antigen levels to undetectable occurred in 2 who were cured, but the third did not respond to antifungal therapy and antigen positivity persisted [114].
Table 5.
Comparison of Diagnostic Tests Utilized in the Diagnosis of Endemic Mycoses
| Diagnostic Test | Sensitivity, % | Specificity, % | Strengths | Limitations |
|---|---|---|---|---|
| Histoplasma | ||||
| Sputum/BAL culture [10, 11, 15, 16, 19, 20] | 15–84 | Inadequate data but presumed ~100 in most studies based on reference standard definitions | More useful in SPH and CPH | Slow growth, 4–8 wk Less useful in APH |
| Cytopathologic examination [8, 16, 17, 19, 20] | 9–50 | Inadequate data available, generally considered fairly specific but presence of Histoplasma in tissue may indicate past rather than current infection; may also be misidentified | Rapid results (hours) More likely to be positive in SPH and CPH |
Sensitivity and specificity vary based on pathologist experience Requires invasive procedures Less useful in pulmonary disease without dissemination |
| Serum antigen [13, 19, 21–23] | 30–87 | 98 | Fast results (days) Improving availability Most useful in APH |
Cross-reacts with other fungi Less useful in SPH and CPH |
| Urine antigen [8, 13, 15, 17, 19, 21, 24] | 40–95 | 95–99 | Fast results (days) Improving availability Most useful in APH |
Cross-reacts with other fungi Less useful in SPH and CPH |
| Antibody [8–10, 13, 14] | 40–95 | 91 | Fast results (days) More useful in SPH and CPH |
Take 4–8 wk to develop antibodies Can be negative in immunocompromised individuals Cross-reacts with other endemic mycoses |
| Blastomyces | ||||
| Sputum/BAL culture [1, 25–28] | 66–90 | Inadequate data but presumed ~100 in most studies based on reference standard definitions | Gold standard for diagnosis Commercial DNA (AccuProbe; GenProbe Inc., San Diego, CA, USA) testing can provide rapid results once there is sufficient growth |
Slow growth, up to 5 wk Diagnostic yield varies based on site DNA probe can cross-react with Paracoccidioides |
| Histologic or cytopathologic examination [26–30] | 38–93 | Inadequate data, generally considered highly specific, but misidentification may occur; presence of Blastomyces in tissue typically indicates active infection |
Rapid results (hours) | Sensitivity varies based on pathologist experience Atypical forms of B. dermatitidis may require special stains |
| Potassium hydroxide smear [25, 27, 28] | 48–90 | No data available, generally considered highly specific but false positives possible |
Rapid results | Varied sensitivity |
| Serum EIA antigen [1, 25, 29, 31, 32] | 36–82 | 99 compared with nonfungal infections or healthy controls but 95.6% cross-reactivity with 90 cases of histoplasmosis | EDTA heat treatment improves sensitivity | Cross-reacts with other fungi Only available at reference labs |
| Urine EIA antigen [1, 27, 29, 31–33] | 76–93 | 79–99 | Can be utilized to monitor response to treatment | Cross-reacts with other fungi Only available at reference labs |
| Antibody testing via complement fixation [28, 34, 35] | 16–77 | 30–100 | Fast results (days) | Difficult to perform, variable performance |
| Antibody testing via immunodiffusion [28, 29, 34, 36] | 32–80 | 100 in 1 study, possibility for cross-reaction remains | Fast results (days) | Can be negative in immunocompromised patients |
| Antibody testing via EIA (BAD-1) [33, 36] | 88 | 94–99 | Low rate of cross-reactivity Increased sensitivity when combined with antigen testing |
May be negative early in infection and in immunocompromised individuals Not commercially available |
| Coccidioides | ||||
| Culture [37] | 56–60 | 100 | Grows well on most media in 2–7 d, specificity | Biohazard to laboratory staff |
| Histologic or cytopathologic examination [37, 38] | 22–55 | 99.6 | Rapid results | Requires invasive procedures Endospores may be mistaken for Histoplasma, Blastomyces, or Cryptococcus |
| Serum antigen [39, 40] | 28–73 | 90–100 | Most useful in immunocompromised and severe disease | Cross-reactivity with Histoplasma and Blastomyces |
| Urine antigen [39, 41, 42] | 50–71 | 90–98 | Most useful in immunocompromised and severe disease | Cross-reactivity with Histoplasma and Blastomyces |
| Immunodiffusion antibody assays (IDTP and IDCF) [42, 43] | 60.2–71 | 98.8 | Quantitative titers correlated to disease severity and can monitor treatment response Commonly used as confirmatory test |
Only available at reference labs Less useful in immunosuppressed patients May be negative early in disease |
| EIA antibody assay [40, 43–49] | 83–100 | 75–98.5 | Commercially available Faster results |
Needs confirmatory testing Not quantitative IgM cross-reacts with other mycoses Less useful in immunosuppressed patients May be negative early in disease |
| Skin testing (Spherusol) [50] | >98 | >98 for prior exposure | Negative test may mean Coccidioides infection less likely |
Only indicates prior exposure, unclear role in active infection |
| Paracoccidioides | ||||
| Culture [51] | 25–44 | 100 | Specificity | Requires 2–4 wk to grow, infrequently used |
| Histologic or cytopathologic examination [52, 53] | 55–97 | Presumed highly specific but inadequate data and misclassification possible |
Gold standard test, results in hours–days | Requires invasive procedures |
| Double immunodiffusion antibody assay [52–56] | 80–90 | >90, inadequate data | Most commonly utilized antibody test |
Cross-reactivity with other fungi Less useful for diagnosis of P. lutzii |
| ELISA antibody assay [52, 57, 58] | 95.7 | 85–100 | Simple to perform Fast results Antibodies can be detected at low concentrations |
Cross-reacts with other fungi Requires confirmatory DID Ab Less useful for diagnosis of P. lutzii |
| Latex agglutination antibody testing [56] | 69.5–84.3 | 81.1 | Simple to perform | Poor reproducibility Limited availability Less useful for diagnosis of P. lutzii |
| Talaromyces | ||||
| Blood culture [59–62] | 72.8–83 | 100 | Gold standard Highly specific May culture other sterile sites as well |
Takes up to 4 wk to grow More likely to be positive in late stages of infection |
| Sputum culture [59, 60, 63] | 11–34 | Inadequate data, presumed highly specific | Highly specific | Takes up to 4 wk to grow |
| Culture from other tissues [59–61] | Inadequate data, presumed highly specific | High specificity, for some tissues high sensitivity | ||
| -Skin | 6–90 | -Yield only accurate if the area is involved, slow growth | ||
| -Bone marrow | 17–100 | -Painful, variable sensitivity–invasive, variable sensitivity | ||
| -Lymph node | 34–100 | -Invasive, small numbers studied | ||
| -Cerebrospinal fluid | 15 | -Invasive, small numbers studied | ||
| -Palatal/pharynx papule | 10 | -Painful, small numbers studied | ||
| -Liver | 5 | -Invasive, small numbers studied | ||
| -Pleural fluid | 5 | -Invasive, small numbers studied | ||
| Cytology [63] | 46 | Inadequate data, presumed highly specific, but misidentification may occur |
Specificity | Small numbers in studies, requires invasive procedures in most cases |
| Lateral flow immunochromatographic antigen assay (4D1) [64] | 87.9 | 100 | Rapid results Easy to perform |
Not commercially available Urine testing only |
| Antigen via EIA (Mp1p antigen) [62] | 86.3 | 98.1 | Rapid results Sensitivity further increased when both urine and serum tested |
Not commercially available |
| Mab 4D1 inhibitory ELISA antigen assay [65] | 100 | 100 | Low cross-reactivity Can be utilized on serum |
Only tested on small sample size (n = 45), results need confirmation |
Adapted from Poplin et al. [159].
Abbreviations: Ab, antibody; APH, acute pulmonary histoplasmosis, CPH, subacute pulmonary histoplasmosis; DID, double immunodiffusion; EIA, enzyme immunoassay; ELISA, enzyme-linked immunosorbent assay; IDCF, immunodiffusion complement fixation; IDTP, immunodiffusion tube precipitin; SPH, subacute pulmonary histoplasmosis.
A more systematic approach to the use of Blastomyces antigen detection to follow the disease course included 19 patients with serial urine antigen tests performed [115]. Antigen levels declined, but time to a negative result varied greatly among 16 patients who were successfully treated. In 3 patients who failed therapy, urine antigen levels increased.
Summary and Recommendations for Surrogate Biomarkers in Blastomycosis
There are insufficient data to recommend biomarkers for the prognosis of blastomycosis or as surrogate end points in clinical trials. Longitudinal change in serum Blastomyces antigen requires further investigation. Surrogate markers for prognosis of the more severe disease manifestations of acute respiratory distress syndrome in blastomycosis are needed.
COCCIDIOIDOMYCOSIS
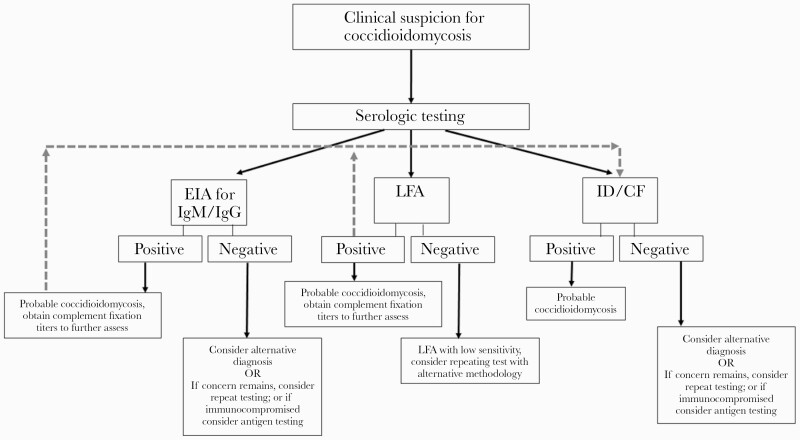
Past studies of coccidioidomycosis have used combined clinical score, radiologic results, and serologic results to determine a composite score for clinical severity [116]. Serologic testing may be performed using a number of methodologies (Figure 2). EIA appears to have the highest sensitivity, although it is prone to false positives and results should be confirmed with immunodiffusion testing [117, 118]. A newly developed lateral flow assay exhibited poor sensitivity (31%) [119]. Coccidioidal complement fixation (CF) testing for quantitative detection of immunoglobulin G has prognostic utility in both serum and CSF samples (Table 5) [120]. Serial serum samples correlate with patient symptoms and are useful for monitoring the course of therapy. It is imperative to recognize the differences in types of coccidioidal infection and the impact this plays in the kinetics of serology in determining a response to therapy, as serologic decline is slower in those with chronic pulmonary or disseminated coccidioidomycosis compared with those with primary pulmonary coccidioidomycosis [120, 121].
Figure 2.
Serologic testing in coccidioidomycosis. Abbreviations: EIA, enzyme immunoassay; IgG, immunoglobulin G; IgM, immunoglobulin M; LFA, lateral flow assay.
Serial serologic testing of CSF in cases of coccidioidal meningitis has shown a correlation with other CSF indices, including cell count, protein, and glucose; however, both serology and routine CSF parameters respond more slowly than clinical signs and symptoms of disease, and clinical decisions should be based on symptoms, CSF parameters, and serology results together [122].
A real-time PCR assay has been developed for detection of Coccidioides in lower respiratory specimens [123]. Coccidioidal antigen testing is also commercially available and may be useful in highly immunocompromised patients who are serologically negative [124]. BDG has been found to have 44% sensitivity and 91% specificity in coccidioidomycosis [125]. Serial antigen and BDG testing of the serum, urine, or CSF are not routinely performed due to a lack of data supporting this approach.
Summary and Recommendations for Surrogate Biomarkers in Coccidioidomycosis
Serologic testing is useful for the diagnosis of coccidioidomycosis, and EIA results should be confirmed by immunodiffusion. Longitudinal assessment using CF serology is useful to monitor the response to antifungal therapy. However, insufficient data exist on their utility as surrogate end points for use in clinical trials.
HISTOPLASMOSIS
Historically, the first prognostic marker for histoplasmosis was duration of positive blood cultures in patients with HIV who were receiving antifungal therapy [126]. In studies performed during the early AIDS era, recurrence of disease was associated with positive or persistently positive blood cultures [127–130]. Blood cultures are infrequently positive in patients who are HIV-negative [131], and the time to positivity (average of >2 weeks) limits their applicability during the care of patients in clinical practice or during the conduct of clinical trials.
The most widely used surrogate marker for histoplasmosis diagnosis is the Histoplasma polysaccharide galactomannan antigen, which can be detected in urine or serum (Figure 3) [132–135]. Histoplasma antigen assays have been developed by MiraVista (Indianapolis, IN, USA) and IMMY (Norman, OK, USA), among others. Only 1 study has compared the MiraVista lateral flow assay and IMMY galactomannan enzyme immunoassay on 391 urine samples (n = 104 proven disseminated histoplasmosis) in persons with HIV; it reported similar sensitivity (90%–91%) and specificity (91%–92%) (Table 5) [136]. The Histoplasma antigen decreases during successful antifungal therapy [137, 138]. Antigenemia decreases more rapidly than antigenuria in successfully treated patients with HIV, although failure of antigen to decline during the first 2 weeks of therapy does not predict treatment failure [133]. Among those with clinical relapse, 86% had a significant increase in their longitudinal antigen levels [139].
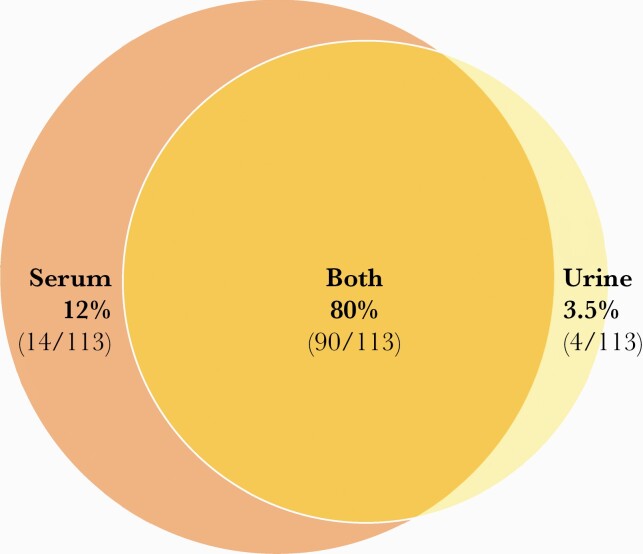
Figure 3.
Diagnostic overlap in histoplasmosis antigen by specimen type. Data are drawn from 113 total cases consisting of 56 disseminated histoplasmosis, 32 acute pulmonary histoplasmosis, 1 subacute pulmonary histoplasmosis, 6 chronic pulmonary histoplasmosis, 1 granulomatous mediastinitis, 15 unknown, and 5 cases not detected in either specimen type [132–135]. Antigen sensitivity is higher in disseminated histoplasmosis and/or in immunocompromised hosts than in acute pulmonary histoplasmosis or in immunocompetent hosts.
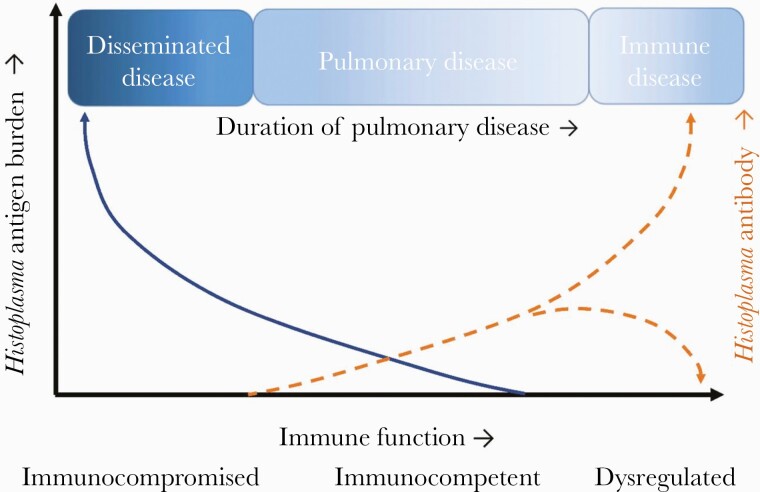
Antibody testing is most useful for nonimmunosuppressed patients with chronic or subacute pulmonary histoplasmosis and may be negative in those with severe immunosuppression (Figure 4). Negative testing should be repeated in 1–2 months if suspicion is high. While antibodies do appear to decay following acute disease, there is no evidence that changes, or lack thereof, in antibody titers predict mortality, treatment success, or recurrence [137].
Figure 4.
Conceptual framework for histoplasmosis. The relationship between host immunosuppression, type of disease, and antigen or antibody titers.
Summary and Recommendations for Surrogate Biomarkers in Histoplasmosis
Urine Histoplasma antigen is well studied in patients with HIV and is useful in diagnosis, patient management, and as a potential end point in clinical trials. In other populations, further studies should be performed to define the usefulness of longitudinal antigen testing to determine prognosis.
PARACOCCIDIOIDOMYCOSIS
Serologic testing is frequently used in the diagnosis of paracoccidioidomycosis and is highly specific (>95%) and sensitive (~80%) [140–142]. Numerous methodologies have been investigated, including in-house assays, although a commercial immunodiffusion test is now available. A previous study found longitudinal serologic assessment useful to monitor response to therapy [143]. This analysis of 43 patients with acute or chronic paracoccidioidomycosis found a decline in antibody levels using counter-immuno-electrophoresis, complement fixation, or enzyme-linked immunosorbent assay testing during a follow-up period of 2 years coincident with patient clinical improvement. Clearance of antibody occurred only in those with single-site disease over 12 months of follow-up. Patients with relapsed disease had an increase in serum titer levels, exhibiting the utility of serial monitoring. However, serology antibody titer in some cases may increase following initiation of antifungal therapy, thereby complicating using serology as a practical surrogate marker.
Antigen testing has also been examined as an adjunct to the diagnosis of paracoccidioidomycosis (Table 5) [142, 144, 145]. In a cohort of 23 patients followed for 8–12 months, antigen decreased during therapy with itraconazole and was undetectable in all patients after 12 months of treatment [144]. Unfortunately, this test is not commercially available.
Summary and Recommendations for Surrogate Biomarkers in Paracoccidioidomycosis
Serologic testing is useful for the diagnosis of paracoccidioidomycosis and is useful to monitor the response to antifungal therapy. However, insufficient data exist on serology as a surrogate end point for use in clinical trials.
SPOROTRICHOSIS
Serologic testing is infrequently used in the diagnosis of sporotrichosis despite the availability of a commercial assay. A latex agglutination assay was previously evaluated to assist with the diagnosis of meningeal sporotrichosis [146]; however, this assay is no longer available, and serologic testing is generally not helpful in other forms of sporotrichosis. Newer assays have been studied and exhibit reasonable sensitivity and specificity but are limited by availability [146–148]. The utility of serology to assess a response to antifungal therapy variably correlates with clinical response, and this test is not routinely recommended or commercially available [149, 150].
Summary and Recommendations for Surrogate Biomarkers in Sporotrichosis
Several assays are in development that may offer promise in the diagnosis of sporotrichosis, and they may be useful biomarkers during future clinical trials following validation. Currently, there are no biomarkers that can be used as a surrogate end point in clinical trials.
TALAROMYCOSIS
The current culture-based diagnosis for talaromycosis is suboptimal due to prolonged incubation (3–14 days) and low yield during the early stage of infection (Table 5) [151]. Several real-time PCR assays performed in blood have shown promising diagnostic performance in small clinical studies [152, 153]. An antigen detection EIA targeting the Talaromyces marneffei–specific cell wall mannoprotein Mp1p is highly sensitive (86%) and specific (98%) in comparison with blood cultures (73% and 86%, respectively) [151, 154, 155]. A commercial Mp1p EIA was approved in China in 2019, and an Mp1p LFA is being developed by IMMY (Norman, OK, USA).
Assessment of therapeutic response in talaromycosis has historically been based on a composite clinical and microbiological end point of resolution of fever, skin lesions, and/or fungemia [156, 157]. Mp1p concentrations have been shown to decline to undetectable levels over 12 to 16 weeks on antifungal therapy (Thuy Le, unpublished), and the role of Mp1p in treatment monitoring and prognosis is a subject of active research.
A new and simple fungal culture technique to serially quantify T. marneffei CFUs in blood was recently developed to measure the quantitative early fungicidal activity (EFA) [158]. The EFA quantified the reduction of blood CFUs/mL/day during the first 14 days. In a multicenter trial of itraconazole vs amphotericin B for talaromycosis, EFA decline was 4-fold faster in the amphotericin B group compared with the itraconazole group, corresponding to an absolute reduction in 24-week mortality from 21.0% to 11.3% (P = .006). Baseline fungal CFUs were an independent predictor of 24-week mortality [158]. These data indicate that EFA in blood is a strong surrogate marker of mortality and is a useful end point to evaluate therapeutic response.
Summary and Recommendations for Surrogate Biomarkers in Talaromycosis
Culture remains the mainstay of diagnosis for talaromycosis. Antigen testing may be useful in future clinical trials following validation, but currently cannot be recommended as a surrogate end point in clinical trials. The longitudinal change in quantitative blood CFUs has been found useful to quantify the EFA in a recent clinical trial and should be a standard metric in future clinical trials.
FUTURE DIRECTIONS
There have been significant advances utilizing surrogate markers of fungal infections to predict patient outcomes. These tests have been a welcome advance; however, numerous challenges remain. The development of affordable, readily available, and rapid in-house diagnostic tests is critically important. Cryptococcosis and talaromycosis are the only fungal infections with established surrogate end points, and further investigation of surrogate markers for other invasive fungal infections is urgently needed to aid in both diagnosis and prognosis. The burden of invasive candidiasis and aspergillosis mandates urgent attention to these diseases. Finally, a concerted effort to evaluate promising diagnostic tests as surrogate markers to measure response to therapy through the conduct of testing well-documented clinical samples is essential to move this field forward and to optimize patient care.
Acknowledgments
Financial support. Existing departmental funding and the Burden Family Gift Fund for Coccidioidomycosis Research supported this work.
Potential conflicts of interest. All authors: no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Jarvis JN, Bicanic T, Loyse A, et al. Determinants of mortality in a combined cohort of 501 patients with HIV-associated cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis 2014; 58:736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Busse O. Uber parasitare Zelleinschlusse und ihre Zuchtung. Zentralbl Bakteriol 1894; 16:175–80. [Google Scholar]
- 3. Boulware DR, Rolfes MA, Rajasingham R, et al. Multisite validation of cryptococcal antigen lateral flow assay and quantification by laser thermal contrast. Emerg Infect Dis 2014; 20:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robertson EJ, Najjuka G, Rolfes MA, et al. Cryptococcus neoformans ex vivo capsule size is associated with intracranial pressure and host immune response in HIV-associated cryptococcal meningitis. J Infect Dis 2014; 209:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen J, Slechta ES, Gates-Hollingsworth MA, et al. Large-scale evaluation of the immuno-mycologics lateral flow and enzyme-linked immunoassays for detection of cryptococcal antigen in serum and cerebrospinal fluid. Clin Vaccine Immunol 2013; 20:52–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binnicker MJ, Jespersen DJ, Bestrom JE, Rollins LO.. Comparison of four assays for the detection of cryptococcal antigen. Clin Vaccine Immunol 2012; 19:1988–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwizera R, Omali D, Tadeo K, et al. Evaluation of the Dynamiker cryptococcal antigen lateral flow assay for the diagnosis of HIV-associated cryptococcosis. J Clin Microbiol. 2021; 59:e02421–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mpoza E, Mukaremera L, Kundura DA, et al. Evaluation of a point-of-care immunoassay test kit “StrongStep” for cryptococcal antigen detection. PLoS One 2018; 13:e0190652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skipper C, Tadeo K, Martyn E, et al. Evaluation of serum cryptococcal antigen testing using two novel semiquantitative lateral flow assays in persons with cryptococcal antigenemia. J Clin Microbiol 2020; 58:e02046–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams DA, Kiiza T, Kwizera R, et al. Evaluation of fingerstick cryptococcal antigen lateral flow assay in HIV-infected persons: a diagnostic accuracy study. Clin Infect Dis 2015; 61:464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tadeo KK, Nimwesiga A, Kwizera R, et al. Evaluation of the diagnostic performance of a semiquantitative cryptococcal antigen point-of-care assay among HIV-infected persons with cryptococcal meningitis. J Clin Microbiol 2021; 59:e0086021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hevey MA, George IA, Rauseo AM, et al. Performance of the lateral flow assay and the latex agglutination serum cryptococcal antigen test in cryptococcal disease in patients with and without HIV. J Clin Microbiol 2020; 58:e01563–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dromer F, Mathoulin-Pelissier S, Launay O, et al. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med 2007; 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh N, Lortholary O, Alexander BD, et al. Antifungal management practices and evolution of infection in organ transplant recipients with Cryptococcus neoformans infection. Transplantation 2005; 80:1033–9. [DOI] [PubMed] [Google Scholar]
- 15. Kabanda T, Siedner MJ, Klausner JD, et al. Point-of-care diagnosis and prognostication of cryptococcal meningitis with the cryptococcal antigen lateral flow assay on cerebrospinal fluid. Clin Infect Dis 2014; 58:113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Powderly WG, Cloud GA, Dismukes WE, Saag MS.. Measurement of cryptococcal antigen in serum and cerebrospinal fluid: value in the management of AIDS-associated cryptococcal meningitis. Clin Infect Dis 1994; 18:789–92. [DOI] [PubMed] [Google Scholar]
- 17. Brouwer AE, Rajanuwong A, Chierakul W, et al. Combination antifungal therapies for HIV-associated cryptococcal meningitis: a randomised trial. Lancet 2004; 363:1764–7. [DOI] [PubMed] [Google Scholar]
- 18. Dyal J, Akampurira A, Rhein J, et al. Reproducibility of CSF quantitative culture methods for estimating rate of clearance in cryptococcal meningitis. Med Mycol 2016; 54:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pullen MF, Hullsiek KH, Rhein J, et al. Cerebrospinal fluid early fungicidal activity as a surrogate endpoint for cryptococcal meningitis survival in clinical trials. Clin Infect Dis 2020; 71:e45–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bicanic T, Muzoora C, Brouwer AE, et al. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis 2009; 49:702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montezuma-Rusca JM, Powers JH, Follmann D, et al. Early fungicidal activity as a candidate surrogate endpoint for all-cause mortality in cryptococcal meningitis: a systematic review of the evidence. PLoS One 2016; 11:e0159727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhein J, Bahr NC, Morawski BM, et al. Detection of high cerebrospinal fluid levels of (1→3)-β-d-glucan in cryptococcal meningitis. Open Forum Infect Dis 2014; 1:ofu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liesman RM, Strasburg AP, Heitman AK, et al. Evaluation of a commercial multiplex molecular panel for diagnosis of infectious meningitis and encephalitis. J Clin Microbiol 2018; 56:e01927–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bridge S, Hullsiek KH, Nerima C, et al. Evaluation of the BioFire(R) FilmArray(R) meningitis/encephalitis panel in an adult and pediatric Ugandan population. J Mycol Med 2021; 31:101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van TT, Kim TH, Butler-Wu SM.. Evaluation of the Biofire FilmArray meningitis/encephalitis assay for the detection of Cryptococcus neoformans/gattii. Clin Microbiol Infect 2020; 26:1375–9. [DOI] [PubMed] [Google Scholar]
- 26. George BJ, Horvath LL, Hospenthal DR.. Effect of inoculum size on detection of Candida growth by the BACTEC 9240 automated blood culture system using aerobic and anaerobic media. J Clin Microbiol 2005; 43:433–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horvath LL, George BJ, Hospenthal DR.. Detection of fifteen species of Candida in an automated blood culture system. J Clin Microbiol 2007; 45:3062–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pliakos EE, Andreatos N, Shehadeh F, et al. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev 2018; 31:e00095–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forrest GN, Mankes K, Jabra-Rizk MA, et al. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol 2006; 44:3381–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altun O, Almuhayawi M, Ullberg M, Ozenci V.. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 2013; 51:4130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salimnia H, Fairfax MR, Lephart PR, et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 2016; 54:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simor AE, Porter V, Mubareka S, et al. Rapid identification of Candida species from positive blood cultures by use of the FilmArray blood culture identification panel. J Clin Microbiol 2018; 56:e01387–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clancy CJ, Nguyen MH.. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin Infect Dis 2013; 56:1284–92. [DOI] [PubMed] [Google Scholar]
- 34. Clancy CJ, Nguyen MH.. Rapid diagnosis of invasive candidiasis: ready for prime-time? Curr Opin Infect Dis 2019; 32:546–52. [DOI] [PubMed] [Google Scholar]
- 35. Hanson KE, Pfeiffer CD, Lease ED, et al. Beta-D-glucan surveillance with preemptive anidulafungin for invasive candidiasis in intensive care unit patients: a randomized pilot study. PLoS One 2012; 7:e42282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sulahian A, Porcher R, Bergeron A, et al. Use and limits of (1-3)-β-d-glucan assay (Fungitell), compared to galactomannan determination (Platelia Aspergillus), for diagnosis of invasive aspergillosis. J Clin Microbiol 2014; 52:2328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He S, Hang JP, Zhang L, et al. A systematic review and meta-analysis of diagnostic accuracy of serum 1,3-beta-D-glucan for invasive fungal infection: focus on cutoff levels. J Microbiol Immunol Infect 2015; 48: 351–61. [DOI] [PubMed] [Google Scholar]
- 38. Karageorgopoulos DE, Vouloumanou EK, Ntziora F, et al. Beta-D-glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin Infect Dis 2011; 52:750–70. [DOI] [PubMed] [Google Scholar]
- 39. Onishi A, Sugiyama D, Kogata Y, et al. Diagnostic accuracy of serum 1,3-beta-D-glucan for Pneumocystis jiroveci pneumonia, invasive candidiasis, and invasive aspergillosis: systematic review and meta-analysis. J Clin Microbiol 2012; 50:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haydour Q, Hage CA, Carmona EM, et al. Diagnosis of fungal infections. A systematic review and meta-analysis supporting American Thoracic Society practice guideline. Ann Am Thorac Soc 2019; 16:1179–88. [DOI] [PubMed] [Google Scholar]
- 41. White SK, Schmidt RL, Walker BS, Hanson KE.. (1-->3)-beta-D-glucan testing for the detection of invasive fungal infections in immunocompromised or critically ill people. Cochrane Database Syst Rev 2020; 7:CD009833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fortun J, Buitrago MJ, Gioia F, et al. Roles of the multiplex real-time PCR assay and beta-D-glucan in a high-risk population for intra-abdominal candidiasis (IAC). Med Mycol 2020; 58:789–96. [DOI] [PubMed] [Google Scholar]
- 43. Fortun J, Meije Y, Buitrago MJ, et al. Clinical validation of a multiplex real-time PCR assay for detection of invasive candidiasis in intensive care unit patients. J Antimicrob Chemother 2014; 69:3134–41. [DOI] [PubMed] [Google Scholar]
- 44. Leon C, Ruiz-Santana S, Saavedra P, et al. Contribution of Candida biomarkers and DNA detection for the diagnosis of invasive candidiasis in ICU patients with severe abdominal conditions. Crit Care 2016; 20:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen MH, Wissel MC, Shields RK, et al. Performance of Candida real-time polymerase chain reaction, beta-D-glucan assay, and blood cultures in the diagnosis of invasive candidiasis. Clin Infect Dis 2012; 54:1240–8. [DOI] [PubMed] [Google Scholar]
- 46. Tissot F, Lamoth F, Hauser PM, et al. Beta-glucan antigenemia anticipates diagnosis of blood culture-negative intraabdominal candidiasis. Am J Respir Crit Care Med 2013; 188:1100–9. [DOI] [PubMed] [Google Scholar]
- 47. Jaijakul S, Vazquez JA, Swanson RN, Ostrosky-Zeichner L.. (1,3)-beta-D-glucan as a prognostic marker of treatment response in invasive candidiasis. Clin Infect Dis 2012; 55:521–6. [DOI] [PubMed] [Google Scholar]
- 48. Avni T, Leibovici L, Paul M.. PCR diagnosis of invasive candidiasis: systematic review and meta-analysis. J Clin Microbiol 2011; 49:665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang SS, Hsieh WH, Liu TS, et al. Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis - a systemic review and meta-analysis. PLoS One 2013; 8:e62323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clancy CJ, Pappas PG, Vazquez J, et al. Detecting Infections Rapidly and Easily for Candidemia Trial, Part 2 (DIRECT2): a prospective, multicenter study of the T2Candida panel. Clin Infect Dis 2018; 66:1678–86. [DOI] [PubMed] [Google Scholar]
- 51. Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 2015; 60:892–9. [DOI] [PubMed] [Google Scholar]
- 52. Bomkamp JP, Sulaiman R, Hartwell JL, et al. Evaluation of a rapid fungal detection panel for identification of candidemia at an academic medical center. J Clin Microbiol 2020; 58:e01408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arendrup MC, Andersen JS, Holten MK, et al. Diagnostic performance of T2Candida among ICU patients with risk factors for invasive candidiasis. Open Forum Infect Dis 2019; 6:ofz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamoth F, Clancy CJ, Tissot F, et al. Performance of the T2Candida panel for the diagnosis of intra-abdominal candidiasis. Open Forum Infect Dis 2020; 7:ofaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mylonakis E, Zacharioudakis IM, Clancy CJ, et al. Efficacy of T2 magnetic resonance assay in monitoring candidemia after initiation of antifungal therapy: the Serial Therapeutic and Antifungal Monitoring Protocol (STAMP) trial. J Clin Microbiol 2018; 56:e01756–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nucci M, Nouer SA, Esteves P, et al. Discontinuation of empirical antifungal therapy in ICU patients using 1,3-beta-d-glucan. J Antimicrob Chemother 2016; 71:2628–33. [DOI] [PubMed] [Google Scholar]
- 57. Posteraro B, Tumbarello M, De Pascale G, et al. (1,3)-beta-d-Glucan-based antifungal treatment in critically ill adults at high risk of candidaemia: an observational study. J Antimicrob Chemother 2016; 71:2262–9. [DOI] [PubMed] [Google Scholar]
- 58. Patch ME, Weisz E, Cubillos A, et al. Impact of rapid, culture-independent diagnosis of candidaemia and invasive candidiasis in a community health system. J Antimicrob Chemother 2018; 73:iv27–30. [DOI] [PubMed] [Google Scholar]
- 59. De Pascale G, Posteraro B, D’Arrigo S, et al. (1,3)-beta-D-glucan-based empirical antifungal interruption in suspected invasive candidiasis: a randomized trial. Crit Care 2020; 24:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kritikos A, Poissy J, Croxatto A, et al. Impact of the beta-glucan test on management of intensive care unit patients at risk for invasive candidiasis. J Clin Microbiol 2020; 58:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Munoz P, Vena A, Machado M, et al. T2MR contributes to the very early diagnosis of complicated candidaemia. A prospective study. J Antimicrob Chemother 2018; 73:iv13–9. [DOI] [PubMed] [Google Scholar]
- 62. Munoz P, Vena A, Machado M, et al. T2Candida MR as a predictor of outcome in patients with suspected invasive candidiasis starting empirical antifungal treatment: a prospective pilot study. J Antimicrob Chemother 2018; 73:iv6–12. [DOI] [PubMed] [Google Scholar]
- 63. Thompson GR 3rd, Young JH.. Aspergillus infections. N Engl J Med 2021; 385:1496–509. [DOI] [PubMed] [Google Scholar]
- 64. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 2020; 71:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hope WW, Kruhlak MJ, Lyman CA, et al. Pathogenesis of Aspergillus fumigatus and the kinetics of galactomannan in an in vitro model of early invasive pulmonary aspergillosis: implications for antifungal therapy. J Infect Dis 2007; 195:455–66. [DOI] [PubMed] [Google Scholar]
- 66. Sheppard DC, Marr KA, Fredricks DN, et al. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin Microbiol Infect 2006; 12:376–80. [DOI] [PubMed] [Google Scholar]
- 67. Mennink-Kersten MA, Donnelly JP, Verweij PE.. Detection of circulating galactomannan for the diagnosis and management of invasive aspergillosis. Lancet Infect Dis 2004; 4:349–57. [DOI] [PubMed] [Google Scholar]
- 68. Maertens J, Buve K, Theunissen K, et al. Galactomannan serves as a surrogate endpoint for outcome of pulmonary invasive aspergillosis in neutropenic hematology patients. Cancer 2009; 115:355–62. [DOI] [PubMed] [Google Scholar]
- 69. Ledoux MP, Guffroy B, Nivoix Y, et al. Invasive pulmonary aspergillosis. Semin Respir Crit Care Med 2020; 41:80–98. [DOI] [PubMed] [Google Scholar]
- 70. Miceli MH, Maertens J.. Role of non-culture-based tests, with an emphasis on galactomannan testing for the diagnosis of invasive aspergillosis. Semin Respir Crit Care Med 2015; 36:650–61. [DOI] [PubMed] [Google Scholar]
- 71. Mercier T, Guldentops E, Lagrou K, Maertens J.. Galactomannan, a surrogate marker for outcome in invasive aspergillosis: finally coming of age. Front Microbiol 2018; 9:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marr KA, Schlamm HT, Herbrecht R, et al. Combination antifungal therapy for invasive aspergillosis: a randomized trial. Ann Intern Med 2015; 162:81–9. [DOI] [PubMed] [Google Scholar]
- 73. Woods G, Miceli MH, Grazziutti ML, et al. Serum Aspergillus galactomannan antigen values strongly correlate with outcome of invasive aspergillosis: a study of 56 patients with hematologic cancer. Cancer 2007; 110:830–4. [DOI] [PubMed] [Google Scholar]
- 74. Nouer SA, Nucci M, Kumar NS, et al. Earlier response assessment in invasive aspergillosis based on the kinetics of serum Aspergillus galactomannan: proposal for a new definition. Clin Infect Dis 2011; 53:671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Neofytos D, Railkar R, Mullane KM, et al. Correlation between circulating fungal biomarkers and clinical outcome in invasive aspergillosis. PLoS One 2015; 10:e0129022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park SH, Choi SM, Lee DG, et al. Serum galactomannan strongly correlates with outcome of invasive aspergillosis in acute leukaemia patients. Mycoses 2011; 54:523–30. [DOI] [PubMed] [Google Scholar]
- 77. Chai LY, Kullberg BJ, Johnson EM, et al. Early serum galactomannan trend as a predictor of outcome of invasive aspergillosis. J Clin Microbiol 2012; 50:2330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chai LY, Kullberg BJ, Earnest A, et al. Voriconazole or amphotericin B as primary therapy yields distinct early serum galactomannan trends related to outcomes in invasive aspergillosis. PLoS One 2014; 9:e90176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koo S, Bryar JM, Baden LR, Marty FM.. Prognostic features of galactomannan antigenemia in galactomannan-positive invasive aspergillosis. J Clin Microbiol 2010; 48:1255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Petraitiene R, Petraitis V, Groll AH, et al. Antifungal activity and pharmacokinetics of posaconazole (SCH 56592) in treatment and prevention of experimental invasive pulmonary aspergillosis: correlation with galactomannan antigenemia. Antimicrob Agents Chemother 2001; 45:857–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Racil Z, Kocmanova I, Toskova M, et al. Galactomannan detection in bronchoalveolar lavage fluid for the diagnosis of invasive aspergillosis in patients with hematological diseases-the role of factors affecting assay performance. Int J Infect Dis 2011; 15:e874–81. [DOI] [PubMed] [Google Scholar]
- 82. Patterson TF, Thompson GR 3rd, Denning DW, et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 2016; 63:e1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hoenigl M, Eigl S, Heldt S, et al. Clinical evaluation of the newly formatted lateral-flow device for invasive pulmonary aspergillosis. Mycoses 2018; 61:40–3. [DOI] [PubMed] [Google Scholar]
- 84. Marr KA, Datta K, Mehta S, et al. Urine antigen detection as an aid to diagnose invasive aspergillosis. Clin Infect Dis 2018; 67:1705–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. White PL, Parr C, Thornton C, Barnes RA.. Evaluation of real-time PCR, galactomannan enzyme-linked immunosorbent assay (ELISA), and a novel lateral-flow device for diagnosis of invasive aspergillosis. J Clin Microbiol 2013; 51:1510–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pazos C, Ponton J, Del Palacio A.. Contribution of (1->3)-beta-D-glucan chromogenic assay to diagnosis and therapeutic monitoring of invasive aspergillosis in neutropenic adult patients: a comparison with serial screening for circulating galactomannan. J Clin Microbiol 2005; 43:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Senn L, Robinson JO, Schmidt S, et al. 1,3-beta-D-glucan antigenemia for early diagnosis of invasive fungal infections in neutropenic patients with acute leukemia. Clin Infect Dis 2008; 46:878–85. [DOI] [PubMed] [Google Scholar]
- 88. Koo S, Baden LR, Marty FM.. Post-diagnostic kinetics of the (1 --> 3)-beta-D-glucan assay in invasive aspergillosis, invasive candidiasis and Pneumocystis jirovecii pneumonia. Clin Microbiol Infect 2012; 18:E122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ellis M, Al-Ramadi B, Finkelman M, et al. Assessment of the clinical utility of serial beta-D-glucan concentrations in patients with persistent neutropenic fever. J Med Microbiol 2008; 57:287–95. [DOI] [PubMed] [Google Scholar]
- 90. White PL, Wingard JR, Bretagne S, et al. Aspergillus polymerase chain reaction: systematic review of evidence for clinical use in comparison with antigen testing. Clin Infect Dis 2015; 61:1293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. White PL, Bretagne S, Caliendo AM, et al. Aspergillus polymerase chain reaction—an update on technical recommendations, clinical applications, and justification for inclusion in the second revision of the EORTC/MSGERC definitions of invasive fungal disease. Clin Infect Dis 2021; 72:S95–101. [DOI] [PubMed] [Google Scholar]
- 92. Arvanitis M, Ziakas PD, Zacharioudakis IM, et al. PCR in diagnosis of invasive aspergillosis: a meta-analysis of diagnostic performance. J Clin Microbiol 2014; 52:3731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cruciani M, Mengoli C, Loeffler J, et al. Polymerase chain reaction blood tests for the diagnosis of invasive aspergillosis in immunocompromised people. Cochrane Database Syst Rev 2015; 9:CD009551. [DOI] [PubMed] [Google Scholar]
- 94. Imbert S, Gauthier L, Joly I, et al. Aspergillus PCR in serum for the diagnosis, follow-up and prognosis of invasive aspergillosis in neutropenic and nonneutropenic patients. Clin Microbiol Infect 2016; 22:562.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Imbert S, Meyer I, Palous M, et al. Aspergillus PCR in bronchoalveolar lavage fluid for the diagnosis and prognosis of aspergillosis in patients with hematological and non-hematological conditions. Front Microbiol 2018; 9:1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lewis RE, Cahyame-Zuniga L, Leventakos K, et al. Epidemiology and sites of involvement of invasive fungal infections in patients with haematological malignancies: a 20-year autopsy study. Mycoses 2013; 56:638–45. [DOI] [PubMed] [Google Scholar]
- 97. Lamoth F, Chung SJ, Damonti L, Alexander BD.. Changing epidemiology of invasive mold infections in patients receiving azole prophylaxis. Clin Infect Dis 2017; 64:1619–21. [DOI] [PubMed] [Google Scholar]
- 98. Lamoth F, Kontoyiannis DP.. Therapeutic challenges of non-Aspergillus invasive mold infections in immunosuppressed patients. Antimicrob Agents Chemother 2019; 63:e01244–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Campo M, Lewis RE, Kontoyiannis DP.. Invasive fusariosis in patients with hematologic malignancies at a cancer center: 1998-2009. J Infect 2010; 60:331–7. [DOI] [PubMed] [Google Scholar]
- 100. Chamilos G, Lewis RE, Kontoyiannis DP.. Delaying amphotericin B-based frontline therapy significantly increases mortality among patients with hematologic malignancy who have zygomycosis. Clin Infect Dis 2008; 47:503–9. [DOI] [PubMed] [Google Scholar]
- 101. Kontoyiannis DP. Rational approach to pulmonary infiltrates in leukemia and transplantation. Best Pract Res Clin Haematol 2013; 26:301–6. [DOI] [PubMed] [Google Scholar]
- 102. Korem M, Polacheck I, Michael-Gayego A, Strahilevitz J.. Galactomannan testing for early diagnosis of Exserohilum rostratum infection. J Clin Microbiol 2013; 51:2800–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nucci M, Barreiros G, Reis H, et al. Performance of 1,3-beta-D-glucan in the diagnosis and monitoring of invasive fusariosis. Mycoses 2019; 62:570–5. [DOI] [PubMed] [Google Scholar]
- 104. Lionakis MS, Lewis RE, Kontoyiannis DP.. Breakthrough invasive mold infections in the hematology patient: current concepts and future directions. Clin Infect Dis 2018; 67:1621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Pickering JW, Sant HW, Bowles CA, et al. Evaluation of a (1->3)-beta-D-glucan assay for diagnosis of invasive fungal infections. J Clin Microbiol 2005; 43:5957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Litvintseva AP, Lindsley MD, Gade L, et al. Utility of (1-3)-beta-D-glucan testing for diagnostics and monitoring response to treatment during the multistate outbreak of fungal meningitis and other infections. Clin Infect Dis 2014; 58:622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dadwal SS, Kontoyiannis DP.. Recent advances in the molecular diagnosis of mucormycosis. Expert Rev Mol Diagn 2018; 18:845–54. [DOI] [PubMed] [Google Scholar]
- 108. Legrand M, Gits-Muselli M, Boutin L, et al. Detection of circulating Mucorales DNA in critically ill burn patients: preliminary report of a screening strategy for early diagnosis and treatment. Clin Infect Dis 2016; 63:1312–7. [DOI] [PubMed] [Google Scholar]
- 109. Gade L, Grgurich DE, Kerkering TM, et al. Utility of real-time PCR for detection of Exserohilum rostratum in body and tissue fluids during the multistate outbreak of fungal meningitis and other infections. J Clin Microbiol 2015; 53:618–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Durkin M, Witt J, Lemonte A, et al. Antigen assay with the potential to aid in diagnosis of blastomycosis. J Clin Microbiol 2004; 42:4873–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Connolly P, Hage CA, Bariola JR, et al. Blastomyces dermatitidis antigen detection by quantitative enzyme immunoassay. Clin Vaccine Immunol 2012; 19:53–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bariola JR, Hage CA, Durkin M, et al. Detection of Blastomyces dermatitidis antigen in patients with newly diagnosed blastomycosis. Diagn Microbiol Infect Dis 2011; 69:187–91. [DOI] [PubMed] [Google Scholar]
- 113. Mongkolrattanothai K, Peev M, Wheat LJ, Marcinak J.. Urine antigen detection of blastomycosis in pediatric patients. Pediatr Infect Dis J 2006; 25:1076–8. [DOI] [PubMed] [Google Scholar]
- 114. Tarr M, Marcinak J, Mongkolrattanothai K, et al. Blastomyces antigen detection for monitoring progression of blastomycosis in a pregnant adolescent. Infect Dis Obstet Gynecol 2007; 2007:89059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Frost HM, Novicki TJ.. Blastomyces antigen detection for diagnosis and management of blastomycosis. J Clin Microbiol 2015; 53:3660–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Catanzaro A, Friedman PJ, Schillaci R, et al. Treatment of coccidioidomycosis with ketoconazole: an evaluation utilizing a new scoring system. Am J Med 1983; 74:64–9. [DOI] [PubMed] [Google Scholar]
- 117. Kaufman L, Sekhon AS, Moledina N, et al. Comparative evaluation of commercial Premier EIA and microimmunodiffusion and complement fixation tests for Coccidioides immitis antibodies. J Clin Microbiol 1995; 33:618–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bays DJ, Thompson GR 3rd. Coccidioidomycosis. Infect Dis Clin North Am 2021; 35:453–69. [DOI] [PubMed] [Google Scholar]
- 119. Donovan FM, Ramadan FA, Khan SA, et al. Comparison of a novel rapid lateral flow assay to enzyme immunoassay results for early diagnosis of coccidioidomycosis. Clin Infect Dis 2021; 73:e2746–53. [DOI] [PubMed] [Google Scholar]
- 120. McHardy IH, Dinh BN, Waldman S, et al. Coccidioidomycosis complement fixation titer trends in the age of antifungals. J Clin Microbiol 2018; 56:e01318–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Galgiani JN, Blair JE, Ampel NM, Thompson GR.. Treatment for early, uncomplicated coccidioidomycosis: what is success? Clin Infect Dis 2020; 70:2008–12. [DOI] [PubMed] [Google Scholar]
- 122. Galgiani JN, Catanzaro A, Cloud GA, et al. Fluconazole therapy for coccidioidal meningitis. The NIAID-Mycoses Study Group. Ann Intern Med 1993; 119:28–35. [DOI] [PubMed] [Google Scholar]
- 123. Saubolle MA, Wojack BR, Wertheimer AM, et al. Multicenter clinical validation of a cartridge-based real-time PCR system for detection of Coccidioides spp. in lower respiratory specimens. J Clin Microbiol 2018; 56:e01277–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Durkin M, Connolly P, Kuberski T, et al. Diagnosis of coccidioidomycosis with use of the Coccidioides antigen enzyme immunoassay. Clin Infect Dis 2008; 47:e69–73. [DOI] [PubMed] [Google Scholar]
- 125. Thompson GR 3rd, Bays DJ, Johnson SM, et al. Serum (1->3)-beta-D-glucan measurement in coccidioidomycosis. J Clin Microbiol 2012; 50:3060–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Baddley JW, Sankara IR, Rodriquez JM, et al. Histoplasmosis in HIV-infected patients in a southern regional medical center: poor prognosis in the era of highly active antiretroviral therapy. Diagn Microbiol Infect Dis 2008; 62:151–6. [DOI] [PubMed] [Google Scholar]
- 127. Wheat LJ, Connolly P, Haddad N, et al. Antigen clearance during treatment of disseminated histoplasmosis with itraconazole versus fluconazole in patients with AIDS. Antimicrob Agents Chemother 2002; 46:248–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wheat LJ, Connolly P, Smedema M, et al. Emergence of resistance to fluconazole as a cause of failure during treatment of histoplasmosis in patients with acquired immunodeficiency disease syndrome. Clin Infect Dis 2001; 33:1910–3. [DOI] [PubMed] [Google Scholar]
- 129. Wheat LJ, Connolly P, Smedema M, et al. Activity of newer triazoles against Histoplasma capsulatum from patients with AIDS who failed fluconazole. J Antimicrob Chemother 2006; 57:1235–9. [DOI] [PubMed] [Google Scholar]
- 130. Wheat J, MaWhinney S, Hafner R, et al. Treatment of histoplasmosis with fluconazole in patients with acquired immunodeficiency syndrome. National Institute of Allergy and Infectious Diseases Acquired Immunodeficiency Syndrome Clinical Trials Group and Mycoses Study Group. Am J Med 1997; 103:223–32. [DOI] [PubMed] [Google Scholar]
- 131. Franklin A, Larson L, Rutjanawech S, et al. 1699. Presentations and outcomes of Histoplasma capsulatum infection vary by immune status: a retrospective cohort study. Open Forum Infect Dis 2019; 6(Suppl 2):S622. [Google Scholar]
- 132. Swartzentruber S, Rhodes L, Kurkjian K, et al. Diagnosis of acute pulmonary histoplasmosis by antigen detection. Clin Infect Dis 2009; 49:1878–82. [DOI] [PubMed] [Google Scholar]
- 133. Hage CA, Kirsch EJ, Stump TE, et al. Histoplasma antigen clearance during treatment of histoplasmosis in patients with AIDS determined by a quantitative antigen enzyme immunoassay. Clin Vaccine Immunol 2011; 18:661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Libert D, Procop GW, Ansari MQ.. Histoplasma urinary antigen testing obviates the need for coincident serum antigen testing. Am J Clin Pathol 2018; 149:362–8. [DOI] [PubMed] [Google Scholar]
- 135. Williams B, Fojtasek M, Connolly-Stringfield P, Wheat J.. Diagnosis of histoplasmosis by antigen detection during an outbreak in Indianapolis, Ind. Arch Pathol Lab Med 1994; 118:1205–8. [PubMed] [Google Scholar]
- 136. Martinez-Gamboa A, Niembro-Ortega MD, Torres-Gonzalez P, et al. Diagnostic accuracy of antigen detection in urine and molecular assays testing in different clinical samples for the diagnosis of progressive disseminated histoplasmosis in patients living with HIV/AIDS: a prospective multicenter study in Mexico. PLoS NeglTrop Dis 2021; 15:e0009215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wheat LJ, Azar MM, Bahr NC, et al. Histoplasmosis. Infect Dis Clin North Am 2016; 30:207–27. [DOI] [PubMed] [Google Scholar]
- 138. Wheat LJ, Connolly-Stringfield P, Blair R, et al. Effect of successful treatment with amphotericin B on Histoplasma capsulatum variety capsulatum polysaccharide antigen levels in patients with AIDS and histoplasmosis. Am J Med 1992; 92:153–60. [DOI] [PubMed] [Google Scholar]
- 139. Wheat LJ, Connolly-Stringfield P, Blair R, et al. Histoplasmosis relapse in patients with AIDS: detection using Histoplasma capsulatum variety capsulatum antigen levels. Ann Intern Med 1991; 115:936–41. [DOI] [PubMed] [Google Scholar]
- 140. Shikanai-Yasuda MA, Telles Filho Fde Q, Mendes RP, et al. Guidelines in paracoccidioidomycosis. Rev Soc Bras Med Trop 2006; 39:297–310. [DOI] [PubMed] [Google Scholar]
- 141. Perenha-Viana MC, Gonzales IA, Brockelt SR, et al. Serological diagnosis of paracoccidioidomycosis through a Western blot technique. Clin Vaccine Immunol 2012; 19:616–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. de Camargo ZP. Serology of paracoccidioidomycosis. Mycopathologia 2008; 165:289–302. [DOI] [PubMed] [Google Scholar]
- 143. Negro G, Pereira CN, Andrade HF, et al. Evaluation of tests for antibody response in the follow-up of patients with acute and chronic forms of paracoccidioidomycosis. J Med Microbiol 2000; 49:37–46. [DOI] [PubMed] [Google Scholar]
- 144. Marques da Silva SH, Queiroz-Telles F, Colombo AL, Blotta MH, Lopes JD, Pires De Camargo Z.. Monitoring gp43 antigenemia in paracoccidioidomycosis patients during therapy. J Clin Microbiol 2004; 42:2419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gomez BL, Figueroa JI, Hamilton AJ, et al. Use of monoclonal antibodies in diagnosis of paracoccidioidomycosis: new strategies for detection of circulating antigens. J Clin Microbiol 1997; 35:3278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Scott EN, Kaufman L, Brown AC, Muchmore HG.. Serologic studies in the diagnosis and management of meningitis due to Sporothrix schenckii. N Engl J Med 1987; 317:935–40. [DOI] [PubMed] [Google Scholar]
- 147. Barros MB, de Almeida Paes R, Schubach AO.. Sporothrix schenckii and sporotrichosis. Clin Microbiol Rev 2011; 24:633–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Bernardes-Engemann AR, Costa RC, Miguens BR, et al. Development of an enzyme-linked immunosorbent assay for the serodiagnosis of several clinical forms of sporotrichosis. Med Mycol 2005; 43:487–93. [DOI] [PubMed] [Google Scholar]
- 149. Macedo PM, Lopes-Bezerra LM, Bernardes-Engemann AR, Orofino-Costa R.. New posology of potassium iodide for the treatment of cutaneous sporotrichosis: study of efficacy and safety in 102 patients. J Eur Acad Dermatol Venereol 2015; 29:719–24. [DOI] [PubMed] [Google Scholar]
- 150. Costa RO, Bernardes-Engemann AR, Azulay-Abulafia L, et al. Sporotrichosis in pregnancy: case reports of 5 patients in a zoonotic epidemic in Rio de Janeiro, Brazil. An Bras Dermatol 2011; 86:995–8. [DOI] [PubMed] [Google Scholar]
- 151. Le T, Wolbers M, Chi NH, et al. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin Infect Dis 2011; 52:945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Hien HTA, Thanh TT, Thu NTM, et al. Development and evaluation of a real-time polymerase chain reaction assay for the rapid detection of Talaromyces marneffei MP1 gene in human plasma. Mycoses 2016; 59:773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Li X, Zheng Y, Wu F, et al. Evaluation of quantitative real-time PCR and Platelia galactomannan assays for the diagnosis of disseminated Talaromyces marneffei infection. Med Mycol 2020; 58:181–6. [DOI] [PubMed] [Google Scholar]
- 154. Thu NTM, Chan JFW, Ly VT, et al. Superiority of a Novel Mp1p antigen detection enzyme immunoassay compared to standard BACTEC blood culture in the diagnosis of talaromycosis. Clin Infect Dis 2021; 73:e330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ly VT, Thanh NT, Thu NTM, et al. Occult Talaromyces marneffei infection unveiled by the novel Mp1p antigen detection assay. Open Forum Infect Dis 2020; 7:ofaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Supparatpinyo K, Chiewchanvit S, Hirunsri P, Uthammachai C, Nelson KE, Sirisanthana T.. Penicillium marneffei infection in patients infected with human immunodeficiency virus. Clin Infect Dis 1992; 14:871–4. [DOI] [PubMed] [Google Scholar]
- 157. Sirisanthana T, Supparatpinyo K, Perriens J, Nelson KE.. Amphotericin B and itraconazole for treatment of disseminated Penicillium marneffei infection in human immunodeficiency virus-infected patients. Clin Infect Dis 1998; 26:1107–10. [DOI] [PubMed] [Google Scholar]
- 158. Le T, Kinh NV, Cuc NTK, et al. A trial of itraconazole or amphotericin B for HIV-associated talaromycosis. N Engl J Med 2017; 376:2329–40. [DOI] [PubMed] [Google Scholar]
- 159. Poplin V, Smith C, Milsap D, Zabel L, Bahr NC.. Diagnosis of pulmonary infections due to endemic fungi. Diagnostics (Basel) 2021; 11:856. [DOI] [PMC free article] [PubMed] [Google Scholar]