Abstract
We propose a novel method for studying the function of specific microbial groups in situ. Since natural microbial communities are dynamic both in composition and in activities, we argue that the microbial “black box” should not be regarded as homogeneous. Our technique breaks down this black box with group-specific fluorescent 16S rRNA probes and simultaneously determines 3H-substrate uptake by each of the subgroups present via microautoradiography (MAR). Total direct counting, fluorescent in situ hybridization, and MAR are combined on a single slide to determine (i) the percentages of different subgroups in a community, (ii) the percentage of total cells in a community that take up a radioactively labeled substance, and (iii) the distribution of uptake within each subgroup. The method was verified with pure cultures. In addition, in situ uptake by members of the α subdivision of the class Proteobacteria (α-Proteobacteria) and of the Cytophaga-Flavobacterium group obtained off the California coast and labeled with fluorescent oligonucleotide probes for these subgroups showed that not only do these organisms account for a large portion of the picoplankton community in the sample examined (∼60% of the universal probe-labeled cells and ∼50% of the total direct counts), but they also are significant in the uptake of dissolved amino acids in situ. Nearly 90% of the total cells and 80% of the cells belonging to the α-Proteobacteria and Cytophaga-Flavobacterium groups were detectable as active organisms in amino acid uptake tests. We suggest a name for our triple-labeling technique, substrate-tracking autoradiographic fluorescent in situ hybridization (STARFISH), which should aid in the “dissection” of microbial communities by type and function.
In situ characterization of the marine picoplankton “black box” has been restricted mostly to determinations of community composition by a number of molecular techniques, including fluorescent in situ hybridization (FISH) (2, 3, 13, 16, 18, 33). In FISH fluorescently labeled oligonucleotide rRNA probes are used to identify and enumerate microorganisms in situ (13, 16). Probe sequences can target organisms at various taxonomic levels. Some probes are universal and target all organisms (18, 32), while others are for the domains Archaea (17, 30, 35) and Bacteria (16, 18); other probes target intermediate levels, such as the α subdivision of the class Proteobacteria (α-Proteobacteria), β-Proteobacteria, and γ-Proteobacteria (29), as well as the Cytophaga-Flavobacterium group (28); and still other probes are specific for species and subspecies (37).
In addition to composition, the marine bacterioplankton community can be characterized by its ecological role in the uptake of dissolved organic matter (DOM) via autoradiography (6, 15, 24, 38). Workers have reported that the bacterioplankton may be responsible for transferring up to 50 to 60% of the local primary production from the DOM pool into higher trophic levels through the microbial loop (7, 14, 15, 20). Standard measurements of in situ nutrient uptake, however, do not discriminate between different prokaryotic groups. Since analysis of 16S rRNA sequences shows that the marine picoplankton community is dynamic and is composed of various diverse organisms (3, 36), while studies of nutrient uptake by pure cultures show that the type and rate of nutrient uptake vary among different strains (22), we can conclude that in situ substrate uptake is dependent on the local picoplankton community composition. Hence, in order to better understand and model DOM uptake in the ocean, microbiologists need to “dissect” the bacterioplankton black box into smaller compartments on the basis of taxonomy and function instead of looking at it as a single homogeneous group.
In situ identification of the different prokaryotic groups in mixed communities, however, is limited by the lack of morphological features (9); this is the prime reason why FISH has become such an important tool in microbial ecology (3). Also, isolation of marine prokaryotes in pure cultures has been demonstrated to be difficult; less than 1% of the total natural community grows under laboratory conditions (39).
Here we suggest a new method in which FISH and microautoradiography are combined to determine in situ nutrient uptake by members of specific picoplankton groups on a single slide. The cells are triple-labeled with a general stain, a fluorescent oligonucleotide probe, and a tritiated substrate. With the same microscopic field it is possible to determine not only the percentage of a specific prokaryotic phylogenetic group in a mixed sample but also the distribution of nutrient uptake within each subgroup. Results obtained with both mixed cultures and natural marine samples are presented below.
MATERIALS AND METHODS
The sequence of events in our triple-labeling technique involved collection of samples, transfer of cells onto emulsion-gelatin-coated slides, emulsion exposure and development, whole-cell hybridization, and epifluorescence counting. All solutions, including the culture media and phosphate-buffered saline (PBS), were filtered with 0.2-μm-pore-size Nuclepore filters, autoclaved, and checked for the presence of prokaryotes by DAPI (4′,6-diamidino-2-phenylindole) staining before they were used throughout the entire procedure. Formalin was filtered with 0.02-μm-pore-size Whatman Anodisc filters before it was used.
Culture preparation.
The following organisms were selected based on their uptake of glucose and morphological characteristics for easy differentiation in nutrient uptake tests and during identification by epifluorescence microscopy and on photomicrographs: Escherichia coli, a short bacillus which can take up glucose; and Moraxella catarrhalis, a coccus (whose cells usually occur in pairs) that does not take up glucose (22). The two main purposes of the culture experiment were to test the protocol under highly controlled conditions and to test whether 16S rRNA, the target molecule for the oligonucleotide probes, could endure the conditions in the photographic solutions.
Replicates of both cultures were grown separately in Luria-Bertani enriched medium at 37°C to the mid-exponential growth phase, as determined with a Klett colorimeter (Klett-Summerson, New York, N.Y.) at an optical density of 525 nm. Cells were concentrated by centrifugation at 2,300 × g at 17°C for 5 min and were washed three times in 1× PBS; this was followed by two additional centrifugation steps, and each time the cells were resuspended in M9 minimal medium (27) lacking glucose.
Natural sample preparation.
The following natural samples were collected in 250-ml acid-washed dark bottles from two sites in southern California: (i) Catalina Island surface water, which was collected on 16 January 1998 at 33°26.71′N and 118°29.07′W; and (ii) water collected in the San Pedro Channel 5 m below the surface on 31 May 1998 at 33°33′N and 118°24′W.
Sample treatment.
Tritiated d-glucose (Dupont) was added at a concentration of 10 nM to two replicate cultures growing in minimal medium. Two killed controls for each culture were prepared by treatment with 10% formalin for 1 h prior to addition of d-[3H]glucose. Cultures were incubated on a rotary shaking platform at 250 rpm at 37°C.
Natural samples were divided into four 40-ml subsamples in sterile conical tubes. Two of the subsamples that were killed by treatment with 10% formalin for 1 h served as controls. All four subsamples were supplemented with a 5 nM 3H-amino acid mixture (Amersham) and were incubated at in situ seawater temperatures.
Nutrient uptake was monitored over time for all of the live and killed cultures, as well as for the natural subsamples, by withdrawing 2-ml aliquots from each culture or subsample, filtering the aliquots onto a 0.2-μm-pore-size 25-mm-diameter Nuclepore polycarbonate filter, rinsing the filter four times with about 2 ml of 1× PBS, and measuring the radioactivity with a scintillation counter by using EcoscintA scintillation cocktail (National Diagnostic, Atlanta, Ga.). The cell concentration was determined with DAPI (34), and the uptake of nutrients was terminated by adding formalin to a concentration of 10% when live samples reached saturation levels of radioactivity. Equal numbers of cells from E. coli and M. catarrhalis cultures were combined in 15-ml polycarbonate tubes prior to slide preparation.
Slide preparation.
In a darkroom, a photographic emulsion (type NTB2; Kodak, Rochester, N.Y.) was melted for 1 h at 43°C in a water bath and then mixed with a 43°C gelatin solution (0.2% [final concentration] gelatin, 0.02% [final concentration] CrKSO4) at a 50:50 (vol/vol) ratio. Using an ITT (Roanoke, Va.) Night Vision scope and a 15-W safelight (model 51/2; Testrine Instrument, Newark, N.J.) placed 2 m away, we coated heavy Teflon glass slides with 10 7-mm-diameter wells (Cel-Line Associates, Inc., Newfield, N.J.) with the emulsion-gelatin solution. The wells on the slides were coated separately by dispensing approximately 20 μl of the emulsion-gelatin solution into each well and immediately withdrawing as much as possible with a micropipette. Coating wells separately was necessary to keep Teflon areas around the wells hydrophobic, which prevented the hybridization buffer (see below) in one well from merging with the buffer in adjacent wells. Also, if the emulsion was not mixed with gelatin, the emulsion tended to peel off the glass slide during emulsion development or during in situ hybridization (see below). Three slides were coated with the emulsion-gelatin solution at a time and were left to dry for 30 min in total darkness while filters with samples were being prepared. This minimized the background exposure of the emulsion in wells. The emulsion was kept in total darkness unless it was used.
Attempts to transfer cells to Teflon slides coated with gelatin followed by in situ hybridization and then by coating the slides with the NTB2 emulsion resulted in very low fluorescence signals from both DAPI and the probes.
Whole-cell prestaining and filtration.
Under room light, 10-μl portions of a formalin-fixed culture containing 6.74 × 107 cells/ml were added to 5 ml of 1× PBS in clean stainless steel filtration towers and filtered down to a volume of 2 ml onto a 0.2-μm-pore-size 25-mm-diameter Nuclepore polycarbonate filter placed over a 0.8-μm-pore-size type AA Millipore filter. For the natural samples 10 ml of the formalin-fixed Catalina Island sample (3.32 × 106 cells/ml) or the San Pedro Channel sample (6.74 × 105 cells/ml) was used and also filtered to 2 ml. The cells in the 2-ml preparation were then stained with 100 μl of a 0.1-μg/μl DAPI solution for 10 min in the dark (obtained by covering the filtration towers with aluminum foil) before the filtration was completed. All of the filters were then rinsed four times with about 2 ml of 1× PBS to remove unincorporated radioactive glucose or amino acid. With clean forceps, the Nuclepore and Millipore filters were placed together with the cell side facing up on a 10-μl drop of 1× PBS in a petri dish, cut into eight equal pieces with a new razor blade, and carried to the darkroom, where they were transferred to Teflon slides that had been precoated with the emulsion-gelatin solution. Only three filters were prepared at a time to prevent the filters from drying out before they were transferred to the emulsion-gelatin-coated slides in the darkroom. Prestaining of cells with DAPI did not result in any noticeable increase in the background silver grain on the emulsion like that reported by Carman (11) when cells were prestained with acridine orange.
Cell transfer to slides, emulsion exposure, and development.
The method used to transfer cells to slides was similar to the microautoradiogram method (version E) by Tabor and Neihof (41). Cells were immediately transferred from the filter to a Teflon-coated slide by peeling each of the eight sections of the 0.2-μm-Nuclepore filter off the 0.8-μm-pore-size Millipore filter in the darkroom and placing it upside down (with the cells facing down) onto a single well coated with the emulsion-gelatin solution. The safelight was placed 2 m away during cell transfer, and the ITT Night Vision scope was used. The slides were left to dry in total darkness for 30 min before they were placed in a light-proof box which was wrapped in aluminum foil, placed inside a cardboard box, and stored at 4°C for 3 days for emulsion exposure. The emulsion was developed by using Kodak specifications, as follows: 2 min in Dektol developer, 10-s stop in deionized water, and 5 min in fixer. The slides were washed in deionized water for 2 min, the pieces of the Nuclepore filters were peeled off, and the slides were dried and prepared for whole-cell hybridization. At this point, the slides could be stored at −80°C for several weeks without noticeable changes in the results.
In situ hybridization.
The procedures used for cell hybridization were the procedures which we described previously (32). Oligonucleotide probes were modified with amino linkers attached at both the 3′ and 5′ termini (Operon Technologies, Inc., Alameda, Calif.) and labeled with cyanine Cy3 monofunctional dye (Amersham) in 0.1 M carbonate buffer (pH 8.5) overnight in the dark at room temperature (19). The method used to separate labeled probes from unlabeled probes was a modification of the method of Amann et al. (2). Briefly, the steps used to purify labeled probes included separating the oligonucleotides from the unreacted Cy3 dye in an STE Select-D G-25 spin column (5′-3′ Prime, Boulder, Colo.). The labeled probes were then separated from the unlabeled probes in a 20% nondenaturing polyacrylamide gel, and the bands containing Cy3-labeled oligonucleotides were visualized on the gel with a hand-held 254-nm UV lamp (model EF-160c; Spectronics Inc., Westbury, N.Y.). The top labeled band was cut out of the gel, and the probe was eluted in low TE (10 mM Tris-hydrochloride, 1 mM EDTA; pH 7.8) at 4°C. The eluted solution was then passed through Nensorb-20 columns (Du Pont, Wilmington, Del.) as described by the manufacturer. The amount of labeled probe was determined by absorbance at 260 nm. The probes were lyophilized and stored dry at −80°C. The probes used for the various samples included a negative control probe (CON), which was not expected to bind to any organism; a universal probe (UNI), which was expected to bind to all organisms; a probe of the domain Bacteria (BAC); and probes for two of the subgroups of the domain Bacteria, a probe for the α-Proteobacteria (αPRT) and a probe for the Cytophaga-Flavobacterium cluster of the Cytophaga-Flavobacterium-Bacteroides phylum (CF). Table 1 shows the probe sequences, E. coli positions, and references.
TABLE 1.
Probe sequence used for FISH
| Target | Sequence (5′-3′)a | Probe nameb | Reference |
|---|---|---|---|
| Control (no group)c | CCTAGTGACGCCGTCGAC | Control | 32 |
| Universal | GWATTACCGCGGCKGCTG | S-*-Univ-0519-a-A-18 | 24a |
| Bacteria | ACCGCTTGTGCGGGCCC | S-Bact-0927-b-A-17 | 18 |
| α-Proteobacteria | CGTTCGYTCTGAGCCAG | S-Sc-aProt-0019-a-A-17 | 29 |
| Cytophaga-Flavobacterium group | TGGTCCGTRTCTCAGTAC | S-*-CF-0319-a-A-18 | 3 |
W = A or T; Y = C or T; R = A or G.
Probe names based on names in the Oligonucleotide Probe Database (31a).
There were more than three mismatches compared with all of the rRNA sequences in the RDP database, and there was no appropriate Oligonucleotide Probe Database name.
In situ hybridization took place in buffer containing 5× SET (1× SET is 150 mM NaCl, 20 mM Tris-HCl [pH 7.8], and 1 mM EDTA), 0.2% bovine serum albumin (acetylated; Sigma), 10% dextran sulfate (molecular weight, 500,000; Pharmacia), 0.01% polyadenylic acid (Sigma), and 0.1% sodium dodecyl sulfate as described by DeLong et al. (13) and Braun-Howland et al. (8); the probe concentration was 5 ng μl−1. The slides were incubated at 43°C for 3 to 16 h, briefly rinsed with distilled water at 43°C, and finally immersed three times in 0.2× SET at 43°C for 10 min each time. After air drying, the slides were mounted in a glycerol-10× SET (50:50, vol/vol) solution and kept at −20°C for at least 1 h before they were observed by fluorescence and transmitted light microscopy.
Autoradiographic and fluorescent-cell counts.
For each microscopic field, the following four types of counts were obtained: (i) total DAPI cell counts with UV excitation (Olympus type U-MWU UV filter; excitation wavelength, 330 to 385 nm; emission wavelength, >420 nm); (ii) probe fluorescence cell counts, which included autofluorescent and probe-labeled cell counts with green light excitation (Chroma type tetramethylrhodamine isothiocyanate [TRITC] U-M41002 filter; excitation wavelength, 535 ± 50 nm; emission wavelength, 565 nm); (iii) microautoradiography counts (cells labeled with one of the tritiated nutrients) under transmitted light; and (iv) counts for cells labeled with a fluorescent probe and simultaneously labeled autoradiographically (overlap between green excitation and transmitted light). The last three types of counts were computed as percentages of the total DAPI cell counts.
All counts were obtained within 2 days of hybridization by using an Olympus Bmax epifluorescence microscope equipped with a UPlanApo objective lens (magnification, ×100), a type HBO 100 Hg vapor lamp, and the filters described above. The images were captured and intensified by using a microchannel plate image intensifier (model COHU intensified charge-coupled device camera), and the background was reduced by image averaging with a model DSP-2000 image processor (Dage-MTI, Inc., Michigan City, Ind.). The images were visualized with a Sony Trinitron color video monitor (model PVM-1353MD). This video system could display cells with fluorescence considerably less than the fluorescence directly detectable by eye.
Comparing counts with FISH counts.
In order to determine whether the treatment of cells during the triple-labeling technique protocol had any effect on the natural picoplankton community composition (from selective transfer of cells to or selective loss of cells from the emulsion-coated slides), the fluorescent probe counts obtained from the triple-labeling experiments were compared to standard FISH counts obtained for the San Pedro Channel sample collected on 31 May 1998. The standard FISH protocol used has been described in detail previously (32), and no chloramphenicol treatment was performed before the cells were preserved.
Photomicrography.
Culture samples were photographed with the Olympus Bmax microscope described above by using Fujifilm Provia 1600 ISO film. The exposure times were 5 s with UV excitation (DAPI-stained cells), about 1 min with green excitation (probe-labeled and autofluorescent cells), and 3 s with transmitted light (autoradiographically labeled cells). The oligonucleotide probe fluorescence from some cells in the natural sample was too dim (compared to that of the intensified video system) to be properly recorded on the photographic film.
RESULTS
Cultures.
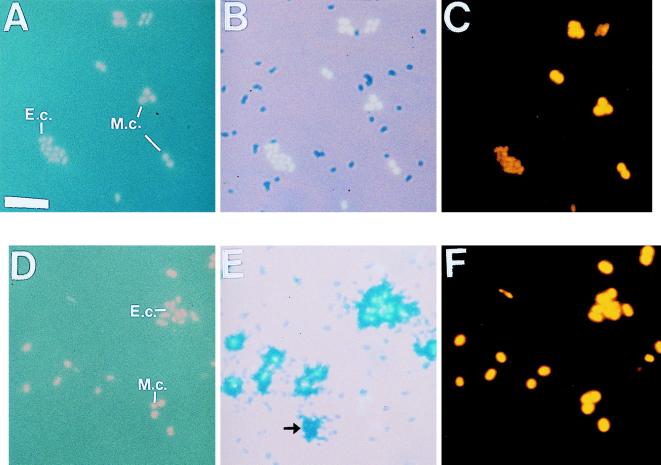
The results of the culture experiment were as expected. From the nutrient uptake curves, we confirmed that the E. coli strain used was capable of taking up glucose but that the M. catarrhalis strain was not. When these strains of E. coli (short bacilli in Fig. 1) and M. catarrhalis (cocci in Fig. 1) were used in the triple-labeling experiment, the autoradiography counts showed an all-or-nothing type of results. While 100% of the live E. coli cells were labeled with [3H]glucose (compare the short bacilli in Fig. 1D with the autoradiographic marks at the same positions in Fig. 1E), the M. catarrhalis cells remained unlabeled (compare the cocci in Fig. 1D with the autoradiographic marks at the same positions in Fig. 1E). None of the killed cells in the control samples took up [3H]glucose (Fig. 1A and 1B).
FIG. 1.
Triple-labeling of mixed E. coli and M. catarrhalis cells radiolabeled with d-[3H]glucose in M9 minimal medium for killed control samples (A through C) and live samples (D through F). (A and D) DAPI-labeled E. coli cells (short bacilli) (E.c.) and M. catarrhalis cells (cocci) (M.c.) with UV excitation. (B and E) Microautoradiograms showing no d-[3H]glucose uptake (B) or d-[3H]glucose uptake only by E. coli cells (E). (C and F) Cy3-conjugated Bacteria probe-labeled cells. The arrow in panel E indicates a silver grain, and no corresponding cell occurred in either the DAPI-stained (D) or probe (F) field; silver grains occurred in less than 2% of the 3H-labeled cells (see text). Bar = 5 μm.
Less than 2% of cells left an autoradiographic record on the emulsion (Fig. 1E) without a corresponding attached cell (Fig. 1D). The silver grain marks left by labeled cells were distinct in shape and size. It is also possible that cells were washed off the emulsion during in situ hybridization, but no tests were done to confirm this.
When cells were excited under green light, none of the cells were visible with no probe (NP) or with the control probe (CON), while all of the cells were visible with the universal probe (UNI) or with the Bacteria probe (BAC) (compare the probe-labeled cells in Fig. 1C and F with the corresponding DAPI-labeled cells in Fig. 1A and B, respectively).
Natural marine samples.
Tritiated amino acid uptake curves for both of the marine samples revealed rapid uptake that reached the saturation level by 3 h. The final corrected levels of radioactivity (live-cell radioactivity minus killed-cell radioactivity) for these two samples were of the same order of magnitude, but the value for the Catalina Island sample (3.14 × 10−2 dpm/cell or 2.8 × 10−19 mol of 3H-amino acid/cell) was two times higher than the value for the San Pedro Channel sample (1.53 × 10−2 dpm/cell or 1.4 × 10−19 mol of 3H-amino-acid/cell). The levels of radioactivity for the killed controls were nearly undetectable (5.6 × 10−5 dpm/cell for the Catalina Island sample control and 8.0 × 10−5 dpm/cell for the San Pedro Channel sample control).
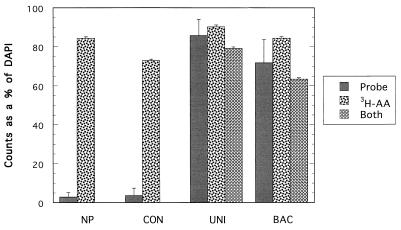
Triple-labeling data confirmed that the killed controls did not take up the 3H-amino acid mixture when either the Catalina Island sample for (Fig. 2) or the San Pedro Channel sample (Fig. 3) was examined. The 3 to 4% live cells detected for either the NP counts (2.8%) or the CON probe counts (3.5%) with the Catalina Island sample were assumed to be autofluorescent counts that probably originated from chlorophyll and phycoerythrin pigments, and they were not detected autoradiographically (Fig. 2). Autofluorescent cells are probably autotrophic, and they did not seem to take up dissolved amino acids in this experiment. With the San Pedro Channel sample the NP or CON probe counts accounted for less than 6% of the live cells, and less than 1% of the cells were simultaneously labeled with probe and by autoradiography (Fig. 3). The differences between the killed and live subsample probe (and NP) counts were not significant for either the Catalina Island sample (P = 0.278, as determined by Student’s t test) or the San Pedro Channel sample (P = 0.128, as determined by Student’s t test).
FIG. 2.
Fluorescent-oligonucleotide probe, 3H-amino acid mixture, and probe plus 3H-amino acid mixture counts as percentages of the total DAPI counts for the Catalina Island (30 km off the coast of Los Angeles) surface sample collected on 16 January 1998. All of the probes, including the control (CON), universal (UNI), and Bacteria domain (BAC) probes, were dually labeled with Cy3 dye. Autofluorescent cell counts were not subtracted from the probe counts. The error bars indicate standard errors. 3H-AA, 3H-labeled amino acids.
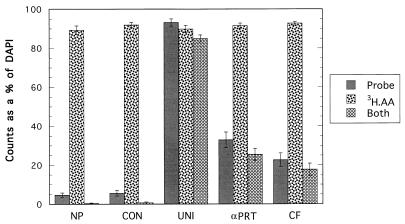
FIG. 3.
Same as Fig. 2, except that the α-Proteobacteria (αPRT) and Cytophaga-Flavobacterium (CF) probes were used instead of the BAC probe for the San Pedro Channel surface sample collected on 31 May 1998. The error bars indicate standard errors. 3H-AA, 3H-labeled amino acids.
The average overall percentage of cells labeled autoradiographically (average autoradiography counts as percentages of DAPI counts for all probe and NP preparations) for the live Catalina Island subsample was 83% (standard error, ± 3.62%) (autoradiography counts divided by DAPI counts) (Fig. 2). The percentage was slightly higher for the San Pedro Channel sample (Fig. 3), 91% (standard error, ± 0.6%). Counts for the Catalina Island sample when cells were simultaneously labeled autoradiographically and with fluorescent probe showed that nearly 80% (standard error, ± 4.0%) of all of the cells were labeled with the universal probe (UNI) and 3H-amino acids and 64% (standard error, ±4.0%) of the cells were labeled with the Bacteria probe (BAC) and 3H-amino acids.
The following two Bacteria subgroup-specific probes were used for the San Pedro Channel sample instead of the BAC probe: the α-Proteobacteria probe (αPRT) (mean ± standard error, 33 ± 4%) and Cytophaga-Flavobacterium probe (CF) (mean ± standard error, 23 ± 3.5%). Labeled amino acid uptake with each of these group-specific probes (computed as the simultaneous probe and autoradiography counts divided by the probe count, including the autofluorescent cell count) showed that 77% of the cells in the San Pedro Channel sample were labeled with the αPRT probe and 78% of the cells labeled with the CF probe were also labeled autoradiographically with the amino acid mixture (Fig. 3).
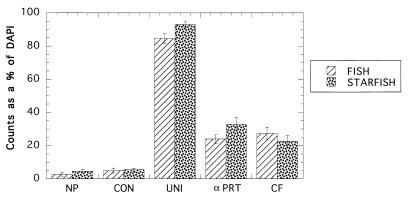
A comparison between the triple-labeling technique and the FISH technique for the San Pedro Channel sample (Fig. 4) showed that there were no significant differences (P = 0.253, as determined by the Student t test) for the NP counts and for all four probe (CON, UNI, αPRT, and CF) counts.
FIG. 4.
Comparison between FISH and STARFISH NP, control (CON), universal (UNI), α-Proteobacteria (αPRT), and Cytophaga-Flavobacterium (CF) probe counts as percentages of total DAPI counts for the San Pedro Channel sample collected on 31 May 1998. The error bars indicate standard errors.
DISCUSSION
Our results show that it is possible to obtain simultaneous in situ measurements for the composition of a bacterioplankton community and the capacity of the community to take up specific organic nutrients. Culture experiments in which E. coli and M. catarrhalis strains were mixed helped demonstrate the feasibility of the triple-labeling technique. Under these highly controlled conditions, culture experiments showed that it is possible to label cells with fluorescent probes even after the cells undergo the additional treatments of the triple-labeling technique. In addition, the culture experiments provided a simple example of one of the applications of the triple-labeling technique. Both E. coli and M. catarrhalis belong to the domain Bacteria, but only E. coli was capable of taking up glucose, supporting our previous statement that in mixed picoplankton communities nutrient uptake is dependent on the local community composition.
Furthermore, results obtained with the triple-labeling technique did not differ significantly from results obtained with FISH alone (P > 0.05) despite the lengthier procedures involved in the former protocol. Hence, any loss of cells from the emulsion-coated slides due to the additional rinse steps or during the development of the emulsion in the triple-labeling technique did not lead to a significant change in the apparent community composition.
Although the oligonucleotide probes used in our study targeted the entire domain Bacteria and two of its subgroups, the α-Proteobacteria and the Cytophaga-Flavobacterium group, other workers have used more group-specific probes (including probes specific at the species level) for in situ studies of mixed microbial communities (3–5, 42). In addition, the tritiated amino acid mixture used in our experiments served as an unspecialized label for easily labeling most or all heterotrophically active organisms. However, more specific substrates, such as simple sugars, polymers, nucleosides, phospholipids, or dissolved inorganic ions, could be used.
With the Catalina Island sample, the Bacteria probe tagged almost all of DAPI-labeled cells, and most of these cells took up detectable levels of amino acids, suggesting that members of most Bacteria subgroups contribute to the uptake of dissolved amino acids at this site. The results obtained with the more specific probes for the San Pedro Channel sample showed that cells labeled with the α-Proteobacteria probe (αPRT) and the Cytophaga-Flavobacterium probe (CF) combined accounted for almost 60% of all of the cells labeled with the UNI probe [(αPRT + CF)/UNI] at that site, assuming that there was no nontarget labeling that resulted in overlap between the two subgroup probes. Such results agree with previous findings which showed that most clones recovered in nearby waters belonged to members of these subgroups (12, 17). Furthermore, most of the cells of members of each of the subgroups (80% of each subgroup) were active in uptake of dissolved amino acids.
It is important to note that probe counts probably underestimate the total number of cells belonging to the probe target group for a given natural sample for two important reasons. First, some naturally occurring prokaryotes seem to have insufficient 16S rRNA for them to be detectable by whole-cell fluorescent hybridization (23–25, 32). These cells may grow too slowly (1, 13, 21), or they may be inactive (43). In contrast to the results of Zweifel and Hagström (43), who found that up to 70% of the cells in the Baltic Sea were inactive, the numbers of inactive cells in our samples ranged from 7 to 15% of the total cells based on probe counts. Furthermore, we showed previously (32) that treatment of natural samples with chloramphenicol at a concentration of 100 μg/ml for 1 h can increase the level of detection of prokaryotes with a universal fluorescent probe by 15% on average to about 95% of the total counts. Although we could not pretreat our samples with chloramphenicol in this study because this antibiotic stops amino acid incorporation, it should be possible to treat samples after uptake is completed.
A second reason for the possible underestimation of the probe counts is related to the fact that the 16S rRNA sequences of only a small portion of the total community have been determined and most likely the probe sequences available do not perfectly match all members of the target groups. Furthermore, because new probe sequences are added to ribosomal databases daily, probes should be checked frequently. Brunk et al. (10) analyzed the specificity of the Bacteria probe used here and reported that it bound to 92% of all Bacteria sequences in the Ribosomal Database Project (RDP) database (26) when 1,484 Bacteria sequences were available. An analysis of the same probe has not been performed with the updated version of the RDP database (9a), which currently contains 5,952 Bacteria sequences (26).
Even though it was not the purpose of this work to evaluate the previously described probe sequences used in our experiments, a preliminary probe check analysis of the RDP database with the αPRT and CF probe sequences confirmed that the sequences of these probes did not perfectly match the sequences of some marine clones in their respective groups. The sequences of clone env.agg41 in the Cytophaga-Flavobacterium subgroup and clones FL1 and FL11 in the α-Proteobacteria subgroup (12), both of which were found in the Santa Barbara Channel near both of our marine sites, did not perfectly match the probe template sequences. The env.agg41 clone sequence had two mismatches (the target sequence had U instead of G at E. coli position 324 and G instead of A at position 326) compared with the CF probe sequence, while the sequences of the α-Proteobacteria clones each had a single mismatch (U instead of A at E. coli position 596) compared with the αPRT probe sequence. Hence, it is possible that the total α-Proteobacteria and Cytophaga-Flavobacterium populations may have been underestimated.
For our calculations, autofluorescent cell counts were not subtracted from the probe counts for the marine samples since autofluorescent cells were most likely cyanobacteria or prochlorophytes and, overall, represented less than 5% of the DAPI counts based on the NP results. The fluorescence of autofluorescent cells (like Synechococcus and Prochlorococcus cells) tends to overlap the fluorescence of the Cy3 dye coupled to the probe. Therefore, autofluorescent cell counts were included in the probe counts. However, since autofluorescent cells did not seem to take up 3H-amino acids, they usually did not contribute to the probe counts plus autoradiography counts, so that the latter counts were always lower in our samples.
The average difference between the probe plus autoradiography counts and the probe counts alone for the Catalina Island and San Pedro Channel samples combined showed that nearly 6% of the cells labeled with the probes were unaccounted for in the probe plus autoradiography counts. In addition, the overall average percentage for autofluorescent cells based on the NP counts for these two samples was 3%. Hence, the data suggest that a small portion (<10%) of the cells in each probe group were inactive or took up amino acids very slowly and were not detected autoradiographically.
We suggest the following name for our triple-labeling technique: substrate-tracking autoradiography fluorescent in situ hybridization (STARFISH), since it is an expansion of the FISH technique. To the best of our knowledge, this is the first time that FISH has been used to aid in the identification of an organism in combination with autoradiography. Autoradiography has been used to study the function of microbes in situ for a long time (9). In previous studies, however, either the organisms of interest (usually limited to a few species) were identifiable by their morphological features (11) or the entire prokaryotic community was studied as a group that was considered homogeneous both in terms of composition and in terms of function in the uptake of DOM (15, 24, 31, 40).
Perhaps due to the unspecialized 3H-amino acid mixture and the general group probes used in our experiments, our results showed that the α-Proteobacteria and the Cytophaga-Flavobacterium subgroups had very similar percentages of cells (80%) involved in uptake of amino acids. Nevertheless, because the members of the α-Proteobacteria were 10% more abundant, their overall significance in DOM uptake may be greater than the significance of members of the Cytophaga-Flavobacterium group, assuming that the average amount of amino acid taken up per cell does not vary significantly between these two prokaryotic subgroups. With further investigation, STARFISH may help break down the bacterioplankton black box into smaller components and may provide a better understanding of the ecological role of various organisms in the uptake of DOM under natural conditions.
ACKNOWLEGMENTS
We are grateful to Markus Karner for help with the initial ideas used in this work; to Louis Bergquist and Martin Huh for the culture strains; to Joel Mefford for suggesting the acronym STARFISH; to Rachel Noble and Alison Davis for reviews and comments; to ZhongDong Huang, Steffanie Gehret, Ximena Hernandez, Julian Herndon, Corey Blake, Albert Perdon, Truc Luu, and Liam Carr for help with various parts of the experiments; to the crew members of the R/V Sea Watch and R/V Yellow Fin; and to workers at the Southern California Marine Institute for sample collection. Special thanks go to Marilene Demasi and Scott Robinson for their support and advice.
This research was funded in part by the USC Sea Grant Program, National Oceanic and Atmospheric Association, under grant NA 86 RG 0054 and by the California State Resources Agency. The other portion of this project was funded by grants OCE 9634028 and DEB 9705523 from the National Science Foundation and by a John and Alice Tyler Environmental Scholarship.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and evironmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angert E R, Clements K D, Pace N R. The largest bacterium. Nature. 1993;362:239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- 6.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 7.Azam F, Smith D C, Stewart G F, Hagström Å. Bacteria-organic matter coupling and its significance for oceanic carbon cycling. Microb Ecol. 1993;28:167–179. doi: 10.1007/BF00166806. [DOI] [PubMed] [Google Scholar]
- 8.Braun-Howland E B, Danielsen S A, Nierzwicki-Bauer S A. Development of a rapid method for detecting bacterial cells in situ using 16S rRNA-targeted probes. BioTechniques. 1992;13:928–933. [PubMed] [Google Scholar]
- 9.Brock T D, Brock M L. Autoradiography as a tool in microbial ecology. Nature. 1966;209:734–736. doi: 10.1038/209734a0. [DOI] [PubMed] [Google Scholar]
- 9a.Brunk, C. Personal communication.
- 10.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carman K R. Radioactive labeling of a natural assemblage of marine sedimentary bacteria and microalgae for trophic studies: an autoradiographic study. Microb Ecol. 1990;19:279–290. doi: 10.1007/BF02017172. [DOI] [PubMed] [Google Scholar]
- 12.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 13.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 14.Fuhrman J A. Bacterioplankton roles in cycling or organic matter: the microbial food web. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 361–383. [Google Scholar]
- 15.Fuhrman J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 16.Fuhrman J A, Lee S H, Masuchi Y, Davis A A, Wilcox R M. Characterization of marine prokaryotic communities via DNA and RNA. Microb Ecol. 1994;28:133–145. doi: 10.1007/BF00166801. [DOI] [PubMed] [Google Scholar]
- 17.Fuhrman J A, Ouverney C C. Marine microbial diversity studied via 16S rRNA sequences: coastal cloning results and counting of native archaea with fluorescent single cell probes. Aquat Ecol. 1998;32:3–15. [Google Scholar]
- 18.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 20.Hagström Å, Larsson U, Horstedt P, Normark S. Frequency of dividing cells, a new approach to the determination of bacterial growth rates in aquatic environments. Appl Environ Microbiol. 1979;37:805–812. doi: 10.1128/aem.37.5.805-812.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. [Google Scholar]
- 23.Hsu D, Shih L, Zee Y C. Degradation of rRNA in Salmonella strains: a novel mechanism to regulate the concentrations of rRNA and ribosomes. J Bacteriol. 1994;176:4761–4765. doi: 10.1128/jb.176.15.4761-4765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, Kemp P F. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr. 1994;39:869–879. [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Fogel K, Woese C R. The RDP (Ribosomal Data Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 30.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novitsky J A. Heterotrophic activity throughout a vertical profile of seawater and sediment in Halifax Harbor, Canada. Appl Environ Microbiol. 1983;45:1753–1760. doi: 10.1128/aem.45.6.1753-1760.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Oligonucleotide Probe Database Website. [Online.] http://www.cme.msu.edu/OPD.
- 32.Ouverney C C, Fuhrman J A. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl Environ Microbiol. 1997;63:2735–2740. doi: 10.1128/aem.63.7.2735-2740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pace N R, Stahl D A, Lane D L, Olsen G J. The analysis of natural microbial populations by rRNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 34.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 35.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rehnstam A S, Backman S, Smith D C, Azam F, Hagström A. Bloom of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol. 1993;102:161–166. [Google Scholar]
- 37.Salama M, Sandine W, Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon M. Specific uptake rates of amino acids by attached and free-living bacteria in a mesotrophic lake. Appl Environ Microbiol. 1985;49:1254–1259. doi: 10.1128/aem.49.5.1254-1259.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 40.Tabor P S, Neihof R A. Direct determination of activities for microorganisms of Chesapeake Bay populations. Appl Environ Microbiol. 1984;48:1012–1019. doi: 10.1128/aem.48.5.1012-1019.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabor P S, Neihof R A. Improved microautoradiographic method to determine individual microorganisms active in substrate uptake in natural waters. Appl Environ Microbiol. 1982;44:945–953. doi: 10.1128/aem.44.4.945-953.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zweifel U L, Hagstrom A. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]