Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), is currently the major global health problem. Still, it continues to infect people globally and up to the end of February 2022, over 436 million confirmed cases of COVID-19, including 5.95 million deaths, were reported to the world health organization (WHO). No specific treatment is currently available for COVID-19, and the discovery of effective therapeutics requires understanding the effective immunologic and immunopathologic mechanisms behind this infection. Type-I interferons (IFN-Is), as the critical elements of the immediate immune response against viral infections, can inhibit the replication and spread of the viruses. However, the available evidence shows that the antiviral IFN-I response is impaired in patients with the severe form of COVID-19. Moreover, the administration of exogenous IFN-I in different phases of the disease can lead to various outcomes. Therefore, understanding the role of IFN-I molecules in COVID-19 development and its severity can provide valuable information for better management of this disease. This review summarizes the role of IFN-Is in the pathogenesis of COIVD-19 and discusses the importance of autoantibodies against this cytokine in the spreading of SARS-CoV-2 and control of the subsequent excessive inflammation.
Keywords: SARS-CoV-2, COVID-19, Type-I interferons, Autoantibody, Inflammation, Immunopathogenesis
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious viral infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Still, it continues to infect people globally and up to the end of February 2022, 436 million confirmed cases of COVID-19, including 5.95 million deaths, were reported to the world health organization (WHO) (Abbasifard and Khorramdelazad, 2020; COVID Worldometer, 2022; Farnoosh et al., 2020; WHO, 2022). COVID-19 was the greatest global health challenge and crisis since the influenza pandemic of 1918 (Cascella et al., 2021). The first case of COVID-19 was reported in Wuhan, China, in late December 2019 and then spread rapidly around the world (Khorramdelazad et al., 2021). Despite widespread vaccination in numerous countries, the new waves of COVID-19 are still catastrophically affecting the health, livelihood, and economy of the world's population (Nesteruk, 2020).
Type-I interferons (IFN-Is), including IFN-Iα and IFN-Iβ, as a distinct class of cytokines, are at the forefront of immunological defense against viruses (Schreiber, 2020). Moreover, these interferons (IFNs) are commonly involved in inflammatory responses, immune system regulation, tumor cell recognition, and functions of T lymphocytes (Lee and Shin, 2020). Based on the available evidence and past pandemic experiences, the administration of IFN-Is as an antiviral drug against SARS and Ebola showed a promising future (Konde et al., 2017; Loutfy et al., 2003; Schreiber, 2020). In this context, it has been revealed that IFN-Is can exert their effects in both autocrine and paracrine manners, inducing expression of the interferon-stimulated genes (ISGs) that triggers antiviral response against infected host cells. It has been disclosed that similar to numerous viruses, SARS-CoV-2 is able to escape the antiviral immune responses, including IFN-I-mediated antiviral episodes (Blanco-Melo et al., 2020). Evaluation of SARS-CoV-2 infected individuals showed that virus-infected bronchial epithelial cells are responsible for the secretion of IFN-Is. However, IFN-I-dependent antiviral responses do not have adequate efficacy to control the viral infection, which may be due to escape mechanisms of the virus (Hadjadj et al., 2020).
The presence of anti–IFN–I autoantibodies in some patients is another challenge in COVID-19 patients (Bastard et al., 2020). These autoantibodies can neutralize IFN-Is in-vitro and in-vivo, and despite 13 different subtypes of IFN-α, autoantibodies against IFN-α2 is more frequently detectable in the bloodstream of COVID-19 patients (Bastard et al., 2020).
It appears that IFN-Is have a vital role in controlling SARS-CoV-2 infection. Moreover, during the initial days of the infection, it was believed that impaired IFN-I-dependent immune responses could lead to increased virus replication, which ultimately leads to the increased production and secretion of inflammatory cytokines and dysregulated pulmonary and systemic inflammation (Zhang et al., 2020).
This review summarizes and describes the key roles of IFN-Is in the immunopathogenesis of COIVD-19 patients, as well as the importance of anti–IFN–I autoantibodies in the spread of SARS-CoV-2 infection and the subsequent excessive inflammation.
2. IFN-Is: production and subtypes
IFN-Is are a group of widely expressed cytokines produced and released by immune and non-immune host cells as the first line of defense against viral infection. In a typical setting, a virus-infected cell releases interferons, causing nearby cells to intensify their antiviral defense (Ivashkiv and Donlin, 2014; Trinchieri, 2010). Human interferons have been classified into three major types, mainly based on the characteristics of their signaling receptors (De Weerd and Nguyen, 2012; Pestka et al., 2004). Genes encoding IFN-Is are located on chromosome 9 in humans and on chromosome 4 in mice (Capobianchi et al., 2015; de Padilla and Niewold, 2016).
The human IFN-Is family encompasses 12 partially homologous IFN-α subtypes (IFN-α1, -α2, -α4, -α5, -α6, -α7, -α8, -α10, -α13, -α14, -α16, and -α17), IFN-β, and several single-gene products, including IFN-ϵ, IFN-τ, IFN-κ, IFN-ω, IFN-δ and IFN-ζ which are not well-defined, yet. IFN-δ and IFN-τ have been described in pig and cattle, respectively, and do not have a human homolog (Walter, 2020).
3. IFN-Is: signaling
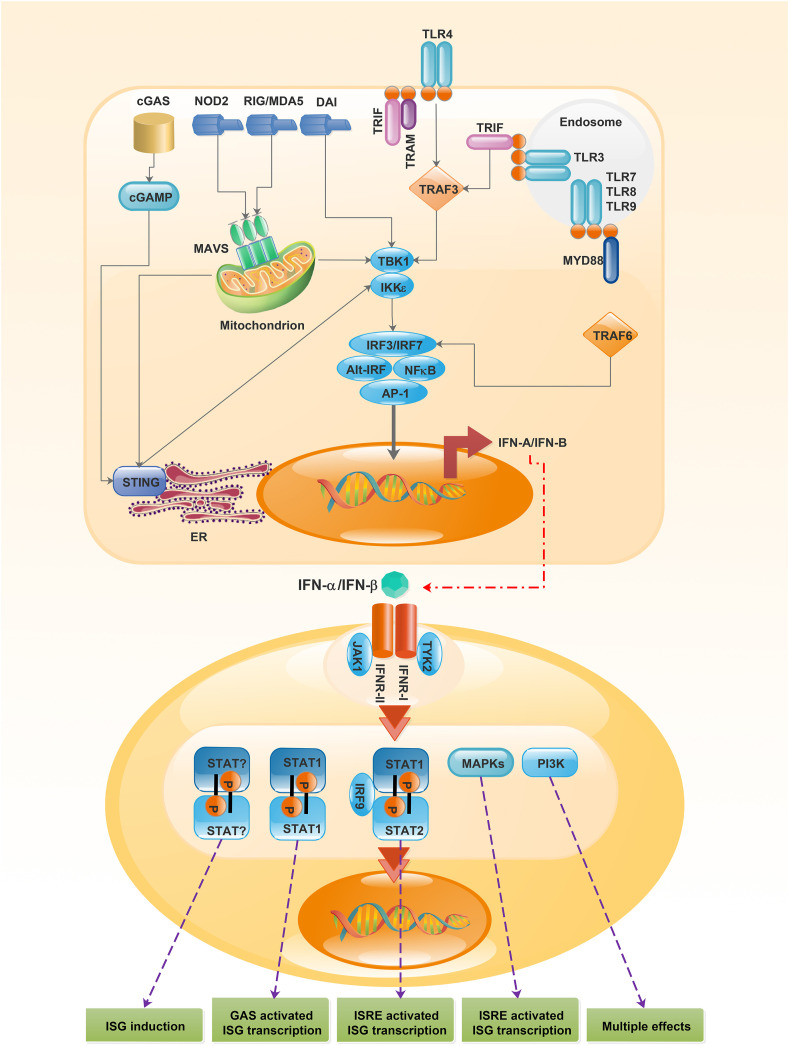
Production of IFN-Is is initiated following the binding of pathogen-associated molecular patterns (PAMPs) to different host pattern recognition receptors (PRRs). The PRRs could be found in various parts of cells, including cell membrane, or within intracellular compartments (McNab et al., 2015; Zahid et al., 2020). IFN-I production and signaling pathways are shown in Fig. 1 .
Fig. 1.
Interferon α/β production and receptor signaling. Recognition of microbial products by a range of PRR on the surface or inside the cell activates several distinct signaling pathways, inducing the expression of the genes encoding IFN α/β. Attachment of IFN-Is to IFN-α receptor stimulates several signaling pathways, creating different biological effects in the cell. The classical pathway contains STAT1–STAT2–IFN-IRF9 signaling complex (also known as ISGF3 complex). This complex binds to ISREs in gene promoters and activates ISGs. Heterodimers and homodimers of some STAT molecules including STAT3, STAT4, and STAT5 may also be activated downstream. MAPKs and the PI3K pathways are STAT independent signaling pathways that can also be involved following recognition of IFN α/β by the receptor. PRR, pattern recognition receptors; IFN α/β, Interferon α/β; IFN-I, type-I interferon; IFNAR, IFN-α receptor; STAT, signal transducer and activator of transcription 1; IRF9, IFN-regulatory factor 9; ISGF3, IFN-stimulated gene factor 3; ISREs, IFN-stimulated response elements; ISGs, IFN-stimulated genes; MAPKs, mitogen-activated protein kinases; PI3K, phosphoinositide 3 kinase; cGAS, cytosolic GAMP synthase; NOD2, NOD-containing protein 2; RIG, retinoic acid-inducible gene; MDA5, melanoma differentiation-associated gene 5; DAI, DNA-dependent activator of IRFs; TLR, toll like receptor; TRIF, TIR domain-containing adaptor protein inducing IFNβ; TRAM, TLR adaptor molecule (also known as TICAM2); TRAF, TNF receptor-associated factor; cGAMP, cyclic di-GMP-AMP; MAVS, mitochondrial antiviral signaling protein; TBK1, TANK-binding kinase 1; IKKε, IκB kinase-ε; MYD88, myeloid differentiation primary response protein 88; IRF, IFN-regulatory factor; Alt-IRF, IRFs other than IRF3 or IRF7; NF-κB, nuclear factor-κB; AP-1, activator protein 1; STING, stimulator of IFN genes; ER, endoplasmic reticulum; TYK2, tyrosine kinase 2; JAK1, Janus Kinase 1; GAS, γ-activated sequence.
The main intercellular receptors that recognize viral products such as RNA fragments are the retinoic acid-inducible gene I (RIG-I; also termed as DExD/H-Box Helicase 58 or DDX58) and melanoma differentiation-associated gene 5 (MDA5) (Goubau et al., 2013; Zahid et al., 2020). The ligation of PAMPs to the mentioned receptors mediates distinct signaling pathways by stimulating the IFN-α/β encoding genes (Platanias, 2005).
In a unique pathway, the RIG-I and MDA5 employ the adaptor mitochondrial antiviral signaling proteins (MAVs) and activate kinases including IκB kinase-ε (IKKε) and TANK-binding kinase 1 (TBK1) (Paludan and Bowie, 2013).
Interestingly, cytosolic nucleotide oligomerization domain-like receptors (NLRs, also called NACHT, LRR, and PYD domain proteins) are expressed in numerous cell types and bind to nucleic acids, resulting in the production and release of IFN-α/β (Kuenzel et al., 2010).
Other DNA-binding receptors involved in IFN-Is production are DNA-dependent activator of IFN-regulatory factors (DAI or ZBP1), cytosolic GAMP synthase, and DExD/H box helicase. In addition to the mentioned receptors, several toll-like receptors (TLRs) can activate IFN-α/β production pathways (Moynagh, 2005; Takeuchi et al., 2004).
All mentioned signals, through different pathways, converge on a key transcription factor named IFN-regulatory factor (IRF). In fact, IFN-α/β gene transcription is mainly induced via IRF3, leading to the transcription of IRF7, which makes a positive feedback loop, triggering the involvement of other IFNα-encoding genes (Moynagh, 2005; Tamura et al., 2008). All IFN-Is could bind to a common cell-surface heterodimeric receptor, known as IFN-α/β receptor (IFNAR) as the inducer of downstream cascades (Ivashkiv and Donlin, 2014).
Receptor-associated protein tyrosine kinases (TYKs) including Janus kinase and signal transducer and activator of transcription (JAK/STAT) are the classical mediators of the IFN-α/β signaling pathway. The binding of IFN-α/β to IFNAR results in the activation of JAK1 and TYK2, which can phosphorylate a couple of cytosolic transcription factors, especially STAT1 and STAT2. Homodimers and heterodimers of STAT1 and STAT2 are translocated to the nucleus and attach to IRF9 and forms IFN-stimulated gene factor 3 (ISGF3), which is regarded as an important transcriptional complex for binding to IFN-stimulated response elements (ISREs) (Ivashkiv and Donlin, 2014). Upon recognizing the ISREs in the ISG promoters, ISGF3 initiates transcription of ISREs, which creates an antiviral state in the cell (Ivashkiv and Donlin, 2014; Platanias, 2005).
Moreover, in mammals, following the binding of IFNα/β to IFNAR, the phosphoinositide 3-kinases (PI3K) can be activated, which is the target of the rapamycin (mTOR) pathway and multiple mitogen-activated protein kinase (MAPK) pathways (Platanias, 2005). Briefly, phosphorylation of the PI3K, downstream of JAKs, can be induced by several IFN-Is, including IFN-α, IFN-β, and IFN-ω, resulting in the activation of the p110 catalytic subunit of PI3K. Finally, PI3K can activate various downstream signaling cascades with diverse biological effects (Platanias, 2005).
Similar to the MAPK pathways, the p38-signaling pathway has a vital role in generating IFN-mediated signals. It was shown that p38α is phosphorylated by IFN-α/β-mediated mechanisms, independent of STAT-mediated pathways and ISRE binding. Moreover, it has been revealed that p38 is also needed for the transcription of the IFN-α/β-driven genes through GAS elements (Platanias, 2005).
Regarding the diversity of IFN-α/β-mediated signaling, widespread biological effects of IFN-α/β are conceivable; therefore, besides activation of specific antiviral genes, they can induce various genes, including pro-and anti-apoptotic molecules (Ivashkiv and Donlin, 2014).
4. Immunologic mechanisms of IFN-Is in viral Infections
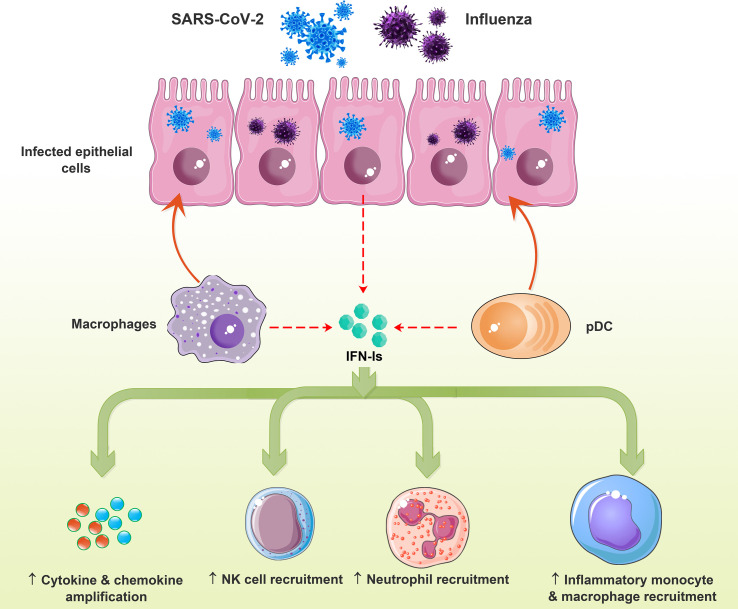
Indeed, in viral infections, protective, immunosuppressive, or immunopathological effects of IFN-Is are determined by the characteristics and context of the pathogen interaction with host cells and exposure timeline with IFN-Is (McNab et al., 2015). For example, IFN-Is contribute to inflammation by recruiting immune cells (Fig. 2 a), which could have beneficial or harmful outcomes for the host, based on immunosuppressive or tissue-damaging effects (da Silva et al., 2021; McNab et al., 2015).
Fig. 2a.
Effects of IFN-I on innate immune cells in the viral diseases. Viral infection of the lung epithelial cells induces IFN-Is that promote cytokine/chemokine production, recruitment of NK cells, neutrophils, and inflammatory macrophage/monocytes, resulting in lung immune-mediated pathology. IFN-I, type-I interferon.
4.1. Mechanisms of the innate immune system
Dendritic cells (DCs), mainly plasmacytoid dendritic cells (pDCs), and macrophages could recognize viral nucleic acids and proteins by their PRRs, inducing the production and release of IFN-Is (Ali et al., 2019; De Weerd and Nguyen, 2012; Ng et al., 2019). IFN-Is upregulate MHC-I expression in multiple cell lineages and enhance antigen presentation, especially in DCs (Murira and Lamarre, 2016; Teijaro, 2016). Exposure of immature DC to IFN-I can result in the upregulation of co-stimulatory molecules, enhanced antigen presentation, increased chemokine and cytokine production, and stimulation of B and T cells (De Weerd and Nguyen, 2012; McNab et al., 2015). Viral infections also enhance TLR expression and production of IFN-Is in neutrophils. Interestingly, IFN-Is cause a positive feedback loop in the formation of neutrophil extracellular traps (NETs) following the binding of viral DNA to TLR9 and enhancing IFN-I production (Stegelmeier et al., 2021).
IFN-Is also enhance natural killer (NK) cell recruitment, cytotoxicity, survival, and functions and protect antiviral CD8+ T cells from the lytic properties of NK cells (Teijaro, 2016). In this regard, Madera et al. showed that expansion and memory cell formation of Ly49H+ NK cells are impaired in IFNAR −/− or STAT1 −/− (as IFNAR downstream signaling molecule) mice following cytomegalovirus (CMV) infection. In addition, IFNAR −/− NK cells were more susceptible to apoptosis than normal NK cells, and the expression of group 2-member D (NKG2D) ligands improved during CMV infection. Eventually, these NK cells demonstrated decreased protection against CMV (Madera et al., 2016). Therefore IFN-I stimulatory or inhibitory effects on IFN-γ production by NK cells depend on the ratio of STAT molecules; for example, increased STAT4 signaling induces IFN-γ production by NK cells (McNab et al., 2015).
4.2. Mechanisms of the adaptive immune system
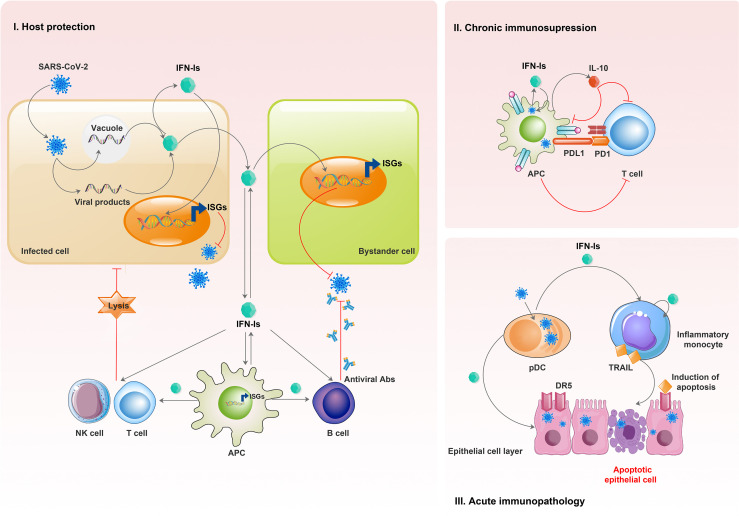
IFN-Is play a pivotal role in the antiviral function of adaptive immune cells (Fig. 2 b). They promote DC maturation, upregulate antigen-presentation of APCs, and orchestrate the adaptive immune responses (Mantlo et al., 2020; Ng et al., 2019). B cells are involved in humoral immune responses against viral infection through the production of neutralizing antibodies. In this context, IFN-Is could stimulate B cell activation, function, and antibody class switching by altering the expression of transcription factors (McNab et al., 2015). Furthermore, following IFN-I-induced maturation and activation of DCs, they release various cytokines and chemokines such as IL-12, which are required for T helper-1 (Th1) differentiation and antiviral activities (McNab et al., 2015). IFN-Is also have different stimulatory impacts on the functionality and characteristics of CD8+ T cells, including the expression level of co-stimulatory molecules, proliferation, survival, and cytotoxicity (Crow and Ronnblom, 2019; Teijaro, 2016).
Fig. 2b.
IFN-I effects on adaptive immune cells in the viral diseases. I) Viral infection of the vertebrate body cells leads to the production of IFN-Is. Infected and bystander cells are affected by the produced IFNs, inducing ISGs, which leads to the blocking of the viral replication process. Also, these interferons are produced by and affect innate immune cells. IFN-Is enhance the antigen-presenting function of APCs. These interferons strengthen the antiviral function of adaptive immune cells, including B cells, T cells, and natural killer (NK) cells. Viral infection can be restricted by producing antibodies (B cells) and cytotoxic responses (T cells and NK cells) of adaptive immune cells. II) During chronic viral infection, produced IFN-Is can induce the production and release of cytokines such as IL-10, which show immunosuppressive effects. Also, during chronic infection, IFN-Is stimulate the expression of ligands such as PDL1, which recognize T cell-inhibitory receptors (such as PD1 and the PDL1 receptor). These events lead to the suppression of T cell function and persistence of the infection. III) Acute viral infections such as influenza result in the production of IFN-Is by myeloid cells, such as pDCs and inflammatory monocytes. IFN-Is upregulate the death ligand TRAIL expression on inflammatory monocytes and the TRAIL receptor DR5 on epithelial cells. Then, inflammatory monocytes expressing TRAIL lead to immunopathological effects by destroying the epithelial cells. IFN-I, type-I interferon; ISGs, IFN-stimulated genes; APCs, antigen-presenting cells; IL-10, interleukin-10; PD1, programmed cell death protein 1; PDL1, programmed cell death 1 ligand 1; pDCs, plasmacytoid dendritic cells; TRAIL, TNF-related apoptosis-inducing ligand; DR5, death receptor 5.
Similar to NK cells, the concentration of IFN-Is and the duration of exposure of CD8+ T cells to IFN-Is can affect the immune response via STAT-mediated signaling pathways (De Weerd and Nguyen, 2012; Teijaro, 2016). However, STAT molecules may demonstrate both positive and negative impacts on IFN-Is production. For instance, in a STAT1 dominant status, such as an increased ratio of STAT1/STAT4 IFN-γ production is inhibited (McNab et al., 2015).
5. IFN-Is in human Coronavirus Infections: common cold, SARS-CoV-1, MERS, and SARS-CoV-2
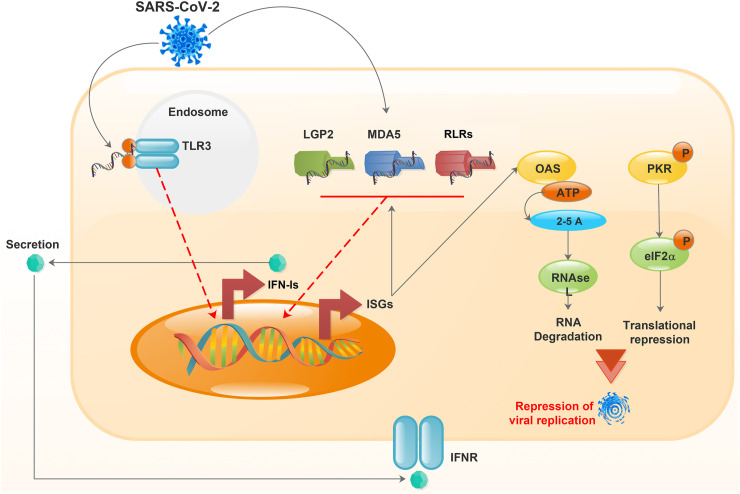
Studies show that IFN-Is exert their antiviral functions through IFN-induced proteins encoded by ISGs. The main IFN-I-induced proteins are protein kinase RNA-activated (PKR), the 2′,5′-oligoadenylate synthetase (OAS), Mx protein GTPase, and Tudor domain-containing protein 7 (TDR7) (Fig. 3 ) (Chattopadhyay and Sen, 2014; Samuel, 2001; Schneider et al., 2014; Subramanian et al., 2018). After binding to viral dsRNA, PKR is autophosphorylated and self-activated, and then it disrupts the translation of viral proteins by inhibiting Eif-2alpha (Fig. 3). However, it seems that this self-activation does not inhibit SARS-CoV-1 infection and is only involved in the apoptosis of infected cells (Krähling et al., 2009; Yim and Williams, 2014). Additionally, the OAS family could be stimulated by viral dsRNA and play a role in breaking viral RNA by activating RNase L (Fig. 3). However, there is no scientific report about the OAS effects on human coronaviruses, yet. Nevertheless, OAS-1 gene polymorphisms can affect the susceptibility to SARS-CoV-1 (Chakrabarti et al., 2011; Hamano et al., 2005; Wreschner et al., 1981). SARS-CoV-2, like other β-coronaviruses, uses similar approaches to induce interferon production and signaling of the downstream pathways. Although this phenomenon was demonstrated in human airway epithelial cells, it was found that following SARS-CoV-2 infection and production of IFN-Is, a relatively poor induction of ISGs occurs (Li et al., 2021; Miorin et al., 2020; Xia et al., 2020). Unlike Middle East respiratory syndrome (MERS), SARS-CoV2 activates PKR and RNase L and can effectively inhibit IFN signaling and the OAS/PKR pathways (Comar et al., 2019; Li et al., 2021; Thornbrough et al., 2016).
Fig. 3.
IFN response induced by viral infection. Virus or viral product recognition by TLR3 or RLRs leads to the activation of TLR3 or RLRs, which finally induce IFN production. The secreted IFN affects the cell through a paracrine or autocrine regulation and stimulates the expression of ISGs. An ISG, either OAS or PKR, activates RNase L via 2′, 5′-oligoadenylate synthetase or phosphorylation of eIF2, resulting in RNA degradation or translational suppression. The IFN response inhibits viral replication and damage to the cell. IFN, interferon; LGP2, laboratory of genetics and physiology 2; MDA5, melanoma differentiation-associated gene 5; TLR3, toll-like receptor 3; RLRs, retinoic acid-inducible gene I (RIG-I)-like receptors; ISGs, FN-stimulated genes; OAS, oligoadenylate synthetase; PKR, protein kinase R; eIF2-α, eukaryotic translation initiation factor 2α; IFNR, interferon receptor.
It has been revealed that the MX1 and MX2 genes are activated in defense against various RNA viruses and provide antiviral effects. In addition, the expression of MX1 and MX2 genes in COVID-19 patients are increased in direct proportion to viral load, and their expression is inversely related to the age of the patients. Considering the lowered expression of MX proteins in elderly patients, it can be hypothesized that the high severity rate of disease in elderly patients is probably associated with decreased expression of MX antiviral proteins (Bizzotto et al., 2020; Verhelst et al., 2012; Zav'yalov et al., 2019).
According to previous investigations, coronavirus members' susceptibility to IFN-Is is significantly different, but there is no explanation for these findings (Kindler et al., 2016). IFN-inducible transmembrane (IFITM) proteins are expressed by the infected cells and show antiviral effects by trapping and destroying the virus in endocytic compartments and inhibiting the virus access to vital parts of the targeted cells (Anafu et al., 2013; Brass et al., 2009; Huang et al., 2006). IFITM1 is found mainly in cellular plasma membranes, and IFITM2 and IFITM3 proteins are principally localized in endo-lysosomal membranes of the cells (Bailey et al., 2014).
The IFITMs play an essential role in defense against many viruses and can be considered one of the main mechanisms of innate immunity and its connection to adaptive immunity (Prelli Bozzo et al., 2021). Nevertheless, some viruses employ the advantages of these proteins. According to the study by Xuesen Zhao et al., the HCoV-OC43 virus, one of the common cold agents, responds oppositely to IFN-Is, and these cytokines increase the possibility of cell infection caused by this virus. An explanation for this contradiction could be traced back to IFITM2 and IFITM3, which can facilitate virus entry into the target cells (Zhao et al., 2014).
The S protein of SARS-CoV-1 mediates virus entry, and similar to influenza A and flaviviruses, this virus requires an acidic pH to enter the target cells (Harrison, 2008; Huang et al., 2011). SARS-COV-1 also requires cathepsin-L to enter the cell by fusion, and IFITM1, 2, and 3 could prevent the entry. The IFITMs can interfere with endosomal trafficking or access to the late endosomal compartment (Huang et al., 2006, 2011). MERS and HCoV-229E, two other coronaviruses, use a similar mechanism to SARS-CoV-1 for entrance to the target cell, and studies show that IFITMs can also prevent the entry of these viruses into the target cells (Bertram et al., 2013; Wrensch et al., 2014). SARS-CoV-2, another member of the coronavirus family, is also sensitive to the IFITMs; however, this susceptibility has been moderately reduced compared to SARS-COV-1 due to polybasic cleavage sites that allow the virus to enter the target cells in a pH-independent manner, resulting in more virus transmission and replication (Winstone et al., 2021). Surprisingly, Prelli Bozzo et al. showed that proper SARS-CoV-2 replication in human respiratory epithelial cells depends on the endogenous IFITMs; therefore, IFITMs could have a dual role in SARS-CoV-2 infection (Prelli Bozzo et al., 2021).
Coronaviruses can serve a variety of strategies to interfere with the synthesis, signaling, and responses of IFN-Is. For example, SARS-CoV-1 enhances the ubiquitination of the subunit 1 of IFNAR and ultimately degrades IFN-Is, leading to impaired innate immunity. Impaired functions of IFN-Is can reduce the activity of other components of the innate immune system such as NK cells and macrophages, as well as the adaptive immune responses and Th1 activation. These defects caused by SARS-COV-1 and MERS eventually worsen the disease condition and increase patient mortality (Dandekar and Perlman, 2005; Minakshi et al., 2009). SARS-COV-2, like other β-coronaviruses, adopts mechanisms to disrupt IFN-I-mediated antiviral processes. It is worth noting that this pathogen, unlike SARS-CoV1, is highly sensitive to IFN-Is. In contrast to SARS-CoV-1, SARS-COV-2 activates the expression of the Th1-related transcription factors and STAT1, which subsequently increases the expression of ISGs (Schreiber, 2020). However, the innate immune system cannot overcome this virus, most likely due to the SARS-CoV-2 inhibitory mechanisms interfering with IFN-I production. For example, both SARS-CoV-1 and SARS-CoV-2 use papain-like proteases (PLpros) to attenuate IFN-I-mediated responses and suppress innate immunity responses (Daczkowski et al., 2017; Schreiber, 2020; Shin et al., 2020).
6. IFN-Is as immunotherapeutic agents in management of COVID-19: benefits and adverse effects
The history of therapeutic and prophylactic use of IFN-I in coronavirus infections dates back to 1983 when Higgins et al. showed that intranasal IFN-α2 administration before and after challenge with coronavirus could reduce virus replication and consequently diminish the incidence and severity of disease in patients (Higgins et al., 1983). In SARS and MERS epidemics, IFN-Is were used along with other treatments to reduce the severity of the disease and to accelerate the recovery process (Loutfy et al., 2003; R Strayer et al., 2014). These promising outcomes encouraged the scientists to use IFN-Is to treat this family's new unwelcome child, SARS-CoV-2.
Probably, a primary mechanism used by SARS-CoV-2 to bypass the innate immune responses is through inhibiting the production of ISGs and IFN-Is, especially IFN-β. Therefore, the IFN-I mediated responses are crucial in antiviral defense, and it appears that they are impaired in COVID-19 patients (Hadjadj et al., 2020). IFN-I-deficient animal models have also demonstrated the critical role of IFN-I in disease recovery (Sun et al., 2020). These findings justified the administration of recombinant external IFN-Is as a treatment for patients with COVID-19, aiming to replace the patient's reduced endogenous IFN-I (Lokugamage et al., 2020; Schreiber, 2020). Furthermore, IFN-I administration is imperative in elder COVID-19 patients that naturally have diminished IFN-I responses (Aricò et al., 2020).
6.1. Safety and efficacy of IFN-Is in clinical trials
Numerous clinical trials have evaluated the safety and efficacy of IFN-Is in COVID-19 patients. These trials used different IFN-Is via various administration routes, including subcutaneous, intranasal, and nebulized respiratory systems. Davoudi-Monfared et al. subcutaneously injected six doses of IFN-β1 in addition to the routine treatments of the COVID-19 patients in a randomized clinical trial study (IRCT20100228003449N28), which significantly lowered the mortality rate compared to the controls (19% vs. 44%) (Davoudi-Monfared et al., 2020). Another clinical trial (NCT04276688) examined the effect of subcutaneous injection of IFN-β with ritonavir, lopinavir, and ribavirin in newly diagnosed patients with COVID-19 and found that IFN-β administration accelerated the recovery, reduced the viral load, and diminished the rate of critically ill patients (Hung et al., 2020). Probably, the subcutaneous injection elicits a systemic response of IFN-Is along with their antiviral functions (Schreiber, 2020).
Because SARS-CoV-2 has tropism to the respiratory tract, numerous clinical trials have used this route of administration to maximize the IFN-Is prophylactic and therapeutic effects (Liu et al., 2020; Monk et al., 2021; Wang et al., 2020; Zhou et al., 2020b). The history of intranasal administration of IFN-I to fight respiratory system infections goes back to the 1970s which aimed to control the replication of rhinoviruses and influenza (Merigan et al., 1973). A study in China on 2944 hospital staff found that IFN-α1 nasal drops were 100% protective against SARS-CoV-2 infection without serious side effects (Meng et al., 2020). Consequently, it seems that this method could be helpful to hinder infection in high-risk individuals during pandemics. However, intranasal administration may unintendedly deliver the substances to the central nervous system (CNS) and result in further neurotoxicity (Thorne et al., 2008). Therefore, some prefer to consider the oral mucosa as the administration route due to rapid absorption of the substances, eliciting mucosal immunity, and less concern about neurotoxicity. It has been shown that the oral administration of low-dose IFN-α promoted immunity against respiratory infections and reduced disease severity (Bennett et al., 2013). Other advantages of mucosal administration are ease of the process, low toxicity, reduced local and systemic effects, and prevention of the entrance of the virus through the entry sites (Aricò et al., 2020).
Another method for mucosal IFN-I administration is using a respiratory nebulizer (Monk et al., 2021). In this method, IFN-I is delivered directly into the infected respiratory epithelial cells with SARS-CoV-2 and replaces the missing endogenous IFN-I (Monk et al., 2021; Zhou et al., 2020b). In a clinical trial (NCT04385095) on 101 patients, the use of SNG001 (IFNβ1a suitable for nebulization) reduced the severity of disease or mortality rate by 79% and increased the chance of recovery by 100% compared to the controls. In this study, the most frequently reported adverse event was headache (Monk et al., 2021). In another study, respiratory administration of IFN-α2b alone or in combination with arbidol (Umifenovir) also diminished the plasma levels of inflammatory mediators such as IL-6 and C-reactive protein (CRP) as well as the presence of the virus in the upper respiratory tract. In this trial, adverse effects of the treatment had not been reported (Zhou et al., 2020a). However, IFN nebulization is currently not applied during standard treatment of COVID-19, because there are some data regarding the risks of transmitting the viral infection by nebulization (Mary et al., 2020; Sethi et al., 2020).
On the other hand, clinical trials investigating effects of recombinant IFN-γ (Type II IFN) and IFN-λ (Type III IFN) administration in COVID-19 are ongoing. In a clinical study by Myasnikov on over 18 years of age patients with moderate COVID-19 infection, daily subcutaneous administration of recombinant interferon gamma (IFN-γ), once a day, during 5 days showed a positive effect on the recovery processes of moderate COVID-19. Patients who received IFN-γ had no progression of respiratory failure and no transfer to intensive care unit was needed (Myasnikov et al., 2021). In another clinical trial (NCT05054114) the efficacy and safety of nasal IFN-γ use in healthy subjects for COVID-19 prevention is under evaluation (ClinicalTrials.gov, 2022). In a clinical trial (NCT04331899) on mild to moderate COVID-19 outpatients subcutaneous administration of Peginterferon Lambda-1a (Lambda) within 72 h of diagnosis didn't improve symptoms, and no effects on viral shedding was found (Jagannathan et al., 2021). In another clinical trial (NCT04354259) subcutaneous administration of Peginterferon Lambda in outpatients with laboratory-confirmed COVID-19 could prevent clinical deterioration and shorten duration of viral shedding (Feld et al., 2021).
6.2. Questions and ambiguities regarding IFN-Is administration
Despite obtaining satisfactory outcomes, there are still several contradictions and questions that limit the administration of IFN-Is in patients with COVID-19. For instance, some studies have reported a negative association between late IFN-I therapy and clinical improvement in COVID-19 (Liu et al., 2020). Another important ambiguity regarding the treatment of COVID-19 using IFN-I is to determine the appropriate time to start the treatment (Aricò et al., 2020; Wang et al., 2020).
A retrospective study compared the efficacy of nebulized IFN-α2 in the initial and advanced phases of COVID-19 and found that early IFN-α2 therapy significantly reduced patient mortality. However, IFN-α2 administration in advanced stages of the disease could not improve the treatment procedure and even delayed the recovery and increased the mortality rate (Wang et al., 2020). Other studies emphasized early initiation of IFN-I treatment and proposed that the ideal administration time could be within the early five days after symptoms onset and could be extended up to maximum ten days after onset of the symptoms (Aricò et al., 2020; Hung et al., 2020; Wang et al., 2020). However, these studies warned about the use of IFN-I treatment in advanced stages of the disease, regarding high mortality due to hyper-inflammation and cytokine storm in late stages (Aricò et al., 2020; Wang et al., 2020). Given that the main therapeutic mechanism of IFN-I is preventing the virus replication, it seems logical that the optimal time of IFN-I usage is the early stages of the disease when the virus is replicating. IFN-I administration at the early stages of the disease can bridge innate and adaptive immunity by stimulating humoral and cellular responses (Aricò et al., 2020). In addition to antiviral functions, IFN-Is also have pro-inflammatory properties. Therefore, if patients enter the hyper-inflammatory phase in which cytokine storm occurs, IFN-I intensifies secretion of the inflammatory cytokines, exacerbating the inflammation and increasing the mortality risk (Aricò et al., 2020). Some studies believe that the severe IFN-I response is one of the main suspects for hyper-inflammatory responses in COVID-19 patients (Wilk et al., 2020; Zhou et al., 2020b). Normally, high levels of tumor necrosis factor (TNF) induce tolerance to TLR signaling in monocytes to prevent more severe inflammation (Park et al., 2017). High levels of IFN-Is abrogate the TLR tolerance to TNF, leading to a maximal response of monocytes and macrophages, which exacerbate the inflammation (Park et al., 2017). Therefore, the timing of IFN-I therapy is of great importance to COVID-19 treatment.
In addition to the time of treatment onset, the duration and frequency of treatment with IFN-I are also questions that must be answered. Preclinical studies on influenza viruses showed that long-term treatment with IFN-I delays the regeneration of lung epithelial cells (Major et al., 2020). Moreover, persistent signaling of IFN-I and IFN-III may disrupt the proliferation and repair of lung epithelial cells by inducing P53 protein (Major et al., 2020). Based on the therapeutic protocols of clinical trials, the frequency of IFN-I administration is usually 2–3 times every week that can be reduced to once a week or once every ten days using PEGylated forms of IFN-I, which has a prolonged half-life (Schreiber, 2020).
In clinical trials, various types of IFN-Is including IFN-α1b, IFN-α2b, IFN-β1a, and IFN-β1b have been used for the treatment of COVID-19 (Davoudi-Monfared et al., 2020; Hung et al., 2020; Meng et al., 2020; Merigan et al., 1973; Monk et al., 2021; Zhou et al., 2020b). However, the question that arises here is which type of IFN-I is the ideal choice for managing and treating of COVID-19?
Considering that SARS-CoV-2 predominantly inhibits IFN-β production, it makes sense that IFN-β might be more effective than IFN-α for the treatment of COVID-19. IFN-β has been longtime prescribed in multiple sclerosis (MS) and has greater anti-inflammatory and immunomodulatory function than IFN-α (Harari et al., 2014). On the other hand, IFN-α induces more robust antiviral responses, leading to use of IFNα2 in viral infections such as hepatitis C (HCV) (Reddy et al., 2002; Rostaing et al., 1998). In the COVIFERON clinical trial, Darazam et al. evaluated the efficacy of IFN-β1a and IFN-β1b in COVID-19 and showed that there was a significant difference between IFN-β1b and the control group in the case of Time to Clinical Improvement (TTCI) (p-value = 0.031), while no significant difference was found between IFN-β1b and the control group (p-value = 0.395). Also, both IFN-βs could relatively improve the clinical responses and reduce the mortality rate from 45% in the control group to 20% in the IFN-β1a group and 30% in the IFN-β1b group but the observed differences were not statistically significant (p-value = 0.231) (Darazam et al., 2021). The results of this study suggest that IFN-β1a might be a more reasonable choice than IFN-β1b for the management of COVID-19 due to a significant reduction in TTCI that was observed using IFN-β1a (Darazam et al., 2021). Surprisingly, although they showed that the treatment efficacy was higher in the early stages than the advanced stages of the disease, serious adverse effects following IFN-I therapy were not observed even in the advanced stages of COVID-19 (Darazam et al., 2021). Despite all the aforementioned studies, the lack of multi-arm multi-stage (MAMS) clinical trials to compare the safety and efficacy of IFN-I in the prevention and treatment of COVID-19 at different stages of the disease is still felt.
Some studies reported the presence of IFN-I neutralizing antibodies in a small proportion of COVID-19 patients, which reduced the effectiveness of IFN-I treatment (Bastard et al., 2020). In these cases, the type of IFN-I recognized by the neutralizing antibody should be determined, and patient treatment should be performed using that type of IFN-I, preferably in anti- IFN-I neutralizing antibody-positive subjects (Bastard et al., 2020).
Taken together, given the long experience of IFN-I therapy and the availability of the drug at an affordable price, IFN-Is could be considered potential candidates for treating COVID-19. However, various clinical questions should be answered before to demonstrate a significant clinical benefit to COVID-19 patients.
7. Anti–IFN–I autoantibody in COVID-19: Immunopathogenesis and clinical findings
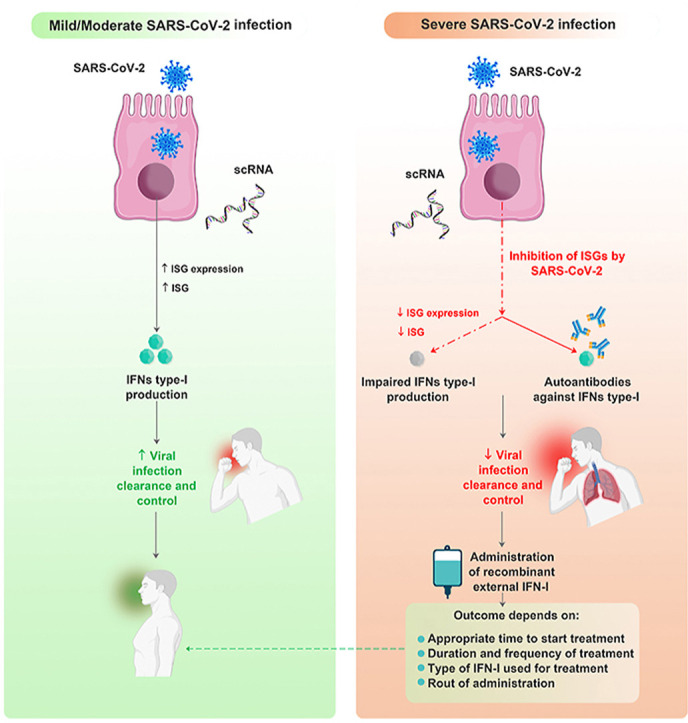
Recently, it has been demonstrated that insufficient antiviral IFN-I responses are the hallmark of COVID-19 infection (Lin and Shen, 2020). Several SARS-CoV-2 proteins, including nucleocapsid (N), membranous (M), and nonstructural (NS) proteins, inhibit the production of IFN-Is (Palermo et al., 2021). On the other hand, the result of multiplexed single-cell epitope and transcriptome sequencing revealed the dysfunction of ISG responses in patients with severe COVID-19 (van der Wijst et al., 2021). Therefore, the production of circulating autoantibodies against IFN-Is could be considered an impairment mechanism of IFN-I responses in critical COVID-19 patients (Goncalves et al., 2021).
It is reasonable to presume that the dysfunction of IFN-I development or production of autoantibodies against it may result in viral spread, subsequent pulmonary and systemic inflammation, as well as COVID-19 predisposition. As summarized in Table-1 , anti–IFN–α autoantibodies were detected in patients recovered from severe COVID-19 (Acosta-Ampudia et al., 2021). Approximately 10% of COVID-19 patients have a remarkable level of autoantibodies against IFN-I and these antibodies are associated with the severity of the infection (Bastard et al., 2020; Zhou and Wang, 2021).
Table 1.
Preclinical findings in COVID-19 patients with positive anti–IFN–I autoantibodies.
| Total number of COVID-19 patients | Total number of normal or healthy subjects | Number or percentage of anti IFN-I autoantibody-positivity in COVID-19 patients | Number or percentage of anti IFN-I autoantibody-positivity in normal or healthy subjects | Reference |
|---|---|---|---|---|
|
105 patients Including 84 critically ill (80%), 10 mild (9.5%) and 11 autoimmune polyendocrinopathy type 1 syndrome (10.5%) patients |
76 | 31/105 (29.5%) | Not Reported (the normal group used to determine a positive cut-off value) | Goncalves et al. (2021) |
|
218 patients Including 135 critical (61.9%), 44 severe (17.9%) and 39 mild/moderate (20.2%) patients |
– | 26/218 (11.9%) including 17/218 (7.8%) or 17/26 (65.3%) autoantibody against both IFN-α and IFN-ω and 1/26 (3.8%) or 1/218 (0.4%) anti IFN-β | – | Abers et al. (2021) |
| 22 COVID-19 patients with APS-1 | 27 | 21/22 (95.5%) autoantibody against both IFN-α and IFN-ω, 1/22 (4.5%) anti IFN- β and 1/22 (4.5%) anti IFN-ω | Not Reported (the normal group used to determine a positive cut-off value) | Bastard et al. (2021b) |
| 47 patients | 118 | 5/47 (10.6%) autoantibody against both IFN-α and IFN-ω | 0% | Troya et al. (2021) |
| 987 patients | 1227 | 101/987 (10.2%) In details 36/987 (3.6%) patients showed anti IFN-α, 13/987 (1.3%) anti IFN-ω and 52/987 (5.2%) had both anti IFN-α and IFN-ω. |
Prevalence of autoantibodies against type I IFNs: 0.33% (0.015–0.67%) | Bastard et al. (2020) |
| 275 patients | – | 49/275 (17.8%) IFN-α2 and/or IFN-ω | – | Solanich et al. (2021) |
| 3595 patients | 1639 | 13.6% | 1% | Bastard et al. (2021a) |
An observational study on more than 980 patients with severe COVID-19 infection has indicated significant autoantibodies against IFN-I, especially both IFN-α2 and IFN-ω (13.7%) compared with unexposed healthy individuals (0.3%) (Bastard et al., 2020). Additionally, a large cohort study in COVID-19 infected patients revealed that 3.5% of these patients harbored some mutations in those eight genes involved in coding the TLR signaling molecules and the IFN-I pathway (Zhang et al., 2020). Abers et al. reported that 26 out of 218 patients with severe COVID-19 had a significant level of anti–IFN–I autoantibodies; 34.7% had anti–IFN–β while 65.3% had anti–IFN–α and anti–IFN–ω (Abers et al., 2021). In another study, 21 out of 84 patients with critical COVID-19 infection (25%) had a significant level of anti–IFN–α2 autoantibodies (Goncalves et al., 2021). In this study, anti–IFN–α2 autoantibodies with neutralizing activity were detected in 15 of the mentioned patients, which made a significant impairment of the IFN-I response (Goncalves et al., 2021). In another study; among 47 patients with severe COVID-19 pneumonia who were hospitalized in Madrid, Spain; more than 10% showed IFN-α and IFN-ω neutralizing autoantibodies compared to asymptomatic control subjects. These autoantibodies were significantly associated with higher CRP levels and lower lymphocyte counts and increased risk of death in patients (Troya et al., 2021). Hence, patients with anti–IFN–I neutralizing autoantibodies might be considered more susceptible to COVID-19 infection (Gupta et al., 2020).
According to the antiviral characteristics of IFN-I, recombinant forms of IFN-I are considered promising candidates for the treatment of severe COVID-19 (Sodeifian et al., 2022). Although plasma exchange has been used to remove autoantibodies against IFN-I, further studies are needed to evaluate the efficacy of this treatment (de Prost et al., 2021). Taken together, the production of IFN-I autoantibodies refers to inborn errors of autoimmune B cells which could benefit as a diagnostic and prognostic criterion.
In addition to anti–IFN–I autoantibodies, COVID-19 patients may produce autoantibodies against other cytokines or their receptors. Wang et al. showed that sever COVID-19 patients have anti-cytokine antibodies targeting IL-1α/β, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF), leptin and IL-18Rβ. Also, autoantibodies targeting chemokines including CXCL1, CXCL7, CCL2, CCL15, CCL16, and the atypical chemokine receptor-1 (ACKR1, or Duffy blood group antigen) were identified in this patients (Wang et al., 2021).
Bastard et al. identified antibodies against cytokines such as GM-CSF, IL-6, IL-10, IL-12p70, IL-17A, IL-17F, IL-22, and TNF-b in COVID-19 patients, among them only autoantibodies targeting IL-6, IL-12p70, and IL-22 showed neutralizing properties (Bastard et al., 2020).
Both Bastard and Wang revealed that a subset of anti-cytokine antibodies inhibit binding of soluble factors to their receptors on the cell surface and are assumed to have a pathogenic role by thwarting protective immune responses against COVID-19 (Chang et al., 2021).
A study by Chang et al. investigated circulating antibodies in hospitalized patients with COVID-19 and demonstrated that about 60–80% of all hospitalized COVID-19 patients had at least one detectable anti-cytokine antibody. In this study many interleukins were identified as autoantibody targets such as IL-1, IL-6, IL-10, IL-15, IL-17A, IL-22, and IL-31, in addition to cytokines with well-known activities such as chemotactic chemokine macrophage inflammatory protein-1 alpha (MIP-1α), leukemia inhibitory factor (LIF), and vascular endothelial growth factor-B (VEGF-B) (Chang et al., 2021).
These anti-cytokine antibodies disturb immune function and impair immunological control of the virus by blocking immunoreceptor signaling and changing composition of the peripheral immune cells, and surrogates of these autoantibodies can worsen disease severity in animal models of SARS-CoV-2 infection (Wang et al., 2021).
8. Therapeutic approaches for the management of Anti-IFN auto-antibodies in COVID-19
Pieces of evidence suggested that the presence of autoantibodies against IFN makes patients vulnerable to the severity of COVID-19. As a result, measurement of the autoantibody levels in patients may help us choose the best treatment protocol and manage the disease more intelligently (Khamsi, 2021).
Several treatment strategies including plasma exchange through plasmapheresis, depletion of plasmablasts by monoclonal antibodies, preventing auto-antibody production, administration of convalescent plasma, recombinant IFN-I, corticosteroids, and intravenous immunoglobulin (IVIG) have been suggested for the management of these autoantibodies in patients with COVID-19. However, still, there is not sufficient data regarding the application of these therapeutic tactics (Casadevall and Pirofski, 2020).
Convalescent plasma is a transfusion method based on passive immunization. In this method, infected patients receive plasma from donors recovered from COVID infection (Selvi, 2020). Early administration of convalescent plasma can decrease the severity of the symptoms (Acosta-Ampudia et al., 2021). Several studies showed that this transfusion has no critical adverse effects on recipients, and they also showed a temporary increase in IgG levels against SARS-CoV-2, which improved symptoms of the disease during 1–3 days after transfusion (Mendoza-Pinto et al., 2021). However, this is not an approved therapeutic procedure and studies did not show reliable and consistent results for its approval as a standard treatment for COVID-19.
Plasma exchange is a therapeutic method aiming to eliminate inflammatory molecules, immunocomplexes, and pathogenic antibodies by separating the plasma from blood and replacing it with solutions with oncotic pressure molecules while maintaining blood volume constant (Mendoza-Pinto et al., 2021). De Prost et al. reported a case series study comprising four patients with destructive COVID-19 and high levels of autoantibodies against IFN-I and SARS-CoV-2. Three of these patients received four sessions of plasma exchange, and one of them got three sessions. Using plasma exchange, the concentration of autoantibodies decreased while anti-SARS-CoV-2 antibodies remained stable. As a result, two patients were discharged from the hospital, while two passed away (de Prost et al., 2021).
Generally, before subcutaneous injection of IFN-I, plasmapheresis is performed to remove autoantibodies and to prevent neutralization of recombinant IFN-I in patients. It was shown that treatment with IFN-I could be effective during the initial phase of COVID-19, while its administration can be lethal when patients' conditions worsen, probably due to cytokine storm and systemic inflammation (Bojdani et al., 2020). A preliminary study by Monk et al. revealed that inhalation of IFN-β can improve the clinical outcomes of COVID-19 patients (Monk et al., 2021). In a cohort study by Troya et al. on 47 patients with severe COVID-19 infection, approximately 10.6% had significantly high levels of autoantibodies against IFN-1α and IFN-ω, while no anti–IFN–β antibody was detected in their plasma samples. They were treated with recombinant IFN-β1b, and there were no significant clinical differences between patients with or without autoantibodies in their response to IFN-β1b therapy (Troya et al., 2021).
IVIG can regulate the immune system response to SARS-COV-2 by blocking pro-inflammatory cytokines such as IL-1, IL-6, and TNF-α, inhibition of Th1 and Th17 cells, neutralization of autoantibodies, and expansion of regulatory T cells (Maddur et al., 2014; Mendoza-Pinto et al., 2021). However, according to the results of IVIG usage in the SARS-CoV-1 outbreak in 2003, IVIG administration can lead to undesirable reactions such as developing venous thromboembolism, which was seen despite using heparin in SARS-CoV-1 patients (Lew et al., 2003).
Plasmablast depletion therapy and inhibition of reactive B-cells through several mechanisms result in the elimination of B-cells harboring inborn abnormality by targeting CD19, CD20, and B-cell activating factor (BAFF), which will prevent the production of autoantibodies. These procedures are used to control autoimmune disorders, including SLE (Lee et al., 2021). These are only some suggestions for future studies, based on plasmablast depletion as currently, there is no registered investigation on this idea.
9. Concluding remarks and future perspectives
It is now obvious that IFN-Is play an important role in the battle between the immune system and SARS-COV-2. Although SARS-COV-2 is highly sensitive to IFN-Is, its inhibitory mechanisms interfere with IFN-I production, and defective functions of IFN-Is could change the outcome of the battle between the immune system and SARS-CoV-2 in favor of the virus.
Despite the promising results achieved by the numerous clinical trials evaluating the effects of exogenous IFN-I administration to overcome defective functions of IFN-Is in COVID-19 patients, some studies have reported a negative association between IFN-I therapy and clinical improvement in COVID-19. It is assumed that several factors influence the outcome of IFN-I therapy in COVID-19 patients such as the time of treatment onset, the duration and frequency of the treatment, the type of IFN-I, and the presence of autoantibodies against IFN-Is.
Therefore, our knowledge regarding different aspects of IFN-I therapy in COVID-19 and its influencing factors is still incomplete.
Also, the available data regarding IFN-Is roles in COVID-19 immunopathology is very limited yet. We need to know more about the mechanisms by which SARS-COV-2 disturbs IFN-Is anti-viral activities. In addition, we need to find strategies that can be applied to overcome these mechanisms. The discovery of these mechanisms can help us to provide better treatment for COVID-19. It is not completely clear yet which of the patient-related factors are involved in the IFN-I impaired responses caused by SARS-COV-2. Could immunodeficiency be the main reason? Why do some individuals have anti–IFN–Is autoantibodies and which mechanisms are involved in the autoantibody production, breakage of B cells’ tolerance, or other similar disorders?
Finally, to overcome defective functions of IFN-Is, we can also explore approaches to induce the production of functional IFN-Is in the patient's body or to find some solutions to protect IFN-Is from undesirable effects of SARS-COV-2.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
All authors participated equally in data gathering, writing, revising of the article. Hossein Khorramdelazad designed pictures.
Declaration of competing interest
There is no conflict of interest to declare.
Acknowledgment
This study was conducted and supported by the Iran University of Medical Sciences, Tehran, Iran.
Abbreviations
- Alt-IRF
IRFs other than IRF3 or IRF7
- AP-1
Activator protein 1
- APCs
Antigen-presenting cells
- APS-1
Autoimmune polyendocrinopathy type 1 syndrome
- BAFF
B-cell activating factor
- cGAMP
cyclic di-GMP-AMP
- cGAS
cytosolic GAMP synthase
- CP
Convalescent plasma
- COVID-19
Coronavirus 2019
- DAI
DNA-dependent activator of IRFs
- DDX58
DExD/H-Box helicase 58
- DCs
Dendritic cells
- DR5
Death receptor 5
- dsRNA
Double-stranded RNA
- eIF2-α
Eukaryotic translation initiation factor 2α
- ER
Endoplasmic reticulum
- GAS
γ-activated site
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- IFNs
Interferons
- IFN-I
Type-I interferons
- IFN-γ
Recombinant interferon gamma
- IFITM
IFN-inducible transmembrane
- IFNAR
IFN alpha-receptor
- IFNR
Interferon receptor
- IRF
IFN-regulatory factor
- ISGs
Interferon-stimulated genes
- ISREs
IFN-stimulated response elements
- IKKε
IκB kinase-ε
- IL-10
Interleukin-10
- IVIG
Intravenous immunoglobulin
- JAK1
Tyrosine kinases Janus kinase 1
- LGP2
Laboratory of genetics and physiology 2
- LIF
Leukemia inhibitory factor
- M
Membrane
- MAPK
Multiple mitogen-activated protein kinases
- MAV
Mitochondrial antiviral signaling protein
- MAVS
Mitochondrial antiviral signaling protein
- MDA5
Melanoma differentiation-associated gene 5
- MIP-1α
Macrophage inflammatory protein-1 alpha
- MS
Multiple sclerosis
- mTOR
Mammalian target of the rapamycin
- MyD88
Myeloid differentiation primary response 88
- N
Nucleocapsid
- NFκB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
Natural killer
- NKG2D
NK group 2 member D
- NLRs
Nucleotide oligomerization domain-like receptors
- NOD2
NOD-containing protein 2
- NS
Nonstructural
- OAS
2′, 5′-oligoadenylate synthetase
- PAMPs
Pathogen-associated molecular patterns
- pDC
Plasmacytoid DC
- PD1
Programmed cell death protein 1
- PDL1
Programmed cell death 1 ligand 1
- PE
Plasma exchange
- PI3K
Phosphoinositide 3-kinases
- PKR
Protein kinase RNA-activated
- PLpros
Papain-like proteases
- PRRs
Pattern recognition receptors
- RA
Rheumatoid arthritis
- RIG
Retinoic acid-inducible gene
- RIG-I
Retinoic acid-inducible gene I
- RLRs
Retinoic acid-inducible gene I (RIG-I)-like receptors
- SAD
Systemic autoimmune diseases
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SLE
Systemic lupus erythematosus
- SSc
Systemic sclerosis
- SS
Sjogren's syndrome
- ssRNA
Single-stranded RNA
- STAT
Signal transducer and activator of transcription
- STING
Stimulator of IFN genes
- TBK1
TANK-binding kinase 1
- TCR
T cell receptor
- TDR7
Tudor domain-containing protein 7
- TLRs
Toll-like receptors
- TNF
Tumor necrosis factor
- TRAM
TLR adaptor molecule
- TRAF
TNF receptor-associated factor
- TRAIL
TNF-related apoptosis-inducing ligand
- TRIF
TIR domain-containing adaptor protein inducing IFNβ
- TTCI
Time to clinical improvement
- TYK2
Tyrosine kinase 2
- VEGF-B
Vascular endothelial growth factor-B
References
- Abbasifard M., Khorramdelazad H. The bio-mission of interleukin-6 in the pathogenesis of COVID-19: a brief look at potential therapeutic tactics. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abers M.S., Rosen L.B., Delmonte O.M., Shaw E., Bastard P., Imberti L., Quaresima V., Biondi A., Bonfanti P., Castagnoli R., Casanova J.L., Su H.C., Notarangelo L.D., Holland S.M., Lionakis M.S. Neutralizing type-I interferon autoantibodies are associated with delayed viral clearance and intensive care unit admission in patients with COVID-19. Immunol. Cell Biol. 2021;99:917–921. doi: 10.1111/imcb.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Ampudia Y., Monsalve D.M., Rojas M., Rodriguez Y., Gallo J.E., Salazar-Uribe J.C., Santander M.J., Cala M.P., Zapata W., Zapata M.I., Manrique R., Pardo-Oviedo J.M., Camacho B., Ramirez-Santana C., Anaya J.M., group C.-C.-. COVID-19 convalescent plasma composition and immunological effects in severe patients. J. Autoimmun. 2021;118 doi: 10.1016/j.jaut.2021.102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Mann-Nüttel R., Schulze A., Richter L., Alferink J., Scheu S. Sources of Type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafu A.A., Bowen C.H., Chin C.R., Brass A.L., Holm G.H. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013;288:17261–17271. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricò E., Bracci L., Castiello L., Gessani S., Belardelli F. Are we fully exploiting type I Interferons in today's fight against COVID-19 pandemic? Cytokine Growth Factor Rev. 2020;54:43–50. doi: 10.1016/j.cytogfr.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Zhong G., Huang I.C., Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annual Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V., Manry J., Shaw E., Haljasmagi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodriguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D'Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., Lab H., Group N.-U.I., Clinicians C., Clinicians C.-S., Imagine C.G., French C.C.S.G., Milieu Interieur C., Co V.C.C., Amsterdam U.M.C.C.-B., Effort C.H.G., Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.Y., Holland S.M., Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D., Abel L., Su H.C., Casanova J.L. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.-H., Eto S., Garcia-Prat M. Autoantibodies neutralizing type I IFNs are present iñ 4% of uninfected individuals over 70 years old and account for∼ 20% of COVID-19 deaths. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Orlova E., Sozaeva L., Levy R., James A., Schmitt M.M., Ochoa S., Kareva M., Rodina Y., Gervais A., Le Voyer T., Rosain J., Philippot Q., Neehus A.L., Shaw E., Migaud M., Bizien L., Ekwall O., Berg S., Beccuti G., Ghizzoni L., Thiriez G., Pavot A., Goujard C., Fremond M.L., Carter E., Rothenbuhler A., Linglart A., Mignot B., Comte A., Cheikh N., Hermine O., Breivik L., Husebye E.S., Humbert S., Rohrlich P., Coaquette A., Vuoto F., Faure K., Mahlaoui N., Kotnik P., Battelino T., Trebusak Podkrajsek K., Kisand K., Ferre E.M.N., DiMaggio T., Rosen L.B., Burbelo P.D., McIntyre M., Kann N.Y., Shcherbina A., Pavlova M., Kolodkina A., Holland S.M., Zhang S.Y., Crow Y.J., Notarangelo L.D., Su H.C., Abel L., Anderson M.S., Jouanguy E., Neven B., Puel A., Casanova J.L., Lionakis M.S. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A.L., Smith D.W., Cummins M.J., Jacoby P.A., Cummins J.M., Beilharz M.W. Low‐dose oral interferon alpha as prophylaxis against viral respiratory illness: a double‐blind, parallel controlled trial during an influenza pandemic year. Influenza Respiratory. Virus. 2013;7:854–862. doi: 10.1111/irv.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzotto J., Sanchis P., Abbate M., Lage-Vickers S., Lavignolle R., Toro A., Olszevicki S., Sabater A., Cascardo F., Vazquez E., Cotignola J., Gueron G. SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. iScience. 2020;23 doi: 10.1016/j.isci.2020.101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojdani E., Rajagopalan A., Chen A., Gearin P., Olcott W., Shankar V., Cloutier A., Solomon H., Naqvi N.Z., Batty N. COVID-19 pandemic: impact on psychiatric care in the United States. Psychiatr. Res. 2020;289 doi: 10.1016/j.psychres.2020.113069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianchi M.R., Uleri E., Caglioti C., Dolei A. Type I IFN family members: similarity, differences and interaction. Cytokine Growth Factor Rev. 2015;26:103–111. doi: 10.1016/j.cytogfr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.-a. In fatal COVID-19, the immune response can control the virus but kill the patient. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:30009–30011. doi: 10.1073/pnas.2021128117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Aleem A., Dulebohn S., Di Napoli R. Treasure Island; StatPearls: 2021. Features, Evaluation, and Treatment of Coronavirus (COVID-19) [PubMed] [Google Scholar]
- Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.E., Feng A., Meng W., Apostolidis S.A., Mack E., Artandi M., Barman L., Bennett K., Chakraborty S., Chang I. New-onset IgG autoantibodies in hospitalized patients with COVID-19. Nat. Commun. 2021;12:1–15. doi: 10.1101/2021.01.27.21250559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Sen G.C. Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 2014;25:533–541. doi: 10.1016/j.cytogfr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrialsgov U.S. National library of medicine. 2022. https://clinicaltrials.gov/ct2/show/NCT05054114/
- Comar C.E., Goldstein S.A., Li Y., Yount B., Baric R.S., Weiss S.R. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS Coronavirus infection. mBio. 2019;10 doi: 10.1128/mBio.00319-19. e00319-00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M.K., Ronnblom L. Type I interferons in host defence and inflammatory diseases. Lupus Sci. Med. 2019;6 doi: 10.1136/lupus-2019-000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva R.P., Goncalves J.I.B., Zanin R.F., Schuch F.B., de Souza A.P.D. Circulating type I interferon levels and COVID-19 severity: a systematic review and meta-analysis. Front. Immunol. 2021;12:657363. doi: 10.3389/fimmu.2021.657363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429:1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darazam I.A., Shokouhi S., Pourhoseingholi M.A., Irvani S.S.N., Mokhtari M., Shabani M., Amirdosara M., Torabinavid P., Golmohammadi M., Hashemi S. Role of interferon therapy in severe COVID-19: the COVIFERON randomized controlled trial. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-86859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoudi-Monfared E., Rahmani H., Khalili H., Hajiabdolbaghi M., Salehi M., Abbasian L., Kazemzadeh H., Yekaninejad M.S. A randomized clinical trial of the efficacy and safety of interferon beta-1a in treatment of severe COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01061-20. e01061-01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Padilla C.M.L., Niewold T.B. The type I interferons: basic concepts and clinical relevance in immune-mediated inflammatory diseases. Gene. 2016;576:14–21. doi: 10.1016/j.gene.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Prost N., Bastard P., Arrestier R., Fourati S., Mahevas M., Burrel S., Dorgham K., Gorochov G., Tandjaoui-Lambiotte Y., Azzaoui I., Fernandes I., Combes A., Casanova J.L., Mekontso-Dessap A., Luyt C.E. Plasma exchange to rescue patients with autoantibodies against type I interferons and life-threatening COVID-19 pneumonia. J. Clin. Immunol. 2021;41:536–544. doi: 10.1007/s10875-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weerd N.A., Nguyen T. The interferons and their receptors—distribution and regulation. Immunol. Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnoosh G., Ghanei M., Khorramdelazad H., Alishiri G., Farahani A.J., Shahriary A., Zijoud S.R.H. Are Iranian sulfur mustard gas-exposed survivors more vulnerable to SARS-CoV-2? Some similarity in their pathogenesis. Disaster Med. Public Health Prep. 2020;14:826–832. doi: 10.1017/dmp.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld J.J., Kandel C., Biondi M.J., Kozak R.A., Zahoor M.A., Lemieux C., Borgia S.M., Boggild A.K., Powis J., McCready J., Tan D.H.S., Chan T., Coburn B., Kumar D., Humar A., Chan A., O'Neil B., Noureldin S., Booth J., Hong R., Smookler D., Aleyadeh W., Patel A., Barber B., Casey J., Hiebert R., Mistry H., Choong I., Hislop C., Santer D.M., Lorne Tyrrell D., Glenn J.S., Gehring A.J., Janssen H.L.A., Hansen B.E. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir. Med. 2021;9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves D., Mezidi M., Bastard P., Perret M., Saker K., Fabien N., Pescarmona R., Lombard C., Walzer T., Casanova J.L., Belot A., Richard J.C., Trouillet-Assant S. Antibodies against type I interferon: detection and association with severe clinical outcome in COVID-19 patients. Clinic. Trans. Immunol. 2021;10:e1327. doi: 10.1002/cti2.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Nakabo S., Chu J., Hasni S., Kaplan M.J. medrxiv; 2020. Association between Anti-interferon-alpha Autoantibodies and COVID-19 in Systemic Lupus Erythematosus. 10.29.20222000. [DOI] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T., Ha L.D., Ban V.V., Matsushita I., Yanai H., Kirikae F., Kirikae T., Kuratsuji T., Sasazuki T., Keicho N. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 2005;329:1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari D., Kuhn N., Abramovich R., Sasson K., Zozulya A.L., Smith P., Schlapschy M., Aharoni R., Koster M., Eilam R., Skerra A., Schreiber G. Enhanced in vivo efficacy of a type I interferon superagonist with extended plasma half-life in a mouse model of multiple sclerosis. J. Biol. Chem. 2014;289:29014–29029. doi: 10.1074/jbc.M114.602474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins P., Phillpotts R., Scott G., Wallace J., Bernhardt L., Tyrrell D. Intranasal interferon as protection against experimental respiratory coronavirus infection in volunteers. Antimicrob. Agents Chemother. 1983;24:713–715. doi: 10.1128/AAC.24.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.-C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan P., Andrews J.R., Bonilla H., Hedlin H., Jacobson K.B., Balasubramanian V., Purington N., Kamble S., de Vries C.R., Quintero O., Feng K., Ley C., Winslow D., Newberry J., Edwards K., Hislop C., Choong I., Maldonado Y., Glenn J., Bhatt A., Blish C., Wang T., Khosla C., Pinsky B.A., Desai M., Parsonnet J., Singh U. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-22177-1. 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamsi R. Rogue antibodies could be driving severe COVID-19. Nat. Commun. 2021;590 doi: 10.1038/d41586-021-00149-1. [DOI] [PubMed] [Google Scholar]
- Khorramdelazad H., Kazemi M.H., Najafi A., Keykhaee M., Emameh R.Z., Falak R. Immunopathological similarities between COVID-19 and influenza: investigating the consequences of Co-infection. Microb. Pathog. 2021;152 doi: 10.1016/j.micpath.2020.104554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V., Weber F. Interaction of SARS and MERS Coronaviruses with the antiviral interferon response. Adv. Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konde M.K., Baker D.P., Traore F.A., Sow M.S., Camara A., Barry A.A., Mara D., Barry A., Cone M., Kaba I., Richard A.A., Beavogui A.H., Gunther S., European Mobile Laboratory C., Pintilie M., Fish E.N. Interferon beta-1a for the treatment of Ebola virus disease: a historically controlled, single-arm proof-of-concept trial. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähling V., Stein D.A., Spiegel M., Weber F., Mühlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J. Virol. 2009;83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel S., Till A., Winkler M., Häsler R., Lipinski S., Jung S., Grötzinger J., Fickenscher H., Schreiber S., Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J. Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Shin E.C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.S.W., Rojas O.L., Gommerman J.L. B cell depletion therapies in autoimmune disease: advances and mechanistic insights. Nat. Rev. Drug Discov. 2021;20:179–199. doi: 10.1038/s41573-020-00092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew T.W., Kwek T.K., Tai D., Earnest A., Loo S., Singh K., Kwan K.M., Chan Y., Yim C.F., Bek S.L., Kor A.C., Yap W.S., Chelliah Y.R., Lai Y.C., Goh S.K. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA. 2003;290:374–380. doi: 10.1001/jama.290.3.374. [DOI] [PubMed] [Google Scholar]
- Li Y., Renner D.M., Comar C.E., Whelan J.N., Reyes H.M., Cardenas-Diaz F.L., Truitt R., Tan L.H., Dong B., Alysandratos K.D., Huang J., Palmer J.N., Adappa N.D., Kohanski M.A., Kotton D.N., Silverman R.H., Yang W., Morrisey E.E., Cohen N.A., Weiss S.R. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2022643118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Shen K. Type I interferon: from innate response to treatment for COVID-19. Pediatric. Invest. 2020;4:275–280. doi: 10.1002/ped4.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Xu A., Zhang Y., Xuan W., Yan T., Pan K., Yu W., Zhang J. Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int. J. Infect. Dis. 2020;95:183–191. doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutfy M.R., Blatt L.M., Siminovitch K.A., Ward S., Wolff B., Lho H., Pham D.H., Deif H., LaMere E.A., Chang M., Kain K.C., Farcas G.A., Ferguson P., Latchford M., Levy G., Dennis J.W., Lai E.K., Fish E.N. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- Maddur M.S., Rabin M., Hegde P., Bolgert F., Guy M., Vallat J.-M., Magy L., Bayry J., Kaveri S.V. Intravenous immunoglobulin exerts reciprocal regulation of Th1/Th17 cells and regulatory T cells in Guillain–Barré syndrome patients. Immunol. Res. 2014;60:320–329. doi: 10.1007/s12026-014-8580-6. [DOI] [PubMed] [Google Scholar]
- Madera S., Rapp M., Firth M.A., Beilke J.N., Lanier L.L., Sun J.C. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med. 2016;213:225–233. doi: 10.1084/jem.20150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., Hartmann R., Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179 doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary A., Hénaut L., Macq P.Y., Badoux L., Cappe A., Porée T., Eckes M., Dupont H., Brazier M. Rationale for COVID-19 treatment by nebulized interferon-β-1b–Literature review and personal preliminary experience. Front. Pharmacol. 2020 doi: 10.3389/fphar.2020.592543. 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Pinto C., Garcia-Carrasco M., Munguia Realpozo P., Mendez-Martinez S. Therapeutic options for the management of severe COVID-19: a rheumatology perspective. Reumatol. Clínica. 2021;17:431–436. doi: 10.1016/j.reumae.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Wang T., Li C., Chen X., Li L., Qin X., Li H., Luo J. An experimental trial of recombinant human interferon alpha nasal drops to prevent coronavirus disease 2019 in medical staff in an epidemic area. medRxiv. 2020 doi: 10.1101/2020.04.11.20061473. [DOI] [Google Scholar]
- Merigan T.C., Reed S.E., Hall T.S., Tyrrell D.A. Inhibition of respiratory virus infection by locally applied interferon. Lancet. 1973;1:563–567. doi: 10.1016/s0140-6736(73)90714-9. [DOI] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., Garcia-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.P., Clark T., Djukanovic R., Wilkinson T.M.A., Inhaled Interferon Beta, C.-S.G Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynagh P.N. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends Immunol. 2005;26:469–476. doi: 10.1016/j.it.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Murira A., Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front. Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikov A.L., Berns S.A., Talyzin P.A., Ershov F.I. [Interferon gamma in the treatment of patients with moderate COVID-19] Prob. Virol. 2021;66:47–54. doi: 10.36233/0507-4088-24. [DOI] [PubMed] [Google Scholar]
- Nesteruk I. medRxiv; 2020. Waves of COVID-19 Pandemic. Detection and SIR Simulations. [DOI] [Google Scholar]
- Ng C.S., Kato H., Fujita T. Fueling type I interferonopathies: regulation and function of type I interferon antiviral responses. J. Interferon Cytokine Res. 2019;39:383–392. doi: 10.1089/jir.2019.0037. [DOI] [PubMed] [Google Scholar]
- Palermo E., Di Carlo D., Sgarbanti M., Hiscott J. Type I interferons in COVID-19 pathogenesis. Biology. 2021;10:829. doi: 10.3390/biology10090829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kang K., Giannopoulou E., Qiao Y., Kang K., Kim G., Park-Min K.H., Ivashkiv L.B. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 2017;18:1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., Walter M.R. Interferons, interferon‐like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Prelli Bozzo C., Nchioua R., Volcic M., Koepke L., Kruger J., Schutz D., Heller S., Sturzel C.M., Kmiec D., Conzelmann C., Muller J., Zech F., Braun E., Gross R., Wettstein L., Weil T., Weiss J., Diofano F., Rodriguez Alfonso A.A., Wiese S., Sauter D., Munch J., Goffinet C., Catanese A., Schon M., Boeckers T.M., Stenger S., Sato K., Just S., Kleger A., Sparrer K.M.J., Kirchhoff F. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Strayer D., Dickey R., A Carter W. Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect. Disord. - Drug Targets. 2014;14:37–43. doi: 10.2174/1871526514666140713152858. [DOI] [PubMed] [Google Scholar]
- Reddy K.R., Modi M.W., Pedder S. Use of peginterferon alfa-2a (40 KD)(Pegasys®) for the treatment of hepatitis C. Adv. Drug Deliv. Rev. 2002;54:571–586. doi: 10.1016/S0169-409X(02)00028-5. [DOI] [PubMed] [Google Scholar]
- Rostaing L., Chatelut E., Payen J.L., Izopet J., Thalamas C., Ton-That H., Pascal J.P., Durand D., Canal P. Pharmacokinetics of alphaIFN-2b in chronic hepatitis C virus patients undergoing chronic hemodialysis or with normal renal function: clinical implications. J. Am. Soc. Nephrol. 1998;9:2344–2348. doi: 10.1681/ASN.V9122344. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G. The role of type I interferons in the pathogenesis and treatment of COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.595739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi V. Convalescent plasma: a challenging tool to treat COVID-19 patients—a lesson from the past and new perspectives. BioMed Res. Int. 2020 doi: 10.1155/2020/2606058. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S., Barjaktarevic I.Z., Tashkin D.P. The use of nebulized pharmacotherapies during the COVID-19 pandemic. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620954366. 1753466620954366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Muller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeifian F., Nikfarjam M., Kian N., Mohamed K., Rezaei N. The role of type I interferon in the treatment of COVID-19. J. Med. Virol. 2022;94:63–81. doi: 10.1002/jmv.27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanich X., Rigo-Bonnin R., Gumucio V.-D., Bastard P., Rosain J., Philippot Q., Perez-Fernandez X.-L., Fuset-Cabanes M.-P., Gordillo-Benitez M.-Á., Suarez-Cuartin G. Pre-existing autoantibodies neutralizing high concentrations of type I interferons in almost 10% of COVID-19 patients admitted to intensive care in barcelona. J. Clin. Immunol. 2021;41:1733–1744. doi: 10.1007/s10875-021-01136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegelmeier A.A., Darzianiazizi M., Hanada K., Sharif S., Wootton S.K., Bridle B.W., Karimi K. Type I interferon-mediated regulation of antiviral capabilities of neutrophils. Int. J. Mol. Sci. 2021;22:4726. doi: 10.3390/ijms22094726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian G., Kuzmanovic T., Zhang Y., Peter C.B., Veleeparambil M., Chakravarti R., Sen G.C., Chattopadhyay S. A new mechanism of interferon's antiviral action: induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J., Li X., Xi Y., Chen R., Alshukairi A.N., Chen Z., Zhang Z., Chen C., Huang X., Li F., Lai X., Chen D., Wen L., Zhuo J., Zhang Y., Wang Y., Huang S., Dai J., Shi Y., Zheng K., Leidinger M.R., Chen J., Li Y., Zhong N., Meyerholz D.K., McCray P.B., Jr., Perlman S., Zhao J. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743 e735. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Hemmi H., Akira S. Interferon response induced by Toll-like receptor signaling. J. Endotoxin Res. 2004;10:252–256. doi: 10.1179/096805104225005896. [DOI] [PubMed] [Google Scholar]
- Tamura T., Yanai H., Savitsky D., Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Teijaro J.R. Type I interferons in viral control and immune regulation. Curr. Opinion. Virol. 2016;16:31–40. doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R., Sims A.C., Baric R.S., Silverman R.H., Weiss S.R. Middle East respiratory syndrome Coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7 doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne R.G., Hanson L.R., Ross T.M., Tung D., Frey W.H., 2nd Delivery of interferon-beta to the monkey nervous system following intranasal administration. Neuroscience. 2008;152:785–797. doi: 10.1016/j.neuroscience.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe? J. Exp. Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troya J., Bastard P., Planas-Serra L., Ryan P., Ruiz M., de Carranza M., Torres J., Martínez A., Abel L., Casanova J.L., Pujol A. Neutralizing autoantibodies to type I IFNs in >10% of patients with severe COVID-19 pneumonia hospitalized in Madrid, Spain. J. Clin. Immunol. 2021;41:914–922. doi: 10.1007/s10875-021-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., Lee D.S., Greenland J.R., Sun Y., Perez R., Ogorodnikov A., Ward A., Mann S.A., Lynch K.L., Yun C., Havlir D.V., Chamie G., Marquez C., Greenhouse B., Lionakis M.S., Norris P.J., Dumont L.J., Kelly K., Zhang P., Zhang Q., Gervais A., Le Voyer T., Whatley A., Si Y., Byrne A., Combes A.J., Rao A.A., Song Y.S., Fragiadakis G.K., Kangelaris K., Calfee C.S., Erle D.J., Hendrickson C., Krummel M.F., Woodruff P.G., Langelier C.R., Casanova J.L., Derisi J.L., Anderson M.S., Ye C.J., consortium U.C. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abh2624. eabh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012;86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M.R. The role of structure in the biology of interferon signaling. Front. Immunol. 2020;11:606489. doi: 10.3389/fimmu.2020.606489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D.J.C.h., microbe . vol. 28. 2020. pp. 455–464. (Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients). e452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.Y., Mao T., Klein J., Dai Y., Huck J.D., Jaycox J.R., Liu F., Zhou T., Israelow B., Wong P. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–288. doi: 10.1101/2020.12.10.20247205. [DOI] [PubMed] [Google Scholar]
- Who WHO Coronavirus (COVID-19) dashboard. 2022. https://covid19.who.int/ 30 February 2022.
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstone H., Lista M.J., Reid A.C., Bouton C., Pickering S., Galao R.P., Kerridge C., Doores K.J., Swanson C.M., Neil S.J.D. The polybasic Cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J. Virol. 2021;95 doi: 10.1128/JVI.02422-20. e02422-02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometer C.O.V.I.D. 2022. https://www.worldometers.info/coronavirus/. coronavirus pandemic 30 February 2022.
- Wrensch F., Winkler M., Pohlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner D.H., McCauley J.W., Skehel J.J., Kerr I.M. Interferon action—sequence specificity of the ppp (A2′p) n A-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H.C., Williams B.R. Protein kinase R and the inflammasome. J. Interferon Cytokine Res. 2014;34:447–454. doi: 10.1089/jir.2014.0008. [DOI] [PubMed] [Google Scholar]
- Zahid A., Ismail H., Li B., Jin T. Molecular and structural basis of DNA sensors in antiviral innate immunity. Front. Immunol. 2020;11:613039. doi: 10.3389/fimmu.2020.613039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zav'yalov V.P., Hamalainen-Laanaya H., Korpela T.K., Wahlroos T. Interferon-inducible myxovirus resistance proteins: potential biomarkers for differentiating viral from bacterial infections. Clin. Chem. 2019;65:739–750. doi: 10.1373/clinchem.2018.292391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Bolze A., Jouanguy E., Zhang S.Y., Effort C.H.G., Cobat A., Notarangelo L.D., Su H.C., Abel L., Casanova J.L. Life-Threatening COVID-19: defective interferons unleash excessive inflammation. Med (N Y) 2020;1:14–20. doi: 10.1016/j.medj.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Wang W. Auto-antibodies against type I IFNs are associated with severe COVID-19 pneumonia. Signal Transduct. Targeted Ther. 2021;6:96. doi: 10.1038/s41392-021-00514-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chen V., Shannon C.P., Wei X.-S., Xiang X., Wang X., Wang Z.-H., Tebbutt S.J., Kollmann T.R., Fish E.N. Interferon-α2b treatment for COVID-19. Front. Immunol. 2020;11:1061. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890 e882. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]