Abstract
A fluorescence in situ hybridization (FISH) technique based on binding of a rhodamine-labelled oligonucleotide probe to 16S rRNA was used to estimate the numbers of ribosome-rich bacteria in soil samples. Such bacteria, which have high cellular rRNA contents, were assumed to be active (and growing) in the soil. Hybridization to an rRNA probe, EUB338, for the domain Bacteria was performed with a soil slurry, and this was followed by collection of the bacteria by membrane filtration (pore size, 0.2 μm). A nonsense probe, NONEUB338 (which has a nucleotide sequence complementary to the nucleotide sequence of probe EUB338), was used as a control for nonspecific staining. Counting and size classification into groups of small, medium, and large bacteria were performed by fluorescence microscopy. To compensate for a difference in the relative staining intensities of the probes and for binding by the rhodamine part of the probe, control experiments in which excess unlabelled probe was added were performed. This resulted in lower counts with EUB338 but not with NONEUB338, indicating that nonspecific staining was due to binding of rhodamine to the bacteria. A value of 4.8 × 108 active bacteria per g of dry soil was obtained for bulk soil incubated for 2 days with 0.3% glucose. In comparison, a value of 3.8 × 108 active bacteria per g of dry soil was obtained for soil which had been air dried and subsequently rewetted. In both soils, the majority (68 to 77%) of actively growing bacteria were members of the smallest size class (cell width, 0.25 to 0.5 μm), but the active (and growing) bacteria still represented only approximately 5% of the total bacterial population determined by DAPI (4′,6-diamidino-2-phenylindole) staining. The FISH technique in which slurry hybridization is used holds great promise for use with phylogenetic probes and for automatic counting of soil bacteria.
For a long time plate counting has been inadequate for estimating the active populations of bacteria in soils (8, 40), and numerous methods have been tested as alternatives. Recently, the frequency of dividing cells (9) and counts of 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyltetrazolium chloride-reducing cells (35), 5-cyano-2,3-ditolyl tetrazolium chloride (CTC)-reducing cells (48), and [3H]thymidine- or [14C]leucine-incorporating cells (3, 15) have been used to determine active bacterial populations. The limitations of these assays include inaccurate conversion factors in the frequency-of-dividing-cell, thymidine, and leucine methods (3, 9, 13, 17), as well as difficulties in using the CTC and thymidine methods with certain soils (15, 48).
The fluorescence in situ hybridization (FISH) method developed recently has great potential for improving determinations of active populations of soil bacteria (2, 14, 20, 42, 49). Oligonucleotide probes are designed based on signature nucleotide positions in the bacterial 16S rRNA and may be used to target either a narrow group of organisms or a broad group of organisms. High levels of rRNA (equivalent to high ribosome numbers), which result in high detection signals with the FISH technique, are observed in active populations of bacteria in which protein synthesis is occurring either in nondividing or dividing cells (12, 30, 37). In contrast, starved bacteria are known to contain low levels of rRNA (18) but might be detected by FISH under some circumstances (44). Similarly, 16S rRNA levels decrease below the detection level in stressed bacteria (44), and moribund bacteria are not expected to contain detectable levels of rRNA.
The majority of soil bacteria are known to be less than 0.5 μm in diameter (6, 7, 8, 17, 27), and it has been suggested that this population of small bacteria consists mainly of gram-positive organisms (32). These bacteria do not grow well under laboratory conditions (7), and their growth rate, as determined by the thymidine technique, may be lower than the growth rate of larger bacteria (4). Still, the polulation dynamics and functional role of the small bacteria are unclear (45). With the FISH technique it may be possible to directly detect the active bacterial populations in the soil and to determine the size class distribution and the responses of the bacteria to different soil conditions.
FISH protocols for bacterial cells that are extracted from soil (20) and bacterial cells that occur in smears (49) are difficult to perform because bacteria associated with soil particles are excluded and because the background fluorescence signals from the soil smears are high, respectively. To determine bacterial numbers in soil without an extraction step, a slurry hybridization protocol followed by membrane filtration and fluorescence microscopy was developed (14). By this procedure, it is possible to include the important fraction of adsorbed bacteria reported to exhibit a high level of activity (5) and to exclude the high background signal from minute clay and organic soil particles by collecting bacterium-sized particles with a filter. It should be noted that the slurry hybridization-membrane filtration method was originally developed with laboratory-grown bacteria added to soil. Hence, when indigenous soil bacteria are counted, some limitations due to nonspecific staining may be encountered. The aims of this investigation were, therefore, to determine the populations of active bacteria belonging to different size groups in soil and to modify the slurry hybridization protocol to compensate for nonspecific staining in soil samples.
MATERIALS AND METHODS
Soil sampling and treatments.
Bulk soil samples were collected from a sandy loam soil (63% sand, 17% silt, 17% clay, 1.1% C, pH [CaCl2] 5.5 [26]) at an experimental farm (Højbakkegård, Tåstrup, Denmark). This freshly collected soil was used during the developmental study to modify the slurry hybridization protocol. In additional experiments, glucose was added or a drying-rewetting cycle was included as described below before the bacterial populations were counted. All of the soil samples used for hybridization (5 to 10 g) were fixed in 100 ml of an ice-cold (pH 7.0-buffered) paraformaldehyde solution (1). After fixation overnight at 4°C, each sample was homogenized three times for 1 min in a Waring blender operated at the maximum speed. The cup containing blended soil was cooled on ice for 3 min between homogenization steps. The blended sample was stored at −20°C after 140 ml of filter-sterilized distilled water, 10 ml of 1 M Tris-HCl buffer (pH 7.5), and 250 ml of 96% ethanol were added. The dry weight of the soil sample was calculated on the basis of the soil moisture content.
Total counts of soil bacteria as determined by DAPI staining.
DAPI (4′,6-diamidino-2-phenylindole) staining was used to determine the total numbers of soil bacteria. Samples corresponding to 0.1 mg (dry weight) were stained in 10 ml of filter-sterilized distilled water containing 1 μg of DAPIs per ml for 10 min (14, 36). Bacteria were collected by filtration through a 0.2-μm-pore-size black polycarbonate filter (diameter, 25 mm; Millipore). The filters were mounted in paraffin oil, and counts were determined with a fluorescence microscope as described below for the filter specimens obtained from in situ slurry hybridization experiments.
In situ slurry hybridization with rhodamine-labelled oligonucleotide probes.
To target the active soil bacteria, 16S rRNA probe EUB338 (for the domain Bacteria) having the nucleotide sequence 5′-TGAGGATGCCCTCCGTCG-3′ (43) was used. A different probe, NONEUB338, having the nucleotide sequence complementary to the nucleotide sequence of EUB338, was used to determine the nonspecific binding of a probe to the soil bacteria. The nucleotide probes were labelled with rhodamine (tetramethyl rhodamine isothiocyanate) (Sigma Chemical Co., St. Louis, Mo.) at a 1:1 ratio by using an amino linker (Applied Biosystems, Foster City, Calif.), and the rhodamine-labelled probes were purified by high-performance liquid chromatography. The staining intensities of different batches of the probes varied slightly because of slight differences in the ratio of rhodamine to oligonucleotide (data not shown).
Sample volumes equivalent to 0.5 mg (dry weight) of soil were taken directly from the blended samples while they were being stirred magnetically. A dehydration process in which a sample was centrifuged (10,000 × g, 5 min) and then resuspended in 80 and 100% ethanol was used (14). Soil pellets were then vacuum dried to evaporate the ethanol and resuspended in 200 μl of standard hybridization buffer (0.9 M NaCl, 20 mM Tris [pH 7.0]) containing 0.1% Nonidet P-40 detergent (Sigma). The samples were centrifuged again, and each pellet was resuspended in 200 μl of hybridization buffer containing 15% formamide and incubated at 37°C for 30 min. The effects of formamide concentrations ranging from 0 to 30% on the cell counts were determined during the developmental study.
Then rhodamine-labelled probe EUB338 or NONEUB338 was added to each slurry at a concentration of 12 ng μl−1. This concentration was found to result in optimal staining, and higher probe concentrations did not result in higher cell numbers (data not shown). The standard protocol developed also included adding unlabelled probes EUB338 and NONEUB338 at a concentration of 36 ng μl−1 together with rhodamine-labelled probes EUB338 and NONEUB338, respectively. The effects of concentrations of unlabelled probe ranging from 0 to 36 ng μl−1 on the cells counts were determined during the developmental study.
Hybridization took place at 37°C for 18 h by using continuous rotation in a hybridization oven. In comparison, a shorter hybridization time (3 h) resulted in lower counts of indigenous bacteria (the counts were reduced 50% or more) (data not shown). After hybridization each slurry was transferred to 10 ml of hybridization buffer containing 15% formamide and incubated at 37°C for 10 min. The significance of this washing procedure was determined during the developmental study by using wash times ranging from 2 to 60 min.
After hybridization, the soil bacteria were collected by filtration onto 0.2-μm-pore-size black polycarbonate filters (diameter, 25 mm; Millipore) in a filtration manifold with stainless steel chimneys which were prewarmed to the hybridization temperature. The filters were rinsed with 20 ml of filter-sterilized distilled water at room temperature (25°C) and stored at 4°C until fluorescence microscopy counting was performed (within 1 week). For longer periods of storage, the filters were frozen at −20°C. Prior to microscopy, the filters were mounted on glass slides in liquid paraffin oil.
Fluorescence microscopy counting and determining the size classes of active soil bacteria.
To count the bacteria in different size classes, a Zeiss Axioplan 2 fluorescence microscope equipped with a Plan-Neofluar objective (magnification, ×100; numerical aperture, 1.3) and a 100-W type HBO high-pressure mercury lamp was used. Zeiss no. 2 and 15 filters were used to visualize bacteria stained with DAPI and rhodamine, respectively. Each bacterium was assigned to one of the following three size classes as defined by spheres of the G12 New Porton graticule (27, 34): small, medium, and large, representing cell widths of 0.25 to 0.5, 0.5 to 0.7, and 0.7 to 1.0 μm, respectively. This classification was chosen since bacteria belonging to the same species predominantly change in cell length rather than in cell width in response to variable growth conditions (24).
For soil samples hybridized with a rhodamine-labelled probe, autofluorescence may sometimes give erroneous results (14, 28). Therefore, a bacterial cell was counted only if autofluorescence was absent, which was ascertained by placing a Zeiss no. 2 filter (normally used for DAPI staining with UV light) immediately behind the rhodamine filter without moving the microscopic field. Autofluorescing cells were never detected with the Zeiss no. 15 filter when samples were prepared for microscopy without the rhodamine probe.
During the developmental study, at least 200 fields of 50 by 50 square units of the G12 New Porton graticule were inspected for each filter. In subsequent studies of treated soils, at least four replicates of hybridization experiments were used, and at least 100 cells were counted for each sample. At low cell densities a total of 600 microscopic fields were inspected, which corresponded to 1% of the sample filter. Statistical differences between counts obtained with different treatments were analyzed by a d test (10). The d test was used because the variance was not the same for all of the data sets compared. Comparable levels of variance are required to perform the t test. The d test follows a Fisher-Behrens distribution, as described by Campbell (10).
For selected samples confocal laser scanning microscopy (CLSM) was used to confirm the distinction of active bacteria from nonactive bacteria or nonbacterial soil particles. The type of confocal microscope used (model TCS4d; Leica Laser Technik, Heidelberg, Germany) and the associated equipment have been described previously by Hansen et al. (23). In this study, we used soil samples pretreated with glucose to stimulate bacterial activity. The filters were first scanned at random by using the optical part of the confocal microscope, and active bacteria were separated from nonactive bacteria and nonbacterial soil particles by using the following criteria: light intensity, morphology, and color. Particles were confirmed to be active bacteria by examining pictures taken by CLSM. Each recording consisted of a stack of images with a vertical distance of 0.3 μm. The stack was subsequently combined into one image by maximum intensity projection performed by the software of the model TCS4d microscope.
RESULTS
Optimization of hybridization conditions.
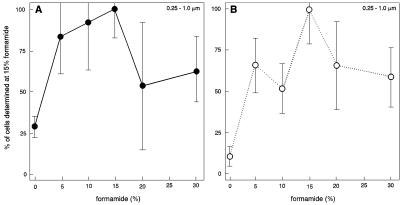
To count indigenous, active soil bacteria, the protocol for in situ slurry hybridization (14) had to be optimized, notably because nonspecific staining was expected to be significant. Addition of formamide to the hybridization buffer is known to decrease the melting point of the DNA double helix (11), and the optimal formamide concentration for hybridization at 37°C was determined to obtain a high-stringency protocol. Figure 1 shows the effects of different formamide concentrations on the hybridization signal for both specific probe EUB338 and nonspecific probe NONEUB338. The total population of cells generally gave the highest counts in the presence of 15% formamide for both probe EUB338 and probe NONEUB338, and this standard concentration (15% formamide in hybridization buffer) was used for all subsequent experiments. All three size classes of bacteria exhibited the same pattern as the total bacterial population (data not shown). Hence, significantly higher numbers were obtained for the small size class of bacteria with the EUB338 probe when 15% formamide was used than when other levels of formamide were used, and for the medium size class significantly higher numbers were obtained in the presence of 5, 10, 15, and 20% formamide than in the presence of 0 and 30% formamide. When the NONEUB338 probe was used, the numbers obtained in the presence of 15% formamide were significantly higher than the numbers obtained in the presence of 0 and 10% formamide for the small size class and significantly higher than the numbers obtained in the absence of formamide for the medium size class.
FIG. 1.
Total numbers of cells (sums of values for all size groups) targeted with specific probe EUB338 (A) or nonspecific probe NONEUB338 (B) at different formamide concentrations. The number of cells obtained with 15% formamide was defined as 100%. The bars indicate standard deviations (n = 3).
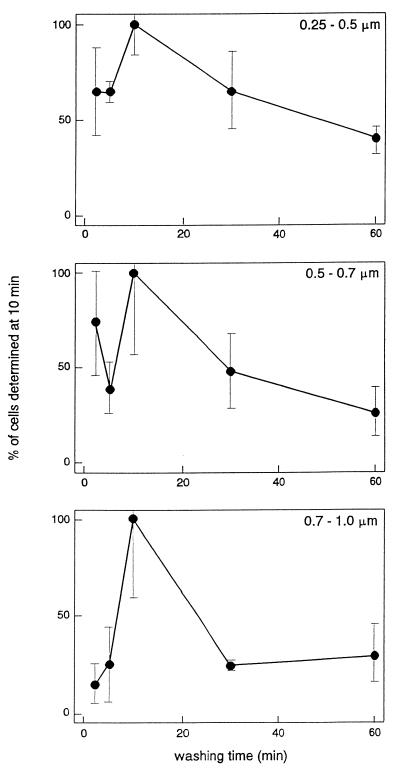
The optimal stringency wash time was determined at a fixed formamide concentration of 15% and a hybridization temperature of 37°C. Figure 2 shows that the highest counts generally were obtained after 10 min of washing. Hence, for the small size class of bacteria, significantly higher numbers were obtained after wash times of 2 and 10 min than after washing for 60 min. For the medium size class, the numbers obtained after 2, 10, and 30 min of washing were significantly higher than the numbers obtained after 5 and 60 min. Finally, for the large size class (and for the total of all size classes), the numbers obtained after 10 min of washing were significantly higher than the numbers obtained after all other wash times. The lower values obtained after 2 and 5 min were probably due to masking of the bacteria by excess stain when the wash time was too short. Hence, the final standard protocol used for in situ slurry hybridization of indigenous active, soil bacteria included a formamide concentration of 15% in the buffer and a reaction time of 18 h at 37°C, followed by 10 min of washing.
FIG. 2.
Numbers of cells in size groups (0.25 to 0.5, 0.5 to 0.7, and 0.7 to 1.0 μm) targeted with specific probe EUB338 after different stringency wash times. The number of cells obtained with a wash time of 10 min was defined as 100%. The bars indicate standard deviations (n = 3).
Finally, probe penetration into gram-positive soil bacteria may be difficult and was investigated for Streptomyces sp. and Frankia sp. by using lysozyme treatment (21, 22). To examine this, we dehydrated soil samples with ethanol and then incubated them in a 1-, 2-, or 3-mg ml−1 lysozyme solution. After this, the dehydration procedure was repeated, and the preparations were washed with 1% toluene in 96% ethanol. The lysozyme treatment had no effect on the numbers of cells targeted by the EUB338 probe (data not shown). This is in agreement with results reported by Roller et al. (39) and Zarda et al. (49), who found that lysozyme did not increase the cell counts of gram-negative bacteria. Treatment with lysozyme was therefore omitted in the final standard protocol for in situ slurry hybridization.
Correction model for nonspecific binding of probe.
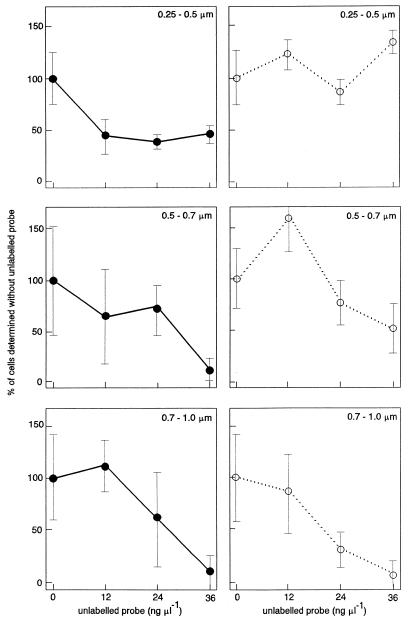
We propose a model for calculating the number of active bacteria in soil (see below) based on the assumption that sufficient unlabelled probe (DNA) should outcompete labelled DNA at both specific and nonspecific DNA binding sites. Figure 3 shows that the cell counts for all three size classes obtained with the EUB338 probe decreased significantly when different amounts of the corresponding unlabelled probe were added. However, no effect of titration with unlabelled probe was observed for the smaller bacteria when they were hybridized with the NONEUB338 probe, indicating that all nonspecific staining in the small cells was due to binding of the rhodamine component of the probe. For medium and large cells, however, the counts obtained with the NONEUB338 probe also decreased as the concentration of unlabelled probe increased. This shows that for larger cells, nonspecific staining caused by DNA binding becomes more important than rhodamine. This is probably due to the lower surface-to-volume ratio of the larger bacteria if it is assumed that rhodamine binding is associated with the cell wall.
FIG. 3.
Numbers of cells in size groups (0.25 to 0.5, 0.5 to 0.7, and 0.7 to 1.0 μm) targeted with specific probe EUB338 (●) or nonspecific probe NONEUB338 (○) in the presence of different concentrations of the corresponding unlabelled probes. The number of cells obtained without unlabelled probes was defined as 100%. The bars indicate standard deviations (n = 3).
The amount of unlabelled probe EUB338 which was sufficient to titrate out binding by DNA of labelled probe EUB338 was defined on basis of the bacteria in the smallest size class because these bacteria were numerically dominant. A constant low number of counts was reached at 12 ng μl−1 (statistically significant), and maximal titration was observed with 36 ng of unlabelled probe per μl for the small size class (Fig. 3). For the medium and large size classes the counts were still decreasing and significantly lower in the presence of 36 ng of unlabelled probe μl−1 than in the presence of 0, 12, and 24 ng of unlabelled probe μl−1; however, at this concentration the counts were only 13 and 9%, respectively, of the counts obtained without unlabelled probe. The numbers of cells in the medium and large size classes represented 7 and 6%, respectively, of the total counts in samples hybridized with labelled EUB338 probe (data not shown). Addition of 36 ng of unlabelled probe μl−1, therefore, reduced the counts to less than 1% of the total counts for the medium and large size classes, and 36 ng of unlabelled probe μl−1 was found to be sufficient to titrate out binding due to the DNA part of the probe.
The model in Table 1 suggests that the numbers of active bacteria determined by in situ slurry hybridization in soil samples can be calculated based on the following four components: (i) homologous binding of the DNA component of the probe to target rRNA; (ii) heterologous binding of the DNA component of the probe to target rRNA; (iii) binding of the DNA component of the probe to nontarget rRNA; and (iv) binding of the rhodamine component of the probe to cell surfaces. In our protocol, the total number of cells determined with labelled, specific probe EUB338 (Nsp) and labelled, nonspecific probe NONEUB338 (Nnon) includes the numbers due to both DNA and rhodamine binding. However, similar samples hybridized with labelled probe plus an excess amount of the corresponding unlabelled probe provide counts (Nsp,unl and Nnon,unl, respectively) resulting from rhodamine binding alone, because the unlabelled probe outcompetes the targets of the labelled probe. To determine the number of cells related to DNA binding alone, the counts related to rhodamine binding are subtracted from the total counts to obtain Nsp − Nsp,unl and Nnon − Nnon,unl, respectively. Finally, the corrected number of active bacteria related to homologous hybridization of the probe to the target rRNA is obtained from (Nsp − Nsp,unl) − (Nnon − Nnon,unl), as indicated in Table 1. While Nsp and Nnon depend on the ratio of rhodamine to DNA in both probe EUB338 and probe NONEUB338, the calculated number of active cells depends only on the labelling of EUB338, because components from nonspecific DNA and rhodamine binding cancel each other out.
TABLE 1.
Model for calculating numbers of actively growing bacteria in soil, as determined by a slurry in situ hybridization protocol performed with rhodamine-labelled oligonucleotide probes targeting rRNA and controls containing excess unlabelled probe
| Designation | Description | Mechanism of binding |
|---|---|---|
| Cell numbers measured | ||
| Nsp | Number of cells hybridized with specific rhodamine-labelled probe (EUB338) | Homologous and heterologous hybridization to rRNA and cell binding by DNA and rhodamine parts of probe |
| Nnon | Number of cells hybridized with nonspecific rhodamine-labelled probe (NONEUB338) | Heterologous hybridization to rRNA and cell binding by DNA and rhodamine parts of probe |
| Nsp,unl | Number of cells hybridized with labelled specific probe and excess of unlabelled specific probe | Cell binding by rhodamine part of probe |
| Nnon,unl | Number of cells hybridized with labelled nonspecific probe and excess of unlabelled nonspecific probe | Cell binding by rhodamine part of probe |
| Cell numbers calculated | ||
| Nsp − Nsp,unl | Homologous and heterologous hybridization to rRNA and cell binding by DNA part of probe | |
| Nnon − Nnon,unl | Heterologous hybridization to rRNA and cell binding by DNA part of probe | |
| (Nsp − Nsp,unl) − (Nnon − Nnon,unl) | Homologous hybridization to rRNA |
Detection limit.
To determine the number of cells targeted by fluorescence-labelled oligonucleotide probes directed against rRNA, the minimum growth rate which can be determined by fluorescence microscopy has to be considered. We investigated the relationship between growth rate and in situ hybridization signal in soil samples which were amended with formalin-killed bacteria obtained from pure cultures of Arthrobacter globiformis DSM 20124T, Pseudomonas putida MM 3 (16), Pseudomonas fluorescens DF 17 (41), and Corynebacterium sp. strain H 19 (13). Bacteria could be detected easily by the EUB338 probe when the generation times were 5 h or less before the bacteria were added to the soil. In contrast, slowly growing bacteria (generation times, 1 to 2 days or more) could not be detected by the probe (data not shown). The range of generation times between 5 h and 1 day was not investigated.
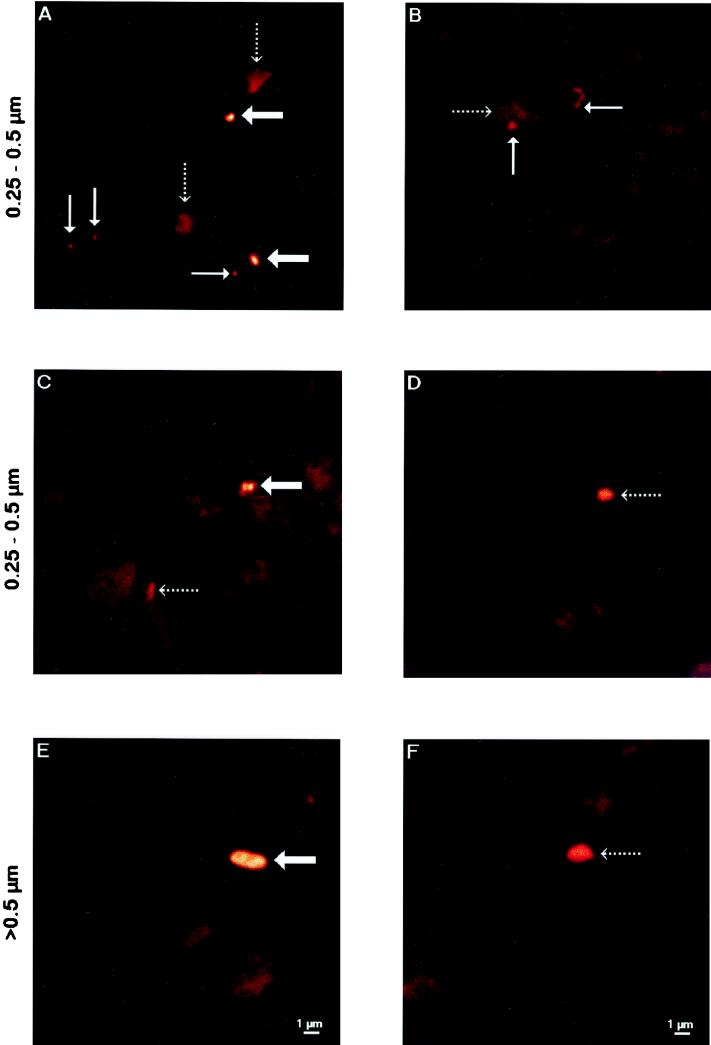
Bacterial cells were counted with the conventional fluorescence microscope based on staining intensity, cell shape, and color. To test our identification of the active bacteria, selected soil samples were investigated by CLSM by using the improved spatial resolution and narrow emission spectrum of this instrument to give high-quality images. Active bacteria, nonactive bacteria, or nonbacterial soil particles were first identified and assigned to size classes by eye by using conventional fluorescence microscopy. Without moving the microscopic field, we then recorded the images by CLSM. As Fig. 4 shows, the particles identified as active bacterial cells by eye had sharper boundaries when they were analyzed by CLSM than the nonbacterial soil particles had, whose shapes were blurred. Furthermore, the light intensity was greater for the active bacteria than for the nonactive bacteria.
FIG. 4.
CLSM images of active bacteria (broad arrows), nonactive bacteria (thin arrows), and nonbacterial soil particles (dotted arrows) preselected randomly by conventional microscopy following in situ slurry hybridization (EUB338 probe). Active and nonactive bacteria representing different size classes (0.25 to 0.5 μm [A through D] and 0.5 to 0.7 μm [E and F]) are shown. The size bars in panels E and F apply to all images.
Case study of pretreated and untreated soils.
The final standard hybridization protocol, including the correction for nonspecific binding, was tested with soil samples representing the following conditions: (i) soil which had been air dried for 2 days and then amended with a glucose solution (final concentration, 3 mg of glucose per g [dry weight] of soil) by rewetting the preparation to field capacity 2 days before hybridization; (ii) soil which had been air dried for 2 days and then rewetted to field capacity 2 days before hybridization; and (iii) soil which had been fixed directly after collection from the field and had a water content of 9.8% on a dry weight basis. The experiment with glucose-treated soil was repeated four times, and the other two experiments were repeated twice. In all repetitions, hybridizations were performed with at least four replicates. The results given below are the means for all experiments for each soil treatment.
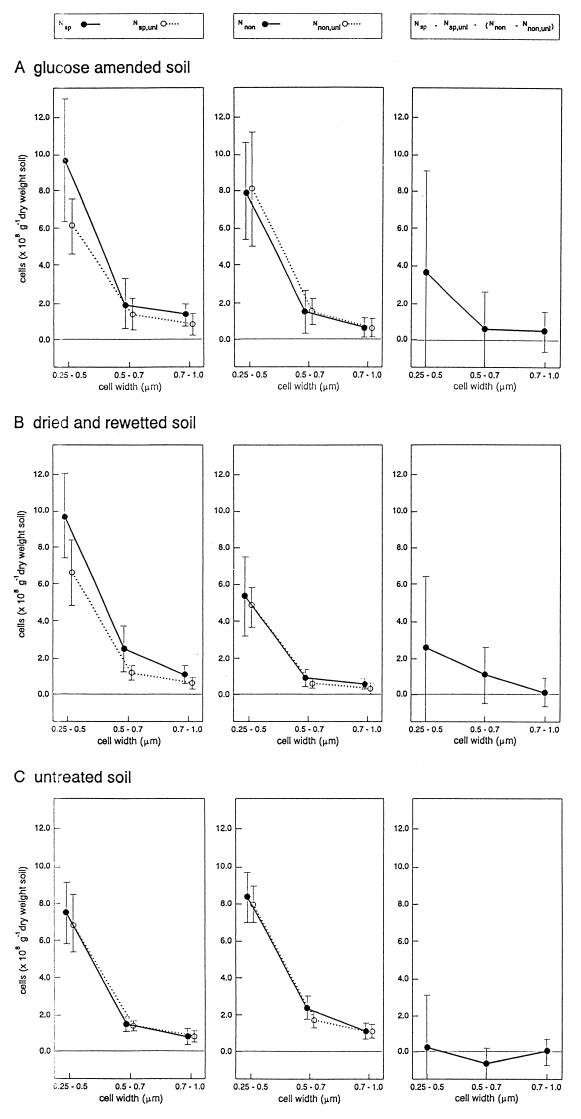
Figure 5 shows that the numbers of specifically stained cells (Nsp) decreased in the following order: glucose-amended soil, air-dried and rewetted soil, untreated soil. After the addition of unlabelled probe, the counts (Nsp,unl) were reduced by 35 and 38% in the glucose-treated and dried and rewetted soils, respectively, but only by 6% in the untreated soil. As Fig. 5 also shows, the counts obtained with the nonspecific NONEUB338 probe rarely decreased when excess unlabelled NONEUB338 probe was added; therefore, the nonspecific counts were interpreted as being mostly due to rhodamine binding. In most cases, the nonspecific staining was thus completely compensated for by adding unlabelled probe, so that the numbers of active cells in these soils could be calculated from the counts obtained with the EUB338 probe after subtraction of the counts obtained with the EUB338 probe plus unlabelled probe.
FIG. 5.
Numbers of active soil bacteria determined by fluorescence in situ slurry hybridization by using rhodamine-labelled specific (Nsp) and nonspecific (Nnon) oligonucleotide probe EUB338. Unlabelled probes were included in order to determine the numbers of cells exhibiting nonspecific binding to the rhodamine part of probe (Nsp,unl, Nnon,unl). The numbers of active bacteria were calculated by compensating for both types of nonspecific labelling ([Nsp − Nsp,unl] − [Nnon − Nnon,unl]). The data represent the results of four experiments performed with glucose-treated soil (A), two experiments performed with air-dried and rewetted soil (B), and two experiments performed with untreated soil (C). Each experiment included four or five independent hybridizations, and the standard deviations thus represent 16, 8, and 9 trials, for the glucose-treated soil, air-dried and rewetted soil, and untreated soil, respectively. The cells were assigned to three size classes, as indicated.
We anticipated that the data obtained for the untreated soil collected under relatively dry conditions would represent the natural low-activity soil conditions under which many small bacteria, but relatively few active bacteria, would be detectable by the in situ slurry hybridization protocol. The results indicate that no active bacteria could be detected in the dry control soil (Fig. 5C), reflecting the detection limit of the method. However, rewetting air-dried soil clearly resulted in higher numbers of active bacteria (3.8 × 108 cells g of soil−1), with the greatest increase occurring in the abundant small-size group of bacteria. Finally, glucose amendment also resulted in a significant increase in the number of active bacteria (4.8 × 108 cells g of soil−1), and the small size group was again the predominant group.
DISCUSSION
The present study shows that the majority of the active soil bacteria, as determined by FISH, are the smallest bacteria (i.e., bacteria with diameters of less than 0.5 μm). The level of activity of these bacteria has not been investigated in detail (45), but the results correspond well with the finding that radiolabelled thymidine and leucine were actively incorporated into such bacteria extracted from soil (4, 5). Addition of a labile carbon source, such as glucose, to soil or rewetting of air-dried soil, resulting in a release of nutrients (38), are both well-known treatments which increase bacterial activity dramatically (8, 9, 13, 47). The increased counts determined after rewetting alone or rewetting plus amendment with glucose are considered evidence that the hybridization protocol and the model for calculating the numbers of active soil bacteria performed correctly.
The rRNA content has been correlated directly with the growth rate for fast-growing bacteria (19, 29), but this relationship may not be valid during slow growth or starvation (12, 30, 44). The relationship between growth rate and sensitivity of the FISH protocol was investigated for laboratory-grown bacteria added to soil. The results demonstrated that cells with generation times of 5 h of less could be easily detected, and the data thus supported the conclusion that the in situ slurry hybridization technique detects only the most active bacteria in soil samples. When flow cytometry was used to quantify light emission by a fluorescein-labelled EUB338 probe, a lower limit of detection was set at generation times of 4 to 7 h for cultures of Escherichia coli and Burkholderia cepacia (46). However, we cannot eliminate the possibility that starved and dormant bacteria may also be detected as active bacteria when their rRNA levels are still relatively high. This has been observed for Salmonella typhimurium (44), and a high level of protein turnover probably accompanied by elevated rRNA levels has been found in starved marine bacteria (31).
In soil samples nonspecific staining may result from binding of either the oligonucleotide (DNA) component or the fluorochrome component of the probe (1, 46). When the FISH technique is used with environmental samples, nonspecifically stained cells are therefore often observed and must be corrected for by subtracting counts obtained by hybridization with a nonspecific probe (28, 33). With soil samples hybridized with labelled probe EUB338 it was not possible to correct for this by directly subtracting counts obtained with the NONEUB338 probe because higher counts were sometimes obtained with the NONEUB338 probe than with the EUB338 probe (Fig. 5). This finding was related to slight differences between the ratios of rhodamine to DNA components in different batches of the probes. To solve this problem, we added different amounts of unlabelled (rhodamine-free) probes to the soil samples in order to titrate out the nonspecific binding of the labelled probe. The contribution of nonspecific binding of DNA was compensated for by subtracting counts obtained with the nonspecific probe from counts obtained with the specific probe after correction for rhodamine binding.
The numbers of active bacteria found in the glucose-treated and the dried and rewetted soil samples represented approximately 5% of the total bacterial counts (5 × 109 to 9 × 109 cells g [dry weight] of soil−1) determined by DAPI staining. This estimate corresponds well with the percentage of active cells determined by the CTC reduction assay performed with bulk soil from the same barley field (48). This is apparently in strong contrast to other reported data, which showed that 41% of the total DAPI counts could be detected with EUB338 probe hybridization by using soil smears (49). However, controls including nonspecific probe NONEUB338 were not included in the previous study, and it is likely that the numbers of bacteria in the samples were overestimated.
The great differences between replicates shows the importance of including at least four replicates with the four combinations of probes in order to obtain consistent results for the different size groups of soil bacteria. The high number of replicates makes the method labor-intensitive and expensive in terms of materials. Additional improvements in the protocol for in situ slurry hybridization should include testing alternative fluorochrome labels for the oligonucleotide probes in order to obtain minimal binding to soil particles and more uniform labelling. To reduce the labor involved, automatic counting procedures based on CLSM, as demonstrated by Bloem et al. (9), could be incorporated into the FISH technique. Despite the obvious limitations at present, including the relatively poor detection limit, our protocol for in situ hybridization to rRNA to determine the number of active soil bacteria is useful and could be improved. The sensitivity for specific groups of bacteria might be increased by performing repeated homogenization-centrifugation extraction for soil bacteria (32). This approach, in combination with soil perturbations, might be used to determine the functions of major soil bacterial groups, such as the Acidobacterium and Verrucomicrobia divisions (25), which have been poorly characterized or not characterized by cultivation methods but are known from 16S rRNA gene sequence comparisons.
ACKNOWLEDGMENTS
Palle Hobolth is acknowledged for synthesizing the oligonucleotide probes. We thank Lars Kongsbak Poulsen and Svend J. Binnerup for their support and advice in developing the in situ hybridization method.
This work was financed by the Danish Center for Microbial Ecology and by the Danish Biotechnology Research Programme 1996-1999 (grant 9502015).
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bååth E. Measurement of protein synthesis by soil bacterial assemblages with the leucine incorporation technique. Biol Fertil Soils. 1994;17:147–153. [Google Scholar]
- 4.Bååth E. Thymidine and leucine incorporation in soil bacteria with different cell size. Microb Ecol. 1994;27:267–278. doi: 10.1007/BF00182410. [DOI] [PubMed] [Google Scholar]
- 5.Bååth E. Thymidine incorporation of bacteria sequentially extracted from soil using repeated homogenization-centrifugation. Microb Ecol. 1996;31:153–166. doi: 10.1007/BF00167861. [DOI] [PubMed] [Google Scholar]
- 6.Bae H C, Cota-Robles E H, Casida L E., Jr Microflora of soil as viewed by transmission electron microscopy. Appl Microbiol. 1972;23:637–648. doi: 10.1128/am.23.3.637-648.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakken L R, Olsen R A. The relationship between cell size and viability of soil bacteria. Microb Ecol. 1987;13:103–114. doi: 10.1007/BF02011247. [DOI] [PubMed] [Google Scholar]
- 8.Bloem J, de Ruiter P C, Koopman G J, Lebbink G, Brussaard L. Microbial numbers and activity in dried and rewetted arable soil under integrated and conventional management. Soil Biol Biochem. 1992;24:655–665. [Google Scholar]
- 9.Bloem J, Veninga M, Shepherd J. Fully automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl Environ Microbiol. 1995;61:926–936. doi: 10.1128/aem.61.3.926-936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell R C. Statistics for biologists. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1981. [Google Scholar]
- 11.Casey J, Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4:1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman S J, Gray R R G. Endogenous metabolism and macromolecular composition of Arthrobacter globiformis. Soil Biol Biochem. 1981;13:11–18. [Google Scholar]
- 13.Christensen H. Conversion factors for the thymidine incorporation technique estimated with bacteria in pure culture and on seedling roots. Soil Biol Biochem. 1993;25:1085–1096. [Google Scholar]
- 14.Christensen H, Poulsen L K. Detection of Pseudomonas in soil by rRNA targeted in-situ hybridization technique. Soil Biol Biochem. 1994;26:1093–1096. [Google Scholar]
- 15.Christensen H, Christensen S. 3H thymidine incorporation technique to determine soil bacterial growth rate. In: Alef K, Nannipieri P, editors. Methods in applied soil microbiology and biochemistry. London, United Kingdom: Academic Press; 1995. pp. 258–261. [Google Scholar]
- 16.Christensen H, Boye M, Poulsen L K, Rasmussen O F. Analysis of fluorescent pseudomonads based on 23S ribosomal DNA sequences. Appl Environ Microbiol. 1994;60:2196–2199. doi: 10.1128/aem.60.6.2196-2199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen H, Rønn R, Ekelund F, Christensen S. Bacterial production determined by [3H]thymidine incorporation in field rhizospheres as evaluated by comparison to rhizodeposition. Soil Biol Biochem. 1995;27:93–99. [Google Scholar]
- 18.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 20.Hahn D, Amann R I, Ludwig W, Akkermans A D L, Schleifer K H. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labelled oligonucleotides. J Gen Microbiol. 1992;138:879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- 21.Hahn D, Amann R I, Zeyer J. Whole-cell hybridization of Frankia strains with fluorescence- or digoxigenin-labeled, 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1993;59:1709–1716. doi: 10.1128/aem.59.6.1709-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn D, Amann R I, Zeyer J. Detection of mRNA in Streptomyces cells by whole-cell hybridization with digoxigenin-labeled probes. Appl Environ Microbiol. 1993;59:2753–2757. doi: 10.1128/aem.59.8.2753-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen M, Kragelund L, Nybroe O, Sørensen J. Early colonization of barley roots by Pseudomonas fluorescens studied by immunofluorescence technique and confocal laser scanning microscopy. FEMS Microbiol Ecol. 1997;23:353–360. [Google Scholar]
- 24.Henrici A T. Morphologic variation and the rate of growth of bacteria. In: Buchanan R E, Fred E B, Waksman S A, editors. Microbiology monographs: general, agricultural, industrial. London, United Kingdom: Bailliere, Tindall and Cox; 1928. pp. 1–194. [Google Scholar]
- 25.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsen I, Nielsen N E. Vesicular-arbuscular mycorrhiza in field-grown crops. I. Mycorrhizal infection in cereals and peas at various times and soil depths. New Phytol. 1983;93:401–413. [Google Scholar]
- 27.Jenkinson D S, Powlson D S, Wedderburn R W M. The effect of biocidal treatments on metabolism in soil. III. The relationship between soil biovolume, measured by optical microscopy, and the flush of decomposition caused by fumigation. Soil Biol Biochem. 1976;8:189–202. [Google Scholar]
- 28.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1417–1431. [Google Scholar]
- 30.Kemp P F, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kjelleberg S, Hermansson M, Mårdén P. The transient phase between growth and nongrowth of heterotrophic bacteria, with emphasis on the marine environment. Annu Rev Microbiol. 1987;41:25–49. doi: 10.1146/annurev.mi.41.100187.000325. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl V, Frostegård Å, Bakken L, Bååth E. Phospholipid fatty acid composition of size fractionated indigenous soil bacteria. Soil Biol Biochem. 1997;29:1565–1569. [Google Scholar]
- 33.Llobet-Brossa E, Roselló-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May K R. A new graticule for particle counting and sizing. J Sci Instrum. 1965;42:500–501. [Google Scholar]
- 35.Norton J M, Firestone M K. Metabolic status of bacteria and fungi in the rhizosphere of ponderosa pine seedlings. Appl Environ Microbiol. 1991;57:1161–1167. doi: 10.1128/aem.57.4.1161-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 37.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powlson D S, Jenkinson D S. The effects of biocidal treatments on metabolism in soil. II. Gamma irradiation, autoclaving, air-drying and fumigation. Soil Biol Biochem. 1976;8:179–188. [Google Scholar]
- 39.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 40.Skinner F A, Jones P C T, Mollison J E. A comparison of a direct- and a plate-counting technique for the quantitative estimation of soil microorganisms. J Gen Microbiol. 1952;6:261–271. doi: 10.1099/00221287-6-3-4-261. [DOI] [PubMed] [Google Scholar]
- 41.Sørensen J, Skouv J, Jørgensen A, Nybroe O. Rapid identification of environmental isolates of Psudomonas aeruginosa, P. fluorescens, and P. putida by SDS-PAGE analysis of whole-cell protein patterns. FEMS Microbiol Ecol. 1992;101:41–50. [Google Scholar]
- 42.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley; 1991. pp. 205–248. [Google Scholar]
- 43.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolker-Nielsen T, Larsen M H, Kyed H, Molin S. Effects of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int J Food Microbiol. 1997;35:251–258. doi: 10.1016/s0168-1605(97)01242-7. [DOI] [PubMed] [Google Scholar]
- 45.Van Elsas J D, Van Overbeek L S. Bacterial responses to soil stimuli. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 55–79. [Google Scholar]
- 46.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 47.West A W, Sparling G P, Grant W D. Relationships between mycelial and bacterial populations in stored, air-dried and glucose-amended arable and grassland soils. Soil Biol Biochem. 1987;19:599–605. [Google Scholar]
- 48.Winding A, Binnerup S J, Sørensen J. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl Environ Microbiol. 1994;60:2869–2875. doi: 10.1128/aem.60.8.2869-2875.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann R A, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]