Abstract
Great differences in capability to detect bacteriophages from urban sewage of the area of Barcelona existed among 115 strains of Bacteroides fragilis. The capability of six of the strains to detect phages in a variety of feces and wastewater was studied. Strains HSP40 and RYC4023 detected similar numbers of phages in urban sewage and did not detect phages in animal feces. The other four strains detected phages in the feces of different animal species and in wastewater of both human and animal origin. Strain RYC2056 recovered consistently higher counts than the other strains and also detected counts ranging from 101 to approximately 103 phages per ml in urban sewage from different geographical areas. This strain detected bacteriophages in animal feces even though their relative concentration with respect to the other fecal indicators was significantly lower in wastewater polluted with animal feces than in urban sewage.
Techniques for distinguishing between human and animal fecal pollution are necessary for assessing the overall protection of water supplies and implementing effective remediation, for epidemiological studies, and even for legal purposes when it is necessary to determine the source of environmental contamination. Chemical (24) and microbiological (11, 20, 23, 25, 26, 30, 35) methods have been proposed for such purposes. Among the microbiological methods, the detection of bacteriophages infecting strain HSP40 of Bacteroides fragilis has very attractive features (7, 12, 15, 21, 33–35). Strain HSP40 detects numbers of phages ranging from 101 to 102 per ml of urban sewage in some geographical areas, such as Southern Europe, South Africa, and Israel (3, 4, 9, 13, 32, 35). However, it recovers lower numbers of phages in sewage from other geographical areas, such as the United States (22). This jeopardizes its usefulness as a universal method.
The purpose of this study was to search for new host strains of B. fragilis that detect more phages than strain HSP40, to assess their usefulness in distinguishing phages originating in the human gut from those originating in the guts of different animal species, and to assess whether they can detect bacteriophages in a wider geographical area than strain HSP40 does.
One hundred and fourteen strains provided by the Microbiology Services of three hospitals in Barcelona and Madrid plus strain HSP40 (ATCC 51477) (34) were used as host strains for bacteriophages infecting B. fragilis. Escherichia coli HS (10), and Salmonella typhimurium WG49 (17) were used to enumerate F-specific bacteriophages. The double-agar layer technique (1) was used for the detection of B. fragilis phages as previously described (36), and for the detection of F-specific bacteriophages using either E. coli HS (10) or S. typhimurium (17) as host strains, all quantifications were done in duplicate. Fecal coliform bacteria were enumerated by standard methods (2). Very diverse samples polluted with human and animal feces were analyzed. Human fecal samples were obtained from 43 healthy volunteers. Fecal samples from cattle, pigs, poultry, horses, and sheep were either liquid manure, lixiviates of recent solid manure, or recent solid manure. Raw urban sewage samples were collected from inflowing waters of sewage treatment plants from the geographical sites indicated below. Samples of wastewater from different slaughterhouses were obtained as samples containing a mixture of feces from many different animals. All samples were collected in sterile bottles and kept in the dark at 4°C until examination. Samples from the Barcelona area were examined within 6 h of collection. The samples from other countries were kept at 4°C or frozen (−20°C) for between 2 days and 1 week before the assay. Previous studies (data not shown) proved that numbers of phages in sewage do not decrease significantly under these conditions.
For fecal analyses, aliquots were suspended in peptone saline at a ratio of 1:10 (wt/vol). The mixture was thoroughly mixed by magnetic stirring for 1 h at 4°C. The large particles were then allowed to sediment for 5 min at 4°C, and the supernatant was decontaminated with chloroform 1/3 (vol/vol) as described elsewhere (1). To analyze fecal coliform bacteria, 1 g of feces was added to 10 ml of 1/4-strength Ringer’s solution. After vigorously shaking the mixture for 10 min, 10-fold dilutions were performed, and bacteria were enumerated as indicated above. For the quantification of phages from sewage, samples were decontaminated by filtration through low-protein-binding polyvinylidene difluoride membrane filters (Millex GV; Millipore) as described elsewhere (36).
Selected strains were phenotypically characterized. The biochemical studies were done with API 20A and API 50CH kits (BioMérieux, Marcy l’Etoile, France). Tests for sensitivity to antibiotics were performed on BPRM agar with filter paper discs from BBL (Cockeysville, Md.), Neosensitabs (Taastrup, Denmark), and Oxoid (Basingstoke, England). The antibiotics tested were amoxicillin (25 μg), ampicillin (30 and 10 μg), chloramphenicol (60 μg), cephalosporin (66 μg), cefazolin (30 μg), ceftriaxone (30 μg), erythromycin (15 and 78 μg), streptomycin (10 and 100 μg), kanamycin (10, 30, and 100 μg), methicillin (5 μg), nalidixic acid (30 μg), neomycin (30 and 120 μg), nitrofurantoin (260 μg), novobiocin (30 μg), oxacillin (1 μg), oxolinic acid (10 μg), penicillin G (5 IU), polymyxin (30 IU), tetracycline (80 μg), tobramycin (10 μg), and netilmicin (30 μg).
To test the capacity of phages isolated on a given strain of B. fragilis to infect other strains, a number of randomly chosen plaques on a determined host were inoculated with a sterile toothpick on a monolayer of the strain to test for sensitivity. After 18 h of incubation at 37°C under anaerobic conditions, an area of lysis could be seen on the monolayer if the host was sensitive to the bacteriophage.
Great variability in the capability of the 115 B. fragilis strains studied to recover bacteriophages from urban sewage from the Barcelona area was observed (Table 1). Only 66 strains recovered bacteriophages from 10-ml samples of sewage, and some strains recovered significantly greater numbers than others. Levels recovered by HSP40, previously shown to be very consistent in urban sewage in the Barcelona area (3), were tested in all samples to guarantee the similarity of the fecal load of the samples. Strain RYC2056 detected the highest numbers, which exceeded by a factor of 1,000 the numbers detected by other strains. This difference is greater than that observed for other fecal bacteria, such as E. coli, in which differences reach values of 10 to 100 (13, 16, 19, 28, 31). However, due to the high numbers of strains already tested, it is not foreseeable that more efficient natural strains will be found for the detection of phages that infect B. fragilis.
TABLE 1.
Levels of bacteriophages recovered in urban sewage from the Barcelona area using different B. fragilis host strains
| No. of strains | Range of PFU/mla | Ratio of range of PFUs to numbers of bacteriophages detected by strain HSP40b |
|---|---|---|
| 49 | 0 | NCc |
| 49 | 0–10 | <0.5 |
| 14 | 10–100 | 0.5–2.0 |
| 3 | >100 | >2.0 |
Average value of at least two tests performed in duplicate.
Phages infecting B. fragilis HSP40 were enumerated in all samples.
NC, not calculable.
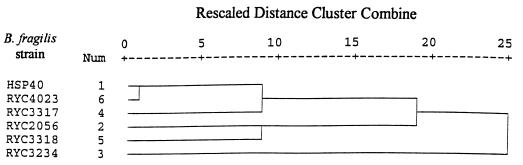
Strains RYC2056, RYC3317, RYC3318, RYC3234, and RYC4023, which gave consistently higher recoveries than strain HSP40 in the preliminary study, plus strain HSP40 were studied further. First, they were characterized phenotypically. From 34 biochemical tests assayed, the maximum differences observed were in the fermentation of inulin, amygdalin, and glycogen. Differences in sensitivity to antibiotics were observed only for ceftriaxone and amoxicillin. Strains HSP40 and RYC4023 presented a high linkage, whereas strain RYC3234 was the most differentiated in a dendrogram (Fig. 1) drawn by the application of hierarchical cluster analysis (SPSS Inc., Chicago, Ill.). However, despite the great phenotypic similarity, major differences in sensitivity to bacteriophage isolates were observed (Table 2). Nearly all the phages isolated with HSP40 could infect RYC4023 and vice versa. However, very few of the phages isolated with the other strains could replicate onto either HSP40 or RYC4023. These data show that strains HSP40 and RYC4023, which do not detect phages in animal feces, as shown below, can support the replication of a very small fraction (less than 1%) of phages present in animal feces. The other four strains showed varied degrees of cross-sensitivity to phages, with percentages of cross-susceptibility ranging from 13 to 87%. Some agreement between the phenotypic characteristics of the different strains and the range of phages infecting them was observed. Thus, strains HSP40 and RYC4023, which are almost identical phenotypically, showed similar patterns regarding sensitivity to bacteriophages, whereas RYC3234 was the most different in terms of both biochemical characteristics and sensitivity to bacteriophages. The great differences in sensitivity to phages of strains of B. fragilis and the narrow host range of phages infecting B. fragilis are in agreement with previous reports for species of Bacteroides (6, 8) and strains of B. fragilis (34).
FIG. 1.
This phenotype-based dendrogram reflects the relationship between the different strains of B. fragilis. The dendrogram was drawn using average linkage (between groups). Analysis was based on the euclidian distance square.
TABLE 2.
Cross-reactivity of B. fragilis phages isolated with different strains from urban sewage
| Strain for which sensi-tivity was tested | Cross-reactivity (%) for B. fragilis strains with which phages were isolateda
|
|||||
|---|---|---|---|---|---|---|
| HSP40 | RYC4023 | RYC3234 | RYC2056 | RYC3317 | RYC3318b | |
| HSP40 | 100 | 0 | 0 | 0 | 0 (0.15) | |
| RYC4023 | 88 | 0 | 0 | 0 | 2 (0.30) | |
| RYC3234 | 1 | 1 | 25 | 28 | 19 | |
| RYC2056 | 0 | 0 | 18 | 59 | 63 | |
| RYC3317 | 0 | 0 | 30 | 73 | 54 | |
| RYC3318 | 0 | 0 | 13 | 87 | 33 | |
These percentages indicate the sensitivity of a strain to detect phages isolated with the other. At least 100 plaques were tested as indicated in the text.
Numbers in parentheses are the percentages of sensitivity of strains HSP40 and RYC4023 to 600 plaques isolated on RYC3318 from abattoir sewage.
Results of detection of bacteriophages infecting strains HSP40, RYC4023, RYC2056, RYC3317, RYC3318, and RYC3234 in animal fecal samples are shown in Table 3. Some, for example RYC2056, detected phages in feces of a range of animals in a percentage of samples similar to the percentage of samples from which F-specific bacteriophages were isolated, and which do not differ significantly from data reported elsewhere regarding the presence in feces of somatic coliphages and F-specific bacteriophages (15, 18, 30). Other strains, such as HSP40 (15, 34) and RYC4023, did not detect phages from animal feces. For human feces, only the presence of bacteriophages infecting RYC2056 was analyzed, and they were isolated in 28% of the samples, which is clearly higher than the percentage described for phages infecting HSP40 (15, 34). Probably due to the great diversity indicated above regarding sensitivity to bacteriophages, the different strains of B. fragilis differ in their capability to support replication of phages present in feces of different animals. The presence of phages in feces depends on the presence in the gut of host strains which are similar in terms of receptors and modification-restriction enzymes to the tested strain and which as a result are able to support phage replication. The great variability in infectivity of bacteriophages infecting B. fragilis is then a consequence of the variability between strains of B. fragilis present in the guts of different animal species. Bacteroides species have evolved in confined environments and have differentiated in accordance with nutrition and other characteristics of the guts of different animal species. The data presented here suggest that phages infecting Bacteroides may be a valuable tool in studying the variability of the Bacteroides present in the microflora of human and animal communities, which have been shown to vary depending on diet, age, etc. (5, 27).
TABLE 3.
Percentage of recovery of bacteriophages of B. fragilis from feces of various animal species using different host strains, F-specific phages, and levels of fecal coliforms
| Host strain | % Positive samples from:
|
||||
|---|---|---|---|---|---|
| Pigs (n = 13) | Poultry (n = 14) | Cows (n = 11) | Sheep and horses (n = 7) | Humans (n = 43) | |
| B. fragilis HSP40 | 0 | 0 | 0 | 0 | 10–13a |
| B. fragilis RYC4023 | 0 | 0 | 0 | 0 | NDc |
| B. fragilis RYC3234 | 31 | 0 | 9 | 0 | NDc |
| B. fragilis RYC2056 | 31 | 29 | 0 | 0 | 28 |
| B. fragilis RYC3317 | 54 | 29 | 18 | 0 | NDc |
| B. fragilis RYC3318 | 46 | 14 | 0 | 14 | NDc |
| E. coli HS(pFamp)Rb | 46 | 50 | 9 | 29 | NDc |
| Fecal coliforms | 100 | 100 | 100 | 100 | NDc |
In an extended study of urban sewage of the area of Barcelona (Table 4), the six strains recovered phages from all the samples. Strain RYC2056 recovered significantly (Student’s t test; P < 0.01) higher numbers of bacteriophages than strains HSP40 and RYC4023 in all samples studied. The other strains recovered significantly (Student’s t test; P < 0.01) higher numbers of phages than strain HSP40 in most of the samples. Average values of F-specific bacteriophages detected in the samples outnumbered those of B. fragilis by factors ranging from 40 to 100 for HSP40 and from 10 to 20 for RYC2056. The average number of fecal coliform bacteria was about 20-fold the number of F-specific coliphages, which is the normal ratio in sewage from different geographical areas (13, 17, 29, 35).
TABLE 4.
Bacteriophages infecting different B. fragilis strains, F-specific phages, and fecal coliforms in different types of sewage from Barcelona area (PFU per ml or CFU per ml)
| Host strain | Bacteriophage result for urban sewage (n = 12)
|
Bacteriophage result for slaughterhouse wastewater (n = 18)
|
||||||
|---|---|---|---|---|---|---|---|---|
| Range (minimum–maximum) | % Positive samples | Arithmetic mean | Median | Range (minimum–maximum) | % Positive samples | Arithmetic mean | Median | |
| B. fragilis HSP40 | 3.2 × 101–1.9 × 102 | 100 | 8.2 × 101 | 7.4 × 101 | 0–7.5 × 100 | 39 | 1 × 100 | 0 |
| B. fragilis RYC4023 | 3.3 × 101–1.8 × 102 | 100 | 9.3 × 101 | 9.6 × 101 | 0–1.6 × 101 | 28 | 1.4 × 100 | 0 |
| B. fragilis RYC3234 | 4.6 × 101–1.9 × 102 | 100 | 9.4 × 101 | 8.8 × 101 | 0–2.3 × 101 | 94 | 5.9 × 100 | 3.9 × 100 |
| B. fragilis RYC2056 | 8.2 × 101–4.4 × 102 | 100 | 2.4 × 102 | 2.1 × 102 | 2.9–2.4 × 102 | 100 | 3.7 × 101 | 2.0 × 101 |
| B. fragilis RYC3317 | 5.9 × 101–1.9 × 102 | 100 | 1.5 × 102 | 1.4 × 102 | 1.4–2.0 × 102 | 100 | 2.2 × 101b | 9.4 × 100 |
| B. fragilis RYC3318 | 4.2 × 101–3.3 × 102 | 100 | 1.7 × 102 | 1.5 × 102 | 0.2–7.8 × 101 | 100 | 1.5 × 101 | 7 × 100 |
| E. coli HS(pFamp)Ra | 1.4 × 103–7.6 × 103 | 100 | 4.1 × 103 | 3.6 × 103 | 1.6 × 102–1.7 × 105 | 100 | 3.7 × 104 | 4.0 × 103 |
| Fecal coliforms | 7.9 × 103–1.9 × 105 | 100 | 6.3 × 104 | 6.0 × 104 | 2.0 × 104–6.8 × 106 | 100 | 9.6 × 105 | 3.0 × 105 |
Host strain for F-specific phages.
n = 17.
The fecal loads of slaughterhouse wastewater studied were higher than those of urban sewage in terms of the values of F-specific bacteriophages and fecal coliforms, which were on average 10 times greater than those of urban sewage (Table 4). In these samples, the ratio of F-specific bacteriophages to fecal coliforms is very similar to the ratio in urban sewage. Of the B. fragilis bacteriophages, only strains RYC2056, RYC3317, and RYC3318 recovered bacteriophages from all samples analyzed. Once again, strain RYC2056 detected numbers significantly higher than the numbers detected by the other strains (Student’s t test; P < 0.01). However, in this case the average values of F-specific bacteriophages outnumbered the average values of phages detected by strain RYC2056 by a factor greater than 103. Strains HSP40 and RYC4023 recovered very low numbers of phages in some of the samples from slaughterhouse sewage, whereas they were not detected in animal feces. But, in this case the average values of F-specific bacteriophages exceeded the average values of phages detected by strain HSP40 by a factor greater than 104.
Since strain RYC2056 recovered more bacteriophages than the others in the area of Barcelona, studies to determine its capability to recover phages from urban (Table 5) and abattoir (Table 6) wastewater samples from different geographical areas were undertaken. Those samples were analyzed for the presence of F-specific bacteriophages and phages infecting B. fragilis HSP40 and RYC2056. Bacterial densities were not determined, since bacterial densities could not be conserved during transportation of the samples. Levels of F-specific phages ranged from 1.2 × 103 to 5.9 × 104 PFU per ml, which are the usual values for urban sewage of developed countries (3, 13, 17, 29). These numbers confirm the properness of the method used for sample transportation. Numbers of phages detected by HSP40 ranged from 0 to 4.5 × 102 PFU per ml. Strain RYC2056 recovered significantly higher (Students’ t test; P < 0.01) and more constant numbers than HSP40, ranging from 2.2 × 101 PFU per ml up to 8.1 × 102 PFU per ml (Table 5). The analyses of bacteriophages from slaughterhouse wastewater of the same geographical areas showed the presence of phages in a few samples when the host used was RYC2056, but no phages were found when HSP40 (Table 6) was the host. F-specific phages were detected in all the samples analyzed, although the numbers were more variable than those of urban sewage. Strain RYC2056 detected a significant number of phages in more areas than HSP40 did.
TABLE 5.
Levels of bacteriophages of B. fragilis and F-specific phages in urban sewage of different countries
| Country | Bacteriophage level (PFU/ml) of:
|
||
|---|---|---|---|
| B. fragilis HSP40 | B. fragilis RYC2056 | S. typhimurium WG49a | |
| The Netherlands | 1 | 4.6 × 102 | 5.1 × 103 |
| The Netherlands | 2.6 | 7.7 × 102 | 7.6 × 103 |
| Ireland | 1.4 | 3.0 × 102 | 4.6 × 103 |
| Ireland | 1.6 | 4.4 × 102 | 6.9 × 103 |
| Austria | 8.5 | 8.1 × 102 | 1.6 × 103 |
| Austria | 0.5 | 6.1 × 102 | 2.2 × 103 |
| Portugal | 0.4 | 1.8 × 102 | 1.8 × 104 |
| Portugal | 0.1 | 1.0 × 102 | 3.8 × 104 |
| Portugal | 0 | 1.0 × 102 | 5.5 × 103 |
| Germany | 2.2 | 7.8 × 102 | 4.8 × 103 |
| Germany | 1.3 | 6.0 × 102 | 2.2 × 103 |
| Sweden | 0.9 | 2.2 × 101 | 1.9 × 103 |
| France | 3.1 × 101 | 2.3 × 102 | 1.2 × 103 |
| South Africa | 1.1 × 102 | 1.8 × 102 | 1.2 × 104 |
| South Africa | 4.5 × 102 | NDb | 5.9 × 104 |
| South Africa | 1.2 × 102 | 5.4 × 102 | 1.7 × 104 |
| South Africa | 2.3 × 102 | 5.0 × 102 | 2.4 × 104 |
Host strain for F-specific bacteriophages.
ND, not done.
TABLE 6.
Levels of bacteriophages of B. fragilis and F-specific phages in wastewater from animal origin of different countries
| Country | Bacteriophage level (PFU/ml) of:
|
||
|---|---|---|---|
| B. fragilis HSP40 | B. fragilis RYC2056 | S. typhimurium WG49a | |
| The Netherlands | 0 | 3.4 × 103 | 3.0 × 104 |
| The Netherlands | 0 | 8.6 × 101 | 3.8 × 103 |
| Ireland | 0 | 0 | 4.8 × 102 |
| Denmark | 0 | 0 | 1.0 × 101 |
| Portugal | 0 | 0.8 | 2.2 × 102 |
| Germany | 0 | 1.3 | 5.7 × 103 |
| South Africa | 0 | 0 | 2.0 × 102 |
| South Africa | 0 | 0 | 5.0 × 103 |
Host strain for F-specific bacteriophages.
When all the quantitative data belonging to samples (mainly polluted with human feces) and those belonging to samples of animal feces and abattoir sewage were grouped, Students’ t test results indicated that the data for F-specific bacteriophages did not differ significantly (P < 0.01) between the two groups of samples, whereas numbers of phages infecting HSP40 and RYC2056 did (P < 0.01). To minimize the effect of the fecal load of the samples studied, the ratios of F-specific bacteriophages to phages infecting HSP40 and F-specific bacteriophages to phages infecting RYC2056 from each sample were calculated. Both ratios significantly (Student’s t test; P < 0.01) differentiated the samples with pollution of human origin from those with pollution of animal origin.
In spite of the fact that strain RYC2056 detects bacteriophages in nonhuman feces, it presents some attractive features to be further assayed as a potential host strain for the detection of bacteriophages infecting B. fragilis, which are a good model of human viruses in the environment. Indeed, it detects good numbers of bacteriophages in a wide range of geographic areas and, from the results of the statistical analysis of the data set presented herein, it may be more useful than other indicators in differentiating human from animal fecal pollution.
Acknowledgments
This work was supported by research grant SMT4-CT95-1603 from the European Commission and GRQ 94-1073 from Generalitat de Catalunya. A.P. was a fellow of the Spanish Ministry of Education.
We give our deepest thanks to K. Mooijman, V. Young, R. Sommer, M. C. Costa, A. Wiedenmann, A. Allard, C. Gantzer, W. O. K. Grabow, and Societat General d’Aigües de Barcelona for the samples. We also thank R. Pericás from Hospital de Sant Pau, J. Vila from Hospital Clínic, and especially M. Reig from Hospital Ramón y Cajal.
REFERENCES
- 1.Adams M H. Bacteriophages. New York, N.Y: John Wiley & Sons, Inc.; 1959. [Google Scholar]
- 2.American Public Health Association. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C: American Public Health Association; 1992. [Google Scholar]
- 3.Araujo R, Puig A, Lasobras J, Lucena F, Jofre J. Phages of enteric bacteria in fresh water with different levels of fecal pollution. J Appl Microbiol. 1997;82:281–286. doi: 10.1046/j.1365-2672.1997.00354.x. [DOI] [PubMed] [Google Scholar]
- 4.Armon R, Kott Y. Distribution comparison between coliphages and phages of anaerobic bacteria (Bacteroides fragilis) in water sources, and their reliability as fecal pollution indicators in drinking water. Water Sci Technol. 1995;31:215–222. [Google Scholar]
- 5.Benno Y, Endo K, Mizutani T, Namba Y, Komori T, Mitsuoka T. Comparison of fecal microflora of elderly persons in rural and urban areas of Japan. Appl Environ Microbiol. 1989;55:1100–1105. doi: 10.1128/aem.55.5.1100-1105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth S J, Van Tassell R L, Johnson J L, Wilkins T D. Bacteriophages of Bacteroides. Rev Infect Dis. 1979;1:325–334. doi: 10.1093/clinids/1.2.325. [DOI] [PubMed] [Google Scholar]
- 7.Chung H, Sobsey M D. Comparative survival of indicator viruses and enteric viruses in seawater and sediment. Water Sci Technol. 1993;27:425–429. [Google Scholar]
- 8.Cooper S W, Szymcza E G, Jacobus N V, Tally F P. Differentiation of Bacteroides ovatus and Bacteroides thetaiotaomicron by means of bacteriophages. J Clin Microbiol. 1984;20:1122–1125. doi: 10.1128/jcm.20.6.1122-1125.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornax R, Moriñigo M A, Paez I G, Muñoz M A, Borrego J J. Application of direct plaque assay for detection and enumeration of bacteriophages of Bacteroides fragilis from contaminated-water samples. Appl Environ Microbiol. 1990;56:3170–3173. doi: 10.1128/aem.56.10.3170-3173.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debartolomeis J, Cabelli V J. Evaluation of Escherichia coli host strain for enumeration of F male-specific bacteriophages. Appl Environ Microbiol. 1991;57:1301–1305. doi: 10.1128/aem.57.5.1301-1305.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geldreich E E. Fecal coliforms and fecal streptococcus density relationships in waste discharge and receiving waters. Crit Rev Environ Control. 1976;6:349–368. [Google Scholar]
- 12.Gironés R, Jofre J, Bosch A. Natural inactivation of enteric viruses in seawater. J Environ Qual. 1989;18:34–39. [Google Scholar]
- 13.Grabow W O K, Holtzhausen C S, De Villiers C J. Report on research project. Research on bacteriophages as indicators of water quality 1990–1992. Pretoria, South Africa: Water Research Commission; 1993. [Google Scholar]
- 14.Grabow W O K, Coubrough P, Nupen E M, Bateman B W. Evaluation of coliphages as indicators of the virological quality of sewage-polluted water. Water S A (Pretoria) 1984;10:7–14. [Google Scholar]
- 15.Grabow W O K, Neubrech T E, Holtzhausen C S, Jofre J. Bacteroides fragilis and Escherichia coli bacteriophages: excretion by humans and animals. Water Sci Technol. 1995;31:223–230. [Google Scholar]
- 16.Havelaar A H, Hogeboom W M. Factors affecting the enumeration of coliphages in sewage and sewage polluted waters. Antonie Van Leeuwenhoek. 1983;49:387–397. doi: 10.1007/BF00399318. [DOI] [PubMed] [Google Scholar]
- 17.Havelaar A H, Hogeboom W M. A method for the enumeration of male-specific bacteriophages in sewage. J Appl Bacteriol. 1984;56:439–447. doi: 10.1111/j.1365-2672.1984.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 18.Havelaar A H, Furuse R, Hogeboom W M. Bacteriophages and indicator bacteria in human and animal feces. J Appl Bacteriol. 1986;60:255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 19.Havelaar A H, Pot-Hogeboom W M, Furuse K, Pot R, Hormann M P. F-specific RNA bacteriophages and sensitive host strains in feces and wastewater of human and animal origin. J Appl Bacteriol. 1990;69:30–37. doi: 10.1111/j.1365-2672.1990.tb02908.x. [DOI] [PubMed] [Google Scholar]
- 20.Hsu F-C, Carol Shien Y S, Van Duin J, Beekwilder M J, Sobsey M D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl Environ Microbiol. 1995;61:3960–3966. doi: 10.1128/aem.61.11.3960-3966.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jofre J, Ollé E, Ribas F, Vidal A, Lucena F. Potential usefulness of bacteriophages that infect Bacteroides fragilis as model organisms for monitoring virus removal in drinking water treatment plants. Appl Environ Microbiol. 1995;61:3227–3231. doi: 10.1128/aem.61.9.3227-3231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kator H, Rodes M. Evaluation of male-specific coliphage as indicators of fecal contamination in point and nonpoint source impacted shellfish growing areas. Gloucester Point, Va: Virginia Institute of Marine Science, College of William and Mary; 1993. [Google Scholar]
- 23.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeming R, Ball A, Ashbolt N, Nichols P. Using fecal sterols from humans and animals to distinguish fecal pollution in receiving waters. Water Res. 1996;30:2893–2900. [Google Scholar]
- 25.Mara D D, Oragui J I. Occurrence of Rhodococcus coprophilus and associated actinomycetes in feces, sewage, and fresh water. Appl Environ Microbiol. 1981;42:1037–1042. doi: 10.1128/aem.42.6.1037-1042.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mara D D, Oragui J I. Sorbitol-fermenting bifidobacteria as specific indicators of human fecal pollution. J Appl Bacteriol. 1983;55:349–357. doi: 10.1111/j.1365-2672.1983.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 27.Moore W E C, Moore L H. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muniesa M, Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl Environ Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieuwstad T J, Mulder E P, Havelaar A H, van Olfen M. Elimination of micro-organisms from wastewater by tertiary precipitation and simultaneous precipitation followed by filtration. Water Res. 1988;22:1389–1397. [Google Scholar]
- 30.Osawa S, Furuse K, Watanabe I. Distribution of ribonucleic acid coliphages in animals. Appl Environ Microbiol. 1981;41:164–168. doi: 10.1128/aem.41.1.164-168.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajala-Mustonen R L, Heinoven-Tanski H. Sensitivity of host strains and host range of coliphages isolated from Finnish and Nicaraguan wastewater. Water Res. 1994;28:1811–1815. [Google Scholar]
- 32.Sun Z P, Levi Y, Kiene L, Dumoutier N, Lucena F. Quantification of bacteriophages of Bacteroides fragilis in environmental water samples of the Seine River. Water Air Soil Pollut. 1997;96:175–183. [Google Scholar]
- 33.Tartera C, Bosch A, Jofre J. The inactivation of bacteriophages infecting Bacteroides fragilis by chlorine treatment and UV-irradiation. FEMS Microbiol Lett. 1988;56:313–316. [Google Scholar]
- 34.Tartera C, Jofre J. Bacteriophages active against Bacteroides fragilis in sewage-polluted waters. Appl Environ Microbiol. 1987;53:1632–1637. doi: 10.1128/aem.53.7.1632-1637.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tartera C, Lucena F, Jofre J. Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl Environ Microbiol. 1989;55:2696–2701. doi: 10.1128/aem.55.10.2696-2701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tartera C, Araujo R, Michel T, Jofre J. Culture and decontaminating methods affecting enumeration of phages infecting Bacteroides fragilis in sewage. Appl Environ Microbiol. 1992;58:2670–2673. doi: 10.1128/aem.58.8.2670-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]