Abstract
Ectonucleoside triphosphate diphosphohydrolase 2 (NTPDase2) hydrolyzes extracellular ATP to ADP, which is the ligand for P2Y1,12,13 receptors. The present study describes the distribution of NTPDase2 in adult rat brains in physiological conditions, and in hippocampal neurodegeneration induced by trimethyltin (TMT). The study also describes the regulation of NTPDase2 by inflammatory mediators in primary astrocytes and oligodendroglial cell line OLN93. In physiological conditions, NTPDase2 protein was most abundant in the hippocampus, where it was found in fibrous astrocytes and synaptic endings in the synaptic-rich hippocampal layers. In TMT-induced neurodegeneration, NTPDase2-mRNA acutely decreased at 2-dpi and then gradually recovered to the control level at 7-dpi and 21-dpi. As determined by immunohistochemistry and double immunofluorescence, the decrease was most pronounced in the dentate gyrus (DG), where NTPDase2 withdrew from the synaptic boutons in the polymorphic layer of DG, whereas the recovery of the expression was most profound in the subgranular layer. Concerning the regulation of NTPDase2 gene expression, proinflammatory cytokines IL-6, IL-1β, TNFα, and IFNγ negatively regulated the expression of NTPDase2 in OLN93 cells, while did not altering the expression in primary astrocytes. Different cell-intrinsic stressors, such as depletion of intracellular energy store, oxidative stress, endoplasmic reticulum stress, and activation of protein kinase C, also massively disturbed the expression of the NTPDase2 gene. Together, our results suggest that the expression and the activity of NTPDase2 transiently cease in neurodegeneration and brain injury, most likely as a part of the acute adaptive response designed to promote cell defense, survival, and recovery.
Keywords: ectonucleoside triphosphate diphosphohydrolase 2 (NTPDase2), hippocampus, trimethyltin-model of neurodegeneration, neuroinflammation, purinergic signaling
Introduction
Extracellular purine and pyrimidine nucleotides, such as ATP and uridine triphosphate (UTP), regulate a wide variety of processes in the central nervous system (CNS), in health and disease (Ipata, 2011). In physiological conditions, nucleotides released from neurons (Fields, 2011; Pankratov et al., 2006; White, 1977), astrocytes (Koizumi, 2010), and microglia (George et al., 2015; Imura et al., 2013), participate in a multitude of processes, including cell proliferation, neurotransmission, and regulation of cell volume, blood-brain barrier permeability and cerebral blood flow (Carman et al., 2011; Narravula et al., 2000). In hypoxic/ischemic conditions, in the metabolic challenge (Juranyi et al., 1999; Melani et al., 2005) or after brain trauma (Davalos et al., 2005; Franke et al., 2006; Wang et al., 2004), ATP released from damaged or dying neurons, functions as an SOS signal for astrocytes and microglia (Bours, Dagnelie, et al., 2011; Bours, Swennen, et al., 2006; Rodrigues et al., 2015). The nucleotide initiates neuroinflammatory responses of microglia (Idzko et al., 2014; Koizumi et al., 2013) and induces the release of IL-1β, essential for morphological and functional activation of astrocytes (Cahill and Rogers, 2008; He et al., 2015). Moreover, the nucleotides produced by enzymatic degradation of ATP, i.e., ADP, AMP, and adenosine, also have a crucial role in the regulation of glial cell responses to an insult and inflammatory status of the tissue (Bours, Swennen, et al., 2006).
The actions of the extracellular purine nucleotides are mediated via P2 receptors, which comprise two families of ligand-gated receptor channels P2X1-7 and G-protein coupled receptors (GPRC) P2Y1−14 (Abbracchio et al., 2009; Khakh et al., 2001). ATP binds to and activates P2X1-P2X7 and a subset of ATP-sensitive P2Y purinoceptors, namely P2Y2 and P2Y11, the latter also being activated by UTP. The subtypes P2Y1, P2Y12, and P2Y13 mediate actions of ADP, while P2Y6 exhibits a preference towards UDP/ADP (Boeynaems et al., 2005). Typically, P2Y1, P2Y2, P2Y4, and P2Y6 are Gq/PLC-coupled membrane receptors, whereas P2Y12, P2Y13, and P2Y14 are the Gi/adenylyl cyclase-coupled receptors (Abbracchio et al., 2009). All purinoceptors, except P2Y4 and P2Y11 (Boehm, 2003) are abundantly expressed in the CNS, at neurons, astrocytes, oligodendrocytes, microglia, epithelial cells of the choroid plexus, and vascular endothelial cells (Burnstock and Knight, 2004), hence, the regulation of ligand availability for P1 and P2 receptor activation has a crucial impact on brain homeostasis and its response to injury (Illes et al., 2020). The second class of purinoceptors, P1, comprises four GPRC subtypes, namely A1, A2A, A2B, and A3, which mediate physiological actions of adenosine (Fredholm, 2007).
The availability of P1 and P2 receptor ligands is determined by the rate of ATP release and its hydrolysis by the ectonucleotidase enzymes. Major ectonucleoside triphosphate phosphohydrolases operating in the CNS are NTPDases1 and NTPDase2 (Robson et al., 2006; Zimmermann et al., 2012). Both ectonucleotidases hydrolyze purine and pyrimidine triphosphate and diphosphate nucleotides while differing in their substrate preference and cell localization (Kukulski and Komoszyński, 2003). Thus, NTPDase1 (CD39) (Kansas et al., 1991), mainly residing in microglia and vascular endothelial cells in the CNS (Langer et al., 2008), hydrolyzes ATP and ADP to AMP. The nucleotide monophosphate is further hydrolyzed to adenosine by ecto-5’-nucleotidase (eN/CD73) (Zimmermann, 1992). Accordingly, simultaneous induction of NTPDase1/eN, often seen in neuropathological conditions (Allard et al., 2017; Dragic et al., 2021b), eliminates P2 receptor ligands and potentiates P1-mediated anti-inflammatory actions of adenosine (Cunha, 2005).
Much less is known about the role of NTPDase2 in pathophysiological conditions (Kirley, 1997; Sévigny et al., 2002). The ectonucleotidase is mostly expressed in astrocytes in vitro (Brisevac, Adzic, et al., 2015; Brisevac, Bajic, et al., 2013; Wink et al., 2006) and in vivo (Jakovljevic et al., 2017). The enzyme catalyzes the removal of the terminal phosphate group from ATP (Kukulski and Komoszyński, 2003), leading to a sustained accumulation of ADP and promotion of the P2Y1,12,13-mediated actions (Heine et al., 1999). The Entpd2 genes in humans, rats, and mice share 83% sequence homology (Chadwick and Frischauf, 1997; Kirley, 1997). The coding regions translate into one 495-residues long polypeptide of about 54.4 kDa (Chadwick and Frischauf, 1997), with six Arg residues which may be fully or partially N-glycosylated (Mateo et al., 2003), resulting in a 70–80 kDa mature glycoprotein (Sévigny et al., 2002). The enzyme is embedded in a cell membrane via two transmembrane segments at N- and C-termini, which flank a large extracellular loop, with five apyrase-conserved regions (ACRs) involved in the catalytic activity, and 11 conserved Cys residues, participating in disulfide bond formation and homo-oligomerization (Heine et al., 1999). Several lines of evidence suggest complex regulation of NTPDase2 at transcriptional, posttranscriptional, and posttranslational levels in physiological and pathological situations. NTPDase2 is negatively regulated by MCP-1/CCL2 in bile duct epithelial cells (Kruglov et al., 2006), by IFNα, IFNβ, IL1α + TNFα + C1q in primary astrocytes (Hasel et al., 2021), and by IL-6 in portal fibroblasts (Yu et al., 2008), where the cytokine directly regulates transcription of Entpd2 via specific negative promoter elements. Alternative splicing may provide two NTPDase2 isoforms (Wang et al., 2005), which are under specific regulation by extracellular signals which activate protein kinase C (Vlajkovic et al., 1999). Finally, NTPDase2 may be modulated at the posttranslational level by alternative glycosylation that affects folding, trafficking, and catalytic activity (Mateo et al., 2003).
In the adult CNS, NTPDase2 exhibits wide distribution, but restricted cellular localization (Gampe, Hammer, et al., 2012; Gampe, Stefani, et al., 2015; Jakovljevic et al., 2017). Available immunohistochemical and in situ hybridization data revealed that NTPDase2 is mostly localized at white matter fibrous astrocytes, particularly in projection tracts (Braun et al., 2003; Jakovljevic et al., 2017), and at specialized astrocytes in distinct brain areas and dorsal root ganglions (Braun et al., 2004). At these locations, NTPDase2 provides ADP for basal signaling between astrocytes, oligodendrocytes, and resting microglia (Amadio et al., 2014; Bianco et al., 2005; Fischer and Krügel, 2007; Kyrargyri et al., 2020). ADP signaling also contributes to progenitor cell homeostasis in the adult neurogenic niches (Gampe et al., 2015; Shukla et al., 2005). Therefore, under basal conditions, the main role of NTPDase2 may be to provide ADP, which functions as a homeostatic regulator of microglial/astrocytes/oligodendrocyte interactions and cell proliferation and expansion (Gampe et al., 2015).
Our interest in NTPDase2 has been kindled by several recent in vivo and in vitro evidence showing acute down-regulation of NTPDase2 in conditions of neuroinflammation and brain injury. Thus, marked down-regulation of NTPDase2 was demonstrated in rat spinal cord during the symptomatic phase of experimental autoimmune encephalomyelitis, with complete recovery at the end of the acute autoimmune disease (Jakovljevic et al., 2017). Down-regulation of NTPDase2 was also demonstrated in primary astrocytes isolated from the SOD1G93A mice model of amyotrophic lateral sclerosis (ALS) (Guttenplan et al., 2020), and in hippocampal astrocytes following ovariectomy (Grkovic et al., 2019). Given that P2Y-mediated ADP signaling is implicated in homeostatic processes and neuroinflammatory responses of astrocytes and microglia (Illes et al., 2020), the present study is designed to complement the existing data on NTPDase2 and its regulation in neuroinflammatory conditions, as a step forward in understanding the role of ADP-mediated signaling in health and disease. To this end, we have analyzed the expression of NTPDase2 in adult rat brains, and in neuroinflammatory conditions in the model of trimethyltin-induced neurodegeneration in vivo and several neuroinflammatory paradigms in vitro.
Materials and Methods
Animal Handling
A total of 44 two-month-old male rats of the Wistar strain were used for in vivo study. Animals were bred in the institutional animal facility, housed (3–4 per cage) under standard conditions of 12 h light/dark regime, constant ambient temperature (22 ± 2 °C), and 40–60% relative humidity, with unlimited access to food and water. All animal procedures were carried out in compliance with the European Communities Council Directive (2010/63/EU) and the National Laboratory Animal Science Association for the protection of animals used for experimental and scientific purposes and were approved by the institutional Ethics Committee.
Trimethyltin-Induced Neurodegeneration
On day zero (0-dpi), 36 animals received single i.p injection of trimethyltin(IV)-chloride (C3H9SnCl, 8 mg/kg) in 0.9% w/v saline. Animals were returned to the cages (3/cage) and kept at standard conditions for 2-, 4-, 7- or 21-dpi (9/per group). From 0-dpi onwards, animals were monitored for signs of the “TMT syndrome“, which include hyperactivity, aggressive behavior, and tremor. Thirteen age-matched animals received an i.p injection of 0.9% saline solution at 0-dpi and served as a control group. Animals were sacrificed by decapitation (Harvard apparatus, Holliston, MA, USA), brains were carefully removed from skulls and processed for subcellular fractionation (4 animals in the control group), total mRNA isolation (5 animals/group), and histology (4 animals/group).
Preparation of Subcellular Fractions
Crude membrane fraction (P2) was prepared by following the protocol of Gray and Whittaker (1962). After isolation, brains were washed in the isolation buffer (0.32 M sucrose, 10 mM Tris-HCl, pH 7.4), dried, and the regions of interest were carefully dissected. The tissue was homogenized in 10 volumes of ice-cold isolation buffer (IB, 0.32 M sucrose in 5 mM Tris–HCl, pH 7.4) with a Teflon/glass homogenizer. Crude nuclear fraction and cell debris were removed by centrifugation at 1,000 × g for 10 min. The resulting supernatants were collected and centrifuged at 16,000 × g for 30 min to obtain crude membrane fraction (P2), which was resuspended in 5 mM Tris–HCl, pH 7.4. All steps were carried out at 4 °C. The samples were aliquoted and stored at −80 °C until use. The P2 fractions were separately isolated from four animals without pooling the tissue.
For purified hippocampal synaptosome (SYN) and gliosome (GLIO) preparations, the P2 fraction was placed on a discontinuous Percoll gradient (2, 6, 10, and 23% v/v Percoll in 0.32 M Tris-buffered sucrose and 1 mM EDTA, pH 7.4), and centrifuged at 35.000 × g for 5 min. The bands containing gliosomal and synaptosomal fractions were separated and recovered from 2- 6% Percoll interface and 10–23% Percoll interface, respectively. Both fractions were resuspended in the IB and washed from the Percoll reagent by double centrifugation at 12.000 × g for 15 min at 4 °C. The purity of the subcellular preparations was tested by immunoblotting, using specific molecular markers directed to GLIO (GFAP) and SYN (synaptophysin, SNAP25). GLIO was enriched in astroglial membrane fragments, while SYN was highly enriched in enclosed presynaptic and postsynaptic membrane fragments (Grkovic et al., 2019, Milanese et al., 2009). The protein content of the isolated subcellular fractions was determined by using bovine serum albumin (BSA) as a standard, as described previously. For functional assays, samples were used immediately after isolation, while for the immunoblotting procedures, samples were kept at −80 °C until use.
Immunohistochemistry/Immunofluorescence and Confocal Microscopy
Serial 25-μm sagittal paraformaldehyde-fixed cryosections were incubated in 0.1% H202 in methanol for 20 min at RT, washed 2 × 5 min in PBS, and blocked with 5% normal donkey serum (NDS) in PBS, for 1h at RT. After the incubation with rabbit anti-rat NTPDase2 antibodies (Ectonucleotidases-ab.com), at 4 °C overnight and 3 × 5 min washing in PBS, sections were incubated for 2h at RT in horseradish peroxidase (HRP)-conjugated secondary antibodies conjugated. The reaction was visualized with HRP chromogen, 3,3′-S-diaminobenzidine-tetrahydrochloride (DAB, Abcam, UK). The reaction was stopped with ddH2O, followed by dehydration in graded ethanol (70%, 95%, and 100%) and clearing in xylene. Sections were mounted with DPX medium (Sigma Aldrich, USA) and left to dry overnight. Sections were analyzed on LEITZ DM RB light microscope (Leica Mikroskopie & Systems GmbH, Wetzlar, Germany), under ×50 and ×200 magnifications, and digital images were captured by LEICA DFC320 CCD camera (Leica Microsystems Ltd., Heerbrugg, Switzerland) and LEICA DFC Twain software (Leica, Germany).
For confocal immunofluorescence, slides were air-dried 30 min, rehydrated in PBS (pH 7.4) for 10 min, and incubated in 5% NDS in PBS for 1h at RT. After incubation with primary antibodies (Table 1), overnight at 4 °C, and 3 × 5 min washes in PBS, sections were incubated with fluorophore-labeled secondary antibodies (Table 1), for 2h in a dark chamber at RT. After 3 × 5 min washing in PBS, sections were mounted with Mowiol (Sigma Aldrich, USA). Microscopic slides were stored at 4 °C until used for confocal laser scanning microscopy (LSM 510, Zeiss), with an argon laser (488 nm) and two helium-neon lasers (543 and 633 nm) at 400× magnification, equipped with a monochrome camera AxioCam ICm1camera (Carl Zeiss GmbH, Germany) and AxioVision software.
Table 1.
List of Primary and Secondary Antibodies.
| Antibody specificity | Source and type | Dilution | Manufacturer |
|---|---|---|---|
| Doublecortin | Goat, polyclonal | 1:100IF | Santa Cruz, sc-8066 RRID: AB_2088494 |
| GFAP | Mouse, monoclonal | 1:100IF | UC Davis/NIH NeuroMab Facility (73–240), RRID: AB_10672298 |
| Microtubule-associated protein 2 | Mouse, monoclonal | 1:200IF | Sigma-Aldrich, M9942 RRID: AB_477256 |
| Myelin basic protein | Mouse, monoclonal | 1:100IF | BioLegend, 801703, RRID: AB_510039 |
| Nestin | Mouse, monoclonal | 1:300IF | Sigma-Aldrich, N5413 RRID: AB_1841032 |
| NG2 | Mouse, monoclonal | 1:50IF | Sigma-Aldrich, N8912, RRID: AB_609907 |
| NTPDase2 | Rabbit, polyclonal | 1:300IF,IHC 1:1000WB |
ectonucleotidases-ab.com. NTPDase2 RRID:AB_2314986 |
| Parvalbumin | Mouse, monoclonal | 1:300IF | Sigma-Aldrich, P3088, RRID:AB_477329 |
| S100 | Mouse monoclonal | 1:200IF | Sigma Aldrich MAB079-1 RRID:AB_571112 |
| Synaptophysin | Rabbit, polyclonal | 1:500IF | Santa Cruz, sc-9116, RRID: AB_2199007 |
| Vimentin | Mouse, polyclonal | 1:300IF | DAKO, M0725, RRID: AB_10013485 |
| Anti-mouse lgG Alexa Fluor 488 | Donkey, polyclonal | 1:400IF | Invitrogen A21202, RRID: AB_ I41607 |
| Anti-goat lgG Alexa Fluor 488 | Donkey, polyclonal | 1:400IF | Invitrogen A-1 1055, RRID: AB_ I42672 |
| Anti-rabbit lgG Alexa Fluor 555 | Donkey, polyclonal | 1:400IF | Invitrogen A-2 1428, RRID: AB_ I41784 |
| Anti-mouse lgG Alexa Fluor 647 | Donkey, polyclonal | 1:400IF | Thermo Fisher Scientific A-31571, RRID: AB_ 162542 |
| Anti-guinea pig lgG Alexa Fluor | Goat, polyclonal | 1:400IF | Invitrogen A-2 1206, RRID: AB_ I41708 |
| Anti-goat HRP-conjugated lgG | Rabbit, polyclonal | 1:200IHC | R&D Systems, HAF017 RRID: AB_56258 |
| Anti-mouse HRP-conjugated lgG | Goat, polyclonal | 1:200IHC | R&D Systems, HAF007 RRID: AB_562588 |
| Anti-goat HRP-conjugated lgG | Rabbit, polyclonal | 1:200IHC | R&D Systems, HAFO 17 RRID: AB_56258 |
Regions of interest were denoted according to The Rat Brain in Stereotaxic Coordinates by Paxinos and Watson (2004). High-resolution digital images (2,088 × 1,550 and 1,024 × 1,024 pixels) were saved as.tiff files. Basic image processing and linear adjustments were performed in ImageJ (U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/, 1997–2018), while composite figures were arranged in Photoshop.
Cell Cultures and Treatments
Primary cortical astrocyte cultures were prepared from 1-day old neonatal rat puppies, as previously described (Adzic and Nedeljkovic, 2018), according to the protocols approved by the institutional Ethics Committee. Briefly, 1-day old rat cerebral cortices were dissociated by mincing and trituration and cells were seeded in a complete medium (Dulbecco's modified Eagle's medium, containing 10% heat-inactivated fetal bovine serum (FBS), 25 mmol/l glucose, 2 mmol/l L-glutamine, 1 mmol/l sodium pyruvate, 100 IU/ml penicillin and 100 μg/ml streptomycin) and cultured until a monolayer was formed (80–90% confluence). Primary microglia and oligodendrocytes were removed by vigorous shaking at 400 rpm for 16–20 h on a plate shaker (Perkin Elmer, Turku, Finland) and additional mechanical washing using the 1 ml pipette if needed. Adherent primary astrocytes were trypsinized (0.25% trypsin and 0.02% EDTA), and replated on 60-mm diameter dishes to multiply cells. Cells were treated after reaching 80–90% confluence with the following inflammatory factors: TNF-α, LPS (100 ng/ml), IL-1β, IL-6, IL-10, IFNγ (10 ng/ml), ATP, and adenosine (1 mM) for 8h (for rt-PCR) or 24h (for immunoblot).
OLN93 is a rat oligodendrocyte progenitor cell line, derived from spontaneously transformed cells in primary rat brain glial cultures (Richter-Landsberg and Heinrich, 1996). Cells were cultured and maintained in complete DMEM, at 37 °C and 5% CO2, as described elsewhere (Teske et al., 2018). The culture medium was changed every 2–3 days at 80–90% confluence. For the experiments, cells were seeded at 4 × 105 cells/well into Poly-D-lysine coated 6-well plates (Sigma-Aldrich, Munich, Germany). OLN93 were treated with TNF-α, IL-1β, IL-6, TGFβ (0.01 μg/ml), IFNγ (100 U/ml), and ATP (0.1 mM). To analyze whether blocking of mitochondrial respiratory chain activity alters the expression of NTPDase2, the following toxins were applied to OLN93 cells: Rotenone (complex-I inhibitor; 1 μM, diluted in dimethyl sulfoxide), antimycin (complex-III inhibitor; 10 μM, diluted in ethanol), sodium azide (complex-IV inhibitor; 10 mM, diluted in H2O), oligomycin (ATPase inhibitor; 10 μM, diluted in ethanol), tunicamycin (N-linked glycosylation and cell cycle arrest agent, 50 μg/ml, diluted in ethanol), all obtained from Sigma-Aldrich, and protein kinase c stimulator, phorbol myristate acetate (100 ng/ml in PBS, diluted from stock solution diluted in DMSO).
For quantitative RT-PCR, cells were collected 8h after the treatment, in TRIzol® (astrocytes) or PeqGold® (OLN93). Total mRNA isolation, concentration determination, reverse transcription, and real-time PCR were conducted as described below. For Western blot analysis, cells were collected 24 h after the treatment, centrifuged at 500 × g for 5 min, and re-suspended in cold RIPA buffer, supplemented with a 0.5% w/v protease inhibitor cocktail. The suspension was centrifuged at 10,000 × g for 10 min, at 4 °C, the supernatant was separated from the pellet, and the protein concentration was determined using the protein assay kit (Pierce BCA protein assay kit).
Gene Expression Analyses
Total RNA was isolated from the hippocampal tissue (5/group) or collected cells, RNA concentrations were measured spectrophotometrically and the purity was determined by determining the A260/A280 and A260/A230 ratios. Volume equivalent to 1 µg of RNA was used for reverse transcription to generate cDNA (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA, US), used for real-time PCR analysis (QuantStudioTM 3 Real-Time PCR System, Applied Biosystems, Foster City, CA, United States). The reaction mixture contained 2 μl cDNA (20 ng/μl), 5 μl QTM SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, US), 0.5 μl primers (100 pmol/μl), and 2 μl RNase-free water (UltraPure, Invitrogen, Germany). The primer sequences were shown in Table 2. The amplification conditions were: 10 min of enzyme activation at 95 °C, 40 cycles of 15 s denaturation at 95 °C, 30 s annealing at 64 °C, 30 s amplification at 72 °C, and 5 s fluorescence measurements at 72 °C. To determine the specificity of the PCR reaction product we performed melting curve analysis and subsequent electrophoretic analysis. The results were expressed as the abundance of NTPDAse2 /CycA-mRNAs at each dpi relative to control (arbitrarily defined as 100%) ± SEM.
Table 2.
List of Primer Pairs for rt-PCR.
| Target gene | Forward | Reverse |
|---|---|---|
| Entpd2 | ccc tca tga cct tct tcc tg | cca aga gac ccg gta tag ca |
| Entpd1 | ccc agc tga aca gcc att at | gat gaa cag ccc tgt gat ga |
| Entpd3 | acg gtt aca gca cca cct tc | aca gct gtg ggt cac cag tt |
| Nt5e | caa atc tgc ctc tgg aaa gc | acc ttc cag aag gac cct gt |
| Enpp1 | cca gaa tca cat tgg cat aat tg | cgg ctg tcc gta cca ca |
| Enpp2 | gac aga tgt ggg gaa gta cga | tgc aga cca ctt ggt agt tgg |
| P2RY1 | ctg gat ctt cgg ggt gtt a | ctg ccc aga gac ttg aga gg |
| P2RY12 | cga aac caa gtc act gag aga | cca gga atg gag gtg gtg ttg |
| P2RY13 | ggc ata acc gtg aag aaat | ttg gaa tca ccg tgt aaa a |
| CycA | ggc aaa tgc tgg acc aaa cac | ggc aaa tgc tgg acc aaa cac |
SDS-PAGE and Immunoblotting
Equivalent amounts of sample proteins (20 µg of P2 subcellular preparations or 10 μg of cell lysates) were resolved by denaturing SDS-PAGE and immunoblotted onto PVDF support membrane (0.45 mm, Millipore, Germany). After thorough washing in TBST (50 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 0.05% Tween 20), the PVDF membrane was blocked (5% non-fat milk/TBST or 5% BSA/TBST), for 1 h at RT. The PVDF membranes were incubated with rabbit anti-rat NTPDase2 antibodies (Table 1), overnight at 4 °C. The membrane was washed 3 × 10 min in TBST and incubated with selected HRP-conjugated secondary antibodies (Table 1), for 1 h at RT. After several rounds of washing in TBST, the membrane was contacted with a peroxide reagent and luminal-containing solution (Bio-Rad Clarity Western ECL substrate), and the protein bands were visualized using chemiluminescence ChemiDoc-It®2 510 Imaging System. Protein bands obtained with the use of anti-β-actin-HRP conjugated antibodies at each membrane were used as a loading control. Densitometric analysis was performed using the ImageJ software package (NIH, Bethesda, USA). The optical density (OD) of the specific band in each lane was normalized for the optical density of the β-actin band in the same lane and the result was expressed relative to the value obtained for control, arbitrarily defined as 100% (±SEM), from n = 3–4 determinations performed on separate preparations.
Statistical Analysis
In the immunoblot and rt-PCR experiments, the abundance of NTPDase2 was normalized to the level of β-actin and CyCA, respectively. The means ± SEM were calculated from n separate determinations, performed in at least two independent sample preparations. Data were tested for normality of distribution and variance homogeneity, by the Shapiro–Wilk test. Based on those outputs, the following statistical tests were applied. In the in vivo study, the mean in each dpi group was compared with the control, and statistical significance was assessed by one-way ANOVA, with a post hoc Tukey's test for multiple comparisons between groups. In the in vitro study, one-way ANOVA was used to compare the mean in each treatment group with the non-treated control, and the values of p < 0.05 were considered statistically significant. Complete statistical analysis was performed in the GraphPad Prism 6 software package (San Diego, CA).
Results
Protein Abundance of NTPDase2 in Distinct Brain Areas
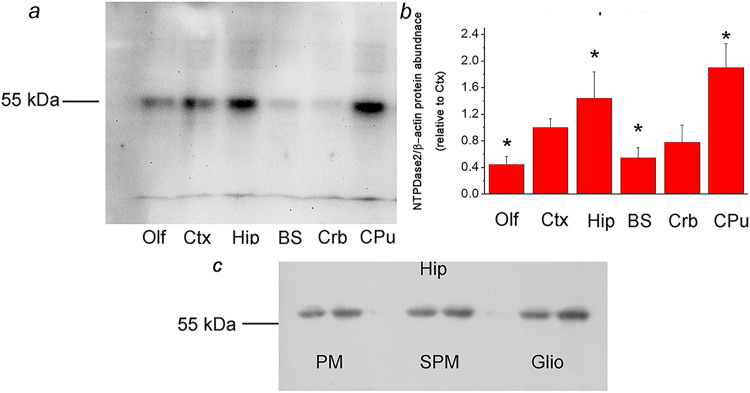
The specificity of rabbit anti-rat NTPDase2 antisera used for immunohistochemistry and immunofluorescence analyses was confirmed by immunoblotting. NTPDase2 was immunodetected in P2 fractions isolated from the olfactory bulb (Olf), cortex (Ctx), hippocampus (Hip), brain stem (BS), cerebellum (Crb), and caudate-putamen (CPu), with 1:500 dilution of the BZ3-4F/rN2-6L antisera. One protein band at about 55-kDa was detected in all tested samples (Figure 1A), while no band was detected in the presence of non-immune rabbit serum (data not shown), confirming the validity of the antisera for the use in NTPDase2-directed immunohistochemistry. The relative abundance of NTPDase2 protein was determined by calculating the OD ratio between the 55-kDa band and the β-actin band in each lane, expressed relative to the same value obtained for Ctx (arbitrarily defined as 1.0) (Figure 1B). The highest relative abundance of NTPDase2 protein was detected in Hip (1.44 ± 0.29, p < 0.05) and CPu (1.90 ± 0.36, p < 0.05), while the lowest was detected in Olf (0.45 ± 0.12, p < 0.05) and BS (0.55 ± 0.15, p < 0.05).
Figure 1.
a: Crude membrane fractions isolated from the olfactory bulb (Olf), cortex (Ctx), hippocampus (Hip), brain stem (BS), cerebellum (Crb), and caudate-putamen (CPu), resolved by SDS-PAGE and probed with rabbit anti-rat NTPDase2 antisera (BZ3-4F/rN2-6L). Dash denotes the position of the specific 55 kDa band. b: Mean NTPDase2/β-actin protein abundance, expressed relative to Ctx (arbitrarily defined as 1.0) ± SEM (from n = 4 determinations performed with two independent P2 preparations). Significance is shown inside the graph: *p < 0.05. c: Immunodetection of NTPDase2 in the plasma membrane (PM), synaptic membrane (SPM), and glial membrane (Glio) fractions isolated from the hippocampal tissue.
NTPDase2 protein was further immunoblotted in the plasma membrane (PM), synaptic membrane (SPM), and glial membrane (Glio) fractions, obtained by density gradient centrifugation of the hippocampal tissue. A single band migrating at about 55-kDa was detected in all samples, suggesting the presence of NTPDase2 protein in astrocyte membranes and synaptic membranes (Figure 1C).
NTPDase2 Immunohistochemistry and Immunofluorescence
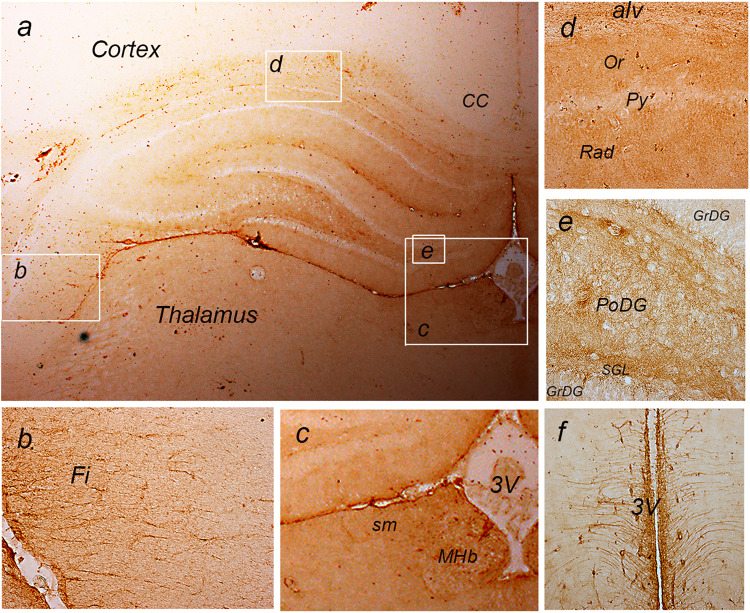
The tissue expression of NTPDase2 in the forebrain is presented in Figure 2. Faint immunoreaction (ir) was observed in deeper cortical layers, while moderate ir-stained fibrous astrocytes in the forebrain projection tracts, including callosal commissure (Figure 2A), fimbria (Figure 2B), stria medullaris (Figure 2C), and alveus (Figure 2D and E),. Moderate to strong ir staining was observed in hippocampal parenchyma, except in principal neuronal cell layers in CA1-CA3 and dentate gyrus (DG), which remained unstained (Figure 2A, D, and E). Prominent ir was observed in ependymal cells lining the third ventricle, medial habenula (MHb) (Figure 2C), tanycytes in the midbrain (Figure 2F), and lateral pial surface (Figure 2A and B).
Figure 2.
a: immunohistochemical labeling of NTPDase2 at coronal forebrain section. The white frames denote areas enlarged at b: The fimbria (fi); c: The third cerebral ventricle; d: Hippocampal CA1 sector; e: Dentate gyrus/Hilus; f: Midbrain tanycytes.
Abbreviations: alv = Alveus; CC = corpus callosum; DG = Dentate gyrus; GrDG = granule cell layer; Fi = Fimbria; MHb = Medial habenula; MoDG = molecular layer; PoDG = Polymorphic layer; Py = Pyramidal cell layer; sm = Stria medullaris; Or = Stratum, oriens; Rad = Stratum radiatum; 3V = Third ventricle. Magnification: ×50 (A) and ×200 (B–F).
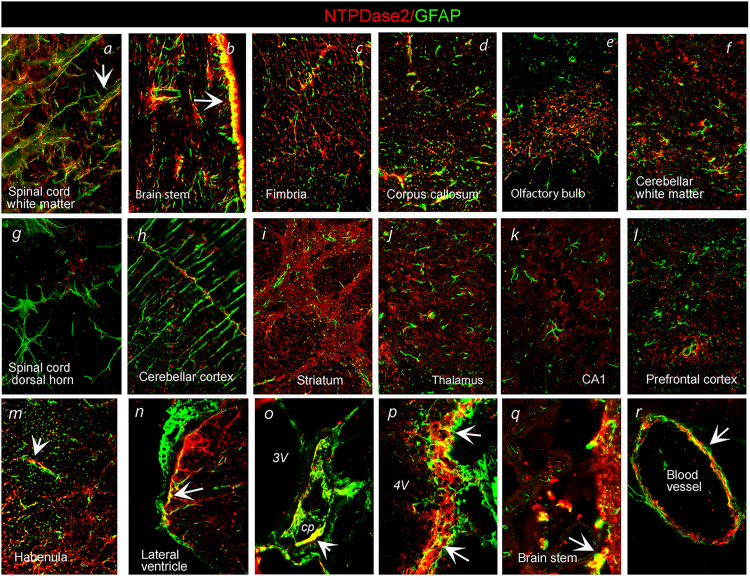
The association of NTPDase2 with glial cells and presynaptic elements was confirmed by double immunofluorescence directed to NTPDase2 in conjunction with astrocyte marker GFAP and presynaptic marker synaptophysin (Syn), respectively. Concerning astrocytic expression, we observed distinct patterns of immunoreactive signals (ir) in the white and gray matter throughout the CNS. In the spinal cord (Figure 3A) and brain stem white matter (Figure 3B), a high degree of GFAP-ir overlap with NTPDase2-ir suggested its dominant expression in fibrous astrocytes. A slightly less degree of overlap was observed within the hippocampal fimbria (Figure 3C), callosal commissure (Figure 3D), and olfactory bulb projection pathways (Figure 3E), whereas the scarce association was found in the cerebellar white matter (Figure 3F). Within gray matter areas, a complete lack of NTPDase2/GFAP-ir overlap suggested the absence of NTPDase2 from typical protoplasmic astrocytes in the spinal cord (Figure 3G), striatum (Figure 3I), thalamus (Figure 3J), hippocampus (Figure 3K), cerebral cortex (Figure 3L) and cerebellar Bergman glia (Figure 3H). The associations between the two signals were observed in non-stellate astrocytes in medial habenula (Figure 3M), ependymal cells lining ventricles (Figure 3N, P, and Q), cells of choroid plexus (Figure 3O) and in perivascular endfeet around surrounding blood vessels (Figure 3R). Sporadic association of NTPDase2 was observed with intermediate filament protein vimentin (Figure 4P) and nestin (Figure 4S), S100 protein (Figure 4T), and OPC cells marker NG2 (Figure 4Q) in the spinal cord and forebrain projection tracts.
Figure 3.
Double immunofluorescence labeling showing immunoreaction corresponding to NTPDase2 (red) and GFAP (green) in distinct brain areas. Arrows point to colocalization of NTPDase2/GFAP-ir (yellow). Abbreviations: cp = choroid plexus; 3V = Third ventricle; 4V = Fourth ventricle. Magnification: ×400 (A–M) and ×600 (N–R).
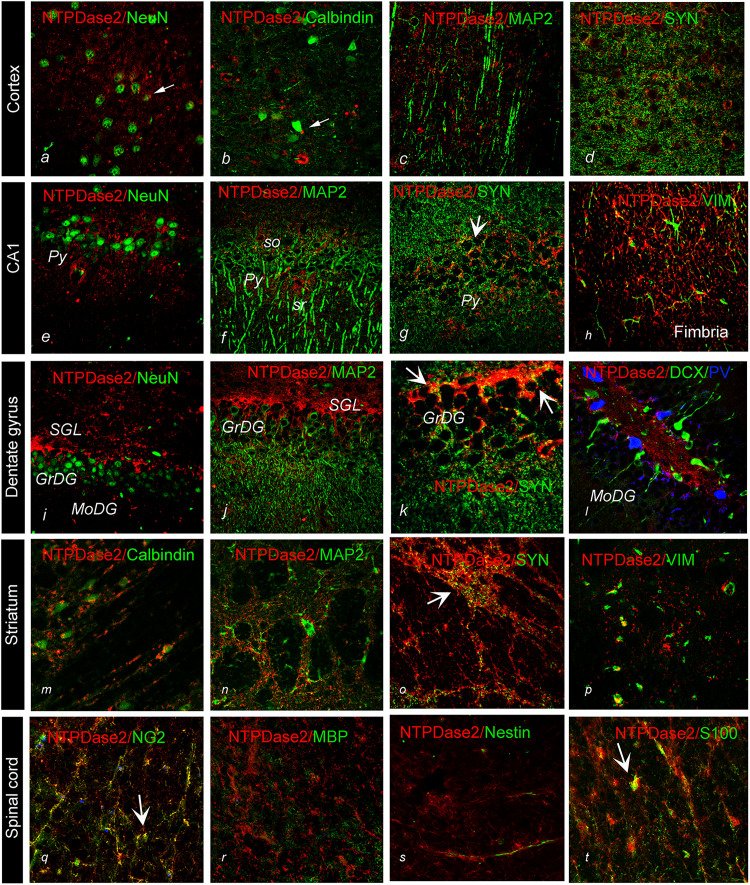
Figure 4.
Double immunofluorescence labeling directed to NTPDase2 (red) and specific cell markers (green) in distinct brain regions. General neuronal marker NeuN; specific neuronal markers, calbindin, MAP2, Doublecortin (DCX); Parvalbumin (PV), Synaptophysin (SYN); glial cell markers, vimentin – VIM, S100, nestin, NG2, myelin basic protein (MBP). Arrows point to colocalization between the signals (yellow).
Abbreviations: GrDG = granule cell layer; fi = Fimbria; MoDG = molecular layer; PoDG = Polymorphic layer; Py = Pyramidal cell layer; Or = Stratum, oriens; Rad = Stratum radiatum. Magnification: ×400.
Concerning the association of NTPDase2 and Syn, significant overlap was found in the cortex, striatum, and hippocampus (Figure 4K). NTPDase2-ir was not found in association with general neuronal marker NeuN (Figure 4A, E, and I), specific neuronal markers calbindin (Figure 4B and M), parvalbumin (Figure 4L), and doublecortin (Figure 4L), and dendritic marker MAP2 (Figure 4C, F, J, and N). Apparent co-localization was also not found with myelin basic protein myelin basic protein (MBP) (Figure 4R).
Expression of NTPDase2 After TMT-Induced Hippocampal Neurodegeneration
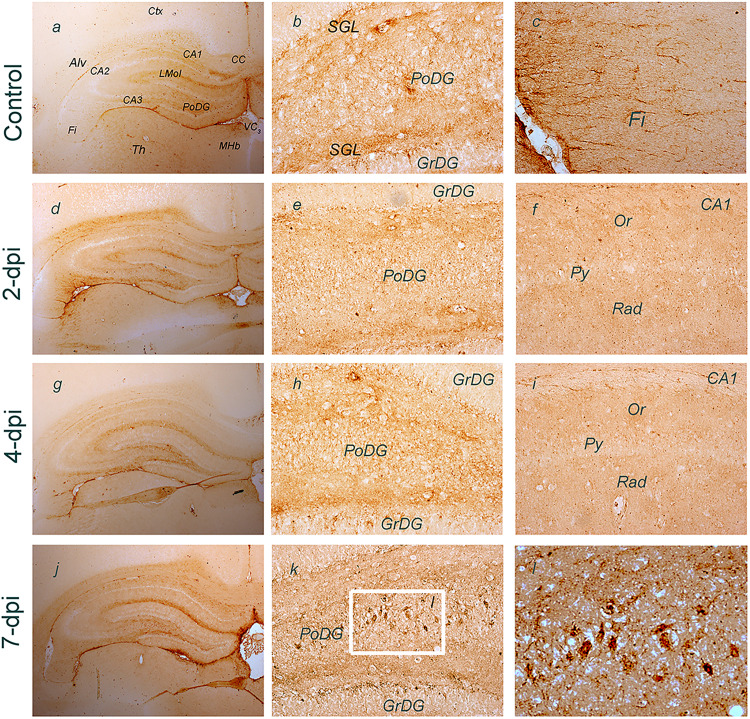
We next used the model of trimethyltin (TMT)-induced neurodegeneration to explore the expression of NTPDase2 in the hippocampal tissue. The neurotoxicant is known to induce progressive hippocampal cell death and astrocyte-driven neuroinflammation and microgliosis (Dragic, Zarić, et al., 2019; Dragic et al., 2021a). Two days after intoxication (2-dpi), NTPDase2-mRNA decreased to about 20% relative to control (p < 0.001), followed by a gradual increase at 4-dpi, and the recovery and overcompensation at 7-dpi at 21-dpi, respectively (Table 3).
Table 3.
Hippocampal Expression of Entpd2 After TMT- Induced Neurodegeneration.
| 2-dpi | 4-dpi | 7-dpi | 21-dpi | |
| NTPD2-mRNA abundance (relative to control) | 0.15 ± 0.34* | 0.69 ± 0.21* | 1.57 ± 0.12 | 1.71 ± 0.23* |
*Significance p < 0.01.
Expression of NTPDase2 was more closely analyzed in the hippocampus at a histological level using NTPDase2-directed immunohistochemistry (Figure 5). The most prominent NTPDase2 labeling could be detected in the DG, particularly in the parenchyma of the polymorphic layer (PoDG), which comprises several types of neurons and axons of granule cells, which constitute the granule cell layer (GrDG). Prominent labeling was also observed in the subgranular layer (SGL), at processes traversing GrDG towards the molecular layer (MoDG), while individual granule neurons remained pale (Figure 5B). Moderate NTPDase2-ir was observed in the fimbria (Fi) and alveus (Alv) (Figure 5C). At 2-dpi sections, the overall intensity of NTPDase2-ir decreased, mostly in LMol and PoDG (Figure 5D–F), and, to a lesser extent, in the hippocampal projection tracts. At 4-dpi, the intensity of NTPDase2-ir further decreased in all the immunoreactive areas (Figure 5G–I). However, the intensity of NTPDase2-ir returned to near control level in LMol at 7-dpi, while conspicuous NTPDase2-ir labeling appeared in PoDG and SGL (Figure 5K). Based on the relative position and cell body size and shape, these prominent NTPDase2-ir elements could be presynaptic boutons of the mossy fiber collaterals ending at giant aspiny stellate cells. These axons and their collaterals form unusually large presynaptic boutons that heavily innervate neurons within PoDG (Amaral, 1978; Amaral et al., 2007).
Figure 5.
Immunohistochemical labeling of NTPDase2 at coronal hippocampal sections after TMT exposure. Abbreviations: alv = Alveus; CC = corpus callosum; DG = Dentate gyrus; GrDG = granule cell layer; fi = Fimbria; MHb = Medial habenula; MoDG = molecular layer; PoDG = Polymorphic layer; Py = Pyramidal cell layer; sm = Stria medullaris; Or = Stratum, oriens; Rad = Stratum radiatum; 3V = Third ventricle. Frame in (K) shows area enlarged in (L). Arrow points to NTPDase2-ir presynaptic boutons terminating within PoDG. Magnification: ×50 (a, d, g, j), ×200 (b, c, e, f, h, i, k, l).
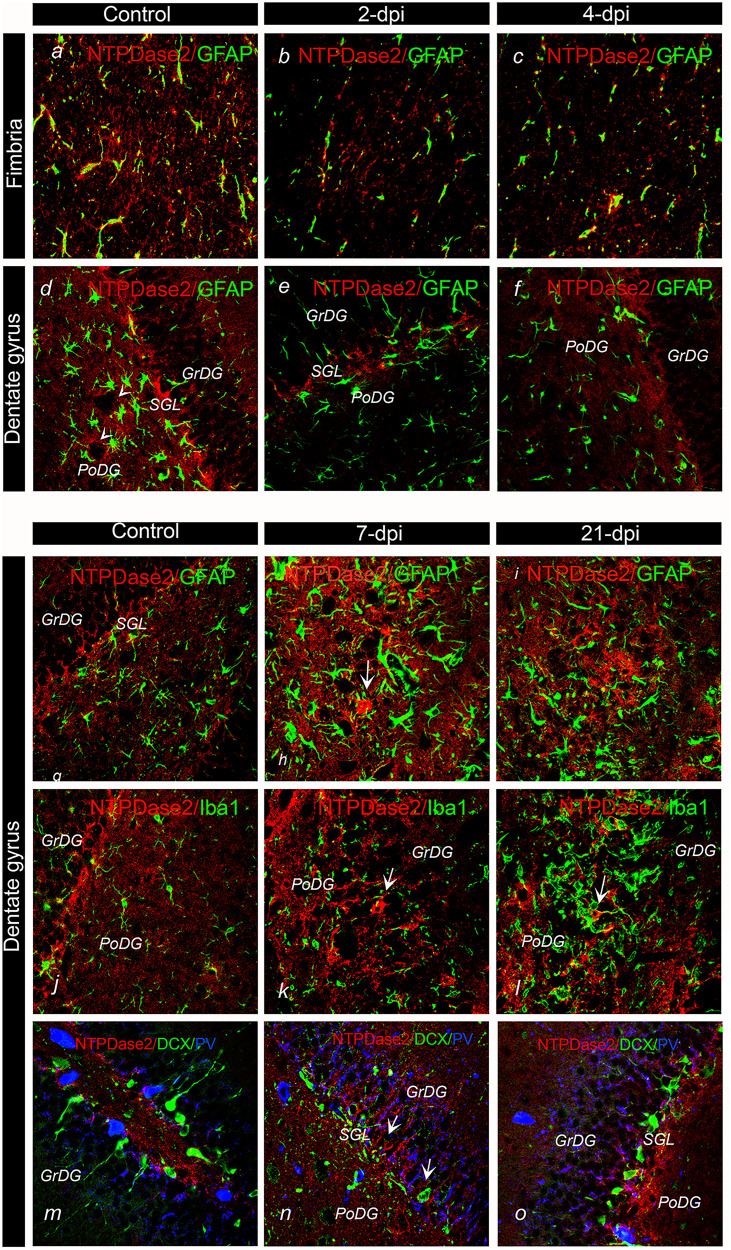
The cell types with altered NTPDase2 expression after TMT exposure were confirmed by double immunofluorescence (Figure 6). Double NTPDase2/GFAP-ir revealed the decrease in NTPDase2-ir intensity at 2- and 4-dpi, mainly at fibrous astrocytes in the fimbria (Figure 6B and C) and in the PoDG parenchyma surrounding conspicuous GFAP-ir astrocytes (Figure 6E and F). As expected, at 7-dpi and 21-dpi, the overall intensity of NTPDase2-ir increased, again mainly in PoDG (Figure 6H and I) and at NTPDase-ir processes traversing the GrDG (Figure 6K and L). Interestingly, although at control sections finely ramified Iba1-ir microglia was surrounded by NTPDase2-ir (Figure 6J), at 7-dpi and 14-dpi, NTPDase2-ir was very sparse in the surroundings of these highly reactive microglial cells (Figure 6L). Because the SGL contains several neuronal cell types, including neuronal progenitor cells at various transition states of the neurogenic pathway (Shukla et al., 2005), we investigated the potential colocalization of NTPDase2 with cell type-specific markers. NTPDase2-ir was not co-localized at parvalbumin (PV)-ir neurons nor doublecortin (DCX)-ir neural stem cells (Figure 6N and O), but was found in apposition with the cell bodies. Based on anatomical and morphological criteria, we concluded that NTPDase2-ir resided at synaptic varicosities of granule cell collaterals ending at mossy cells in the polymorphic layer and astroglial sheats passing through the layer of tightly packed granule cells, where the intensity of NTPDase2-ir increased at 7-dpi and 14-dpi.
Figure 6.
Immunofluorescence labeling directed to NTPDase2 (red) and cell-specific markers in the hippocampus and dentate gyrus 2-7-dpi after TMT exposure. A–L: Double NTPDase2/GFAP immunofluorescence labeling; M–O: NTPDase2/DCX/PV immunofluorescence labeling. Arrows point to NTPDase2-ir elements in the DG.
Abbreviations: GrDG = granule cell layer; Fi = Fimbria; MoDG = molecular layer; PoDG = Polymorphic layer; Py = Pyramidal cell layer; Or = Stratum, oriens; Rad = Stratum radiatum. Magnification: ×400.
Regulation of NTPDase2 by Inflammatory Mediators In Vitro
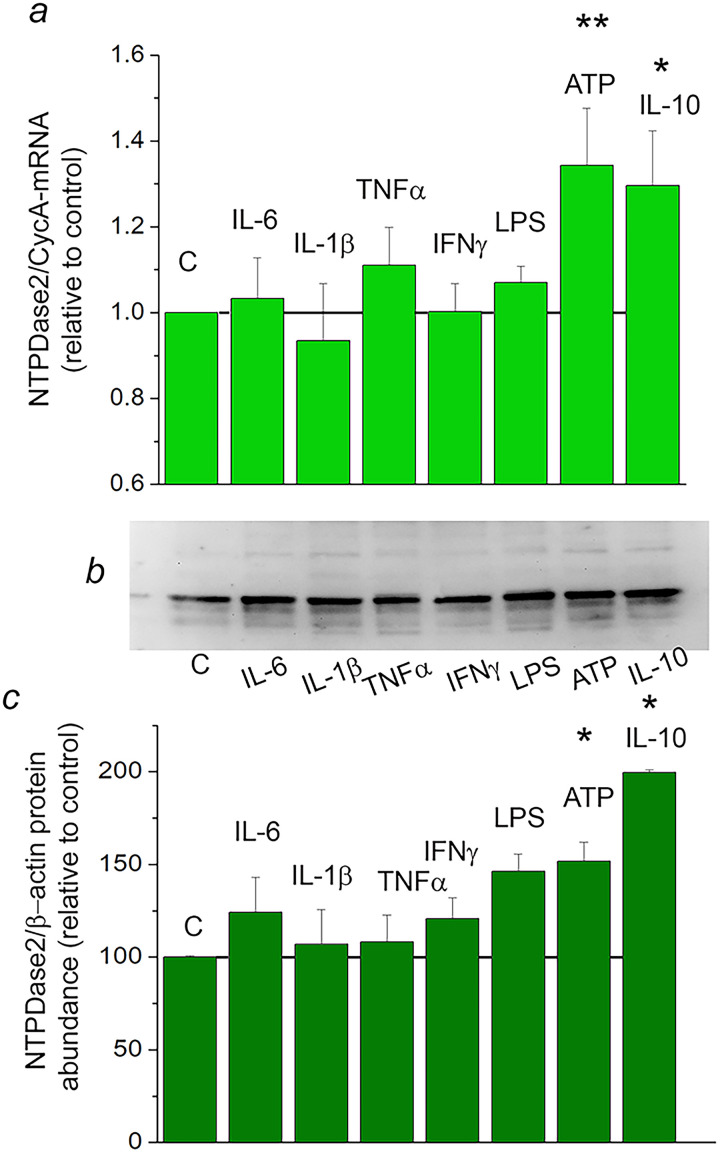
Factors that might regulate transcription of Entpd2 and translation of mature NTPDase2 protein in neuroinflammatory and neurodegenerative conditions were explored in vitro, in rat primary cortical astrocytes (Figure 7) and OLN93 oligodendroglial cell line (Figure 8). The expression of purinergic system components was estimated in the two culture systems by quantitative real-time PCR (Table 4). In both cell types, NTPDase2-mRNA was, by far, the most abundant ectonucleotidase transcript. In primary astrocytes, NTPDase2-mRNA was ∼ 40 times more abundant than NTPDase1-mRNA and ∼ 20 times more abundant than eN-mRNA. Regulation of Entpd2 was further assessed at the mRNA and the protein levels, after exposing primary astrocytes to inflammatory mediators for 8 h and 24 h, respectively (Figure 7A–C). Proinflammatory cytokines IL-6, IL-1β, TNFα, and IFNγ did not affect the expression of Entpd2 in primary astrocytes, either at mRNA (Figure 7A) or the protein levels (Figure 7B and C), while ATP and anti-inflammatory cytokine IL-10 increased the transcript and the protein abundances for ∼50% and ∼90% in respect to non-treated astrocyte culture (p < 0.05). These inflammatory mediators were also evaluated for their potency to alter the expression of P2Y1 and P2Y13. While expression of P2Y1-mRNA remained stable, a several-fold increase in P2Y13-mRNA abundance was demonstrated after exposure of astrocyte cultures to all inflammatory mediators applied (Table 5).
Figure 7.
Regulation of NTPDase2 expression by inflammatory mediators in primary cortical astrocytes. a: NTPDase2/CycA-mRNA abundance in primary astrocytes treated with inflammatory factors for 8 h, expressed relative to non-treated control culture, ± SEM (from n ≥ 3 determinations, performed in two independent culture preparations and mRNA extractions. b: Representative immunoblot membrane and c: quantification of NTPDase2/β-actin protein abundance in astrocytes treated with inflammatory factors for 24 h, expressed relative to non-treated control culture ± SEM (from n = 3 determinations, performed with two independent culture preparations). Significance shown inside the graphs: *p < 0.05.
Figure 8.
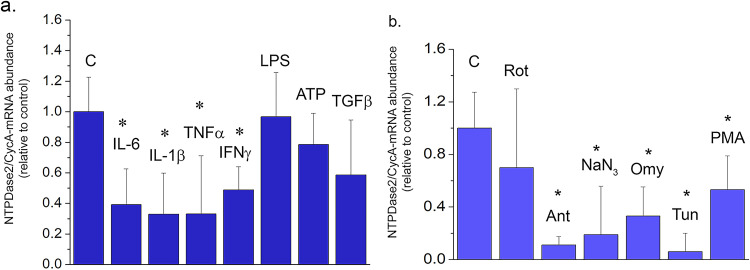
Regulation of NTPDase2 in OLN cell line. NTPDase2/CycA-mRNA abundance in OLN cells treated with (a) inflammatory mediators and (b) cell-intrinsic stressors, for 8 h. Results are expressed relative to non-treated control culture, ± SEM (from n = 3 determinations, performed in two independent culture preparations). Significance is shown inside the graph: *p < 0.01.
Abbreviations: Rot = rotenone, Ant = antimycin, Omy = oligomycin, Tun = tunicamycin, PMA = phorbol myristate ester.
Table 4.
Relative Transcript Abundance (2−ΔCt in Respect to CycA).
| Primary astrocytes | OLN93 cell line | |
|---|---|---|
| Nt5e | 0.00697 | 0.000315 |
| Entpd1 | 0.00364 | n.a. |
| Entpd2 | 0.14063* | 0.001042* |
| Entpd3 | 0.00012 | n.a. |
| Enpp1 | n.a. | n.a. |
| Enpp2 | 0.00106 | n.a. |
| P2Ry1 | 0.00664 | n.a. |
| P2Ry12 | n.a. | n.a. |
| P2Ry13 | 0.00228 | n.a. |
Significance level: *p < 0.05 in respect to control astrocyte and OLN93 culture.
Table 5.
Relative P2Y1R- and P2Y13R Transcript Abundance in Primary Astrocytes (Fold-Change Relative to Untreated Control).
| Treatment | Target gene | |
|---|---|---|
| P2Y1R | P2Y13R | |
| IL6 | 1.06 ± 0.29 | 11.76 ± 1.90 |
| IL-1β | 1.45 ± 0.16 | 5.14 ± 1.33 |
| TNFα | 1.22 ± 0.11 | 4.82 ± 0.57 |
| IFNγ | 1.16 ± 0.06 | 7.95 ± 0.51 |
| ATP | 1.26 ± 0.12 | 7.57 ± 3.39 |
| IL10 | 1.14 ± 0.17 | 6.68 ± 0.67 |
Significance level: *p < 0.05 in respect to control astrocyte culture.
Regulation of NTPDase2 was further assessed in the OLN93 cell line (Figure 8). The data showed that NTPDase2-mRNA and eN-mRNA were the only represented ectonucleotidase transcripts in the OLN93 cell line (Table 4). Exposure of OLN93 cells to pro-inflammatory cytokines IL-6, IL-1β, TNFα, and IFNγ decreased the abundance of NTPDase2-mRNA, whereas ATP and TGFβ did not exert a measurable effect, at least 8h after the exposure (Figure 8A). In light of the known sensitivity of OPC cells to oxidative stress, we also explored the impact of agents that affect cell metabolism and mitochondrial functions on Entpd2 expression (Figure 8B). The results showed that inhibitors of respiratory chain complex I and complex II did not change the abundance of NTPDase2-mRNA, whereas inhibitors of respiratory chain complex III and IV, and ATP synthase markedly decreased the transcript abundance. Reduced abundance of NTPDase2-mRNA was also demonstrated in OLN93 cells treated with tunicamycin which blocks N-linked glycosylation and with phorbol myristate ester, which stimulates protein kinase C.
Discussion
In the present study, we sought to determine how NTPDase2 is affected and regulated under neuroinflammatory conditions in vitro and in vivo. Before that, we qualified the topographic distribution and cellular localization of NTPDase2 in adult rat brains in physiological conditions and demonstrated significant differences at the quantitative level. Namely, the abundance of NTPDase2 protein was the highest in the hippocampus and caudate-putamen and significantly lower in the cerebellum and olfactory bulb. At the cellular and subcellular level, the presence of NTPDase2 protein was immunodetected in the purified plasma membrane, glial membrane, and synaptic membrane fractions, suggesting its prevalent representation in astrocytes and synaptic endings. Succeeding immunofluorescence microscopy confirmed the specific patterns of NTPDase2 expression in the white and gray matter throughout the brain (Braun et al., 2003; Gampe et al., 2012). In the white matter, NTPDase2 was mainly localized at fibrous astrocytes and NG2-expressing OPC cells. At these locations, NTPDase2 is ideally positioned to provide the ligand for homeostatic ADP signaling, mediated via P2Y1, which operates at astrocytes (Fischer and Krügel, 2007), and P2Y12/P2Y13, which operate at oligodendrocytes and quiescent microglia (Amadio et al., 2014; Bianco et al., 2005; Kyrargyri et al., 2020). In the gray matter, NTPDase2 was never observed at typical protoplasmic astrocytes (Braun et al., 2003; Gampe et al., 2012; Jakovljevic et al., 2017). Instead, it was expressed in specialized astrocytes in distinct brain regions, including subependymal and subpial astrocytes, thalamic tanycytes, and laminar astrocytes in the medial habenula. We also found NTPDase2 in association with the presynaptic marker synaptophysin in the synaptic-rich layer of DG. The representation of NTPDase2 in perisomatic nerve endings, suggests its role in the elimination of ATP from the synapses and potentiation of ADP-mediated signaling in physiological conditions (Gampe et al., 2012). We did not observe the expression of NTPDase2 in quiescent or activated microglia, mature myelin, or neuronal elements other than presynaptic membranes. Our immunofluorescence data showing restricted expression of NTPDas2 in specialized astrocytes, synaptic endings, and OPC cells and its absence from mature oligodendrocytes and microglia are following the RNA-seq expression dataset obtained from acutely purified cell populations isolated from mouse cerebral cortex, which demonstrate dominant expression of Entpd2 in astrocytes, and to a much lesser extent, in neurons, OPC, and pericytes, and complete absence from mature oligodendrocytes and microglia (Zhang et al., 2014).
We next explored the NTPDase2 expression profile in the model of TMT-induced hippocampal neurodegeneration. Within the hippocampus as a whole, a significant drop in NTPDase2-mRNA abundance was observed at 2-dpi, followed by a gradual recovery to the control level from 4- to 7-dpi and an overcompensation at 21-dpi. A similar acute decrease in Entpd2 expression in the spinal cord was demonstrated in the rat monophasic EAE model (Jakovljevic et al., 2017). It is of note that TMT inflicts irreversible neuronal damage with progressive neuroinflammation, while EAE represents acute neurodegeneration and self-resolving neuroinflammation. Therefore, we supposed that transient silencing of NTPDase2 occurs as a part of the acute response to injury, irrespective of the nature of the initial insult. The transient decrease followed by full recovery of Entpd2 expression was also demonstrated in isolated rat cortical, hippocampal, and striatal astrocytes in aging (Clarke et al., 2018) and after removal of ovaries (Grkovic et al., 2019).
To address the question of the pathophysiological significance of negative NTPDase2 regulation in neuroinflammatory conditions, we sought to identify cell types that downregulate NTPDase2 and to assess the overall neuroinflammatory context induced by TMT. Given that major changes in the expression of NTPDase2 occurred in DG, we focused the attention on this hippocampal subregion. Within DG, the expression of NTPDase2 decreased in presynaptic boutons innervating neurons in PoDG (Amaral et al., 2007), whereas the recovery occurred in the PoDG, SGL, and at astrocytic sheats traversing the GrDG. As described in several recent papers, exposure to TMT induces selective neuronal death (Ceccariglia et al., 2019) and consequent massive increase in extracellular ATP, which attracts microglia and initiates inflammatory responses of astrocytes (Davalos et al., 2005), mainly via P2X7 receptors (Buffo et al., 2010). From 2-dpi onwards, PoDG became populated with highly reactive astrocytes, which express iNOS, C3, NFkB, and P2X7R, and secrete pro-inflammatory cytokines IL-1β, IL-6, IFN-γ, and TNF-α in the parenchyma (Dragic et al., 2021b; Latini et al., 2010; Little et al., 2012). With a short delay, at 4-dpi numerous amoeboid microglia appear in PoDG, mostly exhibiting a neuroprotective M2 phenotype. The microglial cells massively up-regulate NTPDase1/CD73, which can complete the hydrolysis of ATP to adenosine. At 21-dpi, PoDG microglia reduce the expression of NTPDase1/CD73 (Dragic et al., 2021b), while the surrounding astrocytes retain their inflammatory profile. Within the whole tissue context induced by TMT, down-regulation of NTPDase2 occurs in the robust pro-inflammatory environment induced by neuronal cell death (Ceccariglia et al., 2019), with efficient microglial NTPDase1/eN-catalyzed hydrolysis of ATP and enhanced expression of ATP-sensitive P2X4, P2X7, P2Y1, P2Y6 (Geloso et al., 2011; Dragic et al., 2021a). Thus, down-regulation of Entpd2 most likely attenuates P2Y12/P2Y13-mediated ADP signaling, and switches-on astroglial and neurogenic expansion, which are under the control of ATP-responsive P2X7 and P2Y1 receptors (Franke et al., 2004; Gampe et al., 2015; Quintas et al., 2011). The subsequent recovery of NTPDase2 expression, observed at 21-dpi TMT and in the resolution phase of EAE, most likely re-activates P2Y12- and P2Y13-mediated proliferative brake, and paves the way for immunosuppression and tissue remodeling. This may be particularly relevant in the SGL, where NTPDase2 is associated with a population of astrocyte-like proliferating progenitor cells (Shukla et al., 2005), whereas cells in the transition state lose NTPDase2, express neuronal markers, and move into GrDG. Our study corroborates the existing data, showing the presence of NTPDase2 in small round non-neuronal cell bodies in the SGL and at ventral cellular processes which traverse SGL into the PoDG. It was shown that disruption of NTPDase2 (Gampe et al., 2015) or microglial P2Y13-mediated ADP signaling (Stefani et al., 2018) enhances proliferation in the SGL, confirming the role of ADP in progenitor cell proliferation and expansion. Thus, one direct consequence of the negative regulation of NTPDase2 in neuroinflammatory conditions would be increased proliferation of cells in the SGL, which was already demonstrated in EAE (Schneider et al., 2016), kainic acid-induced neurotoxicity (Gonzales-Reyes et al., 2019), and Alzheimer's disease (Sun et al., 2018).
Previous studies on the molecular mechanism of TMT action in astrocytes have shown that the neurotoxicant increases Ca2+ influx through L-type voltage-gated channels and inflicts oxidative stress and mitochondrial membrane depolarization and enhances the release of pro-inflammatory cytokines, including TNF-α, IFN-γ, IL-1β, and IL-6 (Dragic et al., 2021a; Geloso et al., 2011; Piacentini et al., 2008). Therefore, by using primary astrocytes and OLN93 oligodendroglial cells, in the present study we assessed the regulation of Entpd2 in stress conditions in vitro. Our results show that astrocytes and oligodendroglia may utilize different molecular mechanisms and/or transcription factors to control the expression of Entpd2. The assumption is based on the findings on the selective effects of proinflammatory cytokines TNF-α, IFN-γ, IL-1β, and IL-6 on Entpd2 expression in OLN93 cells and anti-inflammatory cytokine IL-10 and ATP in primary astrocytes. The expression of Entpd2 was also disturbed in OLN93 cells, in the presence of several metabolic inhibitors, known to initiate the integrated stress response (ISR) (Teske et al., 2018). Namely, expression of Entpd2 was severely impaired in the conditions of metabolic stress, oxidative stress, and endoplasmic reticulum stress. The expression was negatively regulated by protein kinase C stimulator, phorbol myristate ester, previously shown to inhibit the activity of NTPDase2 (Wang et al., 2005). It is well-known that stress response initiates intracellular cascades which in a coordinated manner enhance the expression of pro-survival genes while shutting down the expression of others (Pakos-Zebrucka et al., 2016). Among the intracellular pathways activated in conditions of cell stress are NfkB, PKC, and their downstream effector IL-6 (Sanchez et al., 2019). In addition to several important homeostatic and pathophysiological roles performed by IL-6 in neurons, astrocytes, and microglia (Norris et al., 1994; Sparacio et al., 1992), the cytokine acts as a negative regulator of Enpd2 via specific promoter elements (Yu et al., 2008). Therefore, in the present context, one of the potential molecular mediators of TMT-induced down-regulation of Entpd2 may be IL-6, which is massively upregulated in activated astrocytes as a consequence of neuronal death and oxidative stress induced by TMT (Dragic et al., 2021b). We can not exclude the actions of other chemokines and inflammatory mediators, like MCP-1/CCL2, IFN-α and -β, IL-1α + TNFα, and C1q, which altered the expression of Entpd2 in several cell lines including astrocytes (Hasel et al., 2021, Kruglov et al., 2006).
In summary, the present study shows that expression of NTPDase2 is negatively regulated under neuroinflammatory conditions induced by TMT, mainly in fibrous astrocytes and presynaptic ending in PoDG, which result in the reduced formation of ADP and attenuation of P2Y12,13 signaling between astrocyte/oligodendrocytes/microglia. Among the signaling factors that may be responsible for the negative regulation of NTPDase2 gene expression, we have identified proinflammatory cytokines IL-6, IL-1β, TNFα, and IFNγ and signaling induced by cell-intrinsic stressors, including oxidative and endoplasmic reticulum stresses.
Acknowledgments
We would like to express gratitude to Prof. Jean Sevigny for providing rabbit anti-rat NTPDase2 antisera (BZ3-4F/rN2-6L) used in the study.
Footnotes
Author Contributions: All authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article. N.N, I.G., and M.D. conceived and directed the project. M.D. and N.N. designed experiments, D.M and K.M. performed immunohistology, light, and confocal microscopy, M.Z.K. performed immunoblotting, N.M. performed rt-qPCR experiments. M.A., M.J., and M.K. performed the in vitro study in primary astrocytes and OLN93 cells. I.L. and D.L. were involved in immunofluorescence data interpretation. N.N. wrote the manuscript and all authors read and approved the final version. The authors had full access to all of the data in this study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Ministry of Education, Science, and Technological Development of the Republic of Serbia, (grant number 451-03-1/2021-16/14, 451-03-68/2022-14/200178).
ORCID iD: Nadezda Nedeljkovic https://orcid.org/0000-0003-3046-0983
Data Availability: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval: All animal procedures were performed in compliance with the European Communities Council Directive (2010/63/EU) and the National Laboratory Animal Science Association for the protection of animals used for experimental and scientific purposes and were approved by the Institutional Ethics Committee under authorization reference numbers 323-07-02057/2017-05 and EK-BF-2016/05.
References
- Abbracchio M. P., Burnstock G., Verkhratsky A., Zimmermann H. (2009). Purinergic signalling in the nervous system: An overview. Trends in Neurosciences, 32(1), 19–29. PMID: 19008000. 10.1016/j.tins.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Adzic M., Nedeljkovic N. (2018). Unveiling the role of ecto-5’-nucleotidase/CD73 in astrocyte migration by using pharmacological tools. Frontiers in Pharmacology, 9, 153. PMID: 29545748; PMCID: PMC5837971. 10.3389/fphar.2018.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B., Longhi M. S., Robson S. C., Stagg J. (2017). The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunological Reviews, 276(1), 121–144. PMID: 28258700; PMCID: PMC5338647. 10.1111/imr.12528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadio S., Parisi C., Montilli C., Carrubba A. S., Apolloni S., Volonté C. (2014). P2y(12) receptor on the verge of a neuroinflammatory breakdown. Mediators of Inflammation, 2014, 1–15. PMID: 25180027; PMCID: PMC4142314. 10.1155/2014/975849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral D. G. (1978). A Golgi study of cell types in the hilar region of the hippocampus in the rat. The Journal of Comparative Neurology, 182(5), 851–914. PMID: 730852. 10.1002/cne.901820508 [DOI] [PubMed] [Google Scholar]
- Amaral D. G., Scharfman H. E., Lavenex P. (2007). The dentate gyrus: Fundamental neuroanatomical organization (dentate gyrus for dummies). Progress in Brain Research, 163, 3–22. PMID: 17765709; PMCID: PMC2492885. 10.1016/S0079-6123(07)63001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F., Fumagalli M., Pravettoni E., D’Ambrosi N., Volonte C., Matteoli M., Abbracchio M. P., Verderio C. (2005). Pathophysiological roles of extracellular nucleotides in glial cells: Differential expression of purinergic receptors in resting and activated microglia. Brain Research Reviews, 48(2), 144–156. PMID: 15850653. 10.1016/j.brainresrev.2004.12.004 [DOI] [PubMed] [Google Scholar]
- Boehm S. (2003). P2Ys go neuronal: Modulation of Ca2+ and K+ channels by recombinant receptors. British Journal of Pharmacology, 138(1), 1–3. PMID: 12522065; PMCID: PMC1573656. 10.1038/sj.bjp.0705044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems J. M., Communi D., Gonzalez N. S., Robaye B. (2005). Overview of the P2 receptors. Seminars in Thrombosis and Hemostasis, 31(02), 139–149. PMID: 15852217. 10.1055/s-2005-869519 [DOI] [PubMed] [Google Scholar]
- Bours M. J., Dagnelie P. C., Giuliani A. L., Wesselius A., Di Virgilio F. (2011). P2 receptors and extracellular ATP: A novel homeostatic pathway in inflammation. Frontiers in Bioscience, 3(1), 1443–1456. PMID: 21622280. 10.2741/235 [DOI] [PubMed] [Google Scholar]
- Bours M. J., Swennen E. L., Di Virgilio F., Cronstein B. N., Dagnelie P. C. (2006). Adenosine 5’-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacology & Therapeutics, 112(2), 358–404. PMID: 16784779. 10.1016/j.pharmthera.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Braun N., Sévigny J., Mishra S. K., Robson S. C., Barth S. W., Gerstberger R., Hammer K., Zimmermann H. (2003). Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. European Journal of Neuroscience, 17(7), 1355–1364. PMID: 12713638. 10.1046/j.1460-9568.2003.02567.x [DOI] [PubMed] [Google Scholar]
- Braun N., Sévigny J., Robson S. C., Hammer K., Hanani M., Zimmermann H. (2004). Association of the ecto-ATPase NTPDase2 with glial cells of the peripheral nervous system. Glia, 45(2), 124–132. PMID: 14730706. 10.1002/glia.10309 [DOI] [PubMed] [Google Scholar]
- Brisevac D., Adzic M., Laketa D., Parabucki A., Milosevic M., Lavrnja I., Bjelobaba I., Sévigny J., Kipp M., Nedeljkovic N. (2015). Extracellular ATP selectively upregulates ecto-nucleoside triphosphate diphosphohydrolase 2 and ecto-5’-nucleotidase by rat cortical astrocytes in vitro. Journal of Molecular Neuroscience, 57(3), 452–462. PMID: 26080748. 10.1007/s12031-015-0601-y [DOI] [PubMed] [Google Scholar]
- Brisevac D., Bajic A., Bjelobaba I., Milosevic M., Stojiljkovic M., Beyer C., Clarner T., Kipp M., Nedeljkovic N. (2013). Expression of ecto-nucleoside triphosphate diphosphohydrolase1-3 (NTPDase1-3) by cortical astrocytes after exposure to pro-inflammatory factors in vitro. Journal of Molecular Neuroscience, 51(3), 871–879. PMID: 23990338. 10.1007/s12031-013-0088-3 [DOI] [PubMed] [Google Scholar]
- Buffo A., Rolando C., Ceruti S. (2010). Astrocytes in the damaged brain: Molecular and cellular insights into their reactive response and healing potential. Biochemical Pharmacology, 79(2), 77–89. PMID: 19765548. 10.1016/j.bcp.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Burnstock G., Knight G. E. (2004). Cellular distribution and functions of P2 receptor subtypes in different systems. International Review of Cytology, 240, 31–304. PMID: 15548415. 10.1016/S0074-7696(04)40002-3 [DOI] [PubMed] [Google Scholar]
- Cahill C. M., Rogers J. T. (2008). Interleukin (IL) 1beta induction of IL-6 is mediated by a novel phosphatidylinositol 3-kinase-dependent AKT/IkappaB kinase alpha pathway targeting activator protein-1. Journal of Biological Chemistry, 283(38), 25900–25912. PMID: 18515365; PMCID: PMC2533786. 10.1074/jbc.M707692200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman A. J., Mills J. H., Krenz A., Kim D. G., Bynoe M. S. (2011). Adenosine receptor signaling modulates permeability of the blood-brain barrier. Journal of Neuroscience, 31(37), 13272–13280. PMID: 21917810; PMCID: PMC3328085. 10.1523/JNEUROSCI.3337-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccariglia S., Alvino A., Del Fà A., Parolini O., Michetti F., Gangitano C. (2019). Autophagy is activated in vivo during trimethyltin-induced apoptotic neurodegeneration: A study in the rat hippocampus. International Journal of Molecular Sciences, 21(1), 175. PMID: 31881802; PMCID: PMC6982133. 10.3390/ijms21010175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B. P., Frischauf A. M. (1997). Cloning and mapping of a human and mouse gene with homology to ecto-ATPase genes. Mammalian Genome, 8(9), 668–672. PMID: 9271669. 10.1007/s003359900534 [DOI] [PubMed] [Google Scholar]
- Clarke L. E., Liddelow S. A., Chakraborty C., Münch A. E., Heiman M., Barres B. A. (2018). Normal aging induces A1-like astrocyte reactivity. Proceedings of the National Academy of Sciences, 115(8), E1896–E1905. PMID: 29437957; PMCID: PMC5828643. 10.1073/pnas.1800165115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha R. A. (2005). Neuroprotection by adenosine in the brain: From A(1) receptor activation to A (2A) receptor blockade. Purinergic Signalling, 1(2), 111–134. PMID: 18404497; PMCID: PMC2096528. 10.1007/s11302-005-0649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J. V., Zuo Y., Jung S., Littman D. R., Dustin M. L., Gan W. B. (2005). ATP Mediates rapid microglial response to local brain injury in vivo. Nature Neuroscience, 8(6), 752–758. PMID: 15895084. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- Dragić M., Milićević K., Adžić M., Stevanović I., Ninković M., Grković I., Andjus P., Nedeljković N. (2021a). Trimethyltin increases intracellular Ca2+ via L-type voltage-gated calcium channels and promotes inflammatory phenotype in rat astrocytes in vitro. Molecular Neurobiology, 58(4), 1792–1805. PMID: 33394334. 10.1007/s12035-020-02273-x [DOI] [PubMed] [Google Scholar]
- Dragić M., Mitrović N., Adžić M., Nedeljković N., Grković I. (2021b). Microglial- and astrocyte-specific expression of purinergic signaling components and inflammatory mediators in the rat hippocampus during trimethyltin-induced neurodegeneration. ASN Neuro, 13, 17590914211044882. PMID: 34569324; PMCID: PMC8495514. 10.1177/17590914211044882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragić M., Zarić M., Mitrović N., Nedeljković N., Grković I. (2019). Two distinct hippocampal astrocyte morphotypes reveal subfield-different fate during neurodegeneration induced by trimethyltin intoxication. Neuroscience, 423, 38–54. PMID: 31682945. 10.1016/j.neuroscience.2019.10.022 [DOI] [PubMed] [Google Scholar]
- Erta M., Quintana A., Hidalgo J. (2012). Interleukin-6, a major cytokine in the central nervous system. International Journal of Biological Sciences, 8(9), 1254–1266. PMID: 23136554; PMCID: PMC3491449. 10.7150/ijbs.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erta M., Quintana A., Hidalgo J. (2012). Interleukin-6, a major cytokine in the central nervous system.. Int J Biol Sci, 8(9), 1254–1266. Epub 2012 Oct 25. PMID: 23136554. 10.7150/ijbs.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields R. D. (2011). Nonsynaptic and nonvesicular ATP release from neurons and relevance to neuron-glia signaling. Seminars in Cell & Developmental Biology, 22(2), 214–219. PMID: 21320624; PMCID: PMC3163842. 10.1016/j.semcdb.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Krügel U. (2007). P2y receptors: Focus on structural, pharmacological and functional aspects in the brain. Current Medicinal Chemistry, 14(23), 2429–2455. PMID: 17979698. 10.2174/092986707782023695 [DOI] [PubMed] [Google Scholar]
- Franke H., Grummich B., Härtig W., Grosche J., Regenthal R., Edwards R. H., Illes P., Krügel U. (2006). Changes in purinergic signaling after cerebral injury -- involvement of glutamatergic mechanisms? International Journal of Developmental Neuroscience, 24(2-3), 123–132. PMID: 16387466. 10.1016/j.ijdevneu.2005.11.016 [DOI] [PubMed] [Google Scholar]
- Franke H., Krügel U., Grosche J., Heine C., Härtig W., Allgaier C., Illes P. (2004). P2y receptor expression on astrocytes in the nucleus accumbens of rats. Neuroscience, 127(2), 431–441. PMID: 15262333. 10.1016/j.neuroscience.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Fredholm B. B. (2007). Adenosine, an endogenous distress signal, modulates tissue damage and repair. Cell Death & Differentiation, 14(7), 1315–1323. PMID: 17396131. 10.1038/sj.cdd.4402132 [DOI] [PubMed] [Google Scholar]
- Gampe K., Hammer K., Kittel Á, Zimmermann H. (2012). The medial habenula contains a specific nonstellate subtype of astrocyte expressing the ectonucleotidase NTPDase2. Glia, 60(12), 1860–1870. PMID: 22865704. 10.1002/glia.22402 [DOI] [PubMed] [Google Scholar]
- Gampe K., Stefani J., Hammer K., Brendel P., Pötzsch A., Enikolopov G., Enjyoji K., Acker-Palmer A., Robson S. C., Zimmermann H. (2015). NTPDase2 and purinergic signaling control progenitor cell proliferation in neurogenic niches of the adult mouse brain. Stem Cells (Dayton, Ohio), 33(1), 253–264. PMID: 25205248; PMCID: PMC4270857. 10.1002/stem.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geloso M. C., Corvino V., Michetti F. (2011). Trimethyltin-induced hippocampal degeneration as a tool to investigate neurodegenerative processes. Neurochemistry International, 58(7), 729–738. 10.1016/j.neuint.2011.03.009 [DOI] [PubMed] [Google Scholar]
- George J., Gonçalves F. Q., Cristóvão G., Rodrigues L., Meyer Fernandes J. R., Gonçalves T., Cunha R. A., Gomes C. A. (2015). Different danger signals differently impact on microglial proliferation through alterations of ATP release and extracellular metabolism. Glia, 63(9), 1636–1645. PMID: 25847308. 10.1002/glia.22833 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes L. E., Chiang C. C., Zhang M., Johnson J., Arrillaga-Tamez M., Couturier N. H., Reddy N., Starikov L., Capadona J. R., Kottmann A. H., Durand D. M. (2019). Sonic Hedgehog is expressed by hilar mossy cells and regulates cellular survival and neurogenesis in the adult hippocampus. Sci Rep, 9(1), 17402. PMID: 31758070. 10.1038/s41598-019-53192-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E. G., Whittaker V. P. (1962). The isolation of nerve endings from brain: An electron-microscopic study of cell fragments derived by homogenization and centrifugation. Journal oif Anatomy, 96(Pt 1), 79–88. PMID: 13901297; PMCID: PMC1244174. [PMC free article] [PubMed] [Google Scholar]
- Grković I., Mitrović N., Dragić M., Adžić M., Drakulić D., Nedeljković N. (2019). Spatial distribution and expression of ectonucleotidases in rat hippocampus after removal of ovaries and estradiol replacement. Molecular Neurobiology, 56(3), 1933–1945. PMID: 29978426. 10.1007/s12035-018-1217-3 [DOI] [PubMed] [Google Scholar]
- Guttenplan K. A., Weigel M. K., Adler D. I., Couthouis J., Liddelow S. A., Gitler A. D., Barres B. A. (2020). Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nature Communications, 11(1), 3753. PMID: 32719333; PMCID: PMC7385161. 10.1038/s41467-020-17514-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel P., Rose I. V. L., Sadick J. S., Kim R. D., Liddelow S. A. (2021). Neuroinflammatory astrocyte subtypes in the mouse brain. Nature Neuroscience, 24(10), 1475–1487. PMID: 34413515. 10.1038/s41593-021-00905-6 [DOI] [PubMed] [Google Scholar]
- He Y., Jackman N. A., Thorn T. L., Vought V. E., Hewett S. J. (2015). Interleukin-1β protects astrocytes against oxidant-induced injury via an NF-κB-dependent upregulation of glutathione synthesis. Glia, 63(9), 1568–1580. PMID: 25880604; PMCID: PMC4506211. 10.1002/glia.22828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine P., Braun N., Heilbronn A., Zimmermann H. (1999). Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. European Journal of Biochemistry, 262(1), 102–107. PMID: 10231370. 10.1046/j.1432-1327.1999.00347.x [DOI] [PubMed] [Google Scholar]
- Idzko M., Ferrari D., Eltzschig H. K. (2014). Nucleotide signalling during inflammation. Nature, 509(7500), 310–317. PMID: 24828189; PMCID: PMC4222675. 10.1038/nature13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illes P., Rubini P., Ulrich H., Zhao Y., Tang Y. (2020). Regulation of microglial functions by purinergic mechanisms in the healthy and diseased CNS. Cells, 9(5), 1108. PMID: 32365642; PMCID: PMC7290360. 10.3390/cells9051108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura Y., Morizawa Y., Komatsu R., Shibata K., Shinozaki Y., Kasai H., Moriishi K., Moriyama Y., Koizumi S. (2013). Microglia release ATP by exocytosis. Glia, 61(8), 1320–1330. PMID: 23832620. 10.1002/glia.22517 [DOI] [PubMed] [Google Scholar]
- Ipata P. L. (2011). Origin, utilization, and recycling of nucleosides in the central nervous system. Advances in Physiology Education, 35(4), 342–346. PMID: 22139768. 10.1152/advan.00068.2011 [DOI] [PubMed] [Google Scholar]
- Jakovljevic M., Lavrnja I., Bozic I., Savic D., Bjelobaba I., Pekovic S., Sévigny J., Nedeljkovic N., Laketa D. (2017). Down-regulation of NTPDase2 and ADP-sensitive P2 purinoceptors correlate with severity of symptoms during experimental autoimmune encephalomyelitis. Frontiers in Cellular Neuroscience, 11, 333. PMID: 29163045; PMCID: PMC5670145. 10.3389/fncel.2017.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurányi Z., Sperlágh B., Vizi E. S. (1999). Involvement of P2 purinoceptors and the nitric oxide pathway in [3H]purine outflow evoked by short-term hypoxia and hypoglycemia in rat hippocampal slices. Brain Research, 823(1-2), 183–190. PMID: 10095025. 10.1016/S0006-8993(99)01169-5 [DOI] [PubMed] [Google Scholar]
- Kansas G. S., Wood G. S., Tedder T. F. (1991). Expression, distribution, and biochemistry of human CD39. Role in activation-associated homotypic adhesion of lymphocytes. The Journal of Immunology, 146(7), 2235–2244. PMID: 1672348. [PubMed] [Google Scholar]
- Khakh B. S., Burnstock G., Kennedy C., King B. F., North R. A., Séguéla P., Voigt M., Humphrey P. P. (2001). International union of pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacological Reviews, 53(1), 107–118. PMID: 11171941. [PubMed] [Google Scholar]
- Kirley T. L. (1997). Complementary DNA cloning and sequencing of the chicken muscle ecto-ATPase. Homology with the lymphoid cell activation antigen CD39. Journal of Biological Chemistry, 272(2), 1076–1081. PMID: 8995405. 10.1074/jbc.272.2.1076 [DOI] [PubMed] [Google Scholar]
- Koizumi S. (2010). Synchronization of Ca2+ oscillations: Involvement of ATP release in astrocytes. FEBS Journal, 277(2), 286–292. PMID: 19895581. 10.1111/j.1742-4658.2009.07438.x [DOI] [PubMed] [Google Scholar]
- Koizumi S., Ohsawa K., Inoue K., Kohsaka S. (2013). Purinergic receptors in microglia: Functional modal shifts of microglia mediated by P2 and P1 receptors. Glia, 61(1), 47–54. PMID: 22674620. 10.1002/glia.22358 [DOI] [PubMed] [Google Scholar]
- Kruglov E. A., Nathanson R. A., Nguyen T., Dranoff J. A. (2006). Secretion of MCP-1/CCL2 by bile duct epithelia induces myofibroblastic transdifferentiation of portal fibroblasts. American Journal of Physiology-Gastrointestinal and Liver Physiology, 290(4), G765–G771. PMID: 16282363. 10.1152/ajpgi.00308.2005 [DOI] [PubMed] [Google Scholar]
- Kukulski F., Komoszyński M. (2003). Purification and characterization of NTPDase1 (ecto-apyrase) and NTPDase2 (ecto-ATPase) from porcine brain cortex synaptosomes. European Journal of Biochemistry, 270(16), 3447–3454. PMID: 12899702. 10.1046/j.1432-1033.2003.03734.x [DOI] [PubMed] [Google Scholar]
- Kyrargyri V., Madry C., Rifat A., Arancibia-Carcamo I. L., Jones S. P., Chan V. T. T., Xu Y., Robaye B., Attwell D. (2020). P2y13 receptors regulate microglial morphology, surveillance, and resting levels of interleukin 1β release. Glia, 68(2), 328–344. PMID: 31520551; PMCID: PMC6916289. 10.1002/glia.23719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D., Hammer K., Koszalka P., Schrader J., Robson S., Zimmermann H. (2008). Distribution of ectonucleotidases in the rodent brain revisited. Cell and Tissue Research, 334(2), 199–217. Epub 2008 Oct 9. PMID: 18843508. 10.1007/s00441-008-0681-x [DOI] [PubMed] [Google Scholar]
- Latini L., Geloso M. C., Corvino V., Giannetti S., Florenzano F., Viscomi M. T., Michetti F., Molinari M. (2010). Trimethyltin intoxication up-regulates nitric oxide synthase in neurons and purinergic ionotropic receptor 2 in astrocytes in the hippocampus. Journal of Neuroscience Research, 88, 500–509. 10.1002/jnr.22238 [DOI] [PubMed] [Google Scholar]
- Little A. R., Miller D. B., Li S., Kashon M. L., O’Callaghan J. P. (2012). Trimethyltin-induced neurotoxicity: Gene expression pathway analysis, q-RT-PCR and immunoblotting reveal early effects associated with hippocampal damage and gliosis. Neurotoxicology and Teratology, 34(1), 72–82. 10.1016/j.ntt.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Mateo J., Kreda S., Henry C. E., Harden T. K., Boyer J. L. (2003). Requirement of Cys399 for processing of the human ecto-ATPase (NTPDase2) and its implications for determination of the activities of splice variants of the enzyme. Journal of Biological Chemistry, 278(41), 39960–8. PMID: 12888562. 10.1074/jbc.M307854200 [DOI] [PubMed] [Google Scholar]
- Melani A., Turchi D., Vannucchi M. G., Cipriani S., Gianfriddo M., Pedata F. (2005). ATP Extracellular concentrations are increased in the rat striatum during in vivo ischemia. Neurochemistry International, 47(6), 442–448. PMID: 16029911. 10.1016/j.neuint.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Milanese M., Bonifacino T., Zappettini S., Usai C., Tacchetti C., Nobile M., Bonanno G. (2009). Glutamate release from astrocytic gliosomes under physiological and pathological conditions. International Review of Neurobiology, 85, 295–318. PMID: 19607977. 10.1016/S0074-7742(09)85021-6 [DOI] [PubMed] [Google Scholar]
- Narravula S., Lennon P. F., Mueller B. U., Colgan S. P. (2000). Regulation of endothelial CD73 by adenosine: Paracrine pathway for enhanced endothelial barrier function. The Journal of Immunology, 165(9), 5262–5268. PMID: 11046060. 10.4049/jimmunol.165.9.5262 [DOI] [PubMed] [Google Scholar]
- Norris J. G., Tang L. P., Sparacio S. M., Benveniste E. N. (1994). Signal transduction pathways mediating astrocyte IL-6 induction by IL-1 beta and tumor necrosis factor-alpha. The Journal of Immunology, 152(2), 841–850. PMID: 7506738. [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A. M. (2016). The integrated stress response. EMBO Reports, 17(10), 1374–1395. PMID: 27629041; PMCID: PMC5048378. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov Y., Lalo U., Verkhratsky A., North R. A. (2006). Vesicular release of ATP at central synapses. Pflügers Archiv - European Journal of Physiology, 452(5), 589–597. PMID: 16639550. 10.1007/s00424-006-0061-x [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. (2004). The rat brain in stereotaxic coordinates (5th ed.). eBook ISBN: 9780080474120. Elsevier Academic Press. [Google Scholar]
- Piacentini R., Gangitano C., Ceccariglia S., Del Fà A., Azzena G. B., Michetti F., Grassi C. (2008). Dysregulation of intracellular calcium homeostasis is responsible for neuronal death in an experimental model of selective hippocampal degeneration induced by trimethyltin. Journal of Neurochemistry, 105(6), 2109–2121. PMID: 18284612. 10.1111/j.1471-4159.2008.05297.x [DOI] [PubMed] [Google Scholar]
- Quintas C., Fraga S., Gonçalves J., Queiroz G. (2011). Opposite modulation of astroglial proliferation by adenosine 5’-O-(2-thio)-diphosphate and 2-methylthioadenosine-5’-diphosphate: Mechanisms involved. Neuroscience, 182, 32–42. PMID: 21419195. 10.1016/j.neuroscience.2011.03.009 [DOI] [PubMed] [Google Scholar]
- Richter-Landsberg C., Heinrich M. (1996). OLN-93: A new permanent oligodendroglia cell line derived from primary rat brain glial cultures. Journal of Neuroscience Research, 45(2), 161–173. PMID: 8843033. [DOI] [PubMed] [Google Scholar]
- Robson S. C., Sévigny J., Zimmermann H. (2006). The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signalling, 2(2), 409–430. Epub 2006 May 30. PMID: 18404480; PMCID: PMC2254478. 10.1007/s11302-006-9003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues R. J., Tomé A. R., Cunha R. A. (2015). ATP As a multi-target danger signal in the brain. Frontiers in Neuroscience, 9, 148. PMID: 25972780; PMCID: PMC4412015. 10.3389/fnins.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C. L., Sims S. G., Nowery J. D., Meares G. P. (2019). Endoplasmic reticulum stress differentially modulates the IL-6 family of cytokines in murine astrocytes and macrophages. Scientific Reports, 9(1), 14931. PMID: 31624329; PMCID: PMC6797742. 10.1038/s41598-019-51481-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Koop B., Schröter F., Cline J., Ingwersen J., Berndt C., Hartung H. P., Aktas O., Prozorovski T.. (2016). Activation of Wnt signaling promotes hippocampal neurogenesis in experimental autoimmune encephalomyelitis. Mol Neurodegener, 11(1), 53. PMID: 27480121. 10.1186/s13024-016-0117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sévigny J., Sundberg C., Braun N., Guckelberger O., Csizmadia E., Qawi I., Imai M., Zimmermann H., Robson S. C. (2002). Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase 1 (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood, 99(8), 2801–2809. PMID: 11929769. 10.1182/blood.V99.8.2801 [DOI] [PubMed] [Google Scholar]
- Shukla V., Zimmermann H., Wang L., Kettenmann H., Raab S., Hammer K., Sévigny J., Robson S. C., Braun N. (2005). Functional expression of the ecto-ATPase NTPDase2 and of nucleotide receptors by neuronal progenitor cells in the adult murine hippocampus. Journal of Neuroscience Research, 80(5), 600–610. PMID: 15884037. 10.1002/jnr.20508 [DOI] [PubMed] [Google Scholar]
- Sparacio S. M., Zhang Y., Vilcek J., Benveniste E. N. (1992). Cytokine regulation of interleukin-6 gene expression in astrocytes involves activation of an NF-kappa B-like nuclear protein. Journal of Neuroimmunology, 39(3), 231–242. PMID: 1644898. 10.1016/0165-5728(92)90257-L [DOI] [PubMed] [Google Scholar]
- Stefani J., Tschesnokowa O., Parrilla M., Robaye B., Boeynaems J. M., Acker-Palmer A., Zimmermann H., Gampe K. (2018). Disruption of the microglial ADP receptor P2Y13 enhances adult hippocampal neurogenesis. Frontiers in Cellular Neuroscience, 12, 134. PMID: 29867367; PMCID: PMC5966569. 10.3389/fncel.2018.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. L., Li W. W., Zhu C., Jin W. S., Zeng F., Liu Y. H., Bu X. L., Zhu J., Yao X. Q., Wang Y. J. (2018). Clinical research on Alzheimer's disease: Progress and perspectives. Neurosci Bull, 34(6), 1111–1118. Epub 2018 Jun 28. PMID: 29956105. 10.1007/s12264-018-0249-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske N., Liessem A., Fischbach F., Clarner T., Beyer C., Wruck C., Fragoulis A., Tauber S. C., Victor M., Kipp M. (2018). Chemical hypoxia-induced integrated stress response activation in oligodendrocytes is mediated by the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2). Journal of Neurochemistry, 144(3), 285–301. PMID: 29210072. 10.1111/jnc.14270 [DOI] [PubMed] [Google Scholar]
- Vlajkovic S. M., Housley G. D., Greenwood D., Thorne P. R. (1999). Evidence for alternative splicing of ecto-ATPase associated with termination of purinergic transmission. Molecular Brain Research, 73(1-2), 85–92. PMID: 10581401. 10.1016/S0169-328X(99)00244-2 [DOI] [PubMed] [Google Scholar]
- Wang C. J., Vlajkovic S. M., Housley G. D., Braun N., Zimmermann H., Robson S. C., Sévigny J., Soeller C., Thorne P. R. (2005). C-terminal splicing of NTPDase2 provides distinctive catalytic properties, cellular distribution and enzyme regulation. Biochemical Journal, 385(3), 729–736. PMID: 15362980; PMCID: PMC1134748. 10.1042/BJ20040852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Arcuino G., Takano T., Lin J., Peng W. G., Wan P., Li P., Xu Q., Liu Q. S., Goldman S. A., Nedergaard M. (2004). P2x7 receptor inhibition improves recovery after spinal cord injury. Nature Medicine, 10(8), 821–827. PMID: 15258577. 10.1038/nm1082 [DOI] [PubMed] [Google Scholar]
- White T. D. (1977). Direct detection of depolarisation-induced release of ATP from a synaptosomal preparation. Nature, 267(5606), 67–68. PMID: 404573. 10.1038/267067a0 [DOI] [PubMed] [Google Scholar]
- Wink M. R., Braganhol E., Tamajusuku A. S., Lenz G., Zerbini L. F., Libermann T. A., Sévigny J., Battastini A. M., Robson S. C. (2006). Nucleoside triphosphate diphosphohydrolase-2 (NTPDase2/CD39L1) is the dominant ectonucleotidase expressed by rat astrocytes. Neuroscience, 138(2), 421–432. PMID: 16414200. 10.1016/j.neuroscience.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Yu J., Lavoie E. G., Sheung N., Tremblay J. J., Sévigny J., Dranoff J. A. (2008). IL-6 downregulates transcription of NTPDase2 via specific promoter elements. American Journal of Physiology-Gastrointestinal and Liver Physiology, 294(3), G748–G756. PMID: 18202114; PMCID: PMC5239663. 10.1152/ajpgi.00208.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O’Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., Wu J. Q. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience, 34(36), 11929–11947. Erratum in: J Neurosci. 2015;35(2):846-6. PMID: 25186741; PMCID: PMC4152602. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H. (1992). 5’-Nucleotidase: Molecular structure and functional aspects. Biochemical Journal, 285(2), 345–365. PMID: 1637327; PMCID: PMC1132794. 10.1042/bj2850345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H., Zebisch M., Sträter N. (2012). Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signalling, 8(3), 437–502. PMID: 22555564; PMCID: PMC3360096. 10.1007/s11302-012-9309-4 [DOI] [PMC free article] [PubMed] [Google Scholar]