Abstract
Recent studies have revealed that circRNAs can affect tumor DNA damage and repair, apoptosis, proliferation, and invasion and influence the transport of intratumor substances by acting as miRNA sponges and transcriptional regulators and binding to proteins in a variety of ways. However, research on the role of circRNAs in cancer radiotherapy and chemoresistance is still in its early stages. Chemotherapy is a common approach to oncology treatment, but the development of tumor resistance limits the overall clinical efficacy of chemotherapy for cancer patients. The current study suggests that circRNAs have a facilitative or inhibitory effect on the development of resistance to conventional chemotherapy in a variety of tumors, suggesting that circRNAs may serve as a new direction for the study of antitumor drug resistance. In this review, we will briefly discuss the biological features of circRNAs and summarize the recent progression of the involvement of circRNAs in the development and pathogenesis of cancer chemoresistance.
Keywords: circRNA, tumor chemotherapy, drug resistance, therapeutic target, cancer biology
Background
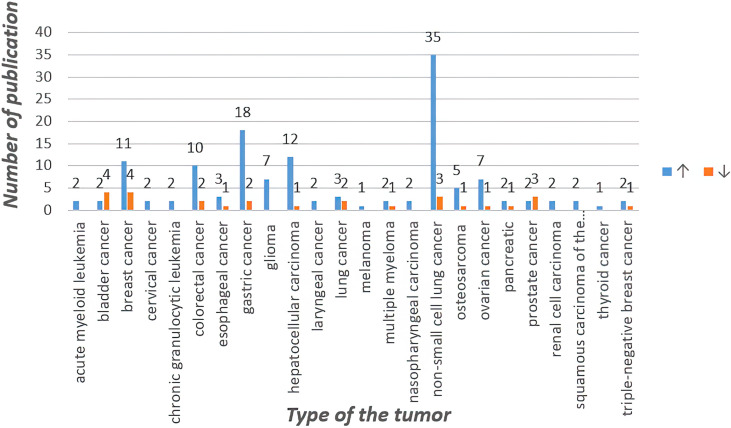
Circular RNA was first found in plant viroid (1), yeast mitochondrial RNA (2). Unlike linear RNA with 5 ‘cap and 3’ tail as terminals, circular RNA is characterized by a covalently closed loop structure with neither 5’to 3 ‘polarity nor polyadenylate tail. This inherent feature led to the widespread underestimation of circular RNA in previous polyadenylated transcriptome analysis. With the advent of specific biochemical and stoichiometric methods, a large number of circular RNA formed by reverse splicing of exons have been found in different cell lines and different species (3). Among them, circular RNA(circRNA), along with their modifiers, have been investigated to play key roles in regulating tumor development and mediating therapy resistance within various cancers, such as hepatocellular carcinoma, breast cancer, lung cancer, gastric cancer, etc. First, almost all of the reported circRNA functions through four classic mechanisms, including:1, miRNA sponge mechanisms 2, sponging of RBPs (RNA-binding proteins)3, regulation of transcription 4, translation into peptides or proteins. In addition, two limitations regarding the generation of tumor drug resistance are raised. First, the existing reports on circRNAs in cancer chemoresistance are limited to a few classical drugs and a few types of cancers. Second, most reported circRNAs, limited to those identified in cancer, remain unstudied in the context of the cancer therapeutic response, particularly radioresistance, and numerous unidentified circRNAs remain to be studied. As these limitations are gradually overcome, potential interventions targeting circRNAs are expected to overcome cancer resistance to radiation and chemotherapy (4, 5). We searched for “circRNA”, “cancer” and “drug resistance” in Pubmed, and obtained a total of 351 relevant papers, and then identified the papers that fit the topic of this paper by preliminary screening of these 351 papers, and the results were 158. According to recent studies, circRNAs mainly (1) upregulate drug efflux transporters or downregulate influx channels, (2) enhance epithelial-mesenchymal transition (EMT) and stemness, (3) alter the copy number of target genes and activation of bypass pathways or downstream signaling, (4) alter endoplasmic reticulum stress, autophagy and phagocytosis, (5) enhance or inhibit DNA repair, (6) impact the tumor microenvironment. Other factors may induce tumor drug resistance but will not be discussed here (4). In this paper, we review the possible tumor drug resistance mechanisms associated with circRNAs and their research progress in the context of chemoresistance by categorizing and summarizing a large number of relevant studies. It also discussed the limitations of available knowledge and future potential directions. The statistics are shown in Figure 1 and Table 1 .
Figure 1.
Statistics of circRNA related to chemotherapy resistance in different tumors in recent five years.
Table 1.
CircRNAs in tumor drug resistance.
| Name of the tumor | CircRNA | Target(s) | Drugs | Alteration | References |
|---|---|---|---|---|---|
| nasopharyngeal carcinoma | hsa_circ_0002346 | miR-422a, FOXQ1 protein | Docetaxel | ↑ | (6) |
| hsa_circ_0004771 | miR-515-5p, IL-25 | DDP | ↑ | (7) | |
| colorectal cancer | hsa_circ_0005963 | miR-122, pyruvate kinase M2 isoform | OXA | ↑ | (8) |
| hsa_circ_0001451 | miR-18b-5p | OXA | ↓ | (9) | |
| hsa_circ_0084443 | miR375, FOXM1 protein, Wnt/β-catenin pathway | 5-Fu | ↑ | (10) | |
| hsa_circ_0000598 | miR-340, the BMI1 protein | Irinotecan | ↑ | (11) | |
| hsa_circ_0032833 | miR-125-5p, MSI1 protein | OXA | ↑ | (12) | |
| hsa_circ_0071589 | miR-526b-3p, KLF12 protein | DDP | ↑ | (13) | |
| hsa_circ_0001806 | miR-944, FZD7 protein | DOX | ↑ | (14) | |
| hsa_circ_0002211 | miR-31-5p, KANK1 protein | 5-Fu | ↓ | (15) | |
| hsa_circ_0007031 | miR-133b, the ABCC5 protein | 5-Fu | ↑ | (16) | |
| hsa_circ_0000338 | miR-217, miR-485-3p | 5-Fu | ↑ | (17) | |
| hsa_circ_0020095 | miR-487a-3p, SOX9 protein | DDP | ↑ | (18) | |
| multiple myeloma | hsa_circ_0007841 | ABCG2 | DOX | ↑ | (19) |
| hsa_circ_0001141 | miR-615-3p, PRKCD protein | BTZ | ↓ | (20) | |
| has_circ_0007841 | miR-129-5p, JAG1 protein | BTZ | ↑ | (21) | |
| non-small cell lung cancer | hsa_circ_0007385 | miR-519D-3p, HMGB1 | DDP | ↑ | (22) |
| hsa_circ_103809 | miR-377-3p, GOT1 | DDP | ↑ | (23) | |
| hsa_circ_0084003 | miR-381-3p, CXCR4 protein | Anti-PD1 | ↑ | (24) | |
| hsa_circ_0082374 | miRNA let-7, PD-L1 | Anti-PD1 | ↑ | (25) | |
| hsa_circ_0031250 | miR-4458, REV3L | DDP | ↑ | (26) | |
| hsa_circ_0096157 | P21 protein, CDK4-cyclin D1, Bcl-2 | DDP | ↑ | (27) | |
| hsa_circ_0004015 | miR-1183, PDPK1 | TKI | ↑ | (28) | |
| hsa_circ_0085131 | miR-654-5p, ATG7 | DDP | ↑ | (29) | |
| hsa_circ_0000079 | FXR1 protein | DDP | ↓ | (30) | |
| hsa_circ_0076305 | miR-296-5p, STAT3 protein | DDP | ↑ | (31) | |
| hsa_circ_0002483 | miR-182-5p, GRB2 protein, FOXO1 protein, FOXO3 protein | PTX | ↓ | (32) | |
| hsa_circ_0002130 | miR-498, GLUT1, HK2, LDHA | Osimertinib | ↑ | (33) | |
| hsa_circ_0011292 | miR-379-5p, TRIM65 protein | PTX | ↑ | (34) | |
| hsa_circ_0014235 | miR-520a-5p, CDK4 | DDP | ↑ | (35) | |
| circMTDH.4 | miR-630, AEG-1 | 5-Fu | ↑ | (36) | |
| hsa_circ_102481 | miR-30a-5p, ROR1 protein | TKI | ↑ | (37) | |
| hsa_circ_0023303 | miR-187-3p, FGF9 protein | DDP | ↑ | (38) | |
| hsa_circ_0003998 | miR-136-5p, CORO1C protein | DTX | ↑ | (39) | |
| hsa_circ_0008928 | miR-488, HK2 protein | DDP | ↑ | (40) | |
| hsa_circ_0005909 | miR-338-3p, SOX4protein | ADM | ↑ | (41) | |
| hsa_circ_103615 | ABCB1 | DDP | ↑ | (42) | |
| hsa_circ_0072083 | miR-195-5p, KPNA4protein | PTX | ↑ | (43) | |
| hsa_circ_0014130 | miR-493-5p, ROCK1protein | DDP | ↑ | (44) | |
| circASXL1 | miR-206 | DDP | ↑ | (45) | |
| hsa_circ_0006404 | miR-543, FOXO3 protein | DDP | ↓ | (46) | |
| hsa_circ_0003222 | miR-527, PHF21B protein, Wnt/β-catenin pathway | Anti-PD1 | ↑ | (47) | |
| hsa_circ_0002874 | miR-1273f, MDM2 protein, p53 protein | PTX | ↑ | (48) | |
| hsa_circ_0001658 | miR-409-3p, TWIST1 protein | Gefitinib | ↑ | (49) | |
| circPRMT5 | miR-138-5p, MYH9 | DDP | ↑ | (50) | |
| hsa_circ_0076305 | miR-186-5p, ABCC1 | DDP | ↑ | (51) | |
| hsa_circ_0014130 | miR-101, ABCC1 | DDP | ↑ | (52) | |
| hsa_circ_0005152 | miR-934, SHP2 protein | Anti-PD1 | ↑ | (53) | |
| hsa_circ_0014130 | miR-545-3p, YAP1 protein | DTX | ↑ | (54) | |
| hsa_circ_0023404 | miR-646, SOX4 protein | DDP | ↑ | (55) | |
| hsa_circ_0001821 | miR-526-5p, GRK5 protein | PTX | ↑ | (56) | |
| hsa_circ_0031608 | miR-137, CXCL12 | DDP | ↑ | (57) | |
| hsa_circ_0079587 | miR-328-3p, miR-3173-5p, PKP3 protein | Anti-PD1 | ↑ | (58) | |
| hsa_circ_100565 | miR-337-3p, ADAM28 protein | DDP | ↑ | (59) | |
| lung cancer | hsa_circ_0000199 | miR-516b-5p, STAT3 protein | DDP | ↑ | (60) |
| hsa_circ_0030998 | miR-558, MMP1 protein, MMP17 protein | PTX | ↓ | (61) | |
| hsa_circ_0008717 | miR-556-3p, AK4 protein | DDP | ↑ | (62) | |
| hsa_circ_0001821 | miR-145-5p, ABCC1 protein | DDP | ↑ | (63) | |
| hsa_circ_0007798 | ASK1 protein | Gefitinib | ↓ | (64) | |
| hepatocellular carcinoma | hsa_circ_0082002 | miR-30-5p, Snail, DPP4 protein, CXCL10 | Anti-PD1 | ↑ | (65) |
| hsa_circ_0048677 | miR-449c-5p, TIM-3 protein | Anti-PD1 | ↑ | (66) | |
| hsa_circ_0087293 | miR-103a-2-5p, miR-660-3p, Wnt/β-catenin pathway | Sorafenib | ↑ | (67) | |
| hsa_circ_0001001 | miR-605, FOXO3 protein, ABCB1 protein | OXA | ↑ | (68) | |
| hsa_circ_0087293 | YBX1 protein | Sorafenib | ↑ | (69) | |
| hsa_circ_0104670 | miR-155-5p, PDK1 protein | DDP | ↑ | (70) | |
| hsa_circ_0031242 | miR-924, POU3F2 protein | DDP | ↑ | (71) | |
| hsa_circ_G004213 | miR-513b-5p, PRPF39 protein | DDP | ↓ | (72) | |
| hsa_circ_0025039 | miR-1324, MECP2 protein | Sorafenib | ↑ | (73) | |
| hsa_circ_0006404 | miR-199a-5p, ABCC1 protein | ADM | ↑ | (74) | |
| hsa_circ_0003998 | miR-218-5p, EIF5A2 | ADM | ↑ | (75) | |
| hsa_circ_0000384 | miR-148a, STX3 protein, PTEN protein | DDP | ↑ | (76) | |
| cervical cancer | hsa_circ_0007874 | miR-6893, S100A1 protein | DDP | ↑ | (77) |
| hsa_circ_0023404 | miR-5047, VEGFA protein | DDP | ↑ | (78) | |
| osteosarcoma | hsa_circ_0000073 | miR-145-5p, miR-151-3p, NRAS protein | MTX | ↑ | (79) |
| hsa_circ_0001821 | ABCB1 protein | ADM | ↑ | (80) | |
| hsa_circ_0003496 | miR-506-3p, SEMA6D protein, Wnt/β-catenin pathway | DDP | ↑ | (81) | |
| hsa_circ_0001141 | miR-524, RASSF6 protein | ADM | ↓ | (82) | |
| hsa_circ_0081001 | miR-494-3p, TGM2 protein | MTX | ↑ | (83) | |
| hsa_circ_0005986 | miR-760, EZH2 | DXR | ↑ | (84) | |
| melanoma | hsa_circ_0020710 | miR-370-3p, CXCL12 | Anti-PD1 | ↑ | (85) |
| laryngeal cancer | hsa_circ_0004507 | miR-873 | DDP | ↑ | (86) |
| hsa_circ_0019340 | miR-376a, ATG2A protein | DDP | ↑ | (87) | |
| acute myeloid leukemia | circPAN3 | Beclin1, p62, AMPK/mTOR pathway | ADM | ↑ | (88) |
| circPAN3 | miR-153-5p, miR-183-5p, XIAP protein | ADM | ↑ | (89) | |
| thyroid cancer | hsa_circ_0060060 | miR-144-3p, TGF-α | DDP | ↑ | (90) |
| glioma | hsa_circ_0102722 | miR-145-5p, ABCG2 protein | TMZ | ↑ | (91) |
| hsa_circ_0005660 | miR-132 | TMZ | ↑ | (92) | |
| hsa_circ_0000284 | miR-421, ZIC5 protein | TMZ | ↑ | (93) | |
| hsa_circ_0000284 | miR-524-5p, KIF2A protein, PI3K/AKT pathway | TMZ | ↑ | (94) | |
| hsa_circ_0072083 | miR-1252-5p, ALKBH5 protein, NANOG protein | TMZ | ↑ | (95) | |
| hsa_circ_0002330 | miR-502-5p, NRAS/MEK1/ERK1-2 signal | TMZ | ↑ | (96) | |
| hsa_circ_0005198 | miR-198, TRIM14 protein | TMZ | ↑ | (97) | |
| squamous carcinoma of the oral cavity | hsa_circ_0109291 | miR-188-3p, ABCB1 protein | DDP | ↑ | (98) |
| hsa_circ_0011946 | miR-338-3p, Lin28B protein | DDP | ↑ | (99) | |
| ovarian cancer | hsa_circ_0001946 | miR-1270, the SCAI factor | DDP | ↓ | (100) |
| hsa_circ_0001741 | miR-1299, NEK2 protein | PTX | ↑ | (101) | |
| hsa_circ_0000714 | miR-370-3p, CDK6 protein, RB protein, RAB17 protein | PTX | ↑ | (102) | |
| hsa_circ_0063809 | miR-1252, FOXR2 protein | PTX | ↑ | (103) | |
| hsa_circ_0008234 | miR-22, miR-150-3p, CEBPG, FMNL3 | DDP | ↑ | (104) | |
| hsa_circ_0002711 | miR-211-5p, HOXC8 protein | PTX | ↑ | (105) | |
| hsa_circ_0001756 | miR-338-3p, CHTOP protein | DDP | ↑ | (106) | |
| hsa_circ_0063809 | miR-149-5p, SIK2 protein | PTX | ↑ | (107) | |
| chronic granulocytic leukemia | hsa_circ_0080145 | miR-326, PPFIA1 protein | IM | ↑ | (108) |
| hsa_circ_0009910 | miR-34a-5p, ULK1 protein | IM | ↑ | (109) | |
| bladder cancer | hsa_circ_0000284 | GEM | ↓ | (110) | |
| hsa_circ_103809 | miR-516a-5p, FBXL18 protein | GEM | ↑ | (111) | |
| hsa_circ_0001785 | DDP | ↑ | (112) | ||
| hsa_circ_0000285 | DDP | ↓ | (113) | ||
| hsa_circ_0072309 | MutSα, ATM, p73 protein | DDP | ↓ | (114) | |
| hsa_circ_0008399 | WTAP protein | DDP | ↓ | (115) | |
| prostate cancer | hsa_circ_0006404 | DTX | ↓ | (116) | |
| hsa_circ_0001427 | miR-181c-5p, ARv7 protein | Enz | ↓ | (117) | |
| hsa_circ_0000735 | miR-7 | DTX | ↑ | (118) | |
| hsa_circ_0001141 | miR-17, Wnt/β-catenin pathway, PI3K/AKT/mTOR pathway | Enz | ↓ | (119) | |
| hsa_circ_0005276 | miR-1182, TPD52 protein | DTX | ↑ | (120) | |
| breast cancer | hsa_circ_0025202 | miR-197-3p, HIPK3 protein | TAM | ↓ | (121) |
| hsa_circ_0025202 | miR-182-5p, FOXO3a protein | TAM | ↓ | (122) | |
| hsa_circ_0001839 | miR-548p, the PBLD protein | ADM | ↓ | (123) | |
| hsa_circ_007874 | EG5 protein, TRAF4 protein | Monastrol | ↓ | (124) | |
| hsa_circ_0008717 | let-7a-5p, the DUSP7 protein | PTX | ↑ | (125) | |
| hsa_circ_0001946 | miR-7, CCNE1 protein | 5-Fu | ↑ | (126) | |
| circ-RNF111 | miR-140-5p, transcription factor E2F3 | PTX | ↑ | (127) | |
| hsa_circ_0001667 | miR-4458, NCOA3 protein | ADM | ↑ | (128) | |
| hsa_circ_0125597 | miR-216b, HMGA2 | 5-Fu | ↑ | (129) | |
| hsa_circ_0001461 | miR-525-5p, SKA1subunit | OXA | ↑ | (130) | |
| hsa_circ_0006528 | miR-1236-3p, CHD4 protein | ADM | ↑ | (131) | |
| hsa_circ_0062558 | miR-153-5p, ANLN protein | Lapatinib | ↑ | (132) | |
| hsa_circ_0085495 | miR-873-5p, integrin β1 | ADM | ↑ | (133) | |
| hsa_circ_0001598 | miR-1184, PD-L1 | Trastuzumab | ↑ | (134) | |
| circ_UBE2D2 | miR-200a-3p | TAM | ↑ | (135) | |
| triple-negative breast cancer | hsa_circ_0005728 | miR-512-3p, CDCA3 protein | ADM | ↓ | (136) |
| hsa_circ_0007503 | miR-142, WWP1 protein, PI3K/AKT pathway | PTX | ↑ | (137) | |
| hsa_circ_005239 | miR-361-5p, TLR4 protein | PTX | ↑ | (138) | |
| renal cell carcinoma | hsa_circ_0035483 | hsa-miR-335, CCNB1 | GEM | ↑ | (139) |
| hsa_circ_0031608 | miR-1184, GPCPD1 protein | Sunitinib | ↑ | (140) | |
| esophageal cancer | hsa_circ_001275 | miR-370-3p, WNT7a protein | DDP | ↑ | (141) |
| hsa_circ_0006168 | miR-194-5p, JMJD1C protein | PTX | ↑ | (142) | |
| circPSMC3 | miR-10a-5p, PTEN protein | Gefitinib | ↓ | (143) | |
| hsa_circ_0000337 | miR-377-3p, JAK2 protein | DDP | ↑ | (144) | |
| gastric cancer | hsa_circ_0026359 | miR-1200, the POLD4 protein | DDP | ↑ | (145) |
| hsa_circ_0006990 | miR-125b-5p, STAT3 protein | DDP | ↑ | (146) | |
| hsa_circ_0032821 | miR-515-5p, SOX9 protein | OXA | ↑ | (147) | |
| hsa_circ_0000234 | miR-142-3p, ROCK2 protein | DDP | ↑ | (148) | |
| hsa_circ_0058147 | miR-182-5p | DDP | ↑ | (149) | |
| hsa_circ_0001313 | miR-618, BCL2protein | DDP | ↑ | (150) | |
| hsa_circ_0000199 | miR-198, PIK3R1 protein, p85α protein, PI3K/AKT pathway | DDP | ↑ | (151) | |
| hsa_circ_0000657 | miR-99a-5p, the MTMR3 protein | DDP | ↓ | (152) | |
| hsa_circ_0001821 | miR-124-3p, ZEB1 protein | PTX | ↑ | (153) | |
| hsa_circ_0004771 | miRNA-138-5p, HIF-1α factor | 5-Fu | ↑ | (154) | |
| hsa_circ_0006089 | miR-330-3p, NT5E protein | DDP | ↑ | (155) | |
| hsa_circ_0004339 | miR-802, BMI1 | DDP | ↑ | (156) | |
| circ MTHFD2 | miR-124, MDR-1 | MTA | ↑ | (157) | |
| hsa_circ_0001546 | miRNA-421, ATM/CHK2/P53 pathway | OXA | ↓ | (158) | |
| hsa_circ_0031452 | miR-137, PBX3 protein | DB | ↑ | (159) | |
| hsa_circ_0000144 | miR-502-5p, ADAM9 protein | OXA | ↑ | (160) | |
| hsa_circ_0000260 | miR-129-5p, MMP11 protein | DDP | ↑ | (161) | |
| hsa_circ_0001821 | miR-152-3p, HDGF/PI3K/AKT pathway | DDP | ↑ | (162) | |
| hsa_circ_0001821 | miR-30a-5p, YAP1protein | DDP | ↑ | (163) | |
| hsa_circ_0002570 | miR-490-3p, HMGA2protein, HNRNPK protein, β-catenin | DDP | ↑ | (164) | |
| pancreatic | hsa_circ_0000284 | miR-330-5p, RASSF1 protein | GEM | ↑ | (165) |
| hsa_circ_0013587 | miR-1227, E-Cadherin | Erlotinib | ↓ | (166) | |
| hsa_circ_0030167 | miR-338-5p, Wif1, Wnt8, β-catenin | GEM | ↑ | (167) |
↑ means increased; ↓ means decreased.
CircRNAs Regulate Tumor Drug Resistance by Affecting Intratumor Drug Concentrations
It has been shown that reduced drug concentration is the main cause of drug resistance. This may be due to drug segregation in intracellular vesicles and compartments, increased drug efflux or decreased drug influx. These changes may be due to the remodeling of drug channels and transporters, which broadly include ATP-binding cassette (ABC) family proteins, solute carriers, and volume-regulated anion channels (VRACs). M Saxena et al. mechanistically demonstrated that the promoters of ABC transporter proteins carry binding sites for several EMT-inducible transcription factors and that EMT-related transcription factors, such as Twist, Snail, and FOXC2, and overexpression of these transcription factors increased the promoter activity of ABC transporter proteins. Furthermore, chromatin immunoprecipitation studies showed that Twist binds directly to the E-box element of the ABC transporter protein. EMT inducers were therefore identified as novel regulators of ABC transporter proteins, thus providing a long-term link between aggressiveness and multidrug resistance. Thus, targeting EMT transcription factors could serve as a novel strategy to inhibit tumor metastasis and associated drug resistance (168–171).
According to current reports, circRNAs can increase drug resistance in tumor cells by targeting miRNAs and down-regulating the expression of genes related to drug concentrations in tumors accordingly. For example, in glioma tissues and cell lines, circRNA CEP128 expression was upregulated, and circRNA CEP128 expression was higher in temozolomide-resistant glioma cells than in their parental cells. In contrast, downregulation of circRNA CEP128 inhibited glioma cell proliferation and reduced temozolomide resistance by decreasing the expression of ATP-binding cassette G superfamily member 2 (ABCG2). In addition, miR-145-5p was expressed at low levels in glioma cells and temozolomide-resistant glioma cells, and miR-145-5p was also identified as a target of circRNA CEP128. miR-145-5p overexpression inhibited the proliferation of U251/temozolomide cells and decreased the expression of ATP-binding cassette superfamily member 2, and circRNA CEP128 overexpression blocked these changes (91).
The mRNA and protein levels of ATP-binding cassette transporter protein G2 (ABCG2) were elevated in myeloma adriamycin chemotherapy-resistant cells. Silencing hsa_circ_0007841 in resistant cells reduces the mRNA and protein levels of ABCG2, and re-expression of hsa_circ_0007841 blocks this reduction. Similarly, overexpression of hsa_circ_0007841 effectively upregulated mRNA and protein levels of ABCG2. Inhibition of ABCG2 blocked hsa_circ_0007841 overexpression-induced cellular chemoresistance. In addition, ABCG2 reduced the difference in the half-maximal inhibitory concentration between parental and resistant cells. These findings suggest that hsa_circ_0007841 can promote a pattern of acquired chemoresistance in myeloma cells through the upregulation of ABCG2, providing a new molecular basis for chemotherapy in multiple myeloma (19).
In laryngeal cancer, knockdown of circ_0004507 reduces the protein levels of multidrug resistance-associated protein-1 (MRP1) and multidrug resistance gene 1 (MDR1), which are associated with cellular cassette transporters and confer multidrug resistance to tumor cells by reducing the uptake of anticancer drugs (86).
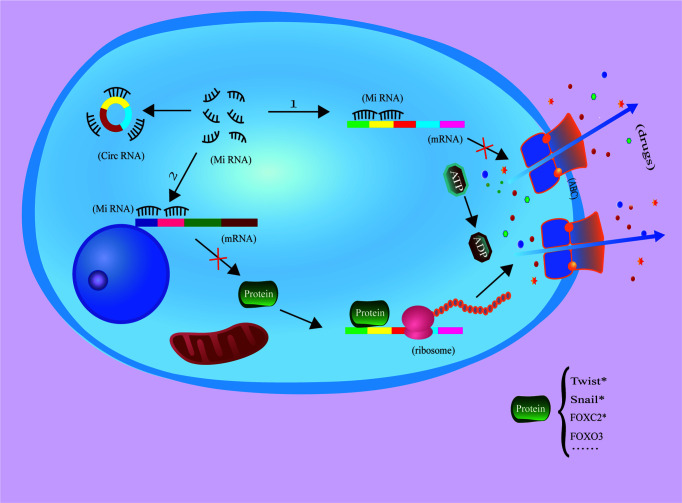
In hepatocellular carcinoma, circRNA PTGR1 regulates drug resistance to 5-fluorouracil (5-FU) and the growth tumor cells by regulating the miR-129-5p/ABCC1 axis (172). Additionally, studies of adriamycin resistance in hepatocellular carcinoma have shown that circFOXO3 promotes resistance to adriamycin in by regulating the miR-199a-5p/ATP binding cassette C family member 1 axis (74). However, circRNA circFBXO11 regulates hepatocellular carcinoma progression and oxaliplatin resistance through the miR-605/FOXO3/ABCB1 axis (68), so further study is needed determine whether circFBXO11 regulates circFOXO3 and its downstream signaling through this axis ( Figure 2 ).
Figure 2.
CircRNAs inhibit miRNA function throug1h the molecular sponging mechanism, increasing ABC expression and drug efflux to increase tumor cell drug resistance. The protein with * has been proven to be an ABC-related transcription factor, but there is no literature to prove that it is regulated by circRNA.
It can be seen that the effect of circRNA on ABC transporter protein is important and diverse in each type of tumor, but there are various factors that affect the concentration of drugs in tumor cells, and ABC transporter protein is only one of the drug channels. However, it is worth thinking why circRNA are mostly reported to regulate ABC transporter protein.
CircRNAs Enhance EMT and Stemness
Briefly, EMT is an epigenetic program by which cells lose their epithelial phenotype and acquire mesenchymal features. Common EMT-inducible transcription factors, such as twist and snail, which contain complementary binding sites, act as promoters of ABC transporters, and they enhance not only EMT but also ABC transporter expression (4, 171). In recent years, it has been shown that circRNAs can influence tumor cell resistance by enhancing EMT; for example, circ_001680 and circ-NOTCH1 are elevated in colorectal cancer (CRC) and gastric cancer (GC), respectively, and both these circRNAs can promote cancer stem cell (CSC) expansion via EMT (11, 173). Chen et al. showed that failure to eradicate cancer cells that acquire stem cell features through EMT activation or senescence is another reason for impaired cancer treatment outcomes (174–177). Second, CSCs are able to remain dormant for a long period of time to escape harmful stresses such as radiotherapy and chemotherapy. Once harmful stress is lifted, CSCs can differentiate, proliferate or metastasize at the primary lesion and subsequently invade other organs, leading to tumor recurrence or metastasis (178, 179). For example, circFOXO3 reduction promotes prostate cancer progression and doxorubicin resistance. circFOXO3 inhibits prostate cancer cell survival, migration, invasion, and doxorubicin resistance, which is associated with circFOXO3-mediated inhibition of Foxo3 and EMT (116).
Nithila A. Joseph et al. showed that in lung adenocarcinoma cells, circRNA CCDC66 increases SUMOlyation modifications related to the HGF-MET signaling pathway and epidermal growth factor receptor (EGFR)-controlled SAE2 through association with EMT and thus affects the resistance of tumor cells to tyrosine kinase inhibitors (TKIs) targeting EGFR (180). Among them, SAE2 is one of the subunits of SUMO-activating enzyme E1, and SUMO-mediated modifications have been shown to be importantly associated with the proliferation, migration and invasion of a variety of tumors (181, 182). We found that circRNA CCDC66 has been reported to be associated with mechanisms of chemoresistance generation not only in lung adenocarcinoma cells but also in gastrointestinal tumors; for example, DHX9 phosphorylation induced by oxaliplatin chemotherapy in colorectal cancer promotes the expression of oncogenic circRNA CCDC66 to affect chemoresistance (183), and circRNA CCDC66 in gastric cancer through the miR-618/BCL2 axis regulates cisplatin resistance in gastric cancer (150). More data is needed to assess the resistance induced by different mechanisms of different drugs yet related to the same circRNA ( Figure 3 ).
Figure 3.
CircRNAs regulate the stem cells of tumor cells by affecting the expression of EMT- and stemness-related proteins, which leads to chemotherapy resistance.
It is thus clear that circRNA also has an important role in the regulation of EMT and stemness. However, EMT and stemness is different from the regulation of drug channel proteins, and changes often imply an all-round overall effcet in EMT and stemness. Therefore, it is clear that since circRNA can enhance drug resistance by regulating EMT and stemness, circRNA must have other regulatory functions while enhancing EMT and stemness. In short, the effect of circRNA on tumors, too, is holistic. For example, the circ-CPA4 regulated cell growth, mobility, stemness and drug resistance in NSCLC cells and inactivated CD8 T cells in the tumor immune microenvironment (25).
CircRNAs Alter the Copy Number of Target Genes and Activate Bypass Pathways or Downstream Signaling
CircRNAs Compete for Endogenous MiRNAs
CircRNAs competitively bind miRNAs (184, 185), so as to serve as intracellular competitive endogenous RNA, suppressing the effects of miRNAs on target genes. For example, the circRNA AKT3 enhances cisplatin resistance in gastric cancer by inhibiting miR-198 and upregulating PIK3R1, which was experimentally shown to be a downstream molecule of miR-198 (151). PIK3R1 encodes a regulatory subunit of PI3K and has a role in positively regulating the PI3K signaling pathway. The autophagic process also requires phosphatidylinositol 3-phosphate (PI3P), and the inhibition of PI3P can inhibit the formation of autophagic vesicles. Whether PI3K can affect tumor cell drug resistance through the regulation of autophagy deserves further study.
In addition, some studies have shown that circRNA can promote its own loop and form positive feedback by up-regulating the expression of downstream transcription factors. For example, in gastric cancer, circFAM73A increases the expression of HMG (high mobility group) A2 through miR-490-3p, HMGA2 plays a dichotomous role in regulating circFAM73A ex-pression, i.e., HMGA2 facilitates the transcriptional acti-vation of FAM73A by E2F1 and elevates the efficiency of circFAM73A circularization by HNRNPL, HMGA2 promotes stem cell-like properties and cell malignancy in GC cells (164). It is worth considering whether this process of positive feedback loops to improve tumor drug resistance is common. With regard to circRNA and its sponge miRNA, through a large number of literature review and summary, we find that there is often more than one miRNA combined with each other under the same circRNA. For example, also in non-small cell lung cancer, circRNAPRMT5 can target miR-381-3p (24) and miR-4458 (26) to regulate the expression of different downstream proteins to promote tumor, and the same circ_007630 can also target miR-296-5p (31) and miR-186-5p (51) respectively. Interestingly, the same miRNA targets different genes in different tumors and regulates downstream pathways, such as miR-338-3p, which promotes the progressive drug resistance of oral squamous cell carcinoma (99) and ovarian cancer (106) at the same time. As we all know, the effect of circRNA on tumor drug resistance is often the result of a variety of mechanisms, so we need to pay attention to the interaction between various mechanisms and pathways, and pay attention to the integration and analysis of various mechanism information. The coincidence targets in these different pathways provide some support for us to analyze the generation of drug resistance of tumor cells as a whole. For example, miR-338-3p can play a role in promoting drug resistance in a variety of tumors according to current reports (99, 106), but the mechanisms of action are different. It is worth thinking whether the regulatory mechanisms of the same miRNAs in different tumor lines overlap, and if not, what determines the target of miRNAs to exert regulatory effects in tumor cells, and if so, whether it is of limited significance to focus on individual pathways according to most of the current literature. Moreover, we found that there are literatures about target overlap at all three levels of circRNA, miRNA and protein (32, 51, 63, 88, 89, 149).
CircRNAs Function as a Reservoir or a Stabilizing Factor for the MiRNA
Studies have shown that in the progress of chemoresistance of prostate cancer, as circRNA17 is positively correlated with the expression level of miR-181c-5p, this circRNA may not function as a sponge to sequester the miRNAs, but likely function as a reservoir or a stabilizing factor for the miR-181c-5p (117). Mechanistically, speculated the binding of miR-181c-5p with circRNA17 might enhance the miR-181c-5p stability by repressing its degradation through nucleases such as Tudor-SN endonuclease (186). In addition, this stabilizing effect appears particularly significant in vivo during tumor growth and less so in cells grown in tissue culture. The exact details of these phenomena remain to be further determined. CircRNA adsorbs miRNA can prevent the degradation of the latter, which reminds us of the need to add deeper thinking to the traditional study of ceRNA(competing endogenous RNA) mechanism.
CircRNAs Bind RNA-Binding Proteins
CircRNAs serve as protein baits or antagonists, thus arrestting the function of proteins, thereby affecting the related progresses (184). For example: circLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73 axis in bladder cancer (114). In addition, the same circRNA can perform different functions in different cells or in the same cell through different pathways. For example, in hepatocellular carcinoma cells, circRNA-SORE can mediate hepatocellular carcinoma resistance to sorafenib by stabilizing YBX1 (67), while N-6-methyladenosine-modified circRNA-SORE can also maintain hepatocellular carcinoma resistance to sorafenib by regulating β-catenin signalin (69). It should be pointed out that the previously mentioned circRNA-SORE mediates the drug resistance of hepatocellular carcinoma to sorafenib by stabilizing the YBX1 protein; in fact, it does not involve gene copy number and activation, but rather, circRNA directly plays a role in the process of protein stability and degradation.YBX1 is a kind of RNA binding protein (RBP). At present, an increasing number of evidences suggest the direct binding of circRNAs to RBPs. Most of them continue to regulate the expression of downstream signaling pathways by affecting the stability or degradation of target proteins, and there is often more than one downstream pathway involved (30, 124). Although the RBP pathway is different from pathways influenced by circRNA regulation of the target gene, in essence, it continues to play its biological function by regulating the expression (by targeting genes) or degradation (by targeting proteins) of related proteins.It is worth mentioning that there are also reports that circRNA directly binds to the DNA promoter to regulate transcription and thus affect the progression of liver cancer (84, 187).
CircRNAs Encode Proteins
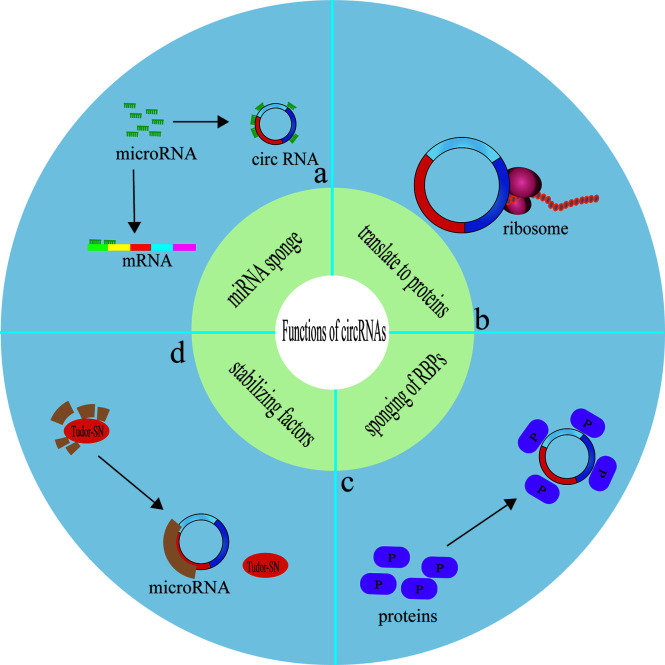
CircRNAs can translate to proteins through a cap-independent manner (184, 188). Yang et al. demonstrated that N-methyladensine(m6A) could promote the initiation of protein translation from circRNAs in human cells (189). For example, PengLi et al. found that circMRPS35 not only regulates the STX3-PTEN axis through sponge miR-148A, but also encodes circMRPS35-168aa, resulting in cisplatin resistance, thus playing a carcinogenic role in hepatocellular carcinoma (76). In addition, in gastric cancer, YuZhang et al. found that circDIDO1 inhibits the progression of gastric cancer by encoding a new DIDO1-529aa protein and regulating the stability of PRDX2 protein (190). At present, there are relatively few articles about circRNA affecting tumor chemotherapy resistance by encoding proteins, and in most of these articles, circRNA plays a role in multiple pathways, For example, even playing the role of molecular sponge also encodes protein, the emergence of this experimental result may be preconceived thinking about the mechanism of circRNA molecular sponge. Whether this preconceived way of thinking limits the study of circRNA multi-mechanism is worthy of further study ( Figure 4 ).
Figure 4.
Functions of circRNAs. (A) circRNAs competitively bind miRNAs, so as to serve as intracellular competitive endogenous RNA, suppressing the effects of miRNAs on target genes. (B) circRNAs can translate to proteins through a cap-independent manner. (C) circRNAs serve as protein baits or antagonists, thus arrestting the function of proteins, thereby affecting the related progresses. (D) circRNA function as a reservoir or a stabilizing factor for the miRNA.
Although this section outlines that circRNAs affect cancer drug resistance through the copy number alteration and activation of downstream genes, circRNA downstream genes are often proto-oncogenes or oncogenes, and the outcomes are mostly achieved by affecting the immune escape of cancer cells and the ability of cancer cells to repair genes or altering autophagy or EMT. For example, in studies of drug resistance in non-small-cell lung cancer (NSCLC), circFGFR1 also promotes NSCLC cell development and immune system escape by sponging miR-381-3p and targeting CXCR4, a gene that has been identified as a key oncogene in several cancers, and knockdown of CXCR4 reverses circFGFR1-induced NSCLC cell proliferation, migration and invasion (24). In addition, circFGFR1 is a sponge for several miRNAs. Additionally, in hepatocellular carcinoma cell resistance-related studies, circMET,can drive hepatocellular carcinoma immunosuppression and anti-PD-1 therapy resistance via the miR-30-5p/Snail/DPP4 axis. circMET also induces hepatocarcinogenesis and immune tolerance via the Snail/DPP4/CXCL10 axis (65).
The role of circRNAs in tumor drug resistance can be broadly classified into two categories according to their role, and in addition to the drug resistance enhancing effect mentioned above, there are also reports of drug resistance inhibiting effects. CircRNA can be negatively correlated with the generation of tumor drug resistance,For example, circ_0000079 induces the ribonucleic acid binding protein FXR1 to block the formation of the FXR1/PrCKI complex and reduces its promoting effects on NSCLC cell invasion and cisplatin resistance; it was shown that circ_0000079 (CiR79) levels were significantly downregulated in NSCLC patients, especially in cisplatin-resistant NSCLC patients, and that reduced circ_0000079 levels were significantly associated with overall survival in NSCLC patients. The results of Cell Counting Kit-8 and Transwell cell invasion assays showed that circ_0000079 overexpression significantly inhibited the proliferation and invasion of cisplatin-resistant NSCLC cells. Functionally and mechanistically, circ_0000079 negatively regulates the FXR1/PRKCI-mediated phosphorylation of glycogen synthase kinase 3β and activator protein 1, thereby decreasing the protein levels of the Snail gene, an important promoter gene that regulates cancer cell growth and EMT (30). This shows that circRNAs may be able to exert opposite effects on the biological behavior of tumor cells through different signaling pathways in different tumor cells.
CircRNAs Regulate Endoplasmic Reticulum Stress, Autophagy and Phagocytosis
Both the activation and inhibition of endoplasmic reticulum stress have been shown to attenuate chemo- and radioresistance in tumors (191–193), and endoplasmic reticulum stress is also a potent trigger of autophagy for controlling cancer sensitivity (194). Autophagy is a process by which cells engulf their own cytoplasmic proteins or organelles and encapsulate them into vesicles that fuses with lysosomes to form autophagic lysosomes, which degrade the contents they encapsulate. However, autophagy is also a double-edged sword. Mild autophagy facilitates cell survival under normal or mild stress conditions, while excessive autophagy can lead to cell death. Smith et al. showed that the development of drug resistance may be attributed to autophagy-mediated oxidative stress and the role of key metabolites provided by autophagy in maintaining cell stemness during dormancy (195–198).
However, in contrast to other mechanisms of drug resistance generation, autophagy and phagocytosis tend to be positively correlated with the generation of drug resistance, i.e., enhanced autophagy generates drug resistance and inhibition of autophagy inhibits drug resistance, presumably related to the specificity of the function of autophagy itself. In cervical cancer, for example, autophagy-related genes are aberrantly regulated in cells overexpressing hsa_circ_0023404 (78). Nevertheless, in studies of drug resistance in cervical cancer, circMTO1 can regulate the expression of the S100A1 protein via miR-6893 and can likewise affect the expression of autophagy-related genes (77).
In laryngeal cancer, circPGAM1 can enhance autophagic signaling during drug resistance by regulating miR-376a, and autophagy can be induced and activated by autophagy-related genes in light chain 3 (LC3). Functionally, autophagy plays a dual role by promoting cell death and cancer cell survival (87).
In acute myeloid leukemia (AML), circPAN3 is involved in AML drug resistance by regulating autophagy. The study suggests that circPAN3 may promote AML drug resistance by regulating autophagy and affecting the expression of apoptosis-related proteins, while AMPK/mTOR signaling plays a key role in circPAN3 regulation of autophagy. Moreover, there is more than one miRNA strongly predicted to be associated downstream of circPAN3 in this example, and regulating autophagy via the AMPK/mTOR pathway may also be only one of these pathways; whether other more precise downstream pathways of circPAN3 exist to influence tumor drug resistance remains to be investigated (88). Similarly, in studies of AML drug resistance, circPAN3 was also found to mediate AML drug resistance via the miR-153-5p/miR-183-5p-XIAP axis (89). Therefore, by summarizing the studies of autophagy-related circRNAs inducing drug resistance, we found that drug resistance induced by the regulation of autophagy and phagocytosis is often not mediated by a single mechanism of action.
Some examples of inhibiting drug resistance by suppressing autophagy are as follows: in a study of drug resistance in gastric cancer, Sun et al. found that circRNA MCTP2 inhibited cisplatin resistance in gastric cancer through miR-99a-5p-mediated MTMR3 expression, and mechanistically, miR-99a-5p directly targeted MTMR3, a myotubulin-associated protein, to inhibit autophagy (152). mTMR3 is a member of the myotubulin family. It is an inositol-like 3-phosphatase that hydrolyzes PtdIns3P (PI3P) (199). PI3P (inositol-like 3-phosphate) is required for the autophagic process, and the inhibitory effect of MTMR3 on PI3P inhibits the formation of autophagic vesicles. In this study, the action of the circRNA MCTP2 molecular sponge led to the inhibition of autophagy through the MTMR3 protein, which then caused changes in related proteins that regulate autophagy, such as p62, related genes, such as BCL-2 and Beclin1. The process of Sun et al. provides ideas to study the regulation of autophagy, but it is unknown whether there is an inverse regulatory relationship between the regulatory proteins or genes and the functional units of autophagy ( Figure 5 ).
Figure 5.
CircRNA regulate autophagy-mediated chemoresistance.
CircRNAs Promotion or Inhibition of DNA Repair
The induction of DNA damage is another major effect of radiotherapy and chemotherapy, usually leading to cell death. However, DNA repair can reduce the sensitivity of cancer cells to treatment (4), so enhancing or suppressing DNA repair can affect the proportion of tumor cells killed by drugs and, in turn, affects drug resistance. For example, in gastric cancer, it has been shown that circ_0026359 is overexpressed and correlated with cisplatin resistance and poor survival in gastric cancer patients, and conversely, deletion of circ_0026359 enhances miR-1200 activity, which reduces the expression level of POLD4 and thus reduces the resistance of gastric cancer cells to cisplatin. In contrast, POLD4 plays a role in cell proliferation and the maintenance of human cell genome stability. Low expression of POLD4 has been reported to impair DNA repair systems, including nucleotide excision repair, and increase the risk of lung cancer formation (145). As an additional example, in triple-negative breast cancer, hsa_circ_0000199 may activate the DNA repair molecule histone family member X (H2AX), leading to cisplatin resistance. This also involves the classical PI3K/AKT pathway (151, 200).
CircRNA AKT3 upregulates PIK3R1 by inhibiting miR-198 to enhance cisplatin resistance (by inhibiting apoptosis and facilitating DNA damage repair) in gastric cancer. Mechanistically, circAKT3 activates the PI3K/AKT signaling pathway in GC cells by sponging miR-198 to eliminate the inhibitory effect of this miRNA on its target gene PIK3R1. Activation of the PI3K/AKT pathway contributes to the upregulation of the DNA repair molecule BRCA1, leading to resistance to cisplatin-based chemotherapy.CircAKT3 affected the DNA damage response (DDR) in GC cells, suggested that circAKT3 may enhance cisplatin resistance through the PI3K/AKT pathway and DDdR mechanism in GC cells (151). In summary, to study the effect of DNA repair ability on tumor drug resistance, it is crucial to determine the connection between circRNA and DNA damage repair genes or proteins, which are commonly associated with DNA damage repair, such as POLD4, H2AX, BRCA1, etc. In addition, these genes or proteins are often linked to some classical signaling pathways, probably because repair involving DNA requires some well-established and stable genes and pathways.
CircRNAs Regulate Tumor Microenvironment (TME)
The TME is a complex ecosystem with microbiota, an acidic pH, inflammatory factors, matrix metalloproteinases, extracellular matrix (ECM), tumor-associated fibroblasts (CAFs), and tumor-associated macrophages (TAMs). The impact of the tumor microenvironment on the tumor is frequent and must be considered, for example, because the TME of the primary tumor and the corresponding metastases differ, which affects the sensitivity of cancer cells (201, 202). It has been demonstrated that external factors such as inflammatory factors, acidity and alkalinity of the microenvironment in which the tumor is located regulate tumor drug resistance by influencing protein synthesis, organelle division, and enzyme function in tumor cells. The TME can both directly and indirectly affect the ability of circRNAs to regulate tumor drug resistance; for example, the inflammatory factor IL-7 can reduce the expression of ABCG2 (ATP-binding cassette family protein) and regulate cisplatin resistance in NSCLC (203). A hypoxic tumor microenvironment can cause excessive mitochondrial division of NK cells, which reduces the immune surveillance function of the organism (204). Direct regulation of circRNAs is also possible; for example, in a study on the previously mentioned circRNA CCDC66 in lung adenocarcinoma resistance, the expression of SAE2 was mainly regulated by EGFR, whereas the expression of circRNA CCDC66 was positively regulated by FAK and c-Met and negatively regulated by nAchR7α. The immediate response to hypoxia can increase phosphorylated c-Met and SAE2 expression and EMT (180). This regulates tumor drug resistance upstream from circRNA. It is thus clear that the TME and the role of circRNAs are closely related, and the effect of the TME on tumor drug resistance may bring new ideas and possibilities for the exploration of mechanisms for drug-resistant tumors in circRNA research.
CircRNAs in Immune Checkpoint Inhibitors Resistance
Chemotherapeutic drugs have no specific target, and their effects are all over the body, and they are extremely sensitive to cells that proliferate rapidly. It can inhibit and kill the rapidly proliferating tumor cells, but also inevitably “accidentally” injure the normal cells of human body, which is a simple and brutal anti-cancer method of “Injure the enemy by 1000 and lose 800 by yourself”. In contrast, there are sites on the surface of targeted drugs that can specifically bind to tumor cells, which can precisely identify the lesion site, and when cancer cells are identified by immune checkpoint inhibitors, they are destroyed. The effect on normal cells is minimal. Immune checkpoint inhibitors (ICIs) have been shown to be highly efficient in the treatment of solid tumors, Therefore, ICIs treatment may be an important tool for tumor treatment in the future. However, limited benefit in terms of response and survival has been reported in many patients because of drug resistance. PD-L1 expression remains the only validated marker in clinics, molecular profiling has brought valuable information, showing that the tumor mutation load and microsatellite instability (MSI) status were associated to higher response rates in nearly all cancer types (205). The main immune checkpoint inhibitors currently in clinical use are anti-CTLA-4 and antibodies against PD-1 and its ligand PD-L1, which can be applied to many types of cancer and have shown significant improvements in patient survival time. However, despite the success of ICIs, resistance to these agents restricts the number of patients able to achieve durable responses, and immune-related adverse events complicate treatment. Due to tumor heterogeneity, current limited research shows that PD-1 or PD-L1 monoclonal antibody drug resistance may be related to the following factors: mutation of tumor antigen and antigen presentation process, multiple immune checkpoint interactions, immune microenvironment changes dynamically, activation of oncogenic pathways, gene mutation and epigenetic changes of key proteins in tumors, tumor competitive metabolism, and accumulation of metabolites, etc, mechanisms of resistance are complex. Therefore, it is the most urgent task to further elucidate the mechanism of immune checkpoint inhibitor resistance, discover multitumor universal biomarkers, and develop new target agents to improve the response rate of immunotherapy in patients (206). It has been shown that there is also a strong link between circRNAs and ICIs treatment resistance. Zhang et al. found circFGFR1 could directly interact with miR-381-3p and subsequently act as a miRNA sponge to upregulate the expression of the miR-381-3p target gene C-X-C motif chemokine receptor 4 (CXCR4), which promoted NSCLC progression and resistance to anti-programmed cell death 1 (PD-1)- based therapy in non-small cell lung cancer cells (24). Interestingly, Hong et al. found that knock-down of circ-CPA4 inhibited intracellular and extracellular PD-L1 by targeting let-7 miRNA. On the one hand, PD-L1 self-regulated NSCLC cell growth, mobility, stemness and chemoresistance to cisplatin treatment. On the other, secreted PD-L1 inactivated CD8 T cells by activating extracellular and intracellular pathways mediated cell death to facilitate immune evasion (25). There are many more reports on circRNAs regulating resistance to ICIs, mainly against PD-1 and PD-L1 in breast cancer and hepatocellular carcinoma (65, 134). CircRNAs regarding anti-CTLA-4 resistance are rare by comparison, but there are reports of circRNAs targeting CTLA4 via ceRNA(competing endogenous RNAs) mechanisms to regulate tumor tumorigenesis (207, 208). Since ICIs treatment is closely related to T cell immunity and tumor microenvironment, and circRNA also plays an important role in this regard, I think the possible relationship of circRNA in future ICIs treatment cannot be ignored.
Conclusion and Perspective
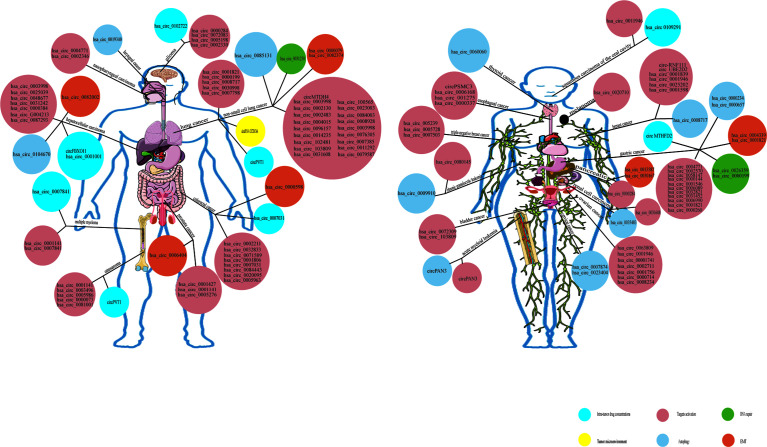
Tumor resistance is now known to be facilitated by factors affecting increased drug efflux transporters or reduced influx channels, enhanced EMT and stem cellularity, alterations in target gene copy number and the activation of bypass pathways or downstream signaling, endoplasmic reticulum stress, the remodeling of autophagy and phagocytosis, enhanced or diminished DNA repair capacity, and alterations in the TME ( Figure 6 ). This paper summarizes the links between the above mechanisms and circRNAs. From the beginning, circRNA was considered to be the “junk” in the cell, Research in recent years has discovered functions of circRNAs ranging from being microRNA (miRNA) sponges and transcription regulators to having protein in teractions and allowing for translation. The function and mechanism of action of circRNAs in tumors is becoming increasingly complex, especially after considering again the function of circ’s encoded proteins, the binding of a previously large number of circRNAs to downstream targets does not seem to be so pure. This also predetermines the diversity of circRNAs functions, in this case, the integration of the same circRNAs function is particularly important. In addition to the functional complexity, the function of the same circRNAs in different tumors or different environments is still worth thinking about whether the function of circRNAs will change with the cellular environment. Secondly, regarding the characterization of circRNAs distribution in cells, researchers habitually take the site where circRNAs saccumulates more (nucleus or cytoplasm) as the site where circRNAs exerts its function, and whether this is reasonable deserves further consideration. Because, we know that the content of circRNAs varies greatly in different tumors, the circRNAs enriched in a certain type of tumor, even if it is relatively less distributed, may still be quite higher after quantification than in other cells where it is more distributed. The role of circRNAs in tumor cells is also becoming increasingly important, and there exists a great potential to become a molecular marker and therapeutic target for tumors in the future. It is believed that, based on the complex mechanism of drug resistance, it is of no clinical significance to simply search for and regulate drug resistance targets, and it may even produce drug resistance again soon. It is speculated that according to the possible tumor characteristics, three types of treatment methods should be combined to change the tumor microenvironment ecology and eliminate various heterogeneous tumor subsets, so as to reduce tumor drug resistance and improve long-term clinical efficacy. Finally, standardizing the nomenclature of circRNAs in publications is imminent. The traditional way of naming circRNAs based on the parent gene has many drawbacks, because it is impossible to distinguish different circRNAs composed of different exons under the same parent gene, and even in many circRNAs related to tumor drug resistance articles do not provide the sequence or location of circRNAs, and the simple parent gene naming makes it impossible for me to target the exact circRNA. Of course, this problem is inevitable, because database inclusion and naming always follows the publication of the researcher’s results. I suggest that the first discovered circRNA can follow the nomenclature of the publisher, but subsequent articles need to specify the sequence and composition of the circRNA to distinguish if it is the same circRNA.
Figure 6.
Overview of functional circRNAs in various types of cancer. The map shows the circRNAs that have been confirmed to function in various types of cancer. As the mechanism of drug resistance is complex, only the main mechanism is listed in this map. Among the number other mechanisms, specific mechanisms such as the regulation of metabolism, cell cycle, apoptosis, RNA binding proteins (RBPs), immune escape,etc, summed up as alterations in target gene copy number and the activation of bypass pathways or downstream signaling.
Author Contributions
SW, LQ, and TC collected the related reports and drafted the manuscript. XH, YX, and LX revised the manuscript. YJ, HH, QF, QL, YW, and JW participated in designing the review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81902515), Excellent Youth Talent Support Program (Key) of Anhui Province (gxyqzd2020029), and Introducing Talents Natural Science Foundation of The First Affiliated Yijishan Hospital of Wannan Medical College (YR202006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
| 5-FU | 5-fluorouracil |
| ABCB10 | ATP binding cassette B family member 10 |
| ADAM28 | ADAM metallopeptidase domain 28 |
| ADAM9 | ADAM metallopeptidase domain 9 |
| ADM | Adriamycin |
| AEG-1 | Astrocyte-elevating protein-1 |
| AK4 | Adenylate kinase 4 |
| AKT | Serine/threonine protein kinase B |
| ALKBH5 | AlkB homolog H5 |
| AML | Acute myeloid leukemia |
| AMPK | Activated protein kinase |
| ANLN | Anillin |
| ARNT2 | Aryl hydrocarbon receptor nuclear translocator 2 |
| ARv7 | The androgen receptor splicing variant 7 |
| ASAP1 | Ankyrin repeat and Pleckstrin homology domain 1 |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| ATG2A | Autophagy-related gene 2A |
| ATG7 | Autophagy Related 7 |
| BCL2 | B-cell lymphoma-2 |
| BCL-2 | B-cell lymphoma-2 |
| BMI1 | B cell-specific Moloney murine leukemia virus integration site 1 |
| BRCA1 | Breast cancer type 1 susceptibility protein |
| BTZ | Bortezomib |
| CAFs | Tumor-associated fibroblasts |
| CCDC66 | Coiled-coil domain containing 66 |
| CCNB1 | Cyclin B1 |
| CCNE1 | Cyclin E1 |
| CDCA3 | Cell division cycle associated 3 |
| CDDP | Cisplatin |
| CDK4 | Cell cycle protein-dependent kinase 4 |
| CDK6 | Cyclin-dependent kinase 6 |
| CEBPG | CCAAT enhancer binding protein gamma |
| CELSR1 | Cadherin EGF LAG seven-pass G-type receptors 1 |
| CHD4 | Chromodomain helicase DNA-binding protein 4 |
| Chk2 | Checkpoint kinase 2 |
| CHTOP | Chromatin target of protein arginine methyltransferase |
| circRNAs | Circular RNAs |
| CORO1C | Coronin 1C |
| CSC | Cancer stem cell |
| CSPP1 | Centrosome and spindle pole associated protein 1 |
| Cul2 | Cullin2 |
| CXCL10 | Chemokine 10 |
| CXCL12 | Chemokine 12 |
| CXCR4 | Chemokine receptor type 4 DB |
| Diosbulbin-B | DCP1A |
| Decapping enzyme 1a | DDP |
| Diamminedichloroplatinum | DDP4 |
| Dipeptidyl peptidase-4 | DDR |
| DNA damage response | DDX17 |
| DEAD-box helicase 17 | DHX9 |
| DExH-box helicase 9 | DONSON |
| Downstream neighbor of SON | DOX |
| Doxorubicin | DPP4 |
| Dipeptidyl peptidase 4 | DTX |
| Docetaxel | DUSP7 |
| Dual specificity phosphatase 7 | E2F3 |
| E2F transcription factor 3 | ECM |
| Extracellular matrix | Eg5 |
| Kinesin-5 | EGFR |
| Epidermal growth factor receptor | EIF5A2 |
| Eukaryotic translation initiation factor 5A-2 | ELP3 |
| Elongator complex protein 3 | EMT |
| Epithelial-mesenchymal transition | Enz |
| Enzalutamide | ERK |
| Extracellular signal-regulated kinase | EZH2 |
| Enhancer of zeste homolog 2 | FAM73A |
| Family with sequence similarity 73 | member A |
| FAT1 | FAT atypical cadherin 1 |
| FBXL18 | F-box and leucine-rich repeat protein 18 |
| FBXL5 | Leucine-rich repeat protein 5 |
| FBXW7 | F-box and WD repeat domain containing 7 |
| FGF9 | Fibroblast growth factor 9 |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FMNL3 | Formin like 3 |
| FN1 | Fibronectin 1 |
| FOXC2 | Forkhead box protein C2 |
| FOXM1 | Forkhead box protein M1 |
| FOXO1 | Forkhead box O1 |
| FOXO3 | Forkhead box O3 |
| FOXO3a | Forkhead box O 3a |
| Foxp1 | Forkhead box protein P1 |
| FOXQ1 | Forkhead box Q1 |
| FOXR2 | Forkhead box 2 |
| FXR1 | Fragile X-Related 1 |
| FZD7 | Frizzled-7 |
| GEM | Gemcitabine |
| GFRA1 | GDNF Family Receptor Alpha 1 |
| GLUT1 | Glucose transporter 1 |
| GOT1 | Glutamate oxaloacetate transaminase 1 |
| GPCPD1 | Glycerophosphocholine phosphodiesterase 1 |
| GRB2 | Growth factor receptor-bound protein-2 |
| GRK5 | G protein-coupled receptor kinase 5 |
| H2AX | Histone family member X |
| HDGF | Hepatoma-derived growth factor |
| HECTD1 | HECT domain E3 ubiquitin ligase 1 |
| HGF | Hepatocyte growth factor |
| HIF-1α | Hypoxia-inducible factor-1α |
| HIPK2 | Homeodomain-interacting protein kinase 2 |
| HIPK3 | Homeodomain-interacting protein kinase 3 |
| HK2 | Hexokinase-2 |
| HMG | High mobility group |
| HMGA2 | High-mobility group AT-hook 2 |
| HMGB1 | High-mobility group box 1 |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K |
| HOXC8 | Homeobox C8 |
| IL-25 | Interleukin -25 |
| IM | Imatinib |
| ITCH | Itchy E3 ubiquitin protein ligase |
| JAG1 | Jagged1 |
| JAK2 | Janus kinase 2 |
| JMJD1C | Jumonji domain containing 1C |
| KANK1 | Kidney ankyrin repeat-containing protein 1 |
| KDM4C | Lysine demethylase 4C |
| KIF2A | Kinesin family member 2A |
| KLF12 | Krüppel-like factor 12 |
| LC3 | Light chain 3 |
| LDHA | Lactate dehydrogenase A |
| LIFR | Leukemia inhibitory factor receptor |
| LIN28B | Lin-28 homolog B |
| m6A | N6-methyladenosine |
| MCTP2 | Multiple C2-domains with two transmembrane regions 2 |
| MDM2 | Mouse double minute 2 |
| MDR1 | Multidrug resistance gene 1 |
| MDR-1 | Multidrug resistance gene-1 |
| MECP2 | Methyl-CpG-binding-protein 2 |
| MEK | Mitogen-activated protein kinase kinase |
| MET | MET proto-oncogene receptor tyrosine kinase |
| miRNAs | MicroRNAs |
| MMP1 | Matrix metalloproteinase 1 |
| MMP11 | Matrix metalloproteinase 11 |
| MMP17 | Matrix metalloproteinase 17 |
| MRP1 | Multidrug resistance-associated protein-1 |
| MSH2 | MutS homolog 2 |
| MSI1 | Musashi1 |
| MTA | Pemetrexed |
| MTMR3 | Myotubularin-related protein 3 |
| MTO1 | Mitochondrial tRNA translation optimization 1 |
| mTOR | Mechanistic target of rapamycin kinase |
| MTX | Methotrexate |
| MutSα | Alpha-melanocyte stimulating hormone |
| MYH9 | Myosin heavy chain 9 |
| nAChRα7 | Nicotinic acetylcholine receptor alpha 7 |
| NANOG | Nanog homeobox |
| NCOA3 | Nuclear receptor coactivator 3 |
| NEK2 | NIMA-related kinase 2 |
| NFIX | Nuclear factor I X |
| NK | Natural killer |
| NRAS | Neuroblastoma RAS |
| NRIP1 | Nuclear receptor-interacting protein 1 |
| NSCLC | Non-small-cell lung cancer |
| NT5E | Ecto-5’-nucleotidase |
| OXA | oxaliplatin |
| PBLD | Problem-based learning discussion |
| PBX3 | Pre-leukemia transcription factor 3 |
| PD-1 | Programmed cell death 1 |
| PDK1 | 3-phosphoinositide-dependent protein kinase 1 |
| PD-L1 | Programmed death-ligand 1 |
| PDPK1 | 3-phosphatidylinositol-dependent protein kinase-1 |
| PDX | Patient-derived xenograft |
| PGAM1 | Phosphoglycerate mutase 1 |
| PI3K | Phosphatidylinositol 3-kinase |
| PI3P | Phosphatidylinositol 3-phosphate |
| PIK3R1 | The PI3K regulatory subunit p85alpha |
| PIP5K1A | Phosphatidylinositol-4-phosphate 5-kinase type 1 alpha |
| PKM2 | The M2 isoform of pyruvate kinase |
| PKP3 | Plakophilin 3 |
| POLD4 | Eukaryote DNA polymeraseδ4 |
| POU3F2 | POU class 3 homeobox 2 |
| PPFIA1 | PTPRF interacting protein alpha 1 |
| PRDM2 | PR/SET domain 2 |
| PRDX2 | Peroxiredoxin 2 |
| PRKCD | Protein kinase C, delta |
| PRPF39 | Pre-mRNA splicing factor 39 |
| PSMC3 | Proteasome 26S ATPase subunit 3 |
| PTEN | Phosphatase and tensin homolog |
| PTGR1 | Effect of prostaglandin reductase 1 |
| PTX | Paclitaxel |
| PVT1 | Plasmacytoma variant translocation 1 |
| RAS | Rat sarcoma |
| RASSF1 | RAS-Association Domain Family 1 |
| RASSF6 | Ras association domain family member 6 |
| RB | Retinoblastoma protein |
| RBP | RNA binding protein |
| REV3L | EV3-like DNA-directed polymerase ζ catalytic subunit |
| RNF | RING finger protein |
| ROCK | Rho-associated coiled-coil-forming protein kinase |
| ROR1 | Receptor tyrosine kinase-like orphan receptor 1 |
| S100A1 | S100A1 calcium-binding protein A1 |
| SAE | SUMO-activating enzyme subunit |
| SAE2 | Activating enzyme subunit 2 |
| SCAI | Suppressor of Cancer Cell Invasion |
| SCMH1 | Sex comb on midleg homolog-1 |
| SEMA6D | Semaphorins 6D |
| SH2 | Src homology region 2 |
| SHP2 | SH2-containing protein tyrosine phosphatase 2 |
| SIK2 | Salt inducible kinase 2 |
| SKA1 | Spindle and kinetochore-associated complex subunit 1 |
| SNAI1 | Snai family zinc finger 1 |
| SNX6 | Sorting nexin 6 |
| SOX4 | SRY-box transcription factor 4 |
| SOX9 | SRY-box transcription factor 9 |
| STAT3 | Signal transducer and activator of transcription 3 |
| STX3 | Syntaxin 3 |
| SUMO | Small ubiquitin-related modifier |
| TAM | Tamoxifen |
| TAMs | Tumor-associated macrophages |
| TGF-α | Transforming growth factor alpha |
| TGM2 | Transglutaminase-2 |
| TIM-3 | T cell immunoglobulin and mucin domain 3 |
| TKI | Tyrosine kinase inhibitor |
| TKIs | Tyrosine kinase inhibitors |
| TLR4 | Toll-like Receptor 4 |
| TME | Tumor microenvironment |
| TMZ | Temozolomide |
| TNPO3 | Transportin3 |
| TPD52 | Tumor protein D52 |
| TRAF4 | Tumor necrosis factor receptor associated factor 4 |
| TRIM14 | Tripartite motif-containing 14 |
| TRIM65 | Tripartite motif-containing protein 65 |
| TWIST1 | Twist family bHLH transcription factor 1 |
| UBAP2 | Ubiquitin associated protein 2 |
| UHRF1 | Ubiquitin like with PHD and ring finger domains 1 |
| ULK1 | Unc-51-like kinase 1 |
| USP7 | Ubiquitin-specific protease-7 |
| VapA | Virulence associated protein A |
| VEGFA | Vascular endothelial growth factor A |
| VRACs | Volume-regulated anion channels |
| WIF1 | Wnt inhibitory factor 1 |
| Wnt7a | Wnt family member 7A |
| WTAP | Wilms tumor 1-associated protein |
| XIAP | X-linked inhibitor of apoptosis protein |
| YAP | Yes-associated protein |
| YAP1 | Yes-associated protein 1 |
| YBX1 | Y box binding protein 1 |
| ZEB1 | Zinc finger E-box binding homeobox 1 |
| ZIC5 | Zinc finger protein of the cerebellum 5 |
References
- 1. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci USA (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some Yeast Mitochondrial RNAs Are Circular. Cell (1980) 19(2):313–9. doi: 10.1016/0092-8674(80)90505-X [DOI] [PubMed] [Google Scholar]
- 3. Chen LL, Yang L. Regulation of circRNA Biogenesis. RNA Biol (2015) 12(4):381–8. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui C, Yang J, Li X, Liu D, Fu L, Wang X, et al. Functions and Mechanisms of Circular RNAs in Cancer Radiotherapy and Chemotherapy Resistance. Mol Cancer (2020) 19(1):58. doi: 10.1186/s12943-020-01180-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu T, Wang M, Jiang L, Ma L, Wan L, Chen Q, et al. CircRNAs in Anticancer Drug Resistance: Recent Advances and Future Potential. Mol Cancer (2020) 19(1):127. doi: 10.1186/s12943-020-01240-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gong J, Luk F, Jaiswal R, George AM, Grau GE, Bebawy M, et al. Circular RNA CRIM1 Functions as a ceRNA to Promote Nasopharyngeal Carcinoma Metastasis and Docetaxel Chemoresistance Through Upregulating FOXQ1. Mol Cancer (2020) 19(1):33. doi: 10.1186/s12943-020-01149-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joyce H, McCann A, Clynes M, Larkin A. CircNRIP1 Modulates the miR-515-5p/IL-25 Axis to Control 5-Fu and Cisplatin Resistance in Nasopharyngeal Carcinoma. Drug Design Dev Ther (2021) 15:323–30. doi: 10.2147/DDDT.S292180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Planells-Cases R, Lutter D, Guyader C, Gerhards NM, Ullrich F, Elger DA, et al. Exosome-Delivered circRNA Promotes Glycolysis to Induce Chemoresistance Through the miR-122-PKM2 Axis in Colorectal Cancer. Mol Oncol (2020) 14(3):539–55. doi: 10.1002/1878-0261.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saxena M, Stephens MA, Pathak H, Rangarajan A. Exosomal Transfer of Circular RNA FBXW7 Ameliorates the Chemoresistance to Oxaliplatin in Colorectal Cancer by Sponging miR-18b-5p. Neoplasma (2021) 68(1):108–18. doi: 10.4149/neo_2020_200417N414 [DOI] [PubMed] [Google Scholar]
- 10. Hua L, Huang L, Zhang X, Feng H, Shen B. Circ-PRKDC Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells by Regulating miR-375/FOXM1 Axis and Wnt/β-Catenin Pathway. Onco Targets Ther (2020) 13:5939–53. doi: 10.2147/OTT.S253468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song Y, Hu N, Song X, Yang J. Hsa_circ_001680 Affects the Proliferation and Migration of CRC and Mediates its Chemoresistance by Regulating BMI1 Through miR-340. Mol Cancer (2020) 19(1):20. doi: 10.1186/s12943-020-1134-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yi X, Chen W, Li C, Chen X, Lin Q, Lin S, et al. Down-Regulation of Circ_0032833 Sensitizes Colorectal Cancer to 5-Fluorouracil and Oxaliplatin Partly Depending on the Regulation of miR-125-5p and MSI1. Cancer Manag Res (2020) 12:11257–69. doi: 10.2147/CMAR.S270123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang W, Wang Z, Cai G, Huang P. Downregulation of Circ_0071589 Suppresses Cisplatin Resistance in Colorectal Cancer by Regulating the MiR-526b-3p/KLF12 Axis. Cancer Manag Res (2021) 13:2717–31. doi: 10.2147/CMAR.S294880 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Xi L, Liu Q, Zhang W, Luo L, Song J, Liu R, et al. Circular RNA Circcspp1 Knockdown Attenuates Doxorubicin Resistance and Suppresses Tumor Progression of Colorectal Cancer via miR-944/FZD7 Axis. Cancer Cell Int (2021) 21(1):153. doi: 10.1186/s12935-021-01855-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren TJ, Liu C, Hou JF, Shan FX. CircDDX17 Reduces 5-Fluorouracil Resistance and Hinders Tumorigenesis in Colorectal Cancer by Regulating miR-31-5p/KANK1 Axis. Eur Rev Med Pharmacol Sci (2020) 24(4):1743–54. doi: 10.26355/eurrev_202002_20351 [DOI] [PubMed] [Google Scholar]
- 16. He X, Ma J, Zhang M, Cui J, Yang H. Circ_0007031 Enhances Tumor Progression and Promotes 5-Fluorouracil Resistance in Colorectal Cancer Through Regulating miR-133b/ABCC5 Axis. Cancer Biomark (2020) 29(4):531–42. doi: 10.3233/CBM-200023 [DOI] [PubMed] [Google Scholar]
- 17. Zhao K, Cheng X, Ye Z, Li Y, Peng W, Wu Y, et al. Exosome-Mediated Transfer of Circ_0000338 Enhances 5-Fluorouracil Resistance in Colorectal Cancer Through Regulating MicroRNA 217 (miR-217) and miR-485-3p. Mol Cell Biol (2021) 41(5):e00517–20. doi: 10.1128/MCB.00517-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Y, Cao Z, Shan J, Gao Y, Liu X, Ma D, et al. Hsa_circ_0020095 Promotes Oncogenesis and Cisplatin Resistance in Colon Cancer by Sponging miR-487a-3p and Modulating Sox9. Front Cell Dev Biol (2020) 8:604869. doi: 10.3389/fcell.2020.604869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song Y, Hu N, Song X, Yang J. Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating Abcg2. Technol Cancer Res Treat (2020) 19:1533033820928371. doi: 10.1177/1533033820928371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J, Du F, Chen C, Li D, Chen Y, Xiao X, et al. CircRNA ITCH Increases Bortezomib Sensitivity Through Regulating the miR-615-3p/PRKCD Axis in Multiple Myeloma. Life Sci (2020) 262:118506. doi: 10.1016/j.lfs.2020.118506 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Lin Q, Song C, Ma R, Li X. Depletion of Circ_0007841 Inhibits Multiple Myeloma Development and BTZ Resistance via miR-129-5p/JAG1 Axis. Cell Cycle (2020) 19(23):3289–302. doi: 10.1080/15384101.2020.1839701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye Y, Zhao L, Li Q, Xi C, Li Y, Li Z, et al. Circ_0007385 Served as Competing Endogenous RNA for miR-519d-3p to Suppress Malignant Behaviors and Cisplatin Resistance of Non-Small Cell Lung Cancer Cells. Thorac Cancer (2020) 11(8):2196–208. doi: 10.1111/1759-7714.13527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu X, Han J, Lan H, Lin Q, Wang Y, Sun X. A Novel Circular RNA Hsa_circRNA_103809/miR-377-3p/GOT1 Pathway Regulates Cisplatin-Resistance in Non-Small Cell Lung Cancer (NSCLC). BMC Cancer (2020) 20(1):1190. doi: 10.1186/s12885-020-07680-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA Circfgfr1 Promotes Progression and Anti-PD-1 Resistance by Sponging miR-381-3p in Non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18(1):179. doi: 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA Circ-CPA4/ Let-7 miRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-Small Cell Lung Cancer (NSCLC). J Exp Clin Cancer Res (2020) 39(1):149. doi: 10.1186/s13046-020-01648-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pang J, Ye L, Zhao D, Zhao D, Chen Q, et al. Circular RNA PRMT5 Confers Cisplatin-Resistance via miR-4458/REV3L Axis in Non-Small-Cell Lung Cancer. Cell Biol Int (2020) 44(12):2416–26. doi: 10.1002/cbin.11449 [DOI] [PubMed] [Google Scholar]
- 27. Lu H, Xie X, Wang K, Chen Q, Cai S, Liu D, et al. Circular RNA Hsa_Circ_0096157 Contributes to Cisplatin Resistance by Proliferation, Cell Cycle Progression, and Suppressing Apoptosis of Non-Small-Cell Lung Carcinoma Cells. Mol Cell Biochem (2020) 475(1-2):63–77. doi: 10.1007/s11010-020-03860-1 [DOI] [PubMed] [Google Scholar]
- 28. Zhou Y, Zheng X, Xu B, Chen L, Wang Q, Deng H, et al. Circular RNA Hsa_Circ_0004015 Regulates the Proliferation, Invasion, and TKI Drug Resistance of Non-Small Cell Lung Cancer by miR-1183/PDPK1 Signaling Pathway. Biochem Biophys Res Commun (2019) 508(2):527–35. doi: 10.1016/j.bbrc.2018.11.157 [DOI] [PubMed] [Google Scholar]
- 29. Kong R. Circular RNA Hsa_Circ_0085131 is Involved in Cisplatin-Resistance of Non-Small-Cell Lung Cancer Cells by Regulating Autophagy. Cell Biol Int (2020) 44(9):1945–56. doi: 10.1002/cbin.11401 [DOI] [PubMed] [Google Scholar]
- 30. Chen C, Zhang M, Zhang Y. Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cell Transplant (2020) 29:963689720961070. doi: 10.1177/0963689720961070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, et al. Circ_0076305 Regulates Cisplatin Resistance of Non-Small Cell Lung Cancer via Positively Modulating STAT3 by Sponging miR-296-5p. Life Sci (2019) 239:116984. doi: 10.1016/j.lfs.2019.116984 [DOI] [PubMed] [Google Scholar]
- 32. Li X, Yang B, Ren H, Xiao T, Zhang L, Li L, et al. Hsa_circ_0002483 Inhibited the Progression and Enhanced the Taxol Sensitivity of Non-Small Cell Lung Cancer by Targeting miR-182-5p. Cell Death Dis (2019) 10(12):953. doi: 10.1038/s41419-019-2180-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma J, Qi G, Li L. A Novel Serum Exosomes-Based Biomarker Hsa_Circ_0002130 Facilitates Osimertinib-Resistance in Non-Small Cell Lung Cancer by Sponging miR-498. Onco Targets Ther (2020) 13:5293–307. doi: 10.2147/OTT.S243214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo C, Wang H, Jiang H, Qiao L, Wang X. Circ_0011292 Enhances Paclitaxel Resistance in Non-Small Cell Lung Cancer by Regulating miR-379-5p/TRIM65 Axis. Cancer Biother Radiopharm (2020) 37(2):84–95. doi: 10.1089/cbr.2019.3546 [DOI] [PubMed] [Google Scholar]
- 35. Xu X, Tao R, Sun L, Ji X. Exosome-Transferred Hsa_Circ_0014235 Promotes DDP Chemoresistance and Deteriorates the Development of Non-Small Cell Lung Cancer by Mediating the miR-520a-5p/CDK4 Pathway. Cancer Cell Int (2020) 20(1):552. doi: 10.1186/s12935-020-01642-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li YH, Xu CL, He CJ, Pu HH, Liu JL, Wang Y, et al. circMTDH.4/miR-630/AEG-1 Axis Participates in the Regulation of Proliferation, Migration, Invasion, Chemoresistance, and Radioresistance of NSCLC. Mol Carcinog (2020) 59(2):141–53. doi: 10.1002/mc.23135 [DOI] [PubMed] [Google Scholar]
- 37. Yang B, Teng F, Chang L, Wang J, Liu DL, Cui YS, et al. Tumor-Derived Exosomal circRNA_102481 Contributes to EGFR-TKIs Resistance via the miR-30a-5p/ROR1 Axis in Non-Small Cell Lung Cancer. Aging (Albany NY) (2021) 13(9):13264–86. doi: 10.18632/aging.203011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geng J, Yang K. Circccnd1 Regulates Oxidative Stress and FGF9 to Enhance Chemoresistance of Non-Small Cell Lung Cancer via Sponging miR-187-3p. DNA Cell Biol (2021) 40(5):675–82. doi: 10.1089/dna.2020.6412 [DOI] [PubMed] [Google Scholar]
- 39. Zhang W, Song C, Ren X. Circ_0003998 Regulates the Progression and Docetaxel Sensitivity of DTX-Resistant Non-Small Cell Lung Cancer Cells by the miR-136-5p/CORO1C Axis. Technol Cancer Res Treat (2021) 20:1533033821990040. doi: 10.1177/1533033821990040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi Q, Ji T, Ma Z, Tan Q, Liang J. Serum Exosomes-Based Biomarker Circ_0008928 Regulates Cisplatin Sensitivity, Tumor Progression, and Glycolysis Metabolism by miR-488/HK2 Axis in Cisplatin-Resistant Nonsmall Cell Lung Carcinoma. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4490 [DOI] [PubMed] [Google Scholar]
- 41. Song HM, Meng D, Wang JP, Zhang XY. circRNA Hsa_Circ_0005909 Predicts Poor Prognosis and Promotes the Growth, Metastasis, and Drug Resistance of Non-Small-Cell Lung Cancer via the miRNA-338-3p/SOX4 Pathway. Dis Markers (2021) 2021:8388512. doi: 10.1155/2021/8388512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liang H, Lin Z, Lin H, Zhao L, Huang W. circRNA_103615 Contributes to Tumor Progression and Cisplatin Resistance in NSCLC by Regulating ABCB1. Exp Ther Med (2021) 22(3):934. doi: 10.3892/etm.2021.10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J, Fan R, Xiao H. Circ_ZFR Contributes to the Paclitaxel Resistance and Progression of Non-Small Cell Lung Cancer by Upregulating KPNA4 Through Sponging miR-195-5p. Cancer Cell Int (2021) 21(1):15. doi: 10.1186/s12935-020-01702-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feng N, Guo Z, Wu X, Tian Y, Li Y, Geng Y, et al. Circ_PIP5K1A Regulates Cisplatin Resistance and Malignant Progression in Non-Small Cell Lung Cancer Cells and Xenograft Murine Model via Depending on miR-493-5p/ROCK1 Axis. Respir Res (2021) 22(1):248. doi: 10.1186/s12931-021-01840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu L, Li J, Peng B, Cai P, Zhao B, Chen Y, et al. CircASXL1 Knockdown Restrains Hypoxia-Induced DDP Resistance and NSCLC Progression by Sponging miR-206. Cancer Manag Res (2021) 13:5077–89. doi: 10.2147/CMAR.S276964 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Zhang Y, Ge P, Zhou D, Xing R, Bai L. Circular RNA FOXO3 Accelerates Glycolysis and Improves Cisplatin Sensitivity in Lung Cancer Cells via the miR-543/Foxo3 Axis. Oncol Lett (2021) 22(6):839. doi: 10.3892/ol.2021.13100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li C, Zhang J, Yang X, Hu C, Chu T, Zhong R, et al. Hsa_Circ_0003222 Accelerates Stemness and Progression of Non-Small Cell Lung Cancer by Sponging miR-527. Cell Death Dis (2021) 12(9):807. doi: 10.1038/s41419-021-04095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J, et al. Overexpression of Hsa_Circ_0002874 Promotes Resistance of Non-Small Cell Lung Cancer to Paclitaxel by Modulating miR-1273f/MDM2/p53 Pathway. Aging (Albany NY) (2021) 13(4):5986–6009. doi: 10.18632/aging.202521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X, Fu X, Zhang X, Bai C, Wang Y. Circ_0001658 Regulates Gefitinib Resistance of Non-Small Cell Lung Cancer Through miR-409-3p/TWIST1 Axis. Anticancer Drugs (2021) 33(2):158–66. doi: 10.1097/CAD.0000000000001257 [DOI] [PubMed] [Google Scholar]
- 50. Xu Y, Zhao R, Wang H, Jiang J, Wang Z, Wang J, et al. Circular RNA PRMT5 Knockdown Enhances Cisplatin Sensitivity and Immune Response in Non-Small Cell Lung Cancer by Regulating miR-138-5p/MYH9 Axis. J buon (2021) 26(5):1850–61. [PubMed] [Google Scholar]
- 51. Wang X, Wang H, Jiang H, Qiao L, Guo C. Circular RNAcirc_0076305 Promotes Cisplatin (DDP) Resistance of Non-Small Cell Lung Cancer Cells by Regulating ABCC1 Through miR-186-5p. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4153 [DOI] [PubMed] [Google Scholar]
- 52. Shao N, Song L, Sun X. Exosomal Circ_PIP5K1A Regulates the Progression of Non-Small Cell Lung Cancer and Cisplatin Sensitivity by miR-101/ABCC1 Axis. Mol Cell Biochem (2021) 476(6):2253–67. doi: 10.1007/s11010-021-04083-8 [DOI] [PubMed] [Google Scholar]
- 53. Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer Cell-Derived Exosomal Circusp7 Induces CD8(+) T Cell Dysfunction and Anti-PD1 Resistance by Regulating the miR-934/SHP2 Axis in NSCLC. Mol Cancer (2021) 20(1):144. doi: 10.1186/s12943-021-01448-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du D, Cao X, Duan X, Zhang X. Blocking Circ_0014130 Suppressed Drug Resistance and Malignant Behaviors of Docetaxel Resistance-Acquired NSCLC Cells via Regulating miR-545-3p-YAP1 Axis. Cytotechnology (2021) 73(4):571–84. doi: 10.1007/s10616-021-00478-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y, Zhai R, Hu S, Liu J. Circular RNA Circ-RNF121 Contributes to Cisplatin (DDP) Resistance of Non-Small-Cell Lung Cancer Cells by Regulating the miR-646/SOX4 Axis. Anticancer Drugs (2022) 33(1):e186–97. doi: 10.1097/CAD.0000000000001184 [DOI] [PubMed] [Google Scholar]
- 56. Liu Y, Li C, Liu H, Wang J. Circ_0001821 Knockdown Suppresses Growth, Metastasis, and TAX Resistance of Non-Small-Cell Lung Cancer Cells by Regulating the miR-526b-5p/GRK5 Axis. Pharmacol Res Perspect (2021) 9(4):e00812. doi: 10.1002/prp2.812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu K, Zhu J, Geng J, Zhang Y, Qin Y, Wang F, et al. Circsnx6 (Hsa_Circ_0031608) Enhances Drug Resistance of Non-Small Cell Lung Cancer (NSCLC) via miR-137. Biochem Biophys Res Commun (2021) 567:79–85. doi: 10.1016/j.bbrc.2021.06.032 [DOI] [PubMed] [Google Scholar]
- 58. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-Methyladenosine-Modified Circigf2bp3 Inhibits CD8(+) T-Cell Responses to Facilitate Tumor Immune Evasion by Promoting the Deubiquitination of PD-L1 in Non-Small Cell Lung Cancer. Mol Cancer (2021) 20(1):105. doi: 10.1186/s12943-021-01398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhong Y, Lin H, Li Q, Liu C, Shen J. CircRNA_100565 Contributes to Cisplatin Resistance of NSCLC Cells by Regulating Proliferation, Apoptosis and Autophagy via miR-337-3p/ADAM28 Axis. Cancer Biomark (2021) 30(2):261–73. doi: 10.3233/CBM-201705 [DOI] [PubMed] [Google Scholar]
- 60. Xu Y, Jiang T, Wu C, Zhang Y. CircAKT3 Inhibits Glycolysis Balance in Lung Cancer Cells by Regulating miR-516b-5p/STAT3 to Inhibit Cisplatin Sensitivity. Biotechnol Lett (2020) 42(7):1123–35. doi: 10.1007/s10529-020-02846-9 [DOI] [PubMed] [Google Scholar]
- 61. Li X, Feng Y, Yang B, Xiao T, Ren H, Yu X, et al. A Novel Circular RNA, Hsa_Circ_0030998 Suppresses Lung Cancer Tumorigenesis and Taxol Resistance by Sponging miR-558. Mol Oncol (2021) 15(8):2235–48. doi: 10.1002/1878-0261.12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu Z, Gong Q, Yu Y, Zhu J, Li W. Knockdown of Circ-ABCB10 Promotes Sensitivity of Lung Cancer Cells to Cisplatin via miR-556-3p/AK4 Axis. BMC Pulm Med (2020) 20(1):10. doi: 10.1186/s12890-019-1035-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zheng F, Xu R. CircPVT1 Contributes to Chemotherapy Resistance of Lung Adenocarcinoma Through miR-145-5p/ABCC1 Axis. BioMed Pharmacother (2020) 124:109828. doi: 10.1016/j.biopha.2020.109828 [DOI] [PubMed] [Google Scholar]
- 64. Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, et al. A Novel Protein Encoded by Circask1 Ameliorates Gefitinib Resistance in Lung Adenocarcinoma by Competitively Activating ASK1-Dependent Apoptosis. Cancer Lett (2021) 520:321–31. doi: 10.1016/j.canlet.2021.08.007 [DOI] [PubMed] [Google Scholar]
- 65. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET Drives Immunosuppression and Anti-PD1 Therapy Resistance in Hepatocellular Carcinoma via the miR-30-5p/Snail/DPP4 Axis. Mol Cancer (2020) 19(1):92. doi: 10.1186/s12943-020-01213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal Circuhrf1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-Methyladenosine-Modified CircRNA-SORE Sustains Sorafenib Resistance in Hepatocellular Carcinoma by Regulating β-Catenin Signaling. Mol Cancer (2020) 19(1):163. doi: 10.1186/s12943-020-01281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li J, Qin X, Wu R, Wan L, Zhang L, Liu R, et al. Circular RNA Circfbxo11 Modulates Hepatocellular Carcinoma Progress and Oxaliplatin Resistance Through miR-605/FOXO3/ABCB1 Axis. J Cell Mol Med (2020) 24(9):5152–61. doi: 10.1111/jcmm.15162 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing YBX1. Signal Transduct Target Ther (2020) 5(1):298. doi: 10.1038/s41392-020-00375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Y, Zhang Y, Zhang S, Huang D, Li B, Liang G, et al. circRNA Circarnt2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the miR-155-5p/PDK1 Axis. Mol Ther Nucleic Acids (2021) 23:244–54. doi: 10.1016/j.omtn.2020.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fan W, Chen L, Wu X, Zhang T. Circ_0031242 Silencing Mitigates the Progression and Drug Resistance in DDP-Resistant Hepatoma Cells by the miR-924/POU3F2 Axis. Cancer Manag Res (2021) 13:743–55. doi: 10.2147/CMAR.S272851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qin L, Zhan Z, Wei C, Li X, Zhang T, Li J. Hsa−circRNA−G004213 Promotes Cisplatin Sensitivity by Regulating Mir−513b−5p/PRPF39 in Liver Cancer. Mol Med Rep (2021) 23(6):421. doi: 10.3892/mmr.2021.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weng H, Zeng L, Cao L, Chen T, Li Y, Xu Y, et al. Circfoxm1 Contributes to Sorafenib Resistance of Hepatocellular Carcinoma Cells by Regulating MECP2 via miR-1324. Mol Ther Nucleic Acids (2021) 23:811–20. doi: 10.1016/j.omtn.2020.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang W, Huang F, Feng C. CircFoxo3 Promotes Adriamycin Resistance Through Regulation of miR-199a-5p/ATP Binding Cassette Subfamily C Member 1 Axis in Hepatocellular Carcinoma. Onco Targets Ther (2020) 13:5113–22. doi: 10.2147/OTT.S243571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li X, He J, Ren X, Zhao H, Zhao H. Circ_0003998 Enhances Doxorubicin Resistance in Hepatocellular Carcinoma by Regulating miR-218-5p/EIF5A2 Pathway. Diagn Pathol (2020) 15(1):141. doi: 10.1186/s13000-020-01056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li P, Song R, Yin F, Liu M, Liu H, Ma S, et al. Circmrps35 Promotes Malignant Progression and Cisplatin Resistance in Hepatocellular Carcinoma. Mol Ther (2022) 30(1):431–47. doi: 10.1016/j.ymthe.2021.08.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen M, Ai G, Zhou J, Mao W, Li H, Guo J, et al. Circmto1 Promotes Tumorigenesis and Chemoresistance of Cervical Cancer via Regulating miR-6893. BioMed Pharmacother (2019) 117:109064. doi: 10.1016/j.biopha.2019.109064 [DOI] [PubMed] [Google Scholar]
- 78. Guo J, Chen M, Ai G, Mao W, Li H, Zhou J, et al. Hsa_circ_0023404 Enhances Cervical Cancer Metastasis and Chemoresistance Through VEGFA and Autophagy Signaling by Sponging miR-5047. BioMed Pharmacother (2019) 115:108957. doi: 10.1016/j.biopha.2019.108957 [DOI] [PubMed] [Google Scholar]
- 79. Li X, Liu Y, Zhang X, Shen J, Xu R, Liu Y, et al. Circular RNA Hsa_Circ_0000073 Contributes to Osteosarcoma Cell Proliferation, Migration, Invasion and Methotrexate Resistance by Sponging miR-145-5p and miR-151-3p and Upregulating NRAS. Aging (Albany NY) (2020) 12(14):14157–73. doi: 10.18632/aging.103423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed Circpvt1, a Potential New Circular RNA Biomarker, Contributes to Doxorubicin and Cisplatin Resistance of Osteosarcoma Cells by Regulating ABCB1. Int J Biol Sci (2018) 14(3):321–30. doi: 10.7150/ijbs.24360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gong X, Li W, Dong L, Qu F. CircUBAP2 Promotes SEMA6D Expression to Enhance the Cisplatin Resistance in Osteosarcoma Through Sponging miR-506-3p by Activating Wnt/β-Catenin Signaling Pathway. J Mol Histol (2020) 51(4):329–40. doi: 10.1007/s10735-020-09883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou W, Liu Y, Wu X. Down-Regulation of circITCH Promotes Osteosarcoma Development and Resistance to Doxorubicin via the miR-524/RASSF6 Axis. J Gene Med (2021) 23(10):e3373. doi: 10.1002/jgm.3373 [DOI] [PubMed] [Google Scholar]
- 83. Wei W, Ji L, Duan W, Zhu J. Circular RNA Circ_0081001 Knockdown Enhances Methotrexate Sensitivity in Osteosarcoma Cells by Regulating miR-494-3p/TGM2 Axis. J Orthop Surg Res (2021) 16(1):50. doi: 10.1186/s13018-020-02169-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yuan J, Liu Y, Zhang Q, Ren Z, Li G, Tian R, et al. CircPRDM2 Contributes to Doxorubicin Resistance of Osteosarcoma by Elevating EZH2 via Sponging miR-760. Cancer Manag Res (2021) 13:4433–45. doi: 10.2147/CMAR.S295147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wei CY, Zhu MX, Lu NH, Liu JQ, Yang YW, Zhang Y, et al. Circular RNA Circ_0020710 Drives Tumor Progression and Immune Evasion by Regulating the miR-370-3p/CXCL12 Axis in Melanoma. Mol Cancer (2020) 19(1):84. doi: 10.1186/s12943-020-01191-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yi X, Chen W, Li C, Chen X, Lin Q, Lin S, et al. Circular RNA Circ_0004507 Contributes to Laryngeal Cancer Progression and Cisplatin Resistance by Sponging miR-873 to Upregulate Multidrug Resistance 1 and Multidrug Resistance Protein 1. Head Neck (2021) 43(3):928–41. doi: 10.1002/hed.26549 [DOI] [PubMed] [Google Scholar]
- 87. Feng B, Chen K, Zhang W, Zheng Q, He Y. Circpgam1 Enhances Autophagy Signaling During Laryngocarcinoma Drug Resistance by Regulating miR-376a. Biochem Biophys Res Commun (2021) 534:966–72. doi: 10.1016/j.bbrc.2020.10.063 [DOI] [PubMed] [Google Scholar]
- 88. Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, et al. CircPAN3 Contributes to Drug Resistance in Acute Myeloid Leukemia Through Regulation of Autophagy. Leuk Res (2019) 85:106198. doi: 10.1016/j.leukres.2019.106198 [DOI] [PubMed] [Google Scholar]
- 89. Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB, et al. CircPAN3 Mediates Drug Resistance in Acute Myeloid Leukemia Through the miR-153-5p/miR-183-5p-XIAP Axis. Exp Hematol (2019) 70:42–54.e3. doi: 10.1016/j.exphem.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 90. Liu F, Zhang J, Qin L, Yang Z, Xiong J, Zhang Y, et al. Circular RNA EIF6 (Hsa_circ_0060060) Sponges miR-144-3p to Promote the Cisplatin-Resistance of Human Thyroid Carcinoma Cells by Autophagy Regulation. Aging (Albany NY) (2018) 10(12):3806–20. doi: 10.18632/aging.101674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hua L, Huang L, Zhang X, Feng H, Shen B. Knockdown of Circular RNA CEP128 Suppresses Proliferation and Improves Cytotoxic Efficacy of Temozolomide in Glioma Cells by Regulating miR-145-5p. Neuroreport (2019) 30(18):1231–8. doi: 10.1097/WNR.0000000000001326 [DOI] [PubMed] [Google Scholar]
- 92. Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, et al. Exosome-Mediated Transfer of circRNA CircNFIX Enhances Temozolomide Resistance in Glioma. Cancer Lett (2020) 479:1–12. doi: 10.1016/j.canlet.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 93. Han C, Wang S, Wang H, Zhang J. Exosomal circ-HIPK3 Facilitates Tumor Progression and Temozolomide Resistance by Regulating miR-421/ZIC5 Axis in Glioma. Cancer Biother Radiopharm (2021) 36(7):537–48. doi: 10.1089/cbr.2019.3492 [DOI] [PubMed] [Google Scholar]
- 94. Yin H, Cui X. Knockdown of Circhipk3 Facilitates Temozolomide Sensitivity in Glioma by Regulating Cellular Behaviors Through miR-524-5p/KIF2A-Mediated PI3K/AKT Pathway. Cancer Biother Radiopharm (2021) 36(7):556–67. doi: 10.1089/cbr.2020.3575 [DOI] [PubMed] [Google Scholar]
- 95. Ding C, Yi X, Chen X, Wu Z, You H, Chen X, et al. Warburg Effect-Promoted Exosomal Circ_0072083 Releasing Up-Regulates NANGO Expression Through Multiple Pathways and Enhances Temozolomide Resistance in Glioma. J Exp Clin Cancer Res (2021) 40(1):164. doi: 10.1186/s13046-021-01942-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J, et al. EIF4A3-Induced Circular RNA ASAP1 Promotes Tumorigenesis and Temozolomide Resistance of Glioblastoma via NRAS/MEK1/ERK1-2 Signaling. Neuro Oncol (2021) 23(4):611–24. doi: 10.1093/neuonc/noaa214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Deng Y, Zhu H, Xiao L, Liu C, Meng X. Circ_0005198 Enhances Temozolomide Resistance of Glioma Cells Through miR-198/TRIM14 Axis. Aging (Albany NY) (2020) 13(2):2198–211. doi: 10.18632/aging.202234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gao F, Han J, Wang Y, Jia L, Luo W, Zeng Y. Circ_0109291 Promotes the Cisplatin Resistance of Oral Squamous Cell Carcinoma by Sponging miR-188-3p to Increase ABCB1 Expression. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3928 [DOI] [PubMed] [Google Scholar]
- 99. Qiu F, Qiao B, Zhang N, Fang Z, Feng L, Zhang S, et al. Blocking Circ-SCMH1 (Hsa_Circ_0011946) Suppresses Acquired DDP Resistance of Oral Squamous Cell Carcinoma (OSCC) Cells Both In Vitro and In Vivo by Sponging miR-338-3p and Regulating LIN28B. Cancer Cell Int (2021) 21(1):412. doi: 10.1186/s12935-021-02110-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol Ther Nucleic Acids (2019) 18:24–33. doi: 10.1016/j.omtn.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101. Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J, et al. Circular RNA Circtnpo3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by miR-1299/NEK2 Signaling Pathway. Mol Ther Nucleic Acids (2020) 21:780–91. doi: 10.1016/j.omtn.2020.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Guo M, Li S, Zhao X, Yuan Y, Zhang B, Guan Y, et al. Knockdown of Circular RNA Hsa_circ_0000714 Can Regulate RAB17 by Sponging miR-370-3p to Reduce Paclitaxel Resistance of Ovarian Cancer Through CDK6/RB Pathway. Onco Targets Ther (2020) 13:13211–24. doi: 10.2147/OTT.S285153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, et al. Circcelsr1 (Hsa_Circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol Ther Nucleic Acids (2020) 19:718–30. doi: 10.1016/j.omtn.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Luo Y, Gui R. Circulating Exosomal Circfoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J Gynecol Oncol (2020) 31(5):e75. doi: 10.3802/jgo.2020.31.e75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li M, Cai J, Han X, Ren Y. Downregulation of Circnrip1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the miR-211-5p/HOXC8 Axis. Cancer Manag Res (2020) 12:9159–71. doi: 10.2147/CMAR.S268872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cao Y, Xie X, Li M, Gao Y. CircHIPK2 Contributes to DDP Resistance and Malignant Behaviors of DDP-Resistant Ovarian Cancer Cells Both In Vitro and In Vivo Through Circhipk2/miR-338-3p/CHTOP ceRNA Pathway. Onco Targets Ther (2021) 14:3151–65. doi: 10.2147/OTT.S291823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wei S, Qi L, Wang L. Overexpression of Circ_CELSR1 Facilitates Paclitaxel Resistance of Ovarian Cancer by Regulating miR-149-5p/SIK2 Axis. Anticancer Drugs (2021) 32(5):496–507. doi: 10.1097/CAD.0000000000001058 [DOI] [PubMed] [Google Scholar]
- 108. Che H, Ding H, Jia X. Circ_0080145 Enhances Imatinib Resistance of Chronic Myeloid Leukemia by Regulating miR-326/PPFIA1 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3600 [DOI] [PubMed] [Google Scholar]
- 109. Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L, et al. Circ_0009910 Promotes Imatinib Resistance Through ULK1-Induced Autophagy by Sponging miR-34a-5p in Chronic Myeloid Leukemia. Life Sci (2020) 243:117255. doi: 10.1016/j.lfs.2020.117255 [DOI] [PubMed] [Google Scholar]
- 110. Xie F, Zhao N, Zhang H, Xie D. Circular RNA CircHIPK3 Promotes Gemcitabine Sensitivity in Bladder Cancer. J Cancer (2020) 11(7):1907–12. doi: 10.7150/jca.39722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Huang W, Lu Y, Wang F, Huang X, Yu Z. Circular RNA circRNA_103809 Accelerates Bladder Cancer Progression and Enhances Chemo-Resistance by Activation of miR-516a-5p/FBXL18 Axis. Cancer Manag Res (2020) 12:7561–8. doi: 10.2147/CMAR.S263083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Su Y, Yang W, Jiang N, Shi J, Chen L, Zhong G, et al. Hypoxia-Elevated Circelp3 Contributes to Bladder Cancer Progression and Cisplatin Resistance. Int J Biol Sci (2019) 15(2):441–52. doi: 10.7150/ijbs.26826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, et al. Downregulation of Hsa_Circ_0000285 Serves as a Prognostic Biomarker for Bladder Cancer and Is Involved in Cisplatin Resistance. Neoplasma (2019) 66(2):197–202. doi: 10.4149/neo_2018_180318N185 [DOI] [PubMed] [Google Scholar]
- 114. Zhang H, Xiao X, Wei W, Huang C, Wang M, Wang L, et al. CircLIFR Synergizes With MSH2 to Attenuate Chemoresistance via Mutsα/ATM-P73 Axis in Bladder Cancer. Mol Cancer (2021) 20(1):70. doi: 10.1186/s12943-021-01360-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, et al. Circ0008399 Interaction With WTAP Promotes Assembly and Activity of the M6a Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res (2021) 81(24):6142–56. doi: 10.1158/0008-5472.CAN-21-1518 [DOI] [PubMed] [Google Scholar]
- 116. Shen Z, Zhou L, Zhang C, Xu J. Reduction of Circular RNA Foxo3 Promotes Prostate Cancer Progression and Chemoresistance to Docetaxel. Cancer Lett (2020) 468:88–101. doi: 10.1016/j.canlet.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 117. Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, et al. Preclinical Study Using Circular RNA 17 and Micro RNA 181c-5p to Suppress the Enzalutamide-Resistant Prostate Cancer Progression. Cell Death Dis (2019) 10(2):37. doi: 10.1038/s41419-018-1048-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gao Y, Liu J, Huan J, Che F. Downregulation of Circular RNA Hsa_Circ_0000735 Boosts Prostate Cancer Sensitivity to Docetaxel via Sponging miR-7. Cancer Cell Int (2020) 20:334. doi: 10.1186/s12935-020-01421-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li S, Yu C, Zhang Y, Liu J, Jia Y, Sun F, et al. Circular RNA Cir-ITCH Is a Potential Therapeutic Target for the Treatment of Castration-Resistant Prostate Cancer. BioMed Res Int (2020) p:7586521. doi: 10.1155/2020/7586521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang H, Li M, Zhang J, Shen Y, Gui Q. Exosomal Circ-XIAP Promotes Docetaxel Resistance in Prostate Cancer by Regulating miR-1182/TPD52 Axis. Drug Des Devel Ther (2021) 15:1835–49. doi: 10.2147/DDDT.S300376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Li H, Li Q, He S. Hsa_circ_0025202 Suppresses Cell Tumorigenesis and Tamoxifen Resistance via miR-197-3p/HIPK3 Axis in Breast Cancer. World J Surg Oncol (2021) 19(1):39. doi: 10.1186/s12957-021-02149-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther (2019) 27(9):1638–52. doi: 10.1016/j.ymthe.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Liang Y, Song X, Li Y, Su P, Han D, Ma T, et al. Circkdm4c Suppresses Tumor Progression and Attenuates Doxorubicin Resistance by Regulating miR-548p/PBLD Axis in Breast Cancer. Oncogene (2019) 38(42):6850–66. doi: 10.1038/s41388-019-0926-z [DOI] [PubMed] [Google Scholar]
- 124. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNA−MTO1 Suppresses Breast Cancer Cell Viability and Reverses Monastrol Resistance Through Regulating the TRAF4/Eg5 Axis. Int J Oncol (2018) 53(4):1752–62. doi: 10.3892/ijo.2018.4485 [DOI] [PubMed] [Google Scholar]
- 125. Yang W, Gong P, Yang Y, Yang C, Yang B, Ren L, et al. Circ-ABCB10 Contributes to Paclitaxel Resistance in Breast Cancer Through Let-7a-5p/DUSP7 Axis. Cancer Manag Res (2020) 12:2327–37. doi: 10.2147/CMAR.S238513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou D, et al. Inhibition of Circular RNA CDR1as Increases Chemosensitivity of 5-FU-Resistant BC Cells Through Up-Regulating miR-7. J Cell Mol Med (2019) 23(5):3166–77. doi: 10.1111/jcmm.14171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zang H, Li Y, Zhang X, Huang G, et al. Circ-RNF111 Contributes to Paclitaxel Resistance in Breast Cancer by Elevating E2F3 Expression via miR-140-5p. Thorac Cancer (2020) 11(7):1891–903. doi: 10.1111/1759-7714.13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Cui Y, Fan J, Shi W, Zhou Z. Circ_0001667 Knockdown Blocks Cancer Progression and Attenuates Adriamycin Resistance by Depleting NCOA3 via Releasing miR-4458 in Breast Cancer. Drug Dev Res (2021) 83(1):75–87. doi: 10.1002/ddr.21845 [DOI] [PubMed] [Google Scholar]
- 129. Zhu M, Wang Y, Wang F, Li L, Qiu X. CircFBXL5 Promotes the 5-FU Resistance of Breast Cancer via Modulating miR-216b/HMGA2 Axis. Cancer Cell Int (2021) 21(1):384. doi: 10.1186/s12935-021-02088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yao Y, Li X, Cheng L, Wu X, Wu B. Circular RNA FAT Atypical Cadherin 1 (Circfat1)/microRNA-525-5p/Spindle and Kinetochore-Associated Complex Subunit 1 (SKA1) Axis Regulates Oxaliplatin Resistance in Breast Cancer by Activating the Notch and Wnt Signaling Pathway. Bioengineered (2021) 12(1):4032–43. doi: 10.1080/21655979.2021.1951929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hao J, Du X, Lv F, Shi Q. Knockdown of Circ_0006528 Suppresses Cell Proliferation, Migration, Invasion, and Adriamycin Chemoresistance via Regulating the miR-1236-3p/CHD4 Axis in Breast Cancer. J Surg Res (2021) 260:104–15. doi: 10.1016/j.jss.2020.10.031 [DOI] [PubMed] [Google Scholar]
- 132. Wu X, Ren Y, Yao R, Zhou L, Fan R. Circular RNA Circ-MMP11 Contributes to Lapatinib Resistance of Breast Cancer Cells by Regulating the miR-153-3p/ANLN Axis. Front Oncol (2021) 11:639961. doi: 10.3389/fonc.2021.639961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Xie H, Zheng R. Circ_0085495 Knockdown Reduces Adriamycin Resistance in Breast Cancer Through miR-873-5p/Integrin β1 Axis. Anticancer Drugs (2021) 33(1):e166–77. doi: 10.1097/CAD.0000000000001174 [DOI] [PubMed] [Google Scholar]
- 134. Huang L, Ma J, Cui M. Circular RNA Hsa_Circ_0001598 Promotes Programmed Death-Ligand-1-Mediated Immune Escape and Trastuzumab Resistance via Sponging miR-1184 in Breast Cancer Cells. Immunol Res (2021) 69(6):558–67. doi: 10.1007/s12026-021-09237-w [DOI] [PubMed] [Google Scholar]
- 135. Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W, et al. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med Sci Monit Int Med J Exp Clin Res (2020) 26:e922253. doi: 10.12659/MSM.922253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, et al. CircUBE2D2 (Hsa_Circ_0005728) Promotes Cell Proliferation, Metastasis and Chemoresistance in Triple-Negative Breast Cancer by Regulating miR-512-3p/CDCA3 Axis. Cancer Cell Int (2020) 20:454. doi: 10.1186/s12935-020-01547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wang L, Zhou Y, Jiang L, Lu L, Dai T, Li A, et al. CircWAC Induces Chemotherapeutic Resistance in Triple-Negative Breast Cancer by Targeting miR-142, Upregulating WWP1 and Activating the PI3K/AKT Pathway. Mol Cancer (2021) 20(1):43. doi: 10.1186/s12943-021-01332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zheng SR, Huang QD, Zheng ZH, Zhang ZT, Guo GL. Circgfra1 Affects the Sensitivity of Triple-Negative Breast Cancer Cells to Paclitaxel via the miR-361-5p/TLR4 Pathway. J Biochem (2021) 169(5):601–11. doi: 10.1093/jb/mvaa148 [DOI] [PubMed] [Google Scholar]
- 139. Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 Sponges hsa-miR-335 to Promote the Gemcitabine-Resistance of Human Renal Cancer Cells by Autophagy Regulation. Biochem Biophys Res Commun (2019) 519(1):172–8. doi: 10.1016/j.bbrc.2019.08.093 [DOI] [PubMed] [Google Scholar]
- 140. Huang KB, Pan YH, Shu GN, Yao HH, Liu X, Zhou M, et al. Circular RNA Circsnx6 Promotes Sunitinib Resistance in Renal Cell Carcinoma Through the miR-1184/GPCPD1/ Lysophosphatidic Acid Axis. Cancer Lett (2021) 523:121–34. doi: 10.1016/j.canlet.2021.10.003 [DOI] [PubMed] [Google Scholar]
- 141. Zou FW, Yang SZ, Li WY, Liu CY, Liu XH, Hu CH, et al. circRNA_001275 Upregulates Wnt7a Expression by Competitively Sponging Mir−370−3p to Promote Cisplatin Resistance in Esophageal Cancer. Int J Oncol (2020) 57(1):151–60. doi: 10.3892/ijo.2020.5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Qu F, Wang L, Wang C, Yu L, Zhao K, Zhong H, et al. Circular RNA Circ_0006168 Enhances Taxol Resistance in Esophageal Squamous Cell Carcinoma by Regulating miR-194-5p/JMJD1C Axis. Cancer Cell Int (2021) 21(1):273. doi: 10.1186/s12935-021-01984-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhu H, Du F, Cao C. Restoration of Circpsmc3 Sensitizes Gefitinib-Resistant Esophageal Squamous Cell Carcinoma Cells to Gefitinib by Regulating miR-10a-5p/PTEN Axis. Cell Biol Int (2021) 45(1):107–16. doi: 10.1002/cbin.11473 [DOI] [PubMed] [Google Scholar]
- 144. Zang R, Qiu X, Song Y, Wang Y. Exosomes Mediated Transfer of Circ_0000337 Contributes to Cisplatin (CDDP) Resistance of Esophageal Cancer by Regulating JAK2 via miR-377-3p. Front Cell Dev Biol (2021) 9:673237. doi: 10.3389/fcell.2021.673237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhang Z, Yu X, Zhou B, Zhang J, Chang J. Circular RNA Circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. BioMed Res Int (2020) 2020:5103272. doi: 10.1155/2020/5103272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Deng P, Sun M, Zhao WY, Hou B, Li K, Zhang T, et al. Circular RNA circVAPA Promotes Chemotherapy Drug Resistance in Gastric Cancer Progression by Regulating miR-125b-5p/STAT3 Axis. World J Gastroenterol (2021) 27(6):487–500. doi: 10.3748/wjg.v27.i6.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA Circ_0032821 Contributes to Oxaliplatin (OXA) Resistance of Gastric Cancer Cells by Regulating SOX9 via miR-515-5p. Biotechnol Lett (2021) 43(2):339–51. doi: 10.1007/s10529-020-03036-3 [DOI] [PubMed] [Google Scholar]
- 148. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. Circcul2 Regulates Gastric Cancer Malignant Transformation and Cisplatin Resistance by Modulating Autophagy Activation via miR-142-3p/ROCK2. Mol Cancer (2020) 19(1):156. doi: 10.1186/s12943-020-01270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Huang XX, Zhang Q, Hu H, Jin Y, Zeng AL, Xia YB, et al. A Novel Circular RNA Circfn1 Enhances Cisplatin Resistance in Gastric Cancer via Sponging miR-182-5p. J Cell Biochem (2020). doi: 10.1002/jcb.29641 [DOI] [PubMed] [Google Scholar]
- 150. Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng A, et al. CircRNACCDC66 Regulates Cisplatin Resistance in Gastric Cancer via the miR-618/BCL2 Axis. Biochem Biophys Res Commun (2020) 526(3):713–20. doi: 10.1016/j.bbrc.2020.03.156 [DOI] [PubMed] [Google Scholar]
- 151. Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 Upregulates PIK3R1 to Enhance Cisplatin Resistance in Gastric Cancer via miR-198 Suppression. Mol Cancer (2019) 18(1):71. doi: 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 Inhibits Cisplatin Resistance in Gastric Cancer by miR-99a-5p-Mediated Induction of MTMR3 Expression. J Exp Clin Cancer Res (2020) 39(1):246. doi: 10.1186/s13046-020-01758-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Liu YY, Zhang LY, Du WZ. Circular RNA Circ-PVT1 Contributes to Paclitaxel Resistance of Gastric Cancer Cells Through the Regulation of ZEB1 Expression by Sponging miR-124-3p. Biosci Rep (2019) 39(12):BSR20193045. doi: 10.1042/BSR20193045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Xu G, Li M, Wu J, Qin C, Tao Y, He H, et al. Circular RNA Circnrip1 Sponges microRNA-138-5p to Maintain Hypoxia-Induced Resistance to 5-Fluorouracil Through HIF-1α-Dependent Glucose Metabolism in Gastric Carcinoma. Cancer Manag Res (2020) 12:2789–802. doi: 10.2147/CMAR.S246272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sun Y, Ma J, Lin J, Sun D, Song P, Shi L, et al. Circular RNA Circ_ASAP2 Regulates Drug Sensitivity and Functional Behaviors of Cisplatin-Resistant Gastric Cancer Cells by the miR-330-3p/NT5E Axis. Anticancer Drugs (2021) 32(9):950–61. doi: 10.1097/CAD.0000000000001087 [DOI] [PubMed] [Google Scholar]
- 156. Liu Y, Xu J, Jiang M, Ni L, Ling Y. CircRNA DONSON Contributes to Cisplatin Resistance in Gastric Cancer Cells by Regulating miR-802/BMI1 Axis. Cancer Cell Int (2020) 20:261. doi: 10.1186/s12935-020-01358-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Xu QY, Xie MJ, Huang J, Wang ZW. Effect of Circ MTHFD2 on Resistance to Pemetrexed in Gastric Cancer Through Regulating Expression of miR-124. Eur Rev Med Pharmacol Sci (2019) 23(23):10290–9. doi: 10.26355/eurrev_201912_19667 [DOI] [PubMed] [Google Scholar]
- 158. Wu Q, Wang H, Liu L, Zhu K, Yu W, Guo J, et al. Hsa_circ_0001546 Acts as a miRNA-421 Sponge to Inhibit the Chemoresistance of Gastric Cancer Cells via ATM/Chk2/p53-Dependent Pathway. Biochem Biophys Res Commun (2020) 521(2):303–9. doi: 10.1016/j.bbrc.2019.10.117 [DOI] [PubMed] [Google Scholar]
- 159. Lu Y, Li L, Li L, Wu G, Liu G. Circular RNA Circhectd1 Prevents Diosbulbin-B-Sensitivity via miR-137/PBX3 Axis in Gastric Cancer. Cancer Cell Int (2021) 21(1):264. doi: 10.1186/s12935-021-01957-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gao H, Xu J, Qiao F, Xue L. Depletion of Hsa_Circ_0000144 Suppresses Oxaliplatin Resistance of Gastric Cancer Cells by Regulating miR-502-5p/ADAM9 Axis. Onco Targets Ther (2021) 14:2773–87. doi: 10.2147/OTT.S281238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Liu S, Wu M, Peng M. Circ_0000260 Regulates the Development and Deterioration of Gastric Adenocarcinoma With Cisplatin Resistance by Upregulating MMP11 via Targeting MiR-129-5p. Cancer Manag Res (2020) 12:10505–19. doi: 10.2147/CMAR.S272324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wang X, Zhang Y, Li W, Liu X. Knockdown of Cir_RNA PVT1 Elevates Gastric Cancer Cisplatin Sensitivity via Sponging miR-152-3p. J Surg Res (2021) 261:185–95. doi: 10.1016/j.jss.2020.12.013 [DOI] [PubMed] [Google Scholar]
- 163. Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis Via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother Radiopharm (2021) 36(4):347–59. doi: 10.1089/cbr.2020.3578 [DOI] [PubMed] [Google Scholar]
- 164. Xia Y, Lv J, Jiang T, Li B, Li Y, He Z, et al. CircFAM73A Promotes the Cancer Stem Cell-Like Properties of Gastric Cancer Through the miR-490-3p/HMGA2 Positive Feedback Loop and HNRNPK-Mediated β-Catenin Stabilization. J Exp Clin Cancer Res (2021) 40(1):103. doi: 10.1186/s13046-021-01896-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X, et al. CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Sponging miR-330-5p and Targets Rassf1. Cancer Manag Res (2020) 12:921–9. doi: 10.2147/CMAR.S239326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Xu H, Chen R, Shen Q, Yang D, Peng H, Tong J, et al. Overexpression of Circular RNA Circ_0013587 Reverses Erlotinib Resistance in Pancreatic Cancer Cells Through Regulating the miR-1227/E-Cadherin Pathway. Front Oncol (2021) 11:754146. doi: 10.3389/fonc.2021.754146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, et al. Exosomal Circ_0030167 Derived From BM-MSCs Inhibits the Invasion, Migration, Proliferation and Stemness of Pancreatic Cancer Cells by Sponging miR-338-5p and Targeting the Wif1/Wnt8/β-Catenin Axis. Cancer Lett (2021) 512:38–50. doi: 10.1016/j.canlet.2021.04.030 [DOI] [PubMed] [Google Scholar]
- 168. Gong J, Luk F, Jaiswal R, George AM, Grau GE, Bebawy M, et al. Microparticle Drug Sequestration Provides a Parallel Pathway in the Acquisition of Cancer Drug Resistance. Eur J Pharmacol (2013) 721(1-3):116–25. doi: 10.1016/j.ejphar.2013.09.044 [DOI] [PubMed] [Google Scholar]
- 169. Joyce H, McCann A, Clynes M, Larkin A. Influence of Multidrug Resistance and Drug Transport Proteins on Chemotherapy Drug Metabolism. Expert Opin Drug Metab Toxicol (2015) 11(5):795–809. doi: 10.1517/17425255.2015.1028356 [DOI] [PubMed] [Google Scholar]
- 170. Planells-Cases R, Lutter D, Guyader C, Gerhards NM, Ullrich F, Elger DA, et al. Subunit Composition of VRAC Channels Determines Substrate Specificity and Cellular Resistance to Pt-Based Anti-Cancer Drugs. EMBO J (2015) 34(24):2993–3008. doi: 10.15252/embj.201592409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Saxena M, Stephens MA, Pathak H, Rangarajan A. Transcription Factors That Mediate Epithelial-Mesenchymal Transition Lead to Multidrug Resistance by Upregulating ABC Transporters. Cell Death Dis (2011) 2(7):e179. doi: 10.1038/cddis.2011.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Li X, Zhang T. Withdrawn: Circular RNA PTGR1 Regulates 5-FU Resistance and Development of Hepatocellular Carcinoma Cells by Modulating miR-129-5p/ABCC1 Axis. Cell Biol Int (2021) 45(11):2391. doi: 10.1002/cbin.11635 [DOI] [PubMed] [Google Scholar]
- 173. Zhao X, Zhong Q, Cheng X, Wang S, Wu R, Leng X, et al. miR-449c-5p Availability Is Antagonized by Circ-NOTCH1 for MYC-Induced NOTCH1 Upregulation as Well as Tumor Metastasis and Stemness in Gastric Cancer. J Cell Biochem (2020) 121(10):4052–63. doi: 10.1002/jcb.29575 [DOI] [PubMed] [Google Scholar]
- 174. Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang Y, et al. circPTN Sponges miR-145-5p/miR-330-5p to Promote Proliferation and Stemness in Glioma. J Exp Clin Cancer Res (2019) 38(1):398. doi: 10.1186/s13046-019-1376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The Epithelial-Mesenchymal Transition Generates Cells With Properties of Stem Cells. Cell (2008) 133(4):704–15. doi: 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Milanovic M, Fan DNY, Belenki D, Däbritz JHM, Zhao Z, Yu Y, et al. Senescence-Associated Reprogramming Promotes Cancer Stemness. Nature (2018) 553(7686):96–100. doi: 10.1038/nature25167 [DOI] [PubMed] [Google Scholar]
- 177. Shibue T, Weinberg RA. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat Rev Clin Oncol (2017) 14(10):611–29. doi: 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Dean M, Fojo T, Bates S. Tumour Stem Cells and Drug Resistance. Nat Rev Cancer (2005) 5(4):275–84. doi: 10.1038/nrc1590 [DOI] [PubMed] [Google Scholar]
- 179. Schulz A, Meyer F, Dubrovska A, Borgmann K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers (Basel) (2019) 11(6):862. doi: 10.3390/cancers11060862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Joseph NA, Chiou SH, Lung Z, Yang CL, Lin TY, Chang HW, et al. The Role of HGF-MET Pathway and CCDC66 cirRNA Expression in EGFR Resistance and Epithelial-to-Mesenchymal Transition of Lung Adenocarcinoma Cells. J Hematol Oncol (2018) 11(1):74. doi: 10.1186/s13045-018-0557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Müller S, Ledl A, Schmidt D. SUMO: A Regulator of Gene Expression and Genome Integrity. Oncogene (2004) 23(11):1998–2008. doi: 10.1038/sj.onc.1207415 [DOI] [PubMed] [Google Scholar]
- 182. Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, Chelbi-Alix MK, et al. Small Ubiquitin-Like Modifier Alters IFN Response. J Immunol (2015) 195(5):2312–24. doi: 10.4049/jimmunol.1500035 [DOI] [PubMed] [Google Scholar]
- 183. Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers (Basel) (2020) 12(3):697. doi: 10.3390/cancers12030697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K, et al. Biogenesis, Functions and Clinical Significance of circRNAs in Gastric Cancer. Mol Cancer (2019) 18(1):136. doi: 10.1186/s12943-019-1069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 186. Elbarbary RA, Miyoshi K, Myers JR, Du P, Ashton JM, Tian B, et al. Tudor-SN-Mediated Endonucleolytic Decay of Human Cell microRNAs Promotes G(1)/S Phase Transition. Science (2017) 356(6340):859–62. doi: 10.1126/science.aai9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Chen Z, Lu T, Huang L, Wang Z, Yan Z, Guan Y, et al. Circular RNA Cia-MAF Drives Self-Renewal and Metastasis of Liver Tumor-Initiating Cells via Transcription Factor MAFF. J Clin Invest (2021) 131(19):e148020. doi: 10.1172/JCI148020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering Circular RNA for Potent and Stable Translation in Eukaryotic Cells. Nat Commun (2018) 9(1):2629. doi: 10.1038/s41467-018-05096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive Translation of Circular RNAs Driven by N(6)-Methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 Inhibits Gastric Cancer Progression by Encoding a Novel DIDO1-529aa Protein and Regulating PRDX2 Protein Stability. Mol Cancer (2021) 20(1):101. doi: 10.1186/s12943-021-01390-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Mazumder A, Lee JY, Talhi O, Cerella C, Chateauvieux S, Gaigneaux A, et al. Hydroxycoumarin OT-55 Kills CML Cells Alone or in Synergy With Imatinib or Synribo: Involvement of ER Stress and DAMP Release. Cancer Lett (2018) 438:197–218. doi: 10.1016/j.canlet.2018.07.041 [DOI] [PubMed] [Google Scholar]
- 192. Wu J, Chen S, Liu H, Zhang Z, Ni Z, Chen J, et al. Tunicamycin Specifically Aggravates ER Stress and Overcomes Chemoresistance in Multidrug-Resistant Gastric Cancer Cells by Inhibiting N-Glycosylation. J Exp Clin Cancer Res (2018) 37(1):272. doi: 10.1186/s13046-018-0935-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Xu Z, Wang H, Wei S, Wang Z, Ji G. Inhibition of ER Stress-Related IRE1α/CREB/NLRP1 Pathway Promotes the Apoptosis of Human Chronic Myelogenous Leukemia Cell. Mol Immunol (2018) 101:377–85. doi: 10.1016/j.molimm.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 194. Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER Stress-Induced Autophagy Overcomes BRAF Inhibitor Resistance in Melanoma. J Clin Invest (2014) 124(3):1406–17. doi: 10.1172/JCI70454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Bristol ML, Di X, Beckman MJ, Wilson EN, Henderson SC, Maiti A, et al. Dual Functions of Autophagy in the Response of Breast Tumor Cells to Radiation: Cytoprotective Autophagy With Radiation Alone and Cytotoxic Autophagy in Radiosensitization by Vitamin D 3. Autophagy (2012) 8(5):739–53. doi: 10.4161/auto.19313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Sheng Y, Sun B, Xie X, Li N, Dong D. DMH1 (4-[6-(4-Isopropoxyphenyl)Pyrazolo[1,5-a]Pyrimidin-3-Yl]Quinoline) Inhibits Chemotherapeutic Drug-Induced Autophagy. Acta Pharm Sin B (2015) 5(4):330–6. doi: 10.1016/j.apsb.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Smith AG, Macleod KF. Autophagy, Cancer Stem Cells and Drug Resistance. J Pathol (2019) 247(5):708–18. doi: 10.1002/path.5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Tian Y, Yang B, Qiu W, Hao Y, Zhang Z, Yang B, et al. ER-Residential Nogo-B Accelerates NAFLD-Associated HCC Mediated by Metabolic Reprogramming of oxLDL Lipophagy. Nat Commun (2019) 10(1):3391. doi: 10.1038/s41467-019-11274-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Walker DM, Urbé S, Dove SK, Tenza D, Raposo G, Clague MJ, et al. Characterization of MTMR3. An Inositol Lipid 3-Phosphatase With Novel Substrate Specificity. Curr Biol (2001) 11(20):1600–5. doi: 10.1016/s0960-9822(01)00501-2 [DOI] [PubMed] [Google Scholar]
- 200. Li H, Xu W, Xia Z, Liu W, Pan G, Ding J, et al. Hsa_circ_0000199 Facilitates Chemo-Tolerance of Triple-Negative Breast Cancer by Interfering With miR-206/613-Led PI3K/Akt/mTOR Signaling. Aging (Albany NY) (2021) 13(3):4522–51. doi: 10.18632/aging.202415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 202. Jiao S, Subudhi SK, Aparicio A, Ge Z, Guan B, Miura Y, et al. Differences in Tumor Microenvironment Dictate T Helper Lineage Polarization and Response to Immune Checkpoint Therapy. Cell (2019) 179(5):1177–90.e13. doi: 10.1016/j.cell.2019.10.029 [DOI] [PubMed] [Google Scholar]
- 203. Ke B, Wei T, Huang Y, Gong Y, Wu G, Liu J, et al. Interleukin-7 Resensitizes Non-Small-Cell Lung Cancer to Cisplatin via Inhibition of ABCG2. Mediators Inflamm (2019) 2019:7241418. doi: 10.1155/2019/7241418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Zheng X, Qian Y, Fu B, Jiao D, Jiang Y, Chen P, et al. Mitochondrial Fragmentation Limits NK Cell-Based Tumor Immunosurveillance. Nat Immunol (2019) 20(12):1656–67. doi: 10.1038/s41590-019-0511-1 [DOI] [PubMed] [Google Scholar]
- 205. Bagchi S, Yuan R, Engleman EG. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol (2021) 16:223–49. doi: 10.1146/annurev-pathol-042020-042741 [DOI] [PubMed] [Google Scholar]
- 206. Wang Z, Wu X. Study and Analysis of Antitumor Resistance Mechanism of PD1/PD-L1 Immune Checkpoint Blocker. Cancer Med (2020) 9(21):8086–121. doi: 10.1002/cam4.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Zhao R, Ni J, Lu S, Jiang S, You L, Liu H, et al. CircUBAP2-Mediated Competing Endogenous RNA Network Modulates Tumorigenesis in Pancreatic Adenocarcinoma. Aging (Albany NY) (2019) 11(19):8484–501. doi: 10.18632/aging.102334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Huang Z, Yao F, Liu J, Xu J, Guo Y, Su R, et al. Up-Regulation of circRNA-0003528 Promotes Mycobacterium Tuberculosis Associated Macrophage Polarization via Down-Regulating miR-224-5p, miR-324-5p and miR-488-5p and Up-Regulating CTLA4. Aging (Albany NY) (2020) 12(24):25658–72. doi: 10.18632/aging.104175 [DOI] [PMC free article] [PubMed] [Google Scholar]