Abstract
Significance:
Mitochondria play a critical role in the physiology of the heart by controlling cardiac metabolism, function, and remodeling. Accumulation of fragmented and damaged mitochondria is a hallmark of cardiac diseases.
Recent Advances:
Disruption of quality control systems that maintain mitochondrial number, size, and shape through fission/fusion balance and mitophagy results in dysfunctional mitochondria, defective mitochondrial segregation, impaired cardiac bioenergetics, and excessive oxidative stress.
Critical Issues:
Pharmacological tools that improve the cardiac pool of healthy mitochondria through inhibition of excessive mitochondrial fission, boosting mitochondrial fusion, or increasing the clearance of damaged mitochondria have emerged as promising approaches to improve the prognosis of heart diseases.
Future Directions:
There is a reasonable amount of preclinical evidence supporting the effectiveness of molecules targeting mitochondrial fission and fusion to treat cardiac diseases. The current and future challenges are turning these lead molecules into treatments. Clinical studies focusing on acute (i.e., myocardial infarction) and chronic (i.e., heart failure) cardiac diseases are needed to validate the effectiveness of such strategies in improving mitochondrial morphology, metabolism, and cardiac function. Antioxid. Redox Signal. 36, 844–863.
Keywords: mitochondrial dynamics, metabolism, bioenergetic dysfunction, oxidative stress, cardiovascular diseases, therapy
Introduction
Cardiac diseases are the major cause of morbidity and mortality worldwide, resulting in the death of 17.9 million people every year and costing ∼$300 billion in the United States (202). The pathophysiology of cardiac diseases is multifactorial and includes genetic (i.e., DNA mutations) and/or environmental factors (i.e., unhealthy lifestyle) (33, 185). More recently, mitochondrial dysfunction has been highlighted as a hallmark of both establishment and progression of cardiac diseases (32, 64, 105, 120).
Mitochondria represent ∼35% of the cardiomyocyte volume, categorized as subsarcolemmal, perinuclear, and intrafibrillar mitochondria. These highly dynamic organelles control many different aspects of the cell, including, but not limited to, ATP production, redox signaling, ion homeostasis, and gene expression (25, 56, 105). Moreover, mitochondrial metabolism regulates the majority of intracellular events in cardiomyocytes, ranging from redox signaling to posttranslational modification of proteins (28, 150).
Under stress conditions, mitochondria play a unique role in triggering and propagating damage throughout the cardiomyocytes (31, 105). As expected, disruption of mitochondrial metabolism and/or morphology negatively affects the viability and survival of cardiomyocytes under physiological and pathological conditions (32, 64). Indeed, genetic defects in mitochondrial DNA (mtDNA) cause inherited mitochondrial diseases and cardiomyopathies (83, 104, 195). Therefore, the tight control of mitochondrial number, shape, and metabolism becomes a promising strategy to tackle cardiac diseases.
This review focuses on the intrinsic mechanisms that control mitochondrial connectiveness and quality control as well as their effect on mitochondrial bioenergetics, oxidative stress, and mtDNA segregation in the heart. We also describe the contribution of such processes to the pathophysiology of cardiac diseases and highlight the main pharmacological approaches targeting mitochondrial dynamics to treat heart diseases.
Mitochondrial Bioenergetics
Mitochondria are double membrane organelles containing over 1300 proteins (69). The outer mitochondrial membrane is more permeable than the inner mitochondrial membrane, due to the existence of large-pore channels termed porins or voltage-dependent anion-selective channels, and molecules with molecular weight less than about 5000 Da are allowed free passage (19, 132). In contrast, the inner mitochondrial membrane is a tightly controlled diffusion barrier where molecules can only cross through specific membrane transport proteins, allowing for two discrete regions to form within the mitochondria (18, 166, 184). Furthermore, the inner mitochondrial membrane folds into the intramitochondrial space, termed mitochondrial cristae, which greatly increases its surface area and allows for the mitochondria to maximize ATP production.
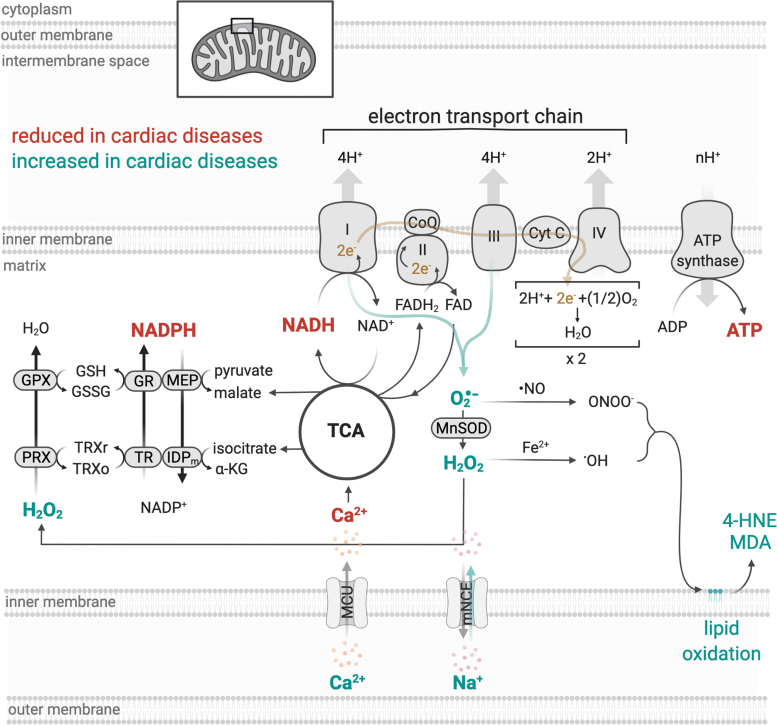
Mitochondrial ATP production results from two closely coordinated metabolic processes, the tricarboxylic acid (TCA) cycle or Krebs cycle, and oxidative phosphorylation (OXPHOS) (23, 103). The TCA cycle involves a series of eight enzymatic steps located in the mitochondrial matrix that oxidizes acetyl coenzyme A from the catabolism of metabolites (e.g., fatty acids, glucose, and proteins) to generate the reduced electron carriers nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2), which transfer electrons to the electron transport chain (ETC) (Fig. 1) (23, 180).
FIG. 1.
Proposed model for impaired mitochondrial bioenergetics and redox balance in cardiac diseases based on previous studies (32, 73, 164), which focused on the interplay between NAD(P)H/NAD(P)+ redox state and mitochondrial ROS generation. 4-HNE, 4-hydroxynonenal; α-KG, α-ketoglutarate; GPX, glutathione peroxidase; GR, glutathione reductase; GSH/GSSG, reduced/oxidized glutathione; H2O2, hydrogen peroxide; IDPm, mitochondrial NADP+-dependent isocitrate dehydrogenase; MCU, mitochondrial Ca2+ uniporter; MDA, malondialdehyde; MEP, malic enzyme; mNCE, Na+/Ca2+ exchanger; Mn-SOD, Mn2+-dependent superoxide dismutase; O2−, anion superoxide; OH, hydroxyl radical; ONOO−, peroxynitrite; PRX, peroxiredoxin; ROS, reactive oxygen species; TCA, tricarboxylic acid; TR, thioredoxin reductase; TRXr/o, reduced/oxidized thioredoxin. Created with BioRender.com
OXPHOS is a process that requires the orchestrated action of four ETC enzyme complexes, complex I–IV, embedded within the mitochondrial cristae of the inner mitochondrial membrane (Fig. 1) (71). Each of these complexes comprised several protein subunits that are associated with a variety of prosthetic clusters with redox potential (103). Complexes I, III, and IV form a “supercomplex,” physically interacting with one another, while complex II represents a point of interaction between TCA cycle and ETC, which occurs through succinate dehydrogenase (3).
During OXPHOS, NADH and FADH2 are oxidized, and their electrons are transferred to complex I (NADH-ubiquinone oxidoreductase) or complex II (succinate-ubiquinone oxidoreductase), respectively, and are transported to complex III (ubiquinol-cytochrome c oxidoreductase) via coenzyme Q. Complex III transfers the electrons from coenzyme Q to the mobile electron carrier, cytochrome c, which delivers electrons to complex IV (cytochrome c oxidase), the terminal complex of the ETC. Once in complex IV, the electrons are transferred from reduced cytochrome c to molecular oxygen, the final electron acceptor, to generate water (103). Recently, Spinelli et al. found that fumarate acts as a terminal electron acceptor when oxygen is not available (hypoxia) (181).
Concomitant with the transfer of electrons between complexes I, III, and IV, protons are pumped from the mitochondrial matrix across the inner mitochondrial membrane, to the intermembrane space using energy from electrons to create an electrochemical proton gradient (Fig. 1). This gradient determines the polarization of the inner mitochondrial membrane (mitochondrial membrane potential), which can be dissipated by the flow of these protons through the ATP synthase or complex V (FoF1-ATP synthase) (2, 134). The proton flow is coupled to the ATP synthesis and leads to the condensation of ADP and inorganic phosphate (Pi) into ATP, which, in turn, is the molecular currency of energy transfer in a cell (2, 81).
The heart has a high demand for ATP synthesis and elevated oxygen uptake rate, therefore relying mainly on mitochondrial OXPHOS to maintain its biochemical and mechanical properties (32, 34). The production of ATP by mitochondria and the use of this energy by cardiomyocytes are highly dynamic. Without oxygen, the mitochondrial-derived ATP supply is only sufficient to power the ATPase required for contraction (e.g., myosin) and relaxation (e.g., sarcoplasmic reticulum calcium ATPase, SERCA) for a few seconds (124, 135, 160). In healthy hearts, fatty acid catabolism, via β-oxidation, is the main substrate utilized for ATP synthesis (up to 70%), which can vary depending on physiological conditions [for review see Bertero and Maack (21)].
In pathological conditions, such as ischemia/reperfusion injury (196) and heart failure (191), glucose becomes the predominant substrate for ATP production. During energy depletion state, the activation of AMP-activated protein kinase (AMPK) stimulates translocation of glucose transporters (mainly GLUT-4) from intracellular vesicles to the sarcolemmal membrane, therefore increasing the rate of glucose uptake (167). In addition, the shift in cardiomyocyte ATP substrate dependence involves the suppression of peroxisome proliferator-activated receptor (PPAR)-α (PPARα) signaling (16) and the activation of both hypoxia-inducible factor 1α (HIF1α) and PPAR, therefore reducing heart exposure to fatty acids (70, 114). This increases glucose uptake and may contribute to cardiac dysfunction and cell death (114).
Moreover, impaired cardiac mitochondrial bioenergetics is associated with defective fatty acid oxidation (120), reduction of mitochondrial supercomplexes (164), and elevated oxidative stress (32, 170), which further leads to cardiomyocyte degeneration and establishment/progression of cardiac diseases in both humans and preclinical models.
In cardiac myocytes, mitochondria represent a major source of reactive oxygen species (ROS). Under physiological conditions, ∼0.12%–2% of the available oxygen is converted into ROS (24, 122, 182), which plays a key role in the cardiac redox signaling and affects several intracellular processes such as protein turnover (26, 29), calcium handling (107, 198), and contractility properties (148). However, under conditions of stress, electrons can prematurely react with oxygen in the ETC and generate excessive ROS, which can interact with different macromolecules (i.e., DNA, lipids, and proteins) (Fig. 1) (24, 78, 207).
Under this scenario, mitochondrial dysfunction-induced oxidative stress can disrupt other intracellular systems and propagate oxidative damage throughout cells, organs, and systems, therefore contributing to the establishment and propagation of cardiac diseases [for review see Bozi et al. (25), Burgoyne et al. (30), and Tsutsui et al. (190)]. To avoid the deleterious effect of excessive oxidative stress, mitochondria contain a variety of enzymatic and nonenzymatic components of the antioxidant defense systems capable of scavenging reactive molecules to nontoxic species, such as the glutathione and thioredoxin systems (Fig. 1) (9) [for details see review Figueira et al. (66)]. Pharmacological inhibition (183) or genetic disruption (85) of thioredoxin system compromises mitochondrial redox balance and cardiac physiology.
Mitochondrial Connectiveness
The mitochondrial network is constantly undergoing fusion/fission cycles, which together control the transport, size, morphology, and turnover of mitochondria (Fig. 2) (37). Collectively, these processes are referred to as mitochondrial dynamics, which directly affect cell viability, proliferation, and ATP production, especially in tissues with high metabolic demand, such as the heart (34, 42, 64). Moreover, mitochondria continuously exchange metabolites, solutes, proteins, DNA, and membranes through mitochondrial fission and fusion processes (169). Therefore, disruption of the mitochondrial fission/fusion balance commonly results in morphological and functional alterations of the mitochondria, which are tightly associated with several chronic disorders, including cardiac diseases (Fig. 3) (31, 177).
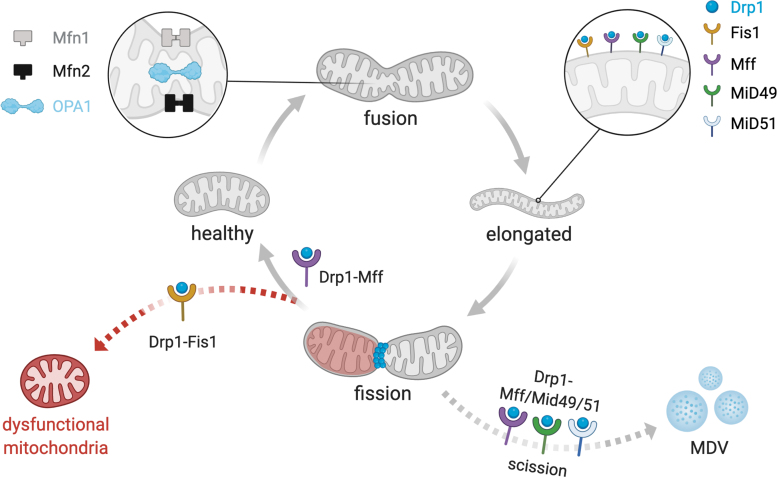
FIG. 2.
Proposed model for mitochondrial fission and fusion. The mitochondrial fission is promoted by Drp1, a cytosolic GTPase that translocates to the mitochondria upon activation and binds to specific proteins located at the outer mitochondrial membrane, including mitochondrial Fis1, Mff, MiD49, and MiD51 (37). Drp1-Mff and Drp1-Fis1 interaction induces two distinct mitochondrial fission signatures and plays a critical role in the mitochondrial physiology and pathology (106). In addition, Mff, MiD49, and MiD51 are essential for Drp1-dependent mitochondrial scission and release of MDV (109). Mitochondrial fusion is coordinated by mitofusins (Mfn1 and Mfn2), located at the outer mitochondrial membrane, and the Opa1, located at the inner mitochondrial membrane. Mitofusins contain a GTPase domain involved in the hydrolysis of GTP, which is required for the mitofusin oligomerization and fusion of the outer mitochondrial membranes of adjacent mitochondria. Drp1, dynamin-related protein 1; Fis1, fission 1 protein; GTPase, guanosine triphosphatase; MDV, mitochondrial-derived vesicles; Mff, mitochondrial fission factor; Mfn1, mitofusin 1; Mfn2, mitofusin 2; MiD49, mitochondrial dynamics protein of 49 kDa; MiD51, mitochondrial dynamics protein of 51 kDa; Opa1, optic atrophy factor 1.
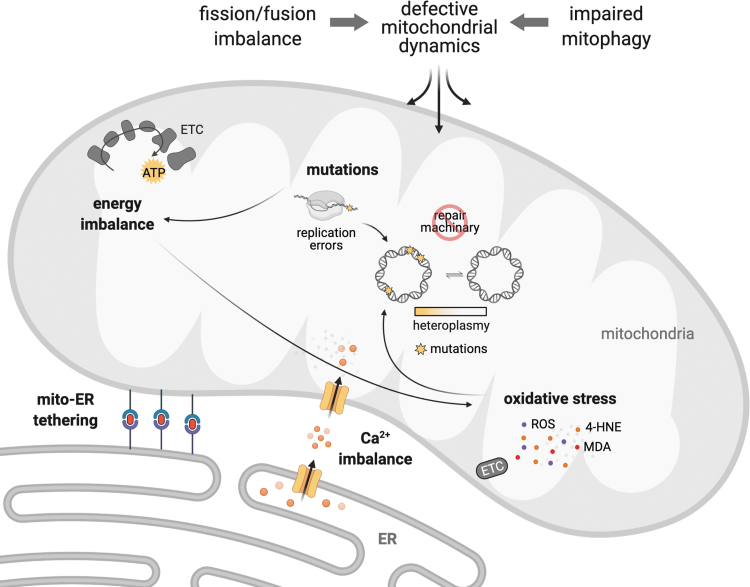
FIG. 3.
Mitochondrial fission/fusion imbalance and impaired mitophagy result in defective mitochondrial dynamics, which negatively affects critical processes in cardiac physiology such as ATP production, redox balance, mtDNA segregation, mitochondrial-ER tethering, and mitochondrial and cytosolic calcium handling. mtDNA, mitochondrial DNA; ER, endoplasmic reticulum.
Mitochondrial fusion
Mitochondrial fusion is fine-tuned by three-membrane guanosine triphosphatases (GTPases), the mitofusins (mitofusin 1 [Mfn1] and mitofusin 2 [Mfn2]), located at the outer mitochondrial membrane, and the optic atrophy factor 1 (Opa1), located at the inner mitochondrial membrane (Fig. 2) (179).
Mitofusins contain a GTPase domain involved in the hydrolysis of GTP, which is required for mitofusin oligomerization and fusion of outer mitochondrial membranes to the adjacent mitochondria (112). Interestingly, mitofusins require membrane curvature for binding, which is promoted by the hydrolysis of phospholipase D-dependent cardiolipin (47, 51). In addition, inner membrane fusion requires a similar process, triggered by Opa1 and coordinated by several other proteins, including prohibitins (131, 179). The interaction between cardiolipin and Opa1 is critical for inner mitochondrial membrane fusion, forming interconnected and elongated mitochondria (14).
Mitofusins are responsible for regulating many cellular processes, including mitochondrial number and morphology (12, 40), biogenesis (152), calcium homeostasis (27), protein turnover (35), and apoptosis (118). Mfn2 regulates cell proliferation (52), oxidative metabolism (20), and autophagy (32, 46), and acts as a mitochondrial antiviral signaling protein (204). Mfn2 is also located at the endoplasmic reticulum and mitochondria contact sites, aiding in the maintenance of cytosolic and mitochondrial calcium homeostasis (27, 139).
The absence of MFN2 in the mitochondria decreases oxygen consumption (40), membrane potential (12, 157), and TCA cycle activity (157), which increase with MFN2 overexpression in embryonic and skeletal muscle cells (12, 157). Finally, Mfn1 and Mfn2 are required for the stability of mtDNA, as mitochondrial fusion prevents the accumulation of mtDNA mutations (40, 138). Of interest, mutations in MFN2 or OPA1 (but not MFN1) cause neurodegenerative diseases (62, 156).
In the context of cardiac physiology, mitofusins are essential for the maintenance of cardiac function and morphology under physiological and pathological conditions. The ablation of both mitofusins leads to mitochondrial fragmentation in cultured myocytes (42) and whole heart (178), resulting in embryonic lethality in mouse. Conditional disruption of MFN1 results in cardiac mitochondrial fragmentation, with no effects on mitochondrial bioenergetics and cardiac function under basal conditions (153). In contrast, MFN2 deletion results in the accumulation of enlarged and pleomorphic cardiac mitochondria, which are associated with a modest mitochondrial bioenergetic dysfunction and pathological cardiac hypertrophy (151).
The differences in cardiac phenotype between MFN1 and MFN2 knockout mice might be explained by their unique biochemical properties and role in mitochondrial dynamics. For example, MFN1-harboring mitochondria (but not MFN2-harboring mitochondria) are efficiently tethered in a GTP-dependent manner (93). Moreover, Mfn1 has higher rate of hydrolysis and a lower affinity for GTP relative to Mfn2 (93). Other individual characteristics of Mfn1 and Mfn2, including posttranslational regulation, protein/protein interaction, and location, seem to explain their distinct role in mitochondria fusion, trafficking, turnover, and contacts with other organelles.
Despite their biochemical and functional singularities, Mfn1 and Mfn2 present a high homology (∼80% sequence identity), therefore suggesting these mitofusins might be functionally redundant under adverse conditions, which might explain the lack of severe cardiac phenotypes of MFN1 or MFN2 deficiency mice. In fact, cardiac-specific ablation of MFN1/MFN2 results in massive accumulation of fragmented and dysfunctional mitochondria, which severely disrupts cardiomyocyte sarcomere architecture and results in left ventricle dysfunction in mice (77, 177). These results suggest a compensatory mechanism, not necessarily functionally redundant, between Mfn1 and Mfn2 in the maintenance of cardiac physiology under baseline conditions. However, the molecular mechanism behind such response remains to be elucidated.
Counterintuitively, individual or combined MFN1 and MFN2 deletion protects against ROS-induced cardiomyocyte death (77, 151, 153). Moreover, MFN2 or MFN1/MFN2 ablation protects mice against in vivo cardiac ischemia/reperfusion injury (77, 151). The role of Mfn1 during in vivo cardiac ischemia/reperfusion injury still needs to be determined. The cardiac compensatory effects triggered by ablation of mitofusins are associated with reduced interaction between the mitochondria and sarcoplasmic reticulum (151), mitochondrial calcium overload (77, 151, 153), oxidative stress (77, 151, 153), and delay in the opening of the mitochondrial permeability transition pore (mPTP) (77, 151, 153).
However, the molecular mechanisms behind such responses are still unknown. The contribution of the mitochondrial permeability transition to the cardioprotective effects triggered by deletion of mitofusins seems unlikely considering the lack of efficacy of cyclosporine, a pharmacological inhibitor of cyclophilin D (a major component of mPTP), in reducing myocardial infarction in patients with ST-segment elevation myocardial infarction (49).
Opa1 plays a critical role in regulating mitochondrial number, cristae organization, and bioenergetic efficiency (57, 94). Cleavage of Opa1 by Yme1l (ATP-dependent zinc metalloprotease) (7) or Oma1 (metalloendopeptidase) (59, 79) is sufficient to induce opposite phenotypes under baseline and stress conditions. Briefly, constitutive Opa1 cleavage by Yme1l results into long forms, which are critical to induce mitochondrial fusion and control mitochondrial crista dynamics (7, 87). On the contrary, stress-induced Opa1 cleavage by Oma1 results in short forms, which triggers mitochondrial fragmentation and cell death (7, 111).
Opa1 levels are reduced in failing human hearts, which is associated with accumulation of smaller mitochondria (43). The contribution of Opa1 to cardiac physiology was previously demonstrated in heterozygous OPA1 mutant mice (44). Aged OPA1 mutant mice display cardiac accumulation of fragmented and dysfunctional mitochondria along with excessive oxidative stress and left ventricle dysfunction when compared with wild-type littermates (homozygous OPA1 mutation is embryonic lethal) (44). Moreover, posttranslational modification of Opa1 through constitutive and inducible proteolytic cleavage by Yme1l and Oma1, respectively, plays a key role in cardiac physiology (199). Cardiac deletion of YMEL1L1 causes accumulation of short forms of Opa1 and fragmented mitochondria, which are followed by dilated cardiomyopathy and heart failure in mice (199).
Interestingly, the ablation of the stress inducible OMA1 is sufficient to neutralize the cardiac negative effects triggered by OPA1 deletion (199). Similarly, deletion of OMA1 reduces the number of fragmented mitochondria and mitigates the progression of heart failure induced by isoproterenol or hypertension in mice (4). These findings provide evidence that preventing mitochondrial fragmentation by deleting OMA1 protects against cell death and heart failure (199).
Mitofusins and Opa1 seem to play a key role in regulating apoptotic cell death. The proapoptotic proteins Bax and Bak colocalize with Mfn2 in the outer mitochondrial membrane, which seems to inhibit its pro-fusion effect and favors the mitochondrial release of cytochrome c and apoptosis-inducing factor (AIF) (141). Indeed, dominant active mutants of MFN2 repress Bax activation and cytochrome c release under proapoptotic conditions (141). Similarly, cells depleted of Opa1 are very sensitive to exogenous apoptosis induction (203). Mechanistically, Opa1 suppresses the proapoptotic action of fission 1 protein (Fis1) (203).
Considering the main role of mitofusins and Opa1 in controlling mitochondrial fusion and cardiomyocyte viability, it is expected that nonpharmacological and pharmacological approaches capable of rescuing the function of these proteins will have a positive impact during cardiac degeneration. In fact, reestablishing mitochondrial fusion through exercise (32, 33) or pharmacological intervention (64) is sufficient to restore mitochondrial bioenergetics and improve the prognosis of heart failure in preclinical studies. Indeed, an 8-week running treadmill exercise reestablishes cardiac mitochondrial morphology and autophagic flux, therefore improving cardiac oxidative capacity in rats with myocardial infarction-induced heart failure (32).
Mechanistically, the synergy between mitochondrial fusion and autophagy drives improved bioenergetic efficiency and cardiovascular fitness induced by exercise. Similarly, pharmacological sustained treatment with a selective antagonist of Mfn1-βIIPKC association (SAMβA) reestablishes Mfn1 GTPase activity, reduces mitochondrial fragmentation in heart failure rats, protects cultured neonatal and adult cardiac myocytes from stress-mediated cell death, and improves cardiac function (64).
Genetic interventions capable of correcting mitochondrial fusion might be another approach to treat inherited heart diseases. A recent article demonstrated that genetic correction of OPA1 variation by using CRISPR-Cas9 technology improves mitochondrial connectiveness, bioenergetics, levels of mtDNA, and resistance to stress in patient-derived OPA1 mutant-induced pluripotent stem cells (176). However, its therapeutic value to treat Opa1-related diseases, such as autosomal dominant optic atrophy and cardiac disorders, remains to be determined. Moreover, the potential use of genetics to correct the mitochondrial fission/fusion imbalance and the consequent mitochondrial dysfunction in cardiac diseases are still uncertain.
Mitochondrial fission
Mitochondrial fission is characterized by splitting one mitochondrion into two smaller mitochondria, which is critical for maintaining mitochondrial quality control and bioenergetics under baseline and stress conditions (Fig. 2) [for review see Chan (37)]. The mitochondrial fission process is orchestrated by the GTPase dynamin-related protein 1 (Drp1). Drp1 is a cytosolic protein that translocates to mitochondria upon activation and binds to specific proteins located at the outer mitochondrial membrane, including mitochondrial Fis1 (136, 205), mitochondrial fission factor (Mff) (145), mitochondrial dynamics protein of 49 kDa (MiD49), and mitochondrial dynamics protein of 51 kDa (MiD51) (123, 149).
Drp1 translocation to mitochondria is induced by many different factors in cardiomyocytes, including β-adrenergic stimulation (97), ischemia/reperfusion injury (53), and exercise (32). Upon Drp1 translocation and binding to aforementioned proteins at constriction sites, it polymerizes around the organelle and through the hydrolysis of GTP, changes its conformation to constrict both the outer and inner membranes, leading to mitochondrial fission and accumulation of smaller and spherical mitochondria (Fig. 2) (53, 92, 129).
Fragmented and dysfunctional mitochondria are not reincorporated into the mitochondrial network, inducing the autophagy-mediated removal of these organelles (Fig. 2). In this scenario, Drp1 plays a key role in cardiac autophagy-mediated mitochondrial removal, also termed mitophagy, and mitochondrial quality control (90, 178). These findings demonstrate the critical role of Drp1 in maintaining cardiac biochemistry homeostasis and contractility properties under physiological conditions.
Under pathological conditions, such as myocardial infarction, a pro-fission phenotype along with impaired mitophagy results in accumulation of fragmented and dysfunctional mitochondria, therefore facilitating the propagation of damage and cardiac degeneration over time (53). Cardiac ablation of DNM1L results in reduction of mitochondrial membrane potential, induction of mPTP (178), impaired mitochondrial respiration (96, 192), and disruption of mitophagy (178), therefore contributing to cardiomyocyte necrosis and dilated cardiomyopathy in mice (90, 177, 178). Mitochondrial translocation of Drp1 is also involved in cardiac ischemic damage, where pharmacological inhibition of Drp1 protects cardiomyocytes in culture and whole heart from ischemia/reperfusion injury (144).
Fis1 is another player involved in mitochondrial fragmentation under baseline and stress conditions (Fig. 2) (89). Interestingly, Fis1 is highly expressed in cardiac cells, therefore suggesting its important role in heart physiology (89).
During ischemia/reperfusion injury, both Fis1 and Drp1 are upregulated in neonatal and adult cardiomyocytes (84, 208). Under this scenario, upstream signaling events favor Drp1 activation and the consequent interaction with Fis1 at the outer mitochondrial membrane (205). This interaction results in an excessive accumulation of fragmented/dysfunctional mitochondria and redox imbalance (53). Fis1 also binds to mitochondrial fusion proteins (Mfn1, Mfn2, and Opa1) and inhibits their GTPase activity, thus blocking mitochondrial fusion in mammalian cells (206). These findings suggest that Fis1 triggers mitochondrial fragmentation also in a Drp1-independent manner. However, this novel role of Fis1 remains to be determined in the context of cardiac physiology.
Mechanistically, there are at least two functionally distinct mitochondrial fission molecular signatures that predict the biogenesis of new mitochondria (midzone division) and the clearance of fragmented and dysfunctional mitochondria (periphery division) (106). Mitochondrial fission in the midzone and periphery occurs through Drp1-Mff interaction and Drp1-Fis1 interaction, respectively (Fig. 2) (106). Moreover, midzone mitochondrial division is marked by endoplasmic reticulum (ER)/mitochondria contact sites (106) and relies on Golgi-derived vesicles (137). The function of these protein/protein interactions to mitochondrial fission, as well as its contribution to physiological and pathological conditions, has only recently been elucidated (100, 110).
The Drp1-Mff interaction plays a critical role during physiological mitochondrial fission and function (110). Disruption of Drp1-Mff interaction, using a rationally designed peptide, is sufficient to reduce ATP levels and impair mitochondrial morphology, leading to behavioral deficits and progression of Huntington's disease in mice (110). On the contrary, Drp1-Fis1 interaction seems to be important for mitochondrial division under adverse conditions such as mitochondrial oxidative stress, impaired calcium homeostasis, and reduced membrane potential (106). This type of mitochondrial fission must be linked to mitochondrial clearance through autophagy. However, autophagy is generally impaired under degenerative conditions, which might result in accumulation of fragmented/dysfunctional mitochondria.
Under these conditions, pharmacological inhibition of excessive Drp1-Fis1 interaction by a cell-permeable peptide termed p110 is sufficient to avoid accumulation of smaller/dysfunctional mitochondria (159) and protect against cardiovascular (53) and neurodegenerative diseases (67, 99, 100, 102). More recently, König et al. demonstrated that Mff, MiD49, and MiD51 are essential for Drp1-dependent mitochondrial scission and release of mitochondrial-derived vesicles (MDV) (Fig. 2) (109).
In summary, a coordinated response involving induction of key mitochondrial fission genes, upstream activation of Drp1 (137), and the bimodal mitochondrial division through Drp1-Mff interaction and Drp1-Fis1 interaction (106) dictates the mitochondrial fission rate in heart physiology (106) from embryonic development (41, 95), cardiomyocyte differentiation (99), to adult cardiomyocytes (53) under baseline and pathological conditions.
Mitophagy
Mitophagy is a process that selectively targets dysfunctional mitochondria for removal and is closely linked to mitochondrial fission and fusion (Fig. 4) [for review see Palikaras et al. (147)].
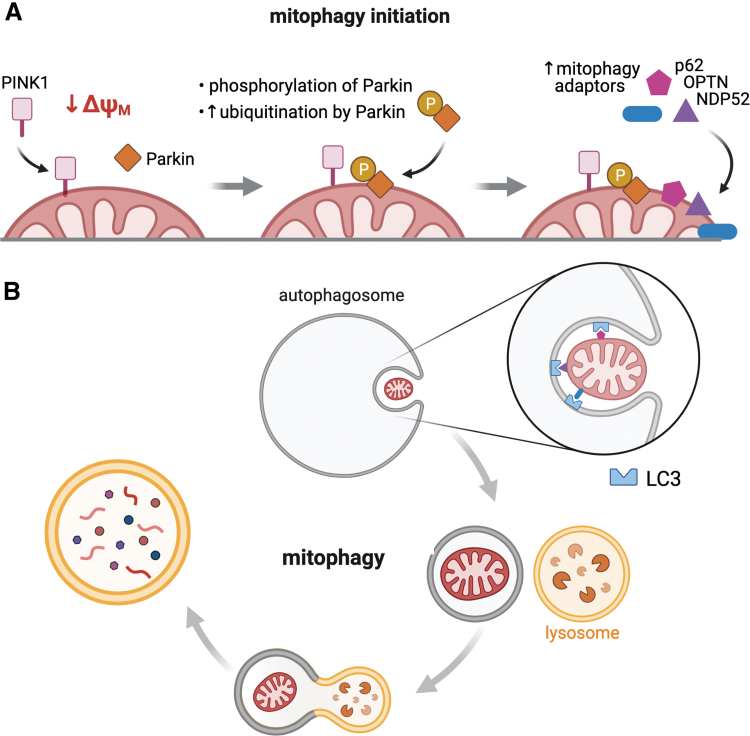
FIG. 4.
Proposed model for mitochondrial mitophagy. (A) Mitophagy selectively targets dysfunctional mitochondria for removal and is closely linked to mitochondrial fission and fusion (147). PINK1-Parkin-mediated mitophagy is triggered by mitochondrial depolarization, which results in PINK1 accumulation in the outer mitochondrial membrane and the subsequent phosphorylation/recruitment of Parkin (a cytosolic E3-ubiquitin ligase) to the mitochondrial surface (133, 171). (B) Parkin-mediated ubiquitination of outer mitochondrial membrane proteins results in the recruitment of autophagy adaptors (p62, OPTN, and NDP52) and initiates autophagosome formation through LC3 binding and the subsequent lysosomal degradation of autophagosome engulfed mitochondria (127, 140). PINK1, PTEN-induced kinase 1.
Briefly, under physiological conditions, PINK1 (PTEN-induced kinase 1) is targeted and imported to the inner mitochondrial membrane to be degraded by mitochondrial proteases (46, 128). However, under stress conditions, when the mitochondrial membrane potential is reduced, this process is disrupted and PINK1 accumulates on the outer mitochondrial membrane. PINK1 then phosphorylates and recruits Parkin (a cytosolic E3-ubiquitin ligase) to the mitochondrial surface (39, 108, 113). Parkin-mediated ubiquitination of outer mitochondrial membrane proteins results in the recruitment of autophagy adaptors (p62, OPTN, and NDP52) and initiates autophagosome formation through LC3 binding and the subsequent lysosomal degradation of autophagosome engulfed mitochondria (Fig. 4) (80, 162).
The PINK1-Parkin pathway affects mitochondrial fission and fusion at different levels. PINK1 indirectly triggers Drp1 translocation to the mitochondrial surface, therefore promoting fission of dysfunctional mitochondria, which enables their removal through autophagy (158). In coordination with Drp1 activation, PINK1-Parkin-mediated phosphorylation and proteasomal degradation of Mfn1 also contribute to mitochondrial fragmentation and removal of dysfunctional organelles (187). The activation of the PINK1-Parkin pathway results in Mfn2 phosphorylation and disruption of ER/mitochondria contact sites, which are critical for the maintenance of calcium handling, bioenergetic profile, and contractility properties of cardiomyocytes (46).
Mitochondrial fission/fusion balance and clearance are also regulated by mitophagy receptors located at the outer mitochondrial membrane, including NIX (NIP3-like protein X) (163), BNIP3 (BCL2 interacting protein 3) (119), and FUNDC1 (FUN14 domain-containing protein 1) (45). The activation of these receptors is critical to control mitochondrial number, size, and metabolism under both health and disease conditions (60). BNIP3 promotes fission and removal of damaged mitochondria in cardiomyocytes through PINK1 stabilization, inhibition of Opa1 assembly, and Drp1 translocation to the mitochondrial surface (119).
There are also other PINK1-Parkin-independent mechanisms of mitophagy [for review see Palikaras et al. (147)]. For example, mitochondrial protein FUNDC1 is located at the outer mitochondrial membrane and has been linked to mitophagy under hypoxia or mitochondrial membrane potential dissipation (45, 121). Phosphorylation of FUNDC1 and the consequent inhibition of FUNCD1-mediated mitophagy result in excessive accumulation of ROS, therefore impairing cardiac redox homeostasis and mitochondrial metabolism (208).
Overall, mitophagy plays a critical role in maintaining the pool of healthy mitochondria under physiological and adverse conditions [i.e., accumulation of mtDNA mutations (192)]. Recent studies provide evidence that dysfunctional mitophagy culminates in the accumulation of damaged mitochondria and degeneration of organs with high demand for ATP synthesis and elevated oxygen uptake rate, such as the heart (22, 32). For example, PINK1 protein levels are reduced in failing hearts (22), and mitophagy is impaired in both aged and failing hearts in mice (86).
There are several findings from preclinical studies supporting the key role of mitophagy in heart physiology. Parkin-deficient mice accumulate dysfunctional mitochondria and have elevated levels of oxidative stress, along with pathological cardiac remodeling and reduced left ventricle fractional shortening (74, 86). Furthermore, genetic disruption of PINK1-PARK2-mediated mitophagy results in lethal heart failure in mice (74), and PARK2-deficient Drosophila heart tubes display progressive cardiomyopathy (46). Lastly, Parkin knockout mice are more sensitive to myocardial infarction, have a higher mortality rate, and a bigger infarct size when compared with wild-type (115).
The PINK1-Parkin pathway is also involved in the cardioprotective effects of ischemic preconditioning. Genetic disruption of the PARK2 pathway abolishes ischemic preconditioning (the benefit of short ischemic events before prolonged ischemia) in mice (88). Pharmacological inhibition of autophagy and the consequent impairment of mitochondria clearance were found to prevent the positive effects of exercise in reestablishing the pool of healthy mitochondria, redox balance, and contractility properties of failing hearts in rats (31, 32).
Counterintuitively, mitophagy overactivation can negatively affect cardiac physiology. The overexpression of mitophagy receptors NIX and BNIP3 maximizes the cardiac damage induced by myocardial infarction (55). Moreover, genetic disruption of BNIP3 is sufficient to decrease apoptosis and improve cardiac remodeling and function in a model of postmyocardial infarction in mice (55).
In summary, functional mitophagy, as well as its interdependence with mitochondria fission and fusion, plays a critical role in the maintenance of cardiac mitochondrial morphology, mtDNA content, bioenergetics, and cell death (Fig. 3) (31), therefore affecting cardiac metabolism, redox homeostasis, and contractility properties in health and disease.
mtDNA Heteroplasmy
The mitochondria of most eukaryotic organisms contain their own circular DNA, which has a high density of genetic information and is maternally inherited (133, 143). In humans, mtDNA is circular, double-stranded, ∼16.6 base pairs in length, and located in the mitochondrial matrix (133). mtDNA encodes 37 genes, of which 13 encode peptide subunits critical for complexes I, III, and IV and ATP synthase, while complex II is encoded only by a nuclear genome (8). In addition, 22 transfer RNAs and 2 ribosome-coding RNAs are encoded by mtDNA (143). Unlike the nuclear genome, mtDNA is not protected by histones—it is organized into nucleoids (an mtDNA-protein complex). Since mtDNA is located in the mitochondrial matrix, it is more vulnerable to ROS oxidation produced during OXPHOS, making the mitochondrial genome more susceptible to mutations (174).
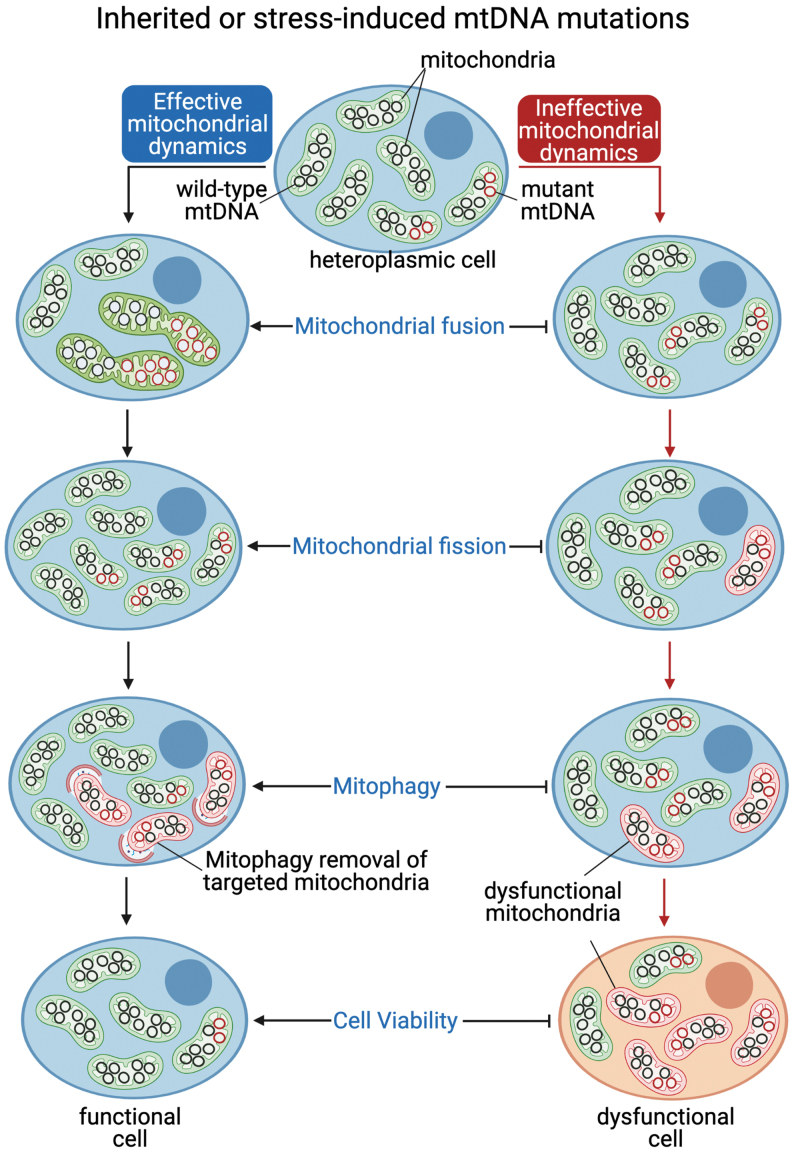
mtDNA is characterized by a high copy number, and the coexistence of wild-type and mutated mtDNA in the cell is defined as heteroplasmy (Fig. 5) (174). The prevalence rate for mtDNA mutations is ∼1 in 5000 individuals (75). In healthy humans, low-level heteroplasmy is usually detected but well tolerated. Most mtDNA mutations are recessive and must accumulate to negatively affect mitochondrial bioenergetics (188). The maximum threshold to tolerate mutations differs among cell types, tissue, or individual (133, 155, 197). Heteroplasmy levels are dynamic and constantly changing, since mtDNA replication occurs throughout the cell cycle. Many point mutations and deletions of mtDNA may arise at embryonic development or over time as heteroplasmy increases (68, 185).
FIG. 5.
Proposed model for mitochondrial segregation under accumulation of inherited or stress-mediated mtDNA mutations. Coexistence of wild-type (black circle) and mutated (red circle) mtDNA in the same cell is defined as mtDNA heteroplasmy, directly regulated and selected by constant events of mitochondrial fission, fusion, and mitophagy (142, 192). Defective mitochondrial dynamics results in accumulation of mtDNA mutations, mitochondrial dysfunction, and the consequent reduction of cell viability (117).
Leber hereditary optic neuropathy (LHON) was the first evidence that mutations of mtDNA maternally inherited could trigger the development of chronic diseases (83), such as cardiomyopathy (82) and diabetes (13). Mothers with high rates of heteroplasmy are more likely to have children with mtDNA mutations. However, the symptoms will only show up according to the percentage of heteroplasmy (197).
In somatic cells, pathogenic mtDNA mutations can arise from replication errors, oxidative stress-induced DNA damage, and/or a defect in the mtDNA repair machinery. Indeed, accumulation of mtDNA mutations (∼60%–90% mutant to wild-type DNA) has a negative impact on mitochondrial bioenergetics, especially in tissues highly dependent on mitochondrial ATP (68, 143, 185). Furthermore, mutations in nuclear genes responsible for mtDNA replication and transcription, such as mitochondrial transcription factor A (TFAM), DNA polymerase POLG, and RNA polymerase (POLRMT), can promote mtDNA mutation and be linked to several chronic diseases (6), including aging phenotypes and reduced life span (189). Therefore, the levels of heteroplasmy are the main determinant for the onset and severity of mitochondrial diseases.
Accumulation of mtDNA mutations in genes that encode subunits of the ETC complex likely results in mitochondrial dysfunction and redox imbalance. This phenotype has a major effect in organs highly dependent on mitochondrial ATP synthesis, including the central nervous system (168), skeletal muscle (197) and heart (195). Clinical studies demonstrate the relationship between cardiac diseases and mtDNA mutations, although the molecular mechanisms are still poorly understood (82, 91, 104, 195). Cardiomyopathy affects ∼20% of individuals with mitochondrial disease and has been related to high heteroplasmy levels in the cardiac tissue and early mortality in children (82, 91).
In a recent study, unique mutations were found to be enriched in stroke and myocardial infarction patients when compared with healthy individuals (195). Briefly, mtDNA mutations (m16145G>A and m.16311T>C) detected in stroke patients were positively associated with potential genetic risk factors. Contrasting, another set of mtDNA mutations (m.72T>C, m.73A>G, and m.16356T>C) found in myocardial infarction individuals was inversely associated with potential genetic risk factors (195). Different mitochondrial mutations have been found in leukocytes from the Russian (m.5178C>A and m.14459G>A) and Mexican (m.13513G>A and m.652insG) populations, and are associated with increased cardiovascular risk factors (104).
mtDNA mutations have also been associated with disrupted mitochondrial morphology, which may contribute to the development of heart failure (11). Taken together, these data suggest that accumulation of mtDNA mutations contributes to the establishment and progression of cardiac diseases. Therefore, evolutionary conserved systems capable of controlling mitochondrial heteroplasmy, such as mitochondrial fission, fusion, and clearance, are important to counteract or mitigate mtDNA mutation-induced heart diseases.
mtDNA Segregation
mtDNA can be segregated along the maternal germ line or in somatic cells and tissues (68, 185). The segregation is highly dynamic, and heteroplasmy levels constantly shift between daughter cells (Fig. 5) (185). During germ line segregation, a reduced number of mtDNA is transmitted from the mother to the offspring within each oocyte, a phenomenon known as a “genetic bottleneck,” which explains the different heteroplasmy levels and clinical disease severity between generations (48, 68). As discussed, mtDNA mutations may also increase during the lifetime through random replication in nondividing (postmitotic) cells, such as cardiomyocytes, a phenomenon that is related to aging (185). For these reasons, both mitochondrial fusion and fission, and mitophagy, are critical to ensure proper selection and/or transmission of mtDNA (1, 143, 192).
mtDNA copy number and turnover, as well as mtDNA segregation rate, are directly affected by changes in mitochondrial number and the continual exchange of mitochondrial content during mitochondrial fission, fusion, and clearance (142, 192). mtDNA is continuously replicated and exchanged between mitochondria during the lifetime through mitochondrial fission and fusion events. This process favors the distribution and propagation of mitochondrial genetic information throughout the cell during mitochondrial network reorganization (38, 42, 200). In the case of dividing cells, mitochondrial dynamics is crucial for the transmission of mtDNA content to daughter cells (Fig. 5).
The effectiveness of mitochondrial dynamics in regulating mtDNA distribution and segregation seems to be tissue- and cell-type-specific, and is affected by the mitochondrial bioenergetic state and redox balance (15, 142, 192). However, the molecular mechanisms regulating these processes remain elusive.
Mitochondrial fission/fusion and mtDNA segregation
Mitochondrial fusion plays a key role in regulating both stability and tolerance of mtDNA mutation in skeletal muscle (42) and mtDNA replication in the heart (161). Patients with mutation in MFN2 and OPA1, responsible for mitochondrial fusion, display increased mtDNA instability (5, 165), which is associated with Charcot-Marie-Tooth type 2A and dominant optic atrophy degenerative diseases, respectively (50, 209). Furthermore, mitofusin-deficient mice show a marked reduction of mtDNA levels and an increase in both mtDNA point mutations and deletions, which are accompanied by mitochondrial myopathy in skeletal muscle. These findings suggest that the maintenance of mtDNA copy number relies on mitochondrial fusion, which is essential to maintain mitochondrial function and muscle contractility properties (42).
Mitochondrial fission is also important in controlling mtDNA number (Fig. 5). Defective mitochondrial fission in DNM1L knockout HeLa cells reduces both mtDNA stability and ATP production, correlating with inhibition of cell proliferation and viability (154). Of interest, mitochondrial fission often occurs adjacent to nucleoids, which suggests a role for mitochondrial division during mtDNA replication and distribution. Indeed, deficient mitochondrial fission results in nucleoid clustering and the consequent dysfunctional mitochondrial bioenergetics (15, 116, 142). Finally, mtDNA segregation seems to be regulated by midzone mitochondrial division through Drp1-Mff interaction, but not Drp1-Fis1 interaction, which is likely associated with peripheral mitochondrial fission and clearance of mitochondrial debris (106).
Muscle-specific DNM1L knockout mice display severe mtDNA nucleoid clustering, which is associated with reduced mitochondrial bioenergetics, dilated cardiomyopathy, and neonatal lethality during heart development (96). In contrast, Jokinen et al. demonstrated that reduced mitochondrial fission in DNM1L mutant animals does not affect cardiac mtDNA segregation (98). This discrepancy may be due to the difference in the time-window that was examined in these studies; in the later study, measurements were conducted before the onset of dilated cardiomyopathy and heart failure. Therefore, whether mitochondrial fission and fusion play a role in cardiac mtDNA segregation under pathological conditions remains to be determined.
Mitophagy and mtDNA segregation
Mitophagy, another critical system involved in mitochondrial quality control, plays an important role in the maintenance and selection of mtDNA. In general, dysfunctional and depolarized mitochondria are identified, targeted, engulfed by autophagosome, and degraded by lysosomes, thereby contributing to selected removal of mtDNA (Fig. 4) (72, 186). It is also expected that a coordinated response between mitochondrial fission/fusion and mitophagy facilitates the clearance of damaged mitochondria, therefore mitigating the propagation of mtDNA mutations and mtDNA heteroplasmy (Fig. 5).
A seminal study by Twig et al. demonstrated that mitochondrial fission followed by selective fusion segregates dysfunctional mitochondria, allowing for elimination through mitophagy (192). In fact, induction of mitochondrial depolarization or overexpression of Parkin optimizes the clearance of deleterious mtDNA in heteroplasmic cells (186). As expected, defective mitophagy increases mtDNA heteroplasmy by accumulating mtDNA mutations, which is related to several genetic diseases, such as hereditary Parkinson's disease (42, 186).
The role of mitophagy in removing deleterious mtDNA has been underexplored in the context of cardiovascular diseases. Induction of mtDNA damage in a transgenic mouse model overexpressing the mutant uracil-DNA glycosylase (mutUNG1) causes a rapid congestive heart failure and premature death (117). These animals display accumulation of dysfunctional mitochondria, therefore suggesting a disruption of the mitochondrial fission/fusion balance and/or mitophagy (117). However, the contribution of mitochondrial dynamics to mutUNG1-induced cardiac phenotype was not addressed in the study.
In another study using a mouse model that accumulates mtDNA damage due to a proofreading-defective mtDNA polymerase γ (POLG), cardiac Parkin protein levels were reduced with aging (201). However, these transgenic mice do not present impaired mitochondrial bioenergetics or cardiac dysfunction compared with wild-type. Under these conditions, neither cardiac-specific overexpression nor global deletion of PARK2 affects the cardiac phenotype of POLG mice (201). These findings suggest that Parkin-mediated mitophagy is not essential for the maintenance of mitochondrial quality control in a preclinical model of cardiac mtDNA damage and aging.
Overall, the mechanisms underlying the clearance of deleterious mtDNA under health and disease conditions are still under debate. However, there is a reasonable amount of evidence suggesting that a coordinated response between mitochondrial fission/fusion and mitophagy plays an important role in controlling mtDNA heteroplasmy, which might have an impact on the onset and progression of cardiac disease.
Potential Therapeutics Targeting Mitochondria Fission and Fusion in Cardiac Diseases
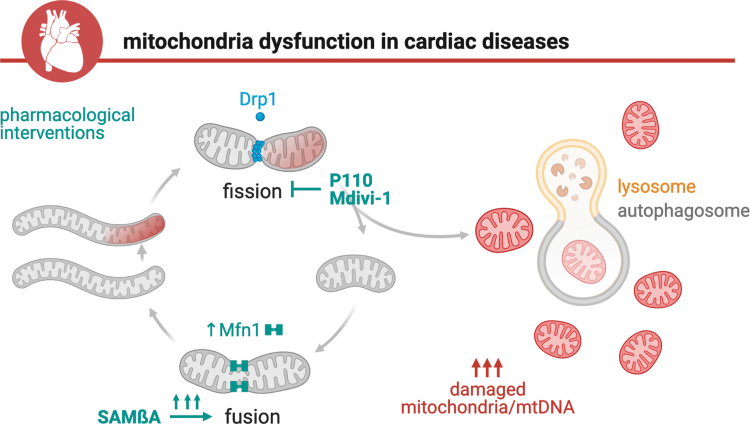
As previously discussed, disruption of mitochondrial quality control through impaired mitochondrial fission, fusion, or defective mitophagy negatively affects mitochondrial number, size, and shape. These changes ultimately result in excessive accumulation of damaged mitochondria, compromised mitochondrial segregation, impaired bioenergetics, and oxidative stress, which together contribute to the establishment and progression of cardiac diseases (Fig. 6). Therefore, it is expected that pharmacological strategies that reestablish mitochondrial connectiveness through inhibition of pathological mitochondrial fission, boosting mitochondrial fusion, or facilitating the clearance of dysfunctional mitochondria will abolish or mitigate cardiac mitochondrial dysfunction and therefore become a promising approach to improve the prognosis of cardiac diseases (193).
FIG. 6.
Proposed model for therapeutics targeting mitochondrial dynamics. Impaired mitochondrial fission/fusion and mitophagy and the consequent accumulation of fragmented and dysfunctional mitochondria are common hallmarks of cardiac diseases. Pharmacological interventions that improve mitochondrial connectiveness and clearance such as P110, Mdivi-1, or SAMβA play a critical role in the maintenance of mitochondrial health, therefore becoming potential novel therapeutic targets for cardiac diseases. SAMβA, selective antagonist of Mfn1-βIIPKC association.
Different strategies targeting mitochondria biology, such as small interfering RNA (siRNA), CRISPR-Cas9 gene editing, small molecules, or short peptides, have been successfully tested in preclinical studies of cardiac diseases. The mitochondrial fission inhibitors mdivi-1 (58, 126), P110 (76), Drp1 siRNA (172), or calcineurin inhibitor (175) efficiently reverse the acute damage induced by ischemia/reperfusion injury in different tissues, including the brain, heart, and kidney. Treatment with mdivi-1, a Drp1 inhibitor, before ischemia attenuates mitochondrial damage and myocardial infarct size in mice subjected to transient coronary artery occlusion (126). Of interest, mdivi-1 poorly inhibits recombinant Drp1 GTPase activity in vitro (58, 125).
A recent study demonstrates that mdivi-1, at the same concentration that reduces mitochondrial fragmentation, reversibly inhibits mitochondrial complex I-dependent oxygen consumption and ROS production, even in the absence of DNM1L (125). Moreover, mdivi-1 induces mitotic spindle abnormalities and mitosis arrest by inhibiting tubulin polymerization in a Drp1-independent manner (61), which might affect fibroblast-mediated cardiac remodeling, function, and inflammation during pathological conditions (194). In summary, Drp1-independent effects of mdivi-1 remain a major issue limiting its use as a promising candidate to treat cardiac diseases.
Inhibition of Drp1-Fis1 interaction by the short peptide P110 (53, 102) reduces Drp1 enzymatic activity and efficiently blocks mitochondrial fragmentation in different cellular and animal models of cardiac diseases (Fig. 6) (53, 76, 101). The administration of P110 at the onset of reperfusion reduces cardiac accumulation of fragmented and dysfunctional mitochondria induced by ex vivo ischemia/reperfusion injury in rats (53). A single dose of P110 peptide at reperfusion is sufficient to inhibit mitochondrial fragmentation and increase mitochondrial size and bioenergetic profile after in vivo transient coronary artery occlusion in animals (53). These P110-mediated changes result in improved cardiac contractility properties when measured 3 weeks after myocardial infarction surgery (53).
Importantly, in contrast to mdivi-1, P110 does not inhibit basal activity of Drp1 in vivo, even after 5 months of treatment (54). P110 blocks pathological Drp1-Fis1 interaction and does not affect Drp1 binding to other adaptor proteins involved in physiological mitochondrial fission (i.e., Mff) (159). In summary, inhibition of DNM1L, specifically by targeting Drp1-Fis1 protein/protein interaction, seems to have a therapeutic potential in preventing ischemia-induced cardiac injury and the subsequent establishment of heart failure (Fig. 6). Similar protective results are observed in other degenerative diseases, including diabetes (173), Huntington's disease (54, 76), amyotrophic lateral sclerosis (102), Parkinson's disease (67, 159), and Alzheimer's disease (101).
The therapeutic use of peptides targeting mitochondrial fission and fusion has promising opportunities due to their high specificity and affinity to their targets as well as ease of modification and high biocompatibility, therefore having relatively few off-target effects.
The use of rationally designed peptides targeting selective protein/protein interaction has been successfully used in clinics to treat cancer [for review see Araste et al. (10)] and is well tolerated in patients with cardiac diseases (17). However, there are still some challenges for peptides as therapeutic entities, including a short half-life and lack of intracellular penetration compared with small molecules. Overall, both opportunities and novel strategies to circumvent the therapeutic challenges place rationally designed peptides targeting mitochondria, such as p110, as promising candidates to treat cardiac diseases.
Another strategy was recently developed to address mitochondrial fragmentation in failing hearts. In this case, the excessive accumulation of fragmented and dysfunctional mitochondria occurs throughout an excessive interaction between protein kinase C βII (PKCβII) and Mfn1 at the outer mitochondrial membrane, which results in PKCβII phosphorylation and the consequent inhibition of Mfn1 in a rodent model of myocardial infarction-induced heart failure (64, 65).
Under this scenario, the design and synthesis of a selective short peptide capable of attenuating PKCβII-Mfn1 interaction, termed SAMβA, are sufficient to counteract excessive mitochondria fragmentation in heart failure (Fig. 6) (64). Targeting intermolecular associations is a promising strategy to mitigate detrimental biological processes in a highly selective manner (63, 64, 146). The development of short peptides derived from protein interfaces that transiently affect intermolecular interaction, and therefore act as competitive inhibitors, becomes a useful tool to counteract the establishment and progression of cardiac diseases (25, 64, 130).
SAMβA protects primary culture of cardiac myocytes against angiotensin II-induced mitochondrial dysfunction and cell death (64). SAMβA treatment also improves heart failure outcome in a rat of model of myocardial infarction-induced heart failure (64). Sustained SAMβA delivery improves both morphological and functional properties of failing hearts compared with vehicle-treated heart failure animals. These changes in SAMβA-treated heart failure animals are paralleled by a series of biochemical modifications in failing hearts, including reduction in the number of fragmented mitochondria, improvement of mitochondrial OXPHOS efficacy, and redox homeostasis.
These findings unravel the contribution of PKCβII-Mfn1 association in disrupting the mitochondrial fission/fusion balance in heart failure, therefore highlighting PKCβII-Mfn1 interaction as a promising target to develop novel pharmacological interventions capable of improving mitochondrial health and heart failure outcome in patients. Note that cardiac overactivation of PKCβII and accumulation of dysfunctional mitochondria are also present in both heart failure with reduced ejection fraction (HFrEF) and heart failure with preserved ejection fraction (HFpEF) patients (36).
Summary and Perspectives
The unique role of mitochondria as intracellular nodes and multieffector players involved in the maintenance of cardiac homeostasis in health and disease has been recently recognized. Under this scenario, extensive work has been performed to decipher the most critical processes involved in mitochondrial dysfunction and its contribution to cardiac diseases. As discussed above, the quality control mechanisms that regulate mitochondrial fission/fusion balance and removal play a critical role in the maintenance of mitochondrial health, therefore becoming potential novel therapeutic targets for cardiac disease. New targets affecting mitochondrial fission/fusion balance and clearance have been recently tested in many preclinical models, therefore paving a new road for the development of interventions capable of improving mitochondrial function to treat cardiac diseases.
Finally, many efforts have been made to identify and validate promising targets affecting mitochondrial quality control. However, the development and improvement of available strategies to increase tissue and cell permeability, as well as drug delivery inside mitochondria, are critical for the translation of these molecules targeting mitochondrial quality control to the clinic. Therefore, combined strategies to approach and treat defective mitochondrial fission/fusion balance and mitophagy seem to be a promising way to design effective interventions and develop new therapies for complex and multifactorial diseases such as cardiac diseases.
Abbreviations Used
- 4-HNE
4-hydroxynonenal
- α-KG
α-ketoglutarate
- BNIP3
BCL2 interacting protein 3
- Drp1
dynamin-related protein 1
- ER
endoplasmic reticulum
- ETC
electron transport chain
- FADH2
flavin adenine dinucleotide
- Fis1
fission 1 protein
- FUNDC1
FUN14 domain-containing protein 1
- GPX
glutathione peroxidase
- GR
glutathione reductase
- GSH/GSSG
reduced/oxidized glutathione
- GTPase
guanosine triphosphatase
- H2O2
hydrogen peroxide
- HFpEF
heart failure with preserved ejection fraction
- HFrEF
heart failure with reduced ejection fraction
- IDPm
mitochondrial NADP+-dependent isocitrate dehydrogenase
- MCU
mitochondrial Ca2+ uniporter
- MDA
malondialdehyde
- MDV
mitochondrial-derived vesicles
- MEP
malic enzyme
- Mff
mitochondrial fission factor
- Mfn1
mitofusin 1
- Mfn2
mitofusin 2
- MiD49
mitochondrial dynamics protein of 49 kDa
- MiD51
mitochondrial dynamics protein of 51 kDa
- mNCE
Na+/Ca2+ exchanger
- Mn-SOD
Mn2+-dependent superoxide dismutase
- mPTP
mitochondrial permeability transition pores
- mtDNA
mitochondrial DNA
- mutUNG1
mutant uracil-DNA glycosylase
- NADH
nicotinamide adenine dinucleotide
- NIX
NIP3-like protein X
- O2−
anion superoxide
- OH
hydroxyl radical
- Oma1
metalloendopeptidase
- ONOO−
peroxynitrite
- Opa1
optic atrophy factor 1
- OXPHOS
oxidative phosphorylation
- PINK1
PTEN-induced kinase 1
- PKCβII
protein kinase C βII
- POLG
proofreading-defective mtDNA polymerase γ
- PPAR
peroxisome proliferator-activated receptor
- PRX
peroxiredoxin
- ROS
reactive oxygen species
- SAMβA
selective antagonist of Mfn1-βIIPKC association
- siRNA
small interfering RNA
- TCA
tricarboxylic acid
- TR
thioredoxin reductase
- TRXr/o
reduced/oxidized thioredoxin
- Yme1l
ATP-dependent zinc metalloprotease
Authors' Contributions
D.L.S. and J.C.B.F. wrote the article. A.A.G., and L.L. designed the figures and contributed to the writing. J.C.B.F. designed the article and D.M-R. and J.C.B.F. revised it. All the coauthors have reviewed and approved the final version of the article.
Author Disclosure Statement
Patents on the design and application of mitochondrial fission peptide inhibitors have been filed. The authors claim that there is no conflict of interest related to this work.
Funding Information
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) numbers 2019/22204-2, 2018/18627-3, 2019/25049-9, 2013/07937-8, and 2015/22814-5, Conselho Nacional de Pesquisa e Desenvolvimento—Brasil (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) Finance Code 001 (to J.C.B.F.), and by NIH R01HL52141 (to D.M-R.).
References
- 1. Abeliovich H, Zarei M, Rigbolt KTG, Youle RJ, and Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun 4: 2789, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abrahams JP, Leslie AGW, Lutter R, and Walker JE. Structure at 2.8 Â resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628, 1994. [DOI] [PubMed] [Google Scholar]
- 3. Acín-Pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, and Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell 32: P529–P539, 2008. [DOI] [PubMed] [Google Scholar]
- 4. Acin-Perez R, Lechuga-Vieco AV, DelMarMuñoz M, Nieto-Arellano R, Torroja C, Sánchez-Cabo F, Jiménez C, González-Guerra A, Carrascoso I, Benincá C, Quiros PM, López-Otín C, Castellano JM, Ruíz-Cabello J, Jiménez-Borreguero LJ, and Enríquez JA. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci Transl Med 10: eaan4935, 2018. [DOI] [PubMed] [Google Scholar]
- 5. Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissière A, Campos Y, Rivera H, De La Aleja JG, Carroccia R, Iommarini L, Labauge P, Figarella-Branger D, Marcorelles P, Furby A, Beauvais K, Letournel F, Liguori R, La Morgia C, Montagna P, Liguori M, Zanna C, Rugolo M, Cossarizza A, Wissinger B, Verny C, Schwarzenbacher R, Martín MÁ, Arenas J, Ayuso C, Garesse R, Lenaers G, Bonneau D, and Carelli V. OPA1 mutations induce mitochondrial DNA instability and optic atrophy “plus” phenotypes. Brain 131: 338–351, 2008. [DOI] [PubMed] [Google Scholar]
- 6. van den Ameele J, Li AYZ, Ma H, and Chinnery PF. Mitochondrial heteroplasmy beyond the oocyte bottleneck. Semin Cell Dev Biol 97: 156–166, 2020. [DOI] [PubMed] [Google Scholar]
- 7. Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, and Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol 204: 919–929, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, and Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981. [DOI] [PubMed] [Google Scholar]
- 9. Aon MA, Stanley BA, Sivakumaran V, Kembro JM, O'Rourke B, Paolocci N, and Cortassa S. Glutathione/thioredoxin systems modulate mitochondrial H2O2 emission: an experimental-computational study. J Gen Physiol 139: 479–491, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Araste F, Abnous K, Hashemi M, Taghdisi SM, Ramezani M, and Alibolandi M. Peptide-based targeted therapeutics: focus on cancer treatment. J Control Release 292: 141–162, 2018. [DOI] [PubMed] [Google Scholar]
- 11. Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, Bellini O, Dal Bello B, Pilotto A, Magrini G, Campana C, Fortina P, Gavazzi A, Narula J, and Viganò M. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am J Pathol 153: 1501–1510, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacín M, Vidal H, Rivera F, Brand M, and Zorzano A. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism: a novel regulatory mechanism altered in obesity. J Biol Chem 278: 17190–17197, 2003. [DOI] [PubMed] [Google Scholar]
- 13. Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, and Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet 1: 11–15, 1992. [DOI] [PubMed] [Google Scholar]
- 14. Ban T, Ishihara T, Kohno H, Saita S, Ichimura A, Maenaka K, Oka T, Mihara K, and Ishihara N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol 19: 856–863, 2017. [DOI] [PubMed] [Google Scholar]
- 15. Ban-Ishihara R, Ishihara T, Sasaki N, Mihara K, and Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proc Natl Acad Sci U S A 110: 11863–11868, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barger PM, Brandt JM, Leone TC, Weinheimer CJ, and Kelly DP. Deactivation of peroxisome proliferator-activated receptor-α during cardiac hypertrophic growth. J Clin Invest 105: 1723–1730, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bates E, Bode C, Costa M, Gibson CM, Granger C, Green C, Grimes K, Harrington R, Huber K, Kleiman N, Mochly-Rosen D, Roe MT, Sadowski Z, Solomon S, and Widimsky P. Intracoronary KAI-9803 as an adjunct to primary percutaneous coronary intervention for acute ST-segment elevation myocardial infarction. Circulation 117: 886–896, 2008. [DOI] [PubMed] [Google Scholar]
- 18. Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, and Mootha VK. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, and Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci U S A 105: 15370–15375, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bell MB, Bush Z, McGinnis GR, and Rowe GC. Adult skeletal muscle deletion of Mitofusin 1 and 2 impedes exercise performance and training capacity. J Appl Physiol 126: 341–353, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bertero E and Maack C. Metabolic remodelling in heart failure. Nat Rev Cardiol 15: 457–470, 2018. [DOI] [PubMed] [Google Scholar]
- 22. Billia F, Hauck L, Konecny F, Rao V, Shen J, and Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A 108: 9572–9577, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Der Bliek AM, Sedensky MM, and Morgan PG. Cell biology of the mitochondrion. Genetics 207: 843–871, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boveris A and Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J 134: 707–716, 1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bozi LHM, Campos JC, Zambelli VO, Ferreira ND, and Ferreira JCB. Mitochondrially-targeted treatment strategies. Mol Aspects Med 71: 100836, 2020. [DOI] [PubMed] [Google Scholar]
- 26. Brennan JP, Bardswell SC, Burgoyne JR, Fuller W, Schröder E, Wait R, Begum S, Kentish JC, and Eaton P. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J Biol Chem 281: 21827–21836, 2006. [DOI] [PubMed] [Google Scholar]
- 27. De Brito OM and Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610, 2008. [DOI] [PubMed] [Google Scholar]
- 28. Budas G, Costa HM, Ferreira JCB, da Silva Ferreira AT, Perales J, Krieger JE, Mochly-Rosen D, and Schechtman D. Identification of ɛPKC targets during cardiac ischemic injury. Circ J 76: 1476–1485, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgoyne JR, Madhani M, Cuello F, Charles RL, Brennan JP, Der ES, Browning DD, and Eaton P. Cysteine redox sensor in PKGIα enables oxidant-induced activation. Science (80-) 317: 1393–1397, 2007. [DOI] [PubMed] [Google Scholar]
- 30. Burgoyne JR, Mongue-Din H, Eaton P, and Shah AM. Redox signaling in cardiac physiology and pathology. Circ Res 111: 1091–1106, 2012. [DOI] [PubMed] [Google Scholar]
- 31. Campos JC, Bozi LHM, Bechara LRG, Lima VM, and Ferreira JCB. Mitochondrial quality control in cardiac diseases. Front Physiol 7: 479, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campos JC, Queliconi BB, Bozi LHM, Bechara LRG, Dourado PMM, Andres AM, Jannig PR, Gomes KMS, Zambelli VO, Rocha-Resende C, Guatimosim S, Brum PC, Mochly-Rosen D, Gottlieb RA, Kowaltowski AJ, and Ferreira JCB. Exercise reestablishes autophagic flux and mitochondrial quality control in heart failure. Autophagy 13: 1304–1317, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campos JC, Queliconi BB, Dourado PMM, Cunha TF, Zambelli VO, Bechara LRG, Kowaltowski AJ, Brum PC, Mochly-Rosen D, and Ferreira JCB. Exercise training restores cardiac protein quality control in heart failure. PLoS One 7: e52764, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Carvalho AETS, Bassaneze V, Forni MF, Keusseyan AA, Kowaltowski AJ, and Krieger JE. Early postnatal cardiomyocyte proliferation requires high oxidative energy metabolism. Sci Rep 7: 15434, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cavellini L, Meurisse J, Findinier J, Erpapazoglou Z, Belgareh-Touzé N, Weissman AM, and Cohen MM. An ubiquitin-dependent balance between mitofusin turnover and fatty acids desaturation regulates mitochondrial fusion. Nat Commun 8: 15832, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaanine AH, Joyce LD, Stulak JM, Maltais S, Joyce DL, Dearani JA, Klaus K, Nair KS, Hajjar RJ, and Redfield MM. Mitochondrial morphology, dynamics, and function in human pressure overload or ischemic heart disease with preserved or reduced ejection fraction. Circ Hear Fail 12: 1–16, 2019. [DOI] [PubMed] [Google Scholar]
- 37. Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46: 265–287, 2012. [DOI] [PubMed] [Google Scholar]
- 38. Chan DC. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol Mech Dis 15: 235–259, 2020. [DOI] [PubMed] [Google Scholar]
- 39. Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RLJ, Hess S, and Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20: 1726–1737, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen H, Chomyn A, and Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280: 26185–26192, 2005. [DOI] [PubMed] [Google Scholar]
- 41. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, and Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, and Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141: 280–289, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen L, Gong Q, Stice JP, and Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res 84: 91–99, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L, Liu T, Tran A, Lu X, Tomilov AA, Davies V, Cortopassi G, Chiamvimonvat N, Bers DM, Votruba M, and Knowlton AA. OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J Am Heart Assoc 1: e003012, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen M, Chen Z, Wang Y, Tan Z, Zhu C, Li Y, Han Z, Chen L, Gao R, Liu L, and Chen Q. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 12: 689–702, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y and Dorn GW. PINK1-phosphorylated mitofusin 2 is a parkin receptor for culling damaged mitochondria. Science (80-) 340: 471–475, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, and Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262, 2006. [DOI] [PubMed] [Google Scholar]
- 48. Cree LM, Samuels DC, De Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HHM, and Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet 40: 249–254, 2008. [DOI] [PubMed] [Google Scholar]
- 49. Cung T-T, Morel O, Cayla G, Rioufol G, Garcia-Dorado D, Angoulvant D, Bonnefoy-Cudraz E, Guérin P, Elbaz M, Delarche N, Coste P, Vanzetto G, Metge M, Aupetit J-F, Jouve B, Motreff P, Tron C, Labeque J-N, Steg PG, Cottin Y, Range G, Clerc J, Claeys MJ, Coussement P, Prunier F, Moulin F, Roth O, Belle L, Dubois P, Barragan P, Gilard M, Piot C, Colin P, De Poli F, Morice M-C, Ider O, Dubois-Randé J-L, Unterseeh T, Le Breton H, Béard T, Blanchard D, Grollier G, Malquarti V, Staat P, Sudre A, Elmer E, Hansson MJ, Bergerot C, Boussaha I, Jossan C, Derumeaux G, Mewton N, and Ovize M. Cyclosporine before PCI in patients with acute myocardial infarction. N Engl J Med 373: 1021–1031, 2015. [DOI] [PubMed] [Google Scholar]
- 50. Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, and Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26: 207–210, 2000. [DOI] [PubMed] [Google Scholar]
- 51. DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, and Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol 186: 793–803, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ding Y, Gao H, Zhao L, Wang X, and Zheng M. Mitofusin 2-deficiency suppresses cell proliferation through disturbance of autophagy. PLoS One 10: e0121328, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Disatnik MH, Ferreira JCB, Campos JC, Gomes KS, Dourado PMM, Qi X, and Mochly-Rosen D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J Am Heart Assoc 2: 1–14, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Disatnik MH, Joshi AU, Saw NL, Shamloo M, Leavitt BR, Qi X, and Mochly-Rosen D. Potential biomarkers to follow the progression and treatment response of Huntington's disease. J Exp Med 213: 2655–2669, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, and Dorn GW. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 117: 2825–2833, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Doenst T, Nguyen TD, and Abel ED. Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 113: 709–724, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Del Dotto V, Mishra P, Vidoni S, Fogazza M, Maresca A, Caporali L, McCaffery JM, Cappelletti M, Baruffini E, Lenaers G, Chan D, Rugolo M, Carelli V, and Zanna C. OPA1 isoforms in the hierarchical organization of mitochondrial functions. Cell Rep 19: 2557–2571, 2017. [DOI] [PubMed] [Google Scholar]
- 58. Duan C, Wang L, Zhang J, Xiang X, Wu Y, Zhang Z, Li Q, Tian K, Xue M, Liu L, and Li T. Mdivi-1 attenuates oxidative stress and exerts vascular protection in ischemic/hypoxic injury by a mechanism independent of Drp1 GTPase activity. Redox Biol 37: 101706, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, and Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187: 1023–1036, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Esteban-Martínez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Mariño G, Seco E, Durand S, Enot D, Graña O, Malumbres M, Cvekl A, Cuervo AM, Kroemer G, and Boya P. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J 36: 1688–1706, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fang CT, Kuo HH, Yuan CJ, Yao JS, and Yih LH. Mdivi-1 induces spindle abnormalities and augments taxol cytotoxicity in MDA-MB-231 cells. Cell Death Discov 7: 118, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Feely SME, Laura M, Siskind CE, Sottile S, Davis M, Gibbons VS, Reilly MM, and Shy ME. MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology 76: 1690–1696, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ferreira JCB, Boer BN, Grinberg M, Brum PC, and Mochly-Rosen D. Protein quality control disruption by PKCβII in heart failure; rescue by the selective PKCβII inhibitor, βIIV5-3. PLoS One 7: e33175, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ferreira JCB, Campos JC, Qvit N, Qi X, Bozi LHM, Bechara LRG, Lima VM, Queliconi BB, Disatnik MH, Dourado PMM, Kowaltowski AJ, and Mochly-Rosen D. A selective inhibitor of mitofusin 1-βIIPKC association improves heart failure outcome in rats. Nat Commun 10: 1–14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ferreira JCB, Koyanagi T, Palaniyandi SS, Fajardo G, Churchill EN, Budas G, Disatnik MH, Bernstein D, Brum PC, and Mochly-Rosen D. Pharmacological inhibition of βIIPKC is cardioprotective in late-stage hypertrophy. J Mol Cell Cardiol 51: 980–987, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Figueira TR, Barros MH, Camargo AA, Castilho RF, Ferreira JCB, Kowaltowski AJ, Sluse FE, Souza-Pinto NC, and Vercesi AE. Mitochondria as a source of reactive oxygen and nitrogen species: from molecular mechanisms to human health. Antioxid Redox Signal 18: 2029–2074, 2013. [DOI] [PubMed] [Google Scholar]
- 67. Filichia E, Hoffer B, Qi X, and Luo Y. Inhibition of Drp1 mitochondrial translocation provides neural protection in dopaminergic system in a Parkinson's disease model induced by MPTP. Sci Rep 6: 1–13, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Floros VI, Pyle A, DIetmann S, Wei W, Tang WWC, Irie N, Payne B, Capalbo A, Noli L, Coxhead J, Hudson G, Crosier M, Strahl H, Khalaf Y, Saitou M, Ilic D, Surani MA, and Chinnery PF. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat Cell Biol 20: 144–151, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Freya TG and Mannellab CA. The internal structure of mitochondria. Trends Biochem Sci 25: 319–324, 2000. [DOI] [PubMed] [Google Scholar]
- 70. Gilde AJ, Van der Lee KAJM, Willemsen PHM, Chinetti G, Van der Leij FR, Van der Vusse GJ, Staels B, and Van Bilsen M. Peroxisome proliferator-activated receptor (PPAR) α and PPARβ/δ, but not PPARγ, modulate the expression of genes involved in cardiac lipid metabolism. Circ Res 92: 518–524, 2003. [DOI] [PubMed] [Google Scholar]
- 71. Gilkerson RW, Selker JML, and Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett 546: 355–358, 2003. [DOI] [PubMed] [Google Scholar]
- 72. Gilkerson RW, De Vries RLA, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, and Schon EA. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet 21: 978–990, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gomes KMS, Campos JC, Bechara LRG, Queliconi B, Lima VM, Disatnik MH, Magno P, Chen CH, Brum PC, Kowaltowski AJ, Mochly-Rosen D, and Ferreira JCB. Aldehyde dehydrogenase 2 activation in heart failure restores mitochondrial function and improves ventricular function and remodelling. Cardiovasc Res 103: 498–508, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, and Dorn GW. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science (80-) 350: aad2459, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, Feeney C, Horvath R, Yu-Wai-Man P, Chinnery PF, Taylor RW, Turnbull DM, and McFarland R. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol 77: 753–759, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, and Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J Clin Invest 123: 5371–5388, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hall AR, Burke N, Dongworth RK, Kalkhoran SB, Dyson A, Vicencio JM, Dorn GW, Yellon DM, and Hausenloy DJ. Hearts deficient in both Mfn1 and Mfn2 are protected against acute myocardial infarction. Cell Death Dis 7: 1–9, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Halliwell B and Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford: Oxford University Press, 1999. [Google Scholar]
- 79. Head B, Griparic L, Amiri M, Gandre-Babbe S, and Van Der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 187: 959–966, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Heo JM, Ordureau A, Paulo JA, Rinehart J, and Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 60: 7–20, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hiroyuki N, Ryohei Y, Masasuke Y, and Kazuhiko K Jr. Direct observation of the rotation of F1-ATPase. Nature 386: 299, 1997. [DOI] [PubMed] [Google Scholar]
- 82. Holmgren D, Wåhlander H, Eriksson BO, Oldfors A, Holme E, and Tulinius M. Cardiomyopathy in children with mitochondrial disease: clinical course and cardiological findings. Eur Heart J 24: 280–288, 2003. [DOI] [PubMed] [Google Scholar]
- 83. Holt IJ, Harding AE, and Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331: 717–719, 1988. [DOI] [PubMed] [Google Scholar]
- 84. Hom J, Yu T, Yoon Y, Porter G, and Sheu SS. Regulation of mitochondrial fission by intracellular Ca2+ in rat ventricular myocytes. Biochim Biophys Acta Bioenerg 1797: 913–921, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Horstkotte J, Perisic T, Schneider M, Lange P, Schroeder M, Kiermayer C, Hinkel R, Ziegler T, Mandal PK, David R, Schulz S, Schmitt S, Widder J, Sinowatz F, Becker BF, Bauersachs J, Naebauer M, Franz WM, Jeremias I, Brielmeier M, Zischka H, Conrad M, and Kupatt C. Mitochondrial thioredoxin reductase is essential for early postischemic myocardial protection. Circulation 124: 2892–2902, 2011. [DOI] [PubMed] [Google Scholar]
- 86. Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, and Matoba S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 4: 2308, 2013. [DOI] [PubMed] [Google Scholar]
- 87. Hu C, Shu L, Huang X, Yu J, Li L, Gong L, Yang M, Wu Z, Gao Z, Zhao Y, Chen L, and Song Z. OPA1 and MICOS regulate mitochondrial crista dynamics and formation. Cell Death Dis 11: 940, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, and Gottlieb RA. Preconditioning involves selective mitophagy mediated by parkin and p62/SQSTM1. PLoS One 6: e20975, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ihenacho UK, Meacham KA, Harwig MC, Widlansky ME, and Hill RB. Mitochondrial fission protein 1: emerging roles in organellar form and function in health and disease. Front Endocrinol (Lausanne) 12: 1–29, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, Abdellatif M, and Sadoshima J. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res 116: 264–278, 2015. [DOI] [PubMed] [Google Scholar]
- 91. Imai-Okazaki A, Kishita Y, Kohda M, Mizuno Y, Fushimi T, Matsunaga A, Yatsuka Y, Hirata T, Harashima H, Takeda A, Nakaya A, Sakata Y, Kogaki S, Ohtake A, Murayama K, and Okazaki Y. Cardiomyopathy in children with mitochondrial disease: prognosis and genetic background. Int J Cardiol 279: 115–121, 2019. [DOI] [PubMed] [Google Scholar]
- 92. Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, and Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol 170: 1021–1027, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ishihara N, Eura Y, and Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117: 6535–6546, 2004. [DOI] [PubMed] [Google Scholar]
- 94. Ishihara N, Fujita Y, Oka T, and Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25: 2966–2977, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto YI, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, and Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11: 958–966, 2009. [DOI] [PubMed] [Google Scholar]
- 96. Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, Mizushima N, Mihara K, and Ishihara N. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol 35: 211–223, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jhun BS, O-Uchi J, Adaniya SM, Mancini TJ, Cao JL, King ME, Landi AK, Ma H, Shin M, Yang D, Xu X, Yoon Y, Choudhary G, Clements RT, Mende U, and Sheu SS. Protein kinase D activation induces mitochondrial fragmentation and dysfunction in cardiomyocytes. J Physiol 596: 827–855, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jokinen R, Marttinen P, Stewart JB, Neil Dear T, and Battersby BJ. Tissue-specific modulation of mitochondrial DNA segregation by a defect in mitochondrial division. Hum Mol Genet 25: 706–714, 2016. [DOI] [PubMed] [Google Scholar]
- 99. Joshi AU, Ebert AE, Haileselassie B, and Mochly-Rosen D. Drp1/Fis1-mediated mitochondrial fragmentation leads to lysosomal dysfunction in cardiac models of Huntington's disease. J Mol Cell Cardiol 127: 125–133, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Joshi AU, Minhas PS, Liddelow SA, Haileselassie B, Andreasson KI, Dorn GW, and Mochly-Rosen D. Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nat Neurosci 22: 1635–1648, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Joshi AU, Saw NL, Shamloo M, and Mochly-Rosen D. Drp1/Fis1 interaction mediates mitochondrial dysfunction, bioenergetic failure and cognitive decline in Alzheimer's disease. Oncotarget 9: 6128–6143, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Joshi AU, Saw NL, Vogel H, Cunnigham AD, Shamloo M, and Mochly-Rosen D. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol Med 10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kennedy EP and Lehninger AL. Oxidation of fatty acids and tricarboxylic acid cycle intermediates by isolated rat liver mitochondria. J Biol Chem 179: 957–963, 1949. [PubMed] [Google Scholar]
- 104. Kirichenko TV, Sobenin IA, Khasanova ZB, Orekhova VA, Melnichenko AA, Demakova NA, Grechko AV, Orekhov AN, Ble Castillo JL, and Shkurat TP. Data on association of mitochondrial heteroplasmy and cardiovascular risk factors: comparison of samples from Russian and Mexican populations. Data Br 18: 16–21, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kiyuna LA, Albuquerque RPE, Chen CH, Mochly-Rosen D, and Ferreira JCB. Targeting mitochondrial dysfunction and oxidative stress in heart failure: challenges and opportunities. Free Radic Biol Med 129: 155–168, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kleele T, Rey T, Winter J, Zaganelli S, Mahecic D, Perreten Lambert H, Ruberto FP, Nemir M, Wai T, Pedrazzini T, and Manley S. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 593: 435–439, 2021. [DOI] [PubMed] [Google Scholar]
- 107. Kohlhaas M and Maack C. Adverse bioenergetic consequences of Na+-Ca2+ exchanger-mediated Ca2+ influx in cardiac myocytes. Circulation 122: 2273–2280, 2010. [DOI] [PubMed] [Google Scholar]
- 108. Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, MacArtney TJ, Deak M, Knebel A, Alessi DR, and Muqit MMK. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2: 120080, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. König T, Nolte H, Aaltonen MJ, Tatsuta T, Krols M, Stroh T, Langer T, and McBride HM. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat Cell Biol 23: 1271–1286, 2021. [DOI] [PubMed] [Google Scholar]
- 110. Kornfeld OS, Qvit N, Haileselassie B, Shamloo M, Bernardi P, and Mochly-Rosen D. Interaction of mitochondrial fission factor with dynamin related protein 1 governs physiological mitochondrial function in vivo. Sci Rep 8: 1–9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Korwitz A, Merkwirth C, Richter-Dennerlein R, Tröder SE, Sprenger HG, Quirós PM, López-Otín C, Rugarli EI, and Langer T. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J Cell Biol 212: 157–166, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, and Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science (80-) 305: 858–862, 2004. [DOI] [PubMed] [Google Scholar]
- 113. Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, and Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510: 162–166, 2014. [DOI] [PubMed] [Google Scholar]
- 114. Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova A, Perriard E, Perriard JC, Larsen T, Pedrazzini T, and Krek W. Activation of a HIF1α-PPARγ axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab 9: 512–524, 2009. [DOI] [PubMed] [Google Scholar]
- 115. Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyans S, Murphy AN, and Gustafsson ÅB. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem 288: 915–926, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Latorre-Pellicer A, Lechuga-Vieco AV, Johnston IG, Hämäläinen RH, Pellico J, Justo-Méndez R, Fernández-Toro JM, Clavería C, Guaras A, Sierra R, Llop J, Torres M, Criado LM, Suomalainen A, Jones NS, Ruíz-Cabello J, and Enríquez JA. Regulation of mother-to-offspring transmission of mtDNA heteroplasmy. Cell Metab 30: 1120.e5–1130.e5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lauritzen KH, Kleppa L, Aronsen JM, Eide L, Carlsen H, Haugen ØP, Sjaastad I, Klungland A, Rasmussen LJ, Attramadal H, Storm-Mathisen J, and Bergersen LH. Impaired dynamics and function of mitochondria caused by mtDNA toxicity leads to heart failure. Am J Physiol Heart Circ Physiol 309: H434–H449, 2015. [DOI] [PubMed] [Google Scholar]
- 118. Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, and Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47: 547–557, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lee Y, Lee HY, Hanna RA, and Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol 301: 1924–1931, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lemieux H, Semsroth S, Antretter H, Höfer D, and Gnaiger E. Mitochondrial respiratory control and early defects of oxidative phosphorylation in the failing human heart. Int J Biochem Cell Biol 43: 1729–1738, 2011. [DOI] [PubMed] [Google Scholar]
- 121. Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, and Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14: 177–185, 2012. [DOI] [PubMed] [Google Scholar]
- 122. Loschen G, Azzi A, Richter C, and Flohé L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett 42: 68–72, 1974. [DOI] [PubMed] [Google Scholar]
- 123. Losón OC, Song Z, Chen H, and Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24: 659–667, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Maack C and O'Rourke B. Excitation-contraction coupling and mitochondrial energetics. Basic Res Cardiol 102: 369–392, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Manczak M, Kandimalla R, Yin X, and Hemachandra Reddy P. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum Mol Genet 28: 177–199, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Maneechote C, Palee S, Kerdphoo S, Jaiwongkam T, Chattipakorn SC, and Chattipakorn N. Differential temporal inhibition of mitochondrial fission by Mdivi-1 exerts effective cardioprotection in cardiac ischemia/reperfusion injury. Clin Sci 132: 1669–1683, 2018. [DOI] [PubMed] [Google Scholar]
- 127. Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, and Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, and Ganley IG. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab 27: 439.e5–449.e5, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, and Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18: 20–27, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Menezes TN, Ramalho LS, Bechara LRG, and Ferreira JCB. Targeting mitochondrial fission-fusion imbalance in heart failure. Curr Tissue Microenviron Rep 1: 239–247, 2020. [Google Scholar]
- 131. Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, Von Kleist-Retzow JC, Waisman A, Westermann B, and Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 22: 476–488, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature 279: 643–645, 1979. [DOI] [PubMed] [Google Scholar]
- 133. Mishra P and Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 15: 634–646, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148, 1961. [DOI] [PubMed] [Google Scholar]
- 135. Mootha VK, Arai AE, and Balaban RS. Maximum oxidative phosphorylation capacity of the mammalian heart. Am J Physiol Heart Circ Physiol 272: H769–H775, 1997. [DOI] [PubMed] [Google Scholar]
- 136. Mozdy AD, McCaffery JM, and Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–379, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nagashima S, Tábara LC, Tilokani L, Paupe V, Anand H, Pogson JH, Zunino R, McBride HM, and Prudent J. Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science (80-) 367: 1366–1371, 2020. [DOI] [PubMed] [Google Scholar]
- 138. Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, and Hayashi JI. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med 7: 934–940, 2001. [DOI] [PubMed] [Google Scholar]
- 139. Naon D, Zaninello M, Giacomello M, Varanita T, Grespi F, Lakshminaranayan S, Serafini A, Semenzato M, Herkenne S, Hernández-Alvarez MI, Zorzano A, De Stefani D, Dorn GW, and Scorrano L. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc Natl Acad Sci U S A 113: 11249–11254, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Narendra D, Tanaka A, Suen DF, and Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Neuspiel M, Zunino R, Gangaraju S, Rippstein P, and McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem 280: 25060–25070, 2005. [DOI] [PubMed] [Google Scholar]
- 142. Nicholls TJ and Gustafsson CM. Separating and segregating the human mitochondrial genome. Trends Biochem Sci 43: 869–881, 2018. [DOI] [PubMed] [Google Scholar]
- 143. Nissanka N and Moraes CT. Mitochondrial DNA heteroplasmy in disease and targeted nuclease-based therapeutic approaches. EMBO Rep 21: e49612, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, and Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 121: 2012–2022, 2010. [DOI] [PubMed] [Google Scholar]
- 145. Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, and Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Palaniyandi SS, Ferreira JCB, Brum PC, and Mochly-Rosen D. PKCβII inhibition attenuates myocardial infarction induced heart failure and is associated with a reduction of fibrosis and pro-inflammatory responses. J Cell Mol Med 15: 1769–1777, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Palikaras K, Lionaki E, and Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20: 1013–1022, 2018. [DOI] [PubMed] [Google Scholar]
- 148. Pallandi RT, Perry MA, and Campbell TJ. Proarrhythmic effects of an oxygen-derived free radical generating system on action potentials recorded from guinea pig ventricular myocardium: a possible cause of reperfusion-induced arrhythmias. Circ Res 61: 50–54, 1987. [DOI] [PubMed] [Google Scholar]
- 149. Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, and Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Palmer JW, Tandler B, and Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977. [PubMed] [Google Scholar]
- 151. Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O'Shea KM, Riley DD, Lugus JJ, Colucci WS, Lederer WJ, Stanley WC, and Walsh K. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol Cell Biol 31: 1309–1328, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, and Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res 111: 1012–1026, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Papanicolaou KN, Ngoh GA, Dabkowski ER, O'Connell KA, Ribeiro RF, Stanley WC, and Walsh K. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am J Physiol Heart Circ Physiol 302: H167–H179, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Parone PA, Da Druz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, and Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One 3: e3257, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Payne BAI, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, and Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 22: 384–390, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pesch UEA, Leo-Kottler B, Mayer S, Jurklies B, Kellner U, Apfelstedt-Sylla E, Zrenner E, Alexander C, and Wissinger B. OPA1 mutations in patients with autosomal dominant optic atrophy and evidence for semi-dominant inheritance. Hum Mol Genet 10: 1359–1368, 2001. [DOI] [PubMed] [Google Scholar]
- 157. Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, Palacín M, and Zorzano A. The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet 14: 1405–1415, 2005. [DOI] [PubMed] [Google Scholar]
- 158. Pryde KR, Smith HL, Chau KY, and Schapira AHV. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. J Cell Biol 213: 163–171, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Qi X, Qvit N, Su YC, and Mochly-Rosen D. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J Cell Sci 126: 789–802, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Qin F, Siwik DA, Lancel S, Zhang J, Kuster GM, Luptak I, Wang L, Tong X, Kang YJ, Cohen RA, and Colucci WS. Hydrogen peroxide-mediated SERCA cysteine 674 oxidation contributes to impaired cardiac myocyte relaxation in senescent mouse heart. J Am Heart Assoc 2: 1–9, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ramos ES, Motori E, Brüser C, Kühl I, Yeroslaviz A, Ruzzenente B, Kauppila JHK, Busch JD, Hultenby K, Habermann BH, Jakobs S, Larsson NG, and Mourier A. Mitochondrial fusion is required for regulation of mitochondrial DNA replication. PLoS Genet 15: 1–28, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, and Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 113: 4039–4044, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Rogov VV, Suzuki H, Marinković M, Lang V, Kato R, Kawasaki M, Buljubašić M, Šprung M, Rogova N, Wakatsuki S, Hamacher-Brady A, Dötsch V, Dikic I, Brady NR, and Novak I. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci Rep 7: 1–12, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, and Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res 80: 30–39, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, Fragaki K, Cano A, Pouget J, Pellissier JF, Procaccio V, Chabrol B, and Paquis-Flucklinger V. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy “plus” phenotype. Brain 135: 23–34, 2012. [DOI] [PubMed] [Google Scholar]
- 166. Ruprecht JJ, King MS, Zögg T, Aleksandrova AA, Pardon E, Crichton PG, Steyaert J, and Kunji ERS. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 176: 435.e15–447.e15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Russell RR, Bergeron R, Shulman GI, and Young LH. Translocation of myocardial GLUT-4 and increased glucose uptake through activation of AMPK by AICAR. Am J Physiol Heart Circ Physiol 277: 643–649, 1999. [DOI] [PubMed] [Google Scholar]
- 168. Scaglia F, Wong LJC, Vladutiu GD, and Hunter J V. Predominant cerebellar volume loss as a neuroradiologic feature of pediatric respiratory chain defects. Am J Neuroradiol 26: 1675–1680, 2005. [PMC free article] [PubMed] [Google Scholar]
- 169. Schrepfer E and Scorrano L. Mitofusins, from mitochondria to metabolism. Mol Cell 61: 683–694, 2016. [DOI] [PubMed] [Google Scholar]
- 170. Sebastiani M, Giordano C, Nediani C, Travaglini C, Borchi E, Zani M, Feccia M, Mancini M, Petrozza V, Cossarizza A, Gallo P, Taylor RW, and D'Amati G. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J Am Coll Cardiol 50: 1362–1369, 2007. [DOI] [PubMed] [Google Scholar]
- 171. Sekine S and Youle RJ. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 16: 2, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Sharp WW, Fang YH, Han M, Zhang HJ, Hong Z, Banathy A, Morrow E, Ryan JJ, and Archer SL. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J 28: 316–326, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Shenouda SM, Widlansky ME, Chen K, Xu G, Holbrook M, Tabit CE, Hamburg NM, Frame AA, Caiano TL, Kluge MA, Duess MA, Levit A, Kim B, Hartman ML, Joseph L, Shirihai OS, and Vita JA. Altered mitochondrial dynamics contributes to endothelial dysfunction in diabetes mellitus. Circulation 124: 444–453, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Sherratt EJ, Thomas AW, and Alcolado JC. Mitochondrial DNA defects: a widening clinical spectrum of disorders. Clin Sci 92: 225–235, 1997. [DOI] [PubMed] [Google Scholar]
- 175. Shirane M and Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol 5: 28–37, 2003. [DOI] [PubMed] [Google Scholar]
- 176. Sladen PE, Perdigão PRL, Salsbury G, Novoselova T, van der Spuy J, Chapple JP, Yu-Wai-Man P, and Cheetham ME. CRISPR-Cas9 correction of OPA1 c.1334G>A: p.R445H restores mitochondrial homeostasis in dominant optic atrophy patient-derived iPSCs. Mol Ther Nucleic Acids 26: 432–443, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Song M, Franco A, Fleischer JA, Zhang L, and Dorn GW. Abrogating mitochondrial dynamics in mouse hearts accelerates mitochondrial senescence. Cell Metab 26: 872–883, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Song M, Mihara K, Chen Y, Scorrano L, and Dorn GW. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab 21: 273–286, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Song Z, Ghochani M, McCaffery JM, Frey TG, and Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell 20: 3525–3532, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Spinelli JB and Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20: 745–754, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Spinelli JB, Rosen PC, Sprenger H-G, Puszynska AM, Mann JL, Roessler JM, Cangelosi AL, Henne A, Condon KJ, Zhang T, Kunchok T, Lewis CA, Chandel NS, and Sabatini DM. Fumarate is a terminal electron acceptor in the mammalian electron transport chain. Science 374: 1227–1237, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. St-Pierre J, Buckingham JA, Roebuck SJ, and Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem 277: 44784–44790, 2002. [DOI] [PubMed] [Google Scholar]
- 183. Stanley BA, Sivakumaran V, Shi S, McDonald I, Lloyd D, Watson WH, Aon MA, and Paolocci N. Thioredoxin reductase-2 is essential for keeping low levels of H 2O 2 emission from isolated heart mitochondria. J Biol Chem 286: 33669–33677, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. De Stefani D, Raffaello A, Teardo E, Szabó I, and Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Stewart JB and Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 16: 530–542, 2015. [DOI] [PubMed] [Google Scholar]
- 186. Suen DF, Narendra DP, Tanaka A, Manfredi G, and Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A 107: 11835–11840, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, and Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367–1380, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Taylor RW and Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 6: 389–402, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gldlöf S, Oldfors A, Wibom R, Törnell J, Jacobs HT, and Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423, 2004. [DOI] [PubMed] [Google Scholar]
- 190. Tsutsui H, Kinugawa S, and Matsushima S. Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol 301: 2181–2190, 2011. [DOI] [PubMed] [Google Scholar]
- 191. Tuunanen H, Engblom E, Naum A, Någren K, Hesse B, Airaksinen KEJ, Nuutila P, Iozzo P, Ukkonen H, Opie LH, and Knuuti J. Free fatty acid depletion acutely decreases cardiac work and efficiency in cardiomyopathic heart failure. Circulation 114: 2130–2137, 2006. [DOI] [PubMed] [Google Scholar]
- 192. Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, and Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ueta CB, Campos JC, Albuquerque RPE, Lima VM, Disatnik MH, Sanchez AB, Chen CH, De Medeiros MHG, Yang W, Mochly-Rosen D, and Ferreira JCB. Cardioprotection induced by a brief exposure to acetaldehyde: role of aldehyde dehydrogenase 2. Cardiovasc Res 114: 1006–1015, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Ugolini GS, Pavesi A, Rasponi M, Fiore GB, Kamm R, and Soncini M. Human cardiac fibroblasts adaptive responses to controlled combined mechanical strain and oxygen changes in vitro. Elife 6: 1–20, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Umbria M, Ramos A, Aluja MP, and Santos C. The role of control region mitochondrial DNA mutations in cardiovascular disease: stroke and myocardial infarction. Sci Rep 10: 2766, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ussher JR, Wang W, Gandhi M, Keung W, Samokhvalov V, Oka T, Wagg CS, Jaswal JS, Harris RA, Clanachan AS, Dyck JRB, and Lopaschuk GD. Stimulation of glucose oxidation protects against acute myocardial infarction and reperfusion injury. Cardiovasc Res 94: 359–369, 2012. [DOI] [PubMed] [Google Scholar]
- 197. Vincent AE, White K, Davey T, Philips J, Ogden RT, Lawess C, Warren C, Hall MG, Ng YS, Falkous G, Holden T, Deehan D, Taylor RW, Turnbull DM, and Picard M. Quantitative 3D mapping of the human skeletal muscle mitochondrial network. Cell Rep 26: 996–1009, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wagner S, Rokita AG, Anderson ME, and Maier LS. Redox regulation of sodium and calcium handling. Antioxid Redox Signal 18: 1063–1077, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Wai T, García-Prieto J, Baker MJ, Merkwirth C, Benit P, Rustin P, Rupérez FJ, Barbas C, Ibañez B, and Langer T. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science (80-) 350: aad0116, 2015. [DOI] [PubMed] [Google Scholar]
- 200. Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884, 2010. [DOI] [PubMed] [Google Scholar]
- 201. Woodall BP, Orogo AM, Najor RH, Cortez MQ, Moreno ER, Wang H, Divakaruni AS, Murphy AN, and Gustafsson ÅB. Parkin does not prevent accelerated cardiac aging in mitochondrial DNA mutator mice. JCI Insight 4: e127713, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. World Health Organization (WHO). Cardiovascular Diseases (CVDs) Fact Sheet. 2017. http://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
- 203. Yang-ja L, Seon-Yong J, Mariusz K, Smith CL, and Richard JY. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell 15: 5001–5011, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata SI, and Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal 2: ra47, 2009. [DOI] [PubMed] [Google Scholar]
- 205. Yoon Y, Krueger EW, Oswald BJ, and McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 23: 5409–5420, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Yu R, Jin S, Lendahl U, Nistér M, and Zhao J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J 38: 1–21, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Zhang D, Li Y, Heims-Waldron D, Bezzerides V, Guatimosim S, Guo Y, Gu F, Zhou P, Lin Z, Ma Q, Liu J, Wang DZ, and Pu WT. Mitochondrial cardiomyopathy caused by elevated reactive oxygen species and impaired cardiomyocyte proliferation. Circ Res 122: 74–87, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Zhou H, Zhu P, Wang J, Zhu H, Ren J, and Chen Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2α-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ 25: 1080–1093, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, De Jonghe P, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battologlu E, Polyakov AV., Timmerman V, Schröder JM, and Vance JM.. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36: 449–451, 2004. [DOI] [PubMed] [Google Scholar]