Abstract
The cpkA gene encoding a second (α) subunit of archaeal chaperonin from Pyrococcus kodakaraensis KOD1 was cloned, sequenced, and expressed in Escherichia coli. Recombinant CpkA was studied for chaperonin functions in comparison with CpkB (β subunit). The effect on decreasing the insoluble form of proteins was examined by coexpressing CpkA or CpkB with CobQ (cobyric acid synthase from P. kodakaraensis) in E. coli. The results indicate that both CpkA and CpkB effectively decrease the amount of the insoluble form of CobQ. Both CpkA and CpkB possessed the same ATPase activity as other bacterial and eukaryal chaperonins. The ATPase-deficient mutant proteins CpkA-D95K and CpkB-D95K were constructed by changing conserved Asp95 to Lys. Effect of the mutation on the ATPase activity and CobQ solubilization was examined. Neither mutant exhibited ATPase activity in vitro. Nevertheless, they decreased the amount of the insoluble form of CobQ by coexpression as did wild-type CpkA and CpkB. These results implied that both CpkA and CpkB could assist protein folding for nascent protein in E. coli without requiring energy from ATP hydrolysis.
Chaperonins have been classified into two distinct groups, I and II (12). Group I chaperonins (the GroEL family) were found in bacteria, chloroplasts, and mitochondria of eukaryotic cells. They are composed of two kinds of subunits, with molecular masses of about 60 and 10 kDa, and form the sevenfold rotational symmetric double-ring structures. Members of group II chaperonins (TCP-1 [t-complex polypeptide-1]; the thermosome family) occur in the cytosol of eukaryotes and archaea. They also form toroidal structures with variations in the numbers of subunits. The eukaryotic cytosolic chaperonin complex (CCT [chaperonin-containing TCP-1]) consists of up to eight or nine kinds of TCP-1 units (15). It has been shown that CCT is involved in the folding of actin, tubulin, and firefly luciferase in an ATPase-dependent manner in vitro (4, 6, 30) and that newly synthesized actin and tubulin monomers are bound by CCT in vivo (25). Archaeal chaperonins, the thermophilic factor 55 (TF55) of Sulfolobus shibatae (27), thermosome of Pyrodictium occultum (20), and Thermococcus strain KS-1 (33), were found to be members of a related family of high-molecular-mass ATPase complexes. They are able to bind several denatured polypeptides in vitro (8, 27, 28) in an ATPase-dependent manner and are ubiquitous in the archaea. Most archaeal chaperonins, except those of methanogens, consist of two kinds of subunits with diverse stoichiometry and rotational symmetry (10). The subunit stoichiometry of the ninefold symmetric chaperonin complex from Sulfolobus solfataricus is reported to be 2:1 (14). Chaperonins from P. occultum and Thermoplasma acidophilum appear to contain two subunits in 1:1 stoichiometry which form two stacked rings with eightfold symmetry. On the other hand, the structures of chaperonins from methanogens seem to be different from those of other archaeal chaperonins. The chaperonin from Methanopyrus kandleri was found to be a homo-oligomer with eightfold symmetry (1). In the case of Methanococcus jannaschii, only one gene was found in the genome (2).
Although the functional consequences of ATP binding and hydrolysis for folding of polypeptide substrate seem to have been conserved between the catalytic cycles of group I and group II chaperonins, the effects of nucleotides on the overall structure of the two chaperonin groups apparently differ. ATP binding drives group II chaperonins from the open, substrate binding conformation into the closed conformation where substrate folds in the central cavity (13). ATP hydrolysis would allow the chaperonin to return to the open conformation with subsequent release of folded substrate. In contrast, GroEL, group I chaperonins, binds its substrates in a compact conformation, and ATP causes its apical domains to move outward to allow binding of GroES and thus closure of the cavity. ATP binding and hydrolysis seem to be essential for chaperonin functions. On the other hand, ATP-independent activity of molecular chaperonin has been reported. Upon incubation with the nonhydrolysis ATP analog AMP-PNP, α-tubulin previously bound to TRiC-CCT undergoes at least partial folding without releasing from the chaperonin (3). In addition, the ATPase activity of chaperonin from M. kandleri was not detected (1). Our previous studies also showed that recombinant β subunit of chaperonin-like protein from Pyrococcus kodakaraensis KOD1 (CpkB) prevents thermal denaturation and enhances thermostability of yeast (Saccharomyces cerevisiae) alcohol dehydrogenase in the absence of ATP, when CpkB is present in excess (31). In the present study, the gene encoding the chaperonin α subunit was cloned from P. kodakaraensis KOD1, expressed in E. coli, and examined for biochemical properties as a molecular chaperone. ATPase-deficient mutant proteins were constructed, and their characteristics were compared with those of wild-type chaperonins.
Cloning and sequencing of the chaperonin gene.
Chaperonins of most microorganisms are known to contain two kinds of subunits. Archaeal chaperonins are likely to form hetero-oligomeric double-ring structures. We previously reported cloning and sequencing analysis of the cpkB gene encoding the β subunit of KOD1 chaperonin (31). In order to obtain the α subunit gene from KOD1, PCR using primers based on the conserved regions of group II chaperonins [primer 1, 5′-GGGNGTACCACNAT(T/A/C)ACNAA(T/C)GA(T/C)GGNGC-3′; primer 2, 5′-GGCATNCC(G/A)AA(G/A)AGGAT(A/T/C) GA(G/A)AA(T/C)GC-3′] was performed. Chromosomal DNA (1 μg) and 200 pmol of each primer in 100 μl of reaction buffer were used for PCR. Southern hybridization was performed using the PCR product as a probe. This probe strongly hybridized with two distinct HindIII fragments whose sizes are 4.2 and 1.8 kb. These fragments were cloned separately into pUC19 and sequenced. Sequence analysis revealed that the 4.2-kb fragment contained the entire cpkB gene. The 1.8-kb fragment possessed an open reading frame encoding a protein with 549 amino acid residues, giving a predicted molecular weight of 59,166.4. The obtained gene was designated cpkA, which stands for α subunit of chaperonin-like protein from P. kodakaraensis KOD1. The constructed plasmid harboring the 1.8-kb fragment was termed pCPA.
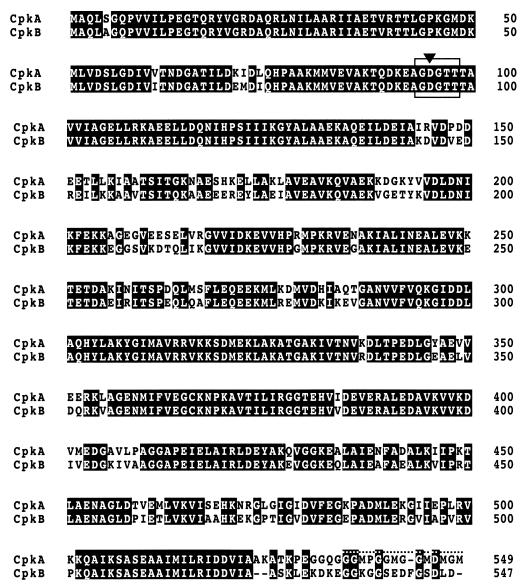
The deduced amino acid sequence from the newly identified open reading frame (cpkA) showed a high degree of similarity (77.1% identity) to that of CpkB (Fig. 1). The most conspicuous difference between the two chaperonins was found at the carboxyl termini. CpkA has a glycine-methionine (G-M) motif, which is typically found in bacterial GroEL chaperonin (18), while the G-M motif was not found in CpkB or other eukaryal chaperonin TCP-1. This region of cpkA might have been transferred from the bacterial counterpart in the course of evolution. Another difference between the two genes was observed in codon usage. When codon usage was examined using 20 cloned genes, some codons were not frequently utilized in KOD1. The typical rare codons for strain KOD1, such as CTT for Leu, AGT for Ser, and GCA for Ala, were found in the 5′ region of the cpkA gene (data not shown). This fact implied that the translational efficiency of cpkA is lower than that of cpkB.
FIG. 1.
Comparison of deduced amino acid sequences of CpkA and CpkB chaperonin subunits from P. kodakaraensis KOD1. White letters on a black background are amino acids identical between CpkA and CpkB. The G-M motif region is indicated by a dotted line above the sequence. The conserved region for nucleotide binding is boxed. The arrowhead indicates Asp95 for site-directed mutagenesis.
In order to determine whether these genes were located close together in chromosomal DNA of strain KOD1, Southern hybridization was performed. A physical map of the KOD1 chromosome was previously constructed for AscI, PmeI, and PacI (5). In order to determine the location of cpkA, cloned HindIII fragment (1,804 bp) was used as a probe for Southern hybridization. Major signals were detected at the AscI-D, PmeI-D, and PacI-D fragments, and minor signals were detected at the AscI-E, PmeI-A, and PacI-A fragments. When the DNA fragment which carries cpkB was used as a probe for Southern hybridization, the AscI-E, PmeI-A, and PacI-A fragments were highlighted. As mentioned above, cpkA shows high sequence identity to cpkB. The weak signals of AscI-E, PmeI-A, and PacI-A detected by the cpkA probe were considered to be due to high sequence identity between cpkA and cpkB. The locus of the cpkA gene was defined at the overlapped region of AscI-D, PmeI-D, and PacI-D. Two genes were located at distinct loci on the physical map.
Expression and purification.
The region for cpkA was amplified by PCR with the following set of forward and reverse primers to introduce restriction enzyme recognition sites: CPAU (5′-TTCCATGGCACAGCTTAGTGGACAGCCG GT-3′) and CPAR′ (5′-ATGGATCCTGCTGGAAGGAAAAGAGAAGTG-3′). The forward and reverse primers possess additional NcoI and BamHI sites at the 5′-terminal regions, respectively, as shown in italic letters in the sequences. The amplified DNA fragment was inserted into the NcoI and BamHI sites of pET-8c. E. coli BL21(DE3) cells were transformed by the recombinant plasmid, and overexpression was performed. However, expressed CpkA was not detected in the cell extract. Some codons of the 5′ terminus are occupied with rare codons which are not efficiently utilized in E. coli. It has been reported that rare codons located near the 5′ end of the gene are negatively effective for an efficient translation (7, 16). In order to achieve the efficient expression, several rare codons for N-terminal amino acids were changed to codons which are frequently used in E. coli. CTT for Leu4, AGT for Ser5, and GGA for Gly6 were replaced by CTG, AGC, and GGC, respectively. Primer CPKU2 (5′-TTCCATGGCACAGCTGAGCGGCACAGCCGGT-3′) was used for PCR instead of CPAU2. The constructed plasmid was designated pCPAE. E. coli BL21(DE3) cells harboring pCPAE efficiently expressed cpkA. The expression plasmid for CpkB carrying cpkB gene was constructed as described by Yan et al. (31). The constructed plasmid was designated pECPK. E. coli BL21(DE3) cells carrying each plasmid were grown at 37°C in NZCYM medium (1% NZ amine, 0.5% NaCl, 0.5% yeast extract, 0.1% Casamino Acids, 0.2% MgSO4 · 7H2O adjusted to pH 7.0 with NaOH) containing ampicillin (50 μg/ml) until the optical density at 660 nm reached 0.4. Chaperonin expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. After harvesting the cells by centrifugation (6,000 × g, 20 min), the pellet was frozen, thawed, and resuspended in buffer A (50 mM Tris-HCl [pH 7.5], 30 mM NaCl, 1 mM dithiothreitol [DTT], 20% [vol/vol] glycerol). The cells were disrupted by sonication in buffer A and then centrifuged at 24,000 × g for 1 h at 4°C. CpkA or CpkB was recovered in a soluble fraction, and the crude extract was treated at 80°C for 20 min followed by centrifugation (24,000 × g, 1 h). Most proteins derived from the host E. coli cell were precipitated and removed as an insoluble inclusion complex.
Relationship between ATP hydrolysis and chaperonin function.
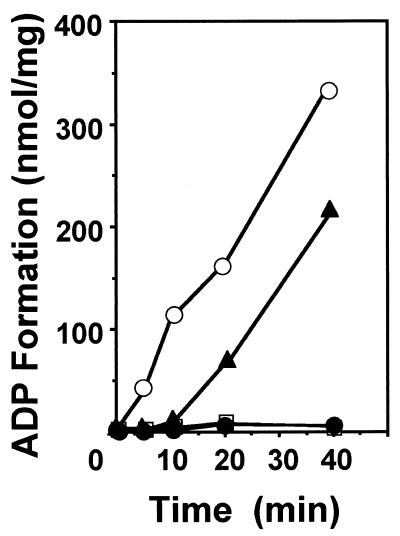
ATP hydrolysis is considered important for the release of folded proteins from chaperonin. Based on the comparative sequence analysis, the cpkA and cpkB genes possess the conserved region which is related to putative nucleotide binding (the GDGTT motif at amino acids 94 to 98; Fig. 1). Recombinant CpkA and CpkB were examined for ATPase activity. An ATPase assay was performed by monitoring ADP formation on polyethylenimine-cellulose thin-layer sheets in accordance with a previously reported procedure (22, 23). As expected, both CpkA and CpkB exhibit ATPase activity at 80°C (Fig. 2). These ATPase activities were maintained even at 90°C (data not shown).
FIG. 2.
ATPase activities of the wild-type and mutant chaperonins at 80°C. ATPase activity was determined in reaction mixtures containing 40 mM HEPES [pH 7.2], 75 mM KCl, 4.5 mM MgCl2, 1.5 mM CaCl2, and 1 mM ATP in a total volume of 20 μl including [α-32P]ATP (400 Ci/mmol). The reaction was started by the addition of the extracts and terminated by rapid cooling to 0°C. ATP hydrolysis was examined by spotting an aliquot (2 μl) on polyethylenimine-cellulose thin-layer sheets (Macherey-Nagel, Duren, Germany). The substrate and products of the reaction were separated by one-dimensional chromatography using 1 M LiCl. The spots were cut out, and the radioactivity was determined by liquid scintillation counting. Symbols: ▴, CpkA; ○, CpkB; ●, CpkA-D95K; □, CpkB-D95K.
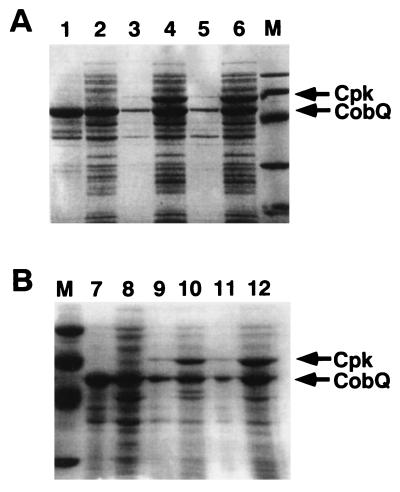
In order to confirm the possible chaperonin function of CpkA, the effect of CpkA on decreasing insoluble foreign protein was examined. When some foreign proteins are overexpressed in E. coli, insoluble inclusion bodies are often formed. It has been observed that molecular chaperone is effective for decreasing insoluble proteins when it is coexpressed (32). The cobQ gene of KOD1, which encodes cobyric acid synthase, forms an insoluble inclusion complex when it is overexpressed in E. coli (31). It is reported that CpkB is functional in vivo and is effective to decrease the amount of the insoluble form of CobQ in E. coli by coexpression. The cobQ gene was previously cloned in pET-8c, and the derivative plasmid was designated pCOB (31). The NruI-ClaI fragments of plasmid pCPAE and pECPK were recloned into the respective sites of plasmid pACYC184, which is compatible with pET-8c, and the constructed plasmids were designated pCPAE2 and pCPK, respectively. Both cobQ and the chaperonin genes are inducible with IPTG by the use of a T7 promoter system. E. coli BL21(DE3) cells were transformed by plasmids pCOB and pCPAE2 or by pCOB and pCPK and were grown in NZCYM medium containing ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml). Overexpression of proteins was induced by the addition of IPTG (1 mM) for 4 h. Cells were then harvested by centrifugation. The pellet was suspended in 1 ml of buffer A. After the cells were disrupted by sonication and centrifuged, the supernatant was rescued as the soluble fraction. The pellet was washed with 1 ml of buffer A and suspended in 1 ml of sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM DTT, 2% sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 10% glycerol), boiled for 5 min, and centrifuged. The supernatant was recovered in an insoluble fraction. Each fraction (20 μl) was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie brilliant blue R-250 staining. When CpkA was coexpressed with CobQ, a significant amount of insoluble CobQ was kept soluble, indicating that CpkA also functions to decrease the insoluble form (Fig. 3A, lanes 3 and 4). CpkA is thought to trap unfolded forms of polypeptides and to correct them in accordance with properly folded ones in E. coli. CpkA and CpkB seem to function to prevent CobQ insolubilization, thus keeping its form soluble.
FIG. 3.
Increased solubility of CobQ protein by coexpression with wild-type (A) and mutant (B) chaperonins in E. coli. Soluble and insoluble forms of CobQ (cobyric acid synthase from P. kodakaraensis) were monitored by SDS-PAGE. Lanes: 1, insoluble fraction from E. coli (pCOBQ); 2, soluble fraction from E. coli (pCOB); 3, insoluble fraction from E. coli (pCPAE2/pCOB): 4, soluble fraction from E. coli (pCPAE2/pCOB); 5, insoluble fraction from E. coli (pCPK/pCOB); 6, soluble fraction from E. coli (pCPK/pCOB); 7, insoluble fraction from E. coli (pCOB); 8, soluble fraction from E. coli (pCOB); 9, insoluble fraction from E. coli (pCPAE2-D95K/pCOB); 10, soluble fraction from E. coli (pCPAE2-D95K/pCOB); 11, insoluble fraction from E. coli (pCPK-D95K/pCOB); 12, soluble fraction from E. coli (pCPK-D95K/pCOB); M, molecular mass markers (94 kDa, rabbit muscle phosphorylase; 67 kDa, bovine serum albumin; 43 kDa, egg white ovalbumin; 30 kDa, bovine erythrocyte carbonic anhydrase; 20.1 kDa, soybean trypsin inhibitor).
In the GroEL-GroES chaperonin system, the binding and releasing of target proteins are associated with ATP hydrolysis. Chaperonin TCP-1 and thermophilic factor TF55 from the thermophilic archaeon S. shibatae also require ATP for refolding of denatured proteins. Several protein folding cycles have been proposed for bacterial chaperonin (17, 19, 24, 26, 29). Quaite-Randall et al. (21) have suggested the conformational cycle of the archaeosome, which includes a change in conformation and in the oligomerization state. According to their model, the binding or hydrolysis of ATP acts as a switch between two conformational forms of chaperonin, open and closed complexes. They suggested that as previously reported for GroEL (26), the thermodynamic barriers separating protein-bound and -free archaeosome states are overcome by ATP hydrolysis. Our previous studies, however, revealed that CpkB functions as a chaperonin in the absence of ATP when it is present in an excess amount. In order to examine whether ATPase activity is necessary for chaperonin functions, ATPase-deficient mutant proteins CpkA-D95K (in which Asp95 was replaced by Lys) and CpkB-D95K (in which Asp95 was replaced by Lys) were constructed by site-directed mutagenesis. For CpkA-D95K construction, primers CPKU2 and DAK2′ (5′-AGACTCAGGACAAGGAGGCCGGTAAAGGTACTACC-3′) were annealed with plasmid pCPA, and an intermediate DNA (about 300 bp) was produced by PCR. Primers CPAR′ and DAK1′ (5′-ATGACGACGGCAGTGGTAGTACCTTTACCGGCCTC-3′) were also annealed with pCPA, and another intermediate DNA (1.5 kbp) was produced by PCR. Those intermediates were joined by PCR with the two outer primers CPKU2 and CPAR′. The synthesized DNA was then digested with NcoI and BamHI and introduced between the NcoI and BamHI sites of pET-8c. The resulting plasmid was named pCPAE-D95K. For CpkB-D95K construction, primers KODHSPF, KODHSPB, DBK1′ (5′-AGACTCAGGACAAGGAGGCCGGTAAAGGAACCACC-3′), and DBK2′ (5′-ATGACAACGGCAGTGGTGGTTCCTTTACCGGCCTC-3′) were replaced with CPKU2, CPAR′, DAK1′, and DAK2′, respectively, using plasmid pECPK (31) as a template for PCR. The construct was designated pECPK-D95K. As shown in Fig. 2, neither mutant did exhibited ATPase activity. In order to confirm their chaperonin function, the coexpression effect on preventing insoluble CobQ formation was examined. The NruI-ClaI fragments of plasmid pCPAE-D95K and pECPK-D95K were recloned into the respective sites of plasmid pACYC184, which is compatible with pET-8c, and the constructed plasmids were designated pCPAE2-D95K and pCPK-D95K, respectively. Although CpkA-D95K and CpkB-D95K exhibited no ATPase activity, both mutants are functional for decreasing insoluble CobQ, as shown in Fig. 3B. This result suggested that CpkA and CpkB are functional in the absence of ATP. The chaperonin function of CpkA-D95K or CpkB-D95K might be cooperative action with a host-derivative chaperonin, such as GroEL.
At present, the exact reason why CpkA and CpkB do not require ATPase activity for their chaperonin functions is unclear. ATP-independent action was also observed for molecular chaperonin from Thermococcus sp. strain KS-1 (32a). ATP is easily degraded at high temperatures (the half-lives for ATP and ADP at 90°C were 115 and 750 min, respectively [11]). At the extremely high temperatures at which hyperthermophiles grow, available ATP would be scarce. In the present study, the functions of CpkA and CpkB were examined only for their solubilization activities for the recombinant protein expressed in E. coli. Further biochemical analysis in vitro is in progress.
Nucleotide sequence accession number.
The cpkA gene produced in this study has been assigned GenBank /EMBL/DDBJ accession no. AB018432.
Acknowledgments
This work was supported by a grant from CREST (Core Research for Evolutional Science and Technology, Japan).
REFERENCES
- 1.Ändra S, Frey G, Nitsch M, Baumeister W, Stetter K O. Purification and structural characterization of the thermosome from the hyperthermophilic archaeum Methanopyrus kandleri. FEBS Lett. 1996;379:127–131. doi: 10.1016/0014-5793(95)01493-4. [DOI] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraster C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Farr G W, Scharl E C, Schumacher R J, Sondek S, Horwich A L. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 4.Frydman J, Nimmesgern E, Erdjument Bromage H, Walls J S, Tempst P, Hartl F U. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara S, Okuyama S, Imanaka T. The world of archaea: genome analysis, evolution and thermostable enzymes. Gene. 1996;179:165–170. doi: 10.1016/s0378-1119(96)00428-3. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Thomas J O, Chow R L, Lee G H, Cowan N J. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 7.Goldman E, Rosenberg A H, Zubay G, Studier F W. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J Mol Biol. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 8.Guagliardi A, Cerchia L, Bertolucci S, Rossi M. The chaperonin from the archaeon Sulfolobus solfatalicus promotes correct refolding and prevents thermal denaturation in vitro. Protein Sci. 1994;3:1436–1443. doi: 10.1002/pro.5560030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemmingsen S M, Woolford C, van der Vies S M, Tilly K, Dennis D T, Georgopoulos C P, Hendrix R W, Ellis R J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 10.Kagawa H K, Osipiuk J, Maltsev N, Overbeek R, Quaite-Randall E, Joachimiak A, Trent J D. The 60 kDa heat shock proteins in the hyperthermophilic archaeon Sulfolobus shibatae. J Mol Biol. 1995;253:712–725. doi: 10.1006/jmbi.1995.0585. [DOI] [PubMed] [Google Scholar]
- 11.Kengen S W M, Stams A J M, de Vos W M. Sugar metabolism of hyperthermophiles. FEMS Microbiol Rev. 1996;18:119–137. [Google Scholar]
- 12.Kim S, Willson K R, Horwich A L. Cytosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 13.Klumpp M, Baumeister W. The thermosome: archetype of group II chaperonins. FEBS Lett. 1998;430:73–77. doi: 10.1016/s0014-5793(98)00541-9. [DOI] [PubMed] [Google Scholar]
- 14.Knapp S, Schmidt-Krey I, Hebert H, Bergman T, Jörnvall H, Landenstein R. The molecular chaperonin TF55 from the thermophilic archaeon Sulfolobus solfataricus. J Mol Biol. 1994;242:397–407. doi: 10.1006/jmbi.1994.1590. [DOI] [PubMed] [Google Scholar]
- 15.Kubota H, Hynes G, Willson K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biol. 1995;230:3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. [DOI] [PubMed] [Google Scholar]
- 16.Makrides S C. Strategies for achieving high-level expression of genes in E. coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin J, Mayhew M, Langer T, Hartl F U. The reaction cycle of GroEL and GroES in chaperonin-assisted protein folding. Nature. 1993;366:228–233. doi: 10.1038/366228a0. [DOI] [PubMed] [Google Scholar]
- 18.McLennan N F, Girshovich A S, Lissin N M, Charters Y, Masters M. The strongly conserved carboxyl-terminus glycine-methionine motif of Escherichia coli GroEL chaperonin is dispensable. Mol Microbiol. 1993;7:49–58. doi: 10.1111/j.1365-2958.1993.tb01096.x. [DOI] [PubMed] [Google Scholar]
- 19.Mendoza J A, Demeler B, Horowitz P M. Alteration of the quaternary structure of cpn60 modulates chaperonin-assisted folding. J Biol Chem. 1994;269:2447–2451. [PubMed] [Google Scholar]
- 20.Phipps B M, Hoffmann A, Stetter K O, Baumeister W. A novel ATPase complex selectively accumulated upon heat shock is a major cellular component of thermophilic archaebacteria. EMBO J. 1991;10:1711–1722. doi: 10.1002/j.1460-2075.1991.tb07695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quaite-Randall E, Trent J D, Josephs R, Joachimiak A. Conformational cycle of the archaeosome, a TCP-1 like chaperonin from Sulfolobus shibatae. J Biol Chem. 1995;270:28818–28823. doi: 10.1074/jbc.270.48.28818. [DOI] [PubMed] [Google Scholar]
- 22.Rothman J E. Polypeptide chain binding proteins: catalysis of protein folding and related processes in cell. Cell. 1989;59:591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 23.Sakoda H, Imanaka T. Cloning and sequencing of the gene coding for alcohol dehydrogenase of Bacillus stearothermophilus and rational shift of the optimum pH. J Bacteriol. 1992;174:1397–1402. doi: 10.1128/jb.174.4.1397-1402.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt M, Rutkat K, Rachel R, Pfeifer G, Jaenicke R, Viitanen P, Lorimer G, Buchner J. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265:656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- 25.Sternlicht H, Farr G W, Sternlicht M L, Driscoll J K, Willson K, Yaffe M B. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd M J, Viitanen P V, Lorimer G H. Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 27.Trent J D, Nimmesgern E, Wall J S, Hartl F U, Horwich A L. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature. 1991;354:490–493. doi: 10.1038/354490a0. [DOI] [PubMed] [Google Scholar]
- 28.Waldmann T, Nimmesgern E, Nitsch M, Peters J, Pfeifer G, Muller S, Kellermann J, Engel A, Hartl F U, Baumeister W. The thermosome of Thermoplasma acidophilum and its relationship to the eukaryotic chaperonin TRiC. Eur J Biochem. 1995;227:848–856. doi: 10.1111/j.1432-1033.1995.tb20210.x. [DOI] [PubMed] [Google Scholar]
- 29.Weissman J S, Kashi Y, Fenton W A, Horwich A L. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 30.Yaffe M B, Farr G W, Miklos D, Horwich A L, Sternlicht M L, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 31.Yan Z, Fujiwara S, Kohda K, Takagi M, Imanaka T. In vitro stabilization and in vivo solubilization of foreign proteins by the β subunit of a chaperonin from the hyperthermophilic archaeon Pyrococcus sp. strain KOD1. Appl Environ Microbiol. 1997;63:785–789. doi: 10.1128/aem.63.2.785-789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasukawa T, Kanei-Ishii C, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of bacterial thioredoxin. J Biol Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- 32a.Yohda, M. Personal communication.
- 33.Yoshida T, Yohda M, Iida T, Maruyama T, Taguchi H, Yazaki K, Ohta T, Odaka M, Endo I, Kagawa Y. Structural and functional characterization of homooligomeric complexes of α and β chaperonin subunits from the hyperthermophilic archaeum Thermococcus strain KS-1. J Mol Biol. 1997;273:635–645. doi: 10.1006/jmbi.1997.1337. [DOI] [PubMed] [Google Scholar]