Abstract
During RNA replication, coronaviruses require proofreading to maintain the integrity of their large genomes. Nsp14 associates with viral polymerase complex to excise the mismatched nucleotides. Aside from the exonuclease activity, nsp14 methyltransferase domain mediates cap methylation, facilitating translation initiation and protecting viral RNA from recognition by the innate immune sensors. The nsp14 exonuclease activity is modulated by a protein co-factor nsp10. While the nsp10/nsp14 complex structure is available, the mechanistic basis for nsp10-mediated modulation remains unclear in the absence of the nsp14 structure. Here, we provide a crystal structure of nsp14 in an apo-form. Comparative analysis of the apo- and nsp10-bound structures explain the modulatory role of the co-factor protein and reveal the allosteric nsp14 control mechanism essential for drug discovery. Further, the flexibility of the N-terminal lid of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nsp14 structure presented in this study rationalizes the recently proposed idea of nsp14/nsp10/nsp16 ternary complex.
Keywords: SARS-CoV-2, nsp14, nsp10, XRD, SAH, coronavirus, methylation, proof-reading, structure, methyltransferase
Graphical abstract
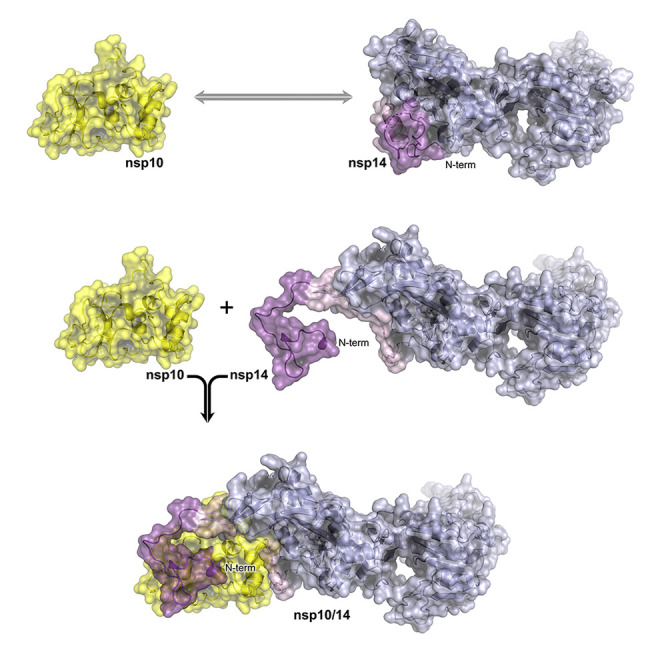
Czarna et al. use X-ray crystallography to determine the structure of SARS-CoV-2 nsp14 methyltransferase with the bound co-factor SAH. The N-terminal “lid” is undefined, revealing a high flexibility of the nsp14 in the absence of its partner nsp10.
Introduction
Upon release into the cytoplasm of the host cell, the coronavirus genome is translated into a single non-functional polyprotein 1a/1ab. Its autoproteolytic processing releases a number of nonstructural proteins (nsps) responsible for viral replication. The activity of a particular nsps is relatively well characterized, but the mechanistic understanding of their interplay is still insufficient (Pyrc et al., 2004; Yadav et al., 2021; Sola et al., 2015).
Nsp14 consists of two domains, each with a distinct catalytic activity. The C-terminal domain is a SAM-dependent methyltransferase, while the N-terminal ExoN domain exhibits 3′-5′ exonuclease activity. Recognition of unmodified RNA molecules constitutes a universal antiviral strategy embedded in the host innate immune defenses. To avoid detection, coronaviral RNA is capped by viral enzymes, which additionally increase its stability and facilitates translation initiation. Capping involves a series of enzymatic reactions. In the penultimate step, the C-terminal domain of nsp14 catalyzes N7-methylation of a GpppA intermediate. Subsequent methylation by nsp16 results in a functional cap (7MeGpppA2'OMe) (Sevajol et al., 2014; Bouvet et al., 2010, 2012; Eckerle et al., 2010; Yan et al., 2021; Chen et al., 2009).
The large coronaviral genome requires replication fidelity to maintain its functionality. While the nsp12-centered polymerase complex is highly processive, it is also error prone. An unaudited replication would result in an excessive accumulation of mutations and the generation of defective progeny virions. Nsp14 associates with the replication complex and removes incorrectly incorporated nucleotides from the 3′ end of the newly formed RNA strand. Nsp14 is associated with nsp10, a protein co-factor that modulates nsp14 exonuclease activity in the replication complex (Smith and Denison, 2013; Eckerle et al., 2010).
The structural and biochemical basis of nsp14 interaction with nsp10 has been elucidated (Baddock et al., 2022; Liu et al., 2021; Lin et al., 2021). We have recently demonstrated that nsp14, nsp10, and nsp16 form a ternary complex, further modulating nsp14 catalytic activity (Matsuda et al., 2022). Despite the detailed structural characterization of the complexes, the mechanistic basis of the modulation of nsp14 exonuclease activity remains elusive. The absence of structural information on nsp14 alone hinders the mechanistic understanding of the consequences of binding with nsp10 and nsp10/nsp16.
Here, we provide the crystal structure of nsp14 in the apo-form (without nsp10), which, together with prior data on nsp10/nsp14 complex, provide insight into the modulatory role of nsp10 on nsp14 exonuclease activity by a set of complex allosteric modifications. Furthermore, the presented structure strongly supports the nsp14/nsp10/nsp16 heterotrimer formation model.
Result
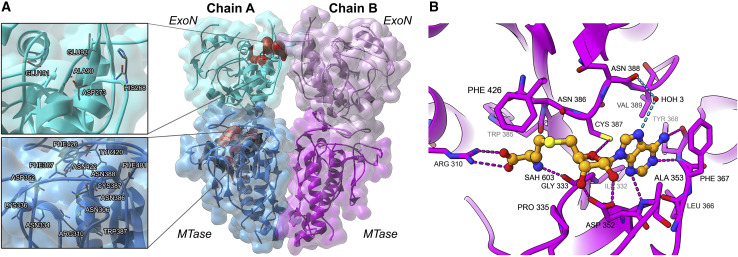
Full-length severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nsp14 exonuclease/methyltransferase was expressed in Escherichia coli and purified. The crystal structure of S-adenosylhomocysteine (SAH)-bound nsp14 was determined at 2.5 Å resolution (Table 1 ). The crystals belonged to P21 space group and contained 2 molecules in the asymmetric unit. Nsp14 is characterized by a bimodular structure corresponding to the two distinct catalytic activities (Figure 1 A). The C-terminal methyltransferase domain of nsp14 (amino acids 287–524) is well defined by the electron density for most of its part save for amino acids 454–465. At the same time, the N-terminal exonuclease domain (encompassing amino acids 1–286) is largely undefined by the electron density, indicating significant flexibility (Figure S1). The flexible regions are between amino acids 1–24, 40–44, 93–103, and 123–154 and are of importance for further discussion. The undefined parts are identical in both chains.
Table 1.
Data collection and refinement statistics
| Data collection statistics | |
| PDB ID | 7R2V |
| Wavelength (Å) | 1.00004 |
| Resolution range | 86.59–2.53 (2.89–2.53) |
| Space group | P 1 21 1 |
| Unit cell | 67.18, 100.31, 90.87 (Å) |
| 90, 107.66, 90 (°) | |
| Data-scaling software | autoPROC, AIMLESS, (STARANISO) |
| Unique reflections | 19,046 (5,869) |
| Rmeas (%) | 18.8 (99.9) |
| Rpim (%) | 8.7 (40.0) |
| Multiplicity | 5 (6.2) |
| CC1/2 | 0.989 (0.664) |
| Mean I/ σ(I) | 6.8 (1.6) |
| Completeness | |
| Spherical (%) | 99.8 (7.7) |
| Elipsoidal (%) | 99.8 (65) |
| Refinement | |
| Rwork/Rfree/test set (%) | 0.196/0.254/4.97 |
| Wilson B factor (Å) | 57.062 |
| Clashscore | 3 |
| Raachandran outliers (%) | 0.5 |
| Sidechain outliers (%) | 5.1 |
| RSRZ outliers (%) | 3.4 |
Values provided in parentheses are for the highest resolution shell. The I/σ(I) > 1.2 was automatically used by the STARANISO server for the highest resolution shell limit. The data was further limited based on analysis of density maps. Rmeas, multiplicity-independent R-factor introduced by Diederichs that indicates the real precision of the measurement, independent of the multiplicity of the reflection; Rpim, precision of the averaged merged intensity measurements; Rwork, working R-factor of the refined structure; Rfree, a residual function calculated during structure refinement in the same way as the conventional R factor, but applied to a small subset of reflections that are not used in the refinement of the structural model. The purpose is to monitor the progress of refinement and to check that the R factor is not being artificially reduced by the introduction of too many parameters; RSRZ, the real-space R-value (RSR) is a measure of the quality of fit between a part of an atomic model (in this case, one residue) and the data in real space. RSR Z-score (RSRZ) is a normalisation of RSR specific to a residue type and a resolution bin.
Figure 1.
Crystal structure of apo-nsp14
(A) Arrangement of the molecules in the asymmetric unit. Model (A) blue and model (B) pink. MTase and ExoN domains within the bimodular structure of nsp14 are distinguished by a primary color shade. Active-sites residues are indicated in red in one molecule, and the detailed arrangements of the active sites are shown in inserts.
(B) Interactions of SAH (orange, stick model) at the active site of nsp14 MTase domain (pink, ribbon). Hydrogen bonds are depicted as dotted lines; waters are indicated as red spheres.
See also Figure S1.
The methyltransferase domain of nsp14 is characterized by a fold atypical to methyltransferases. The structure is built around a central β sheet composed of 5, instead of 7, canonical β strands. Further, the methyltransferase domain of nsp14 contains an atypical peripheral zinc finger stabilized by another element that distinguishes nsp14 from the typical methyltransferase fold, a supplementary C-terminal α-helix. The methyltransferase fold within our structure closely resembles that previously seen in the nsp10/nsp14 complex of SARS-CoV-1 (PDB: 5C8S) with a root-mean-square deviation (RMSD) of 0.57 Å for 197 equiv Cα atoms. A reaction product, SAH, is clearly defined by the electron density in our structure (Figure 1B). The remaining part of the active site has an empty, tubular shape (the methylated reaction product is absent). Interestingly, the active site faces a large, unobstructed water channel. With unobstructed access to the active site, our structure is particularly valuable in terms of inhibitor development (fragment screening and inhibitor soaking).
The SAM/SAH-binding region in our structure (331-DxGxPxA-337) diverges from the classical methyltransferase motif I (E/DxGxGxG; Martin and Mcmillan, 2002) but is consistent with that described in nsp14 of SARS-CoV-1 (Ma et al., 2015). The density in the MTase active site unambiguously shows SAH despite the fact that SAM was added to the protein solution (Figure S2). The purine ring of SAH is positioned via sulfur/π interaction with Cys387; hydrophobic interactions contributed by side chains of Ala353, Val389, Phe367, and direct hydrogen bonds contributed by main chain of Tyr368. A water-mediated hydrogen bond contributed by Asn388 is additionally observed in model B. Ribose is stabilized by hydrogen bonds contributed by the side chain of Asp352. Mainchain carbonyl oxygens of Gly333 and Trp385 contribute hydrogen bonds with the cysteine amine, whereas its carboxyl group forms a salt bridge with Arg310. The identified SAH positioning residues are consistent with the conclusions of prior mutagenesis studies (Ogando et al., 2021). The majority of reported nanomolar MTase inhibitors are SAM competitive (Devkota et al., 2021). By delineating the detailed molecular interactions in the SAH pocket, our study facilitates the development of SAH mimetic inhibitors of SARS-CoV-2 MTase.
A structure of nsp14 from SARS-CoV-2 has been recently deposited at the Protein Data Bank (PDB: 7QGI), which contains an empty active site (no substrates or reaction products). Comparison with our structure demonstrates that the SAM/SAH binding site in 7QGI is occluded by a loop connecting β2' and β3′ strands of a central β sheet (Figure S3). The place where the loop in question is located in SAH-bound structures (nsp10/nsp14 complex from SARS-CoV-1 and this study) is occluded by an adjacent molecule in 7QGI. This may suggest that either the blocked active site in 7QGI is an artifact of crystal packing or, alternatively, it may represent one of the multiple forms found in solution selected by the particular crystal packing. When Ma et al. (2015) analyzed SARS-CoV-1 nsp10/nsp14 complexes in SAH-bound and empty forms, the loop in question had the same conformation in both SAH-bound and empty structures (and comparable to that found in our structure), suggesting indirectly that the open conformation is the prevalent form.
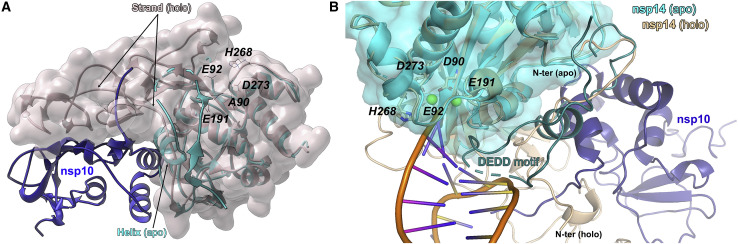
The exonuclease domain of nsp14 is built around a five-stranded, twisted β sheet flanked by α helices. Such an arrangement of the core structure elements follows the overall design of DEDD family exonucleases but contains unique features: the N-terminal region (1–84) and the first zinc finger. The striking features of the exonuclease domain in our structure became evident only upon comparison with the structure of nsp10/nsp14 complex. The N terminus of nsp14 spanning amino acids 1–70 is folded and oriented differently in compared structures (Figures 2A and 2B). The N terminus is well defined by electron density in the nsp10/nsp14 complex but contains few ordered secondary structures. Amino acids 1–56 constitute the first of the two interaction regions with nsp10 (nomenclature of structural features after Ma et al. (2015)). The second interaction region comprises residues 60–67 and 200–202. In apo-nsp14 structure, residues 1–24 are not defined by the electron density. Region 25–70 is completely refolded in the apo-nsp14 structure compared with the nsp10/nsp14 complex. For example, residues Thr50-Met58 form an α helix in the apo-nsp14 structure, whereas residues 51–54 form a β1 strand within β1, β5, and β6 sheets in the nsp10/nsp14 complex. The remaining residues (25–49, 59–67) contribute completely different intermolecular interactions within the apo-nsp14 structure compared with the nsp10/nsp14 complex. In the absence of nsp10, the N-terminal region of nsp14 covers the second nsp10 interaction region, burying, among others, residues 200–202. Thereby, the N terminus of nsp14 acts as a lid, covering the nsp10 binding site in the absence of the protein co-factor. The lid rearranges its structure in the presence of nsp10 to create a significant interaction surface with nsp10. The above conclusion is corroborated by comparing our structure and 7QGI, which show similar arrangements of lid domains irrespective of crystal packing and different from that found in nsp10/nsp14 complex.
Figure 2.
Nsp10 induced refolding of nsp14 ExoN lid subdomain
(A) Nsp10 (navy blue, ribbon)/Nsp14 (gray, surface) complex overlaid on apo-nsp14 (cyan). The N-terminal lid of nsp14 refolds upon binding of nsp10 as exemplified by Thr50-Met58 region assuming a helical structure in apo-nsp14 and forming a strand of a β sheet indicated in nsp10/nsp14 complex. In the apo structure, the lid occludes nsp10-binding site at the surface of nsp14.
(B) The RNA interaction site of nsp10 (navy blue)/nsp14 (beige) complex (7N0B) is sterically occluded by lid subdomain in apo-nsp14 structure (cyan), providing a rationale for the negligible exonuclease activity of nsp14 in the absence of nsp10. RNA (orange). The apo-nsp14 loop, occluding the RNA-binding site, is denoted in dark cyan (dotted line).
See also Figures S2 and S3.
Nsp10 enhances the 3′ exonucleolytic activity of nsp14 but does not significantly affect the methyltransferase activity. Nsp10’s effect on nsp14 was rationalized by the nsp10/nsp14 crystal structure, where nsp10 contributes significant contacts with the ExoN domain of nps14 but does not interact with the N7-methyltransferase domain (Ma et al., 2015). The extensive surface area of interaction suggested that nsp10 maintains the structural stability of ExoN, but the nature and extent of instability of apo-nsp14 were not known in the absence of the apo-nsp14 structure. In the current study, we show that the ExoN domain of nsp14 is partially unfolded in the absence of nsp10. The proximal part of the β2-β3 sheet adjacent to catalytic Glu92 is undefined by electron density (flexible, unstructured) in apo-nsp14. The same is true for the entire region between α3 and β4. Residues constituting α2 and β5-β6 in the nsp10/nsp14 complex structure are not defined by the electron density in the apo-nsp14 structure. Significantly, in apo-nsp14, the catalytic metal ion-binding site within the DEDD motif is partially occluded by the side chain of His188. Further, the RNA-binding site is partially occluded by the lid fragment (Thr35-Lys47), which additionally buries the nsp10-binding site. It has been shown that Lys9 of the nsp14 is critical for exonuclease activity (Moeller et al., 2022). This residue is not visible in our electron density. Thus, the refolded N-terminal part of the protein is not structurally oriented for RNA interaction. This further confirms the role of 1–50 span refolding in exonuclease activity control. Nsp10-induced refolding of the lid removes the steric occlusion and induces a catalytically capable conformation of nsp14 observed in the nsp10/nsp14 complex (Figure 2B).
We have demonstrated earlier that nsp14, nsp10, and nsp16 form a ternary complex centered around nsp10 (Matsuda et al., 2022). However, such complex is not supported by the structural data available on nsp10/nsp14 and nsp10/nsp16 binary complexes. When the binary complexes are overlayed on nsp10, the lid of nsp14 clashes with nsp16, demonstrating that ternary-complex formation would require a structural rearrangement within the lid. The current study demonstrates that the lid of nsp14 is capable of structural morphing, indirectly supporting the mechanism of triplex formation.
Discussion
The current pandemic highlighted the importance of understanding coronavirus physiology. Prior research on other human and animal coronaviruses facilitated an unprecedented pace of vaccine and antiviral development, but the constant genetic drift and the threat of new zoonotic transmission warrant continued effort.
Nsp14 constitutes a valid target in SARS-CoV-2, but the structural information was incomplete. This study provided the structural basis for a mechanistic explanation of nsp10-mediated modulation of nsp14 3′ exonuclease activity, defined the structure of nsp14 N7-methyltransferase in the absence of RNA substrate, and offered crystallization conditions suitable for soaking of small molecules at the active site of nsp14 N7-methyltransferase. Thereby, the study contributes to our understanding of coronavirus physiology and facilitates the effort of anti-coronaviral drug development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21 Rosetta2 (DE3) | Milipore | 71402 |
| Chemicals, peptides, and recombinant proteins | ||
| Terrific Broth medium | Bioshop | TER409 |
| Ampicillin | Sigma-Aldrich | A9393 |
| isopropyl-D-1-thiogalactopyranoside | Sigma-Aldrich | I5502 |
| Tris-HCl | Bioshop | TRS003 |
| NaCl | Bioshop | SOD004 |
| MgCl2 | Bioshop | MAG520 |
| Glycerol | Bioshop | GLY004 |
| β-mercaptoethanol | Sigma-Aldrich | M6250 |
| Imidazole | Bioshop | IMD508 |
| protease inhibitors cocktail | Roche | 11697498001 |
| Benzonase | Millipore | 101654 SAFC |
| tobacco etch virus (TEV) protease | In-house | n.a. |
| S-Adenosyl methionine | New England BioLabs | B9003S |
| Deposited data | ||
| Structure of nsp14 from SARS-CoV-2 in complex with SAH | this paper | PDB ID: 7R2V |
| SARS-CoV-1 nsp14 | (Ma et al., 2015) | PDB ID: 5C8S |
| SARS-CoV-2 nsp14 | (Imprachim et al., 2022) | PDB ID: 7QGI |
| SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex | (Liu et al., 2021) | PDB ID: 7N0B |
| Recombinant DNA | ||
| Uniprot P0DTD1 encoding amino acids 5926 – 6452 | GeneArt | n.a. |
| pETDuet-1 | Milipore | 71146 |
| Software and algorithms | ||
| The STARANISO Server | Global Phasing Limited | https://staraniso.globalphasing.org/cgi-bin/staraniso.cgi |
| CCP4 v7.1 | (Winn et al., 2011) | https://www.ccp4.ac.uk |
| CheckMyBlob server | (Kowiel et al., 2018) | https://checkmyblob.bioreproducibility.org |
| PyMol Molecular Graphics | System Schrodinger, LLC | https://pymol.org/2/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: K.P.: k.a.pyrc@uj.edu.pl
Materials availability
Plasmids produced in this study are available from the authors.
Experimental model and subject details
Recombinant SARS-CoV-2 nsp14 was purified from the expression in E. coli BL21 Rosetta2 (DE3) strain.
Method details
Expression construct
Sequence encoding amino acids 5926 – 6452 of SARS CoV-2 polyprotein 1ab (Uniprot P0DTD1), including D90A and D92A substitutions (Bouvet et al., 2012), flanked by BamHI and NotI cleavage sites and containing a TEV protease cleavage site was codon-optimized for expression in E. coli and obtained by synthesis at GeneArt (sequence available in Table S1). The sequence was subcloned into pETDuet-1 expression vector.
Protein expression and purification
The plasmid was transformed into E. coli BL21 Rosetta2 (DE3) strain. Expression was carried out in Terrific Broth medium (Bioshop) supplemented with 100 μg/mL ampicillin (Sigma-Aldrich). Bacterial cultures were incubated at 37°C until the OD600 value reached 1.2–1.4. The cultures were cooled to 4°C for 20 min, and the recombinant protein expression was induced with 0.5 mM isopropyl-D-1-thiogalactopyranoside (IPTG; Sigma-Aldrich). The bacteria were incubated at 18°C for additional 18 h. Bacterial cells were harvested by centrifugation at 7,000 × g for 30 min at 4°C and stored at −20 °C before the purification.
Bacterial pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.5, 500 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 5 mM β-mercaptoethanol, 10 mM imidazole), supplemented with benzonase (Millipore) and protease inhibitors cocktail (Roche), and lysed by sonication at 80% amplitude for 20 min at 14°C with 3 s on/2 s off cycles. The fraction of the supernatant containing soluble protein was separated from debris by centrifugation at 25,000 × g for 40 min at 4 °C. The clarified supernatant was incubated overnight at 4 °C with HisPur Ni-NTA Resin (Thermo Scientific) pre-equilibrated with the lysis buffer. Resin was sequentially washed with lysis buffer, then with wash buffer 1 (50 mM Tris-HCl pH 8.5, 500 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 5 mM β-mercaptoethanol, 20 mM imidazole) and wash buffer 2 (50 mM Tris-HCl pH 8.5, 500 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 5 mM β-mercaptoethanol, 30 mM imidazole). The immobilized protein was eluted with elution buffer (50 mM Tris-HCl pH 8.5, 250 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 5 mM β-mercaptoethanol, 250 mM imidazole). Fractions containing the protein of interest (as determined by SDS-PAGE) were pooled and supplemented with tobacco etch virus (TEV) protease in a 1:10 molar ratio. TEV cleavage was combined with overnight dialysis into a cleavage buffer (50 mM Tris-HCl pH 8.5, 200 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 5 mM β-mercaptoethanol, 10 mM imidazole). Reverse chromatography on HisPur Ni-NTA resin pre-equilibrated with the dialysis buffer allowed the cleaved His-tag removal. The protein was further purified by size exclusion chromatography on HiLoad 26/600 Superdex 200 prep grade column (GE Healthcare) equilibrated with the working buffer (50 mM Tris-HCl pH 8.5, 150 mM NaCl, 5 mM MgCl2, 1 mM TCEP). The purity of the sample was assessed by SDS-PAGE.
Crystallization and data collection
The purified protein was concentrated to 11.5 mg/mL. The sample was supplemented with 2mM SAM. Crystallization screening was performed at 4°C using the sitting drop vapour diffusion method. Crystals were obtained from a solution containing 15% v/v 2-propanol, 0.2 M imidazole pH 7.6 and 500 mM polypropylene glycol 400 (JBScreen Plus HTS additive screen; Jena Bioscience) and 1:1 protein to mother liquer drops. Further optimization was performed around initial conditions where the crystals generally appeared after 4 days. The crystals were cryoprotected in 30% ethylene glycol in the mother liquor and flash-cooled in liquid nitrogen. The diffraction data were collected at Swiss Light Source (SLS, Villigen, Switzerland) beamline X06DA - PXIII.
Quantification and statistical analysis
The data were indexed and integrated using XDS (Kabsch, 2010). The data were scaled and merged using STARANISO webserver. A molecular replacement solution was found using Phaser (Mccoy, 2007) and the structure of SARS-CoV-1 nsp14 (PDB code: 5C8S) as a search model. SAH was identified in the electron density map using CheckMyBlob server (https://checkmyblob.bioreproducibility.org). Restraints were obtained from ligand builder in Coot (Emsley, 2017). The initial model was subjected to several iterations of manual and automated refinement cycles using COOT and REFMAC5, respectively (Emsley and Cowtan, 2004; Murshudov et al., 2011). Throughout the refinement, 5% of the reflections were used for cross-validation analysis, and the behaviour of Rfree was employed to monitor the refinement strategy. The data collection and refinement statistics are shown in Table 1.
Acknowledgments
This work was supported by a subsidy from the Polish Ministry of Science and Higher Education for research on SARS-CoV-2 and a grant from the National Science Center (UMO-2017/27/B/NZ6/02488; to K.P.) and by Bayerische Forschungsstiftung grant AZ-1453-20C (to G.P.) and Deutsche Forschungsgemeinschaft grant PO 1851/4-1 (to G.P.). K.P. was supported by the Corona Accelerated R&D in Europe (CARE) project funded by the Innovative Medicines Initiative two Joint Undertaking (JU) under grant agreement no. 101005077. The University of Dundee was supported by a grant from the Bill and Melinda Gates Foundation (grant number LO4CVI). We acknowledge the MCB Structural Biology Core Facility (supported by the TEAM TECH CORE FACILITY/2017-4/6 grant from the Foundation for Polish Science) for valuable support.
Author contributions
Conceptualization, A.C. and G.D.; methodology, L.K., A.M., and A.K.; validation, P.G.W., G.D., and K.P.; investigation, J.P., M.P., and P.W.; visualization A.C., A.M., C.R., I.G, and G.P.; writing – original draft, G.D. and K.P.; writing – review & editing, A.C., J.P., C.R., I.G., S.O., and F.C.; formal analysis, J.P and A.C.; project administration, A.C. and P.G.W., supervision, K.P.; funding acquisition, K.P. and G.P.
Declaration of interests
The authors declare no competing interests.
Published: May 23, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.str.2022.04.014.
Supplemental information
Data and code availability
-
•
The atomic coordinates and structure factors of SARS-CoV-2 nsp14 have been deposited to PDB and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table as well as in Table 1.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baddock H.T., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., Swift L.P., Cruz-Migoni A., Fan H., Keown J.R., Walker A.P., et al. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022;50:1484–1500. doi: 10.1093/nar/gkab1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U S A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U S A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota K., Schapira M., Perveen S., Khalili Yazdi A., Li F., Chau I., Ghiabi P., Hajian T., Loppnau P., Bolotokova A., et al. Probing the SAM binding site of SARS-CoV-2 Nsp14 in vitro using SAM competitive inhibitors guides developing selective bisubstrate inhibitors. SLAS Discov. 2021;26:1200–1211. doi: 10.1177/24725552211026261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. Tools for ligand validation in Coot. Acta Crystallogr. D Struct. Biol. 2017;73:203–210. doi: 10.1107/s2059798317003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- Imprachim, Nergis, Yosaatmadja, Yuliana, A N.J. Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development. bioRxiv. 2022 doi: 10.1101/2022.03.11.483836. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowiel M., Brzezinski D., Porebski P.J., Shabalin I.G., Jaskolski M., Minor W. Automatic recognition of ligands in electron density by machine learning. Bioinformatics. 2018;35:452–461. doi: 10.1093/bioinformatics/bty626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Chen H., Chen Z., Yang F., Ye F., Zheng Y., Yang J., Lin X., Sun H., Wang L., et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021;49:5382–5392. doi: 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Shi W., Becker S.T., Schatz D.G., Liu B., Yang Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373:1142–1146. doi: 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U S A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.L., Mcmillan F.M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Matsuda A., Plewka J., Chykunova Y., Jones A.N., Pachota M., Rawski M., Mourão A., Karim A., Kresik L., Lis K., et al. Despite the odds: formation of the SARS-CoV-2 methylation complex. bioRxiv. 2022 doi: 10.1093/nar/gkae165. 2022.01.25.477673 Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/s0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller N.H., Shi K., Demir Ö., Belica C., Banerjee S., Yin L., Durfee C., Amaro R.E., Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. 2022;119 doi: 10.1073/pnas.2106379119. e2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., El Kazzi P., Zevenhoven-Dobbe J.C., Bontes B.W., Decombe A., Posthuma C.C., Thiel V., Canard B., Ferron F., Decroly E., Snijder E.J. Structure-function analysis of the nsp14 N7-guanine methyltransferase reveals an essential role in Betacoronavirus replication. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2108709118. e2108709118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., Van Der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Denison M.R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9:e1003760. doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., Mccoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R., Chaudhary J.K., Jain N., Chaudhary P.K., Khanra S., Dhamija P., Sharma A., Kumar A., Handu S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The atomic coordinates and structure factors of SARS-CoV-2 nsp14 have been deposited to PDB and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table as well as in Table 1.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.