Abstract
Background
Low-energy diets (LEDs) comprise commercially formulated food products that provide between 800 and 1200 kcal/day (3.3–5 MJ/day) to aid body weight loss. Recent small-scale studies suggest that LEDs are associated with marked changes in the gut microbiota that may modify the effect of the LED on host metabolism and weight loss. We investigated how the gut microbiota changed during 8 weeks of total meal replacement LED and determined their associations with host response in a sub-analysis of 211 overweight adults with pre-diabetes participating in the large multicentre PREVIEW (PREVention of diabetes through lifestyle intervention and population studies In Europe and around the World) clinical trial.
Methods
Microbial community composition was analysed by Illumina sequencing of the hypervariable V3-V4 regions of the 16S ribosomal RNA (rRNA) gene. Butyrate production capacity was estimated by qPCR targeting the butyryl-CoA:acetate CoA-transferase gene. Bioinformatics and statistical analyses, such as comparison of alpha and beta diversity measures, correlative and differential abundances analysis, were undertaken on the 16S rRNA gene sequences of 211 paired (pre- and post-LED) samples as well as their integration with the clinical, biomedical and dietary datasets for predictive modelling.
Results
The overall composition of the gut microbiota changed markedly and consistently from pre- to post-LED (P = 0.001), along with increased richness and diversity (both P < 0.001). Following the intervention, the relative abundance of several genera previously associated with metabolic improvements (e.g., Akkermansia and Christensenellaceae R-7 group) was significantly increased (P < 0.001), while flagellated Pseudobutyrivibrio, acetogenic Blautia and Bifidobacterium spp. were decreased (all P < 0.001). Butyrate production capacity was reduced (P < 0.001). The changes in microbiota composition and predicted functions were significantly associated with body weight loss (P < 0.05). Baseline gut microbiota features were able to explain ~25% of variation in total body fat change (post–pre-LED).
Conclusions
The gut microbiota and individual taxa were significantly influenced by the LED intervention and correlated with changes in total body fat and body weight in individuals with overweight and pre-diabetes. Despite inter-individual variation, the baseline gut microbiota was a strong predictor of total body fat change during the energy restriction period.
Trial registration
The PREVIEW trial was prospectively registered at ClinicalTrials.gov (NCT01777893) on January 29, 2013.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-022-01053-7.
Background
To combat the obesity epidemic and its comorbidities such as type 2 diabetes (T2D) [1], energy-restricted diets have been at the forefront of weight management and glucose control. Low-energy diets (LEDs) represent one of the most effective options for weight management [2], with established efficacy for weight loss, but which recently have also proven to be highly successful in normalizing glycaemia in high-risk obese individuals with T2D [3, 4]. However, the success of diet-induced weight loss may vary considerably between individuals and the underlying factors are largely unclear. Mounting evidence suggests that the gut microbiota, one of the most salient features contributing to physiological inter-individual variability [5], is implicated in obesity [6] and influences the host’s metabolic response to diet [7]. In mice, depletion of the gut microbiota nullified the metabolic improvements, especially the decrease in body weight, following energy restriction [8].
The gut microbiota is inextricably linked with the quantity and quality of nutrients in the diet, as gut microbes mainly rely on host diet composition to obtain metabolic substrates [9]. Subsequently, the gut microbiota exerts its impact on host physiology by producing microbial metabolites (e.g., short-chain fatty acids, SCFA) and microbial structural components (e.g., lipopolysaccharides and flagella) that modulate the metabolism of lipids, cholesterol, glucose [10] and inflammatory response [11].
While an association between the gut microbiota and obesity exists, the tripartite interaction between the microbiota, energy restriction and host metabolic response remains little studied in humans. In the context of energy-restricted diets such as LEDs, existing interventions have failed to generate a consensus on changes in the gut microbiota during diet-induced weight loss [12]. For example, Ott and colleagues reported a 4-week LED (3.4 MJ/day) had no effect on the overall gut microbiota [13], whereas another German study by Frost et al. more recently documented distinct shifts of microbiota composition and diversity during a 6-week LED intervention [14]. Variable outcomes regarding specific bacterial taxa affected by LED have been reported in other LED interventions [15–20]. Similarly, the few studies exploring the feasibility of predicting diet-induced weight modulation using baseline features of the gut microbiota arrived at divergent conclusions [7, 21]. This inconsistency is likely due to small sample size (as few as 5 participants), differences in bacterial profiling techniques, population, ethnicity of participants and the intervention diets [12, 22, 23], which preclude generalization of the results. Recently, a re-analysis of omics data collected in two Danish dietary interventions introducing whole grain-rich or low-gluten diets found that inclusion of data on gut microbiota and urine metabolites significantly improved the classification accuracy for weight loss responders [24]. Nevertheless, the definition for weight loss success or responders could vary arbitrarily across studies.
Due to the potential malleability and inter-individual variance in the microbiota, the gut microbiota has increasingly become the focus of precision nutrition, whereby personalized responses to diet predicted and dietary advice tailored to the individual [23]. Therefore, a better understanding of the changes and contribution of the gut microbiota to inter-individual variability in response to a LED may improve the effectiveness of weight-loss interventions.
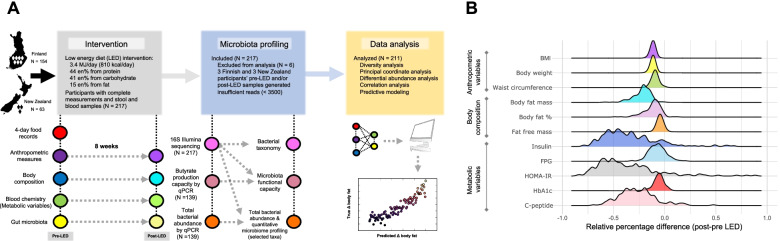
PREVention of diabetes through lifestyle Intervention and population studies in Europe and around the World (PREVIEW) was a multi-centre, 3-year lifestyle intervention in overweight adults with pre-diabetes conducted in 8 countries that aimed to decrease the incidence of T2D. We have previously reported the clinical outcomes from the first phase of PREVIEW, where eligible adult participants followed an 8-week total meal replacement LED [25]. The LED was accompanied by significant weight loss and associated improvements in anthropometry (e.g., body mass index (BMI) and total body fat) and metabolic parameters (e.g., fasting plasma glucose (FPG)), with gender-specific changes [25]. Here we study the gut microbiota from a subset of participants in the PREVIEW trial, from Finland and New Zealand (N = 211), by Illumina sequencing of the 16S rRNA gene. We compared pre- and post-LED differences in the composition and function of the gut microbiota and to determine whether baseline microbiota configuration is predictive of host metabolic response to the LED.
Methods
Study participants and design
PREVIEW was a multi-centre randomized controlled trial (RCT) based on a 3-year lifestyle intervention for T2D prevention across 8 countries, which comprised 2 intervention phases. Phase 1 is an 8-week, weight-loss phase using a formula LED intended to induce weight loss of ≥8% to qualify for the next phase. Phase 2 is a 148-week randomized lifestyle intervention that focuses on diet, physical activity and behaviour modification for maintenance of weight loss [26]. The study population of PREVIEW consisted of 2224 adults with overweight or obesity (BMI≥25 kg/m2) and pre-diabetes (according to the American Diabetes Association (ADA) criteria [27]), aged between 25 and 70 years. Overweight men and women with pre-diabetes were eligible for inclusion. Participants were recruited via advertisements in newspapers and newsletters, radio and television advertisements/interviews and by contacting primary and occupational health care providers [25]. The first participant was enrolled on June 1, 2013, and the last participant was enrolled on February 27, 2015. The last participant visit was in March 2018. Participants self-reported not being engaged in competitive sports, with stable body weight (±5 kg) for at least 2 months prior, and no current glucose medications or changes in prescribed medications for 3 months prior to sample collection. Exclusion criteria included diagnosed T2D, other significant diseases including cardiovascular, liver, gastrointestinal, or kidney disease, malignancy, bariatric or any major surgical procedure in the previous 3 months, pregnancy, or breastfeeding. The PREVIEW primary outcome was incidence of T2D at 3 years; secondary outcomes were incidence of T2D at 2 years, gut microbiota analysis and all relevant clinical and biochemical parameters related to metabolic control, at different time-points (8 weeks, 2 years and 3 years). The analysis described in the present study is based on the LED phase of PREVIEW derived from a multi-ethnic cohort recruited from the University of Auckland (UoA), New Zealand, and the University of Helsinki (HEL), Finland (total N = 217), from whom paired faecal samples were available (baseline/pre- and post-LED; Fig. 1A). None of the participants reported antibiotic use 3 months prior to or during the LED. As the microbiota analysis involved two of the eight PREVIEW study centres, we termed the present study a PREVIEW sub-study herein for clarity. Detailed methods for the LED intervention were described previously [25]. Briefly, all participants were provided with total meal replacement sachets from Cambridge Weight Plan® (Northants, UK) for the 8 weeks duration. In total, the LED provided an estimated 3.4 MJ/day (810 kcal/day), of which 44 energy % (en%) was from protein, 41 en% from carbohydrate and 15 en% from fat. The total dietary fibre content of the LED was 13.3 g/day (participants’ pre-LED intake of dietary fibre was 22.3 g±7.5 (mean±SD)). Participants were advised that psyllium fibre could be used in case of gastrointestinal side effects, mainly constipation. A maximum of 400 g of non-starchy vegetables could be consumed, such as tomatoes, cucumber and lettuce, making the total energy content approximately 4 MJ (1000 kcal). Participants were advised to avoid intense physical activity and maintain current activity levels during the LED intervention. The work of PREVIEW is carried out in full compliance with the relevant requirements of the latest version of the Declaration of Helsinki (59th WMA General Assembly, Seoul, Korea, October 2008), and the ICH-GCP, The International Conference on Harmonisation (ICH) for Good Clinical Practice to the extent that this is possible and relevant. The study protocol was approved by the Ethical Committees of participating countries (Health and Disability Ethics Committee in New Zealand, ref. 14/191 and Medical Ethical Committees of the Hospital District of Helsinki and Uusimaa and HUCH in Finland, ref. 171/13/03/00/2013). All participants provided written informed consent prior to commencing screening procedures in clinic. All information obtained during the trial is handled according to local regulations and the European Directive 95/46/CE (directive on protection of individuals with regard to the processing of personal data and on the free movement of such data). The PREVIEW trial was prospectively registered at ClinicalTrials.gov (NCT01777893). Additional information can be found on the PREVIEW website (https://preview.ning.com/).
Fig. 1.
Overview of the study and variation of host variables. A Schematic overview of the study design. B Density plots showing inter-individual variation in host variables in response to the LED. The density plots display the distribution of the observed data (relative changes in host variables). The density function reflects the estimated underlying continuous probability from which the observed data have been sampled. BMI, body mass index; HOMA, homeostasis model for assessment of insulin resistance; FPG, fasting plasma glucose
Sample collection and clinical outcome measurements
Detailed protocols for clinical sample collection and outcome measurements have been described elsewhere [25, 26]. Before and after the 8-week LED, participants attended the research clinic on clinical investigation days (CIDs) for anthropometry measurements, collection of fasting blood samples and delivery of faecal samples and 4-day food records to assess habitual diet. The faecal samples were collected at home, frozen immediately at – 20 °C in the home freezer and taken in frozen form to the study centres within 1–3 days of collection, then stored at – 80 °C until processing. Body weight and height were measured in duplicate wearing light clothing, without shoes and after voiding the bladder. Body composition including fat mass and fat-free mass was assessed by bioelectrical impedance analysis, BIA (Finland, InBody720 Body Composition Analyzer, Biospace Co., Ltd, Seoul, Korea) and dual-energy X-ray absorptiometry, DXA (New Zealand, iDXA, model DPX+, software version 3.6y, GE-Lunar, Madison, WI). Fasting venous blood samples were collected for laboratory measurements, including FPG, HbA1c, insulin and C-peptide. Laboratory analyses were performed on an Architect ci8200 integrated system (Abbott Laboratories, Abbott Park, Illinois, USA). The Homeostasis Model for Assessment (HOMA) was calculated as a proxy for insulin resistance (IR). The equation used was HOMA-IR = fasting insulin (mU/L) × FPG(mmol/L))/22.5.
DNA extraction and 16S rRNA gene amplicon sequencing
DNA was extracted from all faecal samples with a previously described repeated bead-beating method that efficiently extracts bacterial community DNA including hard-to-lyse Gram-positive bacteria [28]. The quantity of extracted DNA was assessed using the Qubit Fluorometer (Thermo Fisher Scientific). Bacterial community composition was analysed by sequencing the PCR-amplicons of hypervariable V3-V4 regions of the 16S ribosomal RNA (rRNA) gene using primers 341F/785R and Illumina MiSeq (New Zealand, N = 126) or Illumina HiSeq (Finland, N = 308) as previously described [29]. The comparability of sequences generated by the two Illumina platforms was validated by evaluating two artificial communities and nine PREVIEW samples that were sequenced by both MiSeq and HiSeq (Additional file 1: Fig. S1A&B).
Quantification of butyrate production capacity and absolute bacterial abundance by quantitative PCR (qPCR)
Based on sample availability, the butyryl-CoA:acetate CoA-transferase gene and total bacterial abundance were quantified using faecal DNA for a subset of 139 participants (pre- and post-LED sample N = 278) with the degenerate primers BCoATscrF/R and the universal primers 331F/797R by qPCR, respectively. The qPCR assays have been described in detail previously [30, 31] and were performed in triplicate on a BioRad iCycler iQ thermal cycler system (BioRad, Hercules, CA) with HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis BioDyne, Tartu, Estonia). For quantification of the butyryl-CoA:acetate CoA-transferase gene, the mean threshold cycle (Ct) per sample (after excluding triplicates with Ct values that differed >0.5) was used as a proxy for the abundance of the target gene. For quantification of total bacterial abundance, the 10-log-fold standard curves ranging from 102 to 107 copies were produced using full-length amplicons of 16S rRNA gene of Bifidobacterium longum to convert the threshold cycle (Ct) values into the average estimates of target bacterial genomes present in 1 g of faeces (copy numbers/g of wet faeces) in the assays [31]. The absolute abundances of individual bacterial taxa were estimated as previously described [31], adjusting for 16S rRNA gene copy-number variation using the rrnDB database [32].
Data processing and statistical analysis
Demultiplexed reads after adaptor removal were processed using the QIIME2 v.2019.4. pipeline [33]. The high-quality forward reads were truncated to 150 bases and error-corrected using the DADA2 plugin [34] to generate amplicon sequence variants (ASVs). Taxonomic classification was performed using a pre-trained naive Bayes classifier implemented in QIIME2 against the SILVA 132 reference database [35]. Sample pairs from 6 participants (N = 12) were excluded from downstream analysis due to one or both of the samples having low reads after processing (<3500 reads), leaving a total of 211 participants with paired samples (N = 422) for downstream analysis. Samples meeting quality criteria (N = 422) had a mean sequencing depth of 67,453 reads (63,638–71,268, 95% CI). The sequencing files are deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession number PRJEB43667.
To infer the functional contribution of bacterial communities from 16S rRNA gene sequencing data, metagenome prediction was carried out using PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) [36] evaluating KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways [37].
Differential abundance for bacterial taxa or KEGG pathways between time points was identified with the DESeq2 package [38] accounting for sample pairing. DESeq2 employs a generalized linear model of counts based on a negative binomial distribution, scaled by a normalization factor that accounts for differences in sequencing depth between samples. Significance testing was then assessed using the Wald test. Non-count variables (anthropometric and biochemical measurements, microbiota diversity and richness) were analysed with Wilcoxon signed-rank test or paired t-test for non-normally distributed and normally distributed variables, respectively.
Microbiota richness and Shannon diversity index were estimated using the vegan package [39]. Overall microbiota structure was assessed by principle coordinate analysis (PCoA) on beta diversity computed using Bray-Curtis distances, representing the compositional dissimilarity between the samples. Permutational multivariate analysis of variance (PERMANOVA; adonis function in the vegan package [39]) with Bray-Curtis dissimilarities was used to identify factors contributing to variation in microbiota composition. At baseline, variation in the microbiota was significantly associated with gender (P = 0.006), ethnicity (P = 0.004) and age (P = 0.003). Hence, analyses were performed with and without adjustment for gender, ethnicity and age when applicable. Associations between bacterial taxa or KEGG pathways (post–pre-intervention) and clinical measurements (post–pre-intervention) were assessed using Spearman’s correlation as well as a linear mixed-effects model implemented in the mare package [40] for normal and adjusted correlations, respectively.
For prediction of host responses to LED, stepwise regression based on Akaike information criterion (AIC) (PathModel function in the mare package [40]) was used to select baseline features (microbiota, diet, host physiological variables, or a combination of them) that fit parsimonious models for %change in clinical measurements (post–pre-intervention). The PathModel function employs generalized or general linear models (in this case using the function lm in R), identifying first the variables that are most significantly associated with the response variable, combining them all into one model and performing stepwise model reduction using the functions step and stepAIC in the packages stats and MASS to finally arrive at the best model. To assess model performance based on AIC, we adopted the conventional rules of thumb by comparing the difference between the AICs of two models [41]:![]()
where AIC¡ is the AIC of model ¡ with the second lowest AIC, and AICmin is the model with the lowest AIC among the set of models examined. If Δ¡ < 2, there is substantial support for model ¡, whereas models with Δ¡ > 10 have essentially no support. Considering potential non-linear relationships between the microbiota features and the target variables, the prediction based on the baseline microbiota features was also done by Random Forest regression (R package randomForest [42]) using the following parameters: ntree = 10001 and mtry = p/3, where p is the number of input features. We then used repeated cross-validation (5-fold, 10 repetitions) of random forests in the caret package [43] in order to evaluate the R2 of the selected features to predict clinical indices. This method involves repeatedly using a subset of samples as a training set and the remaining samples as the test set to predict the outcome. The importance of each input feature was subsequently ranked according to %increase in mean squared error (%IncMSE). In both approaches, the prediction models based on the microbial features were generated using all prevalent (present in >30% of samples) bacterial genera detected at baseline, Shannon diversity and richness. For the stepwise regression models based on diet, the input features included intake of dietary nutrients; demographic characteristics, anthropometric and metabolic measurements (Table 1) were included in the models based on host physiological variables. For the microbiota and diet-based models, we included potential confounding variables (gender, ethnicity and age) in separate models (adjusted model) to account for confounding effects, since these variables may impact both exposure (e.g., baseline gut microbiota and dietary pattern) and outcome (e.g., change in adiposity).
Table 1.
Demographic characteristics, anthropometric and metabolic measurements of the study participants pre- and post-LED
| Characteristic | pre-LED | post-LED | P value* |
|---|---|---|---|
| Demographic variables | |||
| No. of participants | 211 | – | – |
| Female, No. (%) | 156 (74) | – | – |
| Age (years), mean (95% CI) | 54 (53–55) | – | – |
| Ethnicity, No. (%) | |||
| Caucasian | 194 (92) | – | – |
| Polynesian | 13 (6) | – | – |
| Asian | 3 (1) | – | – |
| Other | 1 (0.4) | – | – |
| Anthropometric variables | |||
| BMI (kg/m2), mean (95% CI) | 34.1 (33.3–34.9) | 30.2 (29.5–30.9) | <0.001 |
| Body weight (kg), mean (95% CI) | 95.8 (93.3–98.2) | 84.7 (82.5–86.9) | <0.001 |
| Waist circumference (cm), mean (95% CI) | 108.6 (106.9–110.2) | 98.4. (96.8–100.0) | <0.001 |
| Body composition | |||
| Body fat mass (kg), mean (95% CI) | 40.1 (38.3–41.8) | 31.8 (30.2–33.5) | <0.001 |
| Body fat (%), mean (95% CI) | 41.8 (407–42.9) | 37.3 (36.0–38.6) | <0.001 |
| Fat-free mass (kg), mean (95% CI) | 55.6 (54.1–57.1) | 52.9 (51.5–54.3) | <0.001 |
| Metabolic variables | |||
| Insulin (mU/L), mean (95% CI) | 13.1 (12.1–14.0) | 7.9 (7.4–8.4) | <0.001 |
| FPG (mmol/L), mean (95% CI) | 6.2 (6.2–6.3) | 5.8 (5.7–5.9) | <0.001 |
| HOMA-IR, mean (95% CI) | 3.6 (3.4–3.9) | 2.0 (1.9–2.2) | <0.001 |
| HbA1c (mmol/mol), mean (95% CI) | 37.0 (36.6–37.5) | 34.8 (34.3–35.2) | <0.001 |
| C-peptide (pmol/L), mean (95% CI) | 908.3 (867.2–949.3) | 648.0 (616.9–679.1) | <0.001 |
FPG fasting plasma glucose, HOMA-IR homeostasis model assessment of insulin resistance
*P value pre-LED vs post-LED was calculated using paired t-test for FPG and body fat % and Wilcoxon signed-rank test for other variables
Statistical analyses were performed with the statistical program R version 3.5.0 and RStudio version 0.99.903. P values were corrected for multiple comparisons by using the Benjamini-Hochberg procedure (FDR) [44]. P values and FDR-adjusted P values <0.05 were considered significant.
Results
Baseline characteristics and post-LED measurements of the 211 participants included in our analysis are summarized in Table 1. Concordant with the findings from the main PREVIEW trial [25, 45], participants lost an average of 11.5% body weight and 22% total body fat during the LED with a significant improvement in all metabolic parameters investigated (Table 1). Seventy-six participants (36%) reverted to normoglycemia (defined as FPG <5.6 mmol/L). Substantial inter-individual variation was found in the LED-induced changes in glucose metabolism-related variables and total body fat (Fig. 1B). Members of the bacterial phylum Firmicutes dominated the baseline (pre-LED) gut microbiota of participants (86%±11; mean±SD), followed by Actinobacteria (9%±9) and Bacteroidetes (2%±3). Verrucomicrobia and Proteobacteria altogether represented on average less than 3% of the microbiota.
Impact of 8-week LED on gut microbiota
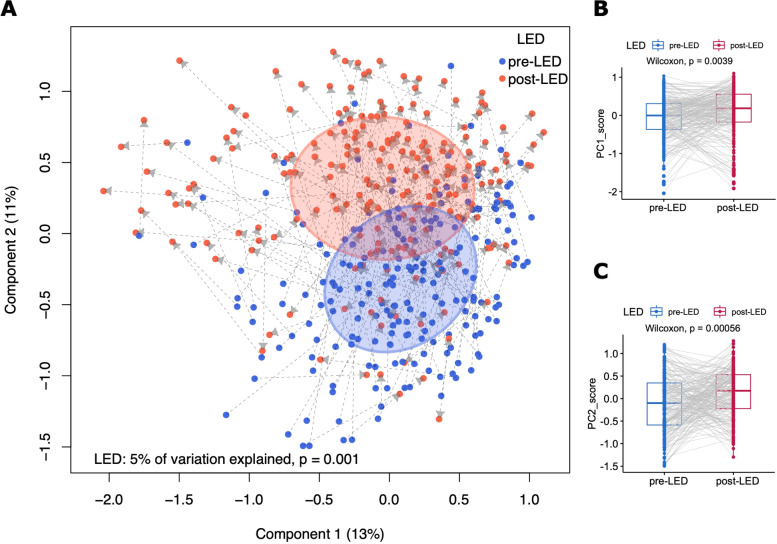
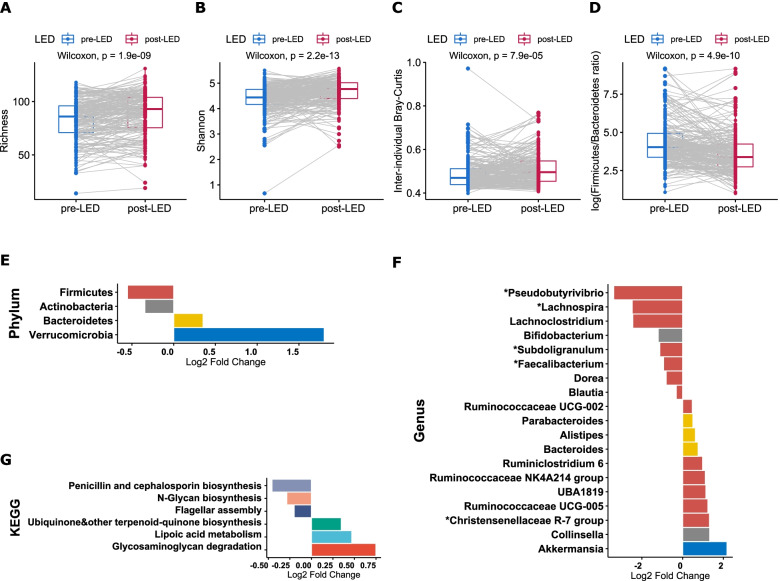
The composition of the gut microbiota changed markedly from pre- to post-LED, visualized in strong clustering of the samples by PCoA and reflected in a significant shift in overall phylogenetic makeup between the two time points (P = 0.001, PERMANOVA) (Fig. 2A). Both the principal component 1 and 2 scores were significantly higher after LED (P < 0.001) (Fig. 2B,C), suggesting a consistent response of the microbiota to the same energy-restricted diet. The LED did not alter total bacterial density, measured by qPCR-based quantification of 16S rRNA gene copies per gram of faeces (Additional file 1: Fig. S2). Microbiota richness and alpha diversity estimated by Shannon index were significantly increased after the intervention (P < 0.001, Fig. 3A,B). Inter-individual Bray-Curtis values, representing how different the microbiota compositions are between participants, were significantly increased after LED weight loss (P < 0.001) (Fig. 3C). A significant decrease was observed in the ratio between Firmicutes and Bacteroidetes (P < 0.001) (Fig. 3D).
Fig. 2.
Principal coordinate analysis (PCoA) of microbiota variation in pre-(blue dots) and post-(red dots) LED samples based on Bray-Curtis distances (A). Arrows link the baseline pre- and post-intervention sample of each individual, indicating direction of change. The blue and red dispersion ellipses represent standard deviations within the groups of pre- and post-intervention samples, respectively. The principal component (PC) scores of PC1 (B) and PC2 (C) are plotted by the sampling time points
Fig. 3.
The LED intervention reshapes the overall microbiota structure, alters relative abundances of individual bacterial taxa and predicted functions. Pre- and post-LED A richness, B diversity within samples (Shannon index), C average dissimilarities (beta diversity) estimated by Bray-Curtis distances between participants, and D Firmicutes to Bacteroidetes ratio. Differentially abundant E phyla, F genera (coloured by respective phyla) and G KEGG modules following the LED ranked by log-fold change are visualized by divergent bar plots. Only the 15 most abundant genera and KEGG modules are shown in F and G. Log2 fold change is calculated as post-LED/pre-LED; only significant results (FDR-P < 0.05) are plotted. The genera known to be able to produce butyrate are marked with an asterisk (*) in I
Having established pre- vs post-LED differences at the level of the entire bacterial community, we sought to identify specific bacteria that were affected by the LED. At the phylum level, the 8-week LED was accompanied by significant increases in Verrucomicrobia and Bacteroidetes (P < 0.001), and concomitant decreases in Actinobacteria and Firmicutes (P < 0.001) (Fig. 3H). At the genus level, in addition to Akkermansia (P < 0.001), the abundances of five genera from family Ruminococcaceae (Ruminococcaceae UCG-002, Ruminiclostridium 6, Ruminococcaceae NK4A214 group, UBA1819, Ruminococcaceae UCG-005) as well as Bacteroides, Alistipes and Christensenellaceae R-7 group, were increased (P < 0.001; Fig. 3I). Christensenellaceae R-7 group appeared to form the hub of the post-LED co-occurrence network with other bacterial groups (Additional file 1: Fig. S3). By contrast, Pseudobutyrivibrio and some other genera belonging to Firmicutes including other butyrate producers such as Lachnospira, Subdoligranulum and Faecalibacterium but also the acetogen Blautia, were significantly decreased (P < 0.001, Fig. 3I). There was also a significant decrease in the abundance of Bifidobacterium (P < 0.001) after the intervention (Fig. 3I). Consistent with the changes in relative abundance, the absolute abundances of Akkermansia and Christensenellaceae R-7 group were significantly increased, and Blautia, Pseudobutyrivibrio and Bifidobacterium significantly decreased after the LED (all P < 0.001) (Additional file 1: Fig. S2).
To understand the functional implications of the observed taxonomic changes, we inferred metagenomes using the PICRUSt2 algorithm. Of 173 KEGG pathways predicted, 6 pathways significantly differed in abundance between the sampling points; these included pathways pertinent to microbial metabolic processes (glycosaminoglycan degradation, lipoic acid metabolism and N-glycan biosynthesis) and the assembly process of flagella (Fig. 3J).
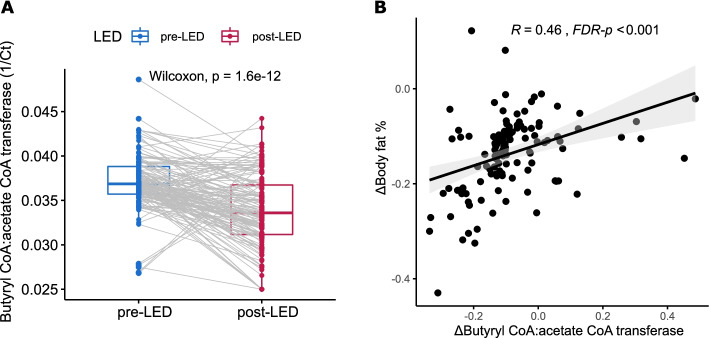
Post–pre-LED changes in butyrate production capacity
As relative abundances of several butyrate-producing bacterial genera were significantly decreased post-LED, we hypothesized that direct quantification of the butyryl-CoA:acetate CoA-transferase gene (responsible for a major route for butyrate production in bacteria) would allow us to gauge changes in butyrate production capacity more precisely. As expected, the post-LED capacity for butyrate production, estimated by qPCR for a subset of participants (N = 139) with available samples, was significantly reduced (P < 0.001, Fig. 4A). Among all the measured clinical indices (Fig. 1B), change in body fat (%) was significantly and positively associated with butyrate production capacity (FDR-P<0.001, Fig. 4B).
Fig. 4.
The LED intervention and body fat (%) reduction associated with reduced capacity for butyrate production in the gut microbiota. A qPCR quantification of the butyryl-CoA:acetate CoA-transferase gene in pre- and post-LED faecal samples. Data are expressed as 1/mean threshold cycle (Ct). B Relative changes (post–pre-LED) in body fat (%) (ΔBody fat %) significantly correlated with relative changes in butyrate production capacity (ΔButyryl CoA:acetate CoA transferase)
Associations between changes in the gut microbiota and clinical variables (post–pre-LED)
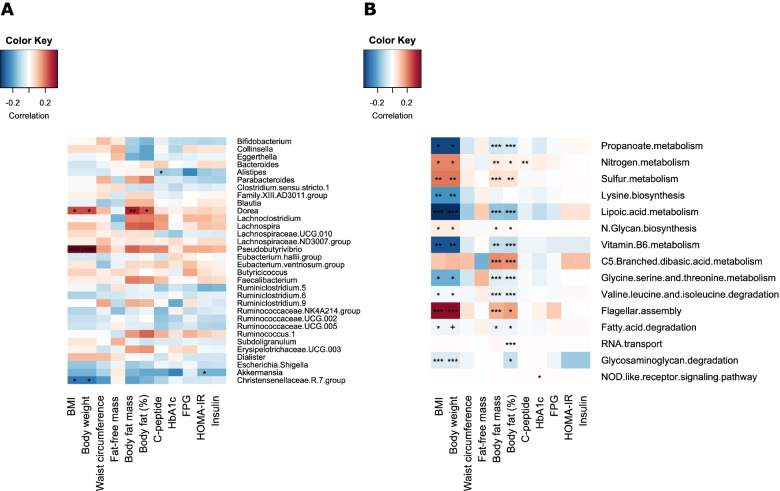
Associations between changes (post–pre diet) in the bacterial genera or predicted functions and clinical indices were first explored by nonparametric correlation analysis to provide an overview of microbiota-host associations (Fig. 5), from which linear mixed-effects models adjusting for demographic variables were fitted to identify significant associations that are potentially generalizable across populations. After the adjustment, changes in BMI and body weight were positively associated with the change in the abundance of Pseudobutyrivibrio (estimate = 0.58, FDR-P<0.01) and negatively with Christensenellaceae R7 group (estimate = − 0.3, FDR-P=0.03). Change in total body fat mass was consistently positively associated with changes in the relative abundances of Pseudobutyrivibrio (estimate=0.16, FDR-P<0.01) and Dorea (estimate=0.04, FDR-P<0.01). These significant associations were not present at baseline (pre-LED) (Additional file 1: Fig. S4). Changes in BMI had a weak but significant negative association with intra-individual Bray-Curtis distance (Additional file 1: Fig. S5), suggesting that the magnitude of microbiota change is associated with individual weight loss.
Fig. 5.
Correlation heatmaps for changes (post–pre-LED) in A bacterial genera and B KEGG functional modules. For readability of the figures, only prevalent bacterial genera (present in >30% of samples) or functional modules that had at least one significant association with changes in clinical measurements are shown. * FDR-P < 0.05; ** FDR-P < 0.01; *** FDR-P < 0.001
With respect to microbiota function, after the adjustment, BMI and body weight remained positively and negatively associated with flagellar assembly (estimate=0.003, FDR-P<0.01) and glycosaminoglycan degradation (estimate = − 0.008, FDR-P<0.01), respectively. These two functional modules were significantly affected by LED as mentioned previously (P < 0.001, Fig. 5B). The changes were attributable to changes in the abundance of Pseudobutyrivibrio (contributing to flagellar assembly) and Akkermansia (contributing to glycosaminoglycan degradation) (Additional file 1: Fig. S6).
Prediction of host responses to LED using baseline microbiota
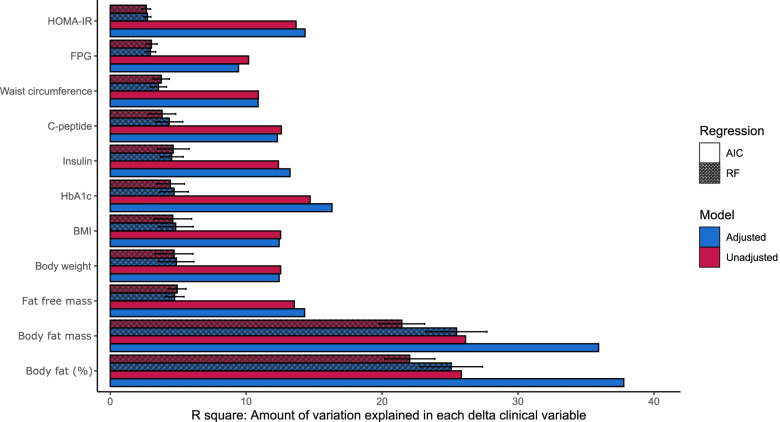
Given the above connection between the microbiota changes and changes in adiposity, we asked whether the extent of host response to LED could be predicted based on an individual’s baseline microbiota. Our results from stepwise and random forest (RF) regressions indicated that baseline features of the gut microbiota explained a significant proportion of variance in both unadjusted and adjusted models (ca. 26–38% in stepwise regression; 22–25% in RF) in %change of total body fat, but not other clinical indices during the LED (Fig. 6). Similar results were obtained by applying the same set of predictors in the Finnish participants only (N = 151) (Additional file 1: Fig. S7). The baseline microbiota features predictive of total body fat change are listed in Table 2 for the stepwise regression model and in Additional file 2: Table S1-S2 for the RF regression models. Erysipelotrichaceae UCG-003 emerged as consistently predictive of total body fat change in both stepwise and RF regressions and had the strongest correlation with changes in total body fat (Additional file 1: Fig. S8). We next constructed predictive models for total body fat change based on 4 sets of baseline host features, including (1) microbiota-only, (2) diet-only, (3) host clinical characteristics (Table 1) and (4) a combination of 1–3. The prediction based on the combined model outperformed the predictions based on all other models, as the difference in the Akaike information criterion (AIC) value between the best (i.e., combined model) and second best model (i.e., host clinical characteristics) was larger than the conventional cutoff of 10 AIC units for significant model support. The predicted and measured total body fat (%) change based on the combined model had Spearman’s R of 0.74, corresponding to 55% of the variance in total body fat (%) change (P < 0.001, Fig. 7). The combined model indicated that the higher the baseline body fat (%), monounsaturated fatty acid intake in the habitual diet, and gut microbiota richness, the less successful was the intervention in terms of fat loss (%). Conversely, a high relative abundance of Clostridium sensu stricto 1, Ruminococcaceae UCG-003 and Parabacteroides at baseline were predictive of increased fat loss (%) during the intervention (Table 2). In the model without adjustment for host characteristics and habitual diet, also Lactococcus and an unclassified genus of Peptostreptococcaceae were significantly associated with a good response, while Erysipelotrichaceae UCG-003 was predictive of poor response. Their effects were, however, explained by the host characteristics, rather than being predictive in their own right.
Fig. 6.
Amount of variation in changes of clinical indices explained by baseline gut microbiota. The bar graph shows the estimated R2 in Random Forest (RF) and stepwise regressions. The error bars show 95% confidence intervals from repeated cross-validation of the random forests to predict the delta (post–pre-LED) clinical indices. In both regression models, the adjusted model includes demographic variables (age, gender and ethnicity) in addition to microbiota features. The unadjusted model is included as a contrast to showcase the predictive power of gut microbiota features for specific clinical indices without conflating the information related to host clinical characteristics
Table 2.
Variables retained in the prediction models for relative change in total body fat (%) based on stepwise selection
| Variable | Coefficient | SE | z | P |
|---|---|---|---|---|
| Combined | ||||
| Body fat (%) | 0.049 | 0.0048 | 10.23 | <0.001 |
| Gut microbiota richness | 0.01 | 0.005 | 1.9 | 0.05 |
| Clostridium sensu stricto 1 | − 0.007 | 0.0045 | − 1.58 | 0.1 |
| Parabacteroides | − 0.008 | 0.0046 | − 1.8 | 0.07 |
| FPG (mmol/L) | − 0.01 | 0.004 | − 2.54 | 0.01 |
| Ruminococcaceae UCG-003 | − 0.01 | 0.0047 | − 2.63 | 0.009 |
| MUFA intake (en%) | 0.01 | 0.0044 | 2.29 | 0.02 |
| Host clinical characteristics | ||||
| Gender | − 0.024 | 0.017 | − 1.443 | 0.15 |
| Waist circumference (cm) | − 0.522 | 0.19 | − 2.73 | 0.007 |
| BMI (kg/m2) | − 0.01 | 0.004 | − 2.37 | 0.02 |
| Body fat (%) | 0.012 | 0.002 | 5 | <0.001 |
| FPG (mmol/L) | − 0.017 | 0.007 | − 2.26 | 0.02 |
| Gut microbiota | ||||
| Ruminococcaceae UCG-003 | − 0.013 | 0.005 | − 2.35 | 0.02 |
| Lactococcus | − 0.01 | 0.006 | − 1.88 | 0.06 |
| Parabacteroides | − 0.013 | 0.005 | − 2.46 | 0.01 |
| Peptostreptococcaceae_unclassified | − 0.021 | 0.006 | − 3.76 | <0.001 |
| Erysipelotrichaceae UCG-003 | 0.014 | 0.006 | 2.56 | 0.04 |
| Diet | ||||
| Total fat intake (en%) | − 11.1 | 6.12 | − 1.82 | 0.07 |
| Total CHO intake (en%) | 0.936 | 0.425 | 2.21 | 0.03 |
| Sugars intake (en%) | − 1.02 | 0.66 | − 1.54 | 0.1 |
| Fibre intake (en%) | − 3.08 | 2.2 | − 1.4 | 0.16 |
| SFA intake (en%) | 11.06 | 7.57 | 1.46 | 0.14 |
| MUFA intake (en%) | 14.2 | 6.43 | 2.21 | 0.03 |
| PUFA intake (en%) | 14 | 8.41 | 1.67 | 0.1 |
BMI body mass index, FPG fasting plasma glucose, CHO carbohydrate, SFA saturated fat, MUFA monounsaturated fat, PUFA polyunsaturated fat
Fig. 7.
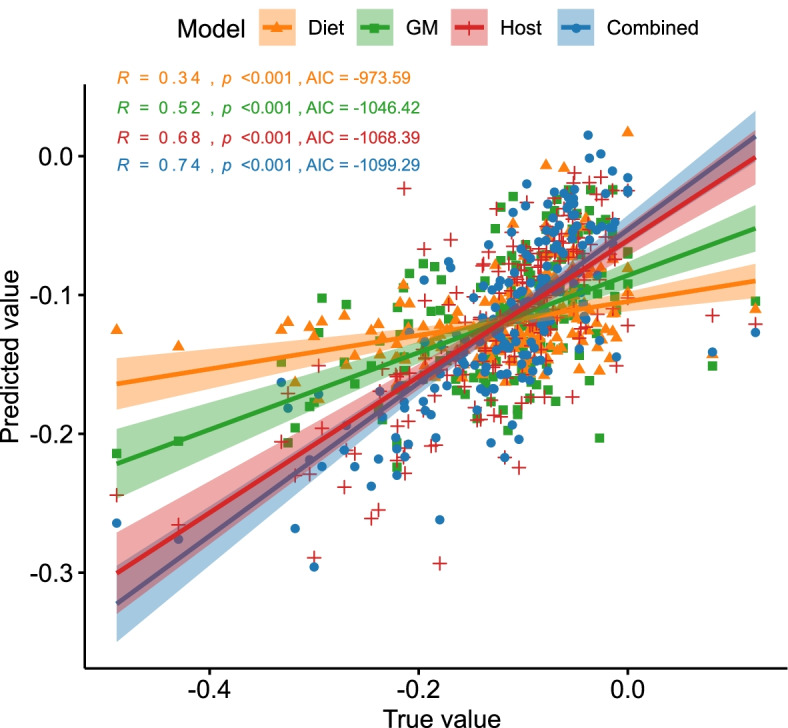
Comparison of the association strengths between the true and predicted total body fat (%) change during the LED based on the four different models. The lines represent the fitted regression lines (Spearman’s rank correlation coefficients displayed at the upper left corner) and the corresponding shaded area represents the 95% confidence intervals for each model. GM, gut microbiota; host, host clinical characteristics including demographic characteristics, anthropometric and metabolic measurements as presented in Table 1
Discussion
Recent preclinical and clinical studies suggest that, along with other host and environmental factors [46, 47], the gut microbiota contributes to individual variability in diet-induced weight loss [7]. Here we present microbiota data derived from PREVIEW, the largest intervention to date in overweight or obese adults with pre-diabetes undertaking an 8-week LED for weight loss. To our knowledge, the participants included in this PREVIEW sub-study represent the largest cohort to date investigating the impact of a commercial total meal replacement LED on the gut microbiota. Our results show that (1) the LED intervention significantly altered the overall community structure, relative and absolute abundances of bacterial taxa and functional potential of the microbiota; (2) changes in the gut microbiota were strongly associated with changes in adiposity-related variables; and ( 3) decrease in body fat during the LED was predicted by the baseline features of the gut microbiota.
We observed drastic shifts in the overall microbiota structure measured by beta diversity after 8-weeks of LED, which is consistent with the previous results from Frost and colleagues’ 6-week energy restriction trial in obese individuals with T2D [14] and Heinsen’s 12-week intervention in obese adults with various chronic diseases [17], both of which used total meal replacements of similar macronutrient composition and energy content (3.4 MJ/day). Similarly, Simões et al. showed significant changes in dominant faecal bacteria following a 6-week very low-energy diet (VLED; providing fewer than 800 kcal/day (3.3 MJ)) in adults with obesity [16]. Moreover, a recent intervention analysing the effects of VLED (2.5 MJ/day) in 30 adults with overweight or obesity and non-alcoholic fatty liver disease (NAFLD) also found that 4 weeks of VLED had a significant impact on the overall gut microbiota [18]. One weight loss study not using total meal replacement products reported that a 6-week high-protein energy-restricted diet significantly increased microbial gene richness that was associated with improved clinical phenotypes in obese and overweight adults [48]. In contrast, many other weight loss interventions with duration ranging from 4 to 12 weeks did not find a significant difference in microbiota structure before and after energy restriction in overweight and obese individuals with a range of metabolic syndrome and NAFLD conditions [13, 15, 19, 20]. Evidence indicates that the microbiota may respond to diet within 1–3 days [49–51], suggesting a 4-week timeframe should have been sufficient to observe changes in the microbiota. The differences are unexpected since most of these trials used a very similar approach to weight loss, namely LED complete meal replacement. The discrepancy may perhaps arise from small differences in macro and micronutrient composition, compliance to the LED, or more likely differences in microbiome methodologies which may be exacerbated in these studies of very small sample size [12, 22]. Interestingly, the microbiota responses to the same LED in the PREVIEW participants were highly consistent, as opposed to personalized microbiota responses to similar foods previously reported in 34 healthy individuals [51].
In contrast to a recent VLED study in 40 post-menopausal women with overweight or obesity [52], we did not observe a reduction in total bacterial density after the 8-week LED. This difference is possibly owing to the different macronutrient compositions of the OPTIFAST® Liquid Diet [52] and the Cambridge Weight Plan® used in our study. Other indices that reflect microbiota structure, namely microbiota richness and alpha diversity, increased significantly following the LED as also reported by previous studies using similar energy-restricted regimes [14, 48]. While greater microbiota diversity does not necessarily imply better health [53], low diversity has been linked to poor metabolic health due to, e.g., loss of metabolically important functional capacity [54]. In comparison with the baseline, the post-LED microbiota can be delineated by (1) reduced fibre-degrading Firmicutes (mainly the Lachnospiraceae family as well as other key butyrate producers), (2) decreased relative and absolute abundance of Bifidobacterium and (3) concomitant increases in Akkermansia and the Christensenellaceae family. Pseudobutyrivibrio in the Lachnospiraceae family was the genus that decreased the most following the LED in our study; this genus has an elusive role as it has been associated with both weight loss [55] and the pro-inflammatory response [56] in animal models.
By inferring metagenomes, we show that reduced Pseudobutyrivibrio may have contributed to lower microbiota capacity in flagellar assembly, the changes in which associated positively to that of body weight and fat mass. This is relevant as flagellins are canonical effectors of Toll-like receptor 5 and have been suggested to contribute to obesity [57]. Reductions in Bifidobacterium and several butyrate-producing bacteria are not surprising considering these bacteria thrive on indigestible polysaccharides that were scarce during the LED. While decreased Bifidobacterium is often associated with metabolic disorders [58], the change in Bifidobacterium did not correlate with clinical measurements investigated here in PREVIEW. The observed small reduction of Blautia is of interest, as Blautia is a highly abundant and acetogenic group and recent studies have shown that the intestinal levels of Blautia spp. are increased in patients with T2D as compared to healthy controls [59]. Akkermansia, previously shown to be inversely correlated with metabolic derangements [60] and recently shown to improve barrier functions and insulinemia in obese individuals [61], was significantly increased following the LED in PREVIEW. This contributed to the elevated microbiota capacity for glycosaminoglycan degradation, in line with the extensive capacity of Akkermansia for genes capable of metabolizing host glycans, e.g., mucins [62] to gain an edge over competitors during scarcity of dietary glycans as also noted in fasting animals [63]. Increases in Christensenellaceae R-7 group were likely partly promoted by the adequate protein content in the LED (43.7 E%, 88 g/day), as this bacterial group specializes in protein fermentation and may produce butyrate [64]. Members of the Christensenellaceae family have been linked to decreased adiposity [65], which is consistent with our correlation analysis. It has been hypothesized that the protective effect of Christensenellaceae against excess adiposity gain involves remodeling the microbial community [64], which is supported by the Christensenellaceae-centred co-occurrence network after the LED as well as in a previous study [66].
Akkermansia was the only genus found to negatively associate with insulin resistance index (HOMA-IR) during LED, but the significance disappeared after adjusting for demographics. While a recent study in ~1500 Swedes strongly associated insulin resistance and glycemic status, specifically impaired glucose tolerance, with the gut microbiota [67], we found few associations between the gut bacteria and changes in glucose metabolism during the LED. A possible explanation is that one key mechanism by which the gut microbiota mediates glucose metabolism is through colonic fermentation of dietary fibre [68], which was however relatively low in the intervention diet. A recent post hoc analysis of a subgroup in Cotillard and colleagues’ calorie restriction (CR) intervention [48], where the diet was rich in total fibre, found that fibre intake and gut metagenomic species that were interconnected in a biological network had the greatest contribution to CR-induced improvement in insulin sensitivity [69].
The capacity for butyrate production in the post-LED gut microbiota, quantified by qPCR rather than inferred metagenomics, was significantly reduced. This mirrors the results from a recent VLED study [52]. Interestingly, we found that reduced butyrate production capacity was proportional to reduced body fat (%). While butyrate is generally considered anti-obesogenic, some studies reported higher concentrations of butyrate in obese humans than in lean individuals [70]. Greater stool calorie loss, a proxy of decreased nutrient absorption, has been linked to lower circulating [71] and faecal [52] levels of butyrate, suggesting decreased gut microbial capacity in processing nutrients. Our findings therefore lend credence to the new working model where weight loss and metabolic improvements in obese individuals induced by caloric restriction are potentially mediated by impaired nutrient absorption that is associated with gut microbiota changes, including reduced butyrate production [52, 71]. This also implies that the effects of butyrate on body weight control likely depend on host health status and/or dietary conditions. While the observations in the present study should not be taken as evidence of causality or causal mechanisms, the role of butyrate in nutrient absorption under different clinical and nutritional conditions warrants further mechanistic investigation. Moreover, the long-term impact of reduced butyrate production capacity induced by the LED on host health requires clarification.
Our previous studies [72, 73] and data from others [74, 75] have suggested that an individual’s metabolic response to different diets depends partly on baseline features of the microbiota. Recently, the baseline ratio between Prevotella and Bacteroides was proposed as a predictive biomarker for weight loss [76, 77]. However, the utility of this ratio may be limited by the sparsity of Prevotella [78]. A recent Chinese study in 83 participants on a 6-month self-managed dieting program suggests that the baseline gut microbiota is predictive of weight loss [21], whereas body composition and other metabolic markers were not measured. Importantly, BMI or body weight per se is an inadequate indicator for metabolic health [79] or the success of any interventions [80]. As the variance in weight change in our PREVIEW cohort was relatively small due to the fully controlled LED and high compliance, we were able to focus on the variables representing the quality of the weight loss, e.g., body fat loss and lean mass loss. By applying predictive modelling in our current study, we show that the baseline microbiota alone was able to explain a significant proportion of variance (up to 38%) in total body fat (%), but not in BMI or body weight, during LED. Since weight loss entails a reduction in both lean (fat-free) and fat mass, the baseline microbiota likely modifies the LED-driven effect on the fat tissue specifically. Indeed, preclinical and clinical studies have consistently shown that the gut microbiota modulates adipose tissue physiology [65, 81–83]. Of note, the cross-sectional results from the large-scale PREDICT1 study (N = 1098) found that visceral fat was more strongly linked to gut microbial composition than BMI [84]. Our findings from the PREVIEW intervention therefore extend on the findings from PREDICT1, highlighting the crucial role of the gut microbiota in adipose tissue physiology. In contrast, the baseline microbiota was not predictive of changes in clinical indices related to glucose metabolism in PREVIEW. Given the lack of association between the gut microbiota and glucose metabolism during weight loss, the LED-driven improvement in glucose metabolism was likely microbiota-independent in our cohort. The baseline relative abundances of Erysipelotrichaceae UCG-003 and Clostridium sensu stricto 1 were selected as important features for prediction of total body fat change by both feature selection techniques, but their abundances were otherwise unchanged during the LED phase. Analogous findings were reported in a recent Danish study [24] and this again suggests that the LED-induced change in body fat was modified by the baseline microbiota as an effect modifier [23]. Erysipelotrichaceae UCG-003 (the top feature in predicting total body fat change in PREVIEW) was recently identified as a key player in a cohort of lean patients with confirmed NAFLD [85], a consequence of increased non-adipose ectopic fat deposition into liver. Members of the Erysipelotrichaceae family have also been repeatedly linked to host lipid and cholesterol phenotypes in humans [86] and experimental animals [87, 88]; likewise, Clostridium sensu stricto 1 has been associated with high-density lipoprotein (HDL) metabolism in a recent population-based study [89].
For prediction of total body fat change during LED in PREVIEW, the combined model with host physiological, dietary and microbial features had better predictive performance than the model with only host physiological features. This suggests that microbial and habitual dietary features modify host responses to dietary change, rather than acting as mere proxies for the bio-clinical features. Notably, several species of Parabacteroides, one of the taxa predictive of fat loss, have been previously shown to reduce obesity in mice [90, 91]. Whether gut microbes influence fat loss via metabolism, or e.g. via eating behaviour [92], is an intriguing question. SCFAs, neurotransmitters and peptides produced by gut microbes are all hypothesized to regulate appetite [93]. Weight loss success during a diet is largely an outcome of behaviour alongside physiology [94]. In this PREVIEW sub-study, we found unexpectedly that the higher the baseline body fat percentage, the poorer the response in terms of fat percentage loss, suggestive of lower compliance and/or more sedentary lifestyle. Taken together, our work and other recent studies [21, 24] demonstrate that data integration using host and microbial features prior to an intervention is able to predict diet-induced metabolic changes over relatively long time spans, while this strategy has only been utilized to successfully predict postprandial responses previously [75, 95].
As inter-individual variation in the gut microbiota is notably high and shaped by various factors such as long-term diet and host genetics [7], small and homogenous cohorts as utilized by previous studies provide little generalizability and applicability. The main strength of our report is therefore a large novel cohort of overweight adults confirmed to be at high risk of T2D from two different. geographical regions and various ethnicities, adhering to a well-controlled and uniform dietary regime for weight loss, rendering the study clinically relevant. Compliance to diet was confirmed by the significant ≥8% body weight loss over 8 weeks. The results presented in this study should nonetheless be interpreted with some limitations in mind, including different methods for total body fat assessment (DXA and BIA) in the two study sites, which may have resulted in differences in absolute body fat mass and fat-free mass. However, we mitigated potential bias to the greatest extent by analysing change from baseline data, making the results from DXA and BIA more comparable [96]. We additionally tested the regression model in the Finnish participants only (body composition assessed using BIA) to ensure consistent findings. While we cannot exclude the possibility that natural temporal fluctuation of the gut microbiota partly contributed to the observed changes, the gut microbiota is known to be rather stable in the absence of extreme external stressors even over a 2-year period [97].
Conclusions
Obesity and its most prevalent co-morbidity, T2D, could affect half of the world’s adult population by 2030 [98]. By identifying the gut microbiota as an important co-determinant of LED-induced reduction in total body fat, our study lays the foundation for pre-intervention assessment and patient stratification using individual microbiota profiles. A recent study suggests that daily pre-diet gut microbiota variability, termed “plasticity,” is associated with diet-induced weight loss [99]. Hence, integration of baseline microbiota variability by daily sampling prior to weight loss might improve the prediction of total body fat change, which warrants investigation in future studies. In conclusion, an 8-week LED weight loss intervention in adults with overweight and pre-diabetes significantly influenced microbiota structure, functional potential and relative abundance of several bacterial taxa. These correlated with favorable changes in adiposity that can be predicted by baseline features of the gut microbiota.
Supplementary Information
Additional file 1. Supplementary Information consisting of eight supporting figures (Figure S1-S8). A short description of the figure and a figure caption for each figure are given within the file.
Additional file 2. Two supporting tables (Table S1-S2) of lists of input predictors for body fat change in random forest regression ranked by feature importance. A table caption for each table is given within the file.
Additional file 3. PREVIEW study protocol.
Acknowledgements
We thank Saara Kettunen and Laura Huuskonen for the faecal DNA extractions of the Finnish samples, Afshan Saleem for performing the qPCR assays for quantification of butyrate production capacity, and Evgenia Dikareva for performing the qPCR assays for total bacterial quantification. We thank Amy Liu, Kelly Storey, Anne-Thea McGill, Lindsay Plank, Katya Volkova, Madhavi Bollineni and Clarence Vivar for assistance in conduct of the Auckland intervention. Also Jouko Sundvall and Laura Råman from National Institute for Health and Welfare THL, Finland for analysis of blood samples.
Abbreviations
- LED
Low-energy diet
- T2D
Type 2 diabetes
- NAFLD
Non-alcoholic fatty liver disease
- BIA
Bioelectrical impedance analysis
- DXA
Dual-energy X-ray absorptiometry
- BMI
Body mass index
- FPG
Fasting plasma glucose
- HOMA-IR
Homeostatic Model Assessment of Insulin Resistance
- HbA1c
Glycated hemoglobin
- SCFA
Short-chain fatty acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- rRNA
Ribosomal RNA
- PCoA
Principal coordinate analysis
- FDR-P
FDR-adjusted P value
- RF
Random forest
Authors’ contributions
CJ processed the sequencing data, analysed, interpreted and visualized the results and wrote the manuscript with contributions from MPS, MWT, SDP, KK, WMdV and AS. KK assisted in the performance and interpretation of predictive modelling. AS, WMdV and MWT coordinated and supervised the processing of faecal samples for sequencing. AR, SDP and MF designed and conceived the clinical study. EJ and MPS coordinated and supervised the intervention, collected and analysed the dietary data and clinical variables. DM and DB processed the NZ faecal samples. All authors read and approved the final manuscript.
Funding
This PREVIEW sub-study was funded by the EU 7th Framework Programme (FP7/2007-2013) under grant agreement # 312057; and the New Zealand Health Research Council (grant #14/191) and University of Auckland Faculty Research Development Fund (grants #3702182, #3706738). The Cambridge Weight Plan® (Northants, UK) kindly donated all low-energy diet products. This study was also supported by grants from the University of Helsinki, the Mary and Georg Ehnrooth Foundation and the Otto A. Malm Foundation.
Availability of data and materials
The sequencing data in this study are available at the European Nucleotide Archive (ENA) under accession number PRJEB43667 (https://www.ebi.ac.uk/ena/browser/view/PRJEB43667) [100]. All codes are fully accessible from the referenced sources in the program R.
Declarations
Ethics approval and consent to participate
The work of PREVIEW is carried out in full compliance with the relevant requirements of the latest version of the Declaration of Helsinki (59th WMA General Assembly, Seoul, Korea, October 2008), and the ICH-GCP, The International Conference on Harmonisation (ICH) for Good Clinical Practice to the extent that this is possible and relevant. The study protocol was approved by the Ethical Committees of participating countries (Health and Disability Ethics Committee in New Zealand, ref. 14/191 and Medical Ethical Committees of the Hospital District of Helsinki and Uusimaa and HUCH in Finland, ref. 171/13/03/00/2013). All participants provided written informed consent prior to commencing screening procedures in clinic. All information obtained during the trial is handled according to local regulations and the European Directive 95/46/CE (directive on protection of individuals with regard to the processing of personal data and on the free movement of such data). The PREVIEW trial was prospectively registered at ClinicalTrials.gov (NCT01777893) on January 29, 2013.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ching Jian and Marta Paulino Silvestre contributed equally to this work.
References
- 1.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.Franz MJ, VanWormer JJ, Crain AL, Boucher JL, Histon T, Caplan W, Bowman JD, Pronk NP. Weight-loss outcomes: a systematic review and meta-analysis of weight-loss clinical trials with a minimum 1-year follow-up. J Am Diet Assoc. 2007;107:1755–1767. doi: 10.1016/j.jada.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Astbury NM, Aveyard P, Nickless A, Hood K, Corfield K, Lowe R, Jebb SA. Doctor Referral of Overweight People to Low Energy total diet replacement Treatment (DROPLET): pragmatic randomised controlled trial. BMJ. 2018;362:k3760. doi: 10.1136/bmj.k3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lean ME, Leslie WS, Barnes AC, Brosnahan N, Thom G, McCombie L, Peters C, Zhyzhneuskaya S, Al-Mrabeh A, Hollingsworth KG, et al. Primary care-led weight management for remission of type 2 diabetes (DiRECT): an open-label, cluster-randomised trial. Lancet. 2018;391:541–551. doi: 10.1016/s0140-6736(17)33102-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruvada P, Leone V, Kaplan LM, Chang EB. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe. 2017;22:589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Hughes RL, Kable ME, Marco M, Keim NL. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models. Part II: Results. Adv Nutr. 2019;10:979–998. doi: 10.1093/advances/nmz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Huang M, You X, Zhao J, Chen L, Wang L, Luo Y, Chen Y. Gut microbiota mediates the anti-obesity effect of calorie restriction in mice. Sci Rep. 2018;8:13037. doi: 10.1038/s41598-018-31353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riedl RA, Atkinson SN, Burnett CML, Grobe JL, Kirby JR. The gut microbiome, energy homeostasis, and implications for hypertension. Curr Hypertens Rep. 2017;19:27. doi: 10.1007/s11906-017-0721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowah SA, Riedl L, Damms-Machado A, Johnson TS, Schübel R, Graf M, Kartal E, Zeller G, Schwingshackl L, Stangl GI, et al. Effects of weight-loss interventions on short-chain fatty acid concentrations in blood and feces of adults: a systematic review. Adv Nutr. 2019;10:673–684. doi: 10.1093/advances/nmy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lorenzo F, De Castro C, Silipo A, Molinaro A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol Rev. 2019;43:257–272. doi: 10.1093/femsre/fuz002. [DOI] [PubMed] [Google Scholar]
- 12.Lane M, Howland G, West M, Hockey M, Marx W, Loughman A, O'Hely M, Jacka F, Rocks T. The effect of ultra-processed very low-energy diets on gut microbiota and metabolic outcomes in individuals with obesity: a systematic literature review. Obes Res Clin Pract. 2020;14:197–204. doi: 10.1016/j.orcp.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Ott B, Skurk T, Hastreiter L, Lagkouvardos I, Fischer S, Büttner J, Kellerer T, Clavel T, Rychlik M, Haller D, Hauner H. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci Rep. 2017;7:11955. doi: 10.1038/s41598-017-12109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost F, Storck LJ, Kacprowski T, Gärtner S, Rühlemann M, Bang C, Franke A, Völker U, Aghdassi AA, Steveling A, et al. A structured weight loss program increases gut microbiota phylogenetic diversity and reduces levels of Collinsella in obese type 2 diabetics: a pilot study. PLoS One. 2019;14:e0219489. doi: 10.1371/journal.pone.0219489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damms-Machado A, Mitra S, Schollenberger AE, Kramer KM, Meile T, Königsrainer A, Huson DH, Bischoff SC. Effects of surgical and dietary weight loss therapy for obesity on gut microbiota composition and nutrient absorption. Biomed Res Int. 2015;2015:806248. doi: 10.1155/2015/806248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simões CD, Maukonen J, Scott KP, Virtanen KA, Pietiläinen KH, Saarela M. Impact of a very low-energy diet on the fecal microbiota of obese individuals. Eur J Nutr. 2014;53:1421–1429. doi: 10.1007/s00394-013-0645-0. [DOI] [PubMed] [Google Scholar]
- 17.Heinsen FA, Fangmann D, Müller N, Schulte DM, Rühlemann MC, Türk K, Settgast U, Lieb W, Baines JF, Schreiber S, et al. Beneficial effects of a dietary weight loss intervention on human gut microbiome diversity and metabolism are not sustained during weight maintenance. Obes Facts. 2016;9:379–391. doi: 10.1159/000449506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chong CYL, Orr D, Plank LD, Vatanen T, O'Sullivan JM, Murphy R. Randomised double-blind placebo-controlled trial of inulin with metronidazole in non-alcoholic fatty liver disease (NAFLD). Nutrients. 2020;12. 10.3390/nu12040937. [DOI] [PMC free article] [PubMed]
- 19.Alemán JO, Bokulich NA, Swann JR, Walker JM, De Rosa JC, Battaglia T, Costabile A, Pechlivanis A, Liang Y, Breslow JL, et al. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J Transl Med. 2018;16:244. doi: 10.1186/s12967-018-1619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PLoS One. 2016;11:e0149564. doi: 10.1371/journal.pone.0149564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jie Z, Yu X, Liu Y, Sun L, Chen P, Ding Q, et al. The baseline gut microbiota directs dieting-induced weight loss trajectories. Gastroenterology. 2021. 10.1053/j.gastro.2021.01.029. [DOI] [PubMed]
- 22.Seganfredo FB, Blume CA, Moehlecke M, Giongo A, Casagrande DS, Spolidoro JVN, Padoin AV, Schaan BD, Mottin CC. Weight-loss interventions and gut microbiota changes in overweight and obese patients: a systematic review. Obes Rev. 2017;18:832–851. doi: 10.1111/obr.12541. [DOI] [PubMed] [Google Scholar]
- 23.Hughes RL, Marco ML, Hughes JP, Keim NL, Kable ME. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models-part I: overview of current methods. Adv Nutr. 2019;10:953–978. doi: 10.1093/advances/nmz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen RL, Helenius M, Garcia SL, Roager HM, Aytan-Aktug D, Hansen LBS, Lind MV, Vogt JK, Dalgaard MD, Bahl MI, et al. Data integration for prediction of weight loss in randomized controlled dietary trials. Sci Rep. 2020;10:20103. doi: 10.1038/s41598-020-76097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen P, Meinert Larsen T, Westerterp-Plantenga M, Macdonald I, Martinez JA, Handjiev S, Poppitt S, Hansen S, Ritz C, Astrup A, et al. Men and women respond differently to rapid weight loss: metabolic outcomes of a multi-centre intervention study after a low-energy diet in 2500 overweight, individuals with pre-diabetes (PREVIEW) Diabetes Obes Metab. 2018;20:2840–2851. doi: 10.1111/dom.13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fogelholm M, Larsen TM, Westerterp-Plantenga M, Macdonald I, Martinez JA, Boyadjieva N, et al. PREVIEW: prevention of diabetes through lifestyle intervention and population studies in Europe and around the world. Design, methods, and baseline participant description of an adult cohort enrolled into a three-year randomised clinical trial. Nutrients. 2017;9. 10.3390/nu9060632. [DOI] [PMC free article] [PubMed]
- 27.2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care. 2020;43:S14–s31. 10.2337/dc20-S002. [DOI] [PubMed]
- 28.Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Luukkonen PK, Sadevirta S, Zhou Y, Kayser B, Ali A, Ahonen L, Lallukka S, Pelloux V, Gaggini M, Jian C, et al. Saturated fat is more metabolically harmful for the human liver than unsaturated fat or simple sugars. Diabetes Care. 2018;41:1732–1739. doi: 10.2337/dc18-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. doi: 10.1128/aem.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jian C, Luukkonen P, Yki-Järvinen H, Salonen A, Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS One. 2020;15:e0227285. doi: 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoddard SF, Smith BJ, Hein R, Roller BR, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–d361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, et al. Package ‘vegan’. Community ecology package. R package version 2.5-6. 2019.
- 40.Korpela K. Mare: microbiota analysis in R easily. R package version 1.0. 2016. Available at: https://github.com/katrikorpela/mare. 10.5281/zenodo.50310.
- 41.Burnham KP, Anderson DR. Multimodel inference:understanding AIC and BIC in model selection. Sociol Methods Res. 2004;33:261–304. doi: 10.1177/0049124104268644. [DOI] [Google Scholar]
- 42.Wiener ALaM. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 43.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28. 10.18637/jss.v028.i05.
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 1995;57:289–300. [Google Scholar]
- 45.Raben A, Vestentoft PS, Brand-Miller J, Jalo E, Drummen M, Simpson L, Martinez JA, Handjieva-Darlenska T, Stratton G, Huttunen-Lenz M, et al. The PREVIEW intervention study: results from a 3-year randomized 2 x 2 factorial multinational trial investigating the role of protein, glycaemic index and physical activity for prevention of type 2 diabetes. Diabetes Obes Metab. 2021;23:324–337. doi: 10.1111/dom.14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heianza Y, Qi L. Gene-diet interaction and precision nutrition in obesity. Int J Mol Sci. 2017;18. 10.3390/ijms18040787. [DOI] [PMC free article] [PubMed]
- 47.Stubbs J, Whybrow S, Teixeira P, Blundell J, Lawton C, Westenhoefer J, Engel D, Shepherd R, McConnon A, Gilbert P, Raats M. Problems in identifying predictors and correlates of weight loss and maintenance: implications for weight control therapies based on behaviour change. Obes Rev. 2011;12:688–708. doi: 10.1111/j.1467-789X.2011.00883.x. [DOI] [PubMed] [Google Scholar]
- 48.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 49.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. Isme J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Walter J, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25:789–802.e785. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 52.von Schwartzenberg RJ, Bisanz JE, Lyalina S, Spanogiannopoulos P, Ang QY, Cai J, Dickmann S, Friedrich M, Liu SY, Collins SL, et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature. 2021;595:272–277. doi: 10.1038/s41586-021-03663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 54.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henning SM, Yang J, Hsu M, Lee RP, Grojean EM, Ly A, Tseng CH, Heber D, Li Z. Decaffeinated green and black tea polyphenols decrease weight gain and alter microbiome populations and function in diet-induced obese mice. Eur J Nutr. 2018;57:2759–2769. doi: 10.1007/s00394-017-1542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Bian G, Sun D, Zhu W, Mao S. Starter feeding supplementation alters colonic mucosal bacterial communities and modulates mucosal immune homeostasis in newborn lambs. Front Microbiol. 2017;8:429. doi: 10.3389/fmicb.2017.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pekkala S, Munukka E, Kong L, Pöllänen E, Autio R, Roos C, Wiklund P, Fischer-Posovszky P, Wabitsch M, Alen M, et al. Toll-like receptor 5 in obesity: the role of gut microbiota and adipose tissue inflammation. Obesity (Silver Spring). 2015;23:581–590. doi: 10.1002/oby.20993. [DOI] [PubMed] [Google Scholar]
- 58.Esteve E, Ricart W, Fernández-Real JM. Gut microbiota interactions with obesity, insulin resistance and type 2 diabetes: did gut microbiote co-evolve with insulin resistance? Curr Opin Clin Nutr Metab Care. 2011;14:483–490. doi: 10.1097/MCO.0b013e328348c06d. [DOI] [PubMed] [Google Scholar]
- 59.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Y, Wang N, Tan HY, Li S, Zhang C, Feng Y. Function of Akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol. 2020;11:219. doi: 10.3389/fmicb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mack I, Penders J, Cook J, Dugmore J, Mazurak N, Enck P. Is the impact of starvation on the gut microbiota specific or unspecific to anorexia nervosa? A narrative review based on a systematic literature search. Curr Neuropharmacol. 2018;16:1131–1149. doi: 10.2174/1570159x16666180118101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waters JL, Ley RE. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019;17:83. doi: 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beaumont M, Goodrich JK, Jackson MA, Yet I, Davenport ER, Vieira-Silva S, Debelius J, Pallister T, Mangino M, Raes J, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016;17:189. doi: 10.1186/s13059-016-1052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Krämer M, Gummesson A, Perkins R, Bergström G, Bäckhed F. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 2020;32:379–390.e373. doi: 10.1016/j.cmet.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 68.Gérard C, Vidal H. Impact of gut microbiota on host glycemic control. Front Endocrinol (Lausanne). 2019;10:29. doi: 10.3389/fendo.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dao MC, Sokolovska N, Brazeilles R, Affeldt S, Pelloux V, Prifti E, Chilloux J, Verger EO, Kayser BD, Aron-Wisnewsky J, et al. A data integration multi-omics approach to study calorie restriction-induced changes in insulin sensitivity. Front Physiol. 2018;9:1958. doi: 10.3389/fphys.2018.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu H, Wang J, He T, Becker S, Zhang G, Li D, Ma X. Butyrate: a double-edged sword for health? Adv Nutr. 2018;9:21–29. doi: 10.1093/advances/nmx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Basolo A, Hohenadel M, Ang QY, Piaggi P, Heinitz S, Walter M, Walter P, Parrington S, Trinidad DD, von Schwartzenberg RJ, et al. Effects of underfeeding and oral vancomycin on gut microbiome and nutrient absorption in humans. Nat Med. 2020;26:589–598. doi: 10.1038/s41591-020-0801-z. [DOI] [PubMed] [Google Scholar]
- 72.Korpela K, Flint HJ, Johnstone AM, Lappi J, Poutanen K, Dewulf E, Delzenne N, de Vos WM, Salonen A. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One. 2014;9:e90702. doi: 10.1371/journal.pone.0090702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jian C, Luukkonen P, Sädevirta S, Yki-Järvinen H, Salonen A. Impact of short-term overfeeding of saturated or unsaturated fat or sugars on the gut microbiota in relation to liver fat in obese and overweight adults. Clin Nutr. 2020. 10.1016/j.clnu.2020.05.008. [DOI] [PubMed]
- 74.Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan M, Avnit-Sagi T, Kosower N, Malka G, Rein M, et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 2017;25:1243–1253.e1245. doi: 10.1016/j.cmet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 75.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Hjorth MF, Blædel T, Bendtsen LQ, Lorenzen JK, Holm JB, Kiilerich P, Roager HM, Kristiansen K, Larsen LH, Astrup A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int J Obes (Lond). 2019;43:149–157. doi: 10.1038/s41366-018-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hjorth MF, Roager HM, Larsen TM, Poulsen SK, Licht TR, Bahl MI, Zohar Y, Astrup A. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int J Obes (Lond). 2018;42:580–583. doi: 10.1038/ijo.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4:15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Denis GV, Obin MS. 'Metabolically healthy obesity': origins and implications. Mol Aspects Med. 2013;34:59–70. doi: 10.1016/j.mam.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernández-Reyes A, Cámara-Martos F, Molina-Luque R, Romero-Saldaña M, Molina-Recio G, Moreno-Rojas R. Changes in body composition with a hypocaloric diet combined with sedentary, moderate and high-intense physical activity: a randomized controlled trial. BMC Womens Health. 2019;19:167. doi: 10.1186/s12905-019-0864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Del Bas JM, Guirro M, Boqué N, Cereto A, Ras R, Crescenti A, Caimari A, Canela N, Arola L. Alterations in gut microbiota associated with a cafeteria diet and the physiological consequences in the host. Int J Obes (Lond). 2018;42:746–754. doi: 10.1038/ijo.2017.284. [DOI] [PubMed] [Google Scholar]
- 82.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le Roy CI, Bowyer RCE, Castillo-Fernandez JE, Pallister T, Menni C, Steves CJ, Berry SE, Spector TD, Bell JT. Dissecting the role of the gut microbiota and diet on visceral fat mass accumulation. Sci Rep. 2019;9:9758. doi: 10.1038/s41598-019-46193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021. 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed]
- 85.Chen F, Esmaili S, Rogers GB, Bugianesi E, Petta S, Marchesini G, Bayoumi A, Metwally M, Azardaryany MK, Coulter S, et al. Lean NAFLD: a distinct entity shaped by differential metabolic adaptation. Hepatology. 2020;71:1213–1227. doi: 10.1002/hep.30908. [DOI] [PubMed] [Google Scholar]
- 86.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martínez I, Perdicaro DJ, Brown AW, Hammons S, Carden TJ, Carr TP, Eskridge KM, Walter J. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl Environ Microbiol. 2013;79:516–524. doi: 10.1128/aem.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Truax AD, Chen L, Tam JW, Cheng N, Guo H, Koblansky AA, Chou WC, Wilson JE, Brickey WJ, Petrucelli A, et al. The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe. 2018;24:364–378.e366. doi: 10.1016/j.chom.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vojinovic D, Radjabzadeh D, Kurilshikov A, Amin N, Wijmenga C, Franke L, Ikram MA, Uitterlinden AG, Zhernakova A, Fu J, et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat Commun. 2019;10:5813. doi: 10.1038/s41467-019-13721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu TR, Lin CS, Chang CJ, Lin TL, Martel J, Ko YF, Ojcius DM, Lu CC, Young JD, Lai HC. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut. 2019;68:248–262. doi: 10.1136/gutjnl-2017-315458. [DOI] [PubMed] [Google Scholar]
- 91.Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, Wang Y, Liu C, Wang W, Wang J, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235.e225. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 92.Alcock J, Maley CC, Aktipis CA. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays. 2014;36:940–949. doi: 10.1002/bies.201400071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van de Wouw M, Schellekens H, Dinan TG, Cryan JF. Microbiota-gut-brain axis: modulator of host metabolism and appetite. J Nutr. 2017;147:727–745. doi: 10.3945/jn.116.240481. [DOI] [PubMed] [Google Scholar]
- 94.Ramage S, Farmer A, Eccles KA, McCargar L. Healthy strategies for successful weight loss and weight maintenance: a systematic review. Appl Physiol Nutr Metab. 2014;39:1–20. doi: 10.1139/apnm-2013-0026. [DOI] [PubMed] [Google Scholar]
- 95.Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf J, Capdevila J, Hadjigeorgiou G, Davies R, Al Khatib H, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26:964–973. doi: 10.1038/s41591-020-0934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frisard MI, Greenway FL, Delany JP. Comparison of methods to assess body composition changes during a period of weight loss. Obes Res. 2005;13:845–854. doi: 10.1038/oby.2005.97. [DOI] [PubMed] [Google Scholar]
- 97.Fu BC, Randolph TW, Lim U, Monroe KR, Cheng I, Wilkens LR, Le Marchand L, Lampe JW, Hullar MAJ. Temporal variability and stability of the fecal microbiome: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev. 2019;28:154–162. doi: 10.1158/1055-9965.Epi-18-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Finkelstein EA, Khavjou OA, Thompson H, Trogdon JG, Pan L, Sherry B, Dietz W. Obesity and severe obesity forecasts through 2030. Am J Prev Med. 2012;42:563–570. doi: 10.1016/j.amepre.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 99.Grembi JA, Nguyen LH, Haggerty TD, Gardner CD, Holmes SP, Parsonnet J. Gut microbiota plasticity is correlated with sustained weight loss on a low-carb or low-fat dietary intervention. Sci Rep. 2020;10:1405. doi: 10.1038/s41598-020-58000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jian C, Silvestre MP, Middleton D, Korpela K, Jalo E, Broderick D, de Vos WM, Fogelholm M, Taylor MW, Raben A, Poppitt SD, Salonen A: Gut microbiota predicts body fat change following a low energy diet: a PREVIEW sub-study. European Nucleotide Archive at EMBL-EBI under accession number PRJEB43667: 2021. Available from: https://www.ebi.ac.uk/ena/browser/view/PRJEB43667. Accessed Mar 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Information consisting of eight supporting figures (Figure S1-S8). A short description of the figure and a figure caption for each figure are given within the file.
Additional file 2. Two supporting tables (Table S1-S2) of lists of input predictors for body fat change in random forest regression ranked by feature importance. A table caption for each table is given within the file.
Additional file 3. PREVIEW study protocol.
Data Availability Statement
The sequencing data in this study are available at the European Nucleotide Archive (ENA) under accession number PRJEB43667 (https://www.ebi.ac.uk/ena/browser/view/PRJEB43667) [100]. All codes are fully accessible from the referenced sources in the program R.