Abstract
Background
Extracellular vesicle (EV) biomarkers have promising diagnosis and screening capacity for several cancers, but the diagnostic value for pancreatic cancer (PC) is controversial. The aim of our study was to review the diagnostic performance of EV biomarkers for PC.
Methods
We performed a systematic review of PubMed, Medline, and Web Of Science databases from inception to 18 Feb 2022. We identified studies reporting the diagnostic performance of EV biomarkers for PC and summarized the information of sensitivity, specificity, area under the curve (AUC), or receiver operator characteristic (ROC) curve) in according to a pre-designed data collection form. Pooled sensitivity and specificity was calculated using a random-effect model.
Results
We identified 39 studies, including 2037 PC patients and 1632 noncancerous, seven of which were conducted independent validation tests. Seventeen studies emphasized on EV RNAs, sixteen on EV proteins, and sixteen on biomarker panels. MiR-10b, miR-21, and GPC1 were the most frequently reported RNA and protein for PC diagnosis. For individual RNAs and proteins, the pooled sensitivity and specificity were 79% (95% CI: 77–81%) and 87% (95% CI: 85–89%), 72% (95% CI: 69–74%) and 77% (95% CI: 74–80%), respectively. the pooled sensitivity and specificity of EV RNA combined with protein panels were 84% (95% CI: 81–86%) and 89% (95% CI: 86–91%), respectively. Surprisingly, for early stage (stage I and II) PC EV biomarkers showed excellent diagnostic performance with the sensitivity of 90% (95% CI: 87–93%) and the specificity of 94% (95% CI: 92–95%). Both in sensitivity and subgroup analyses, we did not observe notable difference in pooled sensitivity and specificity. Studies might be limited by the isolation and detection techniques of EVs to a certain extent.
Conclusions
EV biomarkers showed appealing diagnostic preference for PC, especially for early stage PC. Solving the deficiency of technologies of isolation and detection EVs has important implications for application these novel noninvasive biomarkers in clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-022-09463-x.
Keywords: Extracellular vesicle, Pancreatic cancer, Biomarker, Diagnosis
Introduction
Pancreatic cancer (PC) is the seventh cancer related mortality worldwide, contributing 32, 000 deaths in 2018 [1, 2]. And its incidence and mortality is increasing in the USA and Europe over the years [2, 3]. Despite progresses in therapeutic strategies, the 5-year relative survival rate of PC still below 9%, the 5-year survival rate of patients with distant metastasis even is 2.9% [2, 4]. Surgical intervention, as the only curative strategy for patients with PC so far, is generally estimated to improve the 5-year survival rate to 30 ~ 40% for PC patients diagnosed at an early stage [5]. However, due to most of PC patients diagnosed at an advance stage, less than 20% of the tumors are eligible for surgical resection [6–8]. Imaging-based methods, such as compute tomography, magnetic resonance imaging, and echo-guided ultrasound, have been studied as screening tools only for populations at high risk of PC, which are not only expensive, radiation exposure, or less tolerant, but also have a high rate of false-positive results [9–11]. Current conventional serological biomarkers widely used for PC diagnosis and recurrence, such as carbohydrate antigen 199 (CA199), are neither sensitive nor specific enough to act as accurate early diagnostic strategies [12–16] and also are overexpressed in benign pancreatic diseases [17]. Therefore, novel circulation-based noninvasive biomarkers that can specifically and accurately diagnose PC at early stage in the general population have attracted great interest worldwide.
Extracellular vesicles (EVs), mainly classified exosomes and microvesicles, are as membrane-bound vesicles with biologically active molecules including proteins and nucleic acids and can be released from tumor cells to transport these molecules from parental cells to recipient cells to mediate tumor initiation, progression, and metastasis [18–21]. EVs stably existing in the circulation can protect these functional molecules from impaired by hydrolysis [21]. circulation EV proteins and nucleic acids beneficially classify tumor type for a diagnosis in patients with cancer of unknown primary tumor origin and the concentration of these molecules increases as cancer stage and tumor size [22–25]. Circulation EV proteins and nucleic acids as potential biomarkers for early cancers diagnosis and continuous monitoring have been investigated for a decade [26, 27]. The potential of circulation EV proteins and nucleic acids as biomarkers for PC indicates increasing application and attention. Accumulating evidences have suggested that EV proteins and nucleic acids can be as promising biomarkers for PC diagnosis [28–32]. Two recent prospective studies identified that circulation EV proteins discerned PC patients from individuals with chronic pancreatitis (CP) and healthy individuals, which showed absolute sensitivity and specificity [33, 34]. However, circulation EV biomarkers were also reported as invasive markers for benign pancreatic diseases, such as intraductal papillary mucinous neoplasms (IPMNs) [35] and CP [36, 37]. Yang et al. demonstrated that circulating EV proteins and miRNAs can predict the presence of invasive carcinoma within IPMN [35, 38]. Circulating EV miR-579-3p was identified significant lower expression in CP patients compared to healthy controls [36]. A bioinformatics analysis resulted that exsome miRNAs may be promising markers for early diagnosis and treatment of CP [39]. Therefore, the purpose of this study was to systematically review the characteristics of circulation EV biomarkers for diagnosing PC and further to evaluate the diagnosis value of these biomarkers for distinguishing PC from noncancerous.
Methods
The systematic review and meta-analysis followed a preferred protocol and is reported according with the PRISMA guidelines [40].
Data sources and searches
We performed an electronic search of PubMed, Medline, and Web of Science databases to identify relevant studies assessing circulation EV biomarkers for PC detection up to 18 Feb 2022. The search strategy used the following keywords combination: ([pancreatic OR pancreas] AND [cancer OR carcinoma OR neoplasm OR tumor OR malignancy OR adenocarcinoma OR adenoma] AND [detection OR diagnosis OR biomarker OR marker OR sensitivity OR specificity OR area under the curve] AND [exosome OR Extracellular Vesicles OR exosomal OR membrane vesicles OR intracellular multivesicular endosomes]). Duplicates were removed.
Study selection
We initially screened all titles and abstracts and studies matched any of the following criteria were excluded: (1) non-English articles; (2) non-original articles; (3) not pancreatic cancer articles; (4) non-human studies; (5) not relevant to the topic. Two authors (Erna Jia and Na Ren) reviewed all potentially relevant full texts, the following studies were included: (1) studies that investigated EV biomarkers in plasma, serum, blood, or peripheral blood in PC patients; (2) PC patients were diagnosed as pancreatic ductal adenocarcinoma depending on the cytological or histological examination; (3) studies that reported the diagnostic performance of EV biomarkers for PC had relevant data (such as sensitivity, specificity, area under the curve (AUC), or receiver operator characteristic (ROC) curve); (4) control groups containing healthy people or benign disease. Any discrepancies were resolved by discussion.
Data extraction and quality assessment
The two reviewers independently extracted available information from eligibility studies in according to a pre-designed data collection form and resolved any disagreements by discussion again. We extracted key information on first author, year of publication, country, study design, population characteristics (including sample size, mean age, and gender distribution), type of blood-based specimen, PC stage, population composition of control groups, names or panels of target biomarkers, detection methods of target biomarkers, preparation approaches of EVs, sensitivity, specificity, AUC, and P Value. Risk of bias and application for each study was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) checklist [41]. Publication bias was analyzed and represented by a funnel plot, and funnel plot symmetry was assessed quantitatively with egger’s test through R software (version 3.5.3, R Foundation, Vienna, Austria) [42].
Data synthesis and statistical analysis
We calculated mean age and sex distribution of eligibility studies using statistical software R (version 3.5.3, R Foundation, Vienna, Austria) if relevant data was not obtained but raw data was available. If the values of sensitivity, specificity or AUC were not reported, we estimated these diagnostic indicators based on ROC curve using OriginPro software (version 9.0) according to maximum Youden’s index.
We combined the sensitivity and specificity of EV biomarkers in studies reporting the relevant data to obtain a pooled diagnostic performance for PC using Meta-DiSc version 1.4 by the random-effect model (DerSimonian-Laird method). If noncancerous groups were consisted of healthy controls and/ or benign diseases, we studied them as a whole if the relevant data was reported, or we studied the healthy controls if not. We investigated heterogeneity between studies using Cocharan’s Q test and the inconsistency index (I2 value), with P < 0.05 or I2 > 50% as statistically significant heterogeneity. We performed sensitivity analysis for studies considered at low risk of bias and low concern for applicability to give convincing diagnostic performance of EV biomarkers for PC based on the assessment of QUADAS 2 using Review Manager 5.3. We also applied subgroup analysis to pool the sensitivity and specificity of individual EV miRNAs and individual EV RNAs detected using qPCR for PC diagnosis.
Results
Research results
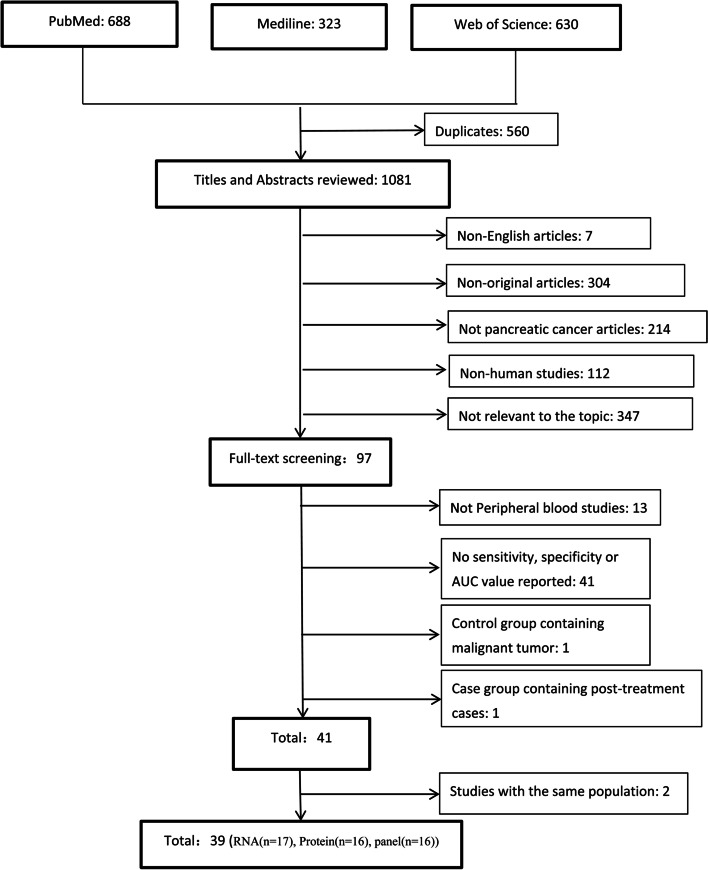
We identified 1641 potentially relevant studies resulted from the above search strategy, with 688 from PubMed, 323 from Medline and 630 from Web of Science (Fig. 1). After deduplication and review of titles and abstracts, we narrowed to 97 studies for full text screening and then a further 56 studies were excluded: the specimens of 13 studies were not peripheral blood; 41 studies had no related information data, such as sensitivity, specificity, AUC value, or ROC curve; the control group of one study contained malignant tumors and the case group of one study contained post-treatment cases. Consequently, we identified 41 studies that were met the inclusion criteria [25, 30, 31, 33, 34, 43–78]. Among these 41 studies, the related information data of four studies was respectively from the same two population cohort, we selected the recently published two studies for analyses [48, 52]. Ultimately, 39 studies identifying EV biomarkers in serum or plasma for diagnosing PC were included in the systematic review [30, 31, 34, 43–57], and 34 of which were included in the meta-analysis [24, 25, 30, 31, 33, 34, 44–47, 53, 59–62, 64, 67–84].
Fig. 1.
Search and selection process
Study characteristics
Across 39 included studies, which involved 2037 PC cases and 1632 noncancerous, 24 studies were conducted in Asia [25, 44, 53, 60–64, 67–72, 74, 76–80, 82–85], eight were in Europe [24, 30, 31, 43, 59, 73, 75, 81], and seven were in North America [33, 34, 45–47, 58, 86]. Sample size of PC cases varied from 6 to 284 (median number, 33) and of noncancerous groups varied from 6 to 152 (median number, 29), respectively. The detail information of mean age, sex distribution, number of cases and controls, detection approaches, and PC stage in included studies were all described in Tables 1, 2 and 3. Seventeen studies focused on the diagnostic performance of individual EV RNAs (microRNAs in nine studies [25, 31, 45, 46, 59, 61, 62, 64, 68, 70, 72, 74, 78, 84], messenger RNAs in two studies [44, 53], small nucleolar RNAs in one study [53], and long noncoding RNAs in one study [80]), one of which was validated by blind external test [44], shown in Table 1; Sixteen focused on individual EV proteins (Membrane proteins (MP) in thirteen studies [24, 30, 33, 34, 47, 58, 60, 62, 75, 76, 79, 82, 83], non-membrane proteins (nMPs) in six studies [47, 58, 67, 69, 81, 82]), three of which were applied independent validation tests [30, 34, 47]. One study reported 446 individual EV proteins with AUC value greater than 0.7 and we selected 156 EV proteins with p value less than 0.01 for analysis [58], shown in Table 2. Sixteen studies focused on EV biomarker panels (miRNA panels in five studies [25, 31, 43, 61, 78], long RNA panels in two studies [63, 85], mRNA panels in one study [71], protein panels in six studies [33, 43, 47, 76, 79, 86], and miRNA combined with protein panels in four study [43, 62, 73, 77]), twelve panels in six studies were conducted independent validation tests [43, 47, 63, 71, 78, 85], one study reported 129 miRNA panels and we selected 119 panels with AUC value greater than 0.70 for analysis [78], shown in Table 3. Nine studies reported the diagnostic performance of EV biomarkers for early stage (stage I and II) PC [30, 34, 46, 53, 60, 75, 84–86].
Table 1.
Diagnostic performance of RNAs in extracellular vesicles for pancreatic cancer
| Study | Country | study design | Cases vs Controls | Specimen | Stage | Status Controls | Detection Method | markers | SEN% | SPE% | AUC | P Value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age | Male (%) | ||||||||||||
| miRNA | ||||||||||||||
| Xu, 2017 [46] | USA | Case–control | 15/15 | 67/48 | 53/27 | Plasma | I-IIA | HC | qPCR | miR-196a | 87e | 73e | 0.81 | < 0.001 |
| miR-1246 | 67e | 80e | 0.73 | 0.019 | ||||||||||
| miR-196b | 67e | 80e | 0.71 | 0.033 | ||||||||||
| Lai, 2017 [45] | USA | Case–control | 29/6 | 67/NA | 52/NA | Plasma | I-IV | HC | qPCR | miR-10b | 100 | 100 | 1.00 | < 0.001 |
| miR-21 | 100 | 100 | 1.00 | < 0.001 | ||||||||||
| miR-30c | 100 | 100 | 1.00 | < 0.001 | ||||||||||
| miR-106b | 62 | 100 | 0.85 | 0.007* | ||||||||||
| miR-20a | 83 | 100 | 0.95 | < 0.001 | ||||||||||
| miR-181a | 100 | 100 | 1.00 | < 0.001 | ||||||||||
| miR-let7a | 100 | 100 | 1.00 | < 0.001 | ||||||||||
| miR-122 | 93 | 100 | 0.99 | < 0.001 | ||||||||||
| Goto, 2018 [84] | Japan | Case–control | 32/22 | 64/58 | 53/64 | Serum | I-IV | HC | qPCR | miR-191 | 72 | 84 | 0.79 | 0.001 |
| miR-21 | 81 | 81 | 0.83 | < 0.001 | ||||||||||
| miR-451a | 66 | 86 | 0.76 | 0.002 | ||||||||||
| 9/22 | NA/58 | NA/64 | Serum | I-IIA | HC | qPCR | miR-191 | 67 | 84 | 0.75 | 0.032 | |||
| miR-21 | 67 | 81 | 0.74 | 0.004 | ||||||||||
| miR-451a | 67 | 86 | 0.74 | 0.044 | ||||||||||
| 23/22 | NA/58 | NA/64 | Serum | IIB-IV | HC | qPCR | miR-191 | 79 | 79 | 0.80 | 0.001 | |||
| miR-21 | 86 | 81 | 0.86 | < 0.001 | ||||||||||
| miR-451a | 70 | 81 | 0.77 | 0.002 | ||||||||||
| Zhou, 2020 [62] | China | Case–control | 30/10 | 60/58 | / | Plasma | I-IV | NCb | 3D mircrofluidic chip | miR-451a | 81e | 100e | 0.93 | / |
| miR-21 | 87e | 100e | 0.94 | / | ||||||||||
| miR-10b | 79e | 99e | 0.88 | / | ||||||||||
| miRNA | ||||||||||||||
| Reese, 2020 [31] | Germany | Case–control | 56/22 | NA/68 | 64/50 | Serum | II-IV | HC | qPCR | miR-200b | 67e | 76e | 0.79 | 0.0001 |
| qPCR | miR-200c | 51e | 90e | 0.67 | 0.0239 | |||||||||
| 56/11 | NA/62 | 64/55 | Serum | II-IV | CP | qPCR | miR-200b | 63e | 86e | 0.77 | 0.0047 | |||
| 56/33 | 64/52 | Serum | II-IV | NCc | qPCR | miR-200b | 85e | 63e | 0.77 | 0.005 | ||||
| 56/22 | NA/68 | 64/50 | II-IV | HC | qPCR | miR-200b | 63e | 68e | 0.69 | 0.0077 | ||||
| Wu, 2020 [61] | China | Case–control | 30/10 | 62/51 | 60/80 | serum | 0-IV | CP | qPCR | miR-21 | 80 | 90 | 0.87 | / |
| miR-210 | 83 | 90 | 0.82 | / | ||||||||||
| Pu, 2020 [25] | China | Case–control | 36/65 | / | / | Plasma | I-IV | HC | cationic lipoplex nanoparticle | miR-21 | 53e | 98e | 0.72 | 0.0003 |
| miR-10b | 42e | 100e | 0.65 | 0.0105 | ||||||||||
| Flammang, 2020 [59]a | Germany | Case–control | 44/12 | / | / | serum | II-IV | HC | qPCR | miR-192-5p | 64e | 99e | 0.83 | 0.0004 |
| Wang, 2021 [64] | China | Case–control | 17/12 | / | / | serum | / | PBT | qPCR | miRNA-1226-3p | 75e | 66e | 0.74 / | |
| Xiao, 2021 [70] | China | Case–control | 10/10 | / | / | Plasma | HC | PNA-functionalized nanochannel sensor | miR-10b | / | / | 0.99 | / | |
| Shao, 2021 [74] | China | Case–control | 63/22 | 60/50 | 56/41 | serum | I-IV | HC | qPCR | miR-483-3p | 82e | 56e | 0.69 | / |
| Wang L, 2021 [72] | China | Case–control | 62/53 | / | / | plasma | HC | qPCR | miR-19b-3p | 85 | 91 | 0.94 | < 0.001 | |
| 62/23 | / | / | CP | 81 | 87 | 0.90 | < 0.001 | |||||||
| 62/30 | / | / | OPT | 94 | 63 | 0.81 | < 0.001 | |||||||
| Chen, 2022 [68] | China | Case–control | 191/90 | 62/57 | 57/52 | serum | I-IV |
HC PBD |
miR-451a | 80 | 87 | 0.90 | / | |
| 191/95 | 62/59 | 57/58 | 71 | 89 | 0.86 | / | ||||||||
| LncRNA | ||||||||||||||
| Takahashi, 2019 [80] | Japan | Case–control | 20/43 | 72/71 | 40/42 | Serum | II-IV | NCd | dPCR | HULC | 80 | 92 | 0.92 | / |
| 20/21 | 72/72 | 40/36 | Serum | II-IV | HC | dPCR | HULC | 80 | 95 | 0.94 | / | |||
| 20/22 | 72/70 | 40/48 | Serum | II-IV | IPMN | dPCR | HULC | 85 | 83 | 0.91 | / | |||
| Guo, 2021 [78]a | China | Case–control | 27/15 | 58/44 | 63/60 | plasma | IB-IV | CP | small RNA sequencing | miR-95-3p | 92e | 94e | 0.91 | |
| miR-199b-3p | / | / | 0.90 | |||||||||||
| miR-3158-3p | / | / | 0.90 | |||||||||||
| miR-199a-3p | / | / | 0.89 | |||||||||||
| miR-4732-3p | / | / | 0.88 | |||||||||||
| miR-10a-5p | / | / | 0.88 | |||||||||||
| miR-145-3p | / | / | 0.87 | |||||||||||
| miR-27b-3p | / | / | 0.86 | |||||||||||
| miR-511-5p | / | / | 0.86 | |||||||||||
| miR-7706 | / | / | 0.85 | |||||||||||
| miR-99b-5p | / | / | 0.85 | |||||||||||
| miR-143-3p | / | / | 0.83 | |||||||||||
| miR-486-3p | / | / | 0.83 | |||||||||||
| miR-99a-5p | / | / | 0.83 | |||||||||||
| miR-223-3p | / | / | 0.82 | |||||||||||
| mRNA | ||||||||||||||
| Hu, 2017 [44]a | China | Case-controld | 20/15 | / | / | Serum | I-IV | HC | LPHN-CHDC biochip | GPC1 mRNA | 95 | 93 | 0.94 | / |
| snoRNA | ||||||||||||||
| Kitagawa, 2019 [53]a | Japan | Case–control | 27/13 | / | 63/31 | Serum | I-III | BGD | qPCR | SNORA14B | 93e | 69e | 0.88 | / |
| SNORA18 | 81e | 84e | 0.88 | / | ||||||||||
| SNORA25 | 92e | 76e | 0.90 | / | ||||||||||
| SNORA74A | 92e | 84e | 0.91 | / | ||||||||||
| SNORD22 | 70e | 93e | 0.86 | / | ||||||||||
| snoRNA | ||||||||||||||
| Kitagawa, 2019 [53] | Japan | Case–control | 27/13 | / | 63/31 | Serum | I-III | BGD | qPCR | CCDC88A | 55e | 92e | 0.72 | / |
| ARF6 | 85e | 92e | 0.94 | / | ||||||||||
| Vav3 | 74e | 85e | 0.84 | / | ||||||||||
| WASF2 | 93e | 84e | 0.94 | / | ||||||||||
| 8/13 | / | NA/31 | Serum | I-IIA | BGD | qPCR | SNORA14B | / | / | 0.86 | / | |||
| SNORA18 | / | / | 0.91 | / | ||||||||||
| SNORA25 | 100e | 78e | 0.91 | / | ||||||||||
| SNORA74A | 100e | 84e | 0.95 | / | ||||||||||
| SNORD22 | / | / | 0.97 | / | ||||||||||
| CCDC88A | / | / | 0.73 | / | ||||||||||
| ARF6 | 100e | 92e | 0.98 | / | ||||||||||
| Vav3 | / | / | 0.89 | / | ||||||||||
| WASF2 | 100e | 84e | 0.97 | / | ||||||||||
| 19/13 | / | NA/31 | Serum | IIB-III | BGD | qPCR | SNORA14B | / | / | 0.88 | / | |||
| SNORA18 | / | / | 0.88 | / | ||||||||||
| SNORA25 | 93e | 77e | 0.90 | / | ||||||||||
| SNORA74A | 93e | 85e | 0.91 | / | ||||||||||
| SNORD22 | / | / | 0.86 | / | ||||||||||
| CCDC88A | / | / | 0.72 | / | ||||||||||
| ARF6 | 85e | 92e | 0.94 | / | ||||||||||
| Vav3 | / | / | 0.84 | / | ||||||||||
| WASF2 | 93e | 84e | 0.94 | / | ||||||||||
SENs, SPEs and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation)
AUC area under the curve, BGD benign gastrointestinal disease, CP chronic pancreatitis, HC healthy control, IPMN intraductal papillary mucinous neoplasm, LPHN-CHDC lipid polymer hybrid nanoparticle-catalyzed hairpin DNA circuit, NC noncancerous, PBT pancretian benign tumor, SEN sensitivity, SPE specificity, OPT other pancreatic tumor (pancreatic neuroendocrine tumor, solid pseudopapillary tumor, serous or mucinous cystadenomas, intraductal papillary mucinous neoplasms, and epithelial cysts), PBD pancreatic benign disease, PNA peptide nucleic acid, OPT pancreatic neuroendocrine tumor, solid pseudopapillary tumor, serous or mucinous cystadenomas, intraductal papillary mucinous neoplasms, and epithelial cysts
arepresent markers extracted from extracellular vesicles
bno history of cancer
cHC and CP
dHC and IPMN
erepresent estimated value
p value indicates p value of AUC
Table 2.
Diagnostic performance of proteins in extracellular vesicles for pancreatic cancer
| Study | Country | study design | Cases vs Controls | Specimen | Stage | Status Controls | Detection Method | markers | SEN% | SPE% | AUC | P Value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age | Male (%) | ||||||||||||
| MPs | ||||||||||||||
| Melo, 2015 [30] | Germany | Case–control | 5/126 | / | / | Serum | 0-I | NCb | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / |
| 18/126 | / | / | Serum | IIa | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / | ||||
| 117/126 | / | / | Serum | IIb | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / | ||||
| 11/126 | / | / | Serum | III | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / | ||||
| 41/126 | / | / | Serum | IV | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / | ||||
| Case–control | 56/26 | 70/NA | 50/NA | Serum | I-IV | Flow cytometry | GPC1 | 100 | 100 | 1.00 | / | |||
| Liang, 2017 [34] a | USA | Case–control | 49/48 | NA/62 | NA/40 | Plasma | I-III | HC | nPES assay | EphA2 | 94 | 85 | 0.96 | < 0.001 |
| 49/48 | NA/52 | NA/52 | Plasma | I-III | CP | nPES assay | EphA2 | 89 | 85 | 0.94 | < 0.001 | |||
| 37/48 | 67/62 | 60/40 | Plasma | I-II | HC | nPES assay | EphA2 | 91 | 85 | 0.96 | < 0.001 | |||
| 37/48 | 67/52 | 60/52 | Plasma | I-II | CP | nPES assay | EphA2 | 86 | 85 | 0.93 | < 0.001 | |||
| Yang, 2017 [47]a | USA | Prospective | 22/21 | / | / | plasma | / | NCc | NPS chip | GPC1 | 82 | 52 | / | / |
| EGFR | 59 | 76 | / | / | ||||||||||
| EPCAM | 45 | 95 | / | / | ||||||||||
| HER2 | 59 | 85 | / | / | ||||||||||
| MUC1 | 36 | 90 | / | / | ||||||||||
| Li, 2018 [82] | China | Case–control | 34/32 | NA/50 | NA/69 | Serum | / | HC | SERS immunosensor | GPC1 | 59 | 58 | 0.67 | / |
| 34/32 | NA/50 | NA/69 | Serum | / | HC | SERS immunosensor | EGFR | 57 | 56 | 0.63 | / | |||
| Jin, 2018 [49, 83] | China | Case–control | 24/46 | 61/49 | 33/50 | Serum | I-IV | HC | ELISA | ZIP4 | 92 | 80 | 0.89 | / |
| 24/32 | 61/44 | 33/28 | Serum | I-IV | NCd | ELISA | ZIP4 | 92 | 84 | 0.89 | / | |||
| 24/32 | 61/63 | 33/53 | Serum | I-IV | BD | ELISA | ZIP4 | 92 | 81 | 0.81 | / | |||
| Xiao, 2019 [79] | China | Case–control | 24/26 | 59/45 | 42/85 | Plasma | / | HC | Flow cytometry | GPC1 | 72* | 84* | 0.89 | / |
| CD82 | 86* | 66* | 0.85 | / | ||||||||||
| 24/6 | 59/73 | 42/83 | Plasma | / | CP | Flow cytometry | GPC1 | 100* | 66* | 0.85 | / | |||
| CD82 | 100* | 66* | 0.90 | / | ||||||||||
| Buscail, 2019 [24, 54, 66] | France | Prospective | 22/28 | 70/58 | 59/28 | Serum-CD63 | I-III | NCe | Flow cytometry | GPC1 | 50 | 90 | 0.68 | / |
| MPs | ||||||||||||||
| Rodrigues, 2019 [33] a | USA | Case–control | 20/12 | / | 45/25 | Serum | / | NCk | nanoparticle-and dyebased fluorescent immunoassay | EpCAM | 75* | 100* | 0.92 | |
| EphA2 | 95* | 83* | 0.93 | |||||||||||
| Zhou, 2020 [62] | China | Case–control | 30/10 | 60/58 | / | Plasma | I-IV | NCf | 3D mircrofluidic chip | EphA2 | 69* | 99* | 0.85 | |
| Wei, 2020 [60] | China | Case–control | 204/74 | 62/NA | / | serum | I-IV | HC | Enzyme-Linked Immunosorbent Assays | EphA2 | 83 | 94 | 0.94 | |
| 204/75 | 62/NA | / | BPD | EphA2 | 81 | 94 | 0.92 | |||||||
| 94/74 | / | / | I-II | HC | EphA2 | 83 | 94 | 0.92 | ||||||
| 94/75 | / | / | BPD | EphA2 | 84 | 88 | 0.90 | |||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | ASGR1 | / | / | 0.89 | 0.0026 |
| ANXA1 | / | / | 0.99 | 0.0000 | ||||||||||
| IL12RB2 | / | / | 0.85 | 0.0083 | ||||||||||
| ADAM9 | / | / | 0.98 | 0.0000 | ||||||||||
| TGFBR2 | / | / | 0.85 | 0.0083 | ||||||||||
| EDA | / | / | 0.85 | 0.0083 | ||||||||||
| BMPR2 | / | / | 0.88 | 0.0034 | ||||||||||
| TYRO3 | / | / | 0.87 | 0.0043 | ||||||||||
| FLRT3 | / | / | 0.87 | 0.0054 | ||||||||||
| SLAMF6 | / | / | 0.95 | 0.0002 | ||||||||||
| CD4 | / | / | 0.87 | 0.0054 | ||||||||||
| RXFP1 | / | / | 0.87 | 0.0054 | ||||||||||
| Li, 2021 [76] a | China | Case–control | 21/29 | 54/62 | 43/55 | plasma | / | HC | AbMB-bioChol paltform | EGFR | 52* | 93* | 0.74 | |
| EpCAM | 94* | 48* | 0.68 | |||||||||||
| GPC1 | 43* | 89* | 0.72 | |||||||||||
| EphA2 | 47* | 86* | 0.64 | |||||||||||
| MPs | ||||||||||||||
| Moutinho-R, 2021 [75] | Portugal | prospective | 60/29 | 67/54 | 55/76 | serum | I-IV | CP | flow cytometry | GPC1 | 98 | 86 | 0.96 | < 0.0001 |
| 15/29 | NA/54 | NA /76 | I | 100* | 96* | 0.97 | ||||||||
| 45/29 | NA /54 | NA /76 | II-IV | 95* | 97* | 0.96 | ||||||||
| nMPs | ||||||||||||||
| Yang, 2017 [47] a | USA | Prospective | 22/21 | / | / | plasma | / | NCc | NPS chip | WNT2 | 64 | 76 | / | / |
| GRP94 | 55 | 71 | / | / | ||||||||||
| Case–control | 22/10 | 68/48 | 36/40 | plasma | II-IV | HC | NPS chip | B7-H3 | 50 | 100 | 0.75 | / | ||
| Li, 2018 [82] | China | Case–control | 71/32 | 60/50 | 54/69 | Serum | I-IV | HC | SERS immunosensor | MIF | 63 | 76 | 0.89 | / |
| Lux, 2019 [54, 81] | Germany | Case–control | 55/34 | 67/NA | 53/NA | Serum | I-IV | NCg | Flow cytometry | c-Met | 70 | 85 | 0.75* | / |
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | IFNGR2 | / | / | 0.87 | 0.0054 |
| TPSG1 | / | / | 0.94 | 0.0003 | ||||||||||
| Yang, 2021 [69] | China | Case–control | 62/42 | / | 48/ NA | plasma | I-IV | NCh | Enzyme-linked immunosorbert assay | ALIX | 53 | 84 | 0.73 | 0.0003 |
| Zheng, 2022 [67] | China | Case–control | 57/84 | / | 31/39 | plasma | II-IV | HC | MALDI-TOF MS | SAA1 | 96 | 28 | 0.61 | / |
| FGG | 40 | 87 | 0.62 | / | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | MAPKAPK2 | / | / | 0.94 | 0.0004 |
| PDXK | / | / | 0.94 | 0.0003 | ||||||||||
| PLCG1 | / | / | 0.94 | 0.0004 | ||||||||||
| INSR | / | / | 0.94 | 0.0004 | ||||||||||
| IL34 | / | / | 0.93 | 0.0006 | ||||||||||
| AIP | / | / | 0.93 | 0.0006 | ||||||||||
| YES1 | / | / | 0.93 | 0.0006 | ||||||||||
| DYNLRB1 | / | / | 0.93 | 0.0006 | ||||||||||
| IDE | / | / | 0.93 | 0.0006 | ||||||||||
| CCL28 | / | / | 0.93 | 0.0006 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | C5 C6 | / | / | 0.92 | 0.0008 |
| CD109 | / | / | 0.92 | 0.0008 | ||||||||||
| CAMK2A | / | / | 0.92 | 0.0008 | ||||||||||
| TNFSF14 | / | / | 0.91 | 0.0011 | ||||||||||
| MMP14 | / | / | 0.91 | 0.0011 | ||||||||||
| PRKCI | / | / | 0.91 | 0.0011 | ||||||||||
| MAP2K1 | / | / | 0.91 | 0.0011 | ||||||||||
| KYNU | / | / | 0.91 | 0.0011 | ||||||||||
| KIF23 | / | / | 0.91 | 0.0011 | ||||||||||
| IL27RA | / | / | 0.91 | 0.0011 | ||||||||||
| IMPDH1 | / | / | 0.91 | 0.0011 | ||||||||||
| TNFRSF13B | / | / | 0.91 | 0.0011 | ||||||||||
| SEMA6B | / | / | 0.91 | 0.0011 | ||||||||||
| AIMP1 | / | / | 0.91 | 0.0011 | ||||||||||
| NMT1 | / | / | 0.90 | 0.0015 | ||||||||||
| EPHA5 | / | / | 0.90 | 0.0015 | ||||||||||
| MSLN | / | / | 0.90 | 0.0015 | ||||||||||
| IBSP | / | / | 0.90 | 0.0015 | ||||||||||
| TNFSF13B | / | / | 0.90 | 0.0015 | ||||||||||
| CTF1 | / | / | 0.90 | 0.0015 | ||||||||||
| BCL2L1 | / | / | 0.90 | 0.0015 | ||||||||||
| IL1A | / | / | 0.90 | 0.0015 | ||||||||||
| MAPK12 | / | / | 0.90 | 0.0015 | ||||||||||
| FABP1 | / | / | 0.90 | 0.0020 | ||||||||||
| CCL17 | / | / | 0.90 | 0.0020 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | SPARCL1 | / | / | 0.90 | 0.0020 |
| LCK | / | / | 0.90 | 0.0020 | ||||||||||
| CCL3L1 | / | / | 0.90 | 0.0020 | ||||||||||
| PDE9A | / | / | 0.90 | 0.0020 | ||||||||||
| PRDX5 | / | / | 0.90 | 0.0020 | ||||||||||
| CD300C | / | / | 0.90 | 0.0020 | ||||||||||
| BCL2L2 | / | / | 0.90 | 0.0020 | ||||||||||
| CSRP3 | / | / | 0.90 | 0.0020 | ||||||||||
| GDF2 | / | / | 0.90 | 0.0020 | ||||||||||
| IL17RB | / | / | 0.90 | 0.0020 | ||||||||||
| LGALS3BP | / | / | 0.90 | 0.0020 | ||||||||||
| SEMA6A | / | / | 0.90 | 0.0020 | ||||||||||
| CLEC11A | / | / | 0.90 | 0.0020 | ||||||||||
| UBC | / | / | 0.89 | 0.0026 | ||||||||||
| HIPK3 | / | / | 0.89 | 0.0026 | ||||||||||
| PARK7 | / | / | 0.89 | 0.0026 | ||||||||||
| MMP13 | / | / | 0.89 | 0.0026 | ||||||||||
| FGFR4 | / | / | 0.89 | 0.0026 | ||||||||||
| FGF5 | / | / | 0.89 | 0.0026 | ||||||||||
| DDR1 | / | / | 0.89 | 0.0026 | ||||||||||
| PDK1 | / | / | 0.89 | 0.0026 | ||||||||||
| CXCL8 | / | / | 0.89 | 0.0026 | ||||||||||
| GDF11 | / | / | 0.89 | 0.0026 | ||||||||||
| IDUA | / | / | 0.89 | 0.0026 | ||||||||||
| CA3 | / | / | 0.89 | 0.0026 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | DYNLL1 | / | / | 0.89 | 0.0026 |
| ACVRL1 | / | / | 0.89 | 0.0026 | ||||||||||
| CXCL1 | / | / | 0.88 | 0.0034 | ||||||||||
| CRLF1/CLCF1 | / | / | 0.88 | 0.0034 | ||||||||||
| DHH | / | / | 0.88 | 0.0034 | ||||||||||
| ENTPD5 | / | / | 0.88 | 0.0034 | ||||||||||
| UBE2G2 | / | / | 0.88 | 0.0034 | ||||||||||
| IFNG | / | / | 0.88 | 0.0034 | ||||||||||
| CHEK2 | / | / | 0.88 | 0.0034 | ||||||||||
| LTA LTB | / | / | 0.87 | 0.0043 | ||||||||||
| FAM107A | / | / | 0.87 | 0.0043 | ||||||||||
| TNFRSF18 | / | / | 0.87 | 0.0043 | ||||||||||
| KLK6 | / | / | 0.87 | 0.0043 | ||||||||||
| PPP3R1 | / | / | 0.87 | 0.0043 | ||||||||||
| CD244 | / | / | 1.00 | 0.0000 | ||||||||||
| ETHE1 | / | / | 0.87 | 0.0043 | ||||||||||
| PDXP | / | / | 0.87 | 0.0043 | ||||||||||
| IFNB1 | / | / | 0.87 | 0.0043 | ||||||||||
| IL36A | / | / | 0.87 | 0.0043 | ||||||||||
| KLK14 | / | / | 0.87 | 0.0043 | ||||||||||
| IL1RN | / | / | 0.87 | 0.0043 | ||||||||||
| F3 | / | / | 0.87 | 0.0043 | ||||||||||
| MAPK11 | / | / | 0.87 | 0.0054 | ||||||||||
| TNFRSF14 | / | / | 0.87 | 0.0054 | ||||||||||
| PRLR | / | / | 0.87 | 0.0054 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | VEGFA | / | / | 0.87 | 0.0054 |
| FABP5 | / | / | 0.87 | 0.0054 | ||||||||||
| CDH3 | / | / | 0.87 | 0.0054 | ||||||||||
| ISG15 | / | / | 0.87 | 0.0054 | ||||||||||
| CKM | / | / | 0.87 | 0.0054 | ||||||||||
| ABL2 | / | / | 0.87 | 0.0054 | ||||||||||
| TNFSF11 | / | / | 0.86 | 0.0067 | ||||||||||
| CA10 | / | / | 0.86 | 0.0067 | ||||||||||
| EPHA2 | / | / | 0.86 | 0.0067 | ||||||||||
| HK2 | / | / | 0.86 | 0.0067 | ||||||||||
| STK16 | / | / | 0.86 | 0.0067 | ||||||||||
| CD47 | / | / | 0.86 | 0.0067 | ||||||||||
| PAK7 | / | / | 0.86 | 0.0067 | ||||||||||
| CDK2 CCNA2 | / | / | 0.86 | 0.0067 | ||||||||||
| ZAP70 | / | / | 0.86 | 0.0067 | ||||||||||
| CXCL5 | / | / | 0.86 | 0.0067 | ||||||||||
| MAPK8 | / | / | 0.86 | 0.0067 | ||||||||||
| IL17F | / | / | 0.86 | 0.0067 | ||||||||||
| PDGFRA | / | / | 0.86 | 0.0067 | ||||||||||
| FGF2 | / | / | 0.86 | 0.0067 | ||||||||||
| AFP | / | / | 0.86 | 0.0067 | ||||||||||
| PECAM1 | / | / | 0.86 | 0.0067 | ||||||||||
| STAT6 | / | / | 0.85 | 0.0083 | ||||||||||
| TOP1 | / | / | 0.85 | 0.0083 | ||||||||||
| CRLF2 | / | / | 0.85 | 0.0083 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | TLR4 | / | / | 0.85 | 0.0083 |
| BCL2 | / | / | 0.85 | 0.0083 | ||||||||||
| TNF | / | / | 0.85 | 0.0083 | ||||||||||
| C1QBP | / | / | 0.85 | 0.0083 | ||||||||||
| CLEC11A | / | / | 0.85 | 0.0083 | ||||||||||
| GDF9 | / | / | 0.85 | 0.0083 | ||||||||||
| ING1 | / | / | 0.85 | 0.0083 | ||||||||||
| BIRC5 | / | / | 0.85 | 0.0083 | ||||||||||
| B2M | / | / | 0.85 | 0.0083 | ||||||||||
| ERP29 | / | / | 0.85 | 0.0083 | ||||||||||
| FLT3LG | / | / | 0.85 | 0.0083 | ||||||||||
| NAPA | / | / | 0.85 | 0.0083 | ||||||||||
| GCKR | / | / | 0.85 | 0.0083 | ||||||||||
| EPHA1 | / | / | 0.85 | 0.0083 | ||||||||||
| HSD17B10 | / | / | 0.85 | 0.0083 | ||||||||||
| CEBPB | / | / | 0.85 | 0.0083 | ||||||||||
| SSRP1 | / | / | 0.85 | 0.0083 | ||||||||||
| SPHK2 | / | / | 0.85 | 0.0083 | ||||||||||
| CCL22 | / | / | 0.85 | 0.0083 | ||||||||||
| GDF5 | / | / | 0.85 | 0.0083 | ||||||||||
| APOD | / | / | 0.85 | 0.0083 | ||||||||||
| POR | / | / | 0.99 | 0.0000 | ||||||||||
| LY9 | / | / | 0.97 | 0.0001 | ||||||||||
| IFNGR1 | / | / | 0.96 | 0.0001 | ||||||||||
| GSK3A/B | / | / | 0.96 | 0.0001 | ||||||||||
| nMPs | ||||||||||||||
| Fahrmann,2020 [58]a | USA | Case–control | 6/21 | / | / | Plasma | IV | HC | SOMAscan assay | FLT3 | / | / | 0.95 | 0.0002 |
| CSNK2A1 | / | / | 0.95 | 0.0002 | ||||||||||
| PIK3CA/R1 | / | / | 0.95 | 0.0002 | ||||||||||
| LIN7B | / | / | 0.95 | 0.0002 | ||||||||||
| METAP1 | / | / | 0.95 | 0.0002 | ||||||||||
| MAPK13 | / | / | 0.90 | 0.0015 | ||||||||||
| FAS | / | / | 0.94 | 0.0003 | ||||||||||
SENs, SPEs and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation)
AUC area under the curve, BD Biliary disease, BPD benign pancreatic disease, CP chronic pancreatitis, HC healthy control, IPMN intraductal papillary mucinous neoplasm, NC noncancerous, nPES nanoplasmon-enhanced scattering, NPS nanoplasmonic sensor, SERS surface-Enhanced Raman Scattering, SOMAscan assay slow off-rate modified DNA aptamer, nMPs nonMembrane proteints, MPs Membrane proteins, SEN sensitivity, SPE specificity, MALDI-TOF MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, AbMB antibody-conjugated magnetic beads
arepresent markers extracted from extracellular vesicles
bBPD and HC
cbenign pancreatic tumor, CP, and HC
dbenign pancreatic tumor and pancreatitis
eHC, CP, and IPMN
fno history of cancer
gserous cyst adenoma and CP
hwell-differentiated pancratic neuroendocrine tumor, pancreatic cystic lesions, choronic pancreatitis, and HC
kliver injury, pancreatitis, and cholangitis
*represent estimated value
p value indicates the p value of AUC
Table 3.
Diagnostic performance of biomarker panels in extracellular vesicles for pancreatic cancer
| Study | Country | study design | Cases vs Controls | Specimen | Stage | Controls Status | Detection Method | Panel | SEN% | SPE% | AUC | P Value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age | Male (%) | ||||||||||||
| Madhavan, 2015 [43] | Germany | Case–control | 75/45 | / | / | Serum | I-IV | NCb | qPCR | panel A | 81 | 93 | 0.94 | / |
| Flow cytometry | panel B | 96 | 100 | 0.99 | / | |||||||||
| qPCR/Flow cytometry | panel A/B | 100 | 93 | / | / | |||||||||
| Yang, 2017 [47]a | USA | prospective | 22/21 | / | / | plasma | / | NCb | NPS chip | panel C | 86 | 86 | / | / |
| panel D | 82 | 90 | / | / | ||||||||||
| panel E | 86 | 81 | / | / | ||||||||||
| panel F | 95 | 81 | / | / | ||||||||||
| Lewis, 2018 [50] | USA | Case–control | 20/11 | 64/NA | / | Plasma | IIA- IIB | HC | ACE Immunoassay | panel G | 94 | 91 | 0.99 | / |
| 20/6 | 64/60 | 70/33 | Plasma | II A- II B | BPD | ACE Immunoassay | 81 | 78 | 0.81 | / | ||||
| Xiao, 2019 [79] | China | Case–control | 24/26 | 59/45 | 42/85 | Plasma | / | HC | Flow cytometry | panel H | 76e | 96e | 0.90 | / |
| 24/6 | 59/73 | 42/83 | Plasma | / | CP | Flow cytometry | 100e | 66e | 0.90 | / | ||||
| Yu, 2019 [85]a | China | Case–control | 95/83 | 61/NA | 57/66 | Plasma | I-IV | NCc | ExLR-seq | panel I | 94 | 92 | 0.94 | / |
| 52/83 | / | / | I-II | 88 | 92 | 0.91 | / | |||||||
| 35/83 | / | / | I | 85 | 92 | 0.90 | / | |||||||
| 17/83 | / | / | II | 94 | 92 | 0.93 | / | |||||||
| 43/83 | / | / | III-IV | 100 | 92 | 0.97 | / | |||||||
| 95/40 | 61/53 | 57/70 | Plasma | I-IV | CP | ExLR-seq | 94 | 90 | 0.95 | / | ||||
| 52/40 | / | / | I-II | 89 | 90 | 0.92 | / | |||||||
| 35/40 | / | / | I | 86 | 90 | 0.91 | / | |||||||
| 17/40 | / | / | II | 94 | 90 | 0.95 | / | |||||||
| 43/40 | / | / | III-IV | 100 | 90 | 0.99 | / | |||||||
| 95/43 | 61/63 | 57/62 | Plasma | I-IV | HC | ExLR-seq | 94 | 93 | 0.92 | / | ||||
| Rodrigues, 2019 [33]a | USA | Case–control | 20/12 | / | 45/25 | serum | NCf | nanoparticle-and dyebased fluorescent immunoassay | panel Q | / | / | 0.95 | / | |
| Reese, 2020 [31] | Germany | Case–control | 56/33 | / | 64/52 | Serum-EpCAM | II-IV | NCc | qPCR | panel J | 64e | 91e | 0.84 | 0.0004 |
| Zhou, 2020 [62] | China | Case–control | 30/10 | 60/58 | / | Plasma | I-IV | NCd | 3D mircrofluidic chip | panel K | 100e | 100e | 1.00 | / |
| Wu, 2020 [61] | China | Case–control | 30/10 | 62/51 | 60/80 | serum | 0-IV | CP | qPCR | panel L | 93 | 80 | / | / |
| Pu, 2020 [25] | China | Case–control | 36/65 | / | / | Plasma | I-IV | HC | cationic lipoplex nanoparticle | panel M | 75 e | 80 e | 0.79 | < 0.0001 |
| Qin, 2021 [63]a | China | Case–control | 44/27 | / | 50/44 | Plasma | I-IV | HC | qPCR | panel N | 75e | 74e | 0.78 | / |
| panel P | 91e | 74e | 0.89 | / | ||||||||||
| 44/40 | / | 50/65 | CP | panel N | 92e | 40e | 0.71 | / | ||||||
| panel P | 64e | 90e | 0.77 | / | ||||||||||
| 44/67 | 50/57 | NCc | panel N | 72e | 63e | 0.70 | / | |||||||
| panel P | 53e | 83e | 0.72 | / | ||||||||||
| Li, 2021 [76]a | China | Case–control | 21/29 | 54/62 | 43/55 | plasma | / | HC | AbMB-bioChol paltform | panel R | 38e | 93e | 0.74 | / |
| Wu, 2021 [71] | China | Case–control | 284/117 | / | 59/62 | plasma | I-IV | HC | ExLR-seq | panel S | 80e | 73e | 0.86 | / |
| 284/100 | / | 59/46 | CP | 64e | 82e | 0.84 | / | |||||||
| 14/32 | / | / | / | HC | RNA-seq | panel T | 100 | 100 | 1.00 | / | ||||
| Verel-Y, 2021 [73] | Germany | Case–control | 72/20 | / | 50/NA | Serum | I-IV | HC | bead-coupled FACS/qPCR | panel U | 100 | 100 | 1.00 | / |
| panel V | 91e | 82e | 0.93 | / | ||||||||||
| Kim, 2021 [77]a | Korea | Case–control | 20/20 | 61/51 | 70/50 | plasma | I-III | CL | qPCR | panel W | 38e | 95e | 0.69 | / |
| panel X | 65e | 99e | 0.77 | / | ||||||||||
| panel Y | 76e | 75e | 0.84 | / | ||||||||||
| panel Z | 85e | 81e | 0.87 | / | ||||||||||
| panel 1 | 56e | 95e | 0.79 | / | ||||||||||
| panel 2 | 81 | 80 | 0.87 | / | ||||||||||
| panel 3 | 76 | 90 | 0.87 | / | ||||||||||
| panel 4 | 76 | 90 | 0.90 | / | ||||||||||
| panel 5 | 81 | 85 | 0.86 | / | ||||||||||
| panel 6 | 76 | 85 | 0.87 | / | ||||||||||
| panel 7 | 76 | 85 | 0.91 | / | ||||||||||
| panel 8 | 86 | 90 | 0.91 | / | ||||||||||
| Kim, 2021 [77]a | Korea | Case–control | 20/20 | 61/51 | 70/50 | plasma | I-III | CL | qPCR | panel 9 | 76 | 90 | 0.93 | / |
| panel 10 | 81 | 85 | 0.91 | / | ||||||||||
| panel 11 | 67 | 90 | 0.81 | / | ||||||||||
| panel 12 | 76 | 90 | 0.90 | / | ||||||||||
| panel 13 | 86 | 90 | 0.94 | / | ||||||||||
| panel 14 | 90 | 90 | 0.95 | / | ||||||||||
| panel 15 | 86 | 90 | 0.94 | / | ||||||||||
| panel 16 | 81 | 85 | 0.92 | / | ||||||||||
| panel 17 | 86 | 90 | 0.91 | / | ||||||||||
| panel 18 | 76 | 90 | 0.94 | / | ||||||||||
| panel 19 | 90 | 85 | 0.94 | / | ||||||||||
| panel 20 | 81 | 90 | 0.96 | / | ||||||||||
| panel 21 | 86 | 90 | 0.94 | / | ||||||||||
| panel 22 | 90 | 90 | 0.96 | / | ||||||||||
| panel 23 | 90 | 85 | 0.95 | / | ||||||||||
| panel 24 | 90 | 90 | 0.96 | / | ||||||||||
| panel 25 | 86 | 90 | 0.95 | / | ||||||||||
| panel 26 | 86 | 85 | 0.96 | / | ||||||||||
| panel 27 | 90 | 90 | 0.97 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 27/15 | 57/43 | 63/60 | plasma | IB-IV | CP | small RNA sequencing | panel 28 | 81 | 93 | 0.88 | / |
| 30/18 | 63/44 | 63/72 | IB-III | panel 29 | / | / | 0.94 | / | ||||||
| panel 30 | / | / | 0.94 | / | ||||||||||
| panel 31 | / | / | 0.94 | / | ||||||||||
| panel 32 | / | / | 0.94 | / | ||||||||||
| panel 33 | / | / | 0.94 | / | ||||||||||
| panel 34 | / | / | 0.94 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 30/18 | 63/44 | 63/72 | plasma | IB-III | CP | small RNA sequencing | panel 35 | / | / | 0.93 | / |
| panel 36 | / | / | 0.93 | / | ||||||||||
| panel 37 | / | / | 0.93 | / | ||||||||||
| panel 38 | / | / | 0.93 | / | ||||||||||
| panel 39 | / | / | 0.93 | / | ||||||||||
| panel 40 | / | / | 0.93 | / | ||||||||||
| panel 41 | / | / | 0.92 | / | ||||||||||
| panel 42 | / | / | 0.92 | / | ||||||||||
| panel 43 | / | / | 0.92 | / | ||||||||||
| panel 44 | / | / | 0.92 | / | ||||||||||
| panel 45 | / | / | 0.92 | / | ||||||||||
| panel 46 | / | / | 0.92 | / | ||||||||||
| panel 47 | / | / | 0.92 | / | ||||||||||
| panel 48 | / | / | 0.92 | / | ||||||||||
| panel 49 | / | / | 0.91 | / | ||||||||||
| panel 50 | / | / | 0.91 | / | ||||||||||
| panel 51 | / | / | 0.91 | / | ||||||||||
| panel 52 | / | / | 0.91 | / | ||||||||||
| panel 53 | / | / | 0.91 | / | ||||||||||
| panel 54 | / | / | 0.91 | / | ||||||||||
| panel 55 | / | / | 0.91 | / | ||||||||||
| panel 56 | / | / | 0.91 | / | ||||||||||
| panel 57 | / | / | 0.91 | / | ||||||||||
| panel 58 | / | / | 0.91 | / | ||||||||||
| panel 59 | / | / | 0.90 | / | ||||||||||
| panel 60 | / | / | 0.90 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 30/18 | 63/44 | 63/72 | plasma | IB-III | CP | small RNA sequencing | panel 61 | / | / | 0.90 | / |
| panel 62 | / | / | 0.90 | / | ||||||||||
| panel 63 | / | / | 0.90 | / | ||||||||||
| panel 64 | / | / | 0.90 | / | ||||||||||
| panel 65 | / | / | 0.90 | / | ||||||||||
| panel 66 | / | / | 0.90 | / | ||||||||||
| panel 67 | / | / | 0.90 | / | ||||||||||
| panel 68 | / | / | 0.90 | / | ||||||||||
| panel 69 | / | / | 090 | / | ||||||||||
| panel 70 | / | / | 0.90 | / | ||||||||||
| panel 71 | / | / | 0.90 | / | ||||||||||
| panel 72 | / | / | 0.90 | / | ||||||||||
| panel 73 | / | / | 0.90 | / | ||||||||||
| panel 74 | / | / | 0.90 | / | ||||||||||
| panel 75 | / | / | 0.90 | / | ||||||||||
| panel 76 | / | / | 0.89 | / | ||||||||||
| panel 77 | / | / | 0.89 | / | ||||||||||
| panel 78 | / | / | 0.89 | / | ||||||||||
| panel 79 | / | / | 0.89 | / | ||||||||||
| panel 80 | / | / | 0.89 | / | ||||||||||
| panel 81 | / | / | 0.89 | / | ||||||||||
| panel 82 | / | / | 0.89 | / | ||||||||||
| panel 83 | / | / | 0.89 | / | ||||||||||
| panel 84 | / | / | 0.89 | / | ||||||||||
| panel 85 | / | / | 0.89 | / | ||||||||||
| panel 86 | / | / | 0.89 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 30/18 | 63/44 | 63/72 | plasma | IB-III | CP | small RNA sequencing | panel 87 | / | / | 0.89 | / |
| panel 88 | / | / | 0.89 | / | ||||||||||
| panel 89 | / | / | 0.89 | / | ||||||||||
| panel 90 | / | / | 0.88 | / | ||||||||||
| panel 91 | / | / | 0.88 | / | ||||||||||
| panel 92 | / | / | 0.88 | / | ||||||||||
| panel 93 | / | / | 0.88 | / | ||||||||||
| panel 94 | / | / | 0.88 | / | ||||||||||
| panel 95 | / | / | 0.88 | / | ||||||||||
| panel 96 | / | / | 0.88 | / | ||||||||||
| panel 97 | / | / | 0.88 | / | ||||||||||
| panel 98 | / | / | 0.88 | / | ||||||||||
| panel 99 | / | / | 0.88 | / | ||||||||||
| panel 100 | / | / | 0.88 | / | ||||||||||
| panel 101 | / | / | 0.88 | / | ||||||||||
| panel 102 | / | / | 0.88 | / | ||||||||||
| panel 103 | / | / | 0.87 | / | ||||||||||
| panel 104 | / | / | 0.87 | / | ||||||||||
| panel 105 | / | / | 0.87 | / | ||||||||||
| panel 106 | / | / | 0.87 | / | ||||||||||
| panel 107 | / | / | 0.87 | / | ||||||||||
| panel 108 | / | / | 0.87 | / | ||||||||||
| panel 109 | / | / | 0.87 | / | ||||||||||
| panel 110 | / | / | 0.86 | / | ||||||||||
| panel 111 | / | / | 0.86 | / | ||||||||||
| panel 112 | / | / | 0.86 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 30/18 | 63/44 | 63/72 | plasma | IB-III | CP | small RNA sequencing | panel 113 | / | / | 0.86 | / |
| panel 114 | / | / | 0.86 | / | ||||||||||
| panel 115 | / | / | 0.85 | / | ||||||||||
| panel 116 | / | / | 0.85 | / | ||||||||||
| panel 117 | / | / | 0.85 | / | ||||||||||
| panel 118 | / | / | 0.84 | / | ||||||||||
| panel 119 | / | / | 0.83 | / | ||||||||||
| panel 120 | / | / | 0.83 | / | ||||||||||
| panel 121 | / | / | 0.83 | / | ||||||||||
| panel 122 | / | / | 0.83 | / | ||||||||||
| panel 123 | / | / | 0.83 | / | ||||||||||
| panel 124 | / | / | 0.82 | / | ||||||||||
| panel 125 | / | / | 0.82 | / | ||||||||||
| panel 126 | / | / | 0.822 | / | ||||||||||
| panel 127 | / | / | 0.82 | / | ||||||||||
| panel 128 | / | / | 0.82 | / | ||||||||||
| panel 129 | / | / | 0.81 | / | ||||||||||
| panel 130 | / | / | 0.81 | / | ||||||||||
| panel 131 | / | / | 0.80 | / | ||||||||||
| panel 132 | / | / | 0.79 | / | ||||||||||
| panel 133 | / | / | 0.77 | / | ||||||||||
| panel 134 | / | / | 0.76 | / | ||||||||||
| panel 135 | / | / | 0.76 | / | ||||||||||
| panel 136 | / | / | 0.76 | / | ||||||||||
| panel 137 | / | / | 0.75 | / | ||||||||||
| panel 138 | / | / | 0.74 | / | ||||||||||
| Guo, 2021 [78]a | China | Case–control | 30/18 | 63/44 | 63/72 | plasma | IB-III | CP | small RNA sequencing | panel 139 | / | / | 0.74 | / |
| panel 140 | / | / | 0.74 | / | ||||||||||
| panel 141 | / | / | 0.72 | / | ||||||||||
| panel 142 | / | / | 0.72 | / | ||||||||||
| panel 143 | / | / | 0.72 | / | ||||||||||
| panel 144 | / | / | 0.71 | / | ||||||||||
| panel 145 | / | / | 0.71 | / | ||||||||||
| panel 146 | / | / | 0.71 | / | ||||||||||
SENs, SPEs and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation)
AUC area under the curve, ACE alternating current electrokinetic, AbMB antibody-conjugated magnetic beads, BPD benign pancreatic disease, CP chronic pancreatitis, CL cholecytitis, HC healthy control, FACS Cartoon of protocol for flow cytometry, NC noncancerous, NPS nanoplasmonic sensor, SEN sensitivity, SPE specificity
arepresent markers extracted from extracellular vesicles
bHC, CP, and benign pancreatic tumor
cHC and CP
dno history of cancer
erepresent 估算值
frepresent liver injury, pancreatitis, and cholangitis
Panel A: miR-1246, miR-4644, miR-3976, miR-4306; Panel B, CD44v6/Tspan8/EpCAM/CD104; panel C, EGFR/EPCAM/HER2/MUC1; Panel D, EGFR/EPCAM/GPC1/WNT2; Panel E, EGFR/EPCAM/MUC1/GPC1/WNT2; Panel F, EGFR/EPCAM/HER2/MUC1/GPC1/WNT2; Panel G, GPC1/CD63; Panel H, GPC1/CD82; Panel I, FGA/KRT19/HIST1H2BK/ITIH2/MARCH2/CLDN1/MAL2/TIMP1; panel Q, EpCAM/EphA2; Panel J, miR-200c/miR-200b; Panel K, miR-451a/21/10b/EphA2; Panel L, miR-21/210; Panel M, miR-21/10b; Panel N, FBXO7/MORF4L1//DDX17/TALDO1//AHNAK/TUBA1B; Panel P, FBXO7/MORF4L1//DDX17/TALDO1//AHNAK/TUBA1B//CD44/SETD3; panel R, EGFR/EpCAM/GPC1/EphA2; panel S, HIST2H2AA3/LUZP6/HLA-DRA; panel T, HIST2H2AA3/HIST1H4K/HLD-DRA/RN7SL1/LUZP6/FAM184B/FGF23/NEUROD2/miR663AHG/GPM6A; panel U, ADAM8/miR-720; panel V, ADAM8/miR-451; panel W, ITGA2/ITGAV/GPC1/miR-10b; panel X, ITGA2/ITGAV/GPC1/miR-21; panel Y, ITGA2/ITGAV/GPC1/miR-155; panel Z, ITGA2/ITGAV/GPC1/miR-429; panel 1, ITGA2/ITGAV/GPC1/miR-1290; panel 2, ITGA2/ITGAV/GPC1/miR-21/miR-155; panel 3, ITGA2/ITGAV/GPC1/miR-21/miR-429; panel 4, ITGA2/ITGAV/GPC1/miR-21/miR-1290; panel 5, ITGA2/ITGAV/GPC1/miR-21/miR-10b; panel 6, ITGA2/ITGAV/GPC1/miR-155/miR-429; panel 7, ITGA2/ITGAV/GPC1/miR-155/miR-1290; panel 8, ITGA2/ITGAV/GPC1/miR-155/miR10b; panel 9, ITGA2/ITGAV/GPC1/miR-429/miR-1290; panel 10, ITGA2/ITGAV/GPC1/miR-429/miR-10b; panel 11, ITGA2/ITGAV/GPC1/miR-1290/miR-10b; panel 12, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-429; panel 13, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-10b; panel 14, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-1290; panel 15, ITGA2/ITGAV/GPC1/miR-21/miR-429/miR-1290; panel 16, ITGA2/ITGAV/GPC1/miR-21/miR-429/miR-10b; panel 17, ITGA2/ITGAV/GPC1/miR-21/miR-1290/miR-10b; panel 18, ITGA2/ITGAV/GPC1/miR-155/miR-429/miR-1290; panel 19, ITGA2/ITGAV/GPC1/miR-155/miR-429/miR-10b; panel 20, ITGA2/ITGAV/GPC1/miR-155/miR-1290/miR-10b; panel 21, ITGA2/ITGAV/GPC1/miR-429/miR-1290/miR-10b; panel 22, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-429/miR-1290; panel 23, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-429/miR-10b; panel 24, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-1290/miR-10b; panel 25, ITGA2/ITGAV/GPC1/miR-21/miR-429/miR-1290/miR-10b; panel 26, ITGA2/ITGAV/GPC1/miR-155/miR-429/miR-1290/miR-10b; panel 27, ITGA2/ITGAV/GPC1/miR-21/miR-155/miR-429/miR-1290/miR-10b; panel 28, miR-95-3p/miR-26b-5p; panel 29, miR-95-3p/miR-3605-3p; panel 30, miR-95-3p/miR-128-3p; panel 31, miR-95-3p/miR-30d-5p; panel 32,miR-95-3p/miR-505-5p; panel 33,miR-95-3p/miR-148b-3p; panel 34, miR-95-3p/miR-342-5p; panel 35,miR-95-3p/miR-532-5p; panel 36, miR-95-3p/let-7 g-5p; panel 37, miR-95-3p/miR-151a-3p; panel 38, miR-95-3p/miR-181a-2-3p; panel 39,miR-95-3p/miR-550a-5p; panel 40, miR-95-3p/let-7b-5p; panel 41, miR-95-3p/miR-191-5p; panel 42,miR-95-3p/miR-92a-3p; panel 43, miR-95-3p/miR-941; panel 44, miR-95-3p/miR-106b-3p; panel 45, miR-95-3p/miR-7706; panel 46, miR-95-3p/miR-183-5p; panel 47,miR-95-3p/miR-25-5p; panel 48,miR-95-3p/miR-486-3p; panel 49,miR-95-3p/miR-3158-3p; panel 50, miR-95-3p/miR-7-5p; panel 51, miR-95-3p/miR-101-3p; panel 52,miR-95-3p/miR-210-3p; panel 53,miR-95-3p/miR-550a-3-5p; panel 54, miR-95-3p/miR-584-5p; panel 55,miR-95-3p/miR-140-3p; panel 56, miR-95-3p/miR-4732-5p; panel 57, miR-95-3p/miR-363-5p; panel 58,miR-95-3p/miR-4326; panel 59, miR-95-3p/miR-1294; panel 60,miR-95-3p/miR-486-5p; panel 61, miR-95-3p/miR-185-3p; panel 62,miR-95-3p/miR-4732-3p; panel 63, miR-95-3p/miR-92b-3p; panel 64,miR-95-3p/miR-423-5p; panel 65,miR-95-3p/miR-503-5p; panel 66, miR-95-3p/miR-1180-3p; panel 67, miR-95-3p/miR-25-3p; panel 68,miR-95-3p/miR-92b-5p; panel 69, miR-95-3p/miR-1284; panel 70,miR-95-3p/miR-17-5p; panel 71, miR-95-3p/miR-2110; panel 72, miR-95-3p/miR-24–2-5p; panel 73,miR-95-3p/miR-339-3p; panel 74,miR-95-3p/miR-660-5p; panel 75,miR-95-3p/miR-6842-3p; panel 76, miR-95-3p/let-7d-5p; panel 77, miR-95-3p/miR-30e-5p; panel 78, miR-95-3p/miR-628-3p; panel 79, miR-95-3p/miR-629-5p; panel 80, miR-95-3p/let-7i-5p; panel 81, miR-95-3p/miR-142-5p; panel 82,miR-95-3p/miR-182-5p; panel 83, miR-95-3p/miR-1908-5p; panel 84, miR-95-3p/miR-425-5p; panel 85,miR-95-3p/miR-942-5p; panel 86, miR-95-3p/miR-93-5p; panel 87, miR-95-3p/miR-363-3p; panel 88, miR-95-3p/miR-18a-3p; panel 89, miR-95-3p/miR-320a; panel 90, miR-95-3p/miR-421; panel 91, miR-95-3p/miR-501-3p; panel 92,miR-95-3p/let-7a-3p; panel 93, miR-95-3p/miR-16–2-3p; panel 94, miR-95-3p/miR-16-5p; panel 95,miR-95-3p/miR-130b-3p; panel 96, miR-95-3p/miR-3613-5p; panel 97, miR-95-3p/miR-451a; panel 98, miR-95-3p/miR-20b-5p; panel 99, miR-95-3p/miR-103a-3p; panel 100, miR-95-3p/miR-1224-5p; panel 101, miR-95-3p/miR-185-5p; panel 102, miR-95-3p/miR-20a-5p; panel 103, miR-95-3p/miR-186-5p; panel 104, miR-95-3p/miR-3615; panel 105, miR-95-3p/miR-7976; panel 106, miR-95-3p/miR-652-3p; panel 107, miR-95-3p/miR-107; panel 108, miR-95-3p/miR-181a-5p; panel 109, miR-95-3p/miR-15b-3p; panel 110, miR-95-3p/let-7b-3p; panel 111, miR-95-3p/miR-10b-5p; panel 112, miR-95-3p/miR-24-3p; panel 113, miR-95-3p/miR-106b-5p; panel 114, miR-95-3p/miR-15a-5p; panel 115, miR-95-3p/miR-197-3p; panel 116, miR-95-3p/miR-32-5p; panel 117, miR-95-3p/miR-450b-5p; panel 118, miR-95-3p/let-7e-5p; panel 119, miR-95-3p/miR-155-5p; panel 120, miR-95-3p/miR-361-5p; panel 121, miR-95-3p/miR-126-3p; panel 122, miR-95-3p/miR-484; panel 123, miR-95-3p/miR-30a-5p; panel 124, miR-95-3p/miR-27a-3p; panel 125, miR-95-3p/miR-29a-3p; panel 126, miR-95-3p/miR-335-5p; panel 127, miR-95-3p/miR-125a-5p; panel 128, miR-95-3p/miR-338-5p; panel 129, miR-95-3p/miR-139-5p; panel 130, miR-95-3p/miR-22-5p; panel 131, miR-95-3p/miR-23a-3p; panel 132, miR-95-3p/miR-382-5p; panel 133, miR-95-3p/miR-543; panel 134, miR-95-3p/miR-499a-5p; panel 135, miR-95-3p/miR-4433b-3p; panel 136, miR-95-3p/miR-1228-5p; panel 137, miR-95-3p/miR-99b-5p; panel 138, miR-95-3p/miR-143-3p; panel 139, miR-95-3p/miR-206; panel 140, miR-95-3p/miR-224-5p; panel 141, miR-95-3p/miR-10a-5p; panel 142, miR-95-3p/miR-223-3p; panel 143, miR-95-3p/miR-134-5p; panel 144, miR-95-3p/miR-485-5p; panel 145, miR-95-3p/miR-760; panel 146, miR-95-3p/miR-199b-3p
The isolation and detection methods of EVs were diversity, 15 studies used the commercial kits [24, 25, 46, 53, 60, 61, 64, 68, 70–72, 74, 83–85], 11 studies used ultracentrifugation [31, 43, 45, 58, 59, 63, 69, 73, 75, 79, 81], one study was not stated the isolation and detection method of EV [80], the remaining studies developed new isolation and detection techniques including ephrin type-A receptor 2/nanoplasmon-enhanced scattering (EphA2-EV-nPES assay) [34], PDA encapsulated antibody-reporter-Ag(shell)-Au(core) multilayer, surface-Enhanced Raman Scattering (chip-exosome-PEARL SERS immunosensor) [82], AC electrokinetic integrated biomarker assay [86], nanoplasmonic sensor assay (NPS chip) [47], lipid-polymer hybrid nanoparticle/catalyzed hairpin DNA circuit (LPHN-CHDC biochip) [44], immunogold transmission electron microscopy [30], Sequential size-exclusion chromatography (SSEC) [67], antibody-conjugated magnetic beads, bivalent cholesterol-modified RNA − DNA duplexes (AbMB-bioChol) platform [76], nanoparticle-and dyebased fluorescent immunoassay [33], immuno-capture using magnetic beads [77], and 3D microfluidic chip [62, 78] (Additional file 1).
Quality assessment
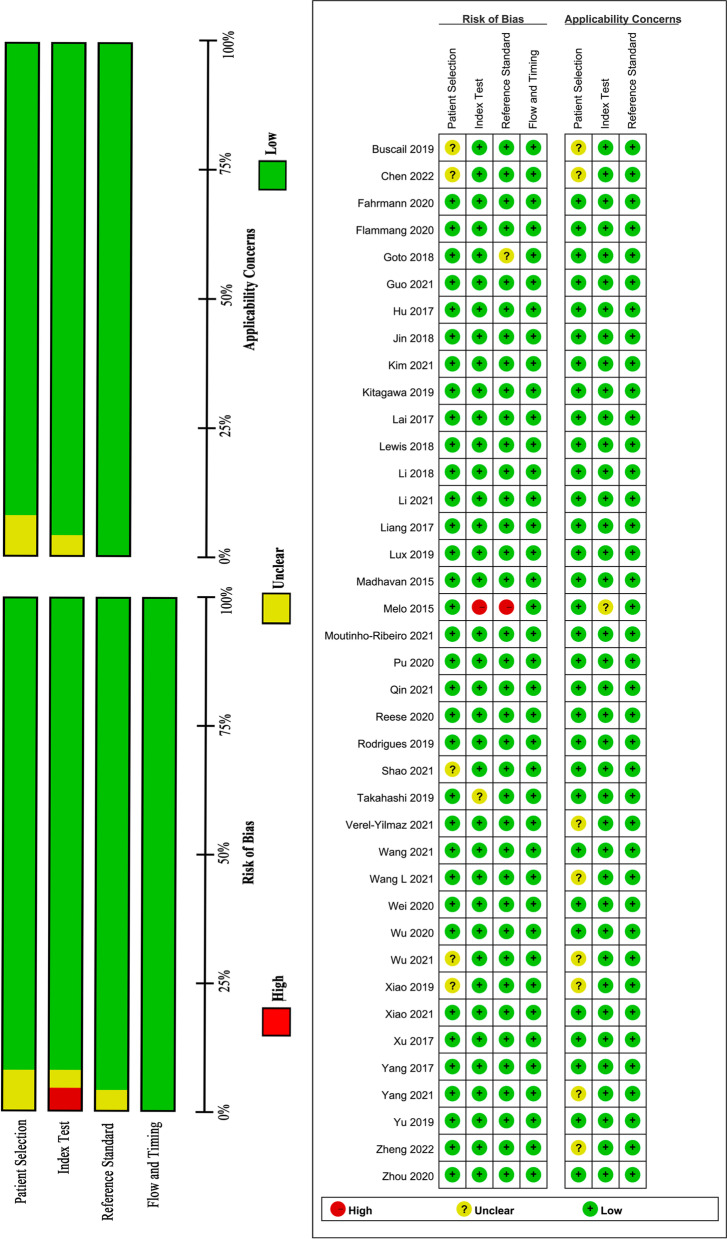
We judged the risk of bias and applicability concerns of the included studies using QUADAS-2 evaluation tool, which were grouped four domains: participant selection, index test, reference standard, flow and timing. The outcomes of our assessment of methodological quality were summarized in Fig. 2. Twenty-seven studies were high quality studies with low risk of bias for all four domains. The risk of bias in the “index test” was high and the applicability concerns of the “index test” was unclear in Melo et al. 2015, as the index test might not be reproduced [30]; the index test of one study had unclear risk due to the unspecified of EVs isolation and detection method [80]; the risk of bias and applicability concerns of the “patient selection” were unclear in 6 studies [52, 56, 68, 71, 74], as the age and gender distribution was significant differences both in PC cases and control groups; The risk of bias and applicability concerns of the “reference standard” were unclear in Goto 2018, as the reference standard in this study was not clarified [84].
Fig. 2.
QUADAS-2 assessment. Risk of bias and applicability concerns summary (A) graph and (B) summary
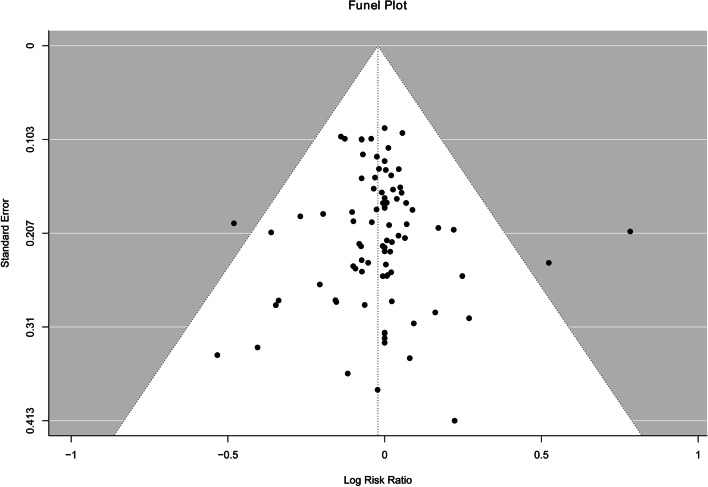
The results of egger’s test (Z = 0.28, P = 0.78) did not provided any evidence of publication bias. The funnel plot showed reasonably symmetrical, which also supports the results of egger’s test (Fig. 3).
Fig. 3.
Funnel plot with 95% confidence limits
Diagnostic performance
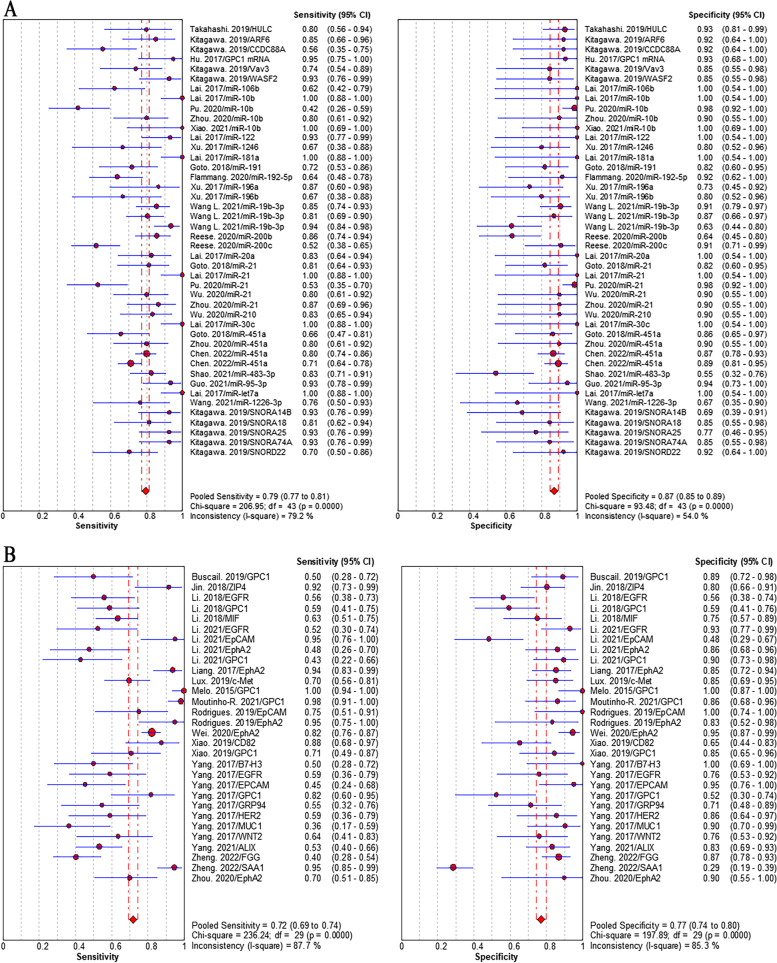
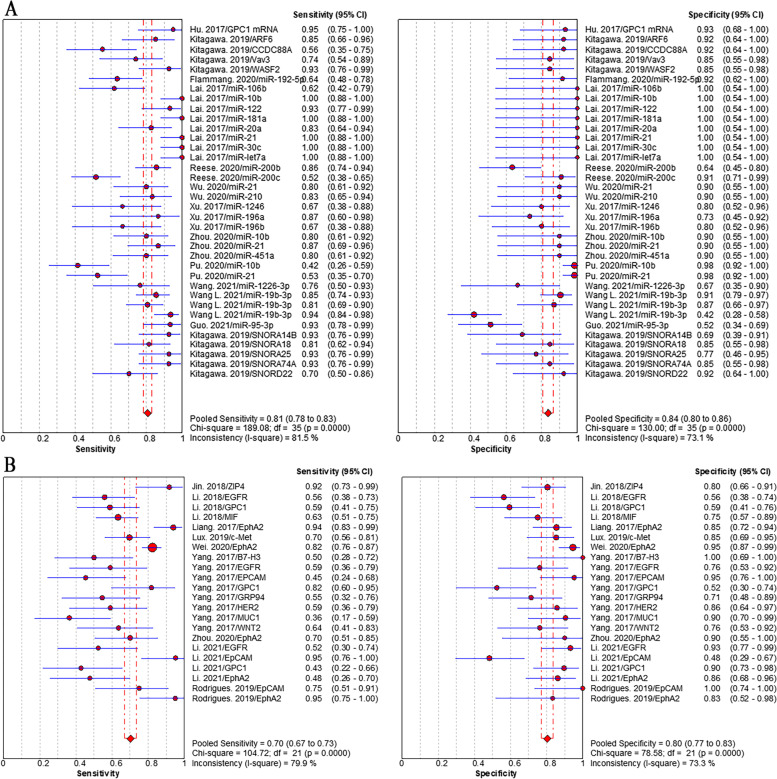
In summary, a total number of 183 EV RNAs in 20 included studies were reported to be statistically significant in PC diagnosis and 161 EV RNAs were included in panels. Nine miRNAs (including miR-10b, miR-21, miR-451a, miR-106b, miR-155, miR-181a, miR-191, miR-1246, and miR-20a) were reported more than one times. Among them, miR-21 and miR-10b were the highest frequently reported RNA which was reported in 6 times. The expression direction of most EV miRNAs were up-regulate, apart from miR-122 [45], miR-let7 [45], miR-1226-3p [64], miR-19b-3p [72], miR-3158-3p [78], miR-4732-3p [78], miR-7706 [78], and miR-486-3p [78] were down-regulate (Additional file 2). The expression directions of miR-192-5p [59] and miR-196b [46] were not reported (Additional file 2). For individual EV RNAs, the median reported sensitivity and specificity were 82% (from 42 to 100%) and 90% (from 63 to 100%), respectively. Both sensitivity and specificity of 52% EV RNAs exceeded 80%. One study conducted validation test by Hu et al., which validated EV GPC1-mRNA as a good diagnostic biomarker for PC, with the sensitivity, specificity, and AUC were 95%, 93% and 0.94, respectively [44].
A total of 177 EV proteins reported in 13 studies were statistically significant for PC diagnosis, 19 EV proteins were included in panels. GPC1 was the most frequently reported EV protein with median sensitivity and specificity of 72% (from 43 to 100%) and 86% (from 52 to 100%), respectively, which was reported in nine times. And the following were EphA2, EGFR, and EPCAM (Additional file 3). For individual EV proteins, the median reported sensitivity and specificity were 64% (36–100%) and 85% (52–100%), respectively. Both sensitivity and specificity of 20% EV proteins exceeded 80%.
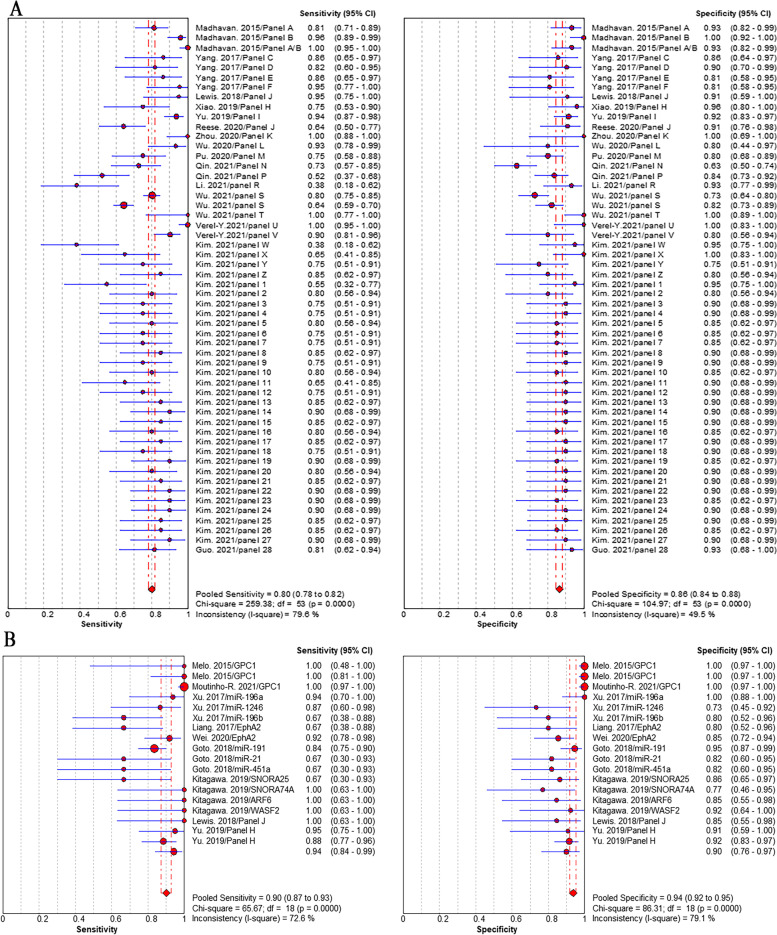
172 EV biomarker panels in 16 studies were reported and 12 panels were verified by independent validation among six studies (Table 3). Most panels showed powerful diagnostic accuracy with median sensitivity and specificity of 82% (from 38 to 100%) and 90% (from 63 to 100%), respectively. Both the sensitivity and specificity of 63% panels exceeded 80%.
Nine studies reported diagnosis performance of EV biomarkers (17 individual biomarkers and 2 panels) for early stage (stage I-II) PC, two of individual biomarkers were conducted independent validation tests. And across these two validation studies, both the sensitivity and specificity of EV biomarkers for early stage (stage I-II) PC was more than 85% [34]. The median reported sensitivity and specificity for early stage PC were 94% (from 67 to 100%) and 86% (from 73 to 100%), respectively. Both sensitivity and specificity of 68% EV biomarkers exceeded 80%.
Results of meta-analysis
Studies that reported individual EV biomarkers for PC diagnosis were performed meta-analyses. Overall, 32 individual EV RNAs investigated in 16 studies and 16 individual EV proteins investigated in 13 studies were included in the meta-analyses. The sensitivity (± 95% confidence intervals) and specificity (± 95% confidence intervals) for each individual EV RNAs and each individual EV proteins were depicted in the corresponding forest plots. The pooled sensitivity and specificity of individual EV RNAs was 79% (95% CI: 77–81%) and 87% (95% CI: 85–89%), respectively (Fig. 4A). The pooled sensitivity and specificity of individual EV proteins was 72% (95% CI: 69–74%) and 77% (95% CI: 74–80%), respectively (Fig. 4B). The pooled sensitivity and specificity of EV biomarker panels were 80% (95% CI: 78–82%) and 86% (95% CI: 84–88%), respectively (Fig. 5A). We separately analyzed the diagnostic performance of EV RNA panels, EV protein panels, and EV RNA combined with protein panels, the results showed that EV RNA combined with protein panels had higher diagnosis value than those of EV RNA panels or EV protein panels. The pooled sensitivity and specificity were 84% (95% CI: 78–82%) and 89% (95% CI: 84–88%) for RNA combined with protein panels, 76% (95% CI: 73–78%) and 82% (95% CI: 79–85%) for RNA panels, and 85% (95% CI: 80–89%) and 91% (95% CI: 86–95%) for protein panels, respectively (addition file 4). Positively, we focused on the diagnosis value of EV biomarkers for early stage (stage I and II) PC. The results showed that the pooled sensitivity and specificity were 90% (95% CI: 87–93%) and 94% (95% CI: 92–95%), respectively (Fig. 5B).
Fig. 4.
Pooled sensitivity and specificity of EV biomarkers for pancreatic cancer diagnosis. A individual EV RNAs, (B) individual EV proteins
Fig. 5.
Pooled sensitivity and specificity of individual EV biomarkers for pancreatic cancer diagnosis. A EV biomarker panels, (B) EV biomarkers for early stage PC
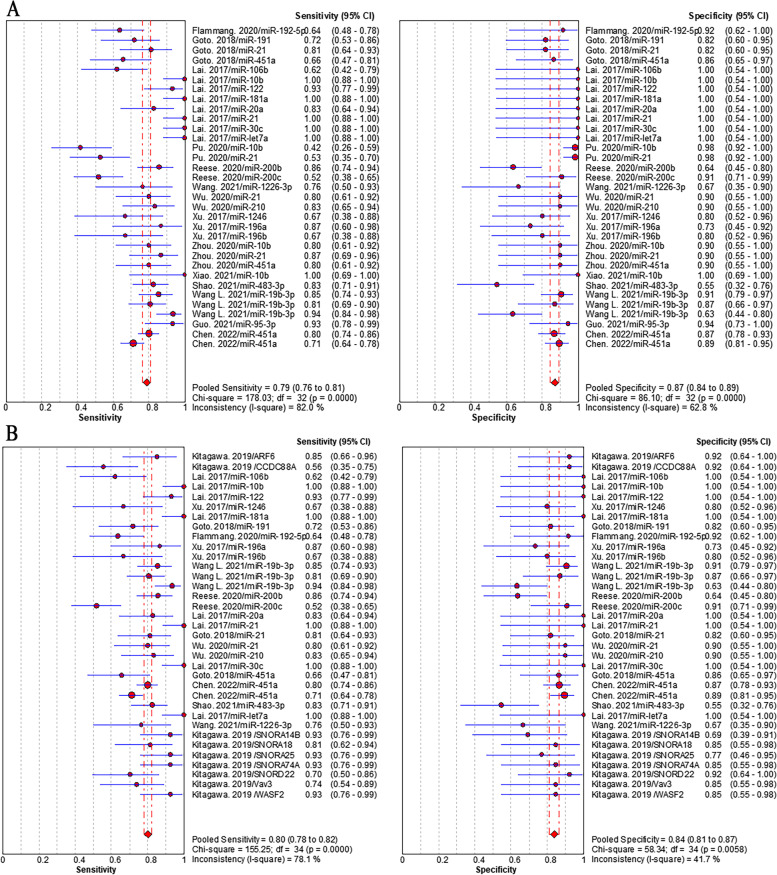
Sensitivity analysis in according to the high quality studies failed to demonstrate a change in the pooled sensitivity and specificity (RNAs: 81% (95%CI: 78–83%) and 84% (95%CI: 80–86%), Fig. 6A; Proteins: 70% (95%CI: 67–73%) and 80% (95% CI: 77–83%), Fig. 6B) for PC diagnosis. In subgroup analysis, we did not observe notable differences in pooled sensitivity and specificity of miRNAs (79% (95% CI: 76–81%) and 87% (95% CI: 84–89%), Fig. 7A) vs those of the whole RNAs (79% (95% CI: 77–81%) and 87% (95% CI: 85–89%)). We also found the similar pooled sensitivity and specificity between RNAs detected by qPCR (80% (95% CI: 78–82%) and 84% (95% CI: 81–87%), Fig. 7B) and those of the whole RNAs. It indicated that the heterogeneity in sensitivity and subgroup analyses was similar to the whole analysis.
Fig. 6.
Pooled sensitivity and specificity of EV biomarkers for pancreatic cancer diagnosis in sensitivity analysis according to high quality studies. A individual EV RNAs, (B) individual EV proteins
Fig. 7.
Pooled sensitivity and specificity of EV biomarkers for pancreatic cancer diagnosis in subgroup analyses. A individual EV miRNAs, (2) individual EV RNAs detected by qPCR
Discussion
A systematic understanding of diagnosis performance of tumor-related moleculars in circulation EVs is critical for PC screening and earliest detection. The present systematic review and meta-analysis has assembled the diagnostic performance of 183 EV RNAs, 177 EV proteins, and 172 EV biomarker panels based on serum/plasma for diagnosing PC in 39 studies from 2015 to 2022. The control groups selected noncancerous population containing healthy population, benign pancreatic disease, IPMN, serious cystadenoma, and nonpancreatic benign diseases. MiR-21 and miR-10b were the highest frequently reported EV RNA for PC diagnosis. GPC1 and EphA2 were mostly reported EV proteins for PC diagnosis. Both the EV RNAs and the EV proteins in high quality studies showed moderate diagnostic values and the EV panels revealed good diagnostic values for PC. Especially, the EV RNA combined protein panels had great diagnostic performances for PC with the pooled sensitivity of 84% and specificity of 89%. Surprisingly, for early stage PC the EV biomarkers showed excellent diagnostic performance, the sensitivity and specificity were 90% and 94%. Respectively. Overall, available studies highlighted the diagnostic performance of EV biomarkers to differentiate PC from noncancerous and further researches will be needed to validate the diagnostic value of EV biomarkers as noninvasive, effective, specific screening tools for PC diagnosis in general population.
EV GPC1 and EV EphA2 were the mostly frequent reported diagnostic proteins for PC in present study. GPC1 was discovered in two prospective studies [47, 75], one of which was conducted a blind validation test [47]. In 2015 Nature, EV GPC1 was demonstrated extremely surprising diagnostic accuracy for diagnosing PC, the AUC value reached 1.00 for all stage PC, and the validation test got consistent results that both the sensitivity and specificity of GPC1 for PC diagnosis was 100% [30]. This conclusion attracted highly attention worldwide at a time, but other researchers found inconsistent results at later [30]. A blind prospective study by Yang et al. reported that EV GPC1 had a good sensitivity for PC diagnosis, but the specificity poorly was 52% [47]. In 2018, Li et al. used exosome-based GPC1 to diagnose PC by an ultrasensitive immunoassay method. Although exosome GPC1 could distinguish PC patients from healthy controls, the diagnosis ability was limited and the sensitivity and specificity was 59% and 58%, respectively [82]. Reccently, Xiao et al. established a simple reproducible analysis method for detection exosome GPC1 for PC screening in chinese cohort, the results showed a excellent diagnostic performance with 100% sepcificity and 100% sensitivity [79]. We speculated that the inconsistent diagnosis performance of EV GPC1 for PC might be related to the non-standardized isolation and detection methods, nonuniform experimental procedure, different sample collection protocol, and various data analysis methods. Although the diagnostic performance of EV GPC1 for PC is varied, EV GPC1 indeed is a promising noninvasive biomarker for early diagnosis PC. Almost all of studies reported the diagnosis performance of EV EphA2 revealed great diagnosis value with AUC value equal or beyond 0.85, except one study with AUC value of 0.64 [76]. The result of a large cohort validated study demonstrated that for both I-II stage PC and all stage PC EV EphA2 showed strongly diagnostic efficiency. Especially, EV EphA2 could accurately distinguish PC patients with I-II stage from healthy controls (AUC = 0.96) or pancreatitis (AUC = 0.93) [34].
EV miRNAs recently have raised more interest as novel noninvasive biomarkers for malignant tumors. In our subgroup analysis EV miRNAs appeared powerful diagnosis capacity for PC with both sensitivity and specificity exceeded 80%, which was consistent with that of the whole EV RNAs. EV miR-21 was the most frequency reported miRNA and was significantly high expressed in PC with consistent direction in all five studies. EV miR-21 performed outstanding specificity for PC diagnosis, excepting in study by Goto et al. the specificity was 81%, the specificity value was all greater than 90% in the remaining studies and even two studies showed a specificity of 100% [25, 45, 61, 62, 84]. Aberrant expressed miR-21 related to the development and metastatic of malignant tumors and impacted signal transducer and activator of transcription 3 (STAT3), epidermal growth factor receptor (EGFR), transforming growth factor-β, and the p53 pathway in malignant tumor progression [87–90]. MiR-10b high expressed in PC and played important role in various malignant cancers, including PC. For example, PC patients had significantly higher expression level of miR-10b than healthy controls or pancreatitis and increased miR-10b expression in PC patients indicated tumor aggressiveness [91–94]; Abnormal expressed miR-10b promoted hepatocellular carcinoma cell proliferation and invasion and high expression level of EV miR-10b was related to advanced tumor [95]. However, there are number of challenges in using EV miRNAs as diagnostic biomarkers for PC, such as racial differences [96], unstandardized sample preparation procession [97], and nonuniform miRNA extraction kits [98]. Continued refinement of EV miRNAs techniques may help identify specific EV miRNAs for screening PC, which offers as promising non-invasive biomarkers for PC diagnosis in clinical practical.
In present study, we found EV biomarker panels showed high diagnosis value for PC, the pooled sensitivity and specificity was 80% and 86%, respectively, which showed a small advantage diagnosis performance than the whole individual EV RNAs and individual EV proteins. In previous studies, compared with the corresponding individual markers the diagnostic accuracy of either protein panels or miRNA panels was not to show any advantage [25, 61, 62, 79, 82]. However, combining EV RNAs with EV proteins, the diagnosis performance for PC was significant greater than that of the corresponding individual biomarkers. For example, both the sensitivity and specificity of EV panel composed by EphA2, miR-451a, miR-21, and miR-10b reached 100% [62]; Another panel consisted by EV miRNAs and EV proteins exhibited great diagnosis efficiency for PC with sensitivity of 100% and specificity of 93% [43]. In present study, we also found the diagnostic value of the EV RNA combined with protein panels showed good diagnostic performance, which was similar to the EV protein panels and a slight greater advantage than the EV RNA panels. Moreover, combined EV RNAs and EV proteins with conventional CA199 also significantly improved the diagnostic accuracy for PC. EV miR-200b and 200c and EpCAM combined with CA199 entailed a diagnostic accuracy of 97%, yielding sensitivity of 92% and specificity of 100% [31]; Therefore, the combine of novel EV biomarkers with conventional CA199 might be explored to increase diagnostic accuracy for PC and can be as noninvasive and low-cost diagnosis methods for PC screening in the future.
Due to high stability and providing specific information from original tumor of RNAs and proteins in EVs, EV RNAs and proteins have attracted considerable attention as noninvasive biomarkers for cancer diagnosis. By far, the analysis of EVs was separated into isolation and detection two steps. Multistep ultracentrifugation is the recommended standard method for EVs isolation, which increased the complexity of the operational process and was time-consuming and expensive, and the centrifugation time and force was nonuniform inducing major difference of EV purity and isolation rate [99]. Most included studies in this systematic review underwent the centrifugation step, but the centrifugation time and force are variable, parts of studies [85] even used once time centrifugation, which greatly affected the isolation concentration of EVs and further impacted the expression level of molecules contained in EVs. For decades, significant progression of EVs isolation and detection techniques has been made and two kinds of microfluidic-based techniques, immunoaffinity-based and size-based platforms, have been reported as most practical solutions to isolate and detect EV biomarkers [34, 44, 47, 62]. Microfluidics-based techniques provide an effective method to integrate the EV isolation and detection into a single chip, which overcome the abovementioned disadvantage [100]. In current study, six studies used microfluidic-based techniques to analysis the expression levels of EV biomarkers, such as 3D microfluidic chip, LPHN-CHDC biochip, ACE integrated biomarker assay, chip-exosome-PEARL SERS immunosensor, surface-Enhanced Raman Scattering, NPS chip, EphA2-EV-nPES assay, SSEC, AbMB-bioChol platform, nanoparticle-and dyebased fluorescent immunoassay, and immuno-capture using magnetic beads. Although microfluidic-based techniques show great advances in EVs isolation and detection, it need sufficient clinical samples to validate the application, accuracy, stability, and reproduction.
The heterogeneity of the present study ranged from mild to severe, neither subgroup analysis nor sensitivity analyses failed to reduce the degree of heterogeneity. Furthermore, we analyzed the pooled diagnostic value of EV biomarkers for early stage PC, the results also did not have impact on heterogeneity. We hypothesized that the sources of heterogeneity might be related to the various methods of EVs isolation and detection, sample selection protocol, and demographic or geographic different of study populations. In addition, the number of samples of case and control groups widely ranged from 6 to 284, which also contributed to the heterogeneity. Although the heterogeneity might limit the reliability of the pooled results in present study, the results of our systematic review and meta-analyses have a certain guide to pick up noninvasive and effective early diagnosis biomarkers for PC.
Conclusions
In this systematic review and meta-analysis, we found circulation based EV biomarkers exhibited considerable diagnosis performance for PC. EV RNAs combined with EV proteins showed appealing higher diagnosis efficiency. Especially, for early stage PC the EV biomarkers revealed excellent diagnostic performance. However, the deficiency of technologies that can effectively isolation and detection EV biomarkers limit the application of EV biomarkers in clinical practice to some extent, highlighting the need for high-quality reproduction researches in this area as well a need for promising accuracy EV biomarkers for PC diagnosis and screening in larger sample prospective cohort.
Supplementary Information
Additional file 1. Protocols of blood exosomes detection.
Additional file 2. Summary of studies reporting significant associations of RNAs in pancreatic cancer.
Additional file 3. Summary of studies reporting significant associations of proteins in pancreatic cancer.
Additional file 4. Pooled sensitivity and specificity of EV biomarker panels for pancreatic cancer diagnosis.
Acknowledgements
Not applicable.
Abbreviations
- EV
Extracellular vesicle
- PC
Pancreatic cancer
- IPMN
Intraductal papillary mucinous neoplasm
- CP
Chronic pancreatitis
- AUC
Area under the curve
- ROC
Receiver operator characteristic
- CA199
Carbohydrate antigen 199
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies 2
- MP
Membrane protein; nMP non-membrane proteins
- CI
Confidence Interval
- EphA2-EV-nPES assay
Ephrin type-A receptor 2/nanoplasmon-enhanced scattering
- chip-exosome-PEARL SERS immunosensor
PDA encapsulated antibody-reporter-Ag(shell)-Au(core) multilayer, surface-Enhanced Raman Scattering
- LPHN-CHDC biochip
Lipid-polymer hybrid nanoparticle/catalyzed hairpin DNA circuit
- NPS chip
Nanoplasmonic sensor assay
- SSEC
Sequential size-exclusion chromatography
- AbMB-bioChol
Antibody-conjugated magnetic beads, bivalent cholesterol-modified RNA − DNA duplexes
Authors’ contributions
All authors read and approved the final manuscript. JX designed the study. EJ, NR, SQ and RZ acquired, analyzed the data and drafted the manuscript. JX, FY and HY contributed to the review and final edition.
Funding
There was no funding received for this article.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
There is no limit to the publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Erna Jia and Na Ren contributed equally to this work and share first authorship.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.(NCI) NCI. Cancer Stat Facts: pancreatic cancer. NCI website 2019,https://seer.cancer.gov/statfacts/html/pancreas.html.
- 5.Ghatnekar O, Andersson R, Svensson M, et al. Modelling the benefits of early diagnosis of pancreatic cancer using a biomarker signature. Int J Cancer. 2013;133:2392–2397. doi: 10.1002/ijc.28256. [DOI] [PubMed] [Google Scholar]
- 6.Buscail L. Commentary: Pancreatic cancer: is the worst to come? Int J Epidemiol. 2017;46:1774–1775. doi: 10.1093/ije/dyx143. [DOI] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 8.Rahib L, Fleshman JM, Matrisian LM, et al. Evaluation of Pancreatic Cancer Clinical Trials and Benchmarks for Clinically Meaningful Future Trials: A Systematic Review. JAMA Oncol. 2016;2:1209–1216. doi: 10.1001/jamaoncol.2016.0585. [DOI] [PubMed] [Google Scholar]
- 9.Force USPST. Owens DK, Davidson KW, et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA. 2019;322:438–444. doi: 10.1001/jama.2019.10232. [DOI] [PubMed] [Google Scholar]
- 10.Prokesch RW, Chow LC, Beaulieu CF, et al. Isoattenuating pancreatic adenocarcinoma at multi-detector row CT: secondary signs. Radiology. 2002;224:764–768. doi: 10.1148/radiol.2243011284. [DOI] [PubMed] [Google Scholar]
- 11.Wiersema MJ, Vilmann P, Giovannini M, et al. Endosonography-guided fine-needle aspiration biopsy: diagnostic accuracy and complication assessment. Gastroenterology. 1997;112:1087–1095. doi: 10.1016/S0016-5085(97)70164-1. [DOI] [PubMed] [Google Scholar]
- 12.Seufferlein T, Mayerle J. Pancreatic cancer in 2015: Precision medicine in pancreatic cancer–fact or fiction? Nat Rev Gastroenterol Hepatol. 2016;13:74–75. doi: 10.1038/nrgastro.2015.215. [DOI] [PubMed] [Google Scholar]
- 13.Chari ST, Kelly K, Hollingsworth MA, et al. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693–712. doi: 10.1097/MPA.0000000000000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herreros-Villanueva M, Bujanda L. Non-invasive biomarkers in pancreatic cancer diagnosis: what we need versus what we have. Ann Transl Med. 2016;4:134. doi: 10.21037/atm.2016.03.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Bamlet WR, Oberg AL, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Sci Transl Med 2017. 9. [DOI] [PMC free article] [PubMed]
- 16.Swords DS, Firpo MA, Scaife CL, et al. Biomarkers in pancreatic adenocarcinoma: current perspectives. Onco Targets Ther. 2016;9:7459–7467. doi: 10.2147/OTT.S100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satake K, Kanazawa G, Kho I, et al. Evaluation of serum pancreatic enzymes, carbohydrate antigen 19–9, and carcinoembryonic antigen in various pancreatic diseases. Am J Gastroenterol. 1985;80:630–636. [PubMed] [Google Scholar]
- 18.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 19.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Liu Y, Liu H, et al. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frampton AE, Prado MM, Lopez-Jimenez E, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006–19013. doi: 10.18632/oncotarget.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino A, Kim HS, Bojmar L, et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020;182(1044–1061):e1018. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buscail E, Chauvet A, Quincy P, et al. CD63-GPC1-Positive Exosomes Coupled with CA19-9 Offer Good Diagnostic Potential for Resectable Pancreatic Ductal Adenocarcinoma. Transl Oncol. 2019;12:1395–1403. doi: 10.1016/j.tranon.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pu X, Ding G, Wu M, et al. Elevated expression of exosomal microRNA-21 as a potential biomarker for the early diagnosis of pancreatic cancer using a tethered cationic lipoplex nanoparticle biochip. Oncol Lett. 2020;19:2062–2070. doi: 10.3892/ol.2020.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JW, Wieckowski E, Taylor DD, et al. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- 27.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartsch DK, Gercke N, Strauch K, et al. The Combination of MiRNA-196b, LCN2, and TIMP1 is a Potential Set of Circulating Biomarkers for Screening Individuals at Risk for Familial Pancreatic Cancer. J Clin Med. 2018;7. [DOI] [PMC free article] [PubMed]
- 30.Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–U182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese M, Flammang I, Yang Z, et al. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2020,12. [DOI] [PMC free article] [PubMed]
- 32.Lan B, Zeng S, Grutzmann R, et al. The Role of Exosomes in Pancreatic Cancer. Int J Mol Sci 2019,20. [DOI] [PMC free article] [PubMed]
- 33.Rodrigues M, Richards N, Ning B, et al. Rapid Lipid-Based Approach for Normalization of Quantum-Dot-Detected Biomarker Expression on Extracellular Vesicles in Complex Biological Samples. Nano Lett. 2019;19:7623–31. [DOI] [PMC free article] [PubMed]
- 34.Liang K, Liu F, Fan J, et al. Nanoplasmonic Quantification of Tumor-derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat Biomed Eng. 2017;1. [DOI] [PMC free article] [PubMed]
- 35.Sato Y, Suzuki R, Takagi T, et al. Circulating extracellular vesicle-encapsulated microRNA as screening biomarkers for intraductal papillary mucinous neoplasm. Oncol Lett. 2020;20:315. doi: 10.3892/ol.2020.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai CS, Khan A, Bellio MA, et al. Characterization of extracellular vesicle miRNA identified in peripheral blood of chronic pancreatitis patients. Mol Cell Biochem. 2021;476:4331–4341. doi: 10.1007/s11010-021-04248-5. [DOI] [PubMed] [Google Scholar]
- 37.Nakamaru K, Tomiyama T, Kobayashi S, et al. Extracellular vesicles microRNA analysis in type 1 autoimmune pancreatitis: Increased expression of microRNA-21. Pancreatology. 2020;20:318–324. doi: 10.1016/j.pan.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Yang KS, Ciprani D, O’Shea A, et al. Extracellular Vesicle Analysis Allows for Identification of Invasive IPMN. Gastroenterology. 2021;160(1345–1358):e1311. [DOI] [PMC free article] [PubMed]
- 39.Sheng LP, Han CQ, Nie C, et al. Identification of potential serum exosomal microRNAs involved in acinar-ductal metaplasia that is a precursor of pancreatic cancer associated with chronic pancreatitis. Medicine (Baltimore) 2021;100:e25753. doi: 10.1097/MD.0000000000025753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 42.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 44.Hu J, Sheng Y, Kwak KJ, et al. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat Commun. 2017;8:1683. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86–93. doi: 10.1016/j.canlet.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu YF, Hannafon BN, Zhao YD, et al. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8:77028–77040. doi: 10.18632/oncotarget.20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang KS, Im H, Hong S, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed]
- 48.Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116. doi: 10.1186/s12885-018-4006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin H. Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Traffic. 2018;109:2946–2956. doi: 10.1111/cas.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis JM, Vyas AD, Qiu Y, et al. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano. 2018;12:3311–20. [DOI] [PubMed]
- 51.Li TD, Zhang R, Chen H, et al. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem Sci. 2018;9:5372–82. [DOI] [PMC free article] [PubMed]
- 52.Buscail E, Chauvet A, Quincy P, et al. CD63-GPC1-Positive Exosomes Coupled with CA19-9 Offer Good Diagnostic Potential for Resectable Pancreatic Ductal Adenocarcinoma. Transl Oncol. 2019;12:1395–1403. doi: 10.1016/j.tranon.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol. 2019;13:212–227. doi: 10.1002/1878-0261.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lux A, Kahlert C, Grutzmann R, et al. c-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed]
- 55.Takahashi K, Ota Y, Kogure T, et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020;111:98–111. doi: 10.1111/cas.14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao D, Dong Z, Zhen L, et al. Combined Exosomal GPC1, CD82, and Serum CA19-9 as Multiplex Targets: A Specific, Sensitive, and Reproducible Detection Panel for the Diagnosis of Pancreatic Cancer. Mol Cancer Res. 2020;18:300–310. doi: 10.1158/1541-7786.MCR-19-0588. [DOI] [PubMed] [Google Scholar]
- 57.Yu S, Li Y, Liao Z, et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69:540–50. [DOI] [PubMed]
- 58.Fahrmann JF, Mao X, Irajizad E, et al. Plasma-Derived Extracellular Vesicles Convey Protein Signatures that Reflect Pathophysiology in Lung and Pancreatic Adenocarcinomas. Cancers (Basel). 2020;12. [DOI] [PMC free article] [PubMed]
- 59.Flammang I, Reese M, Yang Z, et al. Tumor-Suppressive miR-192–5p Has Prognostic Value in Pancreatic Ductal Adenocarcinoma. Int J Cancer. 2020;12. [DOI] [PMC free article] [PubMed]
- 60.Wei Q, Zhang J, Li Z, et al. Serum Exo-EphA2 as a Potential Diagnostic Biomarker for Pancreatic Cancer. Pancreas. 2020;49:1213–1219. doi: 10.1097/MPA.0000000000001660. [DOI] [PubMed] [Google Scholar]
- 61.Wu L, Zhou WB, Zhou J, et al. Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol Lett. 2020;20:1432–1440. doi: 10.3892/ol.2020.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou S, Hu T, Han G, et al. Accurate Cancer Diagnosis and Stage Monitoring Enabled by Comprehensive Profiling of Different Types of Exosomal Biomarkers: Surface Proteins and miRNAs. Small. 2020;16:e2004492. [DOI] [PubMed]
- 63.Qin D, Zhao Y, Guo Q, et al. Detection of Pancreatic Ductal Adenocarcinoma by A qPCR-based Normalizer-free Circulating Extracellular Vesicles RNA Signature. J Cancer. 2021;12:1445–1454. doi: 10.7150/jca.50716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C, Wang J, Cui W, et al. Serum Exosomal miRNA-1226 as Potential Biomarker of Pancreatic Ductal Adenocarcinoma. Onco Targets Ther. 2021;14:1441–51. [DOI] [PMC free article] [PubMed]
- 65.Goto T, Konishi H, Sasajima J, et al. Serum Exosomal MicroRNA-191,-21,-451a are Considered to be Efficient Diagnostic Markers of Pancreatic Neoplasm. Pancreas. 2016;45:1508–1509. doi: 10.1186/s12885-018-4006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buscail E, Alix-Panabieres C, Quincy P, et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers (Basel). 2019:11. [DOI] [PMC free article] [PubMed]
- 67.Zheng H, Zhao J, Wang X, et al. Integrated Pipeline of Rapid Isolation and Analysis of Human Plasma Exosomes for Cancer Discrimination Based on Deep Learning of MALDI-TOF MS Fingerprints. 2022. [DOI] [PubMed]
- 68.Chen J, Yao D, Chen W, et al. Serum exosomal miR-451a acts as a candidate marker for pancreatic cancer. Int J Biol Markers. 2022,37:74–80. [DOI] [PubMed]
- 69.Yang J, Zhang Y, Gao X, et al. Plasma-Derived Exosomal ALIX as a Novel Biomarker for Diagnosis and Classification of Pancreatic Cancer. Front Oncol. 2021;11:628346. doi: 10.3389/fonc.2021.628346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao PP, Wan QQ, Liao T, et al. Peptide Nucleic Acid-Functionalized Nanochannel Biosensor for the Highly Sensitive Detection of Tumor Exosomal MicroRNA. Anal Chem. 2021;93:10966–73. [DOI] [PubMed]
- 71.Wu Y, Zeng H, Yu Q, et al. A Circulating Exosome RNA Signature Is a Potential Diagnostic Marker for Pancreatic Cancer, a Systematic Study. J Cell Mol Med 2021,13. [DOI] [PMC free article] [PubMed]
- 72.Wang L, Wu J, Ye N, et al. Plasma-Derived Exosome MiR-19b Acts as a Diagnostic Marker for Pancreatic Cancer. Cell Commun Signal. 2021;11:739111. doi: 10.3389/fonc.2021.739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verel-Yilmaz Y, Fernández JP, Schäfer A, et al. Extracellular Vesicle-Based Detection of Pancreatic Cancer. Embo J. 2021;9:697939. doi: 10.3389/fcell.2021.697939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao H, Zhang Y, Yan J, et al. Upregulated MicroRNA-483-3p is an Early Event in Pancreatic Ductal Adenocarcinoma (PDAC) and as a Powerful Liquid Biopsy Biomarker in PDAC. Onco Targets Ther. 2021;14:2163–2175. doi: 10.2147/OTT.S288936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moutinho-Ribeiro P, Adem B, Batista I, et al. Exosomal glypican-1 discriminates pancreatic ductal adenocarcinoma from chronic pancreatitis. Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver 2021. [DOI] [PubMed]
- 76.Li P, Wang J, Gao M, et al. Membrane Feature-Inspired Profiling of Extracellular Vesicles for Pancreatic Cancer Diagnosis. Anal Chem. 2021;93:9860–8. [DOI] [PubMed]
- 77.Kim MW, Koh H, Kim JY, et al. Tumor-Specific miRNA Signatures in Combination with CA19-9 for Liquid Biopsy-Based Detection of PDAC. Int J Mol Sci. 2021;22. [DOI] [PMC free article] [PubMed]
- 78.Guo S, Qin H, Liu K, et al. Blood small extracellular vesicles derived miRNAs to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Clin Transl Med. 2021;11:e520. [DOI] [PMC free article] [PubMed]
- 79.Xiao D, Dong Z, Zhen L, et al. Combined Exosomal GPC1, CD82, and Serum CA19-9 as Multiplex Targets: A Specific, Sensitive, and Reproducible Detection Panel for the Diagnosis of Pancreatic Cancer. Mol Cancer Res. 2020;18:300–10. [DOI] [PubMed]
- 80.Takahashi K, Ota Y, Kogure T, et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020;111:98–111. [DOI] [PMC free article] [PubMed]
- 81.Lux A, Kahlert C, Gruetzmann R, et al. c-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed]
- 82.Li T-D, Zhang R, Chen H, et al. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem Sci. 2018;9:5372–5382. doi: 10.1039/C8SC01611A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin H, Liu P, Wu Y, et al. Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 2018;109:2946–2956. doi: 10.1111/cas.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191,-21,-451a of pancreatic neoplasm is considered to be efficient diagnostic marker. Bmc Cancer 2018,18. [DOI] [PMC free article] [PubMed]
- 85.Yu S, Li Y, Liao Z. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. 2019. [DOI] [PubMed]
- 86.Lewis JM, Vyas AD, Qiu Y, et al. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano. 2018;12:3311–3320. doi: 10.1021/acsnano.7b08199. [DOI] [PubMed] [Google Scholar]
- 87.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 89.Ma X, Choudhury SN, Hua X, et al. Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway. Carcinogenesis. 2013;34:1216–1223. doi: 10.1093/carcin/bgt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu HY, Zhang YY, Zhu BL, et al. miR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. Eur Rev Med Pharmacol Sci. 2019;23:4149–4155. doi: 10.26355/eurrev_201905_17917. [DOI] [PubMed] [Google Scholar]
- 91.Sempere LF, Preis M, Yezefski T, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16:4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cote GA, Gore AJ, McElyea SD, et al. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am J Gastroenterol. 2014;109:1942–1952. doi: 10.1038/ajg.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakata K, Ohuchida K, Mizumoto K, et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 94.Ouyang H, Gore J, Deitz S, et al. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-beta actions. Oncogene. 2014;33:4664–4674. doi: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tian XP, Wang CY, Jin XH, et al. Erratum: Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis: Erratum. Theranostics. 2021;11:6522–6523. doi: 10.7150/thno.60140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang RS, Gamazon ER, Ziliak D, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Becker N, Lockwood CM. Pre-analytical variables in miRNA analysis. Clin Biochem. 2013;46:861–868. doi: 10.1016/j.clinbiochem.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 98.El-Khoury V, Pierson S, Kaoma T, et al. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep. 2016;6:19529. doi: 10.1038/srep19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006,Chapter 3:Unit 3 22. [DOI] [PubMed]
- 100.Lin S, Yu Z, Chen D, et al. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small. 2020;16:e1903916. doi: 10.1002/smll.201903916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Protocols of blood exosomes detection.
Additional file 2. Summary of studies reporting significant associations of RNAs in pancreatic cancer.
Additional file 3. Summary of studies reporting significant associations of proteins in pancreatic cancer.
Additional file 4. Pooled sensitivity and specificity of EV biomarker panels for pancreatic cancer diagnosis.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.